Abstract
Blocking the MDM2/X–P53 protein–protein interaction has been widely recognized as an attractive therapeutic strategy for the treatment of cancers. Numerous small-molecule MDM2 inhibitors have been reported since the release of the structure of the MDM2–P53 interaction in 1996, SAR405838, NVP-CGM097, MK-8242, RG7112, RG7388, DS-3032b, and AMG232 currently undergo clinical evaluation for cancer therapy. This review is intended to provide a comprehensive and updated overview of MDM2 inhibitors and proteolysis targeting chimera (PROTAC) degraders with a particular focus on how these inhibitors or degraders are identified from starting points, strategies employed, structure–activity relationship (SAR) studies, binding modes or co-crystal structures, biochemical data, mechanistic studies, and preclinical/clinical studies. Moreover, we briefly discuss the challenges of designing MDM2/X inhibitors for cancer therapy such as dual MDM2/X inhibition, acquired resistance and toxicity of P53 activation as well as future directions.
KEY WORDS: MDM2/X–P53 interaction, MDM2/X inhibitors, PROTAC degraders, Cancer therapy
Graphical abstract
Blocking the MDM2/X–P53 protein–protein interaction is an emerging therapeutic strategy for cancer therapy. In this review, medicinal chemistry efforts and preclinical/clinical studies towards MDM2 inhibitors and PROTAC degraders are extensively summarized.
1. Introduction
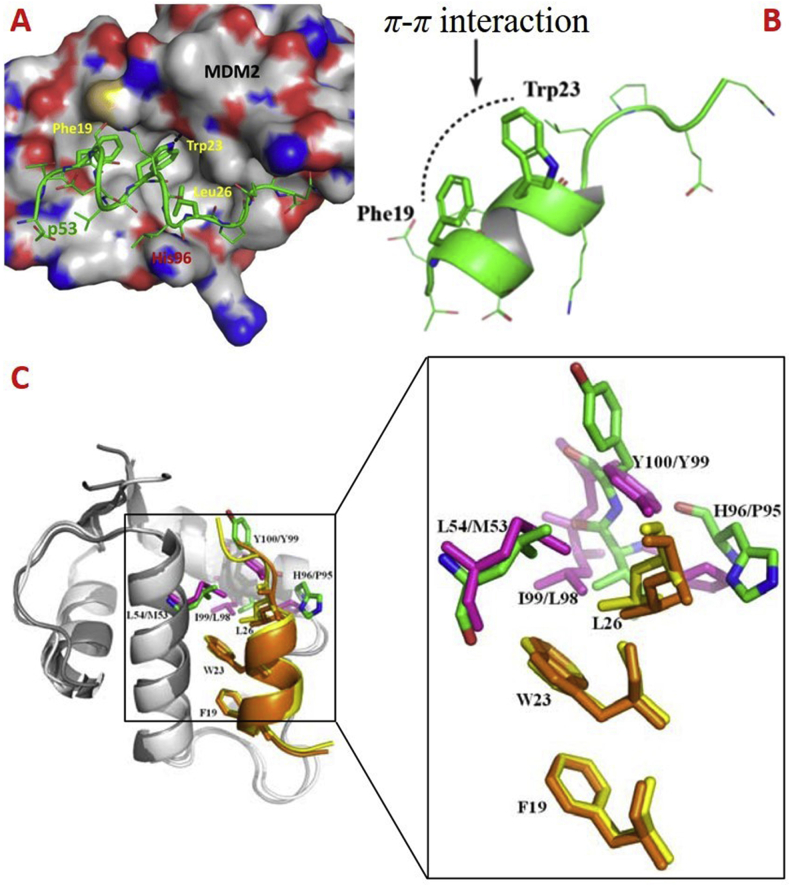
In normal cells, P53, a tumor suppressor, has played crucial roles in regulating cellular processes1, in which the negative feedback loop regulates the P53–MDM2 interactions to maintain the normal functions2,3. This loop operates from P53 activation-initiated MDM2 transcription, thus increasing MDM2 mRNA and expression. The direct interaction between P53 and MDM2 blocks the activity of P53 through diverse mechanisms. In 1996, Kussie et al.4 first uncovered the co-crystal structure of MDM2–P53 complex, showing that P53 maintains the α-helical conformation, and the Phe19, Trp23 and Leu23 residues of the N-terminal domain of P53 primarily mediate the MDM2–P53 interactions5 (Fig. 1A), the π–π interaction between Phe19 and Trp23 maintains the structural stability and thus is crucial for functional roles6 (Fig. 1B). Because of the highly conserved sequence, the spatial orientation of Phe19 and Trp23 residues in P53–MDM2/X complex are highly similar, but the orientation of Leu26 is different7 (Fig. 1C). In structure, the MDMX binding pocket has different shape and is smaller than that of MDM2 due to the Met53 and Tyr99 residues protruding into the hydrophobic cleft of MDMX. The well-defined binding surface of the MDM2/X–P53 complex provides a structural basis for developing new MDM2/X inhibitors8.
Figure 1.
The P53–MDM2/MDMX interactions. (A) MDM2 (surface)–P53 peptide (residues 15–29, colored in green) complex (PDB code: 1YCR). (B) The N-terminal domain of P53 showing π–π interaction between Phe19 and Trp23 (PDB code: 1YCR). (C) The superimposition of the structures of P53–MDM2 (PDB code: 1YCR, shown in cartoon) and P53–MDMX (PDB code: 3DAB, shown in cartoon) complex. The P53 transcriptional activation domain (TAD) fragment (residues 17–29) bound to MDM2 and MDMX is shown in yellow and orange, respectively. Phe19, Trp23 and Leu26 are shown in stick. Leu54, His96, Ile99 and Tyr100 in MDM2 are shown in green and stick, while Met53, Pro95, Leu98 and Tyr99 in MDMX are shown in light magenta.
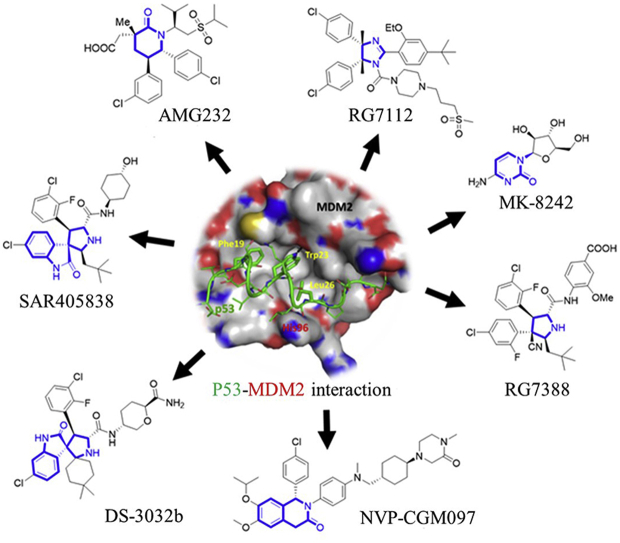
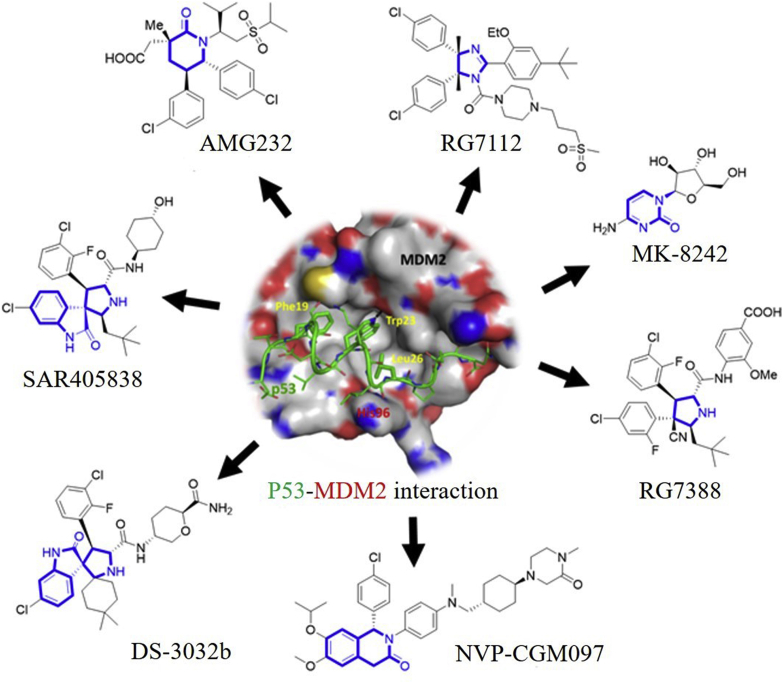
Development of small molecules disrupting the MDM2–P53 interactions has been highly pursued from academia to industry in recent years5,9, 10, 11, 12, 13, 14. Some of these inhibitors including DS-3032 (also known as Milademetan, DS-3032b)15,16, NVP-CGM09717, 18, 19, SAR40583820, 21, 22, RG711223,24, MK-824225,26, RG738827 and AMG 23228, 29, 30 (Fig. 2) are under clinical evaluation for anticancer treatment. The clinical status of these inhibitors is shown in Table 1. This review is intended to provide an overview of MDM2/X inhibitors and MDM2 proteolysis targeting chimera (PROTAC) degraders. We mainly focus on the medicinal chemistry efforts including optimization strategies for improving potency and/or pharmacodynamics (PD)/pharmacokinetics (PK) profiles and structure–activity relationship studies (SARs), binding models, biochemical data, and preclinical/clinical studies. In this review, several types of MDM2 inhibitors such as piperidinones, spirooxindoles, etc. have been previously summarized by our group31 and others11,32, 33, 34, and thus are excluded to avoid duplication. We believe this review, along with previous review articles regarding MDM2/X inhibitors, will offer a comprehensive and updated overview and may be very useful to those devoted to designing small molecules targeting the MDM2/X–P53 interactions.
Figure 2.
MDM2 inhibitors in clinical trials. The core scaffolds are highlighted in blue. The P53 peptide and MDM2 protein are shown in green and gray surface, respectively. Adapted with permission from Ref. 5. Copyright © 2015 American Chemical Society.
Table 1.
Overview of MDM2 inhibitors in clinical trials.
| Drug | Sponsor | Phase | Study title | Trial number | Disease | Status |
|---|---|---|---|---|---|---|
| RG7112 | Hoffmann-La Roche | Phase I | A study of RO5045337 [RG7112] in patients with hematologic neoplasms | NCT00623870 | Hematologic neoplasms | Completed |
| A Study of RO5045337 [RG7112] in patients with advanced solid tumors | NCT00559533 | Advanced solid tumors | ||||
| Icahn School of Medicine at Mount Sinai | Unknown | Tissue banking study—polycythemia vera or essential thrombocythemia (PV & ET) patients | NCT01970930 | Polycythemia vera and essential thrombocythemia | Completed | |
| NVP-CGM097 | Novartis | Phase I | A phase I dose escalation study of CGM097 in adult patients with selected advanced solid tumors | NCT01760525 | Solid tumor with P53 WT status | Active, not recruiting |
| DS-3032 | M.D. Anderson Cancer Center | Phase I | Ascending dose and exploratory expansion study of DS-3032b, an oral MDM2 inhibitor, in subjects with relapsed and/or refractory multiple myeloma | NCT02579824 | Myeloma | Active, not recruiting |
| Phase I Phase II |
Milademetan tosylate and low-dose cytarabine in treating participants with recurrent or refractory acute myeloid leukemia | NCT03634228 | Recurrent acute myeloid leukemia/refractory acute myeloid leukemia/TP53 WT allele | Recruiting | ||
| Daiichi Sankyo | Phase I | A phase I multiple ascending dose study of DS-3032b, an oral murine double minute 2 (MDM2) inhibitor, in subjects with advanced solid tumors or lymphomas | NCT01877382 | Advanced solid tumor Lymphoma |
Recruiting | |
| Study of milademetan in Japanese patients with relapsed or refractory acute myeloid leukemia (AML) | NCT03671564 | Acute myeloid leukemia | Active, not recruiting | |||
| Safety, tolerability and pharmacokinetics of milademetan alone and with 5-azacitidine (AZA) in acute myelogenous leukemia (AML) or high-risk myelodysplastic syndrome (MDS) | NCT02319369 | Acute myelogenous leukemia Myelodysplastic syndrome |
Recruiting | |||
| Milademetan plus quizartinib combination study in FLT3-ITD mutant acute myeloid leukemia (AML) | NCT03552029 | Acute myeloid leukemia | Recruiting | |||
| SAR405838 | Sanofi | Phase I | Phase I safety testing of SAR405838 | NCT01636479 | Neoplasm malignant | Completed |
| A safety and efficacy study of SAR405838 and pimasertib in cancer patients | NCT01985191 | |||||
| MK-8242 | Merck Sharp & Dohme Corp. | Phase I | Study of MK-8242 alone and in combination with cytarabine in participants with acute myelogenous leukemia (P07649) | NCT01451437 | Acute myelogenous leukemia (AML) | Terminated |
| Study of safety and pharmacokinetics of MK-8242 in participants with advanced solid tumors (P07650) | NCT01463696 | Solid tumors | ||||
| AMG 232 | Kartos Therapeutics | Phase I | A phase Ib study evaluating AMG 232 alone and in combination with trametinib in acute myeloid leukemia | NCT02016729 | Advanced malignancy/cancer Oncology/oncology patients/acute myeloid leukemia | Completed |
| National Cancer Institute | Phase I | MDM2 inhibitor AMG-232 in treating patients with recurrent or newly diagnosed glioblastoma | NCT03107780 | Glioblastoma/gliosarcoma/recurrent glioblastoma | Recruiting | |
| MDM2 inhibitor AMG-232, carfilzomib, lenalidomide, and dexamethasone in treating patients with relapsed or refractory multiple myeloma | NCT03031730 | Hypercalcemia Plasmacytoma Recurrent plasma cell myeloma Refractory plasma cell myeloma |
||||
| MDM2 inhibitor AMG-232 and decitabine in treating patients with relapsed, refractory, or newly-diagnosed acute myeloid leukemia | NCT03041688 | Acute myeloid leukemia Recurrent acute myeloid leukemia Refractory acute myeloid leukemia Secondary acute myeloid leukemia |
||||
| MDM2 inhibitor AMG-232 and radiation therapy in treating patients with soft tissue sarcoma | NCT03217266 | Soft tissue sarcoma | ||||
| Amgen | Phase I Phase II |
A phase Ib/2a study evaluating AMG 232 in metastatic melanoma | NCT02110355 | Advanced malignancy, advanced solid tumors, cancer, oncology, oncology patients, tumors, melanoma | Completed | |
| Phase I | A phase I study evaluating AMG 232 in advanced solid tumors or multiple myeloma | NCT01723020 | ||||
| RG7388 | John Mascarenhas | Phase I | Open label study of single agent oral RG7388 in patients with polycythemia vera and essential thrombocythemia | NCT02407080 | Polycythemia vera Essential thrombocythemia |
Completed |
| Mayo Clinic | Phase I Phase II |
Idasanutlin, ixazomib citrate, and dexamethasone in treating patients with relapsed multiple myeloma | NCT02633059 | Loss of chromosome 17p Recurrent plasma cell myeloma | Recruiting | |
| Vanderbilt–Ingram Cancer Center | Phase I Phase II |
Atezolizumab and cobimetinib or idasanutlin in participants with stage iv or unresectable recurrent estrogen receptor positive breast cancer | NCT03566485 | Breast cancer Estrogen receptor-positive HER2/Neu negative |
Recruiting | |
| University Hospital Heidelberg | Phase I Phase II |
NCT neuro master match—N2M2 (NOA-20, N2M2) | NCT03158389 | Glioblastoma, adult | Recruiting | |
| Hoffmann-La Roche | Phase I | A study of idasanutlin in combination with obinutuzumab in relapsed/refractory (R/R) follicular lymphoma (FL) and in combination with rituximab in R/R diffuse large B-cell lymphoma (DLBCL) participants | NCT02624986 | Non-hodgkin's lymphoma | Terminated | |
| Phase I | A study to determine the excretion balance, pharmacokinetics, metabolism and absolute oral bioavailability of a single oral dose of [14C]-labeled idasanutlin and an intravenous tracer dose of [13C]-labeled idasanutlin in a single cohort of participants with solid tumors (malignancies) | NCT02828930 | Solid tumors | Completed | ||
| Phase I | A study to investigate the bioequivalence or relative bioavailability of three new idasanutlin tablet variants following oral administration in participants with solid tumors | NCT03362723 | Solid tumors | Completed | ||
| Phase I Phase II |
A study evaluating the safety and efficacy of idasanutlin in combination with cytarabine and daunorubicin in patients newly diagnosed with acute myeloid leukemia (AML) and the safety and efficacy of idasanutlin in the maintenance of first AML complete remission | NCT03850535 | Acute myeloid leukemia | Recruiting | ||
| Phase I Phase II |
A study evaluating the safety, tolerability, pharmacokinetics, and preliminary activity of idasanutlin in combination with either chemotherapy or venetoclax in the treatment of pediatric and young adult participants with relapsed/refractory acute leukemias or solid tumors | NCT04029688 | Acute myeloid leukemia Acute lymphoblastic leukemia Neuroblastoma Solid tumors |
Recruiting | ||
| Phase I Phase II |
A study of venetoclax in combination with cobimetinib and venetoclax in combination with idasanutlin in patients aged ≥60 years with relapsed or refractory acute myeloid leukemia who are not eligible for cytotoxic therapy | NCT02670044 | Leukemia, myeloid, acute | Recruiting | ||
| Phase I Phase II |
A study of obinutuzumab in combination with idasanutlin and venetoclax in participants with relapsed or refractory (R/R) follicular lymphoma (FL) or rituximab in combination with idasanutlin and venetoclax in participants with R/R diffuse large B-cell lymphoma (DLBCL) | NCT03135262 | Follicular lymphoma Lymphoma, large B-cell, diffuse |
Recruiting | ||
| Phase I Phase II |
A study of multiple immunotherapy-based treatment combinations in participants with metastatic non-small cell lung cancer (morpheus-non-small cell lung cancer, morpheus lung) | NCT03337698 | Non-small-cell lung carcinoma | Recruiting | ||
| Phase I Phase II |
A study evaluating the efficacy and safety of multiple immunotherapy-based treatment combinations in patients with metastatic colorectal cancer (Morpheus-CRC) | NCT03555149 | Colorectal cancer | Recruiting | ||
| Phase II | A study to evaluate the efficacy, safety, pharmacokinetics and pharmacodynamics of idasanutlin monotherapy in participants with hydroxyurea-resistant/intolerant polycythemia vera | NCT03287245 | Polycythemia vera | Recruiting | ||
| Phase III | A study of idasanutlin with cytarabine versus cytarabine plus placebo in participants with relapsed or refractory acute myeloid leukemia (AML) (MIRROS) | NCT02545283 | Leukemia, myeloid, acute | Recruiting |
Data are from ClinicalTrials.gov. Accessed on Oct 16, 2019.
2. Development of MDM2/X inhibitors for cancer therapy
2.1. Nutlins
Among cis-imidazoline series (Nutlin family), a representative example is RG7112 (also known as RO5045337) developed by Hoffmann-La Roche. Phase I clinical trials of RG7112 for the treatment of advanced solid tumors (ClinicalTrials.gov identifier: NCT00559533) and hematologic neoplasms (ClinicalTrials.gov identifier: NCT00623870) were done.
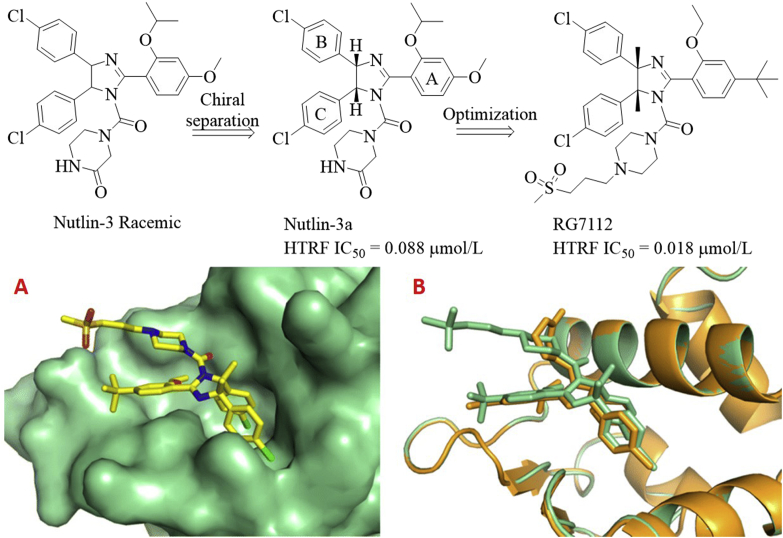
Initially, the Vassilev group8 identified cis-imidazoline analogs (named as Nutlins) after screening a large library of synthetic compounds, which bound to MDM2 with the IC50 values ranging from 100 to 300 nmol/L. Further structural modifications produced racemic Nutlin-3 as illustrated in Fig. 3. Intriguingly, only one enantiomer named Nutlin-3a exhibited potent binding potency to MDM2 (IC50 = 0.088 μmol/L), about 150-fold more potent than another one (IC50 = 13.6 μmol/L). However, Nutlin-3a was not potent enough for clinical development, further optimizations were then carried out while maintaining the key structural features. In the binding models of Nutlin-3a with MDM2, the 4-chlorophenyl groups at the C4 and C5 positions were directed to Trp23 and Leu26 hydrophobic sub-pockets, respectively, while the isopropoxy group was found to fill into the Phe19 pocket. With the structural requirements for optimal binding in mind, further modifications were performed, finally leading to the discovery of RG711223: (a) replacement of isopropoxy group with the ethoxy group to reduce the molecule weight while maintaining comparable binding ability; (b) replacement of metabolically labile methoxy group attached to the A ring with the t-butyl group to enhance metabolic stability; (c) incorporation of methyl group at the C4 and C5 positions to prevent oxidation of imidazoline scaffold to inactive imidazole; substitution with larger ethyl group or monomethylation at the C4 position resulted in reduced potency; (d) variations of the side chain attached to the N-1 position to improve binding potency and PK properties; (e) the absolute configuration plays a pivotal role in potency. The (4S,5R)-enantiomer as presented in RG7112 was about 200 times more potent than (4R,5S)-enantiomer. RG7112 tightly bound to MDM2 (Kd = 10.8 nmol/L) and can displace P53 from the MDM2 surface (IC50 = 18 nmol/L), 4 and 200 times more potent than Nutlin-3a and the inactive enantiomer, respectively24. The crystal structure of RG7112 in complex with MDM2 (PDB code: 4IPF) revealed that two 4-chloro-phenyl rings occupied the Trp23 and Leu26 pockets, while the ethoxy group was projected into the Phe19 pocket (Fig. 3A). Superposition of RG7112 with Nutlin-3a (PDB code: 4J3E, Fig. 3B) showed that introduction of the dimethyl groups had no significant impact on the projection of both 4-chlorophenyl groups.
Figure 3.
Identification of MDM2 inhibitor RG7112 from Nutlin-3. (A) Crystal structure of MDM2 (shown in green surface) in complex with RG7112 (colored in yellow, PDB code: 4IPF). (B) Superposition of the crystal structures of MDM2 in complex with RG7112 (green) and Nutlin-3a (gold, PDB code: 4J3E).
Treatment of cancer cells with RG7112 caused P53 accumulation and activated P21 and MDM2, finally arresting cell cycle at G1 and G2 phase and inducing apoptosis. RG7112 was selectively potent against wild-type (WT) P53 expressed cancer cell lines with the IC50 values ranging from 0.18 to 2.2 μmol/L over those with mutant P53 (IC50 = 5.7–20.3 μmol/L). RG7112 showed favorable plasma exposure in mouse after single oral administration at 50 mg/kg (t1/2 = 8.8 h, Cmax = 15.5 μg/mL, and AUClast = 251.2 μg h/mL) and caused 74% and 69% of tumor growth inhibition (TGI) in SJSA-1 and MHM xenograft models, respectively, at the same dose. Tumor suppression was observed at a dose of 100 mg/kg23. Besides, the synergistic effect was observed in LNCaP when combined with androgen ablation, leading to a significant tumor regression24. Recently, Obrador-Hevia and co-workers35 found that RG7112 can significantly enhance the response of Trabectedin to MDM2-amplified liposarcoma cells. Clinical pharmacological studies showed that high/low-fat food and new formulation can enhance bioavailability; high-dose consecutive daily dosing for 3–5 days can yield required high drug exposures and PD effects for cancer therapy36. However, P53 activation induced by RG7112 caused the thrombocytopenia37, which may restrict its clinical use. Clinical trials of RG7112 in 20 patients indicated that all patients had at least one adverse event, 12 serious adverse events, particularly the neutropenia and thrombocytopenia, were observed in eight patients38.
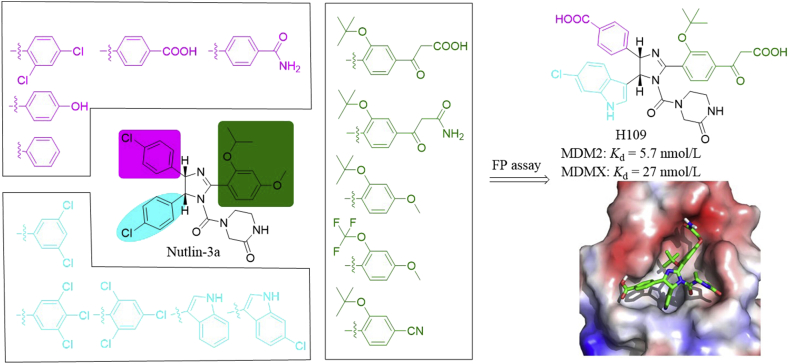
Both Nutlin-3a and RG7112 specifically inhibited MDM2 oncoprotein. The robust antitumor efficacy of Nutlins through MDM2 inhibition has been reported to be compromised by MDMX overexpression39, which has been observed in many cancers40. Therefore, P53 activation by dual inhibition of MDM2 and MDMX is a promising therapeutic option for treating cancers expressing WT P53. Based on the co-crystal structure of MDM2/Nutlin-3a complex and mutational analysis of N-terminus of MDMX, Su and co-workers41 designed a molecular library (216 compounds) sharing the Nutlin-3a core scaffold as shown in Fig. 4. In this design, they focused on introducing the pharmacophores found in U.S. Food and Drug Administration (FDA)-approved drugs. Among these compounds, H109 had high binding affinities to MDMX and MDM2 in the fluorescence polarization (FP) assay (Kd = 27 and 5.7 nmol/L, respectively). The predicted binding model showed that H109 was nicely fitted into the P53 binding surface on N-terminus of MDMX. The 15N–1H heteronuclear NOE data revealed that the flexible region on MDMX became significantly more rigid upon H109 binding. The enhanced hydrophobic interactions of H109 in the Trp23 and Leu26 binding pockets and reduced backbone dynamics in the region around Gln71 and Val92 may be responsible for the enhanced binding affinity of H109 to MDMX.
Figure 4.
Rational design of MDM2/MDMX dual inhibitor H109 and the predicted binding mode of H109 (colored in green) in the MDMX binding site. Adapted with permission from Ref. 41. Copyright © 2014 American Chemical Society.
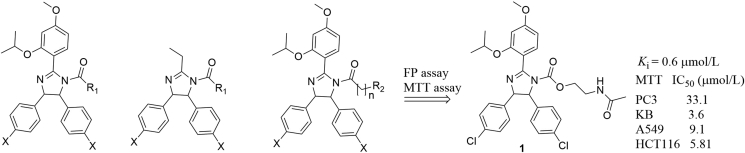
Hu's group42 designed three series of imidazoline analogs and evaluated their binding affinities to MDM2 (Fig. 5). These molecules showed different degrees of binding affinities. Among them, compound 1 bound to MDM2 moderately (Ki = 0.6 μmol/L) in the FR assay, inhibited cell growth of A549 (P53+/+), HCT116 (P53+/+), PC3 (P53−/−) and KB (P53 mutant) cancer cell lines at micromolar levels, and arrested cell growth at G2/M phase. However, compound 1 did not show remarkable differentiation toward cancerous cells with different P53 status.
Figure 5.
Imidazoline-based MDM2 inhibitors developed by Hu's group.
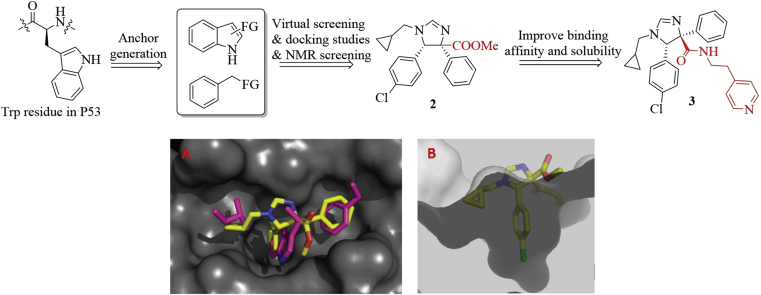
In 2010, Domling et al.43 developed a robust strategy for generating HDM2/P53 antagonists based on computational analysis and the multi-component reaction (MCR) chemistry43. Because of the importance of Trp23 residue in the HDM2–P53 interactions, they chose the indole side chain as the anchor for design. Next, the indole and bioisosteric 4-chlorophenyl groups installed with functional groups were employed as starting materials to access molecular library based on the MCR chemistry (Fig. 6). Virtual screening, followed by constrained docking using REACTOR and Moloc software, respectively generated the prioritized compounds, which mimicked Trp23, Phe19 and Leu26 residues of P53 to occupy the hydrophobic cleft on the surface of HDM2. Seven compounds among this series bound to HDM2 with the Kd values less than 60 μmol/L in the FP assay. NMR spectroscopy was then used to determine their binding ability to HMD2. Syn-2 and anti-2 bond to HDM2 with KdNMR values of 3 and 40 μmol/L, respectively. The binding model of 2 in the active site of HDM2 (PDB code: 1YCR) showed that the methyl ester group was exposed to the solvent region and was not involved in the binding interface (Fig. 6A and B). Replacement of the ester with the amide group produced compound 3, which showed improved binding affinity with a Kd value of 0.8 μmol/L and water solubility44.
Figure 6.
Identification of compound 3 based on computational analysis and MCR chemistry. (A) Predicted binding mode of compound 2 (shown in yellow stick) in the P53 binding site of MDM2 (PDB code: 1YCR, gray surface) and overlapped with the F19, W23, and L26 (pink sticks) in P53. (D) Cut-away view of compound 2 (shown in yellow stick) in the P53 binding site of MDM2, showing the shape and depth of the P53 binding site. Adapted with permission from Ref. 44. Copyright © 2009 ACS publisher.
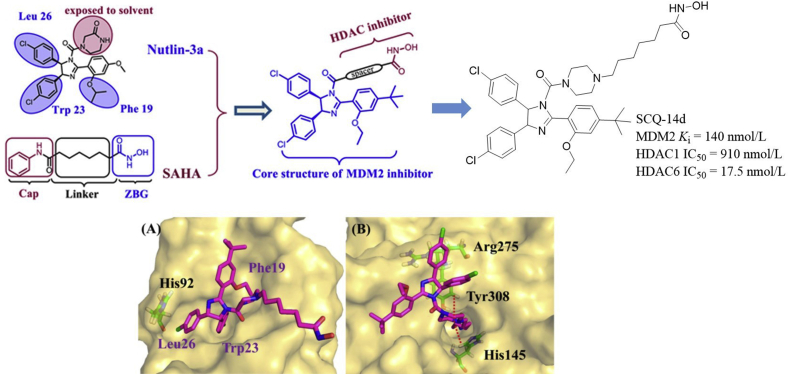
Inspired by the synergistic effects between MDM2 and HDACs, Sheng et al.45 carried out structure-based design of MDM2/HDAC dual inhibitors (Fig. 7). The principle for designing such dual inhibitors was that the zinc binding group (ZBG) of HDAC inhibitor SAHA was introduced to the N3 substitution (exposed to the solvent region) of Nutlin-3a via suitable linkers. Of these compounds, SCQ-14d showed promising activities with the IC50 values of 140, 910 and 17.5 nmol/L against MDM2, HDAC1, and HDAC6, respectively. Docking studies showed that SCQ-14d-2 (4R,5S-enatiomer) fitted well into the MDM2 binding pocket, three phenyl groups were oriented into the sub-pockets normally occupied by Phe19, Trp23, and Leu26 of P53, and the 4-chlorophenyl group formed π–π stacking interaction with His92 (Fig. 7A). While SCQ-14d-2 bound to HDAC1 mainly through the linker (projected into the hydrophobic cavity) and ZBG (Fig. 7B), which chelated with Zn2+ and also formed two H-bonds with Tyr308 and His145. Moreover, the 4-chloropnenyl group in the cap formed π–π stacking interactions with Arg275. Upon oral administration of 100 mg/kg/day of compound SQC-14d, the tumor growth inhibition (TGI) in the A549 xenograft model was 65.4%, higher than that of SAHA and Nutlin at the same dose (TGI = 57.3% and 44.0%, respectively). Besides, compound SQC-14d exhibited reasonable PK properties in Sprague–Dawley (SD) rats [F(%) = 18%, t1/2 = 5.87 h].
Figure 7.
Structure-based design of Nutlin-based MDM2/HDAC dual inhibitors. (A) Binding model of SCQ-14d-2 (red) in the MDM2 binding site (PDB code: 4IPF). (B) Binding mode of SCQ-14d-2 (red) in the active site of HDAC1 (PDB code: 4BKX). Adapted with permission from Ref. 45. Copyright © 2018 American Chemical Society.
2.2. Dihydroisoquinolinones
The dihydroisoquinolinone derivative NVP-CGM097 (also known as CGM097) is an MDM2 inhibitor with high potency and selectivity. NVP-CGM097 is undergoing clinical safety evaluation at phase I for treating advanced solid tumors expressing WT P53 (Table 1)46.
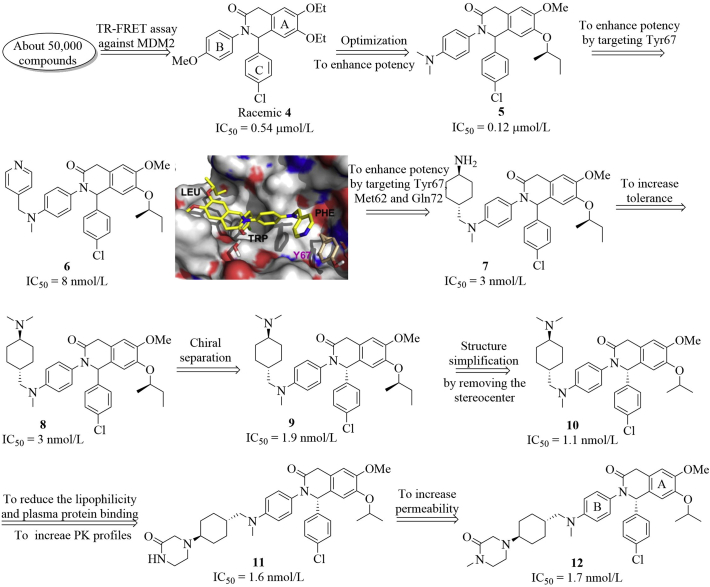
This program was initiated in the Holzer group17 to identify new inhibitors of MDM2–P53 interactions using the three hotspot 2D/3D virtual screening methods. After screening a library containing around 50,000 compounds, they identified an interesting compound 4, which inhibited the MDM2–P53 interactions (IC50 = 0.54 μmol/L) in the TR-FRET assay, albeit with a weak antiproliferative activity against SJSA-1 cells (IC50 = 16.5 μmol/L). Further optimization focused on varying substituents on the A and B phenyl rings (Fig. 8), generating compound 5 with slightly increased binding affinity (IC50 = 0.12 μmol/L). The SARs studies indicated that small aliphatic ether groups on the A ring and a para substituent on the B ring (particularly the dimethyl amino group present in compound 5) were preferred. The co-crystal structure of compound 5 with MDM2 suggested that replacement of one methyl group in the dimethyl amino group of compound 5 with a larger group may offer extra interactions with MDM2 in the Phe19 subpocket. Compound 6 showed significantly improved binding affinity to MDM2, the pyridinyl (Py) group formed a π–π stacking with the phenol group of Tyr67. The molecular modeling showed that additional interactions with Met62 and Gln72 in the Phe19 subpocket through the van der Waals contacts and hydrogen bond, respectively may be helpful to improve the binding ability. Compound 7 with a trans-cyclohexyl amine motif exhibited 2.7-fold improvement in binding affinity and cellular potency against SJSA-1 cells (IC50 = 3 nmol/L, GI50 = 2.85 μmol/L). Methylation of the NH2 group of compound 7 gave compound 8, which showed slightly increased inhibition against SJSA-1 cells (GI50 = 1.13 μmol/L). The X-ray co-crystal structures of this series with MDM2 indicated that the (S)-configuration of the chiral center bearing the para-chlorophenyl group was preferred. Chiral separation of compound 8 yielded (R,S)-9, which showed slightly increased binding potency (IC50 = 1.9 nmol/L) and interesting in vivo ADME properties (logPe = −4.4, clint = 5.6 μL/min/mg, t1/2 = 246 min), albeit with inhibition of CYP450 3A4 isoform and hERG receptor. However, it is worth mentioning that tert-amine compound 9 had a relatively low oral bioavailability and the oral absorption issue. Further structural modifications by changing the chiral isobutyl group to isopropyl group resulted in the identification of compound 10 having similar cellular potency as compound 9 (IC50 = 1.1 nmol/L, GI50 = 0.71 μmol/L toward SJSA-1 cells). To better occupy the Phe19 pocket and to modulate the basicity of basic center, the dimethyl amine group was replaced with other cyclic motifs, generating compound 11 with the piperazin-2-one motif, which showed significantly reduced basicity, around 10-fold reduction of hERG inhibition (IC50 = 16 μmol/L) and comparable cellular potency against SJSA-1 cells (GI50 = 0.35 μmol/L). Besides, compound 11 also showed improved PK profiles and oral absorption but with significantly lower permeability compared to compound 9. Further N-methylation of piperazinone moiety of compound 11 led to the discovery of NVP-CGM097 (compound 12), which possessed significantly improved permeability in the PUMPA assay (logPe = −4.6) and increased bioavailability and oral exposure by a factor of 2–3 compared to compound 11. Further bioisosteric replacement of the phenyl ring (B ring in compound 12) with N-heterocycle (e.g., the pyridine ring) yielded a series of derivatives with increased lipophilicity and comparable binding potency to MDM2. Unfortunately, this series of compounds reduced liver microsomal stability in human and monkey. Interestingly, NVP-CGM097 exhibited species specific binding to MDM2. The binding affinities to MDM2 of different species varied significantly.
Figure 8.
Structure-guided discovery of drug candidate NVP-CGM097. Predicted binding model of 6 (colored in yellow) bound to MDM2, in which the pyridyl ring forms a π–π stacking interaction with the phenol ring of Y67. Adapted with permission from Ref. 17. Copyright © 2015 Elsevier Ltd.
NVP-CGM097 selectively bound to HDM2 (IC50 = 1.7 nmol/L) over MDM4 (IC50 = 2000 nmol/L) and had 4-fold improved potency relative to Nutlin-3a (IC50 = 8 nmol/L). No inhibition against several other protein–protein interactions such as BCL-2/BAD, BCL-2/BAK, MCL-1/NOXA, MCL-1/BAK, c-IAP/BIR3, and XIAP/BIR3 were observed. NVP-CGM097 was well tolerated in inhibiting growth of SJSA-1 cells in nude rat and no histopathology change was observed after daily treatment at 30 mg/kg. The preclinical data of NVP-CGM097 are summarized in Table 2. More interestingly, Porta et al.47 recently identified a gene expression signature consisting of 13 up-regulated P53 downstream target genes, which can be used in both cell lines and in patient-derived tumor xenograft models to predict the sensitivity to NVP-CGM097. This signature may provide a novel patient selection strategy for NVP-CGM097.
Table 2.
The preclinical data of NVP-CGM09719.
| NVP-CGM097 |
MDM2/MDM4 IC50 (nmol/L) |
GI50 (μmol/L) |
CYP3A4 IC50 (μmol/L) |
LogP |
LogPe |
Clint in human LM (μL/min/mg) |
|||
| 1.7/2000 |
0.35 |
17.0 |
5.8 |
−4.6 |
52.3 |
||||
| Dose (mg/kg) |
CL (mL/min/kg) |
VSS (L/kg) |
t1/2term (h) |
AUC p.o. (nmol/L·h/L) |
Cmaxp.o. (nmol/L/L) |
Tmaxp.o. (h) |
BAV (F%) |
PPB (%) |
|
| Mouse | 1/3 | 5 | 2.6 | 6.4 | 3337 | 207 | 4.0 | 71 | >99 |
| Rat | 1/3 | 7 | 6.4 | 12.1 | 2779 | 134 | 4.5 | 85 | >99 |
| Dog | 0.1/0.3 | 3 | 4.2 | 19.8 | 5993 | 251 | 2.7 | 74 | >99 |
| Monkey | 0.1/0.3 | 4 | 2.0 | 8.3 | 3730 | 363 | 1.3 | 57 | >99 |
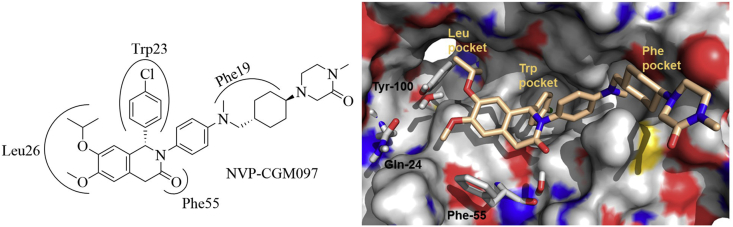
Like other MDM2 inhibitors, NVP-CGM097 mimicked Trp23, Leu26, and Phe19 residues in P53 to occupy the P53 binding interface on HDM2 (Fig. 9). The isopropyl and methyl ether groups filled into the Leu26 subpocket; the ether oxygen atoms formed H-bond interactions with Tyr100 and Gln24. The para-chlorophenyl group was directed to the Trp23 cavity. The carbonyl group of dihydroisoquinolinone core was crucial for the overall binding of NVP-CGM097 to HDM2 by offering a hydrogen bond with Phe55. The conformationally constrained N-aryl group next to the carbonyl group was projected into the Phe19 subpocket. The trans-cyclohexyl group perfectly occupied the central region of Phe19 subpocket, while the N-methyl group filled into a small hydrophobic cavity. The N-methylated piperazinone motif contributed significantly to the binding potency by binding to the exist of Phe19 cavity.
Figure 9.
The co-crystal structure of NVP-CGM097 (colored in wheat) in complex with HDM2 protein (shown in gray surface, PDB code: 4ZYF). Adapted with permission from Ref. 44. Copyright © 2009 American Chemical Society.
2.3. Benzodiazepinediones
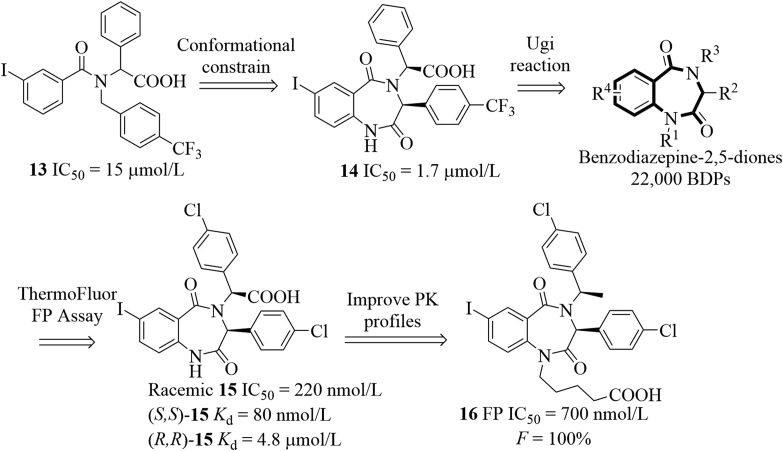
The benzodiazepine-2,5-dione inhibitors that can block the MDM2–P53 interactions were first discovered in 2005 (Fig. 10)48. Initially, they identified compound 13 using the ThermoFluor® assay, which bound to HDM2 with an IC50 value of 15 μmol/L in the FP assay. Further structural rigidification afforded the benzodiazepine-2,5-dione 14, which showed about 10-fold improved potency (FP IC50 = 1.7 μmol/L). Inspired by the promising binding potency, a library of benzodiazepine-2,5-diones (BDPs, 22,000 compounds) were synthesized based on the Ugi multicomponent reactions, which always generate bis-amides from a ketone or aldehyde, an amine, an isocyanide and a carboxylic acid in an efficient manner. This small-molecule library was first screened for the binding affinity to HDM2 using the ThermoFluor® screening technology, the promising hits from the initial screening were further characterized in the FP assay. Extensive structural optimizations49,50, followed by biological characterization led to the discovery of compound 15 with the binding affinity of 220 nmol/L. Interestingly, this kind of molecules bound to HDM2 selectively and stereospecifically51. (S,S)-15 (also known as TDP222669) potently bound to HDM2 with a Kd value of 80 nmol/L, 60-fold more potent than (R,R)-15. (S,S)-15 dose-dependently stabilized P53 and increased P21 and MDM2 expression in JAR choriocarcinoma cells. Besides, (S,S)-15 inhibited growth of JAR P53+/+ cells (IC50 = 30 μmol/L), but was inactive to MDA-MB-231 cells with mutant P5351. The synergistic effect in inhibiting tumor growth in both culture and A375 xenograft model was observed for benzodiazepine-2,4-diones (e.g., TDP665759) when combined with doxorubicin52. Of noted, TDP222669 had poor PK profiles (low bioavailability and rapid clearance) because of the poor permeability and low solubility. To improve the PK profiles, modifications focusing on the carboxylic acid site and substituents attached to the amide nitrogen in TDP222669 were carried out using the structure-based design strategy, ultimately resulting in the discovery of compound 1653. Compound 16 showed desirable early ADME properties and improved PK profiles (Cmax = 1.7 μmol/L after 6 h, t1/2 = 2.5 h, F ∼100%) in mice upon oral administration at 40 mg/kg, albeit with decreased binding potency in the FP assay (IC50 = 0.7 μmol/L), compared to TDP222669 (compound (S,S)-15).
Figure 10.
Discovery of potent benzodiazepine-2,5-dione-based HDM2 inhibitors.
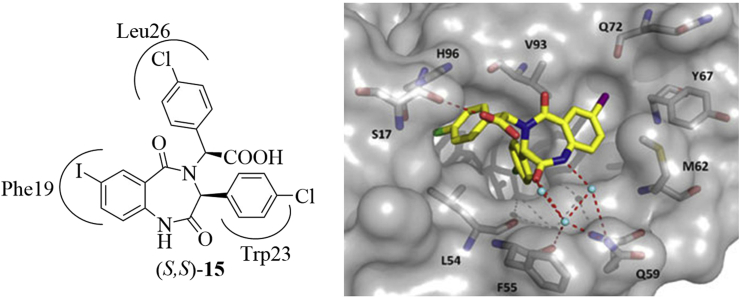
The binding models of (S,S)-15 with HDM2 protein is shown in Fig. 1154. (S,S)-15 filled into the hydrophobic cleft on the surface of HDM2, the 4-chlorophenyl group attached to the C3 position occupied the deep and narrow Trp23 pocket, while the phenyl rings of benzodiazepine scaffold and N-4 substituent were inserted into the Phe19 and Leu26 pockets, respectively. The carboxylic acid and amide formed three hydrogen bonds with H96, F55, and Q59 residues. Compound 16 had a similar binding model with TDP222669.
Figure 11.
The binding models of (S,S)-15 with HDM2 (PDB code: 1T4E). The red dashed lines represent the H-bonds formed with around residues. (S,S)-15 is colored in yellow, while the HDM2 is shown in gray surface. Adapted with permission from Ref. 54. Copyright © 2010 Wiley.
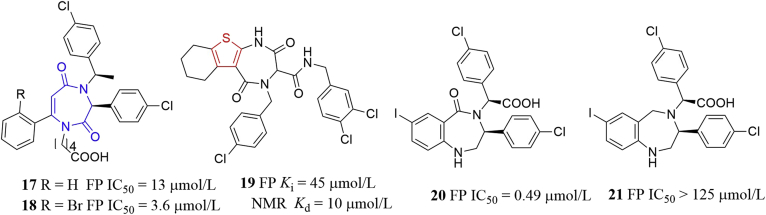
Similarly, the Cummings group48 also identified compounds 17 and 18 (Fig. 12), which could be viewed as seco analogs of benzodiazepines. Compounds 17 and 18 potently displaced P53 peptide from HDM2 protein (IC50 = 13 and 3.6 μmol/L, respectively). Compound 18 showed good permeability in Caco-2 cells (Papp = 2.71 × 10−5 cm/s). Recently, Cummings and coworkers designed and synthesized compound 19, in which the isosteric thiophene ring (highlighted in red) was incorporated instead of the phenyl ring of BDPs. Compound 19 exhibited moderate binding affinity to HDM2 in FP and NMR assays (FP Ki = 45 μmol/L, KdNMR = 10 μmol/L)54. Intriguingly, removal of carbonyl group in (S,S)-16 yielded compounds 20 and 21 showing significantly decreased binding affinity to HDM2 in the FP assay (IC50 = 0.49 and > 125 μmol/L, respectively)55. Taken together, the 1,4-diazepine-2,5-dione scaffold (highlighted in blue in Fig. 12) could mimic the a-helix of natural P53 peptide50, projecting the three halophenyl rings of compound 21 into the regions occupied by the Phe19, Trp23, and Leu26 residues in P53, to antagonize the HMD2–P53 interactions and could serve as a promising template for developing novel small molecule inhibitors of PPIs.
Figure 12.
Benzodiazepine-2,5-dione-derived HDM2 inhibitors.
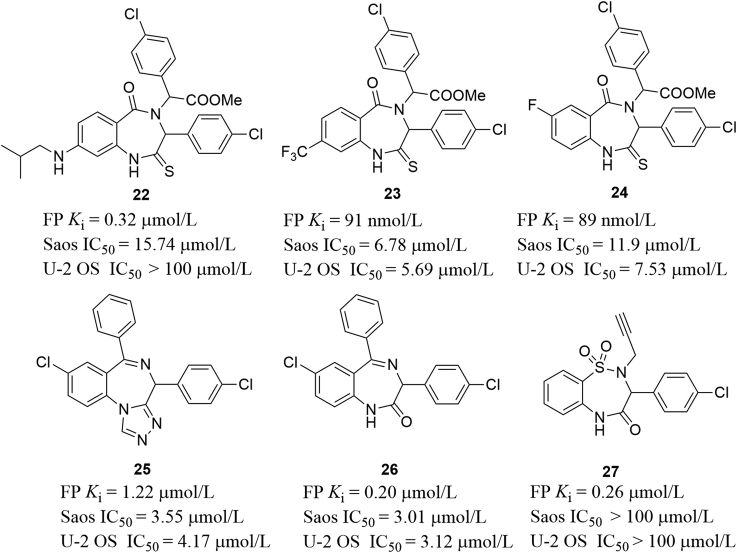
Encouraged by the potential of the benzodiazepine-2,5-dione scaffolds as MDM2 inhibitors, Zhang and co-workers56 designed thio-benzodiazepines that can disrupt the MDM2–P53 interactions based on the bioisosteric replacement principle, in which the oxygen atom of C2 carbonyl was replaced by the sulfur atom (Fig. 13). Among this series, compound 22 bound to MDM2 with a comparable Ki value (Ki = 0.32 μmol/L) with Nutlin-3a (IC50 = 0.23 μmol/L). Further modifications on the aromatic ring of benzodiazepine scaffold were carried out, fluorinated compounds 23 and 24 showed significantly improved binding affinity (Ki = 91 and 89 nmol/L, respectively) as well as enhanced in vitro antitumor activity as shown in Fig. 13 57. Additionally, they also synthesized sulfamide and triazole benzodiazepines and tested their binding affinity to MDM258. Compounds 25 and 26 showed reduced binding activity, albeit with slightly increased in vitro antitumor activity. Compound 27 were found to be inactive toward cancerous cells Saos and U-2 OS, although it represented acceptable binding affinity (Ki = 0.26 μmol/L). However, it should be noted that no selectivity toward Saos P53−/− and U-2 OS P53+/+ cells for all these compounds was observed. Compound 22 was even more sensitive to Saos P53−/− cells.
Figure 13.
Thio-benzodiazepines and related compounds as MDM2–P53 interaction inhibitors.
2.4. Isoindolinones
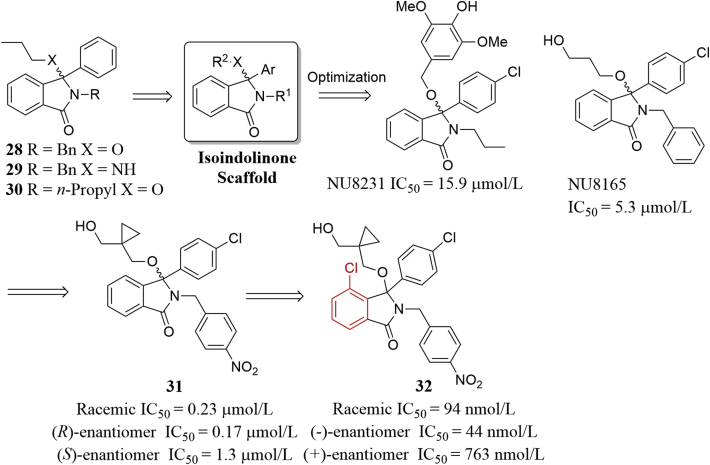
In 2005, the Hardcastle group reported the first isoindolinone-based MDM2 inhibitors59 and finally identified isoindolinone 32 after extensive modifications guided by the virtual screening (Fig. 14). Initially, they identified three isoindolinone-based compounds 28–30 which inhibited the MDM2–P53 interactions moderately (IC50 ∼200 μmol/L)59. Based on the isoindolinone scaffold and the X-ray structure of the MDM2–P53 complex, virtual screening-guided optimization produced NU8231 and NU8165, which inhibited the MDM2–P53 interactions with significantly improved binding affinity (IC50 = 15.9 and 5.3 μmol/L, respectively) and dose-dependently increased expression of P21 and MDM2 in SJSA-1 cells60. Further modifications centered on variations of 3-hydroxypropoxy side chain and substituents on the benzyl group, the introduction of 4-nitro group to the N-benzyl group, coupled with conformational constrain of the alkoxy side chain led to compound 31 with an IC50 of 0.23 μmol/L61. It is believed that the electron-withdrawing character and the lipophilic, directional interactions of nitro group with MDM2 contributed to the observed improved binding potency. The (R)-enantiomer showed slightly enhanced binding potency (IC50 = 0.17 μmol/L). (R)-31 strongly induced expression of MDM2, P53 and P21 and showed comparable cellular activation of P53 to Nutlin-3 in cancer cells expressing P53. Compound 31 is a promising candidate for further development. Next, The Hardcastle group62 paid extra attention to the aromatic ring of isoindolinone and found that replacement of the phenyl group with cyclohexyl or 3,4-dimethyl group resulted in a loss in binging potency. Interestingly, adding a 4-chloro substituent to the aromatic ring resulted in an improved binding potency to MDM2 for almost all other analogs. Racemic 32 potently inhibited the MDM2–P53 interactions (IC50 = 94 nmol/L). (−)-Enantiomer showed about 17-fold potency than (+)-enantiomer. In the SRB assay, both 32 and (−)-32 inhibited growth of SJSA-1 P53+/+ cells with IC50 value less than 6 μmol/L, but less potent than Nutlin-3 and MI-63. Besides, 32 and (−)-32 increased the levels of P53, MDM2 and P21, while (+)-32 failed to activate P53 at the highest concentration.
Figure 14.
Virtual screening-guided identification of isoindolinone MDM2 inhibitors.
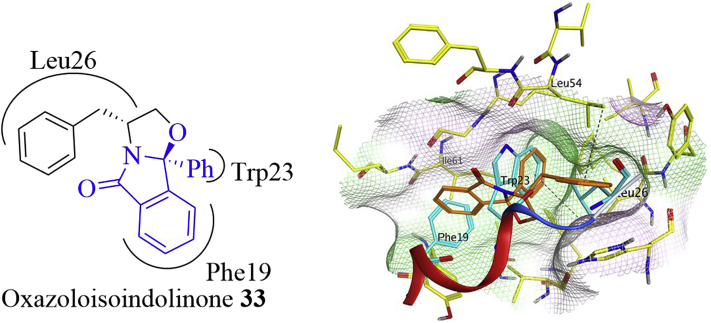
Very recently, Saraiva and co-workers63 identified structurally novel oxazoloisoindolinone analogs that activated P53 pathway by inhibiting MDM2–P53 interactions using a yeast-based screening assay (Fig. 15). Among this series, compound 33 dose-dependently inhibited growth of HCT116 P53+/+ cells through inducing apoptosis and G0/G1 cell cycle arrest. Besides, compound 33 stabilized P53 and increased the expression of P53 transcriptional targets such as P21, MDM2, BAX and PUMA. Docking studies showed that the isoindolinone aromatic ring occupied the Phe19 pocket, while the phenyl and benzyl groups filled into the Trp23 and Leu26 pockets, respectively. A hydrogen–π interaction (shown in black dashed line) between the phenyl ring of benzyl group and Leu54 was also observed. The hydrogen–π interactions refer to the interactions between hydrogen atoms and the conjugate π–systems in organic and biological molecules.
Figure 15.
Oxazoloisoindolinone 33 (colored in brown) and the binding model with MDM2 (PDB code: 3LBL). The Phe19, Trp23, and Leu26 residues are shown in cyan. The hydrogen–π interaction is shown in black dashed line. Adapted with permission from Ref. 63. Copyright © 2015 Elsevier Ltd.
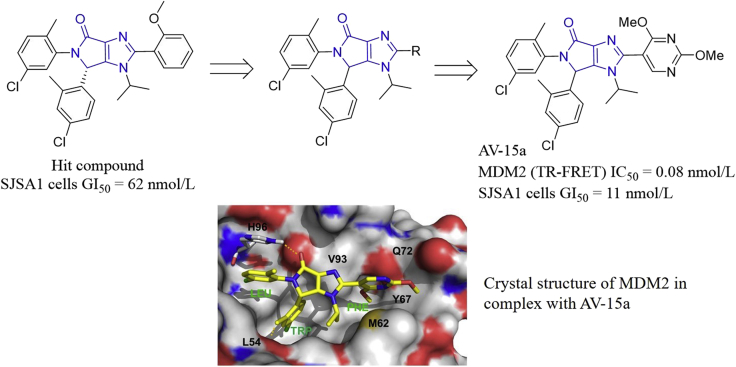
Based on the central valine concept, a pocket-adapted scaffold approach for the de novo design of MDM2 inhibitors, Vaupel and co-authors64 designed a series of bicyclic compounds starting from a previously identified hit compound which potently inhibited cell growth of SJSA1 cells (GI50 = 62 nmol/L). Further modifications around the bicyclic imidazo-pyrrolidinone scaffold gave AV-15a, which effectively inhibited MDM2 with an IC50 value of 0.08 nmol/L in the TR-FRET assay and cell growth of SJSA cells (GI50 = 11 nmol/L, Fig. 16). Crystal structure of MDM2/AV-15a complex (PDB code: 6GGN) revealed that AV-15a had an identical binding model with that of the dihydropyrrolo-pyrazole core (PDB code: 5LN2). The bicyclic core had van der Waals contacts with V93, and the carbonyl oxygen formed an H-bond interaction from H96. Two chlorophenyl groups occupied the Trp23 and Leu19 sub-pockets, and the π–π stacking interaction with H96 was observed in the Leu19 sub-pocket. In the Trp23 sub-pocket, the 2-methyl donated a pseudo H-bond to the backbone-carbonyl of Leu54. The 2,4-dimethoxypyrimidine moiety occupied the Phe19 sub-pocket, where the 4-methoxy group was directed to a small cavity located under the backbone-carbonyl of Q72, and the 2-methoxy substituent formed van der Waals interactions with M62, Y67 and Q72. AV-15a also displayed excellent oral efficacy in the human SJSA1-based xenograft model.
Figure 16.
Identification of MDM2 inhibitor AV-15a based on the central valine concept. The crystal structure (PDB code: 6GGN) of MDM2 in complex with AV-15a (colored in yellow). The dashed line represents the H-bond. The MDM2 sub-pockets accommodating Leu26, Trp23 and Phe19 residues of P53 are labeled LEU, TRP and PHE, respectively. Adapted with permission from Ref. 64. Copyright © 2018 Elsevier Ltd.
2.5. Indoles
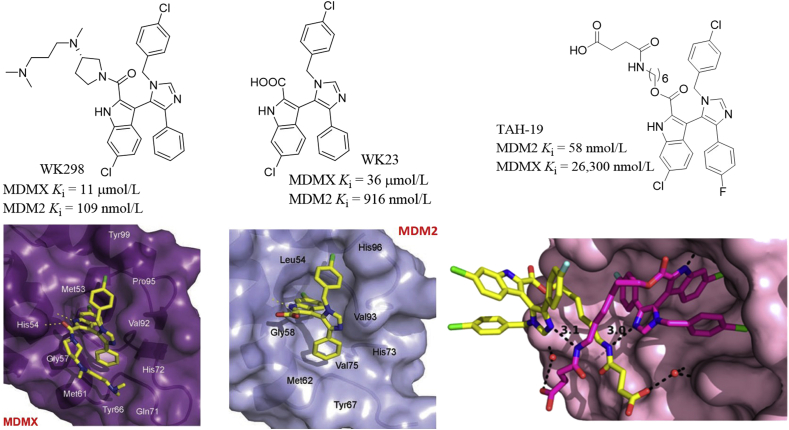
Based on the crystal structure of the octapeptide/MDM2 complex, Furet et al.65 proposed that placing a planar aromatic ring above the MDM2 cleft could project attached substituents to the surrounding subpockets, thereby disrupting the MDM2–P53 interactions. Based on the central valine concept, they designed the imidazolyl indoles that blocked the MDM2–P53 interactions. WK298 (also known as Novartis-101) was the first imidazolyl indole inhibitor of MDMX–P53 interactions, which bound to MDMX with a Ki value of 11 μmol/L in the FP assay, while WK23 lacking the amine side chain exhibited 3-fold potency to MDMX66. This finding showed that the carboxylic acid group attached to the 2-position of indole was beneficial for improving the binding affinity. Both compounds bound to MDM2 at nanomolar levels and showed excellent selectivity to MDM2 over MDMX. WK298 and WK23 displayed very similar bind pose as shown in Fig. 17. The 6-chlorooxindole group of WK298 fitted the key Trp23 pocket, while the 4-phenyl and 1-benzyl groups occupied the Phe19 and Leu26 pockets, respectively. Two H-bonds to His54 and Met53 were formed. The amine side chain contacted with Gly57 and Met61 residues, providing extra hydrophobic protection at the interface of MDMX and P53. For WK23, the imidazole served as a scaffold for projecting the 6-chlorooxindole, phenyl and benzyl substituents to the surrounding Trp23, Phe19 and Leu26 pockets, respectively and also contacted with the Val93 residue. The carboxylic acid group formed a hydrogen bond with Gly58. Interestingly, WK23 did not cause excessive induced-fit changes in MDM2 and therefore could allow for further modifications for improving potency. Holak et al.67 reported a series of 1,4,5-trisubstituted imidazole compounds as P53–MDM2/X antagonists (Fig. 17). Among these compounds, TAH-19 inhibited MDM2 potently with a Ki value of 58 nmol/L in the FP assay, but showed moderate inhibition against MDMX (Ki = 26,300 nmol/L). In P53WT U-2 OS and HCT 116 cells, TAH-19 induced accumulation of P53, P21 and MDM2. TAH-19 inhibited cell cycle progression of U-2 OS cells at G1 phase in a P53-dependent manner. Crystal structures revealed that TAH-19 induced dimerization of MDM2 via its aliphatic linker. Hydrogen-bond interactions between the imidazole ring nitrogens and the amide nitrogens were primary contacts between two TAH-19. Moreover, the oxygen atom of the carboxylic acid group in TAH-19 formed a water-mediated contact with Gln72 and His96 of MDM2 (Fig. 17).
Figure 17.
1,4,5-Trisubstituted imidazole-based P53–MDM2/X antagonists. Imidazo indoles WK298 (A) and WK23 (B) and their binding models in the MDMX (PDB code: 2FEA) and MDM2 (PDB code: 1YCR) active sites, respectively. (C) Crystal structure of TAH-19 in complex with MDM2. Adapted with permission from Refs. 66 and 67. Copyrights © 2010 Taylor & Francis and American Chemical Society, respectively.
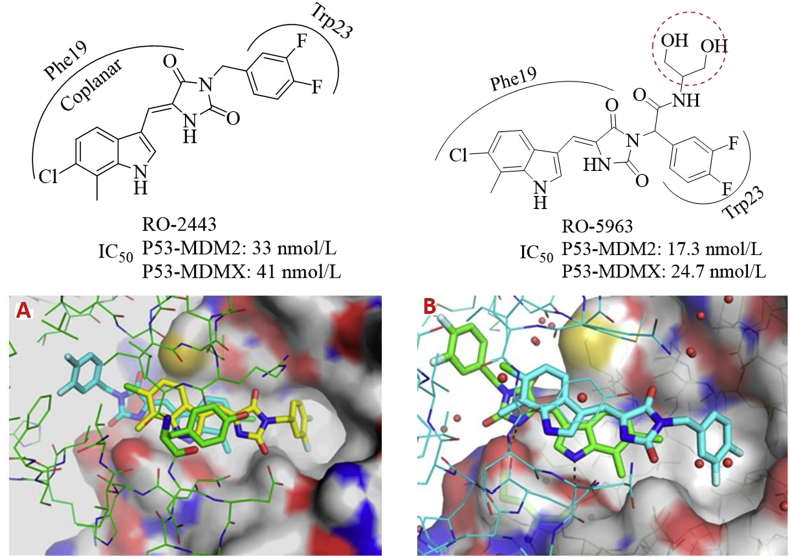
Graves et al.68 reported that the indolyl hydantoins such as RO-2443 and RO-5963 (Fig. 18) dually inhibited MDMX and MDM2 at low nanomolar levels. Interestingly, the coplanar indolyl hydantoin group and difluorophenyl group in both compounds were directed to the Phe19 and Trp23 pockets, respectively, while the Leu26 pocket was not occupied. The unique binding models may provide a structural basis for designing dual inhibitors. The size of Leu26 cavity in MDMX and MDM2 is different, which has posed a great challenge for designing potent dual inhibitors of MDMX and MDM2. The terminal diol group in RO-5963 was beneficial for enhancing the aqueous solubility for further cellular activity assessment. Crystal structures revealed that both compounds induced dimerization of MDM2 and MDMX (Fig. 18).
Figure 18.
MDM2/MDMX dual inhibitors RO-2443 and RO-5963. Shown here are crystal structures of RO-2443 bound to MDM2 (PDB code: 3VBG) and MDMX (PDB code: 3U15).
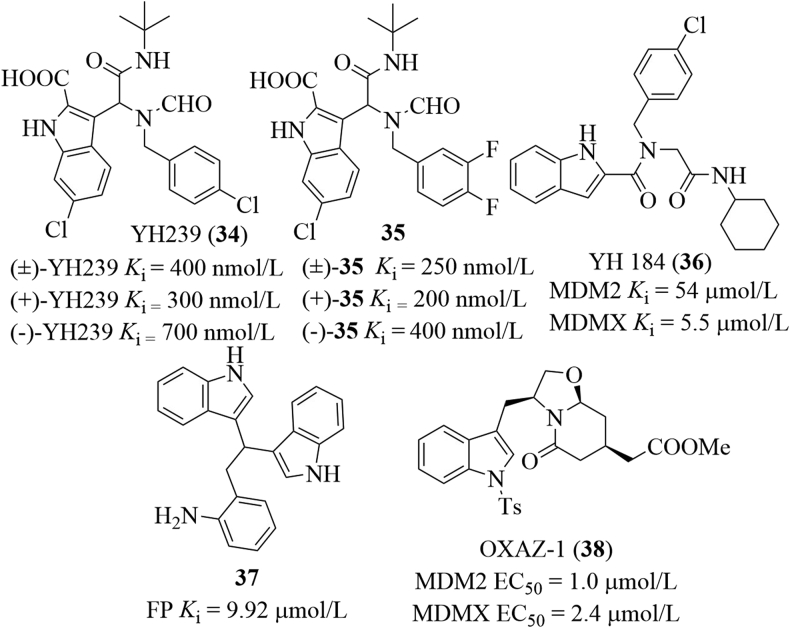
After screening ∼0.5 billion conformer library derived from five million indole-containing compounds for matching the anchor/pharmacophore model, the Domling group69 obtained YH239 (34), which bound to MDM2 with a Ki value of 400 nmol/L in the FP assay and was highly selective to MDM2 over MDMX. Its two enantiomers bound to MDM2 with the Ki values of 300 and 700 nmol/L, respectively (Fig. 19). The ethyl ester of YH239 (known as YH239-EE) concentration-dependently induced apoptosis and cell cycle arrest of AML cells expressing P53 and activated P53 and caspase 3/7. Besides, (+)-YH239-EE was less cytotoxic than Nutlin-3. YH239 mimicked Trp23, Phe19 and Leu26 in P53 to occupy the hydrophobic cleft on MDM2. Among the substituents, the benzyl group formed a π–π stacking with the imidazole group of His96. It is believed that substitution patterns of the fluoro atom on the phenyl ring can modulate the aromatic interactions of small molecules with targets of interest due to the ability of reversing polarization of the fluorine atom70. Based on the ANCHOR.QUERY virtual screening platform, they synthesized 40 derivatives with fluorine atom substituted at the benzyl ring of compound 34 and then performed the fluorine-scanning71. Among this series, racemic compound 35 showed the best binding affinity to MDM2 in the FP assay (Ki = 0.25 μmol/L), about 7 times more potent than compound 34 (Ki = 1.8 μmol/L). (+)-35 and (−)-35 bound to MDM2 with the Ki values of 200 and 400 nmol/L, respectively. The structural basis of the interaction was confirmed by the co-crystal structure. Based on these findings, they also designed a series of peptidomimetic small molecules containing the indole group72. Of this series, YH 184 (36) bound to MDMX with a Ki value of 5.5 μmol/L, showing about 10-fold selectivity to MDMX over MDM2. Further modifications on the indole scaffold or replacement with other aromatic rings failed to give compounds with improved binding affinity. All compounds displayed worse or no binding affinity (Ki > 50 μmol/L). Carlson et al.73 identified compound 37 with a Ki value of 9.92 μmol/L after screening a molecule library containing about 35,000 compounds using the multiple protein structure method. Two indole groups occupied the Trp23 and Leu26 subpockets, while the aminophenyl moiety filled into the Phe19 cavity. Saraiva and co-workers74 identified a dual inhibitor of MDMX/MDM2, namely the (S)-tryptophanol derivative OXAZ-1 (38) after screening a series of oxazolopiperidone lactams derived from tryptophanol using a yeast-based assay. OXAZ-1 inhibited MDMX and MDM2 with the EC50 values of 2.4 and 1.0 μmol/L, respectively. In HCT116 P53+/+ cells, OXAZ-1 stabilized P53, up-regulated MDMX, P21, MDM2, Puma, and BAX, as well as cleaved PARP. Interestingly, OXAZ-1 also exerted comparable cytotoxicity against MCF-7 cells overexpressing MDMX.
Figure 19.
Indole-containing small molecules that inhibit MDMX/MDM2.
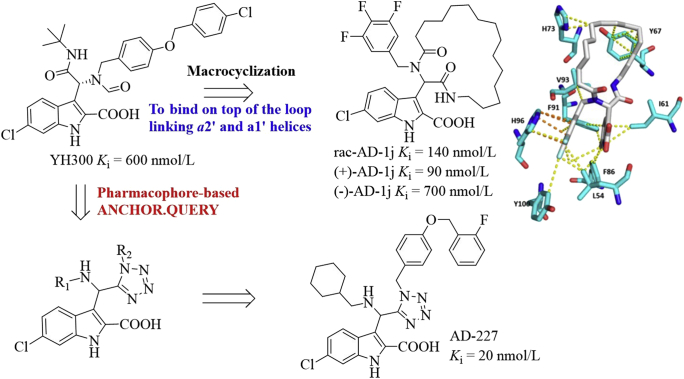
Based on the three and four-point pharmacophore models, Dömling et al.75 designed macrocycles as potent P53–MDM2 inhibitors from YH300 (Ki = 600 nmol/L, Fig. 20). These macrocycles bound on top of the loop linking α2′ and α1′ helices, covering a large hydrophobic surface area formed by Tyr67, Gln72, His73, Val93, and Lys94. Of these macrocycles, compound AD-1j showed the best potency toward MDM2 with the Ki value of 140 nmol/L in the FP assay. Two enantiomers (+)-AD-1j and (−)-AD-1j inhibited the MDM2–P53 interaction with the Ki values of 90 and 700 nmol/L, respectively. Modelling studies revealed the existence of van der Waals interactions of the aliphatic handle with Tyr67 and His73, the 6-chloro-indole moiety and the 3,4,5-trifluorophenyl ring occupied the Trp23 and Leu26 hydrophobic pockets, respectively. Moreover, the 3,4,5-trifluorophenyl fragment formed the π–π interaction with His96 and several van der Waals interactions with surrounding residues such as Leu54, Ile61, Phe86, Phe91, Val93, His96, and Tyr100. Using the pharmacophore based virtual screening ANCHOR.QUERY platform, Dömling et al.76 designed the 1,5-disubstituted tetrazoles as potent MDM2 inhibitors, of which AD-227 inhibited the P53–MDM2 interaction with a Ki value of 20 nmol/L.
Figure 20.
Design of new MDM2 inhibitors based on YH300. Binding model of AD-1j (colored in white) in the MDM2 binding site (PDB code: 3TU1). Key residues are shown in cyan. The van der Waals and π–π interactions are shown in yellow and orange dotted lines, respectively.
2.6. Pyrrolidones
Zhang group77 identified nine compounds through the structure-based virtual screening of the Specs database, these compounds inhibited the MDM2–P53 interactions (Ki 0.57–85.97 μmol/L, six of which possessed the pyrrolidone scaffold. Among them, compound 39 bound to MDM2 at nanomolar levels (Ki = 780 nmol/L, Fig. 21). To improve the binding affinity and physicochemical properties, further modifications on the C4 and C5 phenyl groups as well as the 3-OH group were performed, ultimately leading to the discovery of compounds (R)-40 and 41 (Ki = 150 and 260 nmol/L, respectively), which showed comparable binding affinity with Nutlin-3a (Ki = 230 nmol/L). However, (S)-40 was completely inactive in the FP assay, indicating stereospecific binding with MDM2. (R)-40 potently inhibited cell growth of A549 P53+/+ cells (IC50 = 1.97 μmol/L), about 7-fold more potent than Nutlin-3a (IC50 = 15.12 μmol/L). In A549 cells, (R)-40 dose-dependently increased the expression of MDM2, P21 and P53 proteins. Interestingly, compound 41 also inhibited MDMX and dose-dependently increased the expression of MDMX, P21 and P53 in MCF-7 cells after 24 h treatment. Besides, (R)-40 achieved 40% and 52.53% of TGI, respectively in A549 xenograft model after oral dosing at 200 and 300 mg/kg for 14 days. No significant body weight loss was observed. However, docking studies showed that the imidazole group in compounds 40 and 41 did not contact with Phe19 pocket tightly. Based on the pyrrolidone scaffold, they designed and synthesized a new series of MDM2 inhibitors as represented by compounds 42 and 43 utilizing the molecule fusing strategy78. Compounds 42 and 43 showed favorable binding affinity to MDM2 (Ki = 90 nmol/L). Similarly, compound 42 increased P53 and MDM2 expression in A549 cells. Very recently, they designed a new fluorescence probe 44 that could be used for detecting and imaging the MDM2–P53 interactions by introducing the fluorescent N,N-dimethyl-1,8-naphthalimide79. Probe 44 bound to MDM2 (Ki = 2.29 μmol/L), down-regulated MDM2, but significantly increased expression of MDMX.
Figure 21.
Pyrrolidone-based MDM2 inhibitors.
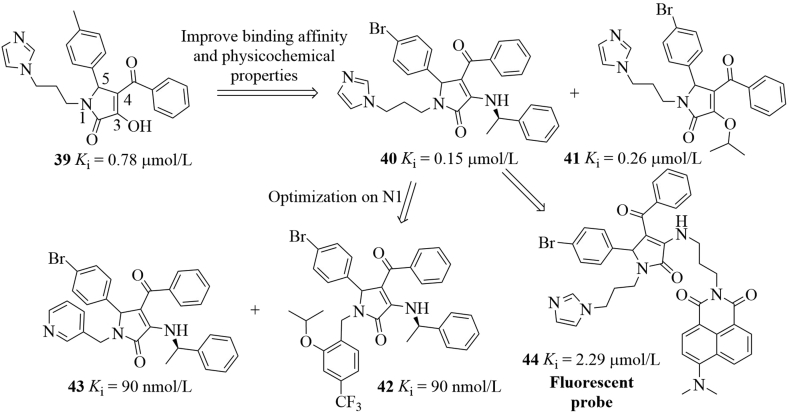
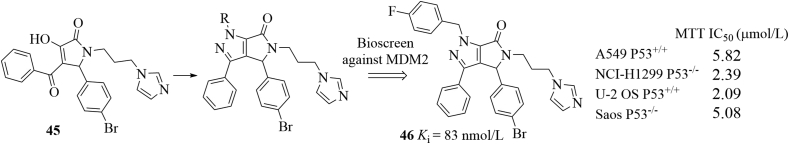
In addition to aforementioned inhibitors, Zhang and co-workers80 rationally designed pyrrolo[3,4-c]pyrazole-based compounds targeting the P53–MDM2 inhibition and the NF-κB pathway for cancer therapy based on the following considerations: (1) pyrrolidone scaffolds could act as promising chemotypes for designing MDM2–P53 interaction inhibitors; (2) the pyrazoles (e.g., celecoxib) have been found to be able to repress NF-κB81; (3) NF-κB proteins have been found to be overexpressed in many cancers and can repress P53 by directly increasing expression of MDM282; (4) many P53 activators have been proved to repress NF-κB83. Therefore, small molecules that activate P53 and inhibit NF-κB simultaneously would have synergistic effect for treating tumors. As shown in Fig. 22, the pyrrolo[3,4-c]pyrazoles can be accessed through the condensation reactions of corresponding hydrazines and 3-hydoxy pyrrolidone 45 and were further screened for their binding affinity toward MDM2. Among them, compound 46 exhibited excellent binding potency (Ki = 83 nmol/L) in the FP assay, which was comparable to that of Nutlin-3 (Ki = 93 nmol/L). Compound 46 showed good antiproliferative activity but with poor selectivity over four cancer cell lines with or without P53. Compound 46 increased P53 expression significantly and dose-dependently. However, decreased expression of MDM2 protein was observed. On the other hand, compound 46 repressed NF-κB activation by inhibiting IκBα phosphorylation, inhibited translocation of NF-κB to nucleus and related kinases of NF-κB pathways. Compound 46 was well tolerated in the A549 xenograft model and achieved 31.86 % inhibition of tumor volume after 14-day oral administration at 200 mg/kg without significant body weight loss. An analog of compound 46, where the fluorine atom was replaced with a hydrogen atom, showed excellent oral bioavailability (F = 72.9%). Interestingly, (R)-46 and (S)-46 showed a synergistic effect by targeting MDM2–P53 interactions and NF-κB, respectively. Specifically, (R)-46 activated P53 by disrupting MDM2–P53 interactions, while the inactive (S)-46 promoted the phosphorylation of IKKs and prevented the IκB phosphorylation, thus inhibiting NF-κB signaling84. Docking studies showed that (R)-46 occupied three key pockets in MDM2, while (R)-46 only filled into the Phe19 and Trp23 pockets.
Figure 22.
Dual inhibitors targeting MDM2–P53 interactions and NF-κB pathway.
2.7. Others
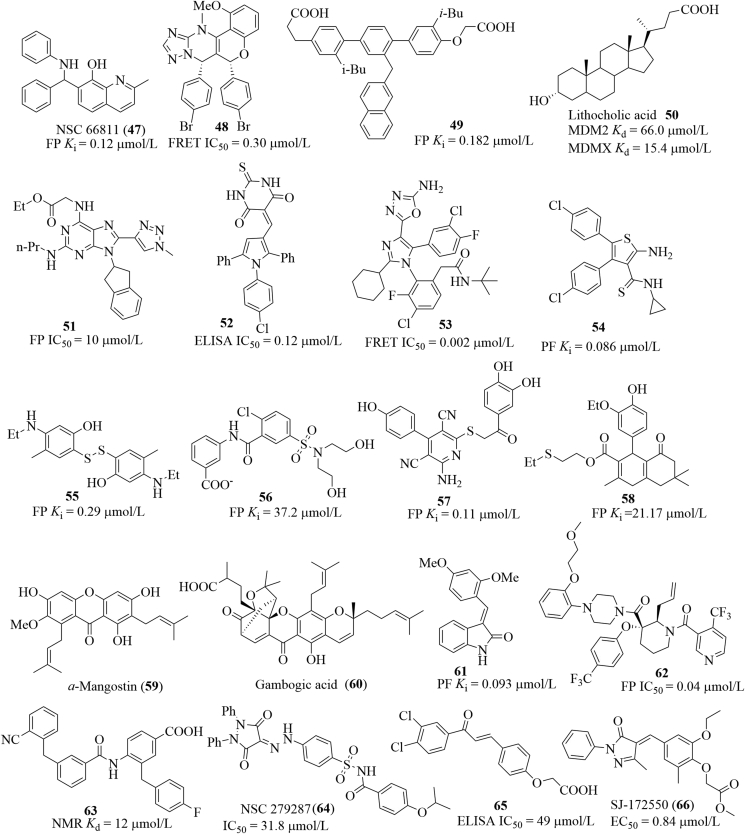
In addition to aforementioned inhibitors, other small molecules have also been reported to be able to inhibit the MDM2/X–P53 interactions at different levels (Fig. 23). Wang et al.85 identified a quinolinol derivative NSC 66811 (47) after evaluating a molecule library containing 150,000 compounds by an integrated virtual screening strategy. NSC 66811 inhibited MDM2 (Ki = 120 nmol/L) and increased expression of P53, P21 and MDM2 in HCT-116 P53+/+ cells, but the result was not observed in HCT-116 P53−/− cells. A high throughput screening of a compound collection (ca. 1.4 million) performed in the Allen group yielded a novel chromenotriazolopyrimidine scaffold, which was proved to be able to disrupt the MDM2–P53 interactions. Further modifications, followed by SAR studies afforded compound 48, which blocked the MDM2–P53 interactions at low micromolar levels (IC50 = 0.30 μmol/L), slightly more potent than its (6R,7S)-enantiomer (IC50 = 0.20 μmol/L)86. Another interesting class is the terphenyl scaffold, which mimicked a-helical structure of P53 and bound to the cleft on the surface of HDM287. Three ortho groups attached to the terphenyl scaffold (e.g., the i-butyl and naphthyl groups in compound 49) are believed to be projected to the three hydrophobic subpockets in MDM2. Compound 49 showed favorable binding affinity to HMD2 in the FP assay (Ki = 0.182 μmol/L). In 2012, Fersht et al.88 reported that lithocholic acid 50 was an endogenous dual inhibitor of MDM2 and MDM4 with the Ki values of 66.0 and 15.4 μmol/L, respectively. On the basis of docking studies and analysis of previously reported inhibitors, Grotli and co-workers89 designed a series of 8-triazolylpurines. Among this series, compound 51 displayed the best inhibitory activity toward the MDM2–P53 interactions in the FP assay (IC50 = 10 μmol/L). After screening a library of 800 structurally diverse compounds from the Cancer Research UK, Hardcastle et al.90 identified the pyrrole scaffold which was capable of targeting the MDM2–P53 interactions. Further SARs studies produced compound 52, which bound to MDM2 potently (IC50 = 0.12 μmol/L), about 35 times selectivity to MDM2 over MDMX (IC50 = 4.2 μmol/L). Compound 52 concentration-dependently induced expression of MDM2, P21 and P53 but exhibited similar cellular activity toward SJSA-1, SN40R2 and MRK-NU-1 cells (GI50 = 2.3, 2.8 and 2.3 μmol/L, respectively) regardless of the MDM2, MDMX and P53 status. By employing the structure-guided approach, Furet et al.91 identified the tetra-substituted imidazole scaffolds, which were used as hit compounds for further structural optimization, the compounds possessed low nanomolar potency in the biochemical FRET assay. Compound 53 inhibited MDM2 potently (IC50 = 0.002 μmol/L) and showed significant inhibition toward SJSA-1 cells expressing P53 (IC50 = 0.5 μmol/L). 3D pharmacophore-based virtual screening in combination with docking studies performed in the Hu group revealed that the aminothiophene scaffold could be used for developing inhibitors of the MDM2–P53 interactions92,93. Further SARs studies yielded compound 54, which bound to MDM2 with a Ki value of 0.086 μmol/L and inhibited cell growth of A549 and PC3 in the SRB assay (IC50 = 8.28 and 22.05 μmol/L, respectively). Carlson et al.73 identified novel scaffolds 55–58 utilizing their own developed multiple protein structure (MPS) technique. All these scaffolds mimicked the key residues of P53 to occupy the cleft on MDM2. Of these scaffolds, compound 59 exhibited favorable binding affinity to MDM2 (Ki = 110 nmol/L). Saraiva et al.94 proved that natural products a-mangostin (60), gambogic acid (61) and the structurally simplified xanthone derivative inhibited growth of several cancer cell lines probably through disrupting the MDM2–P53 interactions based on the yeast approach. Based on the pharmacophore- and structure-based approaches, Li et al.95 identified the (E)-3-benzylidene-indolin-2-one scaffolds as MDM2 inhibitors. Among this series, compound 61 bound to MDM2 potently (Ki = 0.093 μmol/L) and strongly exhibited growth of HCT116 P53+/+ cells with a GI50 value of 13.42 μmol/L. Besides, compound 62 inhibited tumor growth of BALB/c mice bearing CT26, the TGI was 32.4% after administration at 500 mg/kg. After identifying the 3,3-disubstituted piperidine scaffolds as the HMD2/P53 interaction inhibitors96, Bogen et al.97 performed extensive modifications on this scaffold by adding substituents to 2,4,5,6-position of piperidine scaffold, showing that adding substituents to the 4,5,6-position did not lead to a significant improvement in binding potency, while the incorporation of an allyl group to the C2 position resulted in a drastic improvement in binding potency. Compound 63 bound to HMD2 with a Ki value of 0.04 μmol/L. Guy and co-workers98 screened a library of 173 compounds for blocking the MDM2–P53 interactions, which were then characterized by 15N–1H HSQC NMR. The most active Compound 64 exhibited the best binding potency with a Kd value of 12 μmol/L in the NMR assay. Based on the pharmacophore model of MDM2 binding, Abraham et al.99 identified a sulfonamide 65 (also known as NSC 279287) as an inhibitor of MDM2–P53 interactions using 3D database searching. NSC 279287 bound to MDM2 with a moderate binding affinity (IC50 = 31.8 μmol/L) and increased expression of P53 in SJSA-1 cells. Chalcone 66 (SJ-172550) was found to be able to bind to the cleft of MDM2, thereby releasing P53 from the MDM2–P53 complex100. In the ELISA assay, SJ-172550 showed an IC50 value of 49 μmol/L.
Figure 23.
Other small molecules that inhibit the MDM2–P53 interactions.
3. Development of MDM2 PROTAC degraders for cancer therapy
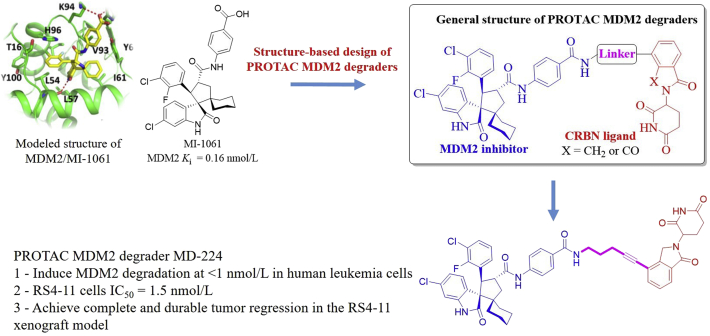
Targeting the P53–MDM2 interaction has become an attractive therapeutic strategy for the treatment of cancers harboring WT P53. To date, several candidate compounds (Fig. 2) have advanced into clinical trials for cancer therapy. However, these small-molecule inhibitors have shown mixed response rates or limited efficacy in some models, probably because of inadequate P53 induction and unwanted toxicities in normal cells and tissues101. Moreover, it is believed that accumulated MDM2 protein in vivo can degrade P53 efficiently and rapidly upon clearance of an MDM2 inhibitor, thus causing reduced therapeutic efficacy of the MDM2 inhibitor102. Therefore, novel strategies to effectively target the MDM2–P53 pathway are greatly needed. Wang et al.103,104 designed the first-in-class MDM2 PROTAC degrader MD-224 starting from a potent MDM2 inhibitor MI-1061 (Ki = 0.16 nmol/L, Fig. 24). Modeled structure of MDM2 in complex with MI-1061 revealed that the carboxylic acid group on the phenyl ring in MI-1061 would be exposed to the solvent region, thus allowing for being linked to CRBN ligands thalidomide and lenalidomide to design bifunctional PROTAC MDM2 degraders. Of these compounds, MD-224 induced rapid MDM2 degradation at <1 nmol/L in human leukemia cells and inhibited growth of RS4-11 cells effectively (IC50 = 1.5 nmol/L). Further studies showed that MD-224 achieved significantly improved efficacy relative to MI-1061, up to 50% tumor regression was observed in the RS4-11 xenograft model following multiple i.v.-dosing of MD-224 at 25 mg/kg every second day without any signs of toxicity.
Figure 24.
Structure-based design of PROTAC MDM2 degrader MD-224. Binding model of MI-1061 (colored in yellow) in the MDM2 binding site (colored in green, PDB ID: 5TRF). The red dashed line represents the hydrogen bond.
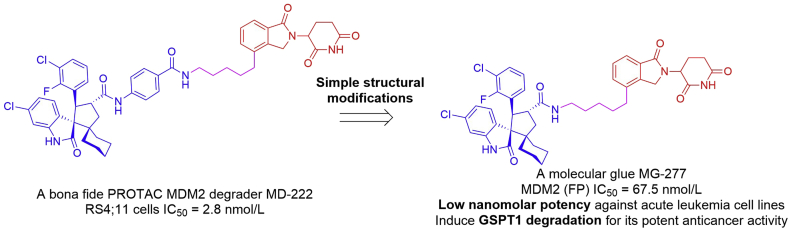
Based on a previously reported MDM2 degrader MD-222 (RS4-11 cells IC50 = 2.8 nmol/L)103, the Wang group105 performed further structural modifications and unexpectedly identified another type of compounds, as exemplified by MG-277 (Fig. 25). MG-277 inhibited the P53–MDM2 interaction potently with an IC50 value of 67.5 nmol/L in the FP assay. In contrast to MD-222, MG-277 displayed high potencies against acute leukemia cell lines with different P53 status and was much less effective in inducing MDM2 degradation. For example, MG-277 inhibited growth of RS4-11, MOLM-13 and MV4-11 with the IC50 values of 1.3, 24.6 and 7.9 nmol/L, respectively. The cellular activity was dependent on cereblon binding, not P53 or MDM2 dependent. Mechanistic studies showed that MG-277 induced degradation of the translation termination factor GSPT1 to achieve its potent anticancer activity. Collectively, MG-277 was a molecular glue, not a PROTAC MDM2 degrader. This study provides the first example that structural modifications can convert a bona fide PROTAC degrader into a molecular glue. For a phthalimide-based degrader, it may be a bona fide degrader for the protein(s) of interest or a molecular glue recruiting neo-substrate protein(s) to CRL4CRBN E3 ligase for ubiquitination and subsequent degradation. Both mechanisms should be considered in designing phthalimide-based PROTAC degraders.
Figure 25.
Discovery of a molecular glue MG-277 from a bona fide PROTAC MDM2 degrader MD-222.
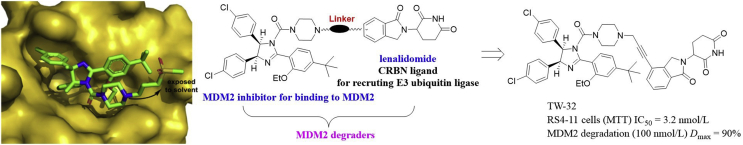
Tang et al.106 also designed a series of new MDM2 PROTAC degraders by conjugating the potent MDM2 inhibitor RG7112 (MDM2 IC50 = 18 nmol/L) to lenalidomide through suitable linkers. The co-crystal structure of MDM2 in complex with RG7112 is shown in Fig. 26, the piperazine ring in RG7112 is exposed to the solvent region, and thus allows for being linked to the E3 ligase ligand lenalidomide for designing bifunctional MDM2 degraders. The most promising compound TW-32 effectively inhibited growth of RS4-11 cells carrying WT P53 (IC50 = 3.2 nmol/L) and achieved 90% of MDM2 degradation (Dmax = 90%) at 100 nmol/L in RS4-11 cells. Notably, TW-32 has C3 (one acetylene and one methylene) unit between lenalidomide and MDM2 inhibitor, representing one of the shortest linkers among all reported PROTACs.
Figure 26.
Structure-based design of MDM2 PROTAC degrader TW-32 from a MDM2 inhibitor RG7112. Co-crystal structure of RG7112 (colored in green) bound to the MDM2 (shown in yellow surface) binding site (PDB code: 4IPF). Adapted with permission from Ref. 106. Copyright © 2019 Elsevier Ltd.
4. Challenges for designing potent MDM2/X antagonists or PROTACs degraders for cancer therapy
Reactivation of P53 by inhibiting its negative suppressors MDM2 and MDMX has proven to an attractive strategy for anti-cancer treatment, significant progress has been witnessed with several drug leads undergoing clinical development. However, challenges for designing MDM2 inhibitors for anticancer treatment in clinic still exist, such as MDM2 and MDMX dual inhibition, acquired resistance and toxicity of P53 activation, etc107.
4.1. MDM2 and MDMX dual inhibition
Like MDM2, MDMX amplification has been also found in many cancers108. MDMX interferes with the ability of P53 to interact with the basal transcription machinery, thus inhibiting P53 transcriptional activity, while MDM2 induces P53 degradation. The MDMX is not regulated by the negative feedback loop. Also, the MDM2–MDMX heterodimers can help degrade P53109. So P53 function can be attenuated by MDMX overexpression. Additionally, MDMX also inhibits MDM2 degradation. Therefore, MDM2/X dual inhibition is highly desirable to achieve full P53 activation for anticancer treatment in clinic. A successful example is the peptide PDI with the sequence of LTFEHYWAQLTS, which has proven to be able to potently inhibit MDM2/X at nanomolar levels and induce apoptosis and cell cycle arrest of cell lines with overexpressed MDM2/X110. The stapled PDIs developed in the same group showed higher inhibitory activity toward MDM2 and MDMX and enhanced cell permeability, albeit with modest in vivo activity111. Peptide-based dual inhibitor ALRN-6924 developed by Aileron Therapeutics has advanced into phase I/II clinical trials for safety and tolerability evaluation in patients with advanced solid tumors retaining WT P53 (ClinicalTrials.gov Identifier: NCT02264613). However, small molecule-induced potent dual inhibition of MDM2/X is quite challenging with very few examples reported. The Graves' group68 reported that RO-2443 and RO-5963 potently bound to MDM2 and MDMX (IC50 < 50 nmol/L). However, MDM2 inhibitors such as Nutlin-3 and spirooxindole derivative MI-219 showed high selectivity to MDM2 over MDMX112. The difficulties for designing potent dual inhibitors are mainly attributed to the difference of Leu26 subpocket size in MDMX and MDM2 (the Leu26 cavity in MDMX is smaller than that in MDM2) and the spatial orientation of key residues close to Leu26 in both proteins, although they share highly similar binding domains113. Increasing the size of hydrophobic group that is designed to occupy the Leu26 pocket can increase binding affinity, while the bulky group may result in decreased binding ability to MDMX114. Interesting, potent dual inhibition of RO-2443 and RO-5963 may be explained by their binding models, where the key Leu26 subpocket is not occupied for both compounds68. Apart from developing MDMX/MDM2 dual inhibitors, another promising strategy is the combination of MDM2 inhibitors with other chemotherapeutic agents (e.g., doxorubicin and irinotecan) and MEK inhibitors (e.g., trametinib) that can effectively down-regulate MDMX115,116. Currently, the combination of SAR405838 or AMG232 with MEK inhibitors (e.g., trametinib and pimasertib) for the treatment of AML or solid tumors is undergoing clinical investigation. Furthermore, the development of specific MDMX inhibitors is also highly needed117. In 2010, after an HTS of chemical library with 285,848 different compounds, Reed and co-workers118 identified the first reversible MDMX inhibitor SJ-172550 (Fig. 23), which effectively inhibited growth of retinoblastoma cells harboring overexpressed MDMX (EC50 = 0.84 μmol/L). An additive effect was also observed when combined with MDM2 inhibitor Nutlin-3a. SJ-172550 can serve as a template for developing more potent MDMX inhibitors and the synergetic effect observed when combined with Nutlin-3a may suggest that the combination of MDMX and MDM2 inhibitors for cancer therapy would be feasible.
4.2. Resistance to P53 activation
Intrinsic or acquired resistance to chemotherapy or molecularly targeted therapies is a major issue in anticancer drug development, making drug treatment less effective119. Because of the tumor suppressing ability of P53, resistance of tumor cells expressing WT P53 may emerge from pre-existing microfoci of P53 mutant cells or through the acquired P53 mutation when treated with MDM2 inhibitors11,120. Recent studies showed that non-genotoxic P53 activation may result in acquired somatic mutations of P53, as illustrated in SJSA-1 cancer cells bearing WT P53 when treated with Nutlin-3121. Different levels of acquired resistance in SJSA-1, RS4-11, and MV4-11 tumor cells to SAR405838 both in vitro and in vivo have also been observed122,123. Therefore, development of new MDM2 inhibitors is highly needed for fighting newly occurred mutations. Combined treatment of MDM2 inhibitors with other therapeutic agents would be a feasible strategy for overcoming the acquired resistance.
4.3. P53 activation induced toxicity to normal tissues
The activation of P53 can induce cell cycle arrest in all proliferative cells, so the toxicity to normal tissues, especially the radiosensitive tissues124, caused by MDM2 inhibitor-induced P53 activation is potentially existed. Restoration of P53 in the absence of MDM2 has been found to cause several pathological damages to radiosensitive mouse tissues or even death125. Well tolerance of Nutlin-3 treatment for three weeks has been observed in nude mice. RG7112 can induce the apoptosis of human megakaryocyte and thrombocytopaenia in patients37. The cell cycle arrest of human normal fibroblasts has also been observed when treated with Nutlin-3126. Therefore, appropriate dose schedules that balance antitumor activity and less toxicity are highly appreciated.
4.4. MDM2 PROTAC degraders
Because of drug resistance, limited efficacy and unwanted toxicities of MDM2 inhibitors, new PROTAC degraders may show promise for cancer therapy127. In contrast to MDM2 inhibitors, MDM2 PROTAC degraders such as MD-224 have shown promise for cancer therapy103,104. Due to the mixed response rates of MDM2 inhibitors in clinical trials, it remains unclear whether the MDM2 PROTAC degraders will possess improved efficacy without substantially increasing toxicity in human cancer patients. The data reported by Wang et al.105 also suggest that for a phthalimide-based degrader, it may be a bona fide degrader for the protein(s) of interest or a molecular glue recruiting neo-substrate protein(s) to CRL4CRBN E3 ligase for ubiquitination and subsequent degradation. Both mechanisms should be considered in designing phthalimide-based PROTAC degraders. Further investigations are needed to reveal precise structural requirements for a phthalimide conjugate to operate either as a bona fide PROTAC degrader or as a molecular glue.
5. Concluding remarks and outlook
In tumor cells, expressions of MDM2 and its homolog MDMX are always upregulated, thereby inhibiting the function of P53. Therefore, restoring P53 function is crucial for inhibiting the growth of tumor cells. Restoring of P53 function by interrupting the MDM2–P53 and MDMX–P53 interactions has been highly pursued to treat cancers. The well-defined structural features of the MDM2–P53 interaction provide a basis for designing new MDM2 inhibitors. To date, many MDM2 inhibitors have been identified since the release of the structure of MDM2–P53 complex. Some of them are currently under clinical investigation for anticancer treatment. However, all these small-molecule inhibitors in clinical trials are highly selective toward MDM2 over MDMX; their inhibitory activity can be compromised by the overexpressed MDMX. Currently, the MDM2 inhibitors are used in clinical trials in combination of other chemotherapeutic agents that can inhibit MDMX. Potent dual inhibition of MDM2 and MDMX seems to be promising but is very challenging as the size of Leu26 subpocket in MDM2 and MDMX is different. It is difficult for small molecules to occupy both pockets. RO-2443 and RO-5963 have been reported to be able to dually inhibit MDM2 and MDMX at nanomolar levels. Interestingly, they did not occupy the Leu26 pocket. Besides, specific potent MDMX inhibitors like SJ-172550 are also rare and highly needed. The drug combination and dual inhibition of MDMX/MDM2 are promising strategies that can help achieve full activation of P53. RO-5963 and SJ-172550 can serve as templates for identifying more potent dual inhibitors and specific MDMX inhibitors, respectively. Although some MDM2 inhibitors are currently undergoing clinical assessment for cancer therapy, challenges such as acquired resistance and toxicity still exist. Appropriate dose schedules balancing antitumor activity and less toxicity would be appreciated. Moreover, due to the mixed response rates of MDM2 inhibitors in clinical trials, MDM2 PROTAC degraders have been highly pursued in recent years, showing improved efficacy for cancer therapy. However, it remains unknown whether the MDM2 PROTAC degraders have substantially increasing toxicity in human cancer patients. It should be kept in mind that a putative phthalimide-based degrader may be a bona fide MDM2 PROTAC degrader or a molecular glue. Precise structural requirements for a phthalimide conjugate to act either as a bona fide PROTAC degrader or as a molecular glue deserves to be further investigated.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 81703326 and 81973177 for Bin Yu, and 81773580 for Guochao Liao), China Postdoctoral Science Foundation (Nos. 2018M630840 and 2019T120641 for Bin Yu), the Open Fund of State Key Laboratory of Pharmaceutical Biotechnology, Nanjing University, China (No. KF-GN-201902 for Bin Yu), Guangdong Key Laboratory for Translational Cancer Research of Chinese Medicine (No. 2018B030322011 for Guochao Liao, China), and Guangdong Province Higher Vocational Colleges and Schools Pearl River Scholar Funded Scheme (No. Guochao Liao, 2019, China).
Author contribution
Bin Yu designed the outline of this article, wrote the abstract and conclusion, revised this review extensively, and submitted this review on behalf of other authors. Yuan Fang and Guochao Liao wrote this review together.
Conflicts of interest
The authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Contributor Information
Guochao Liao, Email: liao@gzucm.edu.cn.
Bin Yu, Email: yubin@zzu.edu.cn.
References
- 1.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of P53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu X., Bayle J.H., Olson D., Levine A.J. The P53–MDM2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 3.Harris S.L., Levine A.J. The P53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 4.Kussie P.H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A.J. Structure of the MDM2 oncoprotein bound to the P53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y., Aguilar A., Bernard D., Wang S. Small-molecule inhibitors of the MDM2-P53 protein–protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J Med Chem. 2015;58:1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saha T., Kar R.K., Sa G. Structural and sequential context of P53: a review of experimental and theoretical evidence. Prog Biophys Mol Biol. 2015;117:250–263. doi: 10.1016/j.pbiomolbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Fu T., Min H., Xu Y., Chen J., Li G. Molecular dynamic simulation insights into the normal state and restoration of P53 function. Int J Mol Sci. 2012;13:9709–9740. doi: 10.3390/ijms13089709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z. In vivo activation of the P53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 9.Zak K., Pecak A., Rys B., Wladyka B., Dömling A., Weber L. MDM2 and MDMX inhibitors for the treatment of cancer: a patent review (2011-present) Expert Opin Ther Pat. 2013;23:425–448. doi: 10.1517/13543776.2013.765405. [DOI] [PubMed] [Google Scholar]
- 10.Wang W., Hu Y. Small molecule agents targeting the P53–MDM2 pathway for cancer therapy. Med Res Rev. 2011;32:1159–1196. doi: 10.1002/med.20236. [DOI] [PubMed] [Google Scholar]
- 11.Vassilev L.T. MDM2 inhibitors for cancer therapy. Trends Mol Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Khoury K., Popowicz G.M., Holak T.A., Domling A. The P53–MDM2/MDMX axis—a chemotype perspective. MedChemComm. 2011;2:246–260. doi: 10.1039/C0MD00248H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popowicz G.M., Dömling A., Holak T.A. The structure-based design of MDM2/MDMX–P53 inhibitors gets serious. Angew Chem Int Ed. 2011;50:2680–2688. doi: 10.1002/anie.201003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tisato V., Voltan R., Gonelli A., Secchiero P., Zauli G. MDM2/X inhibitors under clinical evaluation: perspectives for the management of hematological malignancies and pediatric cancer. J Hematol Oncol. 2017;10:133. doi: 10.1186/s13045-017-0500-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer T., Hong D., Somaiah N., Cai C., Song S., Kumar P. Abstract B27: a phase I dose escalation study of MDM2 inhibitor DS-3032b in patients with hematological malignancies-preliminary results. AACR-NCI-EORTC international conference: molecular targets and cancer; 2015, Nov. 5–9; Boston, MA, USA. Mol Cancer Ther. 2015;14(12 Suppl 2) abstract nr B27. [Google Scholar]
- 16.Bauer T.M., Gounder M.M., Weise A.M., Schwartz G.K., Carvajal R.D., Kumar P. A phase I study of MDM2 inhibitor DS-3032b in patients with well/de-differentiated liposarcoma (WD/DD LPS), solid tumors (ST) and lymphomas (L) J Clin Oncol. 2018;36:11514. [Google Scholar]
- 17.Gessier F., Kallen J., Jacoby E., Chène P., Stachyra-Valat T., Ruetz S. Discovery of dihydroisoquinolinone derivatives as novel inhibitors of the P53–MDM2 interaction with a distinct binding mode. Bioorg Med Chem Lett. 2015;25:3621–3625. doi: 10.1016/j.bmcl.2015.06.058. [DOI] [PubMed] [Google Scholar]
- 18.Valat T., Masuya K., Baysang F., Albrecht G., Buschmann N., Erdmann D. Mechanistic study of NVP-CGM097: a potent, selective and species specific inhibitor of P53–MDM2. Cancer Res. 2014;74:1798. [Google Scholar]
- 19.Holzer P., Masuya K., Furet P., Kallen J., Valat-Stachyra T., Ferretti S. Discovery of a dihydroisoquinolinone derivative (NVP-CGM097): a highly potent and selective MDM2 inhibitor undergoing phase I clinical trials in P53 wt tumors. J Med Chem. 2015;58:6348–6358. doi: 10.1021/acs.jmedchem.5b00810. [DOI] [PubMed] [Google Scholar]
- 20.Wang S., Sun W., Zhao Y., McEachern D., Meaux I., Barrière C. SAR405838: an optimized inhibitor of MDM2–P53 interaction that induces complete and durable tumor regression. Cancer Res. 2014;74:5855–5865. doi: 10.1158/0008-5472.CAN-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Jonge M., de Weger V.A., Dickson M.A., Langenberg M., Le Cesne A., Wagner A.J. A phase I study of SAR405838, a novel human double minute 2 (HDM2) antagonist, in patients with solid tumours. Eur J Cancer. 2017;76:144–151. doi: 10.1016/j.ejca.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 22.de Weger V.A., de Jonge M., Langenberg M.H.G., Schellens J.H.M., Lolkema M., Varga A. A phase I study of the HDM2 antagonist SAR405838 combined with the MEK inhibitor pimasertib in patients with advanced solid tumours. Br J Canc. 2019;120:286–293. doi: 10.1038/s41416-018-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vu B., Wovkulich P., Pizzolato G., Lovey A., Ding Q., Jiang N. Discovery of RG7112: a small-molecule MDM2 inhibitor in clinical development. ACS Med Chem Lett. 2013;4:466–469. doi: 10.1021/ml4000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tovar C., Graves B., Packman K., Filipovic Z., Xia B.H.M., Tardell C. MDM2 small-molecule antagonist RG7112 activates P53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73:2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 25.Kang M.H., Reynolds C.P., Kolb E.A., Gorlick R., Carol H., Lock R. Initial testing (Stage 1) of MK-8242-a novel MDM2 inhibitor by the pediatric preclinical testing program. Pediatr Blood Cancer. 2016;63:1744–1752. doi: 10.1002/pbc.26064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner A.J., Banerji U., Mahipal A., Somaiah N., Hirsch H., Fancourt C. Phase I trial of the human double minute 2 inhibitor MK-8242 in patients with advanced solid tumors. J Clin Oncol. 2017;35:1304–1311. doi: 10.1200/JCO.2016.70.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Q., Zhang Z., Liu J.J., Jiang N., Zhang J., Ross T.M. Discovery of RG7388, a potent and selective P53–MDM2 inhibitor in clinical development. J Med Chem. 2013;56:5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y., Zhu J., Liu J., Chen X., Mihalic J., Deignan J. Optimization beyond AMG 232: discovery and SAR of sulfonamides on a piperidinone scaffold as potent inhibitors of the MDM2–P53 protein–protein interaction. Bioorg Med Chem Lett. 2014;24:3782–3785. doi: 10.1016/j.bmcl.2014.06.073. [DOI] [PubMed] [Google Scholar]
- 29.Sun D., Li Z., Rew Y., Gribble M., Bartberger M.D., Beck H.P. Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2–P53 inhibitor in clinical development. J Med Chem. 2014;57:1454–1472. doi: 10.1021/jm401753e. [DOI] [PubMed] [Google Scholar]
- 30.Canon J., Osgood T., Olson S.H., Saiki A.Y., Robertson R., Yu D. The MDM2 inhibitor AMG 232 demonstrates robust antitumor efficacy and potentiates the activity of P53-inducing cytotoxic agents. Mol Cancer Ther. 2015;14:649–658. doi: 10.1158/1535-7163.MCT-14-0710. [DOI] [PubMed] [Google Scholar]
- 31.Yu B., Liu H.M. The development of new spirooxindoles targeting the P53–MDM2 protein–protein interactions for cancer therapy. In: Sheng C., Georg G., editors. Targeting protein–protein interactions by small molecules. Springer; Singapore: 2018. pp. 213–237. [Google Scholar]
- 32.Liao G., Yang D., Ma L., Li W., Hu L., Zeng L. The development of piperidinones as potent MDM2–P53 protein–protein interaction inhibitors for cancer therapy. Eur J Med Chem. 2018;159:1–9. doi: 10.1016/j.ejmech.2018.09.044. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y., Wang X., Wang G., Yang Y., Yuan Y., Ouyang L. The past, present and future of potential small-molecule drugs targeting P53–MDM2/MDMX for cancer therapy. Eur J Med Chem. 2019;176:92–104. doi: 10.1016/j.ejmech.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 34.Burgess A., Chia K.M., Haupt S., Thomas D., Haupt Y., Lim E. Clinical overview of MDM2/X-targeted therapies. Front Oncol. 2016;6:7. doi: 10.3389/fonc.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obrador-Hevia A., Martinez-Font E., Felipe-Abrio I., Calabuig-Fariñas S., Serra-Sitjar M., López-Guerrero J.A. RG7112, a small-molecule inhibitor of MDM2, enhances trabectedin response in soft tissue sarcomas. Cancer Invest. 2015;33:440–450. doi: 10.3109/07357907.2015.1064534. [DOI] [PubMed] [Google Scholar]
- 36.Patnaik A., Tolcher A., Beeram M., Nemunaitis J., Weiss G.J., Bhalla K. Clinical pharmacology characterization of RG7112, an MDM2 antagonist, in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2015;76:587–595. doi: 10.1007/s00280-015-2830-8. [DOI] [PubMed] [Google Scholar]
- 37.Iancu-Rubin C., Mosoyan G., Glenn K., Gordon R.E., Nichols G.L., Hoffman R. Activation of P53 by the MDM2 inhibitor RG7112 impairs thrombopoiesis. Exp Hematol. 2014;42:137–145. doi: 10.1016/j.exphem.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 38.Ray-Coquard I., Blay J.Y., Italiano A., Le Cesne A., Penel N., Zhi J. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 2012;13:1133–1140. doi: 10.1016/S1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 39.Hu B., Gilkes D.M., Farooqi B., Sebti S.M., Chen J. MDMX overexpression prevents P53 activation by the MDM2 inhibitor nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 40.Danovi D., Meulmeester E., Pasini D., Migliorini D., Capra M., Frenk R. Amplification of MDMX (or MDM4) directly contributes to tumor formation by inhibiting P53 tumor suppressor activity. Mol Cell Biol. 2004;24:5835–5843. doi: 10.1128/MCB.24.13.5835-5843.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qin L., Yang F., Zhou C., Chen Y., Zhang H., Su Z. Efficient reactivation of P53 in cancer cells by a dual MDMX/MDM2 inhibitor. J Am Chem Soc. 2014;136:18023–18033. doi: 10.1021/ja509223m. [DOI] [PubMed] [Google Scholar]
- 42.Hu C., Li X., Wang W., Zhang L., Tao L., Dong X. Design, synthesis, and biological evaluation of imidazoline derivatives as P53–MDM2 binding inhibitors. Bioorg Med Chem. 2011;19:5454–5461. doi: 10.1016/j.bmc.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 43.Czarna A., Beck B., Srivastava S., Popowicz G.M., Wolf S., Huang Y. Robust generation of lead compounds for protein–protein interactions by computational and mcr chemistry: P53/HDM2 antagonists. Angew Chem Int Ed. 2010;49:5352–5356. doi: 10.1002/anie.201001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srivastava S., Beck B., Wang W., Czarna A., Holak T.A., Dömling A. Rapid and efficient hydrophilicity tuning of P53/MDM2 antagonists. J Comb Chem. 2009;11:631–639. doi: 10.1021/cc9000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He S., Dong G., Wu S., Fang K., Miao Z.H., Wang W. Small molecules simultaneously inhibiting P53-murine double minute 2 (MDM2) interaction and histone deacetylases (HDACs): discovery of novel multitargeting antitumor agents. J Med Chem. 2018;61:7245–7260. doi: 10.1021/acs.jmedchem.8b00664. [DOI] [PubMed] [Google Scholar]
- 46.Bauer S., Demetri G., Jeay S., Dummer R., Guerreiro N., Tan D.S. A phase I, open-label, multi-center, dose escalation study of oral NVP-CGM097, a P53/HDM2–protein–protein interaction inhibitor, in adult patients with selected advanced solid tumors. Ann Oncol. 2016;27:114–135. [Google Scholar]
- 47.Jeay S., Gaulis S., Ferretti S., Bitter H., Ito M., Valat T. A distinct P53 target gene set predicts for response to the selective P53–HDM2 inhibitor NVP-CGM097. Elife. 2015;4 doi: 10.7554/eLife.06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raboisson P., Marugán J.J., Schubert C., Koblish H.K., Lu T., Zhao S. Structure-based design, synthesis, and biological evaluation of novel 1,4-diazepines as HDM2 antagonists. Elife. 2005;15:1857–1861. doi: 10.1016/j.bmcl.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 49.Leonard K., Marugan J.J., Raboisson P., Calvo R., Gushue J.M., Koblish H.K. Novel 1,4-benzodiazepine-2,5-diones as HDM2 antagonists with improved cellular activity. Bioorg Med Chem Lett. 2006;16:3463–3468. doi: 10.1016/j.bmcl.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Marugan J.J., Leonard K., Raboisson P., Gushue J.M., Calvo R., Koblish H.K. Enantiomerically pure 1,4-benzodiazepine-2,5-diones as HDM2 antagonists. Bioorg Med Chem Lett. 2006;16:3115–3120. doi: 10.1016/j.bmcl.2006.03.067. [DOI] [PubMed] [Google Scholar]
- 51.Grasberger B.L., Lu T., Schubert C., Parks D.J., Carver T.E., Koblish H.K. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate P53 in cells. J Med Chem. 2005;48:909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 52.Koblish H.K., Zhao S., Franks C.F., Donatelli R.R., Tominovich R.M., LaFrance L.V. Benzodiazepinedione inhibitors of the HDM2:P53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5:160–169. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- 53.Parks D.J., LaFrance L.V., Calvo R.R., Milkiewicz K.L., José Marugán J., Raboisson P. Enhanced pharmacokinetic properties of 1,4-benzodiazepine-2,5-dione antagonists of the HDM2–P53 protein–protein interaction through structure-based drug design. Bioorg Med Chem Lett. 2006;16:3310–3314. doi: 10.1016/j.bmcl.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 54.Huang Y., Wolf S., Bista M., Meireles L., Camacho C., Holak T.A. 1,4-Thienodiazepine-2,5-diones via MCR (I): synthesis, virtual space and P53–MDM2 activity. Chem Biol Drug Des. 2010;76:116–129. doi: 10.1111/j.1747-0285.2010.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parks D.J., LaFrance L.V., Calvo R.R., Milkiewicz K.L., Gupta V., Lattanze J. 1,4-Benzodiazepine-2,5-diones as small molecule antagonists of the HDM2–P53 interaction: discovery and SAR. Bioorg Med Chem Lett. 2005;15:765–770. doi: 10.1016/j.bmcl.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 56.Zhuang C., Miao Z., Zhu L., Zhang Y., Guo Z., Yao J. Synthesis and biological evaluation of thio-benzodiazepines as novel small molecule inhibitors of the P53–MDM2 protein–protein interaction. Eur J Med Chem. 2011;46:5654–5661. doi: 10.1016/j.ejmech.2011.09.043. [DOI] [PubMed] [Google Scholar]
- 57.Guo Z., Zhuang C., Zhu L., Zhang Y., Yao J., Dong G. Structure–activity relationship and antitumor activity of thio-benzodiazepines as P53–MDM2 protein–protein interaction inhibitors. Eur J Med Chem. 2012;56:10–16. doi: 10.1016/j.ejmech.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 58.Yu Z., Zhuang C., Wu Y., Guo Z., Li J., Dong G. Design, synthesis and biological evaluation of sulfamide and triazole benzodiazepines as novel P53–MDM2 inhibitors. Int J Mol Sci. 2014;15:15741–15753. doi: 10.3390/ijms150915741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hardcastle I.R., Ahmed S.U., Atkins H., Calvert A.H., Curtin N.J., Farnie G. Isoindolinone-based inhibitors of the MDM2–P53 protein–protein interaction. Bioorg Med Chem Lett. 2005;15:1515–1520. doi: 10.1016/j.bmcl.2004.12.061. [DOI] [PubMed] [Google Scholar]
- 60.Hardcastle I.R., Ahmed S.U., Atkins H., Farnie G., Golding B.T., Griffin R.J. Small-molecule inhibitors of the MDM2–P53 protein–protein interaction based on an isoindolinone scaffold. J Med Chem. 2006;49:6209–6221. doi: 10.1021/jm0601194. [DOI] [PubMed] [Google Scholar]
- 61.Hardcastle I.R., Liu J., Valeur E., Watson A., Ahmed S.U., Blackburn T.J. Isoindolinone inhibitors of the murine double minute 2 (MDM2)–P53 protein–protein interaction: structure–activity studies leading to improved potency. J Med Chem. 2011;54:1233–1243. doi: 10.1021/jm1011929. [DOI] [PubMed] [Google Scholar]
- 62.Watson A.F., Liu J., Bennaceur K., Drummond C.J., Endicott J.A., Golding B.T. MDM2–P53 protein–protein interaction inhibitors: a-ring substituted isoindolinones. Bioorg Med Chem Lett. 2011;21:5916–5919. doi: 10.1016/j.bmcl.2011.07.084. [DOI] [PubMed] [Google Scholar]
- 63.Soares J., Pereira N.A., Monteiro Â., Leão M., Bessa C., dos Santos D.J. Oxazoloisoindolinones with in vitro antitumor activity selectively activate a P53-pathway through potential inhibition of the P53–MDM2 interaction. Eur J Pharm Sci. 2015;66:138–147. doi: 10.1016/j.ejps.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Vaupel A., Holzer P., Ferretti S., Guagnano V., Kallen J., Mah R. In vitro and in vivo characterization of a novel, highly potent P53–MDM2 inhibitor. Bioorg Med Chem Lett. 2018;28:3404–3408. doi: 10.1016/j.bmcl.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 65.Furet P., Chène P., De Pover A., Valat T.S., Lisztwan J.H., Kallen J. The central valine concept provides an entry in a new class of non peptide inhibitors of the P53–MDM2 interaction. Bioorg Med Chem Lett. 2012;22:3498–3502. doi: 10.1016/j.bmcl.2012.03.083. [DOI] [PubMed] [Google Scholar]
- 66.Popowicz G.M., Czarna A., Wolf S., Wang K., Wang W., Dömling A. Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for P53–MDMX/MDM2 antagonist drug discovery. Cell Cycle. 2010;9:1104–1111. doi: 10.4161/cc.9.6.10956. [DOI] [PubMed] [Google Scholar]
- 67.Twarda-Clapa A., Krzanik S., Kubica K., Guzik K., Labuzek B., Neochoritis C. 1,4,5-Trisubstituted imidazole-based P53–MDM2/MDMX antagonists with aliphatic linkers for conjugation with biological carriers. J Med Chem. 2017;60:4234–4244. doi: 10.1021/acs.jmedchem.7b00104. [DOI] [PubMed] [Google Scholar]
- 68.Graves B., Thompson T., Xia M., Janson C., Lukacs C., Deo D. Activation of the P53 pathway by small-molecule-induced MDM2 and MDMX dimerization. Proc Natl Acad Sci U S A. 2012;109:11788–11793. doi: 10.1073/pnas.1203789109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang Y., Wolf S., Beck B., Köhler L., Khoury K., Popowicz G.M. Discovery of highly potent P53–MDM2 antagonists and structural basis for anti-acute myeloid leukemia activities. ACS Chem Biol. 2014;9:802–811. doi: 10.1021/cb400728e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim C.Y., Chandra P.P., Jain A., Christianson D.W. Fluoroaromatic–fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II. J Am Chem Soc. 2001;123:9620–9627. doi: 10.1021/ja011034p. [DOI] [PubMed] [Google Scholar]
- 71.Huang Y., Wolf S., Koes D., Popowicz G.M., Camacho C.J., Holak T.A. Exhaustive fluorine scanning toward potent P53–MDM2 antagonists. ChemMedChem. 2012;7:49–52. doi: 10.1002/cmdc.201100428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boltjes A., Huang Y., Velde R., Rijkee L., Wolf S., Gaugler J. Fragment-based library generation for the discovery of a peptidomimetic P53–MDM4 inhibitor. ACS Comb Sci. 2014;16:393–396. doi: 10.1021/co500026b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bowman A.L., Nikolovska-Coleska Z., Zhong H., Wang S., Carlson H.A. Small molecule inhibitors of the MDM2–P53 interaction discovered by ensemble-based receptor models. J Am Chem Soc. 2007;129:12809–12814. doi: 10.1021/ja073687x. [DOI] [PubMed] [Google Scholar]
- 74.Soares J., Raimundo L., Pereira N.A.L., Santos D.J.V.A., Pérez M., Queiroz G. A tryptophanol-derived oxazolopiperidone lactam is cytotoxic against tumors via inhibition of P53 interaction with murine double minute proteins. Pharmacol Res. 2015;95–96:42–52. doi: 10.1016/j.phrs.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 75.Estrada-Ortiz N., Neochoritis C.G., Twarda-Clapa A., Musielak B., Holak T.A., Dömling A. Artificial macrocycles as potent P53–MDM2 inhibitors. ACS Med Chem Lett. 2017;8:1025–1030. doi: 10.1021/acsmedchemlett.7b00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Surmiak E., Neochoritis C.G., Musielak B., Twarda-Clapa A., Kurpiewska K., Dubin G. Rational design and synthesis of 1,5-disubstituted tetrazoles as potent inhibitors of the MDM2–P53 interaction. Eur J Med Chem. 2017;126:384–407. doi: 10.1016/j.ejmech.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 77.Zhuang C., Miao Z., Zhu L., Dong G., Guo Z., Wang S. Discovery, synthesis, and biological evaluation of orally active pyrrolidone derivatives as novel inhibitors of P53–MDM2 protein–protein interaction. J Med Chem. 2012;55:9630–9642. doi: 10.1021/jm300969t. [DOI] [PubMed] [Google Scholar]
- 78.Li J., Wu Y., Guo Z., Zhuang C., Yao J., Dong G. Discovery of 1-arylpyrrolidone derivatives as potent P53–MDM2 inhibitors based on molecule fusing strategy. Bioorg Med Chem Lett. 2014;24:2648–2650. doi: 10.1016/j.bmcl.2014.04.063. [DOI] [PubMed] [Google Scholar]
- 79.Liu Z., Miao Z., Li J., Fang K., Zhuang C., Du L. A fluorescent probe for imaging P53–MDM2 protein–protein interaction. Chem Biol Drug Des. 2015;85:411–417. doi: 10.1111/cbdd.12434. [DOI] [PubMed] [Google Scholar]
- 80.Zhuang C., Miao Z., Wu Y., Guo Z., Li J., Yao J. Double-edged swords as cancer therapeutics: novel, orally active, small molecules simultaneously inhibit P53–MDM2 interaction and the NF-κB pathway. J Med Chem. 2014;57:567–577. doi: 10.1021/jm401800k. [DOI] [PubMed] [Google Scholar]
- 81.Shishodia S., Koul D., Aggarwal B.B. Cyclooxygenase (COX)-2 inhibitor celecoxib abrogates TNF-induced NF-κB activation through inhibition of activation of IκBα kinase and Akt in human non-small cell lung carcinoma: correlation with suppression of COX-2 synthesis. J Immunol. 2004;173:2011–2022. doi: 10.4049/jimmunol.173.3.2011. [DOI] [PubMed] [Google Scholar]
- 82.Tergaonkar V., Pando M., Vafa O., Wahl G., Verma I. P53 stabilization is decreased upon NF-κB activation: a role for NF-κB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1:493–503. doi: 10.1016/s1535-6108(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 83.Dey A., Tergaonkar V., Lane D.P. Double-edged swords as cancer therapeutics: simultaneously targeting P53 and NF-κB pathways. Nat Rev Drug Discov. 2008;7:1031–1040. doi: 10.1038/nrd2759. [DOI] [PubMed] [Google Scholar]
- 84.Zhuang C., Sheng C., Shin W.S., Wu Y., Li J., Yao J. A novel drug discovery strategy: mechanistic investigation of an enantiomeric antitumor agent targeting dual P53 and NF-κB pathways. Oncotarget. 2014;15:10830–10839. doi: 10.18632/oncotarget.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lu Y., Nikolovska-Coleska Z., Fang X., Gao W., Shangary S., Qiu S. Discovery of a nanomolar inhibitor of the human murine double minute 2 (MDM2)–P53 interaction through an integrated, virtual database screening strategy. J Med Chem. 2006;49:3759–3762. doi: 10.1021/jm060023+. [DOI] [PubMed] [Google Scholar]
- 86.Allen J.G., Bourbeau M.P., Wohlhieter G.E., Bartberger M.D., Michelsen K., Hungate R. Discovery and optimization of chromenotriazolopyrimidines as potent inhibitors of the mouse double minute 2-tumor protein 53 protein–protein interaction. J Med Chem. 2009;52:7044–7053. doi: 10.1021/jm900681h. [DOI] [PubMed] [Google Scholar]
- 87.Yin H., Lee G.I., Park H.S., Payne G.A., Rodriguez J.M., Sebti S.M. Terphenyl-based helical mimetics that disrupt the P53/HDM2 interaction. Angew Chem Int Ed. 2005;117:2764–2767. doi: 10.1002/anie.200462316. [DOI] [PubMed] [Google Scholar]
- 88.Vogel S.M., Bauer M.R., Joerger A.C., Wilcken R., Brandt T., Veprintsev D.B. Lithocholic acid is an endogenous inhibitor of MDM4 and MDM2. Proc Natl Acad Sci USA. 2012;109:16906–16910. doi: 10.1073/pnas.1215060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pettersson M., Bliman D., Jacobsson J., Nilsson J.R., Min J., Iconaru L. 8-Triazolylpurines: towards fluorescent inhibitors of the MDM2/P53 interaction. PLoS One. 2015;10 doi: 10.1371/journal.pone.0124423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blackburn T.J., Ahmed S., Coxon C.R., Liu J., Lu X., Golding B.T. Diaryl- and triaryl-pyrrole derivatives: inhibitors of the MDM2–P53 and MDMX–P53 protein–protein interactions. Medchemcomm. 2013;4:1297–1304. doi: 10.1039/c3md00161j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vaupel A., Bold G., De Pover A., Stachyra-Valat T., Hergovich Lisztwan J., Kallen J. Tetra-substituted imidazoles as a new class of inhibitors of the P53–MDM2 interaction. Bioorg Med Chem Lett. 2014;24:2110–2114. doi: 10.1016/j.bmcl.2014.03.039. [DOI] [PubMed] [Google Scholar]
- 92.Wang W., Zhu X., Hong X., Zheng L., Zhu H., Hu Y. Identification of novel inhibitors of P53–MDM2 interaction facilitated by pharmacophore-based virtual screening combining molecular docking strategy. MedChemComm. 2013;4:411–416. [Google Scholar]
- 93.Wang W., Lv D., Qiu N., Zhang L., Hu C., Hu Y. Design, synthesis and biological evaluation of novel 3,4,5-trisubstituted aminothiophenes as inhibitors of P53–MDM2 interaction. Part 2. Bioorg Med Chem. 2013;21:2886–2894. doi: 10.1016/j.bmc.2013.03.070. [DOI] [PubMed] [Google Scholar]
- 94.Leão M., Gomes S., Pedraza-Chaverri J., Machado N., Sousa E., Pinto M. α-Mangostin and gambogic acid as potential inhibitors of the P53–MDM2 interaction revealed by a yeast approach. J Nat Prod. 2013;76:774–778. doi: 10.1021/np400049j. [DOI] [PubMed] [Google Scholar]
- 95.Zheng G.H., Shen J.J., Zhan Y.C., Yi H., Xue S.T., Wang Z., Ji X.Y. Design, synthesis and in vitro and in vivo antitumour activity of 3-benzylideneindolin-2-one derivatives, a novel class of small-molecule inhibitors of the MDM2–P53 interaction. Eur J Med Chem. 2014;81:277–288. doi: 10.1016/j.ejmech.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 96.Ma Y., Lahue B.R., Shipps G.W., Jr., Brookes J., Wang Y. Substituted piperidines as HDM2 inhibitors. Bioorg Med Chem Lett. 2014;24:1026–1030. doi: 10.1016/j.bmcl.2014.01.026. [DOI] [PubMed] [Google Scholar]
- 97.Pan W., Lahue B.R., Ma Y., Nair L.G., Shipps G.W., Jr., Wang Y. Core modification of substituted piperidines as novel inhibitors of HDM2–P53 protein–protein interaction. Bioorg Med Chem Lett. 2014;24:1983–1986. doi: 10.1016/j.bmcl.2014.02.055. [DOI] [PubMed] [Google Scholar]
- 98.Lu F., Chi S.W., Kim D.H., Han K.H., Kuntz I.D., Guy R.K. Proteomimetic libraries: design, synthesis, and evaluation of P53−MDM2 interaction inhibitors. J Comb Chem. 2006;8:315–325. doi: 10.1021/cc050142v. [DOI] [PubMed] [Google Scholar]
- 99.Galatin P.S., Abraham D.J. A nonpeptidic sulfonamide inhibits the P53–MDM2 interaction and activates P53-dependent transcription in MDM2-overexpressing Cells. J Med Chem. 2004;47:4163–4165. doi: 10.1021/jm034182u. [DOI] [PubMed] [Google Scholar]
- 100.Stoll R., Renner C., Hansen S., Palme S., Klein C., Belling A. Chalcone derivatives antagonize interactions between the human oncoprotein MDM2 and P53. Biochemistry. 2001;40:336–344. doi: 10.1021/bi000930v. [DOI] [PubMed] [Google Scholar]
- 101.Yang J., Li J., Aguilar A., McEachern A., Przybranowski S., Fernandez-Salas E. Targeted MDM2 degradation as a novel and efficacious cancer therapy. Cancer Res. 2018;78:4870. [Google Scholar]
- 102.Wang S., Sun W., Zhao Y., McEachern D., Meaux I., Barriere C. An optimized inhibitor of MDM2–P53 interaction that induces complete and durable tumor regression. Cancer Res. 2014;74:5855–5865. doi: 10.1158/0008-5472.CAN-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li Y., Yang J., Aguilar A., McEachern D., Przybranowski S., Liu L. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J Med Chem. 2019;62:448–466. doi: 10.1021/acs.jmedchem.8b00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ryan P., Victor J. Targeted degradation of MDM2 as a new approach to improve the efficacy of MDM2–P53 inhibitors. J Med Chem. 2019;62:445–447. doi: 10.1021/acs.jmedchem.8b01945. [DOI] [PubMed] [Google Scholar]
- 105.Yang J., Li Y., Aguilar A., Liu Z.H., Yang C.H., Wang S. Simple structural modifications converting a bona fide MDM2 PROTAC degrader into a molecular glue molecule: a cautionary tale in the design of PROTAC degraders. J Med Chem. 2019;62:9471–9487. doi: 10.1021/acs.jmedchem.9b00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang B., Wu S., Liu J., Yang K., Xie H., Tang W. Development of selective small molecule MDM2 degraders based on Nutlin. Eur J Med Chem. 2019;176:476–491. doi: 10.1016/j.ejmech.2019.05.046. [DOI] [PubMed] [Google Scholar]
- 107.Hoe K.K., Verma C.S., Lane D.P. Drugging the P53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov. 2014;13:217–236. doi: 10.1038/nrd4236. [DOI] [PubMed] [Google Scholar]
- 108.Ramos Y.F., Stad R., Attema J., Peltenburg L.T., van der Eb A.J., Jochemsen A.G. Aberrant expression of HDMX proteins in tumor cells correlates with wild-type P53. Cancer Res. 2001;61:1839–1842. [PubMed] [Google Scholar]
- 109.Linares L.K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. HdmX stimulates HDM2-mediated ubiquitination and degradation of P53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu B., Gilkes D.M., Chen J. Efficient P53 activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 111.Madden M.M., Muppidi A., Li Z., Li X., Chen J., Lin Q. Synthesis of cell-permeable stapled peptide dual inhibitors of the P53–MDM2/MDMX interactions via photoinduced cycloaddition. Bioorg Med Chem Lett. 2011;21:1472–1475. doi: 10.1016/j.bmcl.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shangary S., Qin D., McEachern D., Liu M., Miller R.S., Qiu S. Temporal activation of P53 by a specific MDM2 inhibitor is selectively toxic to tumors and leads to complete tumor growth inhibition. Proc Natl Acad Sci U S A. 2008;105:3933–3938. doi: 10.1073/pnas.0708917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Linke K., Mace P.D., Smith C.A., Vaux D.L., Silke J., Day C.L. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 114.Dickens M.P., Fitzgerald R., Fischer P.M. Small-molecule inhibitors of MDM2 as new anticancer therapeutics. Semin Cancer Biol. 2010;20:10–18. doi: 10.1016/j.semcancer.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 115.Xia M., Knezevic D., Tovar C., Huang B., Heimbrook D.C., Vassilev L.T. Elevated MDM2 boosts the apoptotic activity of P53–MDM2 binding inhibitors by facilitating MDMX degradation. Cell Cycle. 2008;7:1604–1612. doi: 10.4161/cc.7.11.5929. [DOI] [PubMed] [Google Scholar]
- 116.Gilkes D.M., Pan Y., Coppola D., Yeatman T., Reuther G.W., Chen J. Regulation of MDMX expression by mitogenic signaling. Mol Cell Biol. 2008;28:1999–2010. doi: 10.1128/MCB.01633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wade M., Wahl G.M. Targeting MDM2 and MDMX in cancer therapy: better living through medicinal chemistry?. Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Reed D., Shen Y., Shelat A.A., Arnold L.A., Ferreira A.M., Zhu F. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem. 2010;285:10786–10796. doi: 10.1074/jbc.M109.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Holohan C., Van Schaeybroeck S., Longley D.B., Johnston P.G. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 120.Cinatl J., Speidel D., Hardcastle I., Michaelis M. Resistance acquisition to MDM2 inhibitors. Biochem Soc Trans. 2014;42:752–757. doi: 10.1042/BST20140035. [DOI] [PubMed] [Google Scholar]
- 121.Aziz M.H., Shen H., Maki C.G. Acquisition of P53 mutations in response to the non-genotoxic P53 activator Nutlin-3. Oncogene. 2011;30:4678–4686. doi: 10.1038/onc.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hoffman-Luca C.G., Yang C.Y., Lu J., Ziazadeh D., McEachern D., Debussche L. Significant differences in the development of acquired resistance to the MDM2 inhibitor SAR405838 between in vitro and in vivo drug treatment. PLoS One. 2015;10 doi: 10.1371/journal.pone.0128807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hoffman-Luca C.G., Ziazadeh D., McEachern D., Zhao Y., Sun W., Debussche L. Elucidation of acquired resistance to Bcl-2 and MDM2 inhibitors in acute leukemia in vitro and in vivo. Clin Cancer Res. 2015;21:2558–2568. doi: 10.1158/1078-0432.CCR-14-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lowe S.W., Schmitt E.M., Smith S.W., Osborne B.A., Jacks T. P53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 125.Ringshausen I., O'Shea C.C., Finch A.J., Swigart L.B., Evan G.I. MDM2 is critically and continuously required to suppress lethal P53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 126.Miyachi M., Kakazu N., Yagyu S., Katsumi Y., Tsubai-Shimizu S., Kikuchi K. Restoration of P53 pathway by Nutlin-3 induces cell cycle arrest and apoptosis in human rhabdomyosarcoma cells. Clin Cancer Res. 2009;15:4077–4084. doi: 10.1158/1078-0432.CCR-08-2955. [DOI] [PubMed] [Google Scholar]
- 127.Wang Y., Jiang X., Feng F., Liu W., Sun H. Degradation of proteins by PROTACs and other strategies. Acta Pharm Sin B. 2020;10:207–238. doi: 10.1016/j.apsb.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]