Abstract
The motor protein myosin drives muscle and nonmuscle motility by binding to and moving along actin of thin filaments. Myosin binding to actin also modulates interactions of the regulatory protein, tropomyosin, on thin filaments, and conversely tropomyosin affects myosin binding to actin. Insight into this reciprocity will facilitate a molecular level elucidation of tropomyosin regulation of myosin interaction with actin in muscle contraction, and in turn, promote better understanding of nonmuscle cell motility. Indeed, experimental approaches such as fiber diffraction, cryoelectron microscopy, and three-dimensional reconstruction have long been used to define regulatory interaction of tropomyosin and myosin on actin at a structural level. However, their limited resolution has not proven sufficient to determine tropomyosin and myosin contacts at an atomic-level and thus to fully substantiate possible functional contributions. To overcome this deficiency, we have followed a hybrid approach by performing new cryogenic electron microscopy reconstruction of myosin-S1-decorated F-actin-tropomyosin together with atomic scale protein-protein docking of tropomyosin to the EM models. Here, cryo-EM data were derived from filaments reconstituted with α1-actin, cardiac αα-tropomyosin, and masseter muscle β-myosin complexes; masseter myosin, which shares sequence identity with β-cardiac myosin-heavy chain, was used because of its stability in vitro. The data were used to build an atomic model of the tropomyosin cable that fits onto the actin filament between the tip of the myosin head and a cleft on the innermost edge of actin subunits. The docking and atomic scale fitting showed multiple discrete interactions of myosin loop 4 and acidic residues on successive 39–42 residue-long tropomyosin pseudorepeats. The contacts between S1 and tropomyosin on actin appear to compete with and displace ones normally found between actin and tropomyosin on myosin-free thin filaments in relaxed muscle, thus restructuring the filament during myosin-induced activation.
Significance
Mutations in sarcomeric proteins, including mutations detected in actin, tropomyosin, and myosin, are linked to cardiomyopathies and skeletal muscle disease. However, the mechanisms underlying their impact on contraction and relaxation remains unclear because corresponding perturbation of myofilament structure is incompletely defined. In this study, cryoelectron microscopy and protein-protein docking provide a view of tropomyosin-myosin contacts on actin filaments. These interactions are significant in switching between muscle-on and -off states. Indeed, disease-rendering mutations that target such interactions may destabilize the contraction/relaxation cycle. Thus, our results provide a framework for the connection between such molecular insults and pathological progression.
Introduction
During muscle contraction, myosin-motors that project from thick filaments bind to and move along thin filaments formed from filamentous actin (F-actin). To generate requisite force on the actin-based thin filaments for muscle contraction, myosin heads undergo a power stroke, coupled to ATP hydrolysis and product release. In striated muscles, this process is controlled by the regulatory proteins tropomyosin and troponin located on the surface of the actin filaments. The regulatory proteins provide an on-off switching mechanism to control myosin-mediated motility and consequently muscle activity (1, 2, 3, 4). The troponin complex, responding to varied sarcoplasmic Ca2+ levels, as well as myosin-binding itself, bias tropomyosin to occupy one of three distinct regulatory conformations on the actin surface. This involves a steric reconfiguration of the tropomyosin head-to-tail cable to either hinder access or expose myosin-binding sites on actin and thus govern myosin’s interactions with thin filaments and its motor activity (1, 2, 3, 4). At low Ca2+ levels, troponin-I of the troponin complex constrains tropomyosin coiled coils in a “blocked B-state,” in which tropomyosin obstructs myosin-binding to actin, keeping muscle relaxed. With rising Ca2+ levels during muscle activation, Ca2+-occupied troponin-C promotes a “C-state,” in which myosin-binding sites remain partially “closed” but allow weak myosin-binding. Myosin gaining access to actin then “traps” tropomyosins in an otherwise energetically unfavorable “open M-state” position on actin, resulting in strong myosin-binding and actin-activation of the myosin ATPase. Hence, it follows that tropomyosin movement from its C-location into a fully active M-state position on actin requires myosin participation (1, 2, 3, 4). Interestingly, cross-linking studies suggest direct myosin-tropomyosin interaction is possible (5, 6, 7). Thus, myosin does not solely function as a molecular motor during contraction but also acts as a modulator of tropomyosin function, and in many muscles, recruitment of myosin from the surface of the thick filaments also influences activity.
The involvement of troponin in tropomyosin-linked control of thin filament behavior has been investigated in much detail over the past 50 years (reviewed in (3)). By contrast, less is known about regulatory reconfiguration of tropomyosin once myosin binds to thin filaments, particularly when considered from a residue-by-residue structural perspective. For example, it is unclear how myosin displaces tropomyosin from its closed-state position into a default open state (8, 9, 10, 11, 12). As part of this process, attractive and/or repulsive intermolecular chemical forces between myosin and tropomyosin may facilitate the requisite tropomyosin movement. Myosin-tropomyosin interaction, coupled to strong myosin-actin attachment during the cross-bridge cycle is likely to either “push,” “drag” or “chase,” and then trap tropomyosin into the open state. Although cryogenic electron microscopy (cryo-EM) studies by the Raunser, Fujii, and Namba laboratories (10, 11, 12) showed that loop 4 at the tip of the open-state myosin head appears to restrain tropomyosin topologically against a ridge on the extreme inner edge of actin subdomain 3, any further open-state stabilization by myosin-tropomyosin interactions or by actin conformational dynamics is uncertain (10, 11, 12, 13).
In this study, we revisit the structure of M-state thin filaments and address current uncertainties related to myosin-tropomyosin interactions on thin filaments. We define the structure of the myosin motor head on actin-tropomyosin filaments by first performing moderate 4.2 Å resolution cryo-EM of actin-tropomyosin “decorated” with β-myosin subfragment 1 (S1), i.e., with motor heads proteolytically cleaved from full-length myosin. We next apply high-specificity protein-protein-docking protocols to define residue-to-residue contacts made between components and then merge the results of the two methods. We demonstrate periodic interaction between myosin and tropomyosin and provide a more complete paradigm in which myosin-S1 steers tropomyosin away from its default weak C-state interactions with actin subunits along the thin filament, in turn trapping tropomyosin into the M-state position.
Strategy and workflow
The first step in our approach to determine M-state S1-tropomyosin interaction was to generate a three-dimensional (3D) cryo-EM reconstruction of β-myosin-S1-decorated actin-tropomyosin filaments. The objective was to use the reconstruction as a template to fit crystal structures of the filament constituents to generate a hybrid map at high resolution (Fig. 1). Like all recently published cryo-EM structures (10, 11, 12,14,15), a 3–6 Å resolution for actin and S1 is sufficient to match corresponding crystal structures of actin and S1 to densities within these maps (Fig. 1). In our work, tropomyosin resolves as an extremely well-defined coiled coil, yet, like in previous reports (8, 9, 10, 11, 12,14,15), the coiled coil lacks sufficient surface detail or marked asymmetries to readily define the residue-to-residue register of the coiled coil relative to actin and S1. Thus, additional steps were required to determine residue-level longitudinal positioning and rotation of tropomyosin within the EM volume. Here, the programs PIPER and ClusPro (16, 17, 18) were used as computational tools to dock relatively short segments of tropomyosin as ligands to the 4.2 Å resolution actin-S1 model defined by our reconstructions. The docking protocol itself is based on shape complementarity and coiled coil polarity as well as residue-to-residue electrostatic and van der Waals chemical potential. The procedure produced a series of two actin-long acto-S1 cassettes containing tropomyosin fragments that matched respective densities in the EM reconstruction closely, i.e., now the hybrid map contained residue-level tropomyosin assignments in addition to those of actin and S1. The individual cassettes then were stitched together to construct a pseudoatomic model of the M-state filament containing actin, tropomyosin, and S1 components. The composite map was then subjected to energy minimization and molecular dynamics as a means of further refinement (18). It is worth noting that our structural analysis, like that in (12), relied on actin, tropomyosin, and S1 isoforms with sequences found in mammalian muscle.
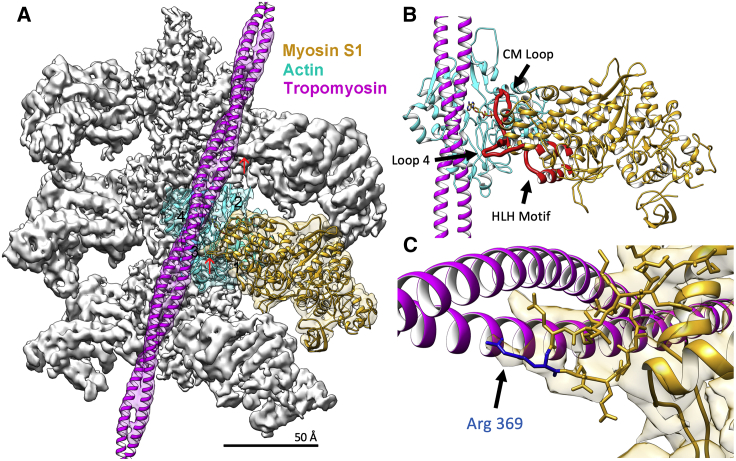
Figure 1.
Cryo-EM reconstruction of S1-decorated actin-tropomyosin filaments. (A) Isosurface rendering of the reconstruction showing atomic models of actin (cyan), myosin (gold), and tropomyosin (magenta) fitted into respective EM densities as described in the Materials and Methods. The pointed end of the filament is facing up; actin subdomains numbered on central actin subunit. Each region of the reconstruction was low-pass filtered to provide better visualization; actin to 4.0 Å, myosin to 5.5 Å, and tropomyosin to 6.5 Å according to local resolution estimates (see Fig. S2). (B) Atomic model of the actin-myosin-tropomyosin complex highlighting the actin-myosin interface. Extensive interactions are formed between actin and the myosin cardiomyopathy loop (CM Loop), loop 4, and the helix-loop-helix (HLH) motif (each highlighted in red). ADP bound to actin is shown. The residue-to-residue contacts between actin and myosin are tabulated in Table S3. (C) Myosin-tropomyosin interaction is visible at myosin loop 4. The cryo-EM density of this region is well resolved, allowing identification of interacting amino acid side chains of myosin, including that of conserved residue Arg369 (blue) making contact with the tropomyosin coil-coil (also noted by red arrows in (A)). To see this figure in color, go online.
Materials and Methods
Proteins used to reconstitute thin filaments
Myosin
β-Myosin was isolated from bovine masseter muscle tissue and S1 then prepared as described in (19). Bovine masseter muscle contains Type 1 slow muscle myosin. In mammals, slow muscle myosin and β-cardiac myosin, the major cardiac muscle isoform, are the same. They are the products of one gene, MyHC7, and have identical heavy chains. The human and bovine isoforms have 96% sequence identity in the motor domain. In the current studies, masseter myosin served as a surrogate for its human cardiac muscle β-myosin counterpart, which is of particular interest because of its dysfunction in many cardiomyopathies. Although these two myosins share virtually identical heavy chains, the isolated and purified masseter myosin and its S1 fragment, unlike the cardiac counterparts, are stable under standard experimental conditions. The masseter S1 can be lyophilized and stored in the presence of 1% (w/v) sucrose before use without loss of activity (19). Also note that loop 4 of mammalian myosin II is highly conserved (see Table S1).
Tropomyosin
Human cardiac α-tropomyosin (Tpm 1.1, also present in significant amounts in skeletal muscles), containing the N-terminal extension Met-Ala-Ser, was expressed and purified as previously (20).
Actin
Skeletal muscle F-actin was prepared by standard methods (21) and used because of its resistance to depolymerization at the low concentrations needed for cryo-EM work; it and cardiac muscle F-actin share the same sequence with conservative substitutions involving four residues.
Sample preparation for cryo-EM
Thin filaments were reconstituted by first mixing F-actin and tropomyosin to final concentrations of 10 μM actin and 7 μM tropomyosin in a buffer consisting 50 mM sodium acetate, 3 mM MgCl2, 1 mM dithiothreitol, 10 mM HEPES buffer at pH 7.0 in a temperature-controlled room at 25°C. We found it essential to replace previously used NaCl or KCl (8) with sodium acetate to prevent tropomyosin dissociation from filaments under cryo-EM conditions. The buffers used throughout preparation were nucleotide-free to maintain “rigor-state” conditions. Excess of tropomyosin was used relative to actin (5:7 mol/mol), rather than a native 1:7 ratio stoichiometry, to ensure full filament saturation. (Although the excess tropomyosin remains unbound, it does not cause noticeable background interference during EM image processing.) Just before applying 1.5 μL actin-tropomyosin to a freshly glow discharged holey-carbon copper grid surface (200 mesh, Quantifoil 2/1; Electron Microscopy Sciences, Hatfield, PA), the surfactant octyl β-D-glucopyranoside (Sigma-Aldrich, St. Louis, MO) was added to the protein solution to a concentration of 12 nM. This facilitated even filament spreading and limited filament adsorption to the air-water interface (22). The grid sample was manually blotted for 1 s at 10°C and 100% humidity, and a 1.5-μL drop of 7.5 μM myosin-S1 subfragment was then applied to the blotted grid sample. The sample was immediately blotted for 4 s and plunge-frozen in liquid ethane using a Vitrobot Mark III system (FEI/Thermo Fisher Scientific, Hillsboro, OR).
Cryo-EM data collection and processing
Samples were examined with a Titan Krios cryo-EM (Thermo Fisher Scientific, Hillsboro, OR) at the Stanford Linear Accelerator Center-National Center for Macromolecular Imaging Cryo-EM facilities (Menlo Park, CA). Micrographs were collected at a magnification of ×130,000 using a 20-eV energy filter. Movies were recorded with a Gatan K2 Summit direct electron detector (Gatan/AMETEK, Berwyn, PA) with each of 35 sequential 0.2 s exposures captured at a dose of 7.5 e−/Å2/s, a defocus of 1.5–2.5 μm and with a pixel size of 1.06 Å. Movie frames were aligned and summed to produce 2496 individual micrographs.
Micrographs were screened to remove images judged to have thick or mottled ice, bundled filaments, or background interference. Image reconstruction was carried out using the RELION 3.0.7 suite of programs, as described by Scheres (23, 24, 25). Briefly, micrographs were first motion corrected using MotioCorr2 and the contrast transfer function (CTF) estimated and normalized by the CtfFind4 subroutines in RELION. Filaments were manually selected and divided into 270 Å long (256 pixels) overlapping segments, each offset by 27.5 Å (90% overlap), and 45,040 segments were extracted from 702 micrographs within the original data. Filament segments were then sorted using reference-free two-dimensional classification. Noisy or empty classes were discarded, resulting in 40,455 segments for processing. To further reduce the number of poorly defined segments to be reconstructed, three-dimensional classification was performed, and class averages inspected for quality, here applying F-actin helical symmetry (166.4° subunit rotation around the F-actin helical axis and a subunit axial translation of 27.5 Å). A final set of 32,158 segments was taken for a preliminary three-dimensional refinement by first aligning these particles to a featureless 200-Å diameter cylinder while applying the above helical parameters. The resulting model generated was low pass filtered to be used as a new reference for a second refinement with a smaller angular sampling of view orientations (3.7°) and incorporating a solvent mask to produce an initial reconstruction at 6.4 Å. We then performed two iterations of CTF refinement and Bayesian “Polishing” (i.e., further electron beam-induced motion correction). Particles underwent CTF refinement with beam tilt estimation and a subset of these particles was evaluated to find optimal polishing parameters. Particles having undergone Bayesian polishing were used in an autorefine run using the 6.4 Å reconstruction, low pass filtered to 60 Å, as a starting model. We additionally employed a soft 6 actin-6 myosin-tropomyosin mask after each iteration to focus the refinement on the central section of the acto-S1 filaments. Our soft solvent mask was created by fitting atomic models of actin, β-myosin-S1, and tropomyosin into the 6.4 Å reconstruction, converting the pdb into an MRC map using the EMAN2 pdb2mrc command (26), and then constructing a mask from this map in RELION. After 24 iterations, this process resulted in a 4.9 Å resolution map (data not shown). The same process was repeated once more to produce a final 4.2 Å resolution map. Further iterations did not improve the resolution. Default processing parameters were used except as noted above. Resolution estimation was made using Fourier shell correlation criteria, FSC0.143, in RELION. (See Figs. S1 and S2; Table S2).
All figures were rendered using UCSF Chimera (27).
Acto-S1 model building
The cryo-EM reconstruction served as a template to build an atomic model of the acto-S1 structure in the rigor state. Using the “Fit In Map” subroutine in UCSF Chimera (27), an F-actin structure taken from the Yamada et al. map (15) (Protein Data Bank, PDB: 6KN8) fitted within its corresponding density of the masseter S1-decorated actin-tropomyosin reconstruction. The von der Ecken actin maps (11,14) (PDB: 5JLH and PDB: 5KLF) also fitted in the volume without any notable difference. It would have been ideal to also directly match the volume of myosin in the reconstruction with a crystal structure of striated muscle S1 and then merge it with the actin fitted in the map. However, there is no complementary high-resolution model of striated muscle “rigor-state” S1, nor structures of the β-cardiac or masseter muscle myosin protein available that are nucleotide-free. Therefore, we built a homology model of the β-myosin-S1 based on the rigor-state crystal structure of the squid muscle myosin-S1 (28) (PDB: 3I5G) (as done previously by Fujii and Namba (12) for the same purpose), here replacing squid residues with corresponding residues in the bovine amino acid sequence (UniProt: Q9BE39). To accomplish this, the SWISS-MODEL suite of programs (29) was used to build the bovine masseter β-myosin-S1 model. Please note that we use the bovine β-myosin sequence numbering throughout the text and in the Supporting Material. The homology model then was fitted to the cryo-EM map using the Fit In Map subroutine in UCSF Chimera (27). The vast majority of Cα-chains in the myosin-S1 motor domain, ranging from residues 216 to 624, matched densities visible in the reconstruction. Myosin loop 1 residues 199–216 and loop 2 residues 624–641 located at the distal ends of the reconstructed S1 were removed from the model because of the lack of definitive density for fitting to these regions. Loop 4 (amino acids 354–380) which varies between muscle and nonmuscle sarcomeric myosins 2, but conserved among muscle myosins (see Table S1) was replaced by the corresponding motif of β-cardiac myosin solved in a postrigor state (30) (PDB: 6FSA). We then refined the models using flexible fitting routines by employing the MDFF routine in the program VMD (31,32), while implementing two plugins, Chirality and Cispeptide, in VMD to prevent possible stereochemical errors and/or overfitting. Each residue in the actin and β-myosin models was assessed and optimized using iterative cycles of real-space refinement with the program PHENIX and manual rebuilding with Coot to diminish Ramachandran outliers, atom clashes, and to optimize rotamer positions to the central EM density (See Figs. S1 and S2; Table S2; (33,34)).
Protein-protein docking
The position of tropomyosin on S1 decorated F-actin was characterized through a global conformational search, using the programs PIPER and ClusPro (16,17) as previously implemented for S1-free F-actin and described in detail (18). In brief, the search was conducted using a series of tropomyosin segments as ligands and acto-S1 as the receptor. Indeed, tropomyosin is a modular protein, which in the muscle isoform consists of seven pseudorepeating elements each represented by tandem α-zones and β-zones; in thin filaments, α-zones lie over subdomains 1 and 3 of actin, and β-zones lie over subdomains 2 and 4 of actin. Each tropomyosin segment here contained of two consecutive α-zones of tropomyosin with an intervening β-zone (58 amino acids in total and ∼85 Å in length). These were docked to atomic model targets comprised of two actin monomers both linked to an S1, using our cryo-EM reconstruction model, where the end of S1 distal to actin was truncated (residues 35–217 and 625–776 were removed) to focus the computation search on the acto-S1 interface. Tropomyosin fragments used for docking were defined as α1β1α2 (residues 8–66), α2β2α3 (residues 47–105), α3β3α4 (residues 86–145), α4β4α5 (residues 127–184), α5β5α6 (residues 165–223), α6β6α7 (residues 205–263) segments, and N- and C-terminal overlapping domains consisting of α1β1α2 (residues 1–66) and β6α7β7 (residues 230–284). The top ranked structures for each of the segments in the docking trials (Fig. 2, A–F) were ascertained based on a combination of van der Waals and electrostatic criteria and they were judged further by visual inspection of protein-protein interactions, polarity relative to the F-actin axis and superimposition onto the above cryo-EM reconstruction with the use of UCSF Chimera (27).
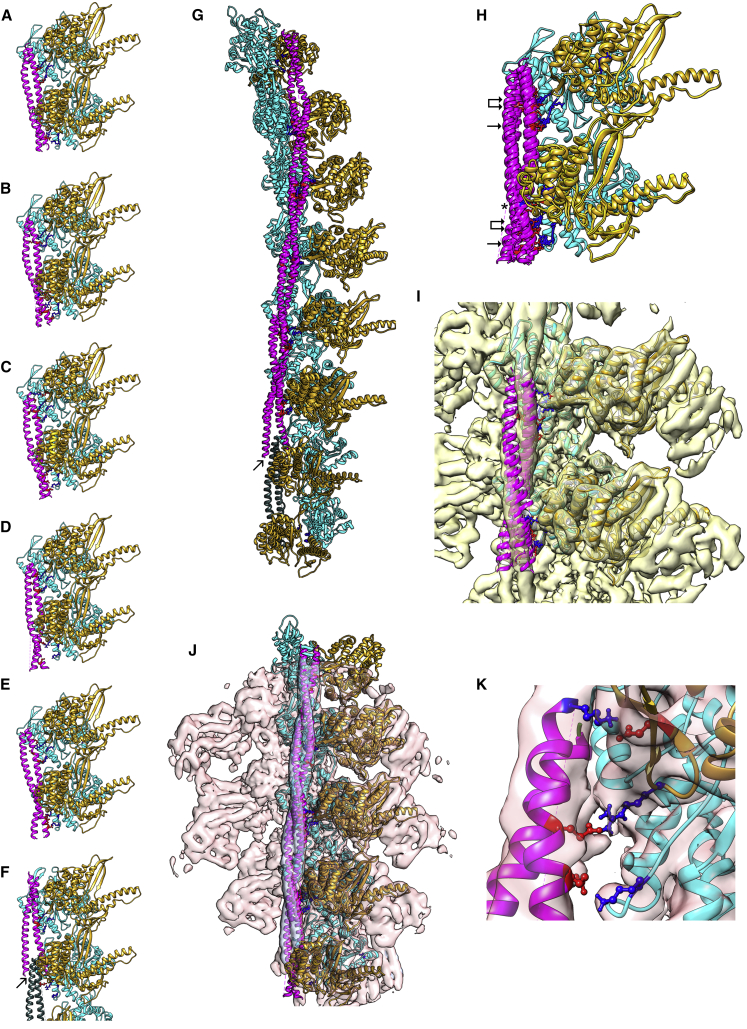
Figure 2.
Docking tropomyosin segments to S1-decorated F-actin. (A–F) Highest scoring αβα-zone tropomyosin segments docked to acto-S1 (tropomyosin, magenta; S1, gold; actin, cyan), showing tropomyosin poses on acto-S1 associated with fragment (A) α2β2α3, (B) α3β3α4, (C) α4β4α5, (D) α5β5α6, (E) α6β6α7, and (F) the overlapping domain (arrow) and surrounding residues, i.e., β6α7β7α1β1α2. Ribbon diagrams are shown with sidechains on actin Lys326 and Lys328 as well as on myosin Arg369 colored blue, along with their interacting partners on tropomyosin in the M-state, colored red. In (A)–(F), segments are offset by one actin subunit and thus, each is related to the next by one common subunit. In (G), the common parts of segments shown in (A)–(F) were superposed to generate a composite “single-stranded” actin-tropomyosin filament; the arrow points to residues in the overlapping domain that form a four-helix bundle in (F). In (H), segments (A)–(E) were directly superposed on each other. Note the superposed motif of actin Lys326 (blue, single arrows) and of myosin Arg369 (blue, double arrows) interacting with sidechains of oppositely charged residues on the various tropomyosin fragments (red); also note occasional interaction of myosin Glu374 and tropomyosin (asterisk). (I) superposed segments fitted within the isosurface of the reconstruction in Fig. 1, using the “Fit in Map” program in UCSF Chimera (27), and similarly in (J), segments α2β2α3, α3β3α4, α4β4α5, and α5β5α6 from the pseudoatomic map in (G) were fitted to the reconstruction. (K) Enlargement of tropomyosin pseudorepeat 4 region in (J), showing side-chain interactions between actin Lys326 and tropomyosin Glu142 (bottom pair), myosin Arg369 and tropomyosin Glu138 (middle pair), and myosin Glu374 and tropomyosin Lys128 (top pair). Please note that the docking in (A)–(F), as well as the superimposition of segments in (G)–(J) and “Fit in Map” alignment to cryo-EM reconstructions in (I) and (K), did not involve any flexible fitting routines or other manipulation of respective coordinates, i.e., only “raw” PIPER/ClusPro output was evaluated. S1 residues 217–623 are shown in the ribbon models displayed because, here, the end of S1 distal to actin was truncated (residues 35–216 and 624–776 were removed) to focus the computation search on the acto-S1 interface. Pointed end of actin was facing up in all panels. To see this figure in color, go online.
Construction and refinement of tropomyosin models
Once tropomyosin docking was completed, the fragments were assembled computationally into a pseudoatomic model by splicing them together to form a continuous tropomyosin model on acto-S1 (Fig. 2 G). The first common α-zone between any two respective fragments were superimposed, and then continuing in this manner an entire chain was created. The resulting models were then subjected to flexible fitting refinement to the cryo-EM volume reference structure (MDFF) (31) and by standard molecular dynamics in explicit solvent (Fig. 3, A and B).
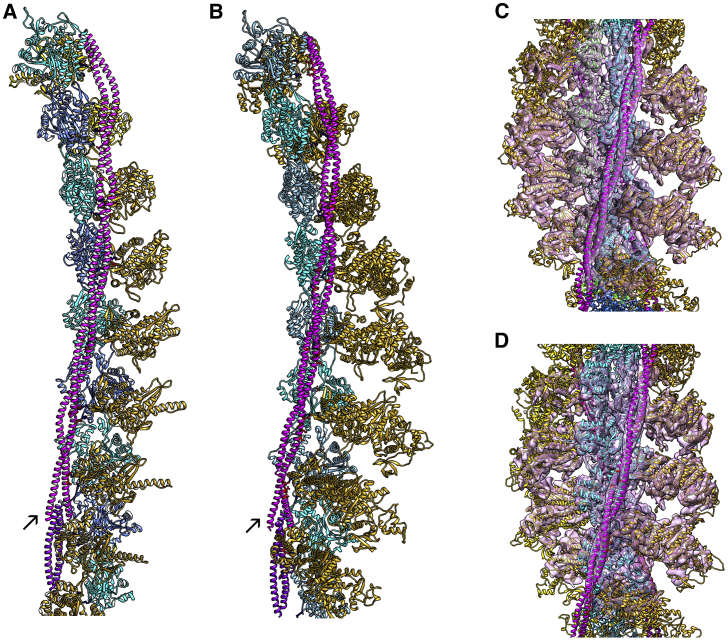
Figure 3.
Annealing docked segments together and molecular dynamics of the M-state model. This figure has the same color coding as in Figs. 1 and 2; in (A) and (B) only a single-helical strand of F-actin and associated proteins are shown. (A) Respective tropomyosin pseudorepeats docked to actin-S1 segments in Fig. 2, A–F shown annealed together by flexible fitting routines. (B) An atomic model of M-state filaments subjected to molecular dynamics in explicit solvent. The MD was started from a canonical model of tropomyosin fitted to the cryo-EM 3D-reconstruction and then threaded into the structure of the tropomyosin overlapping domain complex, i.e., structures described in (15) and (18). The tropomyosin-coiled coil remained intact and its position on actin stable throughout simulation; side-chain interactions from tropomyosin to actin and S1 are noted on Table 1 and highlighted in the figure (red and blue for respective acidic and basic residues). The representative trajectory snapshot shown in (B) was taken from the 30-ns time period of simulation and is a frame that had minimal root mean-square deviation from the average structure over that time period (calculated by backbone atoms superimposition). In (C) and (D), the respective midpieces of the modeled filaments (representing repeats 2–6) shown in (A) and (B) were fitted within densities of the reconstruction of S1-decorated acto-tropomyosin. Note that the construction of starting models in (A) and (C) and in (B) and (D) differed yet yielded virtually the same outcomes. Arrows indicate the position of the tropomyosin overlapping domain in (A) and (B). In (A), S1 residues 217–623 are shown in ribbon format, and in (B)–(D), the full S1 ribbon structure (to residue 777) is shown. To see this figure in color, go online.
To address the tropomyosin head-to-tail overlapping domain interaction with S1, a model of full-length tropomyosin on fully decorated acto-S1 was built starting from an ideal superhelical coiled coil structure (35) that had been optimized by MDFF flexible fitting to the EM density map. A docked overlap domain and surrounding residues were then threaded into the midpiece of the tropomyosin model by combining common residues and the entire structure energy minimized, as done previously (18). The resulting actin-S1-tropomyosin construct was extended to a cable model in a periodic boundary system (18) to simulate tropomyosin dynamics on an infinite long filament during molecular dynamics. Molecular dynamics was carried out using explicit solvent, as previously (18,32) for 30 ns.
Results and Discussion
The following sections detail and appraise results of our stepwise approach in building an atomic model of S1-decorated actin-tropomyosin filaments and in determining the cardiac muscle myosin-head-tropomyosin interaction.
The reconstruction
To evaluate how the binding of myosin affects tropomyosin position, actin-tropomyosin filaments were saturated with myosin-S1 in the absence of ATP, thus reflecting myosin on actin in the “rigor” configuration. Our cryo-EM 3D reconstruction of the S1-decorated filament achieved a resolution of 4.2 Å and shows that myosin-S1 makes extensive contact with the actin subdomains 1 and 2 on the outer aspect of actin subunits (Fig. 1; Table S3). Conversely, tropomyosin lies entirely over the actin subdomains 3 and 4 on the inner aspect of actin. These results are common to all previous reconstructions of M-state thin filaments dating back 30 years (8, 9, 10, 11, 12,36). Despite an ∼13° average rotational translation of tropomyosin around the actin filament and very small longitudinal shift (0.6 Å) over actin to the M-state from the previously delineated C-state position (12,18), the current reconstruction shows relatively little direct contact between myosin and tropomyosin except for a well-resolved extra density emerging from the tip of myosin head. This density, representing loop 4 of myosin, bridges the border between actin subdomains 1 and 3 and extends to the tropomyosin density (Fig. 1, red arrows in Fig. 1 A mark the loop 4 projection, which is most easily viewed by zooming-in to an ∼2-fold magnification).
Fitting actin and S1 to the reconstruction
As mentioned, the well-defined secondary structure as well as side chain density captured by the thin filament reconstruction allows high-resolution structural models to be fitted accurately into corresponding cryo-EM density map. Skeletal muscle F-actin (e.g., PDB: 6KN8) fitted directly to corresponding densities in the reconstruction map without noticeable changes to the structure during follow-up flexible fitting (Fig. 1). Individual actin densities in common with side-chain functional groups in the atomic model were visible in most of the actin structure. The actin subunit configuration in the maps (Electron Microscopy Data Bank ID: EMD-22067; PDB: 6X5Z) are completely consistent with prior solutions irrespective of the actin source (10, 11, 12,14,16).
High-resolution structures of vertebrate muscle myosin-S1 in a nucleotide-free “rigor state” have not been solved and therefore are not available to merge into the cryo-EM map nor to characterize the S1 conformation. After the procedure of Fujii and Namba (12) (see Materials and Methods for details and rationale), a homology model of S1 was therefore built based on a crystal structure of squid muscle S1 in its rigor-like state (PDB: 3I5G, from fast funnel retractor muscle). The model was then fitted into its corresponding density volume in the reconstruction and its complementarity to the map refined by flexible fitting to match residue-level density and further using the programs PHENIX and Coot (33,34). A composite atomic model, containing both fitted actin and S1, shows residue-to-residue contacts at the interface between actin and S1, which are the same as previously proposed (10, 11, 12); namely S1 interacts extensively with the actin surface via the so-called cardiomyopathy loop (residues Thr417 to Pro402), loop 4 (residues Gly360 to Glu379), and the helix-loop-helix motif (residues Ile530 to His556) (Fig. 1; Table S3; Electron Microscopy Data Bank ID: EMD-22067; PDB: 6X5Z; the sequence coding Q9BE39 for MYH7_BOVIN is cited). Our main interest here, however, concerns residue-to-residue interactions between tropomyosin and acto-S1, which as outlined above have remained ambiguous.
The two chains of the tropomyosin coiled coil are easily visible in our reconstruction. The average 137 Å helical pitch of the coiled coil around its own axis is well-characterized and retained in the cryo-EM map. However, because of helical averaging, the short 9–11 residue overlapping domain itself is lost during image processing. In addition, the localized over- and undertwisting of residues in tropomyosin pseudorepeats 1 and 7 (18,37) required during the overlapping domain assembly also are not retained, but can be assessed computationally (see below).
Myosin loop 4 approaches tropomyosin closely
A focus was placed on possible interactions of loop 4 of myosin which interacts closely with tropomyosin. As noted above, in addition to contacting the surface of actin, the loop 4 density protrusion extends from the end of myosin’s upper 50K domain in the direction of tropomyosin. Here, loop 4 reaches over the border between subdomain 1 and 3 of actin and toward the underside of the tropomyosin coiled coil lying on actin (Fig. 1 C). Not only is the loop 4 density well-resolved in the reconstruction, but the homology model fitting identifies the individual residues comprising loop 4 and the adjoining tropomyosin. Moreover, conspicuous extra density is observed to project outward at the very tip of the protruding amino acid loop that represents the side chain of arginine 369 pointing toward tropomyosin. As discussed below, Arg369 of the myosin head may act to reposition tropomyosin from its default C-state position on actin and stabilize it in the M-state location. In the process, C-state tropomyosin interactions with actin residues Lys326 and Lys328 (18) appear to be replaced by ones on myosin Arg369 and more fleeting new links to Lys326.
Molecular level docking of tropomyosin segments to S1-decorated actin
To date, individual tropomyosin residues have not been resolved in cryo-EM maps (8, 9, 10, 11, 12, 13, 14, 15,36), leaving modeling of thin filament regulatory mechanisms uncertain. A bottom-up computational approach was therefore taken here to perform M-state tropomyosin docking onto the S1-decorated actin to determine potential residue-to-residue contacts formed between tropomyosin and myosin. Cardiac tropomyosin (αα-Tpm1.1) was first divided computationally into a series of overlapping segments centered on successive tropomyosin β-zones within individual tropomyosin pseudorepeats. Each β-zone was flanked on both its ends by respective α-zones, thus yielding a series of αiβiαi+1 fragments. We previously found this segment length was the shortest needed for PIPER/ClusPro programs to dock segments of the coiled coil protein onto F-actin with known superhelical orientation and polarity (18). In the current study, the same tropomyosin fragments were docked to the S1-decorated F-actin structure determined above. The docking required a minimum size acto-S1 “receptor” containing two longitudinally linked actin subunits, each associated with S1 motors in a nucleotide-free “rigor” conformation. This arrangement yielded tropomyosin poses with correct superhelical orientation and polarity (Fig. 2, A–G). After docking, all αiβiαi+1 segments examined (i.e., α2β2α3, α3β3α4, α4β4α5, α5β5α6, α6β6α7) produced tropomyosin α-zone structural motifs facing acto-S1, and with individual α-zones in common or entire segments being superposable (Fig. 2, H and I). In all instances, acidic residues on one or both chains of tropomyosin come within 5 Å of the oppositely charged terminal guanidino side chain of Arg369 which extend from myosin loop 4 (Table 1). In fact, Arg369 of loop 4 is buttressed on one side by Gln368 and Arg367, conserved in all mammalian sarcomeric myosin II isoforms, presumably to maintain a positively charged local environment that strengthens tropomyosin contacts (see Table S1). In addition, neighboring acidic residues on tropomyosin (e.g., residues 104, 142, 180) contact Lys326 on successive actin subunits. These contacts appear to form a second set of interactions with tropomyosin (Fig. 2). Thus, it is attractive to consider that myosin-S1 relocates tropomyosin away from its C-state interactions with actin (18), while creating new contacts with actin and tropomyosin. In the process, tropomyosin moves azimuthally ∼13° (9 Å) from its C-state position and less than 1 Å in its longitudinal placement.
Table 1.
Charged Residues that Closely Contact Each Other in the M-State; Tropomyosin and Arginine 369 on Myosin Loop 4; Tropomyosin and Lysine 326 on Actin
| Actin Subunit | Tpm Period | ClusPro Docking |
MD |
||
|---|---|---|---|---|---|
| Myosin Arg369 |
Actin Lys326 |
Myosin Arg369 |
Actin Lys326 |
||
| Tropomyosin Residue | Tropomyosin Residue | ||||
| 7 | 1 | Glu16∗ | – | Glu16∗ | Asp20∗, Glu23∗ |
| 6 | 2 | Glu58 | Glu69 | Glu54∗, Asp58∗ | Glu69 |
| 5 | 3 | Glu97, Glu98 | Glu104∗ | Glu96∗, Glu104 | Glu100∗, Glu104∗ |
| 4 | 4 | Asp137, Glu138∗ | Glu142∗ | Glu131∗, Asp137∗, Glu138∗ | Glu142∗ |
| 3 | 5 | Glu173∗, Asp175 | Glu180∗, Glu181 | Glu173, Asp175∗, Glu180∗ | Glu180∗, Glu181 |
| 2 | 6 | Glu212, Glu218 | Asp219, Glu223∗ | Glu212, Glu218∗ | Asp219, Glu223 |
| 1 | 7 | Asp254, Glu257∗ | Asp258 | Glu250∗, Asp254∗, Glu257∗ | Asp258 |
Residue-to-residue contacts determined for structures displayed in Fig. 3. Acidic residues on tropomyosin within 10 Å of basic ones either on myosin or on actin are listed. Residues underlined in the third and fourth columns were determined by ClusPro docking (a static measurement) to lie less than 5 Å from their oppositely charged partners and those additionally flagged by an asterisk were separated by less than 2 Å. Residues underlined in the fifth and sixth columns were determined by MD (which measures dynamic interactions) to lie “on average” less than 5 Å of oppositely charged partners and those making intermittent contacts with each other by less than 2 Å are marked by asterisks. The docking studies also showed contact made between Glu374 on myosin and Lys128 on tropomyosin (<5 Å apart); during MD, contacts are often noted between Glu374 on myosin and residues Lys12, Lys128, and Lys205 on tropomyosin (<2 Å apart).
Because each tropomyosin fragment docked to acto-S1 was offset from the next by approximately one-half pseudorepeat, their sequences in common could also be superposed together (e.g., α2β2α3 + α3β3α4 to yield α2β2α3β3α4; α2β2α3β3α4 + α4β4α5 to yield α2β2α3β3α4β4α5, etc.). In this way, tropomyosin with its underlying actin and S1 substructure formed a complete pseudoatomic model of an S1-decorated filament (Fig. 2 G). Even though all segment-associated tropomyosin fragments fitted within densities in the cryo-EM reconstruction, periods 2, 3, 4, and 5 of tropomyosin (derived from α2β2α3, α3β3α4, and α4β4α5) aligned best to the EM volume (Fig. 2, I–K). To more faithfully display tropomyosin strand continuity, these actin-tropomyosin-S1 segments were flexibly fitted to the cryo-EM map, which allowed their ends to anneal to form seamless cables on F-actin (Fig. 3, A and C). This was achieved without constraining the respective axial or azimuthal positioning of the tropomyosin. The refined tropomyosin cables were then subjected to MD simulation, maintaining all the same residue-to-residue contacts with actin noted above (Fig. 3, B and D). Thus, the results of the docking and the cryo-EM procedures cross-validate each other.
Thus, the PIPER/ClusPro-derived model of actin, tropomyosin, and S1 fitted well within the EM densities with accuracy already noted for atomic structure fitting shown in Fig. 1. Not only are Asp369 residues on S1 loop 4 particularly well fitted, but the side chains of actin residue Lys326 are also oriented toward oppositely charged tropomyosin targets (Figs. 2, J and K and 3 C) on each pseudorepeat. As noted above, the latter residue is strongly associated with actin-tropomyosin binding in different regulatory states.
Docking the tropomyosin overlapping domain to S1-decorated actin and MD on the full acto-tropomyosin-S1 complex
The tropomyosin overlapping domain docks precisely to S1-decorated actin without clashing with S1 residues and thus does not obstruct the myosin-binding site on actin (Fig. 2 F). Notably, the additional local presence of TnT1, modeled as in (18), also does not obstruct myosin-binding (data not shown). Indeed, PIPER/ClusPro docking indicates that repeats 2–6 as well as the overlapping domain appear to undergo a strict azimuthal sliding from their C-state to their M-state position without obvious conformational reconfiguration or significant longitudinal translation. A preliminary model of full-length tropomyosin in the M-state configuration was therefore constructed computationally by threading pseudorepeats 2–6 to the overlapping domain. MD was carried out, and the composite model remained stable over time. However, loop 4 was seen to be flexible, possibly to optimize Arg369 orientation to favorably target residues on tropomyosin, thus compensating for mismatched thick and thin filament symmetry in vertebrate muscle. Loop 4 flexibility may also contribute to Arg369 promiscuity observed during MD, allowing it the freedom to interact with one or another closely neighboring acidic residues on tropomyosin. Thus, the apparent flexibility of loop 4 may provide a mechanism for myosin heads to adjust to the slight mismatches in orientation and optimize contact of tropomyosin during the cross-bridge cycle on thin filaments. Glu374 on loop 4 also interacts transiently with basic residues on tropomyosin as does Lys326 on actin with acidic residues on tropomyosin, thus bolstering interactions.
Tropomyosin’s roadmap from C to shining M
Cryo-EM reconstruction and tropomyosin docking protocols have identified structures that offer a glimpse of average end-state regulatory structures that reflect blocked, closed, and open biochemical states (see Fig. S3; (8, 9, 10, 11, 12, 13, 14, 15)). In turn, these structures provide insights about the roadmap that myosin must navigate while transitioning between thin filament activation and relaxation. Simply overlaying the M-state S1 configuration over B- or C-state actin-tropomyosin (8, 9, 10, 11, 12, 13, 14, 15) shows that loop 4 of myosin clashes with tropomyosin in prepower stroke myosin-head configurations (for example, see (9, 10, 11)). This steric clash is expected to prevent cleft closure between the upper and lower 50K domains of the myosin head, which is required for strong myosin-actin interaction and productive cross-bridge cycling to occur on actin filaments during muscle contraction (8, 9, 10, 11). Thus, steric interference by tropomyosin in both the low Ca2+-linked blocked B-state and the high Ca2+-linked C-state will inhibit myosin interaction with actin and therefore contraction (8,9). Without tropomyosin displacement from both B- and C-state positions, muscles cannot contract. At low-Ca2+, TnI constrains tropomyosin sufficiently that myosin.ADP.Pi cannot nudge tropomyosin strongly enough to overcome the steric interference imposed by the B-state thin filament (15). It is also possible that localized charge repulsion between loop 4 and TnI-stabilized tropomyosin may further limit the possibility of actin-myosin interaction. However, once tropomyosin is freed of TnI-imposed constraints at high Ca2+, any charge repulsion between loop 4 of weakly bound myosin in the C-state may now promote tropomyosin movement toward the M-state as alluded to by Vibert et al. (8). Alternatively, tropomyosin simply may be propelled toward the M-position by myosin’s competition for residues on actin around the C-state binding site for tropomyosin, effectively pushing tropomyosin away. These alternatives are difficult to decipher energetically or at present by cryo-EM. Equally uncertain is the temporal order of myosin cleft closure, the myosin power stroke, Pi release from myosin after ATP hydrolysis and M-state myosin binding itself or, indeed, if there is a true fixed order. Clearly, weak binding of myosin to C-state thin filaments occurs (2), but when in a kinetic scheme, myosin cleft closure happens has not as yet been revealed by cryo-EM.
In any event, the first myosin bound to C-state actin is expected to have an ∼80% chance of nudging tropomyosin to the M-state position (2,38), accompanied by cleft closure between the myosin upper and lower 50K domains. Then, in a coordinated way, tropomyosin movement from C- to M-states drops myosin loop 4 to a site directly opposed to tropomyosin, likely trapping tropomyosin in the M-state (11), which is coupled to a new local electrostatic energy valley. Little or no additional nudging would be needed to sustain the M-state and promote additional myosin-binding events, given the high persistence length of tropomyosin (39). It is noteworthy that our studies support the premise that myosin-tropomyosin interaction is involved in thin filament regulatory transitions (5, 6, 7).
It is often noted that tropomyosin function is strongly coupled to its unique coiled-coil-structural patterning (40), and this leitmotif is a recurring theme of the current and previous articles (18,37,40,41). It is also worth emphasizing that experimental deletion of the central repeat period regions 3, 4, and 5 greatly reduces tropomyosin affinity for myosin-bound actin filaments but has little effect on tropomyosin interaction with myosin-free thin filaments, suggesting that binding between tropomyosin and myosin on actin is pseudodomain specific (42, 43, 44). Interestingly, our structural work indicates that although all pseudorepeats of tropomyosin contain specific acidic residues likely to interact with myosin loop 4 during the M-state, the central group of tropomyosin pseudorepeats contain relatively larger clusters of acidic residues poised to link to Arg369 on loop 4. Thus, the midpiece of tropomyosin could represent a particularly favorable target for the myosin-head-binding actin, leaving pseudorepeats 1 and 7 with a more restricted role, namely forming the tropomyosin-overlapping domain and, therefore, having lower probability of myosin binding. It is also worth noting that noncanonical d-position tropomyosin residue Asp137 in pseudorepeat 4, which is strictly conserved phylogenetically, may be required to prime the midpiece of the tropomyosin-coiled coil as a hot spot for myosin-binding. The absence of stabilizing electrostatic linkages formed by residues Glu131 and Glu138 between tropomyosin’s coiled coil α-helices (i.e., e-g pairing) is also notable (depicted in Fig. 1 of (41)), possibly serving the same purpose. The functional importance of tropomyosin pseudorepeat 4 is additionally underscored by the observation that few, if any, mutation-linked cardiomyopathies are attributed to this sequence. This suggests that such perturbations might not be compatible with life. These and other possibilities are well-suited for further investigation.
Author Contributions
W.L. thought of the general approach taken and along with M.H.D., M.J.R., E.P., J.R.M., M.R., and M.A.G. formulated the principal concepts presented. M.H.D. performed the cryo-EM, generated the 3D reconstruction, and did the hybrid crystallography; he was trained and advised jointly by W.L., E.B., and M.J.R. E.P. carried out the docking studies. E.P. and M.J.R. performed the molecular dynamics simulations. J.W. prepared the masseter S1 and carried out the myosin sequence analyses. All authors participated in data analysis and interpretation. W.L. and M.H.D. wrote the manuscript. M.H.D., W.L., and J.W. prepared the figures and tables.
Acknowledgments
This work was funded by National Institutes of Health (NIH) Grants R01HL036153 (to W.L.), R01HL123774 (to J.R.M. and W.L.), P30AR074990, RM1GM131981, R01HL128368 (to M.R.), and R01GM029090 (awarded to Leslie A. Leinwand and sponsoring M.A.G.). M.A.G., J.W., and M.R. were also supported by the European Union’s Horizon 2020 Research and Innovation Programme Grant 777204 SILICOFCM. M.H.D. was supported by NIH Training Program Grant T32HL007969 (to Katya Ravid) and by the Boston University Division of Graduate Medical Sciences institutional funds. Computational work was carried out in house and using resources provided by the Massachusetts Green High Performance Computing Center. Preliminary screening of electron microscope samples was carried out in house and supported by NIH Grant S10RR25434 (to E.B.); the rest of the electron microscope work was performed at the Stanford Linear Accelerator Center Cryo-EM Facilities (S2C2) supported by Stanford University and the NIH Common Fund Transformative High-Resolution Cryoelectron Microscopy Program (U24 GM129541) as well as NIH Grant U24GM116787 (to Wah Chiu). Some of this work was performed at the Regional Cryo-EM Consortium, which is also supported by NIH funding (U24GM116787) and the S10 Instrumentation Programs (S10OD021600). The content of the work presented is solely the responsibility of the authors and does not represent the official views of the NIH or the European Union.
Editor: David Thomas.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.07.006.
Supporting Material
References
- 1.Gordon A.M., Homsher E., Regnier M. Regulation of contraction in striated muscle. Physiol. Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 2.Geeves M.A. Thin filament regulation. In: Egelman E.H., Goldman Y.E., Ostap E.M., editors. Volume 4. Academic Press; 2012. pp. 251–267. (Comprehensive Biophysics: Molecular Motors and Motility). [Google Scholar]
- 3.Lehman W. Thin filament structure and the steric blocking model. Compr. Physiol. 2016;6:1043–1069. doi: 10.1002/cphy.c150030. [DOI] [PubMed] [Google Scholar]
- 4.Holmes K.C., Lehman W. Gestalt-binding of tropomyosin to actin filaments. J. Muscle Res. Cell Motil. 2008;29:213–219. doi: 10.1007/s10974-008-9157-6. [DOI] [PubMed] [Google Scholar]
- 5.Lehrer S.S. The regulatory switch of the muscle thin filament: Ca2+ or myosin heads? J. Muscle Res. Cell Motil. 1994;15:232–236. doi: 10.1007/BF00123476. [DOI] [PubMed] [Google Scholar]
- 6.Golitsina N.L., Lehrer S.S. Smooth muscle α-tropomyosin crosslinks to caldesmon, to actin and to myosin subfragment 1 on the muscle thin filament. FEBS Lett. 1999;463:146–150. doi: 10.1016/s0014-5793(99)01589-6. [DOI] [PubMed] [Google Scholar]
- 7.Lehrer S.S. The 3-state model of muscle regulation revisited: is a fourth state involved? J. Muscle Res. Cell Motil. 2011;32:203–208. doi: 10.1007/s10974-011-9263-8. [DOI] [PubMed] [Google Scholar]
- 8.Vibert P., Craig R., Lehman W. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 9.Poole K.J., Lorenz M., Holmes K.C. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 10.Behrmann E., Müller M., Raunser S. Structure of the rigor actin-tropomyosin-myosin complex. Cell. 2012;150:327–338. doi: 10.1016/j.cell.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von der Ecken J., Heissler S.M., Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- 12.Fujii T., Namba K. Structure of actomyosin rigour complex at 5.2 Å resolution and insights into the ATPase cycle mechanism. Nat. Commun. 2017;8:13969. doi: 10.1038/ncomms13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehman W., Orzechowski M., Raunser S. Gestalt-binding of tropomyosin on actin during thin filament activation. J. Muscle Res. Cell Motil. 2013;34:155–163. doi: 10.1007/s10974-013-9342-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von der Ecken J., Müller M., Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamada Y., Namba K., Fujii T. Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat. Commun. 2020;11:153. doi: 10.1038/s41467-019-14008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozakov D., Hall D.R., Vajda S. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13-19. Proteins. 2010;78:3124–3130. doi: 10.1002/prot.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozakov D., Hall D.R., Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavadai E., Lehman W., Rynkiewicz M.J. Protein-protein docking reveals dynamic interactions of tropomyosin on actin filaments. Biophys. J. 2020;119:75–86. doi: 10.1016/j.bpj.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloemink M.J., Adamek N., Geeves M.A. Kinetic analysis of the slow skeletal myosin MHC-1 isoform from bovine masseter muscle. J. Mol. Biol. 2007;373:1184–1197. doi: 10.1016/j.jmb.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coulton A.T., Koka K., Geeves M.A. Role of the head-to-tail overlap region in smooth and skeletal muscle beta-tropomyosin. Biochemistry. 2008;47:388–397. doi: 10.1021/bi701144g. [DOI] [PubMed] [Google Scholar]
- 21.Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 22.Noble A.J., Wei H., Carragher B. Reducing effects of particle adsorption to the air-water interface in cryo-EM. Nat. Methods. 2018;15:793–795. doi: 10.1038/s41592-018-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheres S.H. Processing of structurally heterogeneous cryo-EM data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 25.He S., Scheres S.H.W. Helical reconstruction in RELION. J. Struct. Biol. 2017;198:163–176. doi: 10.1016/j.jsb.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang G., Peng L., Ludtke S.J. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 27.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y., Gourinath S., Cohen C. Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor. Structure. 2007;15:553–564. doi: 10.1016/j.str.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Waterhouse A., Bertoni M., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robert-Paganin J., Auguin D., Houdusse A. Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun. 2018;9:4019. doi: 10.1038/s41467-018-06191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trabuco L.G., Villa E., Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonine P.V., Poon B.K., Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emsley P., Lohkamp B., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenz M., Poole K.J., Holmes K.C. An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin-tropomyosin gels. J. Mol. Biol. 1995;246:108–119. doi: 10.1006/jmbi.1994.0070. [DOI] [PubMed] [Google Scholar]
- 36.Milligan R.A., Whittaker M., Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- 37.Lehman W., Rynkiewicz M.J., Moore J.R. A new twist on tropomyosin binding to actin filaments: perspectives on thin filament function, assembly and biomechanics. J. Muscle Res. Cell Motil. 2020;41:23–38. doi: 10.1007/s10974-019-09501-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKillop D.F., Geeves M.A. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X.E., Holmes K.C., Fischer S. The shape and flexibility of tropomyosin coiled coils: implications for actin filament assembly and regulation. J. Mol. Biol. 2010;395:327–339. doi: 10.1016/j.jmb.2009.10.060. [DOI] [PubMed] [Google Scholar]
- 40.Hitchcock-DeGregori S.E. Tropomyosin: function follows structure. Adv. Exp. Med. Biol. 2008;644:60–72. doi: 10.1007/978-0-387-85766-4_5. [DOI] [PubMed] [Google Scholar]
- 41.Lehman W., Li X., Rynkiewicz M.J. Precise binding of tropomyosin on actin involves sequence-dependent variance in coiled-coil twisting. Biophys. J. 2018;115:1082–1092. doi: 10.1016/j.bpj.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hitchcock-DeGregori S.E., Barua B. Tropomyosin structure, function, and interactions: a dynamic regulator. Subcell. Biochem. 2017;82:253–284. doi: 10.1007/978-3-319-49674-0_9. [DOI] [PubMed] [Google Scholar]
- 43.Landis C., Back N., Tobacman L.S. Effects of tropomyosin internal deletions on thin filament function. J. Biol. Chem. 1999;274:31279–31285. doi: 10.1074/jbc.274.44.31279. [DOI] [PubMed] [Google Scholar]
- 44.Hitchcock-DeGregori S.E., Song Y., Moraczewska J. Importance of internal regions and the overall length of tropomyosin for actin binding and regulatory function. Biochemistry. 2001;40:2104–2112. doi: 10.1021/bi002421z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.