Abstract
Lung cancer has been the focus of attention for many researchers in recent years for the leading contribution to cancer-related death worldwide, in which lung adenocarcinoma (LUAD) is the most common histological type. However, the potential mechanism behind LUAD initiation and progression remains unclear. Aiming to dissect the tumor microenvironment of LUAD and to discover more informative prognosis signatures, we investigated the immune-related differences in three types of genetic or epigenetic characteristics (expression status, somatic mutation, and DNA methylation) and considered the potential roles that these alterations have in the immune response and both the immune-related metabolic and neural systems by analyzing the multi-omics data from The Cancer Genome Atlas (TCGA) portal. Additionally, a four-step strategy based on lasso regression and Cox regression was used to construct the prognostic prediction model. For the prognostic predictions on the independent test set, the performance of the trained models (average concordance index [C-index] = 0.839) is satisfied, with average 1-year, 3-year, and 5-year areas under the curve (AUCs) equal to 0.796, 0.786, and 0.777. Finally, the overall model was constructed based on all samples, which comprised 27 variables and achieved a high degree of accuracy on the 1-year (AUC = 0.861), 3-year (AUC = 0.850), and 5-year (AUC = 0.916) survival predictions.
Keywords: lung adenocarcinoma, tumor immune microenvironment, gene expression, somatic mutation, DNA methylation, prognostic prediction
Graphical Abstract
Liao and colleagues collected multi-omics data on lung adenocarcinoma and performed an integrative analysis. This study dissected the tumor microenvironment, identified the immunity-dependent genetic/epigenetic characteristics (expression status, somatic mutation, and DNA methylation), and finally established a prognosis predictor with great performance based on the identified alterations.
Introduction
Lung cancer represents the peaks of both incidence and mortality among all cancer types, with 2,093,876 (11.6%) new worldwide cases and 1,761,007 (18.4%) deaths out of all cancer types.1 Lung adenocarcinoma (LUAD) is the most common histological type of lung cancer. Many epidemiological investigations and experimental studies have attributed the occurrence and progression of LUAD largely to both environmental factors and genetic alterations.2, 3, 4 Despite the increasing prevalence of health education and the decrease in smoking rates, the incidence trends have not changed considerably. A large number of never-smokers suffering from LUAD overturned the previous theory based only on environmental factors and refocused the researchers’ attention on the in-depth alterations of genetic content. Until now, there are two major types of genetically related therapeutic strategies, i.e., targeted therapy and immunotherapy. Targeted therapy requires a specific gene mutation in cancer patients, and the epidermal growth factor receptor (EGFR) is the most representative mutation in LUAD patients, especially in Asians (51.4%), and can therefore act as a sensitive therapeutic target for the tyrosine kinase inhibitor (TKI).5,6 Unfortunately, there remains a large proportion of LUAD patients trapped in the lack of targetable mutations. Current progress on immunotherapy paves an efficient and safe way to bail them out. Immunotherapy is always realized by the introductions of immune checkpoint blockers (ICBs) such as antibodies targeting programmed death 1 (PD-1) and cytotoxic T lymphocyte antigen-4 (CTLA-4), but this method is also largely affected by the tumor microenvironment (TME) such as T cell abundance, tumor mutation burden, and the regulatory relationships with other biomolecules,6, 7, 8 which highlights the importance and the urgent need of deeply understanding how TME orchestrates the therapeutic and prognostic outcomes.
TME is commonly defined as the environment around a tumor, which comprises the extracellular matrix, blood vessels, and cellular players such as immune cells and neurons, all of which have a strong association with tumor progression and therapeutic outcome.9 Increasing studies have experimentally elucidated the contributive roles of TME infiltrates in the immunotherapy response and resistance in various cancer types, and explored their influence on patient prognosis.10, 11, 12 For example, B7-CD28 family-based signatures were found to have significant prognostic values and demonstrated that the tumor immune landscape plays a role in LUAD.13 Additionally, Song et al.14 established a model comprising of 30 immune-related genes as predictive signatures and attempted to determine their relationship with clinicopathological factors. However, most of the proposed models for LUAD prognosis prediction14, 15, 16 only incorporate expression changes and are therefore not comprehensive enough to reach a satisfying performance.
In the present study, we aim to estimate the TME infiltration patterns, especially the tumor-associated immune system in LUAD based on the expression profiles from The Cancer Genome Atlas (TCGA) portal, and then correlate the immune status with genetic or epigenetic characteristics through analyzing the multi-omics data (RNA sequencing [RNA-seq], whole-exome sequencing [WES], and DNA methylation array), and finally establish a prognosis prediction model from the significant alterations. It is expected that the results from this study could provide a more comprehensive map of the immunogenomic landscape and potentially find a better prognosis predictor for human LUAD.
Results
Construction of the TME in LUAD
Tumor tissues are not simply composed of only tumor cells but also the heterogeneous microenvironment constituents such as fibroblasts, blood vessels, immune cells, stromal cells, and so on, which can be infiltrated by tumor cells and thus equipped with tumor-associated influence. The interactions between the tumor cells and the surrounding infiltrate, especially two major non-tumor constituents (stromal cells and immune cells), are able to orchestrate either tumor progression or inhibition.17
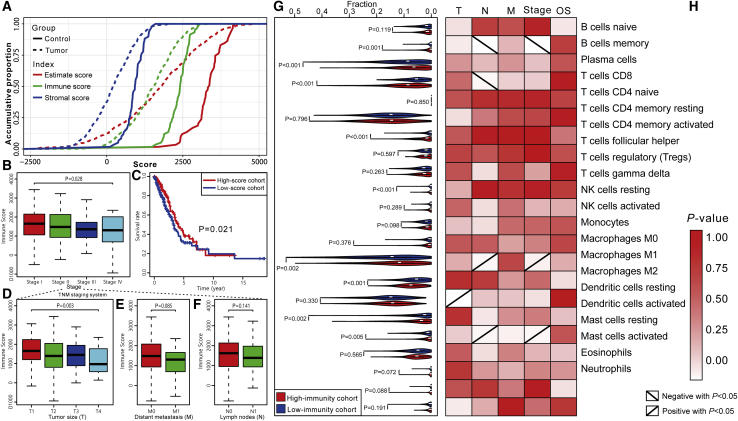
To evaluate the tumor-associated effects of the infiltrating stromal and immune cells, the TME was preliminarily established based on the expression profiles from TCGA portal by using the ESTIMATE (estimation of stromal and immune cells in malignant tumor tissues using expression data) algorithm.18 ESTIMATE generates a stromal score that measures the presence of tumor-associated stroma and an immune score that represents the infiltration level of the immune cells and combines them to produce an index termed “estimate score” that comprehensively infers tumor purity. As shown in Figure 1A, the estimate scores of the LUAD samples were distributed at the significantly lower side, compared with those of the normal samples (Mann-Whitney U test, p < 0.05), as were the stromal scores and immune scores (Mann-Whitney U test, p < 0.05). The difference in the distributions between the tumor and normal cohorts supports the higher tumor purity in the selected LUAD samples.
Figure 1.
Construction of TME in LUAD
(A) Comparison of the distributions of estimate scores, immune scores, and stromal scores between tumor and control cohorts. (B) Comparison of immune scores on the TNM stage. (C) Kaplan-Meier curves show the independent relevance between overall survival time and immune scores. (D–F) Comparisons of immune scores on (D) tumor size, (E) distance metastasis, and (F) lymph nodes. (G) Comparisons of the immune cell members between high-immunity and low-immunity groups. (H) Relevance between clinical factors and immune cell members. The red to white gradient represents the significance level. The positive and negative diagonals in the boxes respectively indicate the significantly positive and negative relationships between cell fractions and the level of clinical factors (p < 0.05). (G) and (H) share the same y axis.
Next, we investigated the relationship between tumor purity and clinical factors. As shown in the results (Figure S1A), the estimate scores were significantly different in tumor size (Kruskal-Wallis test, p < 0.05), distant metastasis (Mann-Whitney U test, p < 0.05), and tumor stage (Kruskal-Wallis test, p < 0.05). Pairwise comparisons (Table S1) show that the samples in stage I have significantly higher estimate scores than do the samples in stages III and IV (Mann-Whitney U test, false discover rate [FDR] < 0.05). Furthermore, patients in the cohort with low estimate scores (below the median) have a poorer prognosis compared with those in the high-score (above the median) cohort (Figure S1A). To separately elaborate the roles of the stromal and immune cells, the relationships between the two types of scores and clinical factors were further studied. From the aspect of the TNM (tumor size, lymph node, and distant metastasis) staging system, the immune scores were significantly different in different tumor sizes (Kruskal-Wallis test, p < 0.05), but there was no difference in lymph nodes and distant metastasis (Figures 1D–1F). As for the integrative TNM stage classification, the immune scores were significantly different between the early stage and advanced stage as shown in Figure 1B (Kruskal-Wallis test, p < 0.05), of which those in stage I were significantly higher than those in stages III and IV (Mann-Whitney U test, FDR < 0.05) (Table S1). In contrast, stromal scores were only significantly related with distant metastasis (Mann-Whitney U test, p < 0.05) rather than tumor size, lymph nodes, and stage (Figure S1B), which conforms to the favorable functions of tumor-associated stroma in tumor metastasis.19,20 A more important finding is that higher immune scores were significantly associated with longer overall survival time (Figure 1C), whereas stromal scores have no significant relevance with patient prognosis (Figure S1B). Extrapolating from all of the above results, we speculated that tumor-infiltrating immune cells have a stronger evidential clinical relevance compared to stromal cells in LUAD. Therefore, the following analyses mainly focused on the tumor immune microenvironment (TIME) and immunity-related genes.
Dissecting the Infiltrating Immune Content in LUAD
Since immune infiltration level and cell composition are strongly related to tumor progression and patient outcome,21 we divided the LUAD samples into a high-immunity cohort and a low-immunity cohort using their median immune score, and further characterized cell composition using CIBERSORT22 to explore the relationship between the immune cell subsets and clinical features. The samples with CIBERSORT22-generated p values greater than 0.05 were removed. A total of 468 samples were reserved, of which 203 belong to the low-immunity cohort and 265 belong to the high-immunity cohort.
Then, the immune content of each sample was dissected into 22 types of immune cell members. The high-immunity cohort had significantly larger fractions of memory B cells, CD8 T cells, activated memory CD4 T cells, M1 macrophages, resting dendritic cells, activated mast cells, and gamma delta T cells and smaller fractions of plasma cells, M0 macrophages, and activated dendritic cells (Mann-Whitney U test, p < 0.05) (Figure 1G), which is consistent with the proposed anti-immune or pro-immune functions of these cell members.23, 24, 25, 26, 27, 28 Of note, some of these immune cell members have potential correlations, such as the positive correlation of activated dendritic cells on CD8 T cell infiltration in lung cancer.29 While considering the relationships between the immune score and the clinical factors detected above, we speculated that different immune cell members may have different contributions to patient outcome, and, as we expected, memory B cells, CD8 T cells, M0 macrophages, M2 macrophages, and activated dendritic cells were detected to have significant differences in TNM stage, tumor size, or lymph nodes (Mann-Whitney U test or Kruskal-Wallis test, p < 0.05), while the other cell subsets appear to be statistically insignificant in all clinical factors (Figure 1H). Furthermore, the single type of cell members contributed little to patient overall survival in LUAD (Figure 1H), although the combination shows discriminative power (Figure 1C).
Identification of the Immune Infiltration-Dependent Differentially Expressed Genes
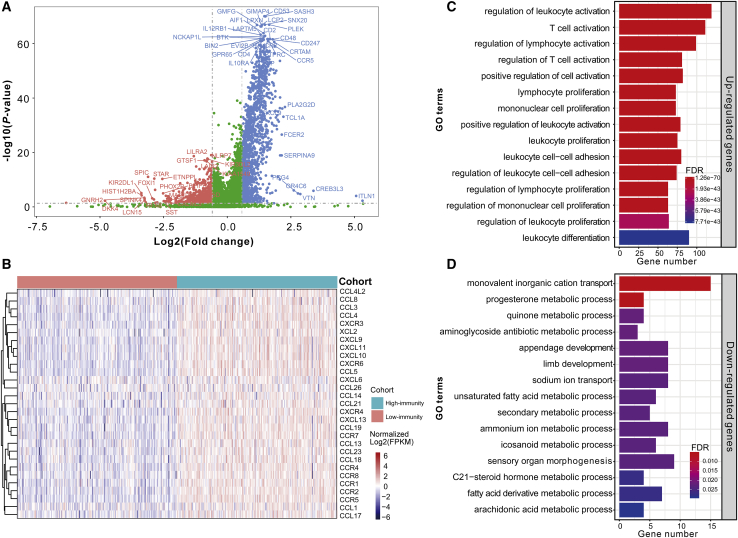
The expression profiles of the LUAD samples from TCGA portal were used to identify the expression change between the high-immunity and low-immunity cohorts. The genes meeting the standard of log2(fold change) >1 and FDR <0.05 (Student t test) were considered to be differentially expressed, of which 611 and 164 genes were, respectively, upregulated and downregulated in the high-immunity cohort (Figure 2A). Note that 29 chemotactic factors such as CXCR4 and CCL8 were found to be significantly upregulated (Figure 2B), which are capable of modulating the recruitment of a wide variety of immune cells to tumor,30
Figure 2.
Investigation of the Immune Infiltration-Dependent Expression Change
(A) Volcano plot showing the differentially upregulated (blue nodes) and downregulated genes (red nodes). (B) Heatmap displaying the expression changes of 29 chemotactic factors. The colors of the top bar and central plot represent the grouping information and gradient of normalized log2FPKM (fragments per kilobase of transcript per million mapped reads), respectively. (C) Bar plot showing the biological processes enriched by the upregulated genes. (D) Bar plot showing the biological processes enriched by the downregulated genes. The y axis in (C) and (D) reflects the name of GO terms. The x axis reflects the overlapped gene numbers between each GO term and query gene set. The color of the bars represents the gradient of adjusted p values (FDR correction).
Then, functional enrichment analyses using clusterProfiler31 were performed to infer the potential functions of these two parts of genes. As we expected, the upregulated genes were found to be enriched in immune-related biological processes such as T cell activation and leukocyte proliferation (Figure 2C), indicating that they have a positive role in the enhancement of tumor-associated immunity. Furthermore, some of the upregulated genes such as LILRB4,32 RUNX3,33,34 and CXCR335 have been experimentally verified in prior studies to regulate T cell activation and support tumor infiltration. Alternatively, the downregulated genes were mainly enriched in the metabolic process (Figure 2D). The metabolic communication between the tumor cells and infiltrating immune cells is a battleground for energy competition and can therefore influence tumor progression.36 Hence, we speculated that some downregulated genes modulate the activities of immune and tumor cells using the metabolic switch; for example, the role of AKR1C1/2/3 in the tumor infiltration level of immune cells and the metabolic process has been partially verified.37
Comparisons of Somatic Mutations under Different Immune Infiltration Levels
After detecting the transcriptional alterations in the above section, we further investigated whether there is evidence of the disparity in the genomic layer between the high-immunity and low-immunity cohorts. Somatic mutations including the single-nucleotide variant (SNV), single-nucleotide polymorphism (SNP), insertion (INS), and deletion (DEL) were analyzed and visualized using the R package maftools38 based on the WES data from TCGA portal in which the mutations had been called by VarScan2.39 As shown in Figure 3A and Figure S3A, most genomic variants were missense mutations in both the high-immunity and low-immunity cohorts (around 60%). Therefore, it was necessary to measure the mutation changes, classify the types of variants, and uncover their potential meanings.
Figure 3.
Landscape of Somatic Mutation in High-Immunity and Low-Immunity Cohorts
(A–D) Boxplots showing the comparisons of mutation frequencies of (A) every mutation type classified by effects, (B) SNV, (C) INDEL and SNP, and (D) the percentage of VAF between high-immunity and low-immunity cohorts. (E) Waterfall plot shows the mutation distribution of the top 15 most frequently mutated genes. The central panel shows the types of mutations in each LUAD sample. The upper panel shows the mutation frequency of each LUAD sample. The bar plots on the left and right side show the frequency and mutation type of genes mutated in the low-immunity and high-immunity cohort, respectively. The lower part shows the clinical features (tumor stage, smoke, resection or biopsy site, sex, and race) and SNV types of each sample. The bottom panel is the legend for mutation types and clinical features. (F) Forest plot displays the top 10 most significantly differentially mutated genes between two cohorts. (G) The lollipop plot illustrates the differential distribution of variants for STK11. (H) Kaplan-Meier curves show the independent relevance between overall survival time and STK11 mutation in high-immunity and low-immunity cohorts. (I) The boxplot shows the expression changes of KRAS with or without SNP rs121913530 between the high-immunity and low-immunity cohorts. (J) The heatmap illustrates the mutually co-occurring and exclusive mutations of the top 25 frequently mutated genes. The color and symbol in each cell represent the statistical significance of the exclusivity or co-occurrence for each pair of genes.
From a global perspective, the samples in the low-immunity cohort hold a significantly larger number of variants (median = 235.00, quartile range = 398.00) than those in the high-immunity cohort (median = 172.50, quartile range = 278.75) (Mann-Whitney U test, p = 0.001) (Figure S2). As for SNVs, a total of 64,344 and 88,708 SNVs across all samples in both high-immunity and low-immunity cohorts were detected, of which C>A was the most common type in both the high-immunity cohort (median frequency = 45, median proportion = 42.65%) and low-immunity cohort (median frequency = 65, median proportion = 42.15%). No matter the type of SNV, the mutation numbers in the low-immunity cohort were significantly higher than those in the high-immunity cohort, as shown in Figure 3B (Mann-Whitney U test, p < 0.05). The ratios between transversion (Tv) and transition (Ti) in all SNVs were approximate 2:1 and remained stable in both cohorts (Figure S3D). Furthermore, Figure 3C shows that the SNPs, INSs, and DELs in the high-immunity cohort (SNP: median = 167.50, quartile range = 270.75; INS: median = 1, quartile range = 3; DEL: median = 5, quartile range = 8) were also outnumbered by those in the low-immunity cohort (SNP: median = 226, quartile range = 386; INS: median = 2, quartile range = 3; DEL: median = 6, quartile range = 9). Note that the samples in the low-immunity cohort have a significantly higher level of variant allele fractions (VAFs) than do those in the high-immunity cohort (Figure 3D), which had been proposed to be associated with tumor progression and a poorer prognosis,40,41 which supports the finding of relatively higher tumor purity and lower heterogeneity in the low-immunity samples. Despite the significant differences in variant number of the four types of somatic mutations between the two immune cohorts, the inner constituent ratio of each mutation type occupied in all variants stayed almost unchanged (Figures S3A–3C), suggesting that the differences observed in mutation number are not caused by type shift.
In the low-immunity cohort, 129 genes were mutated in more than 10% of the samples while only 62 genes met this criterion in the high-immunity cohort, of which there was an overlap of 56 genes. The top 15 most frequently mutated genes in the corresponding cohorts are illustrated in Figure 3E. Interestingly, TP53, TTN, and MUC16 occupy the top three positions in both cohorts, and there were interactions among them that were regulating various tumor-associated biological processes in LUAD,42, 43, 44 which indicates that they may be less involved in the immune infiltration process but are mainly involved in tumor progression. Next, we investigated the co-occurring and exclusive mutations of the top 25 most frequently mutated genes by using the CoMEt algorithm.45 Compared with the pervasive co-occurrence landscape, there are three unique cases in two cohorts (KRAS-TP53, KRAS-TNR, and STK11-TP53) that were exhibiting mutually exclusive mutations (Figure 3J), which suggests their probably redundant effect in the same pathway and the selective advantages between them to keep more than one copy of the mutations. More interestingly, some genes had differential mutation frequencies between the two cohorts. From the results, 268 differentially mutated genes were detected using Fisher’s exact test, which were sorted in ascending order of p value (Table S2), and the top 10 are shown in Figure 3F. Notably, different variants may exert distinct impacts on other genetic alterations and even the clinical outcome of patients. For example, KRAS was detected to hold a differential SNP pattern between high-immunity and low-immunity cohorts (Fisher’s exact test, p < 0.05). The expression levels of KRAS without SNPs were significantly different between the high-immunity and low-immunity cohorts (Mann-Whitney U test, p < 0.05), but the reverse was observed when SNP rs121913530 (C>A) exists (Figure 3I). Furthermore, STK11 is another typical example to demonstrate the different mutation spots between two cohorts (Figure 3G) and the plausible chain reaction of the differences in prognostic impact (Figure 3H).
Depicting DNA Methylation Pattern in TIME of LUAD
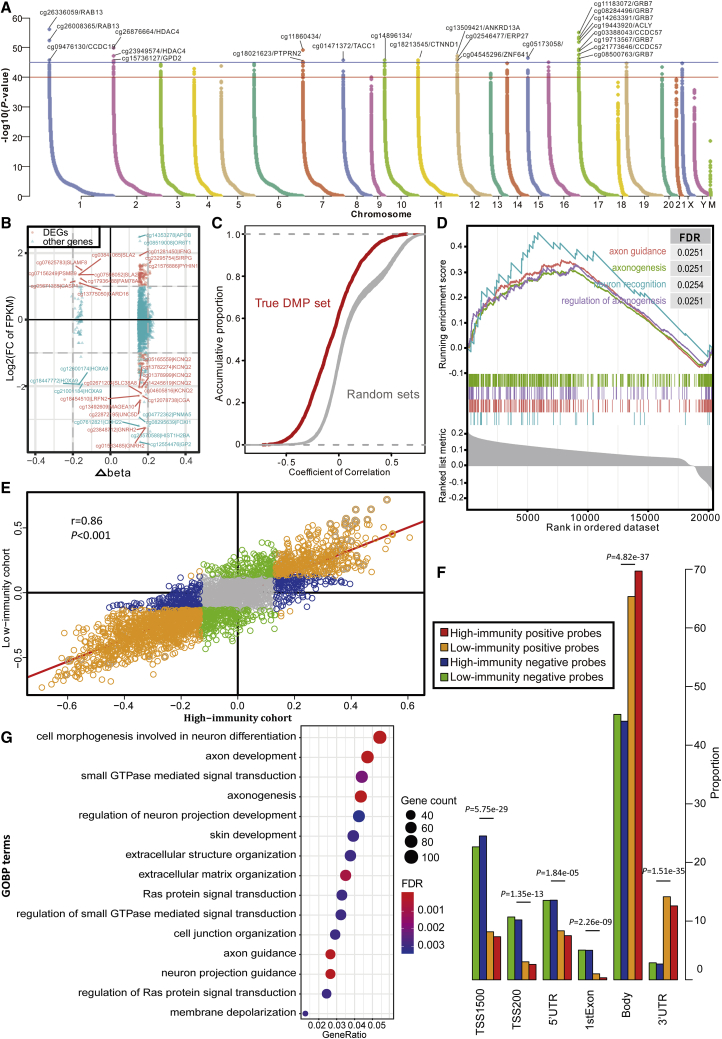
Failure to maintain normal DNA methylation, which includes both hypermethylation in CpG islands and hypomethylation in CpG-poor regions, increases the susceptibility to triggering tumor formation and deterioration.46,47 Therefore, we aimed to detect and compare the effects of DNA methylation patterns in different immune cohorts using Illumina Infinium 450k DNA methylation data from TCGA portal. In this section, 451 samples in which no more than 20% genes have missing beta values were used to detect the differential methylation probes (DMPs) by using ChAMP.48 A total of 5,764 immune-related DMPs were identified according to the standard of Δbeta >0.15 and FDR <0.05 (Figure 4A; Table S3). Compared with the low-immunity cohort, 5,647 (97.97%) hypermethylated positions involving 2,386 genes were detected in the high-immunity cohort, of which 2,221 loci were located on 1,687 CpG islands. In contrast, the hypomethylated positions were drastically outnumbered with only 117 (2.03%) loci related to 68 genes and were located on 56 CpG islands. Therefore, the high-immunity cohort tends to have hypomethylated positions overall, but hypomethylation only occurs in a few genes. In summary, 2,441 DMP-associated genes were composed of 2,393 genes containing either hypermethylated or hypomethylated positions and 13 genes containing both hypermethylated and hypomethylated positions. Additionally, many DMP-associated genes were found to be differentially expressed between the two cohorts (log2FC > 1, p < 0.05). From the 2,386 hypermethylated genes in the high-immunity cohort, there were 63 upregulated and 32 downregulated DEGs (Figure 4B; Table S4). However, from the hypomethylated gene set, only seven upregulated DEGs were detected (Table S5). The functions of the DMP-associated genes were investigated based on the Gene Ontology (GO) analysis. The top 15 enriched GO terms of biological processes with the lowest FDRs indicated their potential roles in cell differentiation and development (Figure 4G). Meanwhile, they are also significantly enriched in various neuron-associated biological processes. In particular, gene set enrichment analysis (GSEA) of the DMP-associated genes shows that the hypermethylated genes with highly positive beta differences have more essential contributions to the tumor-associated neural biological processes such as axon guidance and axonogenesis (Figure 4D), indicating that the aggressive behaviors induced by the aberrant methylations on tumor immunity are through the recognition and participation in the neural pathways. More interestingly is that the hypermethylation-associated upregulated DEGs were found to participate in immune system activation (Figure S4), which suggests that the probable effect of hypermethylation on gene overexpression is to trigger an elevated level of immune cell infiltration.
Figure 4.
DNA Methylation Pattern in TIME
(A) Manhattan plot of the genome-wide DNA differential methylation in high-immunity and low-immunity cohorts. (B) The relationships between expression change and DNA methylation level. The nodes in red represent the DEGs with log2FC of FPKM >2 and Δbeta >0.15. (C) The distributions of Pearson correlation coefficients between gene expression level and beta value in true DMP set and 100 random sets. (D) Gene set enrichment analysis (GSEA) shows the significant enrichment in four neuron-associated biological processes. Genes are ranked by Δbeta. (E) Scatterplot showing the correlations of methylation-expression correlation coefficients between high-immunity and low-immunity cohorts. (F) The histogram shows the regional difference between the positive and negative probes. (G) Bubble plot showing the results of GO biological process (GOBP) enrichment analyses on DMP-associated genes. The x axis (fold enrichment) represents the ratio of two proportions, of which one is the proportion of input genes involved in the given GO term and another is the proportion of genes in the given GO term involved in the background.
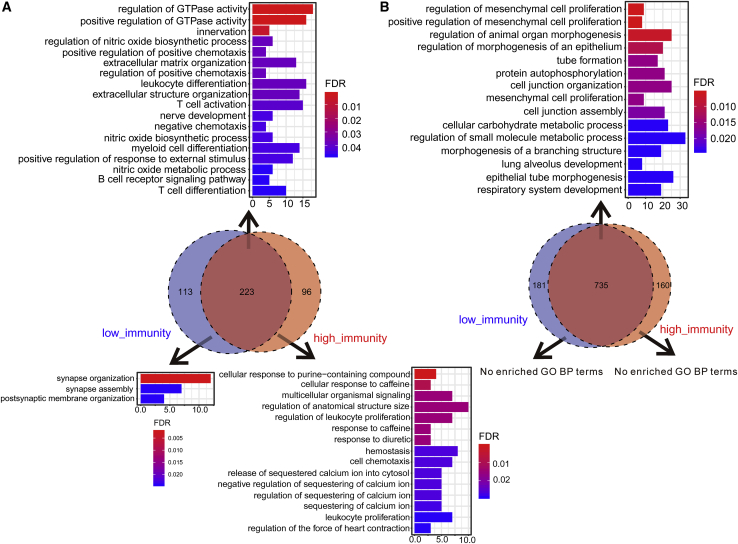
While considering the previous finding about the correlations between DNA methylations and gene expression levels,49 we speculated whether there exists a similar phenomenon in our study and whether the trend is stable across different immune levels. The results showed that out of 2,441 DMP-associated genes, there were 329 positively correlated and 926 negatively correlated genes (Table S6) in the high-immunity cohort, and 346 positively correlated and 939 negatively correlated genes (Table S7) in the low-immunity cohort (p < 0.05, Pearson correlation). Therefore, the preferred negative correlations observed in the DMP-associated genes were as postulated. As expected, the probe signals associated with the DMP-associated genes were prone to be negatively correlated with expression level compared with the relatively balanced distributions of correlation coefficients in 100 random sets, which were constructed using the randomly selected probes (Figure 4C). For example, S100P, a well-studied driver in LUAD,50 was hypomethylated in the low-immunity cohort and had significant correlations between its low methylation signals and high gene expressions (Figure S5). Furthermore, the immunity level did not affect the correlations between methylation levels and expression levels overall, which is supported by the high consistency observed in the correlation coefficients between the two cohorts as shown in Figure 4E (r = 0.86, p < 0.05, Pearson correlation) and the large overlaps in negatively (Venn diagram in Figure 5B) or positively (Venn diagram in Figure 5A) correlated gene sets between two cohorts. These consistently positively and negatively correlated genes in the low-immunity and high-immunity cohorts were respectively enriched in the immune systems and cell proliferation, while the genes with inconsistent trends had some distinctive functions. For example, the uniquely positively correlated genes in low-immunity cohorts could participate in synapse-associated functions (Figure 5A). A previous study that immune synapse proteins produced by tumor cells will suppress the immune system through epigenetic mechanisms, especially DNA methylation,51 provides evidence for this finding. Furthermore, the probes of the positively correlated genes were more frequently located in the gene body and 3′ UTR region (p < 0.001, chi-square test), while the probes of the negatively correlated genes preferred the regions adjacent to a promoter such as transcription start sites (TSSs), the 5′ UTR, and the first exon (Figure 4F), indicating that there are regional differences in the effect of DNA methylation on expression.
Figure 5.
Functional Enrichment Analyses of the Positively and Negatively Correlated Genes
(A) Positively correlated genes. (B) Negatively correlated genes. The results of the consistently correlated genes between the high-immunity and low-immunity cohorts are shown at the top of the Venn diagram, and the results of the uniquely correlated genes are shown on the bottom. The x axis reflects the overlapped gene numbers between each GO term and query gene set. The color of the bars represents the gradient of adjusted p values (FDR correction).
Multi-omics Characteristics Provide an Accurate Prognostic Prediction
From the results obtained in the above sections, some significant immune-related alterations in the multi-omics characteristics, which include expression change, somatic mutation, and DNA methylation, were identified. For expression change, a total of 611 upregulated and 164 downregulated genes were detected in the high-immunity cohort. For somatic mutation, 129 and 62 frequently mutated genes were detected in the low-immunity and high-immunity cohorts, respectively. For DNA methylation, 3,858 out of 5,764 DMPs were located at the regions of 2,442 annotated genes and were differentially methylated.
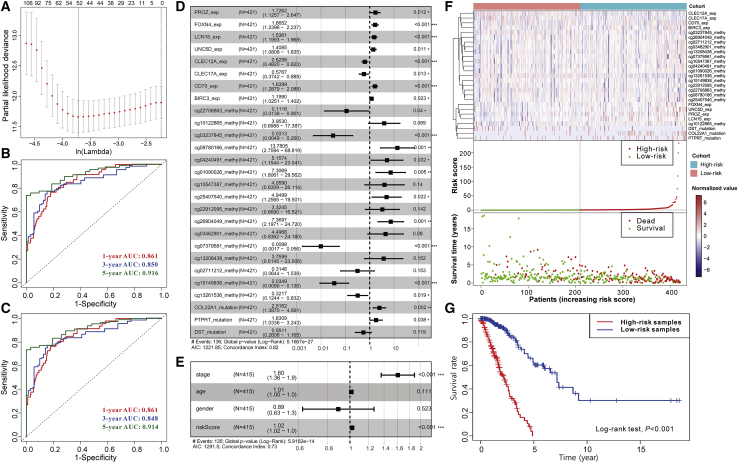
To identify the immune-related prognostic signatures from the numerous genetic alterations, a four-step strategy (see Materials and Methods) based on lasso regression and Cox proportional hazards regression was adopted. Additionally, the joint and separate effects of the three types of alterations were investigated separately to determine which model had the best performance. On the one hand, for joint effect, all genetic alterations were combined, of which 337 items composed of DEGs, 5 mutations, and 217 DMPs were identified to have a significantly independent effect on the overall survival time of patients using a univariate Cox proportional hazards model. Due to the large number of significant alterations and the possible interactions among them, a lasso regression model was adopted to crop the less contributive variables. Under the optimal parameter ln(lambda) = −3.7 (Figure 6A), 52 variables were reserved (Table S8) and used to establish the multivariate Cox proportional hazards regression model (stepwise method). For the lack of the matching multi-omics data from other sources, we randomly divided TCGA samples into a training and independent test set. Each third of the samples (n = 142) took turns to be the independent test set, and the other two-thirds served as the training set, thus constructing three pairs of sample sets. In the results (Table 1; Figure S6), the performance of the trained models is satisfied with the average concordance index (C-index) equal to 0.839. Next, the risk score for each sample was calculated based on the established models, which has great discriminative power on survival status. The average AUC values of 1-year, 3-year, and 5-year prognosis predictions on training sets reached 0.871, 0.875, and 0.928. With regard to the predictions on the test sets, the performance exhibited a slight decline with the average AUC values of 1-year, 3-year, and 5-year survival equal to 0.796, 0.786, and 0.777. Moreover, the samples were classified into high-risk and low-risk cohorts by median risk score (Table S9; Figure 6F). Kaplan-Meier survival analyses (Figure 6G) show that the high-risk cohort had a poorer overall survival compared with the low-risk cohort (p < 0.001).
Figure 6.
Establishing a Prognostic Model for LUAD
(A) Identification of the optimal penalization coefficient lambda in the Lasso regression model. (B) ROC curves of the risk score for predicting 1-year, 3-year, and 5-year survival. (C) ROC curves of the risk score combined with clinical factors for predicting 1-year, 3-year, and 5-year survival. (D) Forest plot of the prognostic impact of 27 variables. (E) Forest plot of the prognostic impact of risk score and clinical factors. (F) Heatmap in the top panel shows the gradient of 27 variables. The color in the heatmap represents the normalized values of log2FPKM, log2Δbeta, and mutation status (0 for non-mutated and 1 for mutated). The scatterplots in the middle and bottom panels respectively illustrate the distribution of risk score and survival status of LUAD patients. (G) Kaplan-Meier curves show the independent relevance between overall survival time and risk scores.
Table 1.
The AUC Values and C-indexes of Three Trained Models
| Index | Model 1 |
Model 2 |
Model 3 |
Average |
||||
|---|---|---|---|---|---|---|---|---|
| Training Set 1 | Test Set 1 | Training Set 2 | Test Set 2 | Training Set 3 | Test Set 3 | Training Set | Test Set | |
| 1-Year AUC | 0.881 | 0.775 | 0.869 | 0.862 | 0.863 | 0.752 | 0.871 | 0.796 |
| 3-Year AUC | 0.854 | 0.787 | 0.873 | 0.811 | 0.897 | 0.761 | 0.875 | 0.786 |
| 5-Year AUC | 0.943 | 0.723 | 0.909 | 0.793 | 0.933 | 0.814 | 0.928 | 0.777 |
| C-index | 0.841 | 0.839 | 0.836 | 0.839 | ||||
Considering the great robustness and validity of the above-constructed models, we then combined all TCGA samples and generated an overall prediction model comprising of 27 variables (Figure 6D), of which some such as the DMPs cg10122865 and cg13208438 seemed to be statistically insignificant initially (p > 0.05) but are likely associated with other signatures and outcome. The contributions of 27 variables on the overall model are shown in Figure S7. In brief, the expression levels of PROZ, FOXN4, LCN15, UNC5D, CD70, and BIRC3, the methylation levels of cg08780166, cg04240491, cg01090026, cg25407540, and cg26904049 probes, and the mutation of COL22A1 and PTPRT have significantly positive contributions to a poorer prognosis, while the expression levels of CLEC12A and CLEC17A and the methylation levels of cg22706883, cg03237845, cg07379581, cg15149938, and cg13261536 probes played opposite roles. Moreover, in line with our above findings, the prognosis of the high-risk cohort is poorer compared to the low-risk cohort according to the results of the survival analysis (Figures 5F and 5G). Furthermore, no matter whether it is the 1-year, 3-year, or 5-year survival rates, the risk score has high discriminative power, with respective AUC values equal to 0.861, 0.850, and 0.916 (Figure 6B; Figure S7). On the other hand, we adopted the same strategy as mentioned above to determine whether the separate effect of each type of genetic alterations has equal or even superior power and whether the differential mutations between the high-immunity and low-immunity cohorts can replace the role of frequently mutated genes in the prediction model. From the results shown in Figure S9, no matter the expression change, somatic mutations, and differential DNA methylation, no single characteristic could provide a powerful enough prognostic prediction compared to the joint model. Additionally, when the frequent somatic mutations were replaced by differential mutations, a 23 variable-based prediction model was obtained, but the performance had no improvement (Figure S10). Also, given the relatively low frequency of these differential mutations, this model was eventually not reserved.
In addition to the genetic alterations, some clinical factors may also have predictive values for the overall survival time. Therefore, the risk score of the joint model was further combined with three clinical and demographic factors (stage, sex, and age) to construct another Cox model. The contributions of the risk score and clinical factors are as shown in Figure S8. Although the stage was significantly associated with overall survival time, the discrimination ability of the new model that incorporated the clinical factor was not improved as shown in Figure 6E (C-index = 0.73), compared with the above overall model constructed only based on multi-omics alterations (C-index = 0.82). Moreover, this new model did not achieve a better performance on its 1-year (AUC = 0.861), 3-year (AUC = 0.848), and 5-year (AUC = 0.914) survival predictions (Figure 6C). We speculated that these clinical factors were highly correlated with and reflected by the genetic characteristics. Hence, only multi-omics characteristics comprising the above 27 alterations already could produce an accurate prognostic prediction.
Discussion
Significant effort and attention have been invested to explore the sophisticated mechanism of LUAD, but the current understanding, especially on TME, therapeutic target, and prognostic factors, remains far from satisfactory. In the present study, we first used gene expression profiles to construct the TME of LUAD and conducted a further study on the immune infiltration landscape. As expected, some immune member cells had distinct fractions between the high-immunity and low-immunity cohorts, which had been partially reported before. For instance, memory B cells, a kind of antigen-presenting cell, can drive the expansion and memory formation of T cells, thus exerting their anti-tumor functions in tumors,52 which conforms to our findings that memory B cells hold a higher fraction in the high-immunity study and are negatively correlated with tumor stage and lymph nodes in LUAD. Then, we turned to investigate the genetic or epigenetic alterations under different immunity infiltration levels and to infer their potential functions. Apart from the canonical immune biological processes such as T cell activation and lymphocyte proliferation, some altered genes are likely able to modulate the immune-related metabolism and neural systems. The genes with downregulated expression levels may have a role in the metabolic communication between tumor cells and infiltrating immune cells. Additionally, the differentially methylated genes may be associated with axon guidance and axonogenesis, thus modulating cell migration and tumor angiogenesis and further promoting LUAD progression. Several studies had reported similar findings in various cancer types.53, 54, 55, 56 Finally, according to the detected genetic or epigenetic alterations, a prognosis model was constructed and was found to have a superior performance with a higher C-index or AUC value compared to the previous models.14, 15, 16 To our surprise, the trained models showed higher accuracy of 5-year survival prediction on the training sets compared with the result from the 1-year survival prediction, whereas the superiority was lost when the prediction was done using the test set. This is likely because the immune-related genetic characteristics have increasingly amplified but uncertain alterations during cancer progression. The uncertainty may be the increasingly different compositions of tumor-infiltrating immune cells for long-term cancer patients. For example, high levels of tumor-infiltrating T cells and macrophages are respectively correlated with better and worse prognosis,57 which caused higher individual variation, thus making the model unstable when projecting long-term patient survival.
Although this study provides a more comprehensive view into the TIME of LUAD and establishes a powerful model for prognostic prediction, there are still two major drawbacks that require further study. The first drawback, that is, because matching multi-omics data and clinical information are required, we were limited to data solely from TCGA portal and cannot cover other data sources. This hindered us from testing the robustness of the model when used for other data. The second drawback was that the application of the prognostic prediction model required three kinds of omics data, including RNA-seq, WES, and DNA methylation array data, which is cost-intensive and not easy to realize in practical use. Even so, the rapid development of biological technologies will hopefully produce a three-in-one toolkit and thereby blaze the way for its realization and popularization. Yet, despite having such limitations, there is no denying that our study provides significant clues for the elucidation of TIME in LUAD. Moreover, our prognostic prediction model can potentially exhibit compelling clinical value that may lead to the improvement of overall survival and even for the development of new therapeutic strategies for LUAD patients.
Materials and Methods
Data Source
The data of 535 LUAD samples and 59 normal control samples of RNA-seq profiles, 561 LUAD samples of WES data, and 504 profiles of the Illumina 450k DNA methylation array were collected from TCGA portal.
TME Construction
First, the ESTIMATE18 algorithm was used to construct the TME and generate an estimate score, stromal score, and immune score based on the expression profiles. A lower estimate score, stromal score, and immune score respectively represent the higher tumor purity and lower infiltration levels of stromal and immune cells in tumor tissue. Next, the LUAD samples of expression profiles were divided into a high-immunity cohort (n = 268) and low-immunity (n = 267) cohort by the median immune score. The infiltrating immune content for each sample was dissected using the software CIBERSORT.22 Samples with a p value greater than 0.05 were filtered out, which means that there is no cell type in the signature matrix that can reflect on the given gene expression profile mixture. The fraction levels of all 22 immune cell members between the high-immunity and low-immunity cohorts were compared using the Mann-Whitney U test. For the LUAD samples of the WES and methylation profiles, we also constructed a high-immunity cohort and a low-immunity cohort by mapping the sample IDs of the RNA-seq profiles.
Multi-omics Data Analyses
In this study, we aimed to investigate the differences in gene expression, somatic mutations, and DNA methylation between the high-immunity cohort and low-immunity cohort, respectively, based on the data obtained from TCGA portal. For gene expression change, the differentially expressed genes (DEGs) between the high-immunity cohort (n = 268) and low-immunity cohort (n = 267) were identified based on the RNA-seq data, which should meet the standard of expression fold change greater than 2 and p value adjusted by FDR correction lower than 0.05 (Student’s t test). For somatic mutations, WES data of both high-immunity (n = 250) and low-immunity cohorts (n = 257) were used to detect the SNVs, SNPs, and INDELs using the software VarScan2.39 Fisher’s exact test was used to identify the differential mutation pattern, and genes with a p value lower than 0.05 were defined as differentially mutated genes. The co-occurrence and mutually exclusive mutations were identified using the CoMEt algorithm.45 R package maftools38 was used to create the visualization of the somatic mutations. For DNA methylation, the R package ChAMP was used to process the Illumina Infinium 450k DNA methylation array data. Samples with more than 20% missing values were filtered out and 451 samples were used, which were further divided into the high-immunity cohort with 219 samples and the low-immunity cohort with 232 samples. The remaining missing values were statistically imputed using ChAMP’s imputation function. The beta values were normalized using peak-based correction (PBC). Furthermore, the differential methylation probes and regions were respectively identified using the limma package58 and Bumphunter algorithm. The correlation between the probe signal and gene expression level was investigated using the Pearson correlation, and the same numbers of probes as in the true DMP set were randomly selected from all probes to construct 100 random sets.
Functional Enrichment Analysis
GO analyses of the DEGs and DMP-associated genes and GSEA were performed using the R package clusterProfiler.31
Clinical Relevance Investigation
Clinical factors include the TNM staging system, integrative stage classification, and overall survival. The differences of the scores generated by ESTIMATE and the fractions of immune cell members in distant metastasis and lymph nodes were evaluated using the Mann-Whitney U test, while those in integrative stages and tumor size were evaluated using the Kruskal-Wallis test. Moreover, the contributions to overall survival were investigated by Kaplan-Meier survival analysis (log-rank test).
Establishing and Evaluating the Prognosis Prediction Model
Survival time and status were used to evaluate the prognosis of LUAD patients. We only considered cases that included their overall survival. Next, 494, 490, and 430 cases were reserved to construct the prognosis prediction models based on gene expression values, gene mutation, and methylation probe signals, respectively. For the model with integrating characteristics from multi-omics data, we only considered cases with all of the characteristics being studied, and 421 same cases were obtained. The workflow to explore the prognostic signatures from multiple immunity-dependent genetic or epigenetic alterations in LUAD consists of four steps: (1) univariate Cox proportional hazards regression was used to assess the individual effect of every alteration using the “survival” R package, and then the features with a p value less than 0.05 were selected for further analysis; (2) a lasso regression model was adopted to filter out some less informative variables using the R package glmnet; (3) multivariate Cox proportional hazards regression with a stepwise procedure was used to obtain the genetic variables-based prediction model and to generate the risk scores for all samples using the R package survival. In model construction, TCGA datasets (n = 423) were randomly divided into three equal parts, of which each part took turns to serve as the independent testing set and the remaining two parts to serve as the training set to obtain the optimal variable combination. (4) The risk score of each patient was predicted based on the training model by using the “predict” function included in the survival R package. The risk scores were combined with three other clinical and demographic characteristics (stage, sex, and age) and further introduced into the multivariate Cox proportional hazards regression model to assess the overall effect. Then, the patients were divided into a high-risk group (above the median) and low-risk group (below the median) by the median risk score. We conducted the survival analyses of LUAD patients with respect to the risk level and drew a 1-year, 3-year, and 5-year receiver operating characteristic (ROC) curve using the R package timeROC.59 The AUC value of the survival ROC curve and C-index were calculated to evaluate the performance of the prognosis prediction model.
Author Contributions
Y.Z. and Q.L. performed most of the analyses and wrote the manuscript. Q.L. and G.Z. conceived and designed the study. G.Z., M.Y., and H.Z. performed some of the analyses and edited the manuscript. D.M.N, M.H., S.H., and T.Y. contributed to additional analyses in the revision procedure.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant no. 31970630); the Fundamental Research Funds for the Provincial Universities of Zhejiang (grant no. SJLZ2021001); Ningbo Health Branding Subject Fund (grant no. PPXK2018-05), Natural Science Foundation of Ningbo (grant no. 2017A610154); the Zhejiang Key Laboratory of Pathophysiology (grant no. 201812); The Scientific Innovation Team Project of Ningbo (grant nos. 2016C51001 and 2017C110019); and by the K.C. Wong Magna Fund in Ningbo University.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.07.024.
Contributor Information
Guofang Zhao, Email: guofzhao@hotmail.com.
Qi Liao, Email: liaoqi@nbu.edu.cn.
Supplemental Information
The Results of Pairwise Comparisons of Estimate Score and Immune Score on Tumor Size and Stage
The List of the Differentially Mutated Genes between High-Immunity and Low-Immunity Cohorts
The List of the Differentially Methylated Probes
The List of the Genes with Expression-Methylation Correlations in High-Immunity Cohort
The List of the Genes with Expression-Methylation Correlations in Low-Immunity Cohort
The Table Shows the Contributions of 27 Variables and Risk Score for Each Sample
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Vaz M., Hwang S.Y., Kagiampakis I., Phallen J., Patil A., O’Hagan H.M., Murphy L., Zahnow C.A., Gabrielson E., Velculescu V.E. Chronic cigarette smoke-induced epigenomic changes precede sensitization of bronchial epithelial cells to single-step transformation by KRAS mutations. Cancer Cell. 2017;32:360–376.e6. doi: 10.1016/j.ccell.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ortega-Gómez A., Rangel-Escareño C., Molina-Romero C., Macedo-Pérez E.O., Avilés-Salas A., Lara-García A., Alanis-Funes G., Rodríguez-Bautista R., Hidalgo-Miranda A., Arrieta O. Gene-expression profiles in lung adenocarcinomas related to chronic wood smoke or tobacco exposure. Respir. Res. 2016;17:42. doi: 10.1186/s12931-016-0346-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song M.A., Benowitz N.L., Berman M., Brasky T.M., Cummings K.M., Hatsukami D.K., Marian C., O’Connor R., Rees V.W., Woroszylo C., Shields P.G. Cigarette filter ventilation and its relationship to increasing rates of lung adenocarcinoma. J. Natl. Cancer Inst. 2017;109 doi: 10.1093/jnci/djx075. djx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi Y., Au J.S., Thongprasert S., Srinivasan S., Tsai C.M., Khoa M.T., Heeroma K., Itoh Y., Cornelio G., Yang P.-C. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J. Thorac. Oncol. 2014;9:154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446–454. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 7.Sharma P., Hu-Lieskovan S., Wargo J.A., Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168:707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He J., Hu Y., Hu M., Li B. Development of PD-1/PD-L1 pathway in tumor immune microenvironment and treatment for non-small cell lung cancer. Sci. Rep. 2015;5:13110. doi: 10.1038/srep13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., Coussens L.M., Gabrilovich D.I., Ostrand-Rosenberg S., Hedrick C.C. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariathasan S., Turley S.J., Nickles D., Castiglioni A., Yuen K., Wang Y., Kadel E.E., III, Koeppen H., Astarita J.L., Cubas R. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee K., Hwang H., Nam K.T. Immune response and the tumor microenvironment: how they communicate to regulate gastric cancer. Gut Liver. 2014;8:131–139. doi: 10.5009/gnl.2014.8.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentles A.J., Bratman S.V., Lee L.J., Harris J.P., Feng W., Nair R.V., Shultz D.B., Nair V.S., Hoang C.D., West R.B. Integrating tumor and stromal gene expression signatures with clinical indices for survival stratification of early-stage non-small cell lung cancer. J. Natl. Cancer Inst. 2015;107 doi: 10.1093/jnci/djv211. djv211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng S., Luo X., Dong C., Zheng D., Xie J., Zhuge L., Sun Y., Chen H. A B7-CD28 family based signature demonstrates significantly different prognoses and tumor immune landscapes in lung adenocarcinoma. Int. J. Cancer. 2018;143:2592–2601. doi: 10.1002/ijc.31764. [DOI] [PubMed] [Google Scholar]
- 14.Song Q., Shang J., Yang Z., Zhang L., Zhang C., Chen J., Wu X. Identification of an immune signature predicting prognosis risk of patients in lung adenocarcinoma. J. Transl. Med. 2019;17:70. doi: 10.1186/s12967-019-1824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yue C., Ma H., Zhou Y. Identification of prognostic gene signature associated with microenvironment of lung adenocarcinoma. PeerJ. 2019;7:e8128. doi: 10.7717/peerj.8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong Y., Whitford C.M., Robertsen H.L., Blin K., Jørgensen T.S., Klitgaard A.K., Gren T., Jiang X., Weber T., Lee S.Y. Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proc. Natl. Acad. Sci. USA. 2019;116:20366–20375. doi: 10.1073/pnas.1913493116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higareda-Almaraz J.C., Ruiz-Moreno J.S., Klimentova J., Barbieri D., Salvador-Gallego R., Ly R., Valtierra-Gutierrez I.A., Dinsart C., Rabinovich G.A., Stulik J. Systems-level effects of ectopic galectin-7 reconstitution in cervical cancer and its microenvironment. BMC Cancer. 2016;16:680. doi: 10.1186/s12885-016-2700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morales M., Planet E., Arnal-Estape A., Pavlovic M., Tarragona M., Gomis R.R. Tumor-stroma interactions a trademark for metastasis. Breast. 2011;20(Suppl 3):S50–S55. doi: 10.1016/S0960-9776(11)70294-6. [DOI] [PubMed] [Google Scholar]
- 20.Guo S., Deng C.X. Effect of stromal cells in tumor microenvironment on metastasis initiation. Int. J. Biol. Sci. 2018;14:2083–2093. doi: 10.7150/ijbs.25720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coussens L.M., Zitvogel L., Palucka A.K. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Selitsky S.R., Mose L.E., Smith C.C., Chai S., Hoadley K.A., Dittmer D.P., Moschos S.J., Parker J.S., Vincent B.G. Prognostic value of B cells in cutaneous melanoma. Genome Med. 2019;11:36. doi: 10.1186/s13073-019-0647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savelyeva N., Allen A., Chotprakaikiat W., Harden E., Jobsri J., Godeseth R., Wang Y., Stevenson F., Ottensmeier C. Linked CD4 T cell help: broadening immune attack against cancer by vaccination. Curr. Top. Microbiol. Immunol. 2017;405:123–143. doi: 10.1007/82_2016_500. [DOI] [PubMed] [Google Scholar]
- 25.Yang L., Carbone D.P. Tumor-host immune interactions and dendritic cell dysfunction. Adv. Cancer Res. 2004;92:13–27. doi: 10.1016/S0065-230X(04)92002-7. [DOI] [PubMed] [Google Scholar]
- 26.Navegantes K.C., de Souza Gomes R., Pereira P.A.T., Czaikoski P.G., Azevedo C.H.M., Monteiro M.C. Immune modulation of some autoimmune diseases: the critical role of macrophages and neutrophils in the innate and adaptive immunity. J. Transl. Med. 2017;15:36. doi: 10.1186/s12967-017-1141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg A.S., Pariser A.R., Diamond B., Yao L., Turka L.A., Lacana E., Kishnani P.S. A role for plasma cell targeting agents in immune tolerance induction in autoimmune disease and antibody responses to therapeutic proteins. Clin. Immunol. 2016;165:55–59. doi: 10.1016/j.clim.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Ryzhov S., Goldstein A.E., Matafonov A., Zeng D., Biaggioni I., Feoktistov I. Adenosine-activated mast cells induce IgE synthesis by B lymphocytes: an A2B-mediated process involving Th2 cytokines IL-4 and IL-13 with implications for asthma. J. Immunol. 2004;172:7726–7733. doi: 10.4049/jimmunol.172.12.7726. [DOI] [PubMed] [Google Scholar]
- 29.Goc J., Germain C., Vo-Bourgais T.K., Lupo A., Klein C., Knockaert S., de Chaisemartin L., Ouakrim H., Becht E., Alifano M. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 2014;74:705–715. doi: 10.1158/0008-5472.CAN-13-1342. [DOI] [PubMed] [Google Scholar]
- 30.Zheng X., Hu Y., Yao C. The paradoxical role of tumor-infiltrating immune cells in lung cancer. Intractable Rare Dis. Res. 2017;6:234–241. doi: 10.5582/irdr.2017.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng M., Gui X., Kim J., Xie L., Chen W., Li Z., He L., Chen Y., Chen H., Luo W. LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature. 2018;562:605–609. doi: 10.1038/s41586-018-0615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manandhar S., Lee Y.M. Emerging role of RUNX3 in the regulation of tumor microenvironment. BMB Rep. 2018;51:174–181. doi: 10.5483/BMBRep.2018.51.4.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W.N., Du Y.F., Bai J., Song X.M., Zheng Y., Yuan H., Zhang W., Zhang Z.D., Wu Y.N. RUNX3 plays a tumor suppressor role by inhibiting cell migration, invasion and angiogenesis in oral squamous cell carcinoma. Oncol. Rep. 2017;38:2378–2386. doi: 10.3892/or.2017.5857. [DOI] [PubMed] [Google Scholar]
- 35.Li K., Zhu Z., Luo J., Fang J., Zhou H., Hu M., Maskey N., Yang G. Impact of chemokine receptor CXCR3 on tumor-infiltrating lymphocyte recruitment associated with favorable prognosis in advanced gastric cancer. Int. J. Clin. Exp. Pathol. 2015;8:14725–14732. [PMC free article] [PubMed] [Google Scholar]
- 36.Ho P.C., Liu P.S. Metabolic communication in tumors: a new layer of immunoregulation for immune evasion. J. Immunother. Cancer. 2016;4:4. doi: 10.1186/s40425-016-0109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu M., Zhong Y., Yang F., Liu K., Zhuang B. Identification and validation of AKR1C1/2/3 as hepatocellular carcinoma risk modules via an integrated strategy. Research Square. 2020 doi: 10.21203/rs.2.24543/v1. [DOI] [Google Scholar]
- 38.Mayakonda A., Lin D.C., Assenov Y., Plass C., Koeffler H.P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sallman D.A., Padron E. Integrating mutation variant allele frequency into clinical practice in myeloid malignancies. Hematol. Oncol. Stem Cell Ther. 2016;9:89–95. doi: 10.1016/j.hemonc.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Shin H.T., Choi Y.L., Yun J.W., Kim N.K.D., Kim S.Y., Jeon H.J., Nam J.Y., Lee C., Ryu D., Kim S.C. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat. Commun. 2017;8:1377. doi: 10.1038/s41467-017-01470-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi W., Qiao D., Martinez J.D. Caffeine induces TP53-independent G1-phase arrest and apoptosis in human lung tumor cells in a dose-dependent manner. Radiat. Res. 2002;157:166–174. doi: 10.1667/0033-7587(2002)157[0166:citigp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 43.Cheng X., Yin H., Fu J., Chen C., An J., Guan J., Duan R., Li H., Shen H. Aggregate analysis based on TCGA: TTN missense mutation correlates with favorable prognosis in lung squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2019;145:1027–1035. doi: 10.1007/s00432-019-02861-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lakshmanan I., Salfity S., Seshacharyulu P., Rachagani S., Thomas A., Das S., Duan R., Li H., Shen H. MUC16 regulates TSPYL5 for lung cancer cell growth and chemoresistance by suppressing p53. J. Cancer Res. Clin. Oncol. 2017;145:1027–1035. doi: 10.1158/1078-0432.CCR-16-2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leiserson M.D., Wu H.T., Vandin F., Raphael B.J. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 2015;16:160. doi: 10.1186/s13059-015-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soozangar N., Sadeghi M.R., Jeddi F., Somi M.H., Shirmohamadi M., Samadi N. Comparison of genome-wide analysis techniques to DNA methylation analysis in human cancer. J. Cell. Physiol. 2018;233:3968–3981. doi: 10.1002/jcp.26176. [DOI] [PubMed] [Google Scholar]
- 47.Koch A., Joosten S.C., Feng Z., de Ruijter T.C., Draht M.X., Melotte V., Smits K.M., Veeck J., Herman J.G., Van Neste L. Analysis of DNA methylation in cancer: location revisited. Nat. Rev. Clin. Oncol. 2018;15:459–466. doi: 10.1038/s41571-018-0004-4. [DOI] [PubMed] [Google Scholar]
- 48.Morris T.J., Butcher L.M., Feber A., Teschendorff A.E., Chakravarthy A.R., Wojdacz T.K., Beck S. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinformatics. 2014;30:428–430. doi: 10.1093/bioinformatics/btt684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenberg M.V.C., Bourc’his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 50.Chien M.H., Lee W.J., Hsieh F.K., Li C.F., Cheng T.Y., Wang M.Y., Chen J.-S., Chow J.-M., Jan Y.-H., Hsiao M. Keap1-Nrf2 interaction suppresses cell motility in lung adenocarcinomas by targeting the S100P protein. Clin. Cancer Res. 2015;21:4719–4732. doi: 10.1158/1078-0432.CCR-14-2880. [DOI] [PubMed] [Google Scholar]
- 51.Berglund A., Mills M., Putney R.M., Hamaidi I., Mulé J., Kim S. Methylation of immune synapse genes modulates tumor immunogenicity. J. Clin. Invest. 2020;130:974–980. doi: 10.1172/JCI131234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuen G.J., Demissie E., Pillai S. B lymphocytes and cancer: a love-hate relationship. Trends Cancer. 2016;2:747–757. doi: 10.1016/j.trecan.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang H., Wei P., Duell E.J., Risch H.A., Olson S.H., Bueno-de-Mesquita H.B., Gallinger S., Holly E.A., Petersen G., Bracci P.M. Axonal guidance signaling pathway interacting with smoking in modifying the risk of pancreatic cancer: a gene- and pathway-based interaction analysis of GWAS data. Carcinogenesis. 2014;35:1039–1045. doi: 10.1093/carcin/bgu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang D., Qu F., Cai H., Chuang C.H., Lim J.S., Jahchan N., Grüner B.M., S Kuo C., Kong C., Oudin M.J. Axon-like protrusions promote small cell lung cancer migration and metastasis. eLife. 2019;8 doi: 10.7554/eLife.50616. e50616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nasarre P., Potiron V., Drabkin H., Roche J. Guidance molecules in lung cancer. Cell Adhes. Migr. 2010;4:130–145. doi: 10.4161/cam.4.1.10882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang X., Deng Y., He R.Q., Li X.J., Ma J., Chen G., Hu X.H. Upregulation of HOXA11 during the progression of lung adenocarcinoma detected via multiple approaches. Int. J. Mol. Med. 2018;42:2650–2664. doi: 10.3892/ijmm.2018.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blanche P., Dartigues J.F., Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013;32:5381–5397. doi: 10.1002/sim.5958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Results of Pairwise Comparisons of Estimate Score and Immune Score on Tumor Size and Stage
The List of the Differentially Mutated Genes between High-Immunity and Low-Immunity Cohorts
The List of the Differentially Methylated Probes
The List of the Genes with Expression-Methylation Correlations in High-Immunity Cohort
The List of the Genes with Expression-Methylation Correlations in Low-Immunity Cohort
The Table Shows the Contributions of 27 Variables and Risk Score for Each Sample