Abstract
During T cell development in mice, thymic negative selection deletes cells with the potential to recognize and react to self-antigens. In human T cell-dependent autoimmune diseases such as Type 1 diabetes, multiple sclerosis, and rheumatoid arthritis, T cells reactive to autoantigens are thought to escape negative selection, traffic to the periphery and attack self-tissues. However, physiological thymic negative selection of autoreactive human T cells has not been previously studied. We now describe a human T-cell receptor-transgenic humanized mouse model that permits the study of autoreactive T-cell development in a human thymus. Our studies demonstrate that thymocytes expressing the autoreactive Clone 5 TCR, which recognizes insulin B:9–23 presented by HLA-DQ8, are efficiently negatively selected at the double and single positive stage in human immune systems derived from HLA-DQ8+ HSCs. In the absence of hematopoietic expression of the HLA restriction element, negative selection of Clone 5 is less efficient and restricted to the single positive stage. To our knowledge, these data provide the first demonstration of negative selection of human T cells recognizing a naturally-expressed tissue-restricted antigen. Intrathymic antigen presenting cells are required to delete less mature thymocytes, while presentation by medullary thymic epithelial cells may be sufficient to delete more mature single positive cells. These observations set the stage for investigation of putative defects in negative selection in human autoimmune diseases.
Keywords: T cell selection, Type 1 diabetes, PD-1, HLA, Tolerance, Autoimmunity
Highlights
-
•In the presence of the HLA-restriction element on hematopoietic stem cells (HSCs) and thymus.
-
•Thymocytes bearing insulin peptide-reactive TCRs express markers of thymic negative selection.
-
•Insulin peptide-reactive T cells are efficiently negatively selected in the human thymus.
-
•The few transgenic T cells that escape negative selection largely express endogenous TCRs.
-
•
-
•
HLA-restriction element on HSCs is required for efficient negative selection.
1. Introduction
Developing T cells pass through stringent positive and negative selection processes to ensure that cells entering the periphery express T-cell receptors (TCRs) tailored to recognize antigens presented on self-MHC molecules but do not react to self-antigens presented by MHC. TCR-transgenic and TCR signaling reporter mouse models have demonstrated that negative selection is driven by exposure to antigens expressed endogenously by medullary thymic epithelial cells (mTECs) expressing the autoimmune regulator (AIRE) and Fezf2 transcription factors, which drive production of tissue-restricted self-antigens [[1], [2], [3]]. mTECs and intrathymic hematopoietic stem cell (HSC)-derived antigen presenting cells (APCs) that acquire self-antigens from the periphery or expressed by mTECS [4,5] present them to thymocytes. Thymocytes that react with moderate strength to these antigens become T regulatory cells [6] while those that receive strong TCR stimulation are deleted. Deletion involves a signaling cascade that upregulates a negative selection gene expression profile including PD-1 [7]. This cascade ultimately leads to apoptosis of those thymocytes [8], purging the T-cell repertoire of cells that could potentially cause autoimmunity by reacting to self-antigens in the periphery.
While these mouse models are extremely valuable, thymic selection of autoreactive TCRs has been less accessible to study in human immune systems. Previous studies of human TCR selection involved non-physiological conditions such as enforced expression of antigen or superantigen during selection [9,10]. Although useful in delineating the steps in human thymic negative selection, these models do not recapitulate naturally occurring selection. The importance of negative selection in humans is underscored by the occurrence of Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy (APECED), a severe multi-organ autoimmune disease caused by mutations in AIRE [11,12]. Understanding negative selection is vital to understanding multigenic T-cell-mediated autoimmune diseases. Despite the negative selection checkpoint during T cell maturation, autoreactive T cells escape into the periphery of patients with T-cell-mediated autoimmune diseases [[13], [14], [15]] and attack self-tissues. There is evidence to suggest that negative selection could be defective in patients with autoimmune disease and defects in this process have been demonstrated in the murine non-obese diabetic (NOD) model of autoimmune diabetes [[16], [17], [18], [19], [20]]. Tools to study human thymocyte selection are needed to determine whether genetic variants associated with human autoimmune disease result in impaired negative selection and whether these autoreactive T cells initiate or drive the disease process. To enable such studies, we asked whether a human autoreactive TCR normally undergoes negative selection in a human thymus. We examined the role of APCs expressing the HLA restriction element that presents the cognate peptide in this process. Specifically, we developed a novel TCR-transgenic humanized mouse model to study the selection of Clone 5, an insulin B:9–23 peptide-reactive TCR restricted by HLA-DQ8 and isolated from the blood of a Type 1 diabetic [21,22], in an immune system reconstituted with HLA-DQ8+ or HLA-DQ8- HSCs. Insulin B:9–23 is an essential antigen for the development of diabetes in NOD mice [23] and Clone 5-transduced peripheral T cells were shown to be capable of initiating beta cell destruction and autoimmune diabetes development when injected into HLA-DQ8 transgenic humanized mice [24]. Previous work has demonstrated the presence of all human APC lineages in human thymus grafts of TCR-transgenic humanized mice generated similarly to the model used in this study [10]. We show here that Clone 5-expressing T cells going through thymic selection are enriched for phenotypic markers of negative selection. Developing Clone 5 T cells in DQ8+ HSC mice are also enriched among T cell precursors that have not undergone thymic selection. Clone 5 T cells are absent from populations of more mature thymocytes and are rarely found in the periphery of DQ8+ HSC TCR-transgenic humanized mice. Interestingly, negative selection of Clone 5 T cells in immune systems containing DQ8- HSCs is less efficient than in those with DQ8+ HSCs and is limited to single positive (SP) thymocytes. To our knowledge, this is the first evidence of negative selection of a TCR recognizing a natural tissue-restricted antigen (TRA) in a human immune system. These studies provide the groundwork for characterization of human T cell negative selection and an opportunity to investigate possible negative selection deficiencies in multiple autoimmune diseases.
2. Materials and methods
2.1. Mice
NOD/SCID IL2Rγko (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, NSG, Jax strain 005557 [25]) and NSG mice transgenic for human HLA-DQ8, Jax strain 026561, were obtained from The Jackson Laboratory (Bar Harbor, ME). NSG mice transgenic for porcine IL-3, Granulocyte-macrophage colony-stimulating factor (GM-CSF), and Stem cell factor (SCF) (pig cytokine transgenic, or PCT, mice) [26,27] were regenerated from frozen embryos at The Jackson Laboratory (Bar Harbor, ME). Mice were bred, maintained, and housed in microisolator cages in a Helicobacter, Pasteurella pneumotropica and Specific Pathogen Free (HPPF) animal facility. All animal procedures were approved by the Columbia University Medical Center (CUMC) Institutional Animal Care and Use Committee.
2.2. Human tissues
All human fetal tissues (gestational age 17–21 weeks) were obtained from Advanced Bioscience Resources (Alameda, CA). Fetal thymus and liver were prepared as previously described [27]. Briefly, fetal thymi were processed to 2 × 2 × 2 mm fragments and cryopreserved in 10% DMSO and 90% human AB serum (Gemini Bio-Products, Sacramento, CA). Single cell suspensions were generated from fetal livers by Liberase digestion (Sigma-Aldrich, St. Louis, MO). CD34+ cells were isolated using positive selection by magnetic-activated cell sorting (MACS) with anti-human CD34+ microbeads according to the manufacturer’s instructions (Miltenyi Biotec, Bergish Gladbach, Germany). Allele-level molecular human leukocyte antigen (HLA) typing of fetal tissue was performed using Sanger sequencing to identify HLA-DQB1∗03:02:01 (HLA-DQ8+) fetal liver and thymus tissue. The use of human tissues and cells was approved by the CUMC Institutional Review Board.
2.3. Transduction of fetal liver CD34+ cells
cDNA of the Clone 5 TCR was obtained from a T-cell clone that was isolated from a Type 1 diabetic patient and reactive with insulin B9-23 [13]. The Clone 5 TCR plasmid was generously provided to us by Yong-Guang Yang [24] and was subcloned into a pHR-EF1α_IRES_GFP_SIN (pHR) second-generation lentiviral backbone [28] then modified to express the EF1α promoter. The final plasmid construct contains the Clone 5 TCR alpha sequence followed by a P2A site then the TCR beta sequence followed by an F2A site then the GFP sequence. The use of the EF1α promoter results in constitutive expression of GFP in transduced cells and expression of the TCR alpha and beta on the cell surface only when CD3 is also present. The packaging vectors consisted of pDelta R8.2 and the pVSV-G pseudotyping vector.
Virus was produced in HEK293 FT cells by standard procedures. HEK293 FT cells were transfected with viral plasmids using Lipofectamine 2000 according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA) and resulting supernatants were harvested and concentrated 100x by ultracentrifugation. Concentrated supernatants were flash frozen and stored at −80 °C prior to use. Viral titer was determined by transducing HEK293 FT cells with serial dilutions of virus and measuring GFP expression by FCM.
Lentiviral transduction of human fetal liver CD34+ cells was performed as previously described [29]. Briefly, human fetal liver CD34+ cells were thawed and pre-stimulated for 3 h in retronectin-coated plates (25 μg/mL, Clontech, Mountain View, CA) in Stemline II medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10 μg/mL protamine sulfate (Sigma-Aldrich) and 60 ng/mL, 300 ng/mL and 150 ng/mL recombinant human IL-3, SCF and Flt3 Ligand (PeproTech, Rocky Hill, NJ), respectively. Cells were transduced overnight (at least 13 h) at a multiplicity of infection (MOI) of 50 for Clone 5 TCR lentivirus and 5 MOI for GFP-only lentivirus. All incubations were performed at 37 °C and 5% CO2. Cells were harvested after transduction and prepared for intratibial injection. A small number of transduced CD34+ cells were cultured in stem cell medium with cytokines but without protamine sulfate for 96 h, then assessed for transduction efficiency by flow cytometry.
2.4. TCR-transgenic humanized mouse transplantation
Mice were surgically thymectomized as described [30] and allowed to recover for at least two weeks, then irradiated with 1 Gy total body irradiation (TBI) by an X-Ray irradiator (RS-2000, Rad Source Technologies, Inc., Suwanee, GA). Mice were then transplanted with 2-5x105 lentivirally-transduced CD34+ cells intratibially. In some cases, more CD34+ cells were transduced than could be injected intratibially and additional cells were transplanted intravenously. Mice were also transplanted with a fragment of human fetal thymus under the kidney capsule. Prior to transplantation, thymus fragments were extensively pipetted to deplete passenger thymocytes that might otherwise be co-transplanted, as previously described [31,32]. To ensure that only de novo development of thymocytes in the transplanted thymus was considered in selection studies, mice were injected intravenously with 0.4 μg of the rat anti-human CD2 monoclonal antibody LoCD2b [33] on the day of thymus transplantation and seven days post-transplantation to deplete remaining passenger T cells emerging from the grafted thymus, as previously described [32].
2.5. Monitoring peripheral blood reconstitution
Human reconstitution in peripheral blood of transplanted mice was monitored every two weeks, beginning 3–6 weeks post-transplantation. Approximately 100 μL of whole blood was collected by tail vein puncture. Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood by density gradient separation using Histopaque 1077 (Sigma Aldrich) for flow cytometric (FCM) analysis using the antibodies listed in Supplementary Table 1.
2.6. Thymus and peripheral tissue processing
TCR-transduced humanized mice were euthanized at 16 weeks post-transplant. This time-point was chosen for optimal analysis of thymopoiesis. Human T cells typically first emerge in transplanted animals between 10 and 14 weeks post-transplant, and therefore we expect peak thymopoiesis at 12–16 weeks post-transplant. At harvest, blood was collected by cardiac puncture and spleens, grafted thymus, residual anterior mediastinum and bone marrow were collected. Spleens were crushed to make a single cell suspension, which was passed through a 70 μm nylon filter and treated with Ammonium-Chloride-Potassium (ACK) erythrocyte lysis buffer (Life Technologies, Carlsbad, CA) prior to counting and staining. Single-cell bone marrow suspensions from the tibia and femur were prepared by crushing using a mortar and pestle, counted, and stained for FCM using the antibodies listed in Supplementary Table 1. Grafted thymi were dissected from kidneys and crushed to single cell suspension, and residual tissue in the anterior mediastinum was collected and processed to single cell suspension to verify removal of the mouse native thymus. Animals that showed residual thymopoiesis in the mouse native thymus were excluded from peripheral analysis. As has been previously reported [10], some transplanted mice showed poor development of grafted thymus tissue. Therefore, only animals with robust thymopoiesis, defined as the presence of CD4 and CD8 double positive (DP) and SP thymocytes indicative of thymopoiesis at the time of sacrifice, were included in analysis. Total numbers of animals grafted for each group are shown in Supplementary Table 2.
2.7. Flow cytometric (FCM) analysis
FCM data were acquired in the Columbia Center for Translational Immunology Flow Core. Data were collected using a BD FACSCanto, a BD LSRII or a BD Fortessa cytometer and analyzed using FlowJo (TreeStar, Ashland, OR) or FCS Express (De Novo Software).
2.8. Statistical analysis
Statistical analysis was performed using GraphPad Prism Version 7 (La Jolla, CA) as described. Groups were considered significantly different if the p value according to the appropriate test was <0.05.
3. Results
3.1. Human immune reconstitution in TCR-transgenic humanized mice
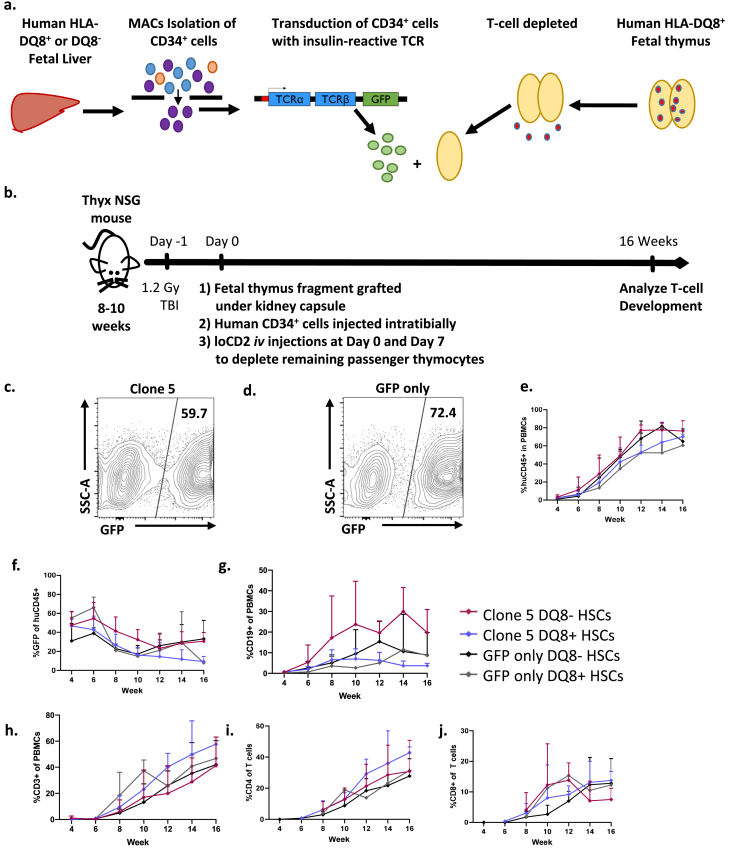
To assess how the diabetogenic Clone 5 TCR is selected in a normal human immune system, we generated TCR-transgenic humanized mice using HLA-DQ8+ fetal thymus tissue and HLA-DQ8+ autologous CD34+ fetal liver cells (FLCs) from 1 donor or HLA-DQ8- FLCs from another donor transduced with a lentivirus containing the Clone 5 TCR (Fig. 1a and b). The transduction protocol generated similar transduction efficiencies across different CD34+ FLCs in different experiments in both the Clone 5 and GFP-only groups. Representative transductions are shown for both Clone 5 and GFP-only transduced FLCs (Fig. 1 c, d). Confirmation of Clone 5 TCR expression on developing thymocytes was demonstrated by high levels of TCR Vβ11+ cells in thymic grafts of Clone 5 mice (Supplemental Fig. 1). In this model, the TCR-transduced cells only express TCR on the surface when CD3 is expressed concurrently, preventing forced T-cell lineage commitment or expression of the TCR on other immune cell lineages, such as B cells. It is important to note that not all CD34+ FLCs acquire the transgene during transduction, providing a population of untransduced immune cells within each mouse that will develop normally and can be used as an internal control.
Fig. 1.
TCR-transgenic humanized model for the study of human T-cell selection. a. CD34+ cells are MACS-sorted from HLA-DQ8+ or HLA-DQ8- human fetal liver tissue, then lentivirally transduced with an insulin B:9-23-reactive TCR containing a GFP reporter or with vector containing a GFP reporter alone. HLA-DQ8+ human fetal thymus tissue is isolated and T-cell-depleted by freeze-thawing. b. Thymectomized 8–10 week old NSG mice receive TBI on day −1. Next, the mice are injected intratibially with transduced CD34+ cells from (A). Mice are co-transplanted with a fragment of fetal thymus from (A) under the kidney capsule. Mice are injected with anti-CD2 antibody on days 1 and 7 to deplete any T cells escaping from the thymus graft. The human immune system reconstitution in peripheral blood is then monitored every two weeks until sacrifice. c, d. Representative FCM analysis of lentiviral transduction efficiency of FLCs measured by GFP expression. e-j. Peripheral blood reconstitution of human immune cell subsets in mice transplanted with Clone 5 or GFP-only virus is shown over time post-transplantation. e. Proportions of huCD45+ cells f. % GFP+ of huCD45+, g. Proportions of human B cells; h. Proportions of human T cells, i. Proportions of human CD4+ T cells, and j. Proportions of human CD8+ T cells. Data are a compilation of n = 4 Clone 5 and n = 3 GFP-only mice transplanted with CD34+ and thymus tissue from a single HLA-DQ8+ fetal donor and n = 4 Clone 5, n = 4 GFP-only mice transplanted with HLA-DQ8- CD34+ cells and HLA-DQ8+ thymus.
Sixteen weeks post-transplant, the CD34+ FLCs engrafted in the mice had reconstituted all immune cell lineages. Overall, total human cell reconstitution and GFP+ PBMC levels were similar in animals transplanted with GFP-only and Clone 5-transduced DQ8+ or DQ8- cells (Fig. 1e and f). The levels of B cell reconstitution were similar between the four groups (Fig. 1g). We did not observe differences in total CD4 or CD8 T cell reconstitution between any groups (Fig. 1h–j).
3.2. Phenotypic characteristics of Clone 5 thymocytes suggest they are efficiently intrathymically negatively selected in the presence of HLA-DQ8+ HSCs
Sixteen weeks post-transplantation, developing thymocytes from the human thymic grafts were analyzed to characterize the selection of the Clone 5 TCR compared to GFP-only thymocytes expressing only endogenous TCRs. GFP was used to identify transgenic thymocytes in both Clone 5 and GFP-only mice because surface TCR expression is not detectable at all stages of development and varies in intensity over the course of development. Three of the recipients of HLA-DQ8- HSCs were transgenic for HLA-DQ8, allowing the potential for mouse APCs entering the human thymus graft to interact with developing human T cells [32]. Although this could impact human thymocyte selection, there was no significant difference observed in mice with and without the HLA transgene as described below. These three mice are marked on each figure for clarity.
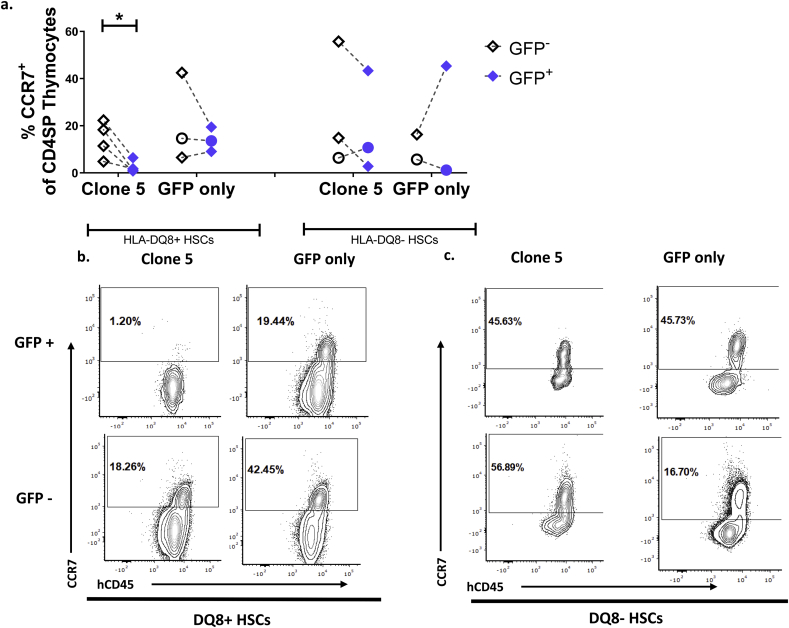
Because antigen recognition by Clone 5 is restricted by an MHC Class II molecule, we would expect to see positive selection of the Clone 5 TCR towards the CD4SP fate. CCR7 is upregulated on thymocytes after positive selection and is required for migration to the thymic medulla, which is classically considered the main site for production and presentation of tissue-associated self-antigens to thymocytes for negative selection [34,35]. We detected robust populations of CCR7+ cells among GFP- CD4 SP thymocytes in all groups of mice (Fig. 2 b,c). However, CCR7+ cells were absent among GFP+ CD4SPs in Clone 5 DQ8+ HSC mice (Fig. 2 a-c). When comparing GFP+ and GFP- populations within each mouse, we detected a significant decrease in CCR7+ CD4SPs in GFP+ populations only in Clone 5 DQ8+ HSC mice (Fig. 2a). This striking decrease in Clone 5 medullary thymocytes compared to non-transgenic thymocytes likely reflects negative selection of Clone 5 T cells in DQ8+ immune systems. There was no significant difference in the percentage of CCR7+ CD4SP cells among GFP+ versus GFP- thymocytes in mice with and without the HLA-DQ8 transgene in the Clone 5 HLA-DQ8- HSC group (Fig. 2a).
Fig. 2.
Developing Clone 5 thymocytes in animals constructed with DQ8+ HSCs show phenotypic characteristics of negative selection. Thymocytes were analyzed by FCM. Within each sample the cells are first gated into GFP+ and GFP- populations. All further analysis is conducted on both populations separately, treating the GFP- population as an internal control within each mouse. a. Comparison of the percentage of CCR7+ CD4SP in total GFP+ and GFP- thymocytes within the same mouse transplanted with Clone 5 or GFP-only transduced cells. b., c. Representative staining of grafted thymi of mice transplanted with b. DQ8+ HSCs c. DQ8- HSCs. Cells are gated on GFP expression, then on CD4SP prior to analysis of CCR7 expression. n = 3 Clone 5 DQ8-, n = 4 Clone 5 DQ8+, n = 2 GFP-only DQ8-, n = 3 GFP-only DQ8+. ∗ = p < 0.05 by paired t-test. Mice marked with a circle are transgenic for HLA-DQ8.
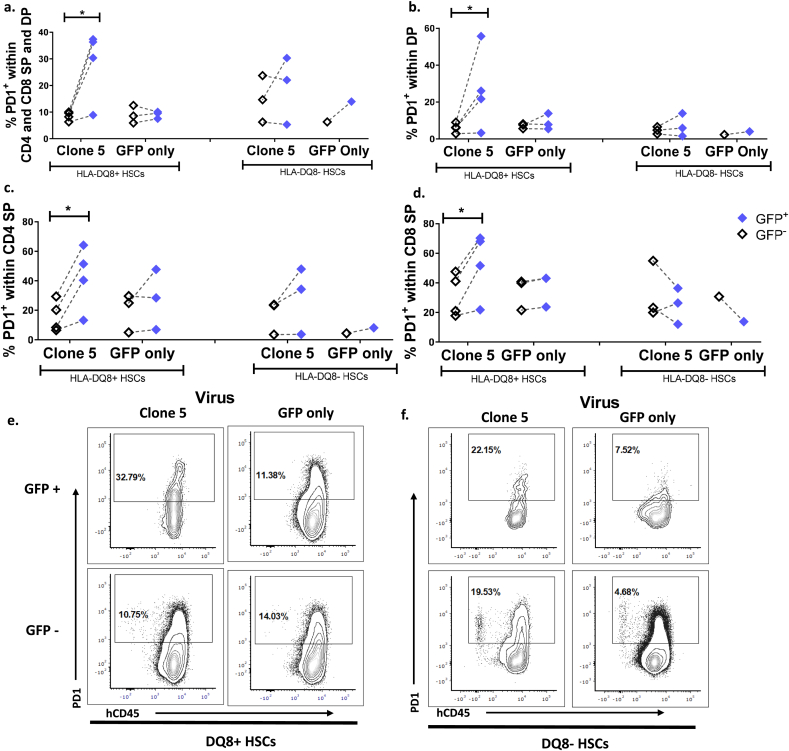
Additionally, surface expression of PD-1, a marker associated with thymocytes undergoing negative selection [10], was measured on all CD4/CD8 DP and SP developing thymocytes. The percentage of PD-1+ thymocytes was significantly higher in the GFP+ cells than in GFP- cells in Clone 5 DQ8+ HSC mice but not in Clone 5 DQ8- HSC mice or DQ8+ and DQ8- HSC GFP-only mice (Fig. 3 a, e, f). Analysis of individual thymocyte subsets revealed increased PD-1 expression on the DP subset of thymocytes in GFP+ cells in Clone 5 DQ8+ HSC mice compared to GFP- DP cells in the same mice. This increase was not seen in Clone 5 DQ8- HSC mice or GFP-only mice (Fig. 3b). In the CD4 SP subset there were increased proportions of PD-1+ GFP+ compared to GFP- cells in Clone 5 DQ8+ HSC mice, and in 2 of 3 Clone 5 DQ8- HSC mice (Fig. 3c). There was also an increased proportion of PD-1+ GFP+ cells in the CD8 SP subset of Clone 5 DQ8+ HSC mice that was not seen in the other conditions (Fig. 3d). These results suggest that HSC-derived intrathymic APCs with the HLA restriction element for a given TCR are responsible for negative selection at the DP stage, and in their absence negative selection may still proceed at the SP stage, although less efficiently. Furthermore, thymocytes containing the Clone 5 TCR transgene developing in an immune system with DQ8+ APCs were negatively selected at a higher rate than thymocytes with only endogenous TCRs (no transgene) in the same mice.
Fig. 3.
Increased PD-1 expression by all subsets of Clone 5 thymocytes in DQ8+ HSC mice. Quantification of PD-1 expression on a. CD4 and CD8 SP and DP GFP+ or GFP− cells, b. DP GFP+ or GFP- cells, c. CD4 SP GFP+ or GFP- cells, d. CD8 SP GFP+ or GFP- cells. n = 3 Clone 5 DQ8-, n = 4 Clone 5 DQ8+, n = 1 GFP-only DQ8-, n = 3 GFP-only DQ8+ ∗ = p < 0.05 by paired t-test. e, f. Representative contour plots of PD-1 expression in thymocytes expressing CD4, CD8, or both markers from thymus grafts of mice transplanted with e. DQ8+ HSCs f. DQ8- HSCs are shown.
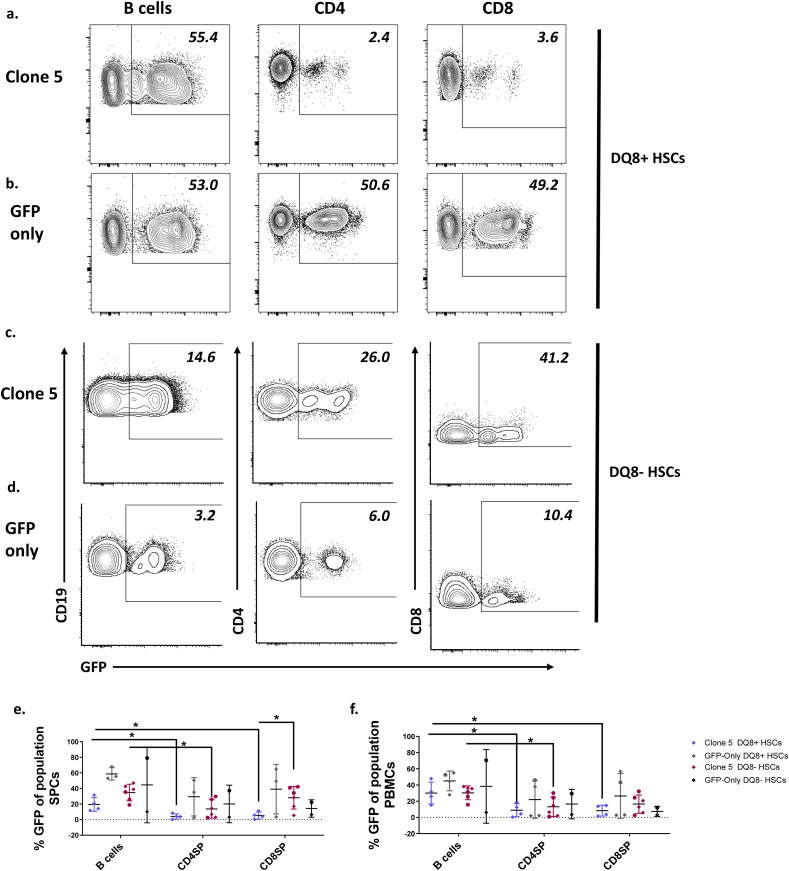
3.3. Peripheral T cells show reduced percentages of GFP+ cells compared to B cells in Clone 5 but not GFP-only animals
We assessed the percentage of GFP+ cells in peripheral populations. The percentage of GFP+ B cells was high in the blood and spleen of mice receiving Clone 5-transduced DQ8+ (Fig. 4a, e, f) and DQ8- (Fig. 4c, e, f) cells, but the percentages of GFP+ CD4 T cells were significantly lower than percentages of GFP+ B cells in Clone 5 mice receiving DQ8+ or DQ8- HSCs. Percentages of GFP+ CD4 T cells were higher in spleens of 3 of 6 Clone 5 DQ8- HSC mice than in spleens of Clone 5 DQ8+ HSC mice, in which the percentages were uniformly low (Fig. 4 e, f), possibly reflecting more escape from negative selection of Clone 5 T cells in mice lacking DQ8+ HSC-derived APCs. Additionally, a significant decrease in percentages of CD8 T cells expressing GFP was observed only in Clone 5 DQ8+ HSC and not in Clone 5 DQ8- HSC mice, consistent with deletion at the double positive stage of development only when DQ8+ thymic APCs are present. In contrast, mice receiving GFP-only DQ8+ (Fig. 4b, e, f) and DQ8- (Fig. 4d, e, f) HSCs showed similar percentages of GFP+ B and T cells and no significant difference in the percentage of GFP+ cells was observed between lineages. In Clone 5 mice with HLA-DQ8- HSCs, there was no significant difference in the percentage of GFP+ B cells, CD4 T cells, or CD8 T cells in the spleen or PBMCs in mice with and without the HLA-DQ8 transgene (Fig. 4e and f). These results are consistent with the interpretation that the decrease in GFP+ T cells is the result of increased negative selection of the transduced Clone 5 TCR compared to endogenously rearranged TCRs and that this deletion occurs at the double positive stage of thymocyte development only when HLA-DQ8+ human HSCs are present.
Fig. 4.
Reduced percentage of GFP+T cells compared to B cells in peripheral compartment in Clone 5 but not GFP-only mice. Representative FCM analysis of splenocytes (SPCs) of mice transplanted with a. Clone 5 DQ8+, b. GFP-only DQ8+, c. Clone 5 DQ8-, or d. GFP-only DQ8- HSCs. e, f. Percentage GFP+ of B cells, CD4, and CD8 T cells in e. spleen and f. blood of mice transplanted with Clone 5 or GFP-only transduced cells. n = 6 Clone 5 DQ8-, n = 4 Clone 5 DQ8+, n = 2 GFP-only DQ8-, n = 3 GFP-only DQ8+ ∗ = p < 0.05 by One-way ANOVA followed by Dunnett’s multiple comparisons test. Mice marked with a circle are transgenic for HLA-DQ8.
3.4. GFP+ CD4 T cells in Clone 5 mice are mainly Clone 5 TCR negative
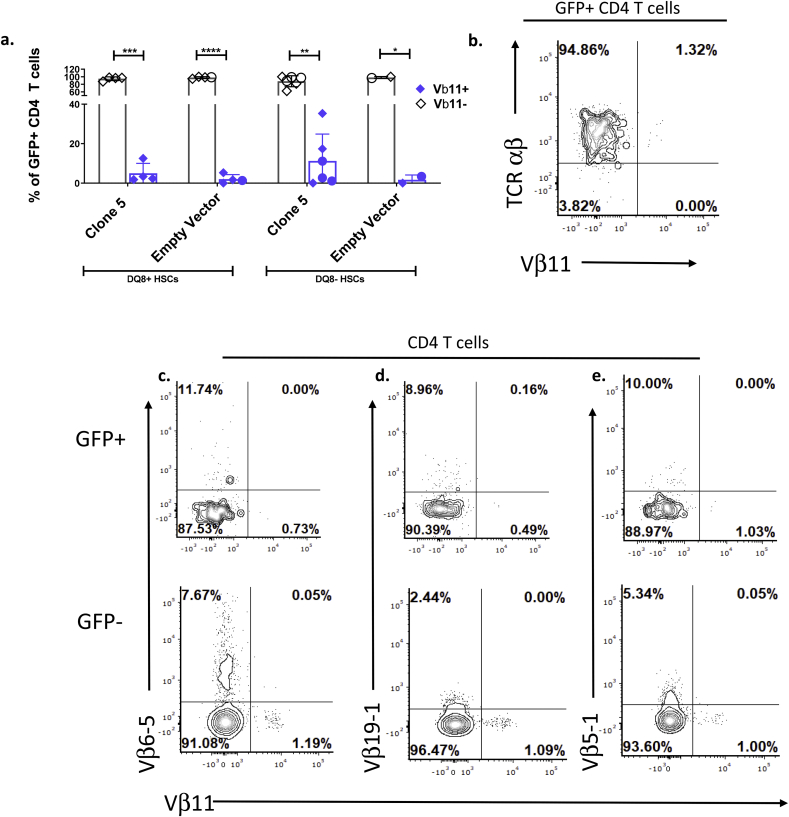
The small number of GFP+ CD4 T cells detected in the PBMCs of Clone 5 DQ8+ mice largely lacked expression of TCR Vβ11, the TCR Vβ chain used by the Clone 5 TCR, but still expressed TCR αβ (Fig. 5a and b). Expression of Vβ11 by GFP+ CD4 T cells in 3 of 4 Clone 5 DQ8+ HSC mice was similar to that of GFP+ CD4 T cells in GFP-only mice, which express Vβ11 as a result of its use in endogenous TCRs (Fig. 5a). The GFP+ peripheral CD4 cells that lacked expression of the Clone 5 TCR in Clone 5 DQ8+ mice expressed TCR αβ and CD3, indicating the presence of an expressed TCR (Fig. 5b, Supplemental Fig. 2a and b). To determine if these cells expressed other TCRs, the cells were stained with mAbs recognizing some common Vβ chains (Fig. 5c–e) [36]. The GFP+ CD4 T cells in Clone 5 DQ8+ HSC mice expressed these additional Vβ chains at similar levels as GFP- CD4 T cells within the same mouse. An increased percentage of Vβ11+ cells was detected in GFP hi CD4+ T cells compared to GFP lo cells (Supplemental Fig. 2c) suggesting that GFP lo cells may have lower expression of the transgene, allowing expression of endogenous TCR and escape from negative selection. There was no significant difference in the percentage of Vβ11+ GFP+ CD4 T cells in HLA-DQ8-transgenic mice and non-transgenic mice in the Clone 5 group receiving HLA-DQ8- HSCs (Fig. 5a).
Fig. 5.
GFP+ CD4 T cells in the periphery of Clone 5 DQ8+ mice mainly express alternative TCRs. a. Quantification of Vβ11+ and Vβ11- cells within total GFP+ CD4 T cells in peripheral blood of all mice at sacrifice. n = 6 Clone 5 DQ8-, n = 4 Clone 5 DQ8+, n = 2 GFP-only DQ8-, n = 3 GFP-only DQ8+b-e. Representative flow cytometry plot of b. TCR c. Vβ6-5, d. Vβ19-1, e. Vβ5-1, and Vβ11-expressing cells within GFP+ and GFP- CD4 T cells in the peripheral blood of a Clone 5 DQ8+ mouse. ∗∗ = p < 0.01, ∗ = p < 0.05, ∗∗∗ = p < 0.001, ∗∗∗∗ = P < 0.0001by paired t-test. Mice marked with a circle are transgenic for HLA-DQ8.
Thus, although small populations of GFP+ CD4 T cells were detectable in the periphery of Clone 5 mice, almost all of those T cells are likely to express endogenous TCRs. These results suggest that the small number of GFP+ CD4 cells that evade negative selection and appear in the periphery of these animals may do so by failing to express the Clone 5 transgenic TCR and thereby arranging endogenous TCR β chains in addition to α chains. Among mice receiving Clone 5 DQ8- HSCs, Vβ11 was expressed in peripheral CD4 cells at a higher frequency than the mean in Clone 5 DQ8+ mice in 3 of 6 animals(Fig. 5a), consistent with decreased negative selection efficiency and increased escape into the periphery of Clone 5 thymocytes when HLA-DQ8 is not expressed by HSCs.
4. Discussion
Our novel TCR-transgenic humanized mouse model provides a tool to study human thymic selection in a “natural” human thymic environment without exogenously-provided antigen. We report the first demonstration of negative selection of human autoreactive T cells to a naturally-expressed TRA. We also report that human HSC-derived intrathymic APCs are required for complete deletion of autoreactive T cells and mainly carry out this selection at the DP stage. In the absence of human HSCs expressing the required HLA molecule, negative selection can also occur at the CD4 SP stage, possibly as a result of HLA/antigen expression on mTECs.
Within the human thymus, evidence for negative selection included a striking absence of more mature medullary (CCR7+) Clone 5 thymocytes in mice receiving HLA-DQ8+ HSCs. In contrast, robust populations of CCR7+ medullary thymocytes were observed in Clone 5 transgene negative cells in Clone 5 HLA-DQ8+ mice and in transgene positive and negative cells in Clone 5 DQ8- HSC mice and GFP-only mice. Developing human thymocytes that expressed the Clone 5 TCR transgene in immune systems with HLA-DQ8+ HSCs included an increased percentage of cells expressing PD-1, a key marker of negative selection, in DP and SP thymocyte subsets compared to developing thymocytes lacking the transgene within the same mice. Collectively, these results are consistent with negative selection at both the DP and SP stages of development when the HLA-DQ8 presenting molecule is present on hematopoietically-derived APCs. Strong reliance on these hematopoeitcially-derived APCs is also seen in murine models, where half of total negative selection events rely on them [37]. Since insulin is expressed only by mTECs in the thymus under control of the AIRE transcription factor [1,38], the deletion of DP thymocytes, which reside predominantly in the thymic cortex, recognizing a TRA that is produced only by medullary thymocytes, is somewhat unexpected. While a murine study demonstrated that most thymocytes are negatively selected in the cortex, T cells specifically recognizing TRAs required trafficking to the medulla for complete deletion [37,39]. Our observation in a human immune system that most clonal deletion of TRA-specific thymocytes occurs in the cortex is therefore somewhat surprising. It is possible that APCs that pick up and process peripheral insulin in the pancreas migrate to the thymic cortex, where they interact with DP thymocytes and induce clonal deletion [40]. Trafficking to the cortex of medullary DCs picking up TRAs from mTECs could have a similar effect. Murine studies have demonstrated the capacity of APCs to pick up TRAs produced by mTECs and mediate negative selection in the cortex [39]. Alternatively, free insulin peptides produced by the pancreas may migrate to the thymic cortex, as immunogenic products of catabolized insulin have been detected in multiple lymphoid tissues of mice [41]. Comparison of mice with and without the HLA transgene that received HLA-DQ8- HSCs did not reveal a statistically significant impact of the transgene on selection of Clone 5 thymocytes. However, a larger study would be needed to definitively determine whether or not HLA-transgenic mouse APCs can influence selection of human thymocytes with this TRA specificity.
In contrast to these results, there was no increase in PD-1 expression on Clone 5 thymocytes in HLA-DQ8- HSC mice in the combined DP and SP subsets or the DP subset. Two of 3 animals did show an increase in PD-1 expression among Clone 5 GFP+ compared to GFP- CD4 SP thymocytes, suggesting that negative selection may take place at the SP but not the DP stage. Taken together, these phenotypic characteristics reflect the partial negative selection of Clone 5 T cells in the presence of HLA-DQ8- HSCs. Our studies do not distinguish whether this partial negative selection is mediated strictly by medullary TECs, which are known to produce insulin under control of the AIRE gene [1,38] and express the HLA-DQ8 presenting molecule in our system, or may be mediated by residual HLA-DQ8+ APCs that are carried in the fetal thymus graft. The evidence that these cells are deleted at the CD4 SP stage, later than the DP stage when Clone 5 cells in HLA-DQ8+ mice are deleted, and the presence of Clone 5 thymocytes expressing the medullary CCR7 marker only in mice with DQ8- HSCs, is consistent with the notion that, in the absence of HLA-DQ8+ APCs, mTECs that produce insulin are responsible for directly presenting the insB9-23 peptide on their HLA-DQ8 molecules, resulting in clonal deletion. Consistently, murine studies have demonstrated negative selection of TRA-specific thymocytes directly by mTEC presentation of the antigen [42].
In contrast to the analyses of thymocytes and peripheral T cells presented here, analysis of the relative size of each thymic subset (DN, DP, and SP) (data not shown) was relatively uninformative with respect to thymocyte selection, likely due to inherent variability of the efficiency of thymopoiesis between individual humanized mice and because thymic grafts in humanized mice eventually “burn out”, showing increasingly poor thymopoiesis at different late times post-transpalnt. These biological variables make it impossible to demonstrate group differences in thymocyte subsets as a result only of selection. In contrast, the thymic readout parameters presented here, paired with the peripheral data, seem to provide an accurate measure of selection outcomes independent of these biological variables.
Peripheral evidence for intrathymic deletion of Clone 5 T cells included reduced percentages of GFP+ CD4 T cells in the spleen and peripheral blood of Clone 5 DQ8+ and DQ8- HSC animals compared to GFP+ B cells in the same mice. This decrease was not as marked in Clone 5 DQ8- HSC mice, consistent with the less complete negative selection. No such difference was seen in GFP-only mice, supporting the conclusion that these differences were due to negative selection of Clone 5 T cells. Clone 5 DQ8+ HSC mice lacked both CD8 and CD4 T cells expressing the Clone 5 TCR in the peripheral tissues. This observation is consistent with negative selection of Clone 5 T cells at the DP stage when HLA-DQ8+ HSC-derived intrathymic APCs are present, as also suggested by the observed upregulation of PD-1 in DP transgenic cells compared to non-transgenic cells in the same mice.
Within the small population of transgenic GFP+ CD4 T cells that were found in the periphery of Clone 5 DQ8+ and DQ8- HSC mice, only a low percentage expressed the Vβ chain used by the Clone 5 TCR. Although still significantly decreased compared to Vβ11- cells in the same mice, levels of Vβ11+ cells in some Clone 5 DQ8- HSC mice were higher than those in Clone 5 DQ8+ HSC mice. The GFP+ CD4 T cells in the periphery of Clone 5 DQ8+ HSC mice were found to express other TCR Vβ chains. Expression of an alternative TCRα chain in TCR-transgenic humanized mice has been previously reported [10], but there is no precedent for alternative β chain expression in TCR Tg humanized mice. Although β chains are known to be subject to allelic exclusion in mice, so that endogenous TCR β genes do not rearrange in animals with a transgenic β chain, exceptions to this rule in mice have been reported [43]. It is conceivable in our model that alternative TCR-expressing GFP+ CD4 T cells in Clone 5 mice form a majority of the peripheral population because GFP+ CD4 T cells expressing the Clone 5 TCR are so efficiently negatively selected before egressing into the periphery. These data suggest that a subset of TCR transgenic human thymocytes is able to bypass allelic exclusion of the TCRβ chain and it is currently unclear whether this is a “natural” phenomenon or an artifact of TCR-transgenic systems.
The model TCR used in this study, Clone 5, is an insulin B:9–23 peptide-autoreactive TCR isolated from the peripheral blood of a T1D patient [21,22]. Clone 5 T cells are capable of initiating autoimmune diabetes in humanized mice when injected into the periphery of HLA-DQ8 transgenic mice with polyclonal human T cell repertoires under conditions of islet stress and peptide immunization [24]. Further, the insulin B:9–23 antigen, which is conserved between mice and humans, is absolutely required for development of diabetes in NOD mice [23]. The B:9–23 antigen is known to be presented in multiple registers, some weakly bound, on HLA-DQ8 [44]. This multiplicity of registers and weak binding to HLA molecules is characteristic of other autoantigens and potentially allows autoreactive T cells to escape negative selection in autoimmune-prone individuals [45,46]. Nevertheless, we have demonstrated efficient negative selection of Clone 5 in healthy human immune systems derived from thymi and HSCs expressing the HLA restriction element of the TCR.
Our studies demonstrate that human insulin-reactive T cells exhibit markers of thymic negative selection previously described. PD-1 is upregulated on human insulin-reactive thymocytes, indicative of their negative selection to a natural ligand. CCR7, a marker of medullary thymocytes [34,35], was also used to delineate the negative selection observed here. We have also demonstrated that HSC-derived intrathymic APCs are required for complete negative selection of human TRA-specific TCRs at the DP stage and in their absence, partial clonal deletion likely occurs in the thymic medulla. It is quite possible that the negative selection efficiency of autoreactive T cells differs depending on the antigen specificity and source of antigen.
The importance of negative selection in immune system development makes knowledge of this process in humans key to understanding autoimmune disease. While the human immune systems used in these studies here are of fetal origin, analysis of the selection of known autoreactive TCRs in the context of immune systems from patients with autoimmune diseases will allow us to see if central tolerance is defective in such patients. The Personalized Immune (PI) mouse model, which allows for de novo generation of immune systems derived from transplantation of adult bone marrow-derived CD34+ cells and a human fetal thymus fragment into immunodeficient mice [32], will allow assessment of HSC-intrinsic genetic abnormalities that may impair negative selection in patients with autoimmune diseases.
Author contributions
GN and MS designed the experiments. GN and CB established and optimized the experimental systems. GN, RM, ND, CB, MK, HL, and EC performed the experiments. GN and RM analyzed the data. RM, GN, and MS wrote the manuscript. RJC, MN and BR provided resources and expertise for the project. MS supervised the project.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the following NIH grants: R01 DK103585, UC4 DK104207, R21 AI146828, and U01 DK123559. Research reported in this publication was performed in the CCTI Flow Cytometry Core and supported in part by the Office of the Director, National Institutes of Health under awards S10RR027050. GN was supported by NIH T32 5T32AI106711. RM was supported by the NIH TL1 5TL1TR001875-03. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The authors also thank Dr. Robert Winchester and Jing Bi for their invaluable assistance performing Sanger allele-level HLA typing for fetal tissue.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtauto.2020.100061.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Anderson M.S., Venanzi E.S., Klein L., Chen Z., Berzins S.P., Turley S.J. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 2.Liston A., Lesage S., Wilson J., Peltonen L., Goodnow C.C. Aire regulates negative selection of organ-specific T cells. Nat. Immunol. 2003;4:350–354. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 3.Takaba H., Morishita Y., Tomofuji Y., Danks L., Nitta T., Komatsu N. Fezf2 orchestrates a thymic program of self-antigen expression for immune tolerance. Cell. 2015;163:975–987. doi: 10.1016/j.cell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Brocker T. The role of dendritic cells in T cell selection and survival. J. Leukoc. Biol. 1999;66:331–335. doi: 10.1002/jlb.66.2.331. [DOI] [PubMed] [Google Scholar]
- 5.Brocker T., Riedinger M., Karjalainen K. Targeted expression of major histocompatibility complex (MHC) class II molecules demonstrates that dendritic cells can induce negative but not positive selection of thymocytes in vivo. J. Exp. Med. 1997;185:541–550. doi: 10.1084/jem.185.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee T., Sprouse M.L., Banerjee P., Bettini M., Bettini M.L. Ectopic expression of self-antigen drives regulatory T cell development and not deletion of autoimmune T cells. J. Immunol. 2017;199:2270–2278. doi: 10.4049/jimmunol.1700207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldwin T.A., Hogquist K.A. Transcriptional analysis of clonal deletion in vivo. J. Immunol. 2007;179:837–844. doi: 10.4049/jimmunol.179.2.837. [DOI] [PubMed] [Google Scholar]
- 8.Klein L., Hinterberger M., Wirnsberger G., Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat. Rev. Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 9.Baccala R., Vandekerckhove B.A., Jones D., Kono D.H., Roncarolo M.G., Theofilopoulos A.N. Bacterial superantigens mediate T cell deletions in the mouse severe combined immunodeficiency-human liver/thymus model. J. Exp. Med. 1993;177:1481–1485. doi: 10.1084/jem.177.5.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Teteloshvili N., Tan S., Rao S., Han A., Yang Y.G. Humanized mice reveal new insights into the thymic selection of human autoreactive CD8(+) T cells. Front. Immunol. 2019;10:63. doi: 10.3389/fimmu.2019.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagamine K., Peterson P., Scott H.S., Kudoh J., Minoshima S., Heino M. Positional cloning of the APECED gene. Nat. Genet. 1997;17:393–398. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 12.Cheng M.H., Shum A.K., Anderson M.S. What’s new in the Aire? Trends Immunol. 2007;28:321–327. doi: 10.1016/j.it.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Michels A.W., Landry L.G., McDaniel K.A., Yu L., Campbell-Thompson M., Kwok W.W. Islet-derived CD4 T cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2017;66:722–734. doi: 10.2337/db16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jingwu Z., Medaer R., Hashim G.A., Chin Y., van den Berg-Loonen E., Raus J.C. Myelin basic protein-specific T lymphocytes in multiple sclerosis and controls: precursor frequency, fine specificity, and cytotoxicity. Ann. Neurol. 1992;32:330–338. doi: 10.1002/ana.410320305. [DOI] [PubMed] [Google Scholar]
- 15.Law S.C., Street S., Yu C.H., Capini C., Ramnoruth S., Nel H.J. T-cell autoreactivity to citrullinated autoantigenic peptides in rheumatoid arthritis patients carrying HLA-DRB1 shared epitope alleles. Arthritis Res. Ther. 2012;14:R118. doi: 10.1186/ar3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zucchelli S., Holler P., Yamagata T., Roy M., Benoist C., Mathis D. Defective central tolerance induction in NOD mice: genomics and genetics. Immunity. 2005;22:385–396. doi: 10.1016/j.immuni.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Kishimoto H., Sprent J. A defect in central tolerance in NOD mice. Nat. Immunol. 2001;2:1025–1031. doi: 10.1038/ni726. [DOI] [PubMed] [Google Scholar]
- 18.Habib T., Funk A., Rieck M., Brahmandam A., Dai X., Panigrahi A.K. Altered B cell homeostasis is associated with type I diabetes and carriers of the PTPN22 allelic variant. J. Immunol. 2012;188:487–496. doi: 10.4049/jimmunol.1102176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugliese A., Zeller M., Fernandez A., Jr., Zalcberg L.J., Bartlett R.J., Ricordi C. The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat. Genet. 1997;15:293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 20.Schuster C., Gerold K.D., Schober K., Probst L., Boerner K., Kim M.J. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity. 2015;42:942–952. doi: 10.1016/j.immuni.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eerligh P., van Lummel M., Zaldumbide A., Moustakas A.K., Duinkerken G., Bondinas G. Functional consequences of HLA-DQ8 homozygosity versus heterozygosity for islet autoimmunity in type 1 diabetes. Gene Immun. 2011;12:415–427. doi: 10.1038/gene.2011.24. [DOI] [PubMed] [Google Scholar]
- 22.Michels A.W., Ostrov D.A., Zhang L., Nakayama M., Fuse M., McDaniel K. Structure-based selection of small molecules to alter allele-specific MHC class II antigen presentation. J. Immunol. 2011;187:5921–5930. doi: 10.4049/jimmunol.1100746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakayama M., Abiru N., Moriyama H., Babaya N., Liu E., Miao D. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature. 2005;435:220–223. doi: 10.1038/nature03523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan S., Li Y., Xia J., Jin C.H., Hu Z., Duinkerken G. Type 1 diabetes induction in humanized mice. Proc. Natl. Acad. Sci. U. S. A. 2017;114:10954–10959. doi: 10.1073/pnas.1710415114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa F., Yasukawa M., Lyons B., Yoshida S., Miyamoto T., Yoshimoto G. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain(null) mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan P., Wang L., Diouf B., Eguchi H., Su H., Bronson R. Induction of human T-cell tolerance to porcine xenoantigens through mixed hematopoietic chimerism. Blood. 2004;103:3964–3969. doi: 10.1182/blood-2003-10-3697. [DOI] [PubMed] [Google Scholar]
- 27.Lan P., Tonomura N., Shimizu A., Wang S., Yang Y.G. Reconstitution of a functional human immune system in immunodeficient mice through combined human fetal thymus/liver and CD34+ cell transplantation. Blood. 2006;108:487–492. doi: 10.1182/blood-2005-11-4388. [DOI] [PubMed] [Google Scholar]
- 28.Breckpot K., Dullaers M., Bonehill A., van Meirvenne S., Heirman C., de Greef C. Lentivirally transduced dendritic cells as a tool for cancer immunotherapy. J. Gene Med. 2003;5:654–667. doi: 10.1002/jgm.400. [DOI] [PubMed] [Google Scholar]
- 29.Nauman G., Borsotti C., Danzl N., Khosravi-Maharlooei M., Li H.-W., Chavez E. Reduced positive selection of a human TCR in a swine thymus using a humanized mouse model for xenotolerance induction. Xenotransplantation. 2019 doi: 10.1111/xen.12558. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khosravi-Maharlooei M., Hoelzl M., Li H., Madley R., Waffarn E., Danzl N. Rapid thymectomy of NSG mice to analyze the role of native and grafted thymi in humanized mice. Eur. J. Immunol. 2019 doi: 10.1002/eji.201948205. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalscheuer H., Onoe T., Dahmani A., Li H.W., Holzl M., Yamada K. Xenograft tolerance and immune function of human T cells developing in pig thymus xenografts. J. Immunol. 2014;192:3442–3450. doi: 10.4049/jimmunol.1302886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalscheuer H., Danzl N., Onoe T., Faust T., Winchester R., Goland R. A model for personalized in vivo analysis of human immune responsiveness. Sci. Transl. Med. 2012;4:125ra30. doi: 10.1126/scitranslmed.3003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dehoux Jp T.S., Dewolf N., Otsuka M., Oike F., Famar F., de la Parra B., Latinne D., Bazin H., Gianello P. Effects on human and nonhuman primate immune response of a new rat anti-CD2 monoclonal antibody. Transplantation. 2000;69:2622–2633. doi: 10.1097/00007890-200006270-00024. [DOI] [PubMed] [Google Scholar]
- 34.Kurobe H., Liu C., Ueno T., Saito F., Ohigashi I., Seach N. CCR7-dependent cortex-to-medulla migration of positively selected thymocytes is essential for establishing central tolerance. Immunity. 2006;24:165–177. doi: 10.1016/j.immuni.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 35.Kwan J., Killeen N. CCR7 directs the migration of thymocytes into the thymic medulla. J. Immunol. 2004;172:3999–4007. doi: 10.4049/jimmunol.172.7.3999. [DOI] [PubMed] [Google Scholar]
- 36.Khosravi-Maharlooei M., Obradovic A., Misra A., Motwani K., Holzl M., Seay H.R. Crossreactive public TCR sequences undergo positive selection in the human thymic repertoire. J. Clin. Invest. 2019;129:2446–2462. doi: 10.1172/JCI124358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breed E.R., Watanabe M., Hogquist K.A. Measuring thymic clonal deletion at the population level. J. Immunol. 2019;202:3226–3233. doi: 10.4049/jimmunol.1900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabater L., Ferrer-Francesch X., Sospedra M., Caro P., Juan M., Pujol-Borrell R. Insulin alleles and autoimmune regulator (AIRE) gene expression both influence insulin expression in the thymus. J. Autoimmun. 2005;25:312–318. doi: 10.1016/j.jaut.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Nitta T., Nitta S., Lei Y., Lipp M., Takahama Y. CCR7-mediated migration of developing thymocytes to the medulla is essential for negative selection to tissue-restricted antigens. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17129–17133. doi: 10.1073/pnas.0906956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bonasio R., Scimone M.L., Schaerli P., Grabie N., Lichtman A.H., von Andrian U.H. Clonal deletion of thymocytes by circulating dendritic cells homing to the thymus. Nat. Immunol. 2006;7:1092–1100. doi: 10.1038/ni1385. [DOI] [PubMed] [Google Scholar]
- 41.Wan X., Zinselmeyer B.H., Zakharov P.N., Vomund A.N., Taniguchi R., Santambrogio L. Pancreatic islets communicate with lymphoid tissues via exocytosis of insulin peptides. Nature. 2018;560:107–111. doi: 10.1038/s41586-018-0341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hubert F.X., Kinkel S.A., Davey G.M., Phipson B., Mueller S.N., Liston A. Aire regulates the transfer of antigen from mTECs to dendritic cells for induction of thymic tolerance. Blood. 2011;118:2462–2472. doi: 10.1182/blood-2010-06-286393. [DOI] [PubMed] [Google Scholar]
- 43.Auger J.L., Haasken S., Steinert E.M., Binstadt B.A. Incomplete TCR-beta allelic exclusion accelerates spontaneous autoimmune arthritis in K/BxN TCR transgenic mice. Eur. J. Immunol. 2012;42:2354–2362. doi: 10.1002/eji.201242520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J., Chow I.T., Sosinowski T., Torres-Chinn N., Greenbaum C.J., James E.A. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14840–14845. doi: 10.1073/pnas.1416864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith K.J., Pyrdol J., Gauthier L., Wiley D.C., Wucherpfennig K.W. Crystal structure of HLA-DR2 (DRA∗0101, DRB1∗1501) complexed with a peptide from human myelin basic protein. J. Exp. Med. 1998;188:1511–1520. doi: 10.1084/jem.188.8.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y., Huang Y., Lue J., Quandt J.A., Martin R., Mariuzza R.A. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24:2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.