Main Text
Proteins from the tubulin and actin superfamilies self-assemble, forming dynamic filaments that are essential for DNA segregation, cell division, cytoplasmic organization, and motility. These filaments translocate (treadmill) fueled by nucleotide hydrolysis to perform their functions, even without motor proteins, growing from one end, whereas shortening from the other. Tubulin and bacterial FtsZ assembly machines adopt two main conformations: the free unassembled proteins are in a low self-association affinity relaxed (R) structure, whereas the subunits in polymers (microtubules and FtsZ filaments) are held in a high self-association affinity tense (T) form. Switching from the R to the T state is fundamentally coupled to the formation of a tight longitudinal association interface between consecutive subunits in the filament rather than to the nucleotide state. The assembly switch entails a rotation of the GTPase-activating domain with respect to the GTP-binding domain (FtsZ: open interdomain cleft = T vs. closed cleft = R; tubulin: straight subunits = T vs. curved subunits = R). The local interactions made by the nucleotide γ-phosphate chemically enhance the affinity of the longitudinal association interface rather than induce large structural changes. Tubulin makes additional lateral contacts with neighbor protofilaments in microtubules.
The cooperative assembly of single-stranded FtsZ filaments and their treadmilling mechanism were puzzles, for which the solutions can also illuminate the dynamic mechanisms of more complex cytomotive filaments. Assembly models in which FtsZ monomers switch between low- and high-affinity conformations explain nucleated condensation polymerization of linear filaments (1,2) and the directional filament treadmilling upon GTP hydrolysis at the association interfaces (3). In this issue of Biophysical Journal, Corbin and Erickson present a timely numerical model of FtsZ filament assembly, nucleotide hydrolysis, and treadmilling employing Monte Carlo methods (4). Interestingly, the tubulin assembly switch possibly enables both microtubule treadmilling and dynamic instability. Actin filament treadmilling appears to involve an analogous mechanism, in which G-actin may be identified as an R state and F-actin as a T state.
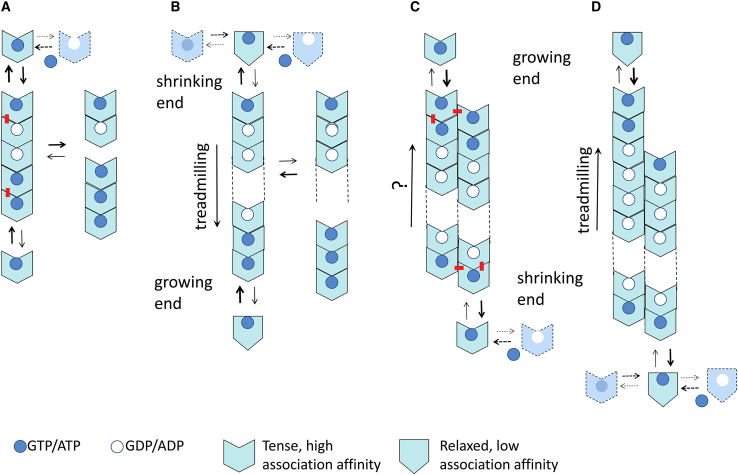
To analyze possible treadmilling mechanisms, let us first consider a GTP-hydrolyzing single-stranded protein filament (Fig. 1, Model A). A polar filament can, in principle, treadmill with nucleotide hydrolysis (5). However, three problems can be identified that prevent the formation of a treadmilling single-stranded filament by this mechanism: 1) oligomers are formed, with a smaller proportion of larger species, rather than long filaments because the association affinity is constant at each association step; 2) oligomers fragment at the interfaces, where the nucleotide has been hydrolyzed at a rate similar to the dissociation rate at a diphosphate-containing end; and 3) the monomer association reactions at both oligomer ends are identical and thus have the same rate.
Figure 1.
(Model A) Linear isodesmic protein self-association with nucleotide hydrolysis. The nucleotide γ-phosphate at the association interface provides a chemical signal increasing the affinity of subunit addition. Hydrolysis stochastically takes place after the formation of each association contact at an intrinsic average rate, inducing subunit dissociation; nucleotide triphosphate (in excess) is assumed to replace nucleotide diphosphate in monomers and at the exposed subunit at the top. The pentamer in the model represents the product of adding one subunit at each end of a trimer. The newly formed top and bottom interfaces are marked with a red dash. Notice that the penultimate subunit at the top is GDP bound when releasing the top subunit but becomes GTP bound when exposed. (Model B) Linear nucleated polymerization with nucleotide hydrolysis and treadmilling is shown. The protein monomers switch between a state with low self-association affinity when unassembled (relaxed; R) and a state with high-association affinity (tense; T) that forms tight interfaces in the filament. This is the mechanism thought to work for FtsZ. (Model C) Multistranded condensation polymerization with nucleotide hydrolysis (no switch) is shown. In the double-stranded filament exemplified by the model it can be appreciated how filament elongation involves longitudinal and lateral contacts, stabilizing the filament against fragmentation, whereas formation of a hypothetical dimer nucleus involves one type of contact only. (Model D) Multistranded condensation polymerization with nucleotide hydrolysis and assembly switch in combination of Models B and C is shown. This type of mechanism may apply to microtubule and actin assembly. To see this figure in color, go online.
For the association rates to be different at the top and bottom ends of a single-stranded filament, the association reactions at each end must have different initial, transition, or end states. Such differences might, in principle, be induced by the nucleotide γ-phosphate (in monomers or in polymer) or by the intermolecular protein contacts in the polymer. In the second model (Fig. 1, Model B), the protein monomers autosterically switch (6) between a state with low self-association affinity when unassembled (R) and a state with high-association affinity (T) when forming part of the polymer.
Three main features can be identified in Model B: 1) the unfavorable switching from R to T subunits explains nucleated condensation polymerization of linear filaments with a cooperative behavior (1,2,7, 8, 9) driven by the tight association contacts in the filament (10) rather than forming shorter oligomers as in model A. In support of this model, FtsZ T subunits relax to R subunits when isolated during atomistic molecular dynamics simulations starting from the crystal filament T structure ((11) and references therein) and also relax upon FtsZ disassembly during solution experiments, as monitored with fluorescent reporters (12,13). Moreover, it has been possible to determine both T and R crystal structures from the same FtsZ protein (3,14). Notice that the structures of diverse unassembled FtsZ proteins correspond to the R state, irrespective of the bound nucleotide. On the other hand, the available crystal structures of T-FtsZ filaments contain GDP and are still missing a filament structure with a bound GTP analog. 2) A second feature is that the fragmentation rate is slower than dissociation from the end because an inner subunit is held in the T state by the contacts to the surrounding subunits along the filament. 3) Importantly, the structural switch between R and T makes both ends kinetically different. It creates different initial pairs of surfaces for association at each end so that the association rates become different, and the single filament can thus treadmill with nucleotide hydrolysis (3). This creates a translocating filament, a linear motor able to move and generate force when reversibly attached to a suitable stator. The end additions in Model B really encompass both switching and association reactions in a nonspecified kinetic pathway. Also, the kinetic growth (plus) and shrinkage (minus) ends can, in principle, be at the structural top (exposed nucleotide) and bottom ends, or vice versa, the growing end can be at the bottom, and the shrinking end can be at the top. This second possibility, with the filament treadmilling downward, has been chosen in Model B, according to the kinetic polarity deduced from the effects of top and bottom mutations in FtsZ (15).
Thanks to the Monte Carlo model of Corbin and Erickson (4), we can now quantitatively understand how treadmilling of single filaments works. The available MATLAB (The MathWorks, Natick, MA) code and user-friendly application permit testing of various values for the switch equilibrium constant, the rates of subunit addition, dissociation from plus and minus ends, and the GTP hydrolysis rate. We now know how both the structural switch and GTP hydrolysis are required for treadmilling. The model captures the birth and dynamic behavior of numerous simulated individual filaments. The bulk properties of simulated FtsZ solutions, including the monomer concentration, filament length distribution, GTP hydrolysis, and exchange of monomers between filaments, are in good agreement with experimental measurements from the Erickson lab on various FtsZ proteins. The model also incorporates sequestering and capping proteins. Interestingly, the treadmilling velocity depends on the GTPase activity, similar to FtsZ treadmilling in bacterial cells. Adding a reversible surface association feature to the model simulations might more closely simulate membrane-tethered FtsZ filaments in cells and in synthetic models systems.
Let us now turn to multistranded polymers with nucleotide hydrolysis (Fig. 1, Model C). Bidimensional polymerization is typically cooperative, proceeding above a critical protein concentration for elongation (16). A multistranded polymer with nucleotide hydrolyzing subunits treadmills with a head-to-tail polymerization mechanism analytically demonstrated by Wegner in his landmark study (5). Notice that the treadmilling direction in Model C has been drawn opposite to Model B to make it compatible with that of actin filaments and microtubules, which were previously thought to treadmill according to this type of mechanism.
This model posits that the transition states, and therefore the rate constants of subunit addition and dissociation at the ends of a polar polymer, need not be equal, even in the absence of nucleotide hydrolysis, in which the equilibrium association constants are identical at both ends. Clearly, this is not the case for a single-stranded oligomer, in which the protein contacts are identical at both ends (Fig. 1, Model A), but in a multistranded polymer the association interfaces at each end may be somehow different. In Model C the longitudinal and lateral interactions made by one monomer at the top and bottom ends are similar, although with opposite handedness (see contacts marked by red dashes in Fig. 1, Model C). It might be debated whether this could be enough for different encounter kinetics at each filament end or whether a monomer structural switch is actually involved (6), similar to the case of single-stranded filaments. A key feature of this model is that different critical concentrations at each end are created by nucleotide hydrolysis. Notice that, in the model, the nucleotide forms part of the axial association interface, such as GTP/GDP in FtsZ and tubulin, and thus the association affinity may be directly reduced by hydrolysis at the shrinking end. However, in an actin-like filament, the ATP/ADP nucleotide binds into the monomer core rather than at an association interface. It may thus be argued that for a Wegner-type Model C to work for actin, it must contain some structural change transmitted from the association interfaces for internal nucleotide hydrolysis and that nucleotide hydrolysis has to allosterically reduce the association affinity at the shrinking end.
A mechanism of multistranded polymerization with nucleotide hydrolysis and assembly switch (Fig. 1, Model D) may be proposed for actin and microtubule assembly, supported by current results. This model is a logical combination of Models B and C. It should treadmill as the single-stranded FtsZ filament B can, and it is further stabilized by the lateral interactions in the polymer lattice, similar to C.
The concept of a polymerization-driven structural switch in microtubule assembly was proposed to explain the curved (R) structures of unassembled tubulins, irrespective of the bound nucleotide, in contrast with straight (T) tubulin in microtubules ((17,18) and references therein); the tubulin assembly switch has been further supported by biochemical and structural studies. Indeed, recent cryo-EM structures of undecorated microtubules with different nucleotides have revealed similar tubulin subunit structures, with subtle structural changes mainly taking place at the longitudinal association interface (19). However, the exact role of the lateral interactions in microtubule assembly is still debated. Observations of individual curved protofilaments at growing microtubule ends (20) suggest that the longitudinal interactions might primarily induce tubulin affinity switching or propagate from the lattice end, followed by protofilament straightening as the lateral interactions are formed. Extension of the numerical simulation model of Corbin and Erickson (4) to multistranded filaments should be helpful in analyzing the interplay of the structural switch, longitudinal and lateral interactions in microtubule treadmilling, and dynamic instability.
Interestingly, actin appears to have converged to a conceptually similar assembly-switch mechanism, in which G-actin may be identified as an R state and F-actin as a T state. Indeed, recent cryo-EM structures of actin filaments with different bound nucleotides have confirmed a large structural change (monomer flattening) with interdomain rotation and translation that takes place upon actin monomer incorporation into filaments, rather than by the bound nucleotide, and have also revealed the small structural changes responsible for ATP hydrolysis and for subunit dissociation after phosphate release (21,22). Further investigation should reveal whether assembly switches are a constitutive feature of all treadmilling protein filaments.
Editor: John Correia.
References
- 1.Huecas S., Llorca O., Andreu J.M. Energetics and geometry of FtsZ polymers: nucleated self-assembly of single protofilaments. Biophys. J. 2008;94:1796–1806. doi: 10.1529/biophysj.107.115493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miraldi E.R., Thomas P.J., Romberg L. Allosteric models for cooperative polymerization of linear polymers. Biophys. J. 2008;95:2470–2486. doi: 10.1529/biophysj.107.126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagstaff J.M., Tsim M., Löwe J. A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. MBio. 2017;8:e00254-17. doi: 10.1128/mBio.00254-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbin L., Erickson H.P. A unified model for treadmilling and nucleation of single-stranded FtsZ protofilaments. Biophys. J. 2020;119:792–805. doi: 10.1016/j.bpj.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wegner A. Head to tail polymerization of actin. J. Mol. Biol. 1976;108:139–150. doi: 10.1016/s0022-2836(76)80100-3. [DOI] [PubMed] [Google Scholar]
- 6.Caspar D.L. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys. J. 1980;32:103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michie K.A., Löwe J. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- 8.Dajkovic A., Lutkenhaus J. Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 2006;11:140–151. doi: 10.1159/000094050. [DOI] [PubMed] [Google Scholar]
- 9.Lan G., Dajkovic A., Sun S.X. Polymerization and bundling kinetics of FtsZ filaments. Biophys. J. 2008;95:4045–4056. doi: 10.1529/biophysj.108.132837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsui T., Han X., Tanaka I. Structural change in FtsZ Induced by intermolecular interactions between bound GTP and the T7 loop. J. Biol. Chem. 2014;289:3501–3509. doi: 10.1074/jbc.M113.514901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramírez-Aportela E., López-Blanco J.R., Chacón P. Understanding nucleotide-regulated FtsZ filament dynamics and the monomer assembly switch with large-scale atomistic simulations. Biophys. J. 2014;107:2164–2176. doi: 10.1016/j.bpj.2014.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y., Erickson H.P. Conformational changes of FtsZ reported by tryptophan mutants. Biochemistry. 2011;50:4675–4684. doi: 10.1021/bi200106d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artola M., Ruíz-Avila L.B., Huecas S. The structural assembly switch of cell division protein FtsZ probed with fluorescent allosteric inhibitors. Chem. Sci. (Camb.) 2017;8:1525–1534. doi: 10.1039/c6sc03792e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita J., Harada R., Matsumura H. Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus. J. Struct. Biol. 2017;198:65–73. doi: 10.1016/j.jsb.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Du S., Pichoff S., Lutkenhaus J. FtsZ filaments have the opposite kinetic polarity of microtubules. Proc. Natl. Acad. Sci. USA. 2018;115:10768–10773. doi: 10.1073/pnas.1811919115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oosawa F., Asakura S. Academic Press; London and New York: 1975. Thermodynamics of the Polymerization of Protein. [Google Scholar]
- 17.Buey R.M., Díaz J.F., Andreu J.M. The nucleotide switch of tubulin and microtubule assembly: a polymerization-driven structural change. Biochemistry. 2006;45:5933–5938. doi: 10.1021/bi060334m. [DOI] [PubMed] [Google Scholar]
- 18.Rice L.M., Montabana E.A., Agard D.A. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. USA. 2008;105:5378–5383. doi: 10.1073/pnas.0801155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang R., LaFrance B., Nogales E. Separating the effects of nucleotide and EB binding on microtubule structure. Proc. Natl. Acad. Sci. USA. 2018;115:E6191–E6200. doi: 10.1073/pnas.1802637115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIntosh J.R., O’Toole E., Gudimchuk N. Microtubules grow by the addition of bent guanosine triphosphate tubulin to the tips of curved protofilaments. J. Cell Biol. 2018;217:2691–2708. doi: 10.1083/jcb.201802138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merino F., Pospich S., Raunser S. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol. 2018;25:528–537. doi: 10.1038/s41594-018-0074-0. [DOI] [PubMed] [Google Scholar]
- 22.Chou S.Z., Pollard T.D. Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. USA. 2019;116:4265–4274. doi: 10.1073/pnas.1807028115. [DOI] [PMC free article] [PubMed] [Google Scholar]