Graphical abstract
Abbreviations: 2,4-D, 2,4-dichlorophenoxy acetic acid; COW, Correlation Optimized Warping; CGA, chlorogenic acid; DV1, first derivatives; DV2, second derivatives; DW, dry weight; DHQ, dihydroquercetin; FT-NIR, fourier transform near-infrared spectroscopy; FW, fresh weight; GAE, gallic acid equivalents; KT, Kinetin; MeJA, Methyl jasmonate; MVA, multivariate analysis; OPLS-DA, orthogonal partial least square-discriminant analysis; PC, phenolic compounds; PCA, principal component analysis; PLS, partial least square-discriminant analysis; RP-HPLC, reversed phase-high performance liquid chromatography; SA, salicylic acid; SG, Savitzky Golay; SH, Schenk and Hildebrandt; SNV, Standard Normal Variate
Keywords: Thevetia peruviana, Plant cell culture, FT-NIR, RP-HPLC, Multivariate analysis
Highlights
-
•
Near infrared spectroscopy was used for the detection of phenolic content in plant cell cultures.
-
•
Multivariate analysis applied to HPLC data was satisfactory to determine changes in the phenolic profile.
-
•
Dihydroquercetin increased significantly in T. peruviana cultures treated with SA/MeJA.
-
•
Chlorogenate and dihydroquercetin are possible biomarkers of the MeJA effects in T. peruviana.
Abstract
Plant cell suspension culture of T. peruviana is a feasible biotechnological platform for the production of secondary metabolites with anti-proliferative/cytotoxic activity, as phenolic compounds (PC); however, different in in vitro growth conditions may affect the production, demanding strategies to increase the metabolite biosynthesis, as well as the development of sensitive and rapid analytical methods for metabolite monitoring. The Fourier transform near-infrared (FT-NIR) spectroscopy and Reversed-phase high-performance liquid chromatography (RP-HPLC) combined with Multivariate analysis (MVA) were used to detect significant differences in the PC production in cultures treated with two elicitors. The results suggest that the FT-NIR-MVA is useful for discriminating samples according to the treatment, showed significant influence of the PC signal. RP-HPLC-MVA showed that the elicitor effect occurs at 72 h post-elicitation. Detection of dihydroquercetin (maximum concentration = 12.59 mg/L), a flavonoid with anti-cancer properties, is highlighted. Future studies will be aimed at scaling this culture to increase the productivity of dihydroquercetin.
1. Introduction
In vitro plant cell suspension cultures are a biotechnological strategy widely used to increase the production of biomass and secondary metabolites of pharmaceutical value [1,2]. Thevetia peruviana (Pers.) K. Schum is an ornamental shrub belonging to the Apocynaceae family grown in Central and South America [3]. This plant has important biological properties, which include cardiac [4], anti-cancer [5,6], antimicrobial [7], antioxidant [8] and antiviral activity [9]. Many of these biological activities have been attributed to the presence of secondary metabolites such as cardiac glycosides and phenolic compounds (PC); nevertheless, obtaining these metabolites from in vivo plants implies problems such as availability, productivity and reproducibility; therefore, it becomes necessary to have a continuous source of quality biological material for extraction, purification and development of potential therapeutic agents. Consequently, plant cell suspension culture of T. peruviana was established in our laboratory [10], demonstrating stability in kinetic parameters of cell growth (duplication time, specific growth and substrate consumption rates) over the years [11].
On the other hand, phytohormones as salicylic acid (SA) y and methyl jasmonate (MeJA) have been used as exogenous elicitors in in vitro plant cell cultures [12]. These signaling molecules can trigger defense responses at a low concentration, promoting the biosynthesis of secondary metabolites of biotechnological interest such as PC through the activation of specific metabolic pathways (e.g. shikimate pathway and phenyl propanoids) [13,14]. A recently study showed that cell suspension culture of T. peruviana respond to SA and MeJA treatment producing changes in the content and chromatographic profile of PC [15]. However, analysis of complex samples (e.g. plant extracts) using univariate analysis techniques have limitations, in terms of the biological information that can be extracted, hindering slight changes detection between treatments; in addition, the quantitative analysis is restricted to a few compounds of known identity, leading unknown analytes, that could be important markers of the cellular response to treatment, being ignored.
One solution to these drawbacks is the application of multivariate analysis (MVA) to the data. Currently, it is common to use chromatographic or spectroscopic analytical techniques combined with MVA to extract relevant biological information from living organisms [16,17]. This approach is widely used in plant research; for example, in classification studies according to geographical origin [18], species [19] and cultivars discrimination [20], metabolic evaluation of genetically modified plants [21] and prediction of chemical quality components [22], among others. In the same way, the PC chromatographic profile, combined with MVA, has been used in plant classification studies according to infection susceptibility [23] and quality of medicinal plants [24], proving to be an effective tool for these purposes.
On the other hand, non-destructive analytical techniques, such as Fourier transform infrared spectroscopy (FT-IR) [25,26], have been used together with chemometric analysis to discriminate and classify plants. Although these techniques have lower sensitivity, compared with chromatographic ones, they offer the advantage of being quick and economical, since no laborious preparation of samples is required, nor is the use of reagents or solvents, therefore, if the objective is to perform a rapid discriminant analysis, these techniques should be considered. In our knowledge, the application of infrared spectroscopy to the investigation of plant cell suspension cultures has focused on the quantification of products [27] and the monitoring of kinetic parameters of the culture, such as substrate consumption rate [28].
The purpose of the present study was to evaluate the utility of FT-NIR spectroscopy and RP-HPLC combined with MVA, as an alternative method to determinate the elicitation effect on PC production in cell suspension cultures of T. peruviana.
2. Materials and methods
2.1. Cell suspension cultures and elicitor treatments
Cell suspensions were established from 10 g of fresh friable callus (g FW) in 100 mL of Schenk and Hildebrandt (SH) sterile medium [29] supplemented with 2 mg/L 2,4-Dichlorophenoxyacetic acid (2,4-D), 0.5 mg/L kinetin (KT), 30 g/L sucrose, and 1 g/L myo-inositol (pH 5.8) in 250 mL flasks. Cell suspension cultures were maintained in an orbital shaker at 110 rpm (New Brunswick™ Innova® 2300), natural photoperiod (12 h light/12 h dark) and 25 °C. Sub-cultures were made every 2 weeks [10].
In order to increase and stabilize the production of PC, cell suspension cultures were treated with two elicitors, SA and MeJA. Elicitor concentrations (300 μM SA, 3 μM MeJA and the combination 300 μM SA/3 μM MeJA), as well as elicitor addition time (day 4 of culture) were previously established [15]. The elicitation experiments were carried out using 6-day old cell suspensions in 250 mL flasks with 100 mL of SH liquid sterile medium, under the same conditions described above. Elicitors were prepared in an aqueous solution of 50 % ethanol (v/v) and filtered through Millipore membranes of 0.45 μm (Minisart, Sartorius, Germany) before being added to cell suspensions. Control (suspension cells without elicitor) and treatments with SA, MeJA, SA/MeJA were harvested at 24, 48, 72, 96 and 120 h post-elicitation for a total of 20 treatments. All the experiments were carried out in triplicate. The harvested cells were vacuum filtered and subjected to drying in a convection oven at 60 °C for 48 h, then was pulverized using a mortar, stored and protected from light at −20 °C.
2.2. FT-NIR analysis
This analysis was used as an exploratory method to visualize changes in the spectral profile of the cell suspension cultures of T. peruviana associated with metabolic responses to treatment with different elicitors.
2.2.1. Samples
Dry biomass was analyzed in an infrared spectrometer (BUCHI NIR Master™ FT-NIR Spectrometer). Spectra were obtained by scanning at 4000 - 10,000 cm−1 intervals at 4 cm−1 resolution. Sixty-four scans were obtained per reading and averaged into one spectrum.
2.2.2. Processing of spectral data
Absorption at wavenumber > 7000 cm−1 were negative, so the region between 4000 and 7000 cm−1 was selected to construct a two-way array raw data, whose dimensions were 20 observations (row) x 750 variables (columns). Prior to the MVA, the spectral data were pre-processed with the purpose to remove spectral variations that are unrelated to the samples such as background noise, solving superimposed signals and suppress effects of light scattering caused by the heterogeneity of the samples; also to enhance subtle differences between samples, improving discriminating features [30]. Pre-processing was performed as implemented in SIMCA 14.1.0.2047 software (MKS U-metrics, Sweden) using first and second derivatives (DV1 and DV2) and numerical algorithms such as Savitzky-Golay (SG) and SNV (Standard Normal Variate). The quality of each model was determined by the goodness of fit parameter (R2) and prediction parameter (Q2). After selecting the best processing model, the data from the X matrix was normalized and scaled using the Pareto method.
2.2.3. MVA of spectral data
From the processed data, a principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA) and orthogonal partial least squares discriminant analysis (OPLS-DA) were performed. PCA was used to establish the presence of relevant and interpretable patterns in the data and to detect outliers through Hottelling T2 analysis (95 % of the confidence interval), while PLS-DA and OPLS-DA were used to recognize possible patterns that maximize the separation between samples [31]. PCA, PLS-DA and OPLS-DA models were generated using SIMCA software. Score plot will be used to quickly visualize the discrimination between treatments, while the loading plot will allow identifying variables with the greatest value for discriminating the treatments.
2.3. RP-HPLC analysis
In order to improve the sensitivity and selectivity in the PC analysis, samples were also analyzed by liquid chromatography. RP-HPLC is one of the most widely used methods to identify and quantify of PC in plants and naturals products [32]. In addition, chemometrics analysis of chromatographic data could provide more biological information about the effect of elicitors on the PC production.
The PCs were extracted from the biomass using a previously described protocol [15]. Briefly, dried and pulverized biomass was extracted using 50 % ethanol solution with 50:1 solvent: sample ratio, in an ultrasonic bath (frequency 40 kHz) at 30 °C for 30 min, followed by centrifugation at 3000 rpm for 10 min. One milliliter of each supernatant was diluted to 5 mL with ethanol solution and then filtered through 0.45 μm membranes. Chromatographic profiling was performed in a Shimadzu Prominence HPLC (Shimadzu Corporation, Kyoto, Japan), equipped with an isocratic pump LC-20 AT, an auto- sampler SIL-20AC, and a diode array detector SPD-M20 A (190−800 nm). The column used was a LiChrospher® 250−4 RP-18 (5 μm) (Merck S.A). Analysis conditions were: 28 °C, flow rate of 1.5 mL min−1 and injection volume of 20 μL. The mobile phase consisted of: phase A (formic acid 1% in water) and B (formic acid 1% in acetonitrile). The elution gradient was as follows: 0 min (80 % A + 20 % B); 7 min (75 % A + 25 % B); 13 min (70 % A + 30 % B); 7 min (65 % A + 35 % B); 3 min (80 % A + 20 % B). Data were collected every 640 milliseconds (msec). The monitoring wavelength selected was 280 nm.
2.3.1. Chromatographic and MAV data processing
Absorbance at 280 nm as function of time was exported as 2D ASCII files for each sample and a matrix of 20 observations x 4686 variables were built. Prior to the MVA, the HPLC data were preprocessed in order to correct differences in the profile of the chromatograms related to aspects such as retention time drift or changes in peak shape. These changes usually occur due to gradual contamination of the column during the consecutive analysis of many samples or due to differences in operating conditions [33]. The alignment of the chromatographic peaks was performed using the Correlation Optimized Warping algorithm (COW) from Warping Toolbox provided by The Quality and Technology Webs [34]; this algorithm was developed in MATLAB R2014a 8.3.0.532 software (The MathWorks, Inc., Natick, Massachusetts, USA). Then, chromatographic data were imported into SIMCA 14.1.0.2047 software (MKS U-metrics, Sweden) for normalization and scaling using the Pareto method. PCA, PLS-DA and OPLS-DA analyzes were then applied.
2.3.2. Identification and quantification of phenolic and flavonoid compounds
Time retention (tR) and the UV–vis absorbance of the peaks present in samples and those obtained from some analytical standards of phenolic and flavonoid compounds previously reported in T. peruviana [35] and others available in our laboratory were compared. Quantification of the identified compounds was performed, using calibration curves with the reference standards. The results obtained were presented as values of the mean ± standard deviation (SD). The differences between treatments were evaluated with one-way ANOVA, with a level of significance of 0.05. Also, multiple comparisons were made (in pairs) using Tukey's Honestly Significant Differences (HSD) method. These analyzes were carried out with the R studio version 1.1.383.
3. Results and discussion
3.1. FT-NIR and MVA of the spectral data
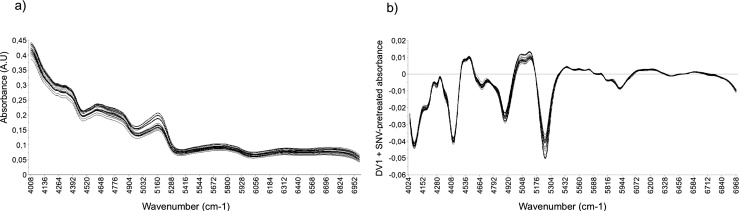
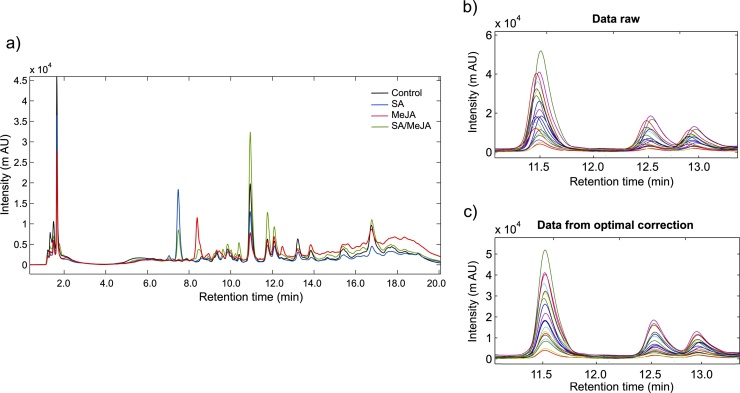
Fig. 1a shows the overlap of the NIR spectra average of all T. peruviana samples treated with different elicitors and the control (raw data). The spectra superposition allow to visualize the great similarity in the spectral profile of the different samples, as well as the presence of regions where there are variations in bands intensity; however, some of these variations may be caused by artifacts or imperfections of the spectrum. In order to remove the spectral variations not attributable to samples, data was pre-processed using different mathematical algorithms. This step allows the generation of reliable models that are interpretable using MVA techniques. The combined use of DV1 and SNV was the best method for processing the spectra (Fig. 1b), generating a PCA model that reduced data dimensionality to seven main components (R2X (cum) = 0.985 and Q2 (cum) = 0.962). SNV is a common method of infrared spectrum processing [30,36], its objective is to correct the dispersion effects of signals, being used in multivariate regression studies, in classification models and exploratory analyzes. In our case, the application of the SNV algorithm to the data helped correct the spectra baseline, while DV1 improved the separation of possible superimposed signals.
Fig. 1.
FT-NIR spectra of T. peruviana samples treated with SA, MeJA, SA/MeJA and control. (a) Raw NIR spectra. (b) NIR spectra pretreated by the 1 st derivative (DV 1) and subsequent Standard Normal Variate (SNV) method.
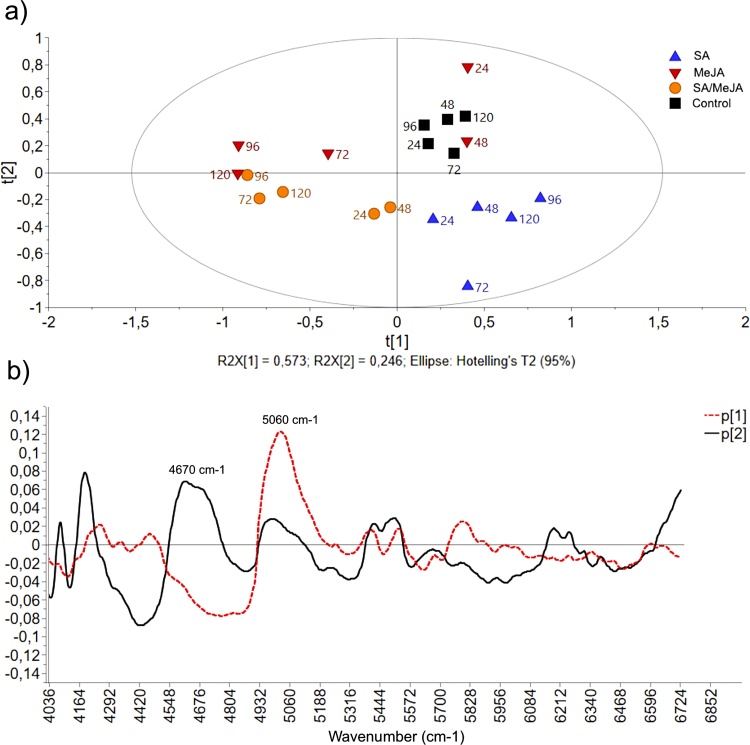
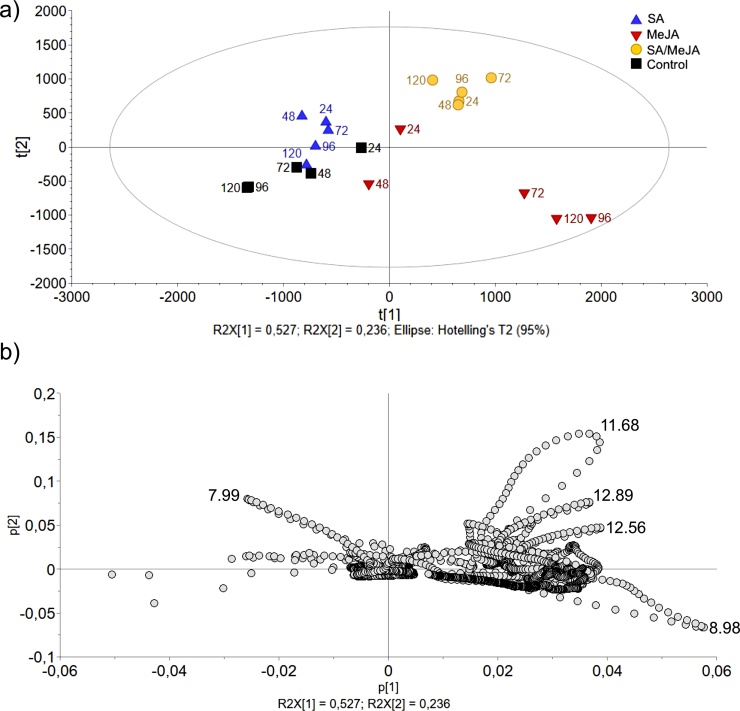
Fig. 2a shows the score plot using the first two principal components, which represent 57.3 % and 24.6 % of the total variation of data, respectively. The existence of a distribution pattern of the scores according to the treatment was observed. The samples corresponding to treatment with SA/MeJA and the control group were separated by the first t[1] component; while, samples corresponding to treatments with SA and SA/MeJA were separated from the control group by the second t[2] component. Additionally, the scores within the MeJA treatment exhibited a broad distribution limit; thus the samples corresponding at 24 h and 48 h were grouped close to the control, while samples harvested at 72, 96 and 120 h were segregated in a different group, which suggests that the elicitor effect of MeJA occurs after 72 h.
Fig. 2.
PCA of FT-NIR data for T. peruviana biomass collected at different times (24, 48, 72, 96 and 120 h) after the treatment with SA, MeJA, SA/MeJA and control. (a) PCA score plot indicating variations in the spectra profiles of samples, R2X [1] and R2X [2] correspond to the fractions of the variation of the X variables explained by the model on the first and second component, respectively. The ellipse represents Hotelling's T2, with 95 % confidence; (b) Loadings plot of the two principal components of PCA (p[1] and p[2]), most significant spectral variables for the samples separation in the PCA are located at 5060 cm−1 and 4670 cm−1.
The loadings plot of PCA (Fig. 2b) showed that the most significant spectral variables for the samples separation in PCA were distributed mainly in the region of 4500 - 5200 cm−1. The first main component (57.3 %) emphasized the spectral signal centered at 5060 cm−1, corresponding to the combination of stretch and flexion of the −OH of water molecules; specifically, this signal has been assigned to the formation of hydrogen bonds of water to carbonyl oxygen in the central skeleton of proteins [37]. The second main component (24.6 %) highlighted a spectral signal centered at 4670 cm−1, this signal is characteristic of the CH bond of phenolic species [38]. This result confirms that treatments with SA and MeJA produces qualitative and quantitative changes in phenolic metabolism of cell suspension cultures of T. peruviana; an effect on the metabolism of proteins is also suggested, which should be confirmed by metabolomic studies.
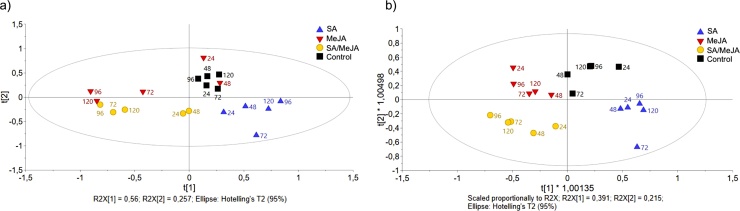
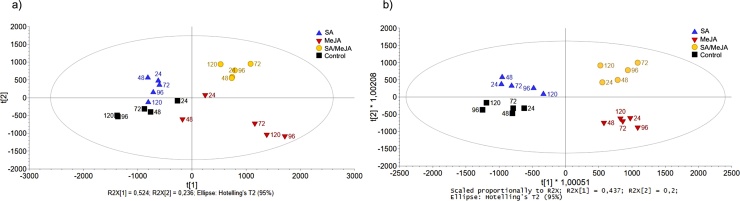
A subsequent analysis of the spectral data using the supervised PLS-DA method supports what is observed in the PCA. The PLS-DA model reduced the dimensionality of the spectral data to five main components that explained 99.8 % of the variance (R2X = 0.958, R2Y = 0.867 and Q2X = 0.712). The score plot (Fig. 3a) of PLS-DA showed a better grouping of samples according to the treatment and confirmed the relevance of the second main component for the separation of samples treated with SA and MeJA. In addition, OPLS-DA generated a model (R2X = 0.941, R2Y = 0.826 and Q2R = 0.681) that reduced the dimensionality of data to three predictive components and one orthogonal component (3 + 1+0). The two-dimensional score sequence showed that the model reduces the effect of the elicitor time exposure, which explains the lower segregation of samples within the same treatment (Fig. 3b). Both PLS-DA and OPLS-DA are powerful statistical modeling tools commonly used in classification and discrimination studies between experimental groups; however, these techniques can force the separation of groups, generating erroneous statistical interpretations; especially when the PCA does not expose separation of groups [39]. In the present study, PCA showed the existence of a separation pattern of the samples according to the treatment, so it is assumed that results of the supervised analyzes are reliable.
Fig. 3.
Two-dimensional score sequences for FT-NIR spectra for T. peruviana biomass collected at different times (24, 48, 72, 96 and 120) after the treatment with elicitors. (a) PLS-DA score plot and (b) OPLS-DA score plot. R2X [1] and R2X [2] correspond to the fractions of the variation of the X variables explained by the model on the first and second component, respectively. The ellipse represents the Hotelling T2 with 95 % confidence.
NIR spectroscopy characterizes the material based on its absorption in a range of ∼ 4000–12,500 cm−1, this range corresponds to overtones and combinations of absorption bands of the infrared fundamental vibrations; so that, the NIR spectrum of a given sample is considered to behave as a ‘fingerprint’ of the sample [40]. The bands that are observed in a NIR spectrum arise mainly from the stretching of OH, CH, and NH, so this technique has been used for qualitative and quantitative analysis of natural products [41]. However, a difficulty of NIR spectroscopy is its low molecular selectivity, attributed to the widening of the bands and the omnipresence of OH, CH and NH bond in organic molecules; this problem can be solved through the chemometrics analysis of the data [42].
MVA is a powerful statistical tool that has been used in qualitative and quantitative studies to extract the required information from the NIR spectra. Conzzolino et al. [43] used NIR spectroscopy combined with MVA in a metabolic study to differentiate strains of Saccharomyces cerevisiae, proving its applicability as a screening tool for both discriminating between yeast strains and grouping strains with deletions in genes that disturb similar metabolic pathways. Combination of chemometrics and visible/NIR spectroscopy was also used for red wine fermentation monitoring in a pilot scale, obtaining a correct classification of the samples, regardless of the variety or time of fermentation [44]. These studies prove the potential of NIR spectroscopy combined with MVA in classification studies of complex samples.
MVA has also been used for the development of predictive methods for the content of PC [45,46] and flavonoids [47] in plants from NIR spectral data, evidencing the predictive value of the phenolic band at 4670 cm−1. In the present study, the application of MVA to FT-NIR data was only used as a discriminant method, demonstrating its usefulness as an exploratory analysis of the effect of elicitors on the phenolic metabolism of T. peruviana cell suspension cultures. Future studies should be aimed at developing a predictive method for the content of phenolic compounds in these cultures.
3.2. Profiling RP-HPLC and MVA of chromatographic data
RP-HPLC profiles were monitored at 280 nm in order to detect phenolic-related compounds. Fig. 4a corresponds to the superposition of representative chromatograms of cell suspension cultures under elicitor treatments. This figure shows differences in both profile and intensity of some peaks; however, to improve the comprehensive analysis of all samples and generate reproducible and comparable results, MVA was applied to the chromatographic data, previously pre-processed to correct the observed instrumental drift (difference in retention time and changes in peak shape). The COW algorithm largely corrected inter-sample variability affecting the peak alignment (Fig. 4b and c). The processed chromatographic data were subsequently normalized and scaled using the Pareto method.
Fig. 4.
Chromatograms obtained with UV detection at 280 nm from T. peruviana samples. (a) Chromatograms of representative samples treated with elicitors and control (72 h post-elicitation); (b) raw data and (c) processed data with Correlation Optimized Warping (COW) algorithm. The alignment of three peaks present in all samples is shown.
The PCA model (R2X = 0.954; Q2X = 0.867) reduced the dimensionality of the data to six main components. The first two components could explain 52.7 % and 23.6 %, respectively, of the total variance of the data. The PCA score plot (Fig. 5a) shows explicit differentiation between samples according to the treatment. The separation was higher between SA/MeJA and control than SA and MeJA with respect to control, this separation was influenced by the elicitor time exposure. The loadings graph of PCA (Fig. 5b) was used to determine the chemical compounds responsible for the variance of data. It was observed that compounds with tR of 11.68–12.52 and 12.89 min influence the separation of the group of samples treated with SA/MeJA; while compounds with tR of 7.99 and 8.98 appear to be important for the separation of groups of samples treated only with SA and only with MeJA (at 72, 96 and 120 h post-elicitation), respectively.
Fig. 5.
PCA of RP-HPLC data for extracts of T. peruviana biomass collected at different times (24, 48, 72, 96 and 120 h) after the treatment with elicitors. (a) PCA score plot indicating variations in the chromatographic profiles of samples, R2X [1] and R2X [2] correspond to the fractions of the variation of the X variables explained by the model on the first and second component, respectively. The ellipse represents Hotelling's T2, with 95 % confidence; (b) Loadings plot of the two principal components of PCA (p[1] and p[2]), most significant chromatographic variables (tR) for the samples separation in the PCA are show.
The application of PLS-DA analysis to chromatographic data improved the separation of samples according to the treatment. The PLS-DA model reduced the dimensionality of the spectral data to six main components that explained 99.8 % of the variance (R2X = 0.954, R2Y = 0.977 and Q2X = 0.839). The score plot (Fig. 6a) shows that the second main component t[2] separates the groups of SA and SA/MeJA from MeJA and the control; while the first main component separates the groups of MeJA and SA/MeJA from SA and the control. This graph also indicates that the separation of MeJA was influenced by the time of post-elicitation harvest, observing a significant difference at 72 h of exposure to MeJA. Fig. 6b presents OPLS-DA analysis results. This model reduced the dimensionality of the chromatographic variables to three predictive components and two orthogonal components (3 + 2+0), evidencing reduction of the effect of post-elicitation harvest time of cells, which improved the separation of the groups of samples according to the treatment. The same was observed with FT-NIR data.
Fig. 6.
Two-dimensional score plot for RP-HPLC data for extracts of T. peruviana biomass collected at different times (24, 48, 72, 96 and 120 h) after the treatment with elicitors. (a) PLS-DA score plot and (b) OPLS-DA score plot. R2X [1] and R2X [2] correspond to the fractions of the variation of the X variables explained by the model on the first and second component, respectively. The ellipse represents the Hotelling T2 with 95 % confidence.
Studies of the elicitor effect on a particular metabolic pathway are often based on univariate statistical analysis, where p-values are calculated to determine if there are differences between two groups or treatments. This univariate approach has limitations, one of them is the inability to evaluate interactions between variables that may be correlated; in these cases, MVA is more appropriate. In the present study, combination of RP-HPLC and MVA enabled the discrimination of the T. peruviana samples based on their phenolic profile, establishing correlation between the PC content and the exposure time to the elicitor in cell suspension cultures treated with MeJA, this is of great relevance in operational terms since it is possible to establish the optimal harvest time of cells.
3.3. Identification and quantification of phenolic and flavonoid compounds
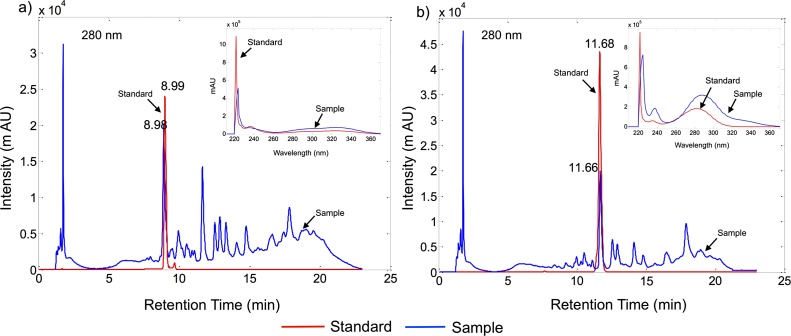
Table 1 presents the tR at 280 nm and the maximum absorbance of standard used in the present study. Two of these compounds were tentatively identified in cell suspension cultures, one with tR =11.68 min corresponding to dihydroquercetin (DHQ) and another with tR =8.99 min corresponding to chlorogenic acid (CGA) (Fig. 7).
Table 1.
Retention time (tR) and maximum absorbance wavelength (UV–vis) for standard compounds.
| Compounds | Molecular Formula |
tR (min) |
Maximum Absorbance (nm) |
|---|---|---|---|
| Chlorogenic acid | C16H18O9 | 8.99 | 228/321/635 |
| Caffeic acid | C9H8O4 | 9.44 | 232/317/622 |
| Trans-sinapic acid | C11H12O5 | 9.91 | 222/237/292 |
| Dihydroquercetin | C15H12O7 | 11.68 | 221/288/237 |
| Coumarin | C9H6O2 | 14.54 | 225/274/309 |
| Quercetin | C15H10O7 | 16.37 | 226/253/369 |
| Hesperidin | C16H14O6 | 18.65 | 223/237/286 |
| Kaempferol | C15H10O6 | 18.99 | 228/266/356 |
Fig. 7.
Chromatograms obtained with UV detection at 280 nm from T. peruviana samples compared with the standards (a) Chlorogenic acid and (b) Dihydroquercetin. UV/Vis absorption spectra are shown in the top right of the figure.
Table 2 shows the amounts of DHQ and CGA produced in cell suspension cultures of T. peruviana, under different treatments and post-elicitation harvest time.
Table 2.
Intracellular concentration of dihydroquercetin and chlorogenic acid in cell suspension cultures of T. peruviana treated with SA, MeJA and SA/MeJA.
| Treatment (hour) | Dihydroquercetin (mg/L of culture) | Chlorogenic acid (mg/L of culture) |
|---|---|---|
| Control (p value = 0.014) | ||
| 24 | 4.71 ±0.38a | ND |
| 48 | 3.30 ± 0.63a | ND |
| 72 | 4.47 ± 1.36ab | ND |
| 96 | 2.42 ± 1.34ab | ND |
| 120 | 1.86 ± 0.29b | ND |
| SA (p value = 0.379) | ||
| 24 | 4.42 ± 0.87a | ND |
| 48 | 5.67 ± 1.46a | ND |
| 72 | 5.89 ± 1.22a | ND |
| 96 | 4.12 ± 1.81a | ND |
| 120 | 3.96 ± 1.66a | ND |
| MeJA (p value = 0.0017) | ||
| 24 | 5.70 ± 0.94a | ND |
| 48 | 2.89 ± 1.25ab | 1.82 ± 0.04b |
| 72 | 3.67 ± 0.48ab | 14.79 ± 2.57a |
| 96 | 2.31 ± 0.75b | 17.52 ± 2.19a |
| 120 | 2.03 ± 1.02b | 12.15 ± 4.56a |
| SA/MeJA (p value = 0.00598) | ||
| 24 | 7.62 ± 1.46b | ND |
| 48 | 8.01 ± 1.29ab | ND |
| 72 | 12.59 ± 1.75a | ND |
| 96 | 9.87 ± 1.04ab | ND |
| 120 | 12.41 ± 1.48ab | ND |
Values of the mean ± SD of 3 independent trials. For each elicitor, values with different letters are significantly different (p < 0.05). ND = Not detected.
DHQ was detected in all cell suspensions however the highest concentrations were produced with SA/MeJA (12.59 ± 1.75 mg/L) at 72 h post-elicitation; this result was statistically different with respect to the control (4.47 ± 1.36 mg/L) (p value = 0.0000). On the other hand, CGA was only produced in cell suspensions treated with MeJA, obtaining a maximum concentration of 17.52 ± 2.19 mg/L at 96 h post-elicitation. These results suggest that MeJA directs the phenolic metabolism of T. peruviana cells towards the biosynthesis of polyphenols such as CGA. However, the treatment of cells with SA seems to annul the MeJA-inducing effect in the biosynthesis of CGA, when these elicitors are used in combination. To verify this hypothesis, future studies will be aimed at transcriptomic and metabolomic studies.
Both DHQ and CGA are secondary metabolites of great pharmaceutical interest. DHQ, also called taxifolin, is a type of flavonoid that has a wide range of biochemical and pharmacological effects [48], including antioxidant [49,50], anti-cancer [51], antidiabetic [52], hepatoprotective [53], cardioprotective [54] and inhibitory of the formation/accumulation of β-amyloid oligomers activity [55,56]. Commercially, DHQ is part of complex pharmaceutical preparations such as Venoruton®, which is indicated for the relief of symptoms related to mild venous insufficiency of the lower extremities [57]. For its part, CGA is a caffeic and quinic acid ester. This compound has antispasmodic [58], antioxidant [59] and anti-mutagenic [60] properties and act as an inhibitor of HIV-1 integrase [61] and there is also evidence that it exhibits anti-obesity potential and improves lipid metabolism [62].
4. Conclusions
The present study shows the significance of the application of FT-NIR spectroscopy and RP-HPLC combined with MVA to examine the qualitative and quantitative differences in PC production and footprint as well as to detect the effect of the elicitor treatment in cell suspension cultures of T. peruviana. This analysis allowed the tentative identification of two molecules CGA and DHQ, with important biological application, the latter not previously reported in T. peruviana cultures. This strategy could improve the current protocols of PC analysis in complex mixtures, making it possible to carry out quickly and accurately exploratory comparisons, in a high volume of samples, reducing reagent consumption. Finally, this method represents a viable alternative to ease the evaluation of elicitor effects on the phenolic metabolism of in vitro cell cultures of T. peruviana and could be applied in other plant cell systems. Future studies will be aimed at deepening our understanding of the metabolic responses of T. peruviana cells using more sensitive and selective metabolomic platforms such as mass spectrometry and increase the DHQ production yield in pilot-scale culture.
Funding
This study was supported by Patrimonio Autónomo Fondo Nacional de Financiamiento para la Ciencia, la Tecnología y la Innovación Francisco José de Caldas - Departamento Administrativo de Ciencia, Tecnología e Innovación de Colombia – COLCIENCIAS [Grant No. FP44842-006-2018].
CRediT authorship contribution statement
Dary Mendoza: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Writing - original draft, Visualization. Juan Pablo Arias: Conceptualization, Methodology, Investigation, Writing - review & editing. Olmedo Cuaspud: Conceptualization, Methodology, Investigation, Writing - review & editing, Visualization. Orlando Ruiz: Resources, Writing - review & editing, Supervision. Mario Arias: Resources, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgments
FT-NIR analyzes were performed in the Laboratorio de Suelos, Universidad Nacional de Colombia, Medellín- Colombia.
References
- 1.Ramachandra Rao S., Ravishankar G.A. Plant cell cultures: chemical factories of secondary metabolites. Biotechnol. Adv. 2002;20:101–153. doi: 10.1016/S0734-9750(02)00007-1. [DOI] [PubMed] [Google Scholar]
- 2.Whitaker R.J., Hobbib G.C., Steward L.A. Biogeneration of Aromas. 1986. Production of secondary metabolites in plant cell cultures; pp. 347–362. [DOI] [Google Scholar]
- 3.Bandara V., Weinstein S.A., White J., Eddleston M. A review of the natural history, toxinology, diagnosis and clinical management of Nerium oleander (common oleander) and Thevetia peruviana (yellow oleander) poisoning. Toxicon. 2010;56:273–281. doi: 10.1016/j.toxicon.2010.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Kohls S., Scholz-Böttcher B.M., Teske J., Zark P., Rullkötter J. Cardiac glycosides from Yellow Oleander (Thevetia peruviana) seeds. Phytochemistry. 2012;75:114–127. doi: 10.1016/j.phytochem.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Haldar S., Karmakar I., Chakraborty M., Ahmad D., Haldar P.K. Antitumor potential of Thevetia peruviana on Ehrlich’s ascites carcinoma-bearing mice. J. Environ. Pathol. Toxicol. Oncol. 2015;34:105–113. doi: 10.1615/jenvironpatholtoxicoloncol.2015012017. [DOI] [PubMed] [Google Scholar]
- 6.Ramos-Silva A., Tavares-Carreón F., Figueroa M., De la Torre-Zavala S., Gastelum-Arellanez A., Rodríguez-García A., Galán-Wong L.J., Avilés-Arnaut H. Anticancer potential of Thevetia peruviana fruit methanolic extract. BMC Complement. Altern. Med. 2017;17(1):241. doi: 10.1186/s12906-017-1727-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassan M.M., Saha A.K., Khan S.A., Islam A., Mahabub-Uz-Zaman M., Ahmed S.S.U. Studies on the antidiarrhoeal, antimicrobial and cytotoxic activities of ethanol-extracted leaves of yellow oleander (Thevetia peruviana) Open Vet. J. 2011;1(1):28–31. [PMC free article] [PubMed] [Google Scholar]
- 8.Dixit A., Singh H., Sharma R.A., Sharma A. Estimation of antioxidant and antibacterial activity of crude extracts of Thevetia peruviana (Pers.) K. Schum. Int. J. Pharm. 2015;7:55–59. [Google Scholar]
- 9.Tewtrakul S., Nakamura N., Hattori M., Fujiwara T., Supavita T. Flavanone and flavonol glycosides from the leaves of Thevetia peruviana and their HIV-1 reverse transcriptase and HIV-1 integrase inhibitory activities. Chem. Pharm. Bull. (Tokyo) 2002;50:630–635. doi: 10.1248/cpb.50.630. [DOI] [PubMed] [Google Scholar]
- 10.Arias M., Angarita M., Restrepo J.M., Caicedo L.A., Perea M. Elicitation with methyl-jasmonate stimulates peruvoside production in cell suspension cultures of Thevetia peruviana. Vitr. Cell. Dev. Biol. – Plant. 2010;46:233–238. doi: 10.1007/s11627-009-9249-z. [DOI] [Google Scholar]
- 11.Villegas-Quiceño A.P., Arias-Echeverri J.P., Aragón-Mena D., Ochoa-Cáceres S., Arias-Zabala M.E. Multi-objective optimization in biotechnological processes: application to plant cell suspension cultures of Thevetia peruviana. Rev. Fac. Ing. Univ. Antioquia. 2018:35–40. doi: 10.17533/udea.redin.n87a05. [DOI] [Google Scholar]
- 12.Reymond P., Farmer E.E. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1998;1:404–411. doi: 10.1016/S1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- 13.Khan M.I.R., Khan N.A. Salicylic acid and Jasmonates: approaches in abiotic stress tolerance. J. Plant Biochem. Physiol. 2013;1:e113. doi: 10.4172/2329-9029.1000e113. [DOI] [Google Scholar]
- 14.Rejeb I., Pastor V., Mauch-Mani B. Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants. 2014;3:458–475. doi: 10.3390/plants3040458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza D., Cuaspud O., Arias J.P., Ruiz O., Arias M. Effect of salicylic acid and methyl jasmonate in the production of phenolic compounds in plant cell suspension cultures of Thevetia peruviana. Biotechnol. Rep. 2018;19 doi: 10.1016/j.btre.2018.e00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granato D., Putnik P., Kovačević D.B., Santos J.S., Calado V., Rocha R.S., Cruz A.G., Jarvis B., Rodionova O.Y., Pomerantsev A.L. Trends in chemometrics: food authentication, microbiology, and effects of processing. Compr. Rev. Food Sci. Food Saf. 2018;17:663–677. doi: 10.1111/1541-4337.12341. [DOI] [PubMed] [Google Scholar]
- 17.Siebert K.J. Chemometrics in brewing—a review. J. Am. Soc. Brew. Chem. 2001;59:147–156. doi: 10.1094/ASBCJ-59-0147. [DOI] [Google Scholar]
- 18.Li J., Zhang J., Zhao Y.L., Huang H.Y., Wang Y.Z. Comprehensive quality assessment based specific chemical profiles for geographic and tissue variation in Gentiana rigescens using HPLC and FTIR method combined with principal component analysis. Front. Chem. 2017;22(5):125. doi: 10.3389/fchem.2017.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X., Gu N., Xue C., Li B. Plant flavonoid taxifolin inhibits the growth, migration and invasion of human osteosarcoma cells. Mol. Med. Rep. 2017;17(2):3239–3245. doi: 10.3892/mmr.2017.8271. [DOI] [PubMed] [Google Scholar]
- 20.Guo Y., Ding X., Ni Y. The combination of NIR spectroscopy and HPLC chromatography for differentiating lotus seed cultivars and quantitative prediction of four main constituents in lotus with the aid of chemometrics. Anal. Methods. 2017;9:6420–6429. doi: 10.1039/C7AY02021J. [DOI] [Google Scholar]
- 21.Moreira I., Scarminio I.S. Chemometric discrimination of genetically modified Coffea arabica cultivars using spectroscopic and chromatographic fingerprints. Talanta. 2013;107:416–422. doi: 10.1016/j.talanta.2013.01.053. [DOI] [PubMed] [Google Scholar]
- 22.Zareef M., Chen Q., Ouyang Q., Kutsanedzie F.Y.H., Hassan M.M., Viswadevarayalu A., Wang A. Prediction of amino acids, caffeine, theaflavins and water extract in black tea using FT-NIR spectroscopy coupled chemometrics algorithms. Anal. Methods. 2018;10:3023–3031. doi: 10.1039/C8AY00731D. [DOI] [Google Scholar]
- 23.Diabaté S., De Franqueville H., Adon B., Coulibaly O., Ake S. The role of phenolic compounds in the determination of wilt disease tolerance of oil palm (Elaeis guineensis JACQ) Afr. J. Biotechnol. 2009;8:5679–5690. [Google Scholar]
- 24.Liu Z., Wang D., Li D., Zhang S. Quality evaluation of Juniperus rigida Sieb. et Zucc. based on phenolic profiles, bioactivity, and HPLC fingerprint combined with chemometrics. Front. Pharmacol. 2017;8:198. doi: 10.3389/fphar.2017.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borille B.T., Marcelo M.C.A., Ortiz R.S., Mariotti K.C., Ferrão M.F., Limberger R.P. Near infrared spectroscopy combined with chemometrics for growth stage classification of cannabis cultivated in a greenhouse from seized seeds. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017;173:318–323. doi: 10.1016/j.saa.2016.09.040. [DOI] [PubMed] [Google Scholar]
- 26.Christou C., Agapiou A., Kokkinofta R. Use of FTIR spectroscopy and chemometrics for the classification of carobs origin. J. Adv. Res. 2018;10:1–8. doi: 10.1016/j.jare.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashimoto A., Yamanaka A., Kanou M., Nakanishi K., Kameoka T. Simple and rapid determination of metabolite content in plant cell culture medium using an FT-IR/ATR method. Bioprocess Biosyst. Eng. 2005;27(2):115–123. doi: 10.1007/s00449-004-0388-7. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto A., Nakanishi K., Motonaga Y., Kameoka T. Sugar metabolic analysis of suspensions of plant cells using an FT-IR/ATR method. Biotechnol. Prog. 2001;17(3):560–564. doi: 10.1021/bp010013w. [DOI] [PubMed] [Google Scholar]
- 29.Schenk R.U., Hildebrandt A.C. Medium and techniques for induction and growth of monocotyledonous and dicotyledonous plant cell cultures. Can. J. Bot. 1972;50:199–204. doi: 10.1139/b72-026. [DOI] [Google Scholar]
- 30.Bi Y., Yuan K., Xiao W., Wu J., Shi C., Xia J., Chu G., Zhang G., Zhou G. A local pre-processing method for near-infrared spectra, combined with spectral segmentation and standard normal variate transformation. Anal. Chim. Acta. 2016;909:30–40. doi: 10.1016/j.aca.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Bartel J., Krumsiek J., Theis F.J. Statistical methods for the analysis of high-throughput metabolomics data. Comput. Struct. Biotechnol. J. 2013;4 doi: 10.5936/csbj.201301009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoddami A., Wilkes M.A., Roberts T.H. Techniques for analysis of plant phenolic compounds. Molecules. 2013;18(2):2328–2375. doi: 10.3390/molecules18022328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rusilowicz M., Dickinson M., Charlton A., O’Keefe S., Wilson J. A batch correction method for liquid chromatography–mass spectrometry data that does not depend on quality control samples. Metabolomics. 2016;12(3):56. doi: 10.1007/s11306-016-0972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomasi G., Skov T., Van den Berg F. Dynamic time warping (DTW) and correlation optimized warping (COW) [WWW document] Spectrosc. Chemom. Sect. 2004 http://www.models.life.ku.dk/dtw_cow_more (accessed 10.1.19) [Google Scholar]
- 35.Department of Agriculture Dr. Duke’s phytochemical and ethnobotanical databases [WWW document] Agric. Res. Serv. 1992 doi: 10.15482/USDA.ADC/1239279. (accessed 12.6.18) [DOI] [Google Scholar]
- 36.Rinnan Å., Berg F., Engelsen S.B., Rinnan Å., Berg F.V.D. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009;28:1201–1222. doi: 10.1016/j.trac.2009.07.007. [DOI] [Google Scholar]
- 37.Iwamoto R. Infrared and Near-Infrared study of the interaction of amide C O with water in ideally inert medium. J. Phys. Chem. A. 2010;114:7398–7407. doi: 10.1021/jp102479t. [DOI] [PubMed] [Google Scholar]
- 38.Li W., Qu H. Rapid quantification of phenolic acids in Radix Salvia Miltrorrhiza extract solutions by FT-NIR spectroscopy in transflective mode. J. Pharm. Biomed. Anal. 2010;52:425–431. doi: 10.1016/j.jpba.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Worley B., Powers R. PCA as a practical indicator of OPLS-DA model reliability. Curr. Metabolomics. 2016;4(2):97–103. doi: 10.2174/2213235X04666160613122429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cordella C., Moussa I., Martel A.-C., Sbirrazzuoli N., Lizzani-Cuvelier L. Recent developments in food characterization and adulteration detection: technique-oriented perspectives. J. Agric. Food Chem. 2002;50(7):1751–1764. doi: 10.1021/jf011096z. [DOI] [PubMed] [Google Scholar]
- 41.Cozzolino D. Near infrared spectroscopy in natural products analysis. Planta Med. 2009;75(7):746–756. doi: 10.1055/s-0028-1112220. [DOI] [PubMed] [Google Scholar]
- 42.Blanco M., Villarroya I. NIR spectroscopy: a rapid-response analytical tool. TrAC Trends Anal. Chem. 2002;21(4):240–250. doi: 10.1016/S0165-9936(02)00404-1. [DOI] [Google Scholar]
- 43.Cozzolino D., Flood L., Bellon J., Gishen M., De Barros Lopes M. Combining near infrared spectroscopy and multivariate analysis as a tool to differentiate different strains of Saccharomyces cerevisiae: a metabolomic study. Yeast. 2016;23(14–15):1089–1096. doi: 10.1002/yea.1418. [DOI] [PubMed] [Google Scholar]
- 44.Cozzolino D., Parker M., Dambergs R.G., Herderich M., Gishen M. Chemometrics and visible-near infrared spectroscopic monitoring of red wine fermentation in a pilot scale. Biotechnol. Bioeng. 2016;95(6):1101–1107. doi: 10.1002/bit.21067. [DOI] [PubMed] [Google Scholar]
- 45.Frizon C.N.T., Oliveira G.A., Perussello C.A., Peralta-Zamora P.G., Camlofski A.M.O., Rossa Ü.B., Hoffmann-Ribani R. Determination of total phenolic compounds in yerba mate (Ilex paraguariensis) combining near infrared spectroscopy (NIR) and multivariate analysis. LWT - Food Sci. Technol. 2015;60:795–801. doi: 10.1016/j.lwt.2014.10.030. [DOI] [Google Scholar]
- 46.Hassan H., Fan M., Zhang T., Yang K. Prediction of Total phenolics and flavonoids contents in chinese wild rice (Zizania latifolia) Using FT-NIR Spectroscopy. Am. J. Food Technol. 2015;10:109–117. doi: 10.3923/ajft.2015.109.117. [DOI] [Google Scholar]
- 47.Baiyi L., Jianyang C., Weisu H., Di W., Wei X., Qing X., Xiao Y., Lanjuan L. Determination of flavonoids and phenolic acids in the extract of bamboo leaves using near-infrared spectroscopy and multivariate calibration. Afr. J. Biotechnol. 2011;10:8448–8455. doi: 10.5897/AJB11.320. [DOI] [Google Scholar]
- 48.Sunil C., Xu B. An insight into the health-promoting effects of taxifolin (dihydroquercetin) Phytochemistry. 2019;166 doi: 10.1016/j.phytochem.2019.112066. [DOI] [PubMed] [Google Scholar]
- 49.Topal F., Nar M., Gocer H., Kalin P., Kocyigit U.M., Gülçin İ., Alwasel S.H. Antioxidant activity of taxifolin: an activity–structure relationship. J. Enzyme Inhib. Med. Chem. 2016;31:674–683. doi: 10.3109/14756366.2015.1057723. [DOI] [PubMed] [Google Scholar]
- 50.Xie X., Feng J., Kang Z., Zhang S., Zhang L., Zhang Y., Li X., Tang Y. Taxifolin protects RPE cells against oxidative stress-induced apoptosis. Mol. Vis. 2017;23:520–528. [PMC free article] [PubMed] [Google Scholar]
- 51.Chen P., Luthriau D., Harringtonohio Pde B., Harnly J.M. Discrimination among Panax species using spectral fingerprinting. J. AOAC Int. 2011;94:1411–1421. doi: 10.5740/jaoacint.10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun X., Chen R.C., Yang Z.H., Sun G.B., Wang M., Ma X.J., Yang L.J., Sun X.B. Taxifolin prevents diabetic cardiomyopathy in vivo and in vitro by inhibition of oxidative stress and cell apoptosis. Food Chem. Toxicol. 2014;63:221–232. doi: 10.1016/j.fct.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 53.Vargas-Mendoza N., Madrigal-Santillán E., Morales-Gonzales A., Esquivel-Soto J., Esquivel-Chirino C., Garcia-Luna M., Gonzales-Rubio M., Gayosso-de-Lucio J.A., Morales-Gonzales J.A. Hepatoprotective effect of silymarin. World J. Hepatol. 2014;6:144. doi: 10.4254/wjh.v6.i3.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo H., Zhang X., Cui Y., Zhou H., Xu D., Shan T., Zhang F., Guo Y., Chen Y., Wu D. Taxifolin protects against cardiac hypertrophy and fibrosis during biomechanical stress of pressure overload. Toxicol. Appl. Pharmacol. 2015;287:168–177. doi: 10.1016/j.taap.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 55.Park S.Y., Kim H.Y., Park H.J., Shin H.K., Hong K.W., Kim C.D. Concurrent treatment with taxifolin and cilostazol on the lowering of β-amyloid accumulation and neurotoxicity via the suppression of P-JAK2/P-STAT3/NF-κB/BACE1 signaling pathways. PLoS One. 2016;11 doi: 10.1371/journal.pone.0168286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito S., Yamamoto Y., Maki T., Hattori Y., Ito H., Mizuno K., Harada-Shiba M., Kalaria R.N., Fukushima M., Takahashi R., Ihara M. Taxifolin inhibits amyloid-β oligomer formation and fully restores vascular integrity and memory in cerebral amyloid angiopathy. Acta Neuropathol. Commun. 2017;5(1):26. doi: 10.1186/s40478-017-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weidmann A.E. Dihydroquercetin: more than just an impurity? Eur. J. Pharmacol. 2012;684(1-3):19–26. doi: 10.1016/j.ejphar.2012.03.035. [DOI] [PubMed] [Google Scholar]
- 58.Farah A., de Paulis T., Trugo L.C., Martin P.R. Effect of roasting on the formation of chlorogenic acid lactones in Coffee. J. Agric. Food Chem. 2005;53:1505–1513. doi: 10.1021/jf048701t. [DOI] [PubMed] [Google Scholar]
- 59.Kweon M.H., Hwang H.J., Sung H.C. Identification and antioxidant activity of novel Chlorogenic acid derivatives from Bamboo (Phyllostachys edulis) J. Agric. Food Chem. 2001;49:4646–4655. doi: 10.1021/jf010514x. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y.J., Zhou C.Y., Qiu C.H., Lu X.M., Wang Y.T. Chlorogenic acid induced apoptosis and inhibition of proliferation in human acute promyelocytic leukemia HL-60 cells. Mol. Med. Rep. 2013;8:1106–1110. doi: 10.3892/mmr.2013.1652. [DOI] [PubMed] [Google Scholar]
- 61.Kwon H.C., Jung C.M., Shin C.G., Lee J.K., Choi S.U., Kim S.Y., Lee K.R. A new caffeoyl quinic acid from Aaster scaber and its inhibitory activity against Human Immunodeficiency Virus-1(HIV-1) Integrase. Chem. Pharm. Bull. (Tokyo) 2000;48(11):1796–1798. doi: 10.1248/cpb.48.1796. [DOI] [PubMed] [Google Scholar]
- 62.Cho A.S., Jeon S.M., Kim M.J., Yeo J., Seo K.I., Choi M.S., Lee M.K. Chlorogenic acid exhibits anti-obesity property and improves lipid metabolism in high-fat diet-induced-obese mice. Food Chem. Toxicol. 2010;48(3):937–943. doi: 10.1016/j.fct.2010.01.003. [DOI] [PubMed] [Google Scholar]