Abstract
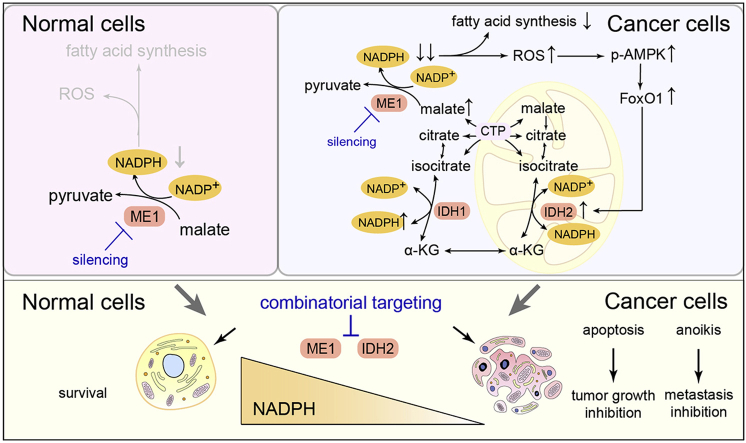
NADPH is a pivotal cofactor that maintains redox homeostasis and lipogenesis in cancer cells and interference with NADPH production is a promising approach for treating cancer. However, how normal and cancer cells differentially exploit NADPH-producing pathways is unclear, and selective approaches to targeting NADPH are lacking. Here, we show that the assayed cancer cell lines preferentially depend on ME1-mediated NADPH production. ME1 knockdown increases intracellular ROS levels and impairs lipogenesis in cancer cells, leading to retarded proliferation and increased anoikis, while sparing normal cells. Notably, ME1 interference ultimately resulted in adaptive upregulation of mitochondrial IDH2 dependent of AMPK-FoxO1 activation to replenish the NADPH pool and mitigate cytosolic ROS. Combining ME1 ablation and IDH2 inhibition drastically reduces intracellular NADPH and prevents resistance to ME1 interference, resulting in increased apoptosis and impeded tumor growth and metastasis. This study demonstrates that cytosolic ME1 integrated with mitochondrial IDH2 is essential for tumor growth and metastasis, thereby highlighting the blockade of metabolic compensation by disrupting mitochondrial-cytosol NADPH transport as a promising approach to selectively targeting NADPH in cancer cells that rely on NADPH-driven antioxidant systems.
Keywords: NADPH, ROS, ME1, IDH2, Reductive carboxylation, Metabolic adaptability
Graphical abstract
Highlights
-
•
NADPH is vital in mitigating ROS stress and supporting lipogenesis in cancer cells.
-
•
Certain cancer cells preferentially depend on ME1-mediated NADPH production route.
-
•
ME1 knockdown adaptively upregulates IDH2 dependent of AMPK-FoxO1 axis.
-
•
Compensatory IDH2 contributes to replenish the NADPH pool and mitigates ROS.
-
•
Combined targeting ME1 and IDH2 depletes NADPH and inhibits tumor growth and metastasis.
Abbreviations
- 2-HG
2-hydroxyglutarate
- 6PGD
6-phosphogluconate dehydrogenase
- α-KG
α-ketoglutarate
- AICAR
acadesine
- AMPK
Adenosine 5'-monophosphate activated protein kinase
- CRC
colorectal cancer
- CTP
citrate transporter protein
- ECM
extracellular matrix
- EGF
epidermal growth factor
- FoxO1
forkhead box O1
- G6PD
glucose-6-phosphate dehydrogenase
- GBM
glioblastoma
- GLS
glutaminase
- GSH
reduced glutathione
- IDH
isocitrate dehydrogenase
- ME
malic enzyme
- MTHFD
methylenetetrahydrofolate dehydrogenase
- NAC
N-acetylcysteine
- NADP
nicotinamide adenine dinucleotide phosphate
- NADPH
reduced nicotinamide adenine dinucleotide phosphate
- oxPPP
oxidative pentose phosphate pathway
- PDAC
pancreatic ductal adenocarcinoma
- PDX
patient-derived xenograft
- ROS
reactive oxygen species
1. Introduction
Cancer cells are characterized by rapid proliferation, anchorage-independent cell growth and migration [[1], [2], [3]]. During the process of rapid proliferation and metastasis, cancer cells encounter a drastic increase in the production of reactive oxygen species (ROS) [[4], [5], [6]]. To survive the increased ROS levels, cancer cells have evolved precise machinery for maintaining redox homeostasis, which is important for supporting tumor growth and metastasis [7]. NADPH is one of the most important cofactors that scavenges ROS by replenishing the reduced glutathione (GSH) and thioredoxin pools [8]. In addition, NADPH is essential and is likely a rate-limiting factor for fueling lipid biosynthesis [9].
Because of the contribution of NADPH to redox homeostasis and lipogenesis, cells have evolved multiple pathways and enzymes to produce NADPH in both the cytosol and mitochondria. At least four metabolic pathways are involved in NADPH production; these pathways include those controlled by glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD), which directly produces NADPH from glucose via the oxidative pentose phosphate pathway (oxPPP); isocitrate dehydrogenase (IDH1 and IDH2), which catalyze isocitrate decarboxylation; methylenetetrahydrofolate dehydrogenase (MTHFD1 and MTHFD2), which couple NADPH production to methylene tetrahydrofolate oxidation; and malic enzyme (ME1, ME2 and ME3), which catalyze the oxidative decarboxylation of malate to pyruvate [9,10]. Disrupting NADPH production by modulating the activity or expression of these enzymes has been proposed as a viable strategy of exploiting cancer metabolic vulnerabilities [11,12]. For instance, G6PD inhibition reduced NADPH production and enhanced oxaliplatin-induced apoptosis in colorectal cancer (CRC) xenografts and in a patient-derived xenograft (PDX) model [13]. Genetic and pharmacological suppression of IDH1 activity inhibits glioblastoma (GBM) cell growth and prolongs survival in an animal model of GBM in vivo [14], whereas MTHFD2 inhibition suppresses colorectal cancer growth [15]. In addition, pancreatic ductal adenocarcinoma (PDAC) patients who harbor homozygous deletion of SMAD4 and ME2 become metabolically vulnerable to ME3 depletion due to collateral depletion of NADPH [16]. Another line of evidence indicating that targeting NADPH production is a promising strategy for cancer therapy is the finding that several key oncogenes and tumor suppressor genes function to control tumor growth and metastasis at least partially by modulating NADPH-producing pathways [[17], [18], [19], [20], [21]].
Although accumulating evidence indicates that targeting NADPH is a promising strategy for cancer therapy [11], two critical questions that represent a bottleneck for the ultimate translation of NADPH targeting strategies to clinical cancer therapies remain unaddressed. First, since NADPH is essential in both normal and cancer cells, the mechanisms by which cancer cells and normal cells differentially exploit metabolic enzymes to produce NADPH and the approaches by which we can harness this distinction to selectively kill cancer cells while sparing normal cells remain unclear. Second, a primary concern is that targeting a single enzyme may not sufficiently inhibit NADPH production, considering that at least four pathways are involved in NADPH production and that several paralogous genes are involved even in a single pathway [9,22]. The metabolic plasticity of cancer cells may thus induce other enzymes to replenish NADPH production when certain enzyme is genetically deleted or pharmacologically inhibited [10]. Even in normal cells, adipocytes reveal a hypoxia-induced switch of the major NADPH producer from ME1 to the oxPPP [23]. Projecting forward, collateral targeting of the paralogous isoform ME3 in patients harboring genomic deletion of ME2 has been proposed as a desirable approach to treat PDAC. However, patients with genomic deletion of paralogous isozymes often account for only a small fraction of the population [16]. We thus reasoned that it is tempting to target NADPH for cancer therapy based on understanding of the mechanism by which certain subtypes of tumor cells differentially exploit NADPH-producing pathways compared with normal cells and may utilize the compensatory mechanism to replenish NADPH.
Here, we find that the assayed cancer cells but not normal cells preferentially depend on ME1-catalyzed NADPH production. Interference with ME1 pronouncedly reduces NADPH and increases ROS levels, thereby slowing tumor growth and facilitating detachment-induced death of cancer cells while sparing normal cells. Notably, ME1 interference alone is insufficient to trigger cancer cell apoptosis, and acquired resistance is particularly identified during prolonged cell culture. Mechanistically, ME1 silencing leads to adaptive activation of mitochondrial IDH2, which mediates NADPH compensation in an AMPK-forkhead box O1 (FoxO1)-dependent manner. Combinatorial targeting of cytosolic ME1 and mitochondrial IDH2 is thus proposed and demonstrated to synergistically impede tumor growth and metastasis both in vitro and in vivo.
2. Materials and methods
2.1. Cell lines and cell culture
Human mammary epithelial cells MCF-10A, human breast carcinoma cells MCF-7, and human lung adenocarcinoma cells A549 were obtained from the American Type Culture Collection (ATCC). Human embryonal lung fibroblast MRC-5 and human colorectal cancer cells HCT116 were obtained from the Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). Human colon mucosal epithelial cells NCM460 was obtained from Jining Shiye (Shanghai, Chnia). MCF-10A and MRC-5 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 1 μg/mL streptomycin. MCF-7, NCM460 and A549 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% FBS, 100 U/mL penicillin and 1 μg/mL streptomycin. HCT116 cells was cultured in McCoy's 5A medium supplemented with 10% FBS, 100 U/mL penicillin and 1 μg/mL streptomycin. All cells were incubated at 37 °C with 5% CO2 and the medium was changed every other day.
2.2. Human breast cancer tissue specimens
Female breast cancer tissue specimens were collected after obtaining written informed consent from all the patients. The scientific use of clinical specimens was conducted in compliance with the Declaration of Helsinki and approved by the ethics committee of The First Hospital of Jilin University. 12 female patients with luminal A breast cancer (1 patient), luminal B breast cancer (9 patients) and triple negative breast cancer (2 patients) with ages between 41 and 76. The available clinical characters of these patients are summarized in Supplementary Table 1. Each pair of tumor specimens and adjacent noncancerous tissues was obtained from the same patient. The collected tissue specimens were stored frozen at −80 °C until required for protein extraction.
2.3. SiRNA transfection
Scrambled small interfering RNA (siRNA) and siRNA targeting G6PD and ME1 were purchased from Invitrogen. The targeted sequence siRNA sequences for G6PD is 5′-ACG AGC UGA UGA AGA GAG UGG GUU U-3′, and the sequences for ME1 #1 and #2 siRNAs are 5′-GAGAAGAGUAAGAGGUUCUGAAUAU-3′ and 5′-UGACCAGAUUCUACCUGAUUGUUAU-3′, respectively. SiRNA targeting ME2 (cat. no. sc-75764), IDH1 (cat. no. sc-60829), IDH2 (cat. no. sc-62487), MTHFD1 (cat. no. sc-61082), MTHFD2 (cat. no. sc-75937) and CTP (cat. no. sc-77046) were purchased from Santa Cruz. Cells were transfected using lipofectamine™ RNAiMAX regent (Invitrogen, cat. no.13778030) for 48 h according to the manufacturer's instructions and the efficiency of silencing was confirmed by immunoblotting. Unless otherwise specified, the ME1 siRNA used in this study is a mixture of ME1 #1 and ME1 #2.
2.4. Metabolic tracing
Isotopic tracers used in this study were all purchased from Cambridge Isotope Laboratories. For metabolic tracing experiments, all cells were cultured in the identical medium (RPMI 1640) for at least 3 passages. For NADPH tracing, cells were cultured in glucose-free RPMI 1640 containing 10 mM [1-2H] glucose, 10 mM [4-2H] glucose and 2 mM [2,3,3-2H] aspartate for indicated duration, respectively. For fatty acids synthesis tracing, cells were cultured in glucose-free RPMI 1640 containing 10 mM [U-13C] glucose for 72 h. For glutamine tracing, cells were cultured in glutamine-free RPMI 1640 containing 2 mM [U-13C] glutamine for 24 h. The labeling medium was replaced 2 h before cell harvest.
2.5. Metabolomic sample preparation
Cells (approximately 3 × 105/well) were seeded, cultured in 6-well plates and harvested after indicated treatments. For metabolomic sample preparation, cells were permeabilized with 1 mL ice-cold 80% methanol aqueous buffer containing 1.5 μg/mL 4-chloro-phenylalanine as an internal standard (IS), followed by incubation at −80 °C for 20 min. The cell lysates were centrifuged at 14,000 g for 10 min at 4 °C, and the resultant supernatants were evaporated to dryness. The samples were reconstituted with HPLC-grade water in 100 μL and subjected to analysis. Notably, samples involving NADPH measurement must be processed within 6 h prior to analysis to avoid NADPH degradation. As for fatty acid measurement, cells were washed with cold PBS twice, collected and sonicated in 100 μL water on ice. Then, 800 μL ethyl acetate containing 0.5 μg/mL heptadecanoic acid as an internal standard (IS) was added into the samples followed by vortexing for 10 min. The resultant lysates were centrifuged at 16,000 g for 10 min at 4 °C, and the supernatants were evaporated to dryness. The samples were reconstituted with 200 μL HPLC-grade methanol and subjected to analysis.
2.6. LC-MS/MS-based metabolite analysis
Metabolites were analyzed using an LC-30A Shimadzu LC system (Kyoto, Japan) coupled to a TripleTOF 5600 system (AB SCIEX, Foster City, CA, USA). An XBridge BEH Amide HPLC column (100 mm × 4.6 mm, i,d. 3.5 μm) (Water, Milford, MA, USA) was used for metabolite separation with a column temperature of 40 °C. The mobile phase consisted of solvent A (95% 5 mM ammounium acetate buffer, pH adjusted to 9, 5% acetonitrile) and solvent B (acetonitrile). The gradient was set as follows: 0-3 min, 85% B; 3-6 min, 85-30% B; 6-15 min, 30-2% B; 15-18 min, 2%B; 18-19 min, 2-85% B; 19-26 min, 85% B. The flow rate was set at 0.4 mL/min. Following separation, MS data was acquired in the negative mode, and collected in a data-dependent acquisition mode. The following parameters were used: time-of-flight MS scan, m/z 50-1000 Da; product ion scan, m/z 50-1000 Da; ion source gas 1 (gas 1), 50 psi; ion source gas 2 (gas 2), 30 psi; curtain gas, 30 psi; source temperature, 500 °C; ion spray voltage floating, −4500 V; declustering potential (DP), −100 V; collision energy (CE), −35 V and CE spread, 10 V. The accurate mass was calibrated by Calibration Delivery System (AB SCIEX) and automatic calibration was performed every five samples. Mass isotopologue distribution for assayed metabolites was obtained by dividing the intensity of each isotopologue by the summed intensities of all the detected isotopomers with correction for natural isotope abundance when 13C-labeling tracers were used; detected isotopologues using 2H-labeling tracers were not corrected due to negligible natural abundance of 2H [9,24].
Fatty acids were analyzed using an Acquity H class UPLC system coupled with a Synapt G2 Si system (Waters, Milford, MA, USA). An BEH C18 column (50 mm × 2.1 mm, i.d. 1.7 μm) (Waters, Milford, MA, USA) was used for lipid separation with a column temperature of 40 °C. The mobile phase consisted of solvent A (water) and solvent B (80% acetonitrile, 20% isopropyl alcohol, v/v). The gradient was set as follows: 0-2 min, 70% B; 2-5 min, 70-75% B; 5-10 min, 75-80% B; 10-13 min, 80-90% B; 13-16 min, 90-99% B; 16-21 min, 99%; 21-22 min, 99-70% B and 22-25 min, 70% B. The flow rate was set at 0.4 mL/min. MS data was acquired in the negative mode and electrospray ionization was used as the ionization source. The following parameters were used: capillary voltage, 2.5 kV; sample cone, 55 V; extraction cone, 4 V; desolvation temperature, 450 °C; source temperature, 150 °C; desolvtion gas flow, 650 L/h; cone gas flow, 50 L/h. Mass accuracy was automatically calibrated by lockspray. Fatty acids were identified by matching with reference standards. Peak areas of targeted compounds were integrated and quantified by QuanLynx™ (Waters, Milford, MA, USA), corrected for natural isotope abundance and normalized to TIC (total ion chromatogram) algorithm.
2.7. Immunoblotting
Collected cells or tissue homogenates were lysed in RIPA lysis buffer (Beyotime Biotechnology, cat. no. P0013B) with protease inhibitor cocktail (APExBio, cat. no. K1007). The protein concentrations were then determined by the bicinchoninic acid (BCA) Protein Assay (Beyotime Biotechnology, cat. no. P0011). The lysates were diluted by 4x XT Sample Buffer (Bio-Rad, cat. no. 1610791) followed by boiling for at least 5 min. Approximately 50-70 μg proteins were then loaded onto and separated by 6%–12% SDS-PAGE gels, transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad, cat. no. 1620177) and incubated with primary antibodies. Subsequently, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at 37 °C. The immunoblotted bands were detected by addition of HRP substrate (Bio-Rad, cat. no. 170-5601), captured on a ChemDoc XRS+ System (Bio-rad, Hercules, USA), and analyzed by ImageLab software. The primary antibodies used in this study include antibodies against ME1 (1:1000, 5% non-fat milk, Abcam, cat. no. ab97445), G6PD (1:1000, 5% non-fat milk, Abcam, cat no. ab993), β-actin (1:1000, 5% BSA, Cell Signaling Technology, cat. no. 8457), IDH1 (1:1000, 5% non-fat milk, Cell Signaling Technology, cat. no. 8137), IDH2 (1:1000, 5% BSA, Cell Signaling Technology, cat. no. 56439), AMPK (1:1000, 5% BSA, Cell Signaling Technology, cat. no. 5831), p-AMPK (1:1000, 5% BSA, Cell Signaling Technology, cat. no. 2535), FoxO1 (1:1000, 5% non-fat milk, Cell Signaling Technology, cat. no. 2880), ME2 (1:200, 5% non-fat milk, Santa Cruz Biotechnology, cat no. 514850), MTHFD1 (1:500, 5% non-fat milk, Proteintech, cat. no. 10794-1-AP), MTHFD2 (1:1000, 5% non-fat milk, Proteintech, cat. no. 12270-1-AP) and β-Tubulin (1:3000, 5% non-fat milk, Bioworld, cat. no. AP0064). The raw data for immunoblotting has been uploaded to Mendeley Data.
2.8. ROS measurement
Cells were incubated with serum-free medium containing 10 μM 2′,7′-dichlorodihydrofluorescein diacetate (DHFC-DA, Beyotime Biotechnology, cat. no. S0033) for intracellular ROS measurement and 10 μM MitoSox solution (Invitrogen, cat.no. M36008) for mitochondrial ROS measurement at 37 °C for 30 min, respectively. Afterwards, cells were washed twice with cold phosphate buffered saline (PBS) and resuspended in PBS. Fluorescence was immediately measured using BD Accuri C6 Flow Cytometer (BD Biosciences, San Jose, USA).
2.9. Proliferation and colony formation assay
For cell proliferation assay, cells (approximately 2.5 × 103 cells/well) were seeded in 96-well plates and cultured in medium containing 10% FBS. The medium was changed every day. At indicated time points, cells were stained with Hoechst 33342 (Beyotime Biotechnology, cat. no. C1022) and the number of cells with stained nuclei was counted by LionHeart FX (BioTek, Winooski, VT, USA). For colony formation assay, ~500 cells were seeded per well in 6-well plates and were transfected with siRNAs or control. After 2 weeks, cells were fixed with 4% paraformaldehyde for 20 min and stained with crystal violet solution (Beyotime Biotechnology, cat. no. C0121) for 20 min. The images were captured using Leica DMI 3000B light microscope (Leica, Wetzlar, Germany) in a blinded manner.
2.10. Cell proliferation and cell viability
For EdU cell proliferation assay, cells (approximately 1 × 105 cells/well) were seeded and cultured in 12-well plates. After indicated treatments, cells were incubated with EdU solution (Beyotime Biotechnology, cat. no. C0071S) for 2 h. EdU incorporation was measured according to the manufacturer's instructions. To monitor cell viability, a cell counting kit (CCK-8, Dojido Molecular Technologies, cat. no. DJDB4000X) was used by following the manufacturer's instructions. Briefly, cells (approximately 7.5 × 103/well) were seeded and cultured in 96-well plates. After treatment, cells were washed with PBS twice, and incubated with 100 μL non-phenol red medium containing 10% CCK-8 solution at 37 °C for 1-4 h in each well. The absorbance was measured at 450 nm using an automatic microplate reader (Synergy H1, Bio-tek, Winooski, VT, USA).
2.11. Migration assay
To evaluate cell migration capacity, wound healing assay and transwell migration assay were employed. For wound healing assay, cells (approximately 2 × 105 cells/well) were seeded in 12-well plates. After reaching ~95% confluence, cells monolayers were scratched by the tip of a 200 μL pipette, washed with serum-free medium and cultured in serum-free medium for 24 h. The images of wounds were captured at 0 h and 24 h with at least 3 fields per well using a Leica DMI 3000B light microscope (Leica, Wetzlar, Germany). The relative migration distances [(width of 0 h–width of 24 h)/width of 0 h × 100%] were calculated using Image J (National Institutes of Health, Version 1.51). For transwell migration assay, 24-transwell chambers (8.0 μm pore size, Corning, NY, USA) were used. Briefly, approximately 2 × 105/well cells were seeded in the upper chamber and cultured with serum-free medium at 37 °C with 5% CO2. After 24 h, medium containing 10% FBS was added to the lower chamber. Then, cells adhering to the lower chamber were fixed with 4% paraformaldehyde and stained with crystal violet solution for 20 min. Five randomly selected fields were counted for each filter (n = 3).
2.12. Spheroid cell culture
Cells (approximately 7.5 × 103 cells/well) were seeded in 96-well ultra-low attachment spheroid microplates (Corning, USA), and cultured in RPMI 1640 supplemented with 10% FBS, 100 U/mL penicillin and 1 μg/mL streptomycin at 37 °C with 5% CO2 for 7 days. The medium was changed every other day.
2.13. Flow cytometric analysis of cell apoptosis
For cellular apoptotic rate analysis, cells were treated with 0.25% typsin solution without EDTA, centrifuged at 3000 rpm for 5 min, washed once with PBS and stained using Annexin V-FITC and Propidium iodide (PI) solution (BD Pharminge, cat. no. 556547) for 15 min at room temperature (avoid light). The percentage of apoptotic cells for each sample was subsequently evaluated by BD Accuri C6 flow cytometer (BD Biosciences, San Jose, USA).
2.14. RNA extraction and quantitative RT-PCR
Total RNA was extracted from cells by RNAiso Plus reagent (Takara, Japan) according to the manufacturer's instructions. About 500 ng of total RNA was used to generate complementary DNA (cDNA) using Superscript II reverse transcriptase (Takara, cat. no. RR047A). Quantitative RT-PCR (qRT-PCR) was performed using SYBR qPCR Master Mix (Vazyme Biotech, cat. no. Q311-02) on a real-time PCR cycler (Step One Plus, Applied Biosystems, CA, USA). Endogenous β-actin gene was used as the internal control for normalizing target gene expression changes. The primer Sequences used in this study are listed in Supplementary Table 2.
2.15. Measurement of mitochondrial membrane potential
The mitochondrial membrane potential was determined using JC-1 mitochondrial membrane potential assay kit (Beyotime Biotechnology, cat no. C2006) according to the manufacturer's instructions. Briefly, approximately 1 × 104/well cells were seeded in 96-well plates. After treatment, cells were incubated with 100 μM JC-1 probe solution in 37 °C for 30 min and followed by washing with wash solution according to the instructions. JC-1 was detected by a fluorescent reader (Synergy H1, Bio Tek, Winooski, VT, USA) with Ex 490/Em 530 nm for monomer detection and Ex 525/Em 590 nm for aggregate detection. The ratio of aggregates/monomers indicates the dissipation of mitochondrial transmembrane potential.
2.16. shRNA knockdown
Short hairpin RNA (shRNA) directed against ME1 (GCATCTGAACTCTGACTTTGA) was cloned into PHY-315 vector and packaged into lentivirus (Hanyin Biotechnology, Shanghai, China). Approximately 5 × 104/well cells were seeded in 6-well plates and cultured for 24 h, followed by transfection with lentivirus encoding ME1 shRNA. After 2 days of transfection, cells were selected in 1 μg/ml puromycin for 5 days. Knockdown efficiency was confirmed by immunoblotting.
2.17. In vivo tumorigenesis and metastasis models
All animal experiments were performed in accordance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) at China Pharmaceutical University. For in vivo tumorigenesis study, A549 cells (5 × 106) were injected subcutaneously into the flank of 5-week-old male BABL/c nude mice (Model Animal Research Center of Nanjing University, Nanjing, China). Mice were randomized before being allocated to cages for treatment. When the tumor volumes reached approximately 50 mm3, the tumor-bearing mice were randomly assigned into 4 groups (n = 6). For tumoral knockdown of ME1, ME1 siRNAs (#1, 5 nmol/mouse) were multi-point injected intratumorally every other day. For IDH2 inhibition, AGI-6780 (60 mg/kg) was intragastrically administered every day. Tumor size was measured every other day, and tumor volume was calculated using a formula of length × width2/2. After 18 days, mice were euthanized and tumors were removed, weighed and photographed. For the metastasis model, A549 cells (4 × 106) were injected into the tail veins of four groups of mice (n = 6). AGI-6780 (60 mg/kg) was intragastrically administered every day. Mice were subjected to intraperitoneal injection of D-luciferin 60 days post-injection, and lung colonization was monitored using a PerkinElmer IVIS Spectrum bioluminescent imaging system (Waltham, MA, USA). Then, the animals were sacrificed and lungs were removed for examination of the metastatic nodules followed by staining with hematoxylin and eosin (H&E).
2.18. Immunohistochemical staining and TUNEL
After sacrifice, the tumor tissues of mice were harvested, fixed with 4% polyformaldehyde and embedded in paraffin and subjected to immunohistological (IHC) analysis as previously described. In brief, tumor sections were deparaffinized, rehydrated and heated at a subboiling temperature in sodium citrate buffer (pH 6.0) for antigen retrieval. The sections were then incubated in 3% hydrogen peroxide to inactivate endogenous peroxidase activity and stained with indicated primary antibodies at 4 °C overnight. The primary antibodies used in this study include antibodies against ME1 (Abcam, 97445, 1:200), Ki-67 (Abcam, ab16667, 1:200) and IDH2 (Cell Signaling Technology, 56439, 1:200). After overnight incubation, the sections were washed, incubated with secondary antibodies (DAKO, cat. no. K5007) and stained with DAB substrate. Counterstaining was lastly carried out with hematoxylin.
The terminal deoxynucleotidyl transferase-mediated nick end labeling (TUNEL) assay was carried out using a TUNEL kit (Roche, cat no.11684817910) according to the manufacturer's instructions. The staining images were captured on a Leica DMI 3000B light microscope (Leica, Wetzlar, Germany) in a blinded manner.
2.19. Promoter reporter assay
The dual reporter plasmid expressing firefly luciferase under the human IDH2 promoter and Renilla luciferase under the SV40 promoter was constructed. MCF-7 cells were seeded in 12-well plates 24 h before transfection and transiently transfected with 500 ng of the reported plasmid using PolyJet (SignaGen, cat. no. SL100688). The luciferase activity was determined according to the manufacturer's instructions (GeneCopoeia, cat. no. LF004).
2.20. Quantification and statistical analysis
The metabolomic analysis and isotope tracing experiments were performed once with multiple replicates as indicated in related figure legends. Animal studies were performed once with each group containing 6 mice to minimize type I/II errors. No sample was excluded from data analysis. Data in the remaining figure panels represent at least two independent experiments, unless stated otherwise, and the contents are representive of the results drawn based on experimental replicates. Statistical analyses were performed using GraphPad Prism 7 software and statistical significance was determined by Student's t-test (unpaired, two-tailed). In all figures, error bars represent standard deviation and the p values of less than 0.05 were considered significant; *p < 0.05, **p < 0.01 and ***p < 0.001.
3. Results
3.1. Cancer cells and normal cells show different NADPH pathway usages
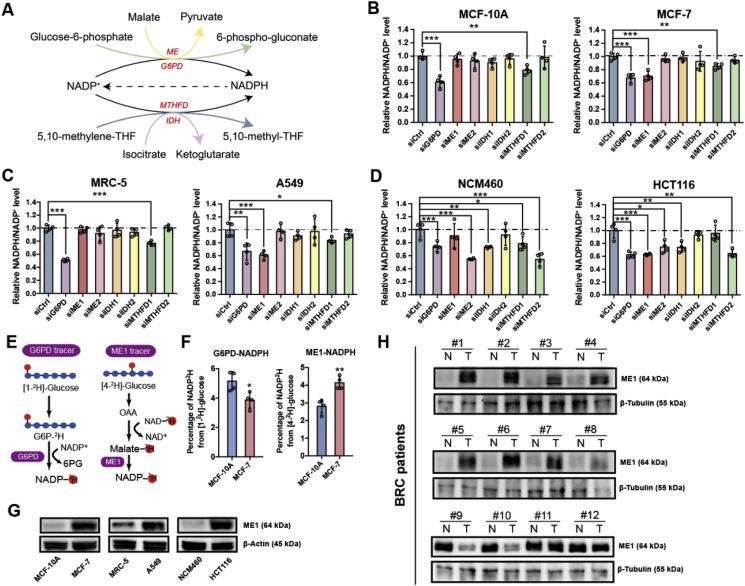
Tumor cells are characterized by enhanced production of ROS during rapid proliferation and metastasis and by an amplified demand for lipid biosynthesis [5]. NADPH is thus a critical factor required for cancer cell proliferation and metastasis due to its contribution to both redox defense and biosynthesis [25]. Four major intracellular pathways are responsible for producing NADPH to meet the demand for reducing equivalent generation: the oxPPP, folate pathway, ME pathway and IDH pathway (Fig. 1A). Since cancer cells require more NADPH than normal cells to support their uncontrolled growth and metastasis, we reasoned that cancer cells exploit these pathways differently from normal cells. To test this hypothesis, we used siRNAs to knock down the major NADPH-producing enzymes (Supplementary Fig. 1A) and measured the NADPH/NADP+ ratio in three human non-transformed cell lines (MCF-10A, MRC-5 and NCM460) and established human cancer cell lines (MCF-7, A549 and HCT116). As shown in Fig. 1B, in MCF-10A cells, silencing ME1, ME2, IDH1, IDH2 and MTHFD2 did not significantly affect the NADPH/NADP+ ratio, whereas knocking down G6PD or MTHFD1 significantly decreased the NADPH/NADP+ ratio with G6PD contributing more to NADPH production than MTHFD1. In contrast, in MCF-7 cells, ME1 knockdown, in addition to G6PD and MTHFD1 knockdown, decreased the NADPH/NADP+ ratio. Similarly, both G6PD and MTHFD1 knockdown markedly influenced the NADPH/NADP+ ratio in the non-transformed cell lines MRC-5 and NCM460, whereas ME1 knockdown decreased the NADPH/NADP+ ratio specifically in the cancer cell lines A549 and HCT116 (Fig. 1C and D).
Fig. 1.
Cancerous but not normal cells rely on ME1-mediated NADPH production.
A. Major NADPH-producing pathways.
B. Effect of NADPH-producing enzyme knockdown on the relative NADPH/NADP+ ratio in MCF-10A and MCF-7 cells (n = 4).
C. Effect of NADPH-producing enzyme knockdown on the relative NADPH/NADP+ ratio in MRC-5 and A549 cells (n = 4).
D. Effect of NADPH-producing enzyme knockdown on the relative NADPH/NADP+ ratio in NCM460 and HCT116 cells (n = 4).
E. Schematic illustration of the tracing of G6PD- and ME1-mediated NADPH production using [1-2H] glucose and [4-2H] glucose.
F. Comparison of G6PD- and ME1-mediated NADPH production in MCF-10A cells and MCF-7 cells after labeling with 10 mM [1-2H] glucose or 10 mM [4-2H] glucose for 60 min (n = 4).
G. Immunoblot analysis of ME1 expression in the indicated cell lines. β-Actin was used as the loading control.
H. Immunoblot analysis of ME1 expression in eight human breast cancer tissue biopsies (T) and adjacent noncancer tissues (N). β-Tubulin was used as the loading control.
Data are represented as mean ± SD. The experiments in B, C, D and F were conducted once, and all other experiments were repeated at least twice. Statistical significance was determined by Student's t-test. *p < 0.05, **p < 0.01, ***p < 0.001. See also Supplementary Fig. 1.
To validate whether the assayed cancer cells and normal cells prefer different NADPH production routes, we employed deuterium tracers to directly compare redox-active hydrogen labeling flux of NADPH mediated by the two major NADPH producers, G6PD and ME1, in MCF-7 and MCF-10A (Fig. 1E) [9,23,26]. Considering the redox-active hydrogen labeling of NADPH occurs very quickly (Supplementary Fig. 1B) and may undergo enzyme-catalyzed hydrogen-deuterium exchange with water during prolonged incubation [27], we performed a short-term labeling experiment and examined the labeled NADPH mass isotopomers when NADP+ remains unlabeled. Fig. 1F showed the preferential dependence of MCF-7 cells on ME1 and MCF-10A cells on G6PD for NADPH production. This distinction may be explained by the observation that the ME1 protein level was much higher in these representative cancer cells than in the assayed normal cells, while the protein levels of the other NADPH-producing enzymes showed negligible or inconsistent differences between the cancer and normal cells (Fig. 1G and Supplementary Fig. 1C). In addition, the ME1 protein level mostly increased (8 out of 12 biopsies) in examined breast cancer tissue biopsies compared with the adjacent normal tissues (Fig. 1H). Collectively, these data suggest that certain cancer cells preferentially depend on ME1-catalyzed NADPH production compared to the assayed normal cells, and such specific NADPH pathway usage can be exploited to target cancer cells.
3.2. ME1 is pivotal for supporting cancer cell growth
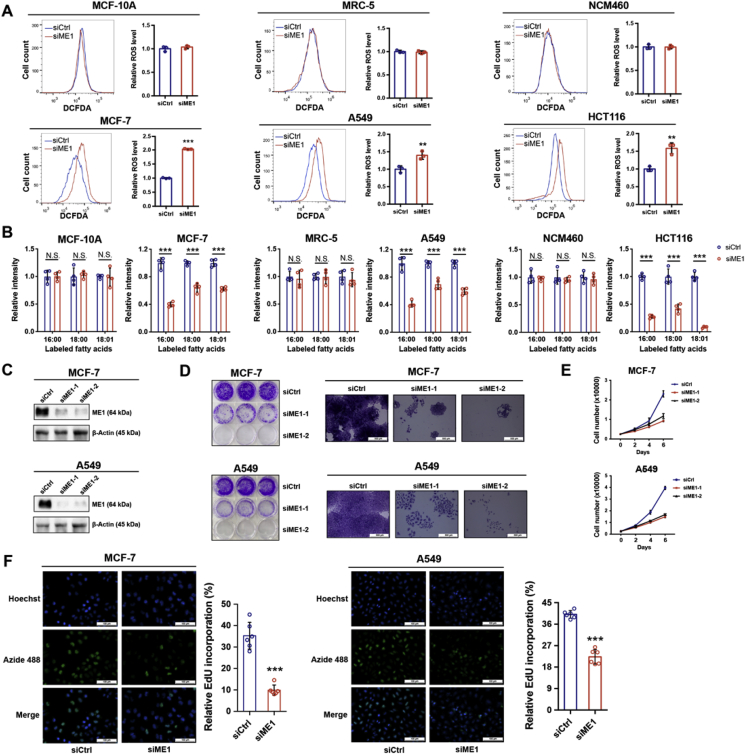
The metabolic tracing results revealed a stronger dependence of certain cancer cells on ME1 for NADPH production. It is thus anticipated that ME1 interference may selectively disrupts redox homeostasis and lipid biosynthesis in these cancer cells while sparing the normal cells. As expected, ME1 knockdown resulted in increased intracellular ROS levels in cancer cells including MCF-7, A549 and HCT116 but not in the examined non-transformed cells (Fig. 2A). NADPH is also important for de novo synthesis of fatty acids. Consistently, both saturated and unsaturated fatty acid synthesis was impaired in cancer cells but not in normal cells with ME1 knockdown as revealed by [U-13C] glucose tracing, implying the diminished fatty acid synthesis due to ME1-mediated NADPH loss in the examined cancer cells (Fig. 2B). These observations indicated an unmet demand for NADPH by rapidly proliferating cancer cells to withstand redox stress and support lipogenesis when ME1 was interfered.
Fig. 2.
ME1 is pivotal for supporting cancer cell growth.
A. Intracellular ROS levels in MCF-10A and MCF-7 cells, MRC-5 and A549 cells and NCM460 and HCT116 cells after ME1 knockdown (n = 3).
B. Effect of ME1 knockdown on labeled fatty acid levels in MCF-10A and MCF-7 cells, MRC-5 and A549 cells, and NCM460 and HCT116 cells cultured in medium containing 10 mM [U-13C] glucose for 72 h (n = 4).
C. Immunoblot analysis of ME1 knockdown efficiency using two specific siRNAs in MCF-7 and A549 cells.
D. Colony formation assay of MCF-7 and A549 cells with control transfection or ME1 knockdown. Scale bar, 500 μm.
E. Growth curve of MCF-7 and A549 cells with control transfection or ME1 knockdown (n = 3).
F. EdU incorporation assay of MCF-7 and A549 cells with control transfection or ME1 knockdown (n = 6). Scale bar, 100 μm.
Data are represented as mean ± SD. The experiments in B and F were conducted once, and all other experiments were repeated at least twice. Statistical significance was determined by Student's t-test. *p < 0.05, **p < 0.01, ***p < 0.001; N.S., nonsignificant. See also Supplementary Fig. 2.
In agreement with the above results, ME1 silencing (Fig. 2C) drastically inhibited the colony formation and impeded the proliferation of both MCF-7 and A549 cells (Fig. 2D and E). This was further confirmed by the EdU incorporation assay in MCF-7 and A549 cells (Fig. 2F) but not in the assayed normal cells (Supplementary Fig. 2A and B). Meanwhile, the diminished proliferation of MCF-7 and A549 cells after ME1 knockdown can be rescued by exogenous supplementation with fatty acids (Supplementary Fig. 2C). To further validate the role of ME1 in supporting the rapid proliferation of cancer cells, EGF was administered to promote proliferation [28]. ROS levels increased rapidly in an EGF dose-dependent manner, supporting that rapid cell proliferation is associated with increased production of intracellular ROS levels (Supplementary Fig. 2D and E). Importantly, ME1 was adaptively upregulated upon EGF treatment (Supplementary Fig. 2E), and ME1 silencing abrogated the EGF-promoted proliferation of both MCF-7 and A549 cells (Supplementary Fig. 2F). Collectively, these data indicate that ME1 is critical in maintaining NADPH levels for redox homeostasis and lipid biosynthesis, and thereby supports cancer cell proliferation.
3.3. ME1 regulates cancer cell migration and anchorage-independent survival
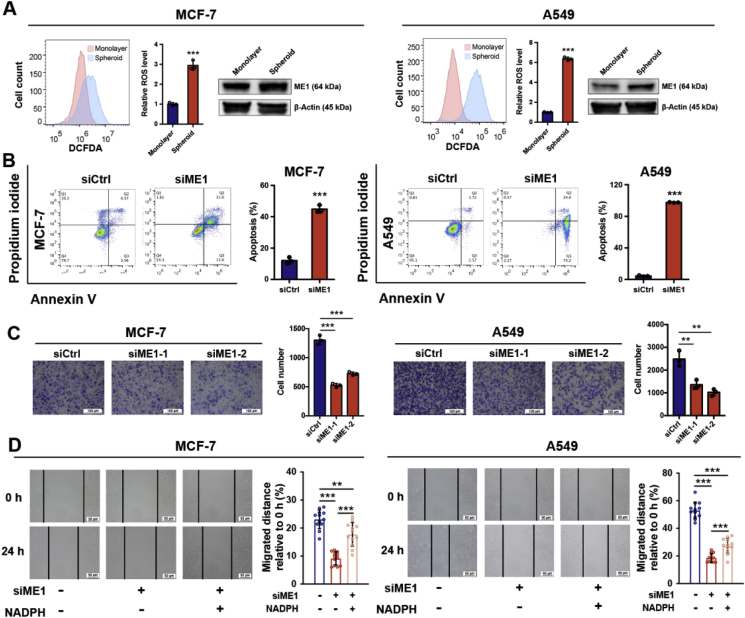
Metastasis is the major factor enhancing cancer malignancy and is often responsible for the failure of anticancer treatment [29,30]. During tumor metastasis, cancer cells must detach from the extracellular matrix (ECM) and overcome the increased ROS generated during this process to avoid anoikis [[31], [32], [33]]. Hence, anchorage-independent growth is a hallmark of cancer cells with migratory capacity. Thus, we hypothesized that ME1 might also be vital for the anchorage-independent survival and growth of cancer cells by maintaining redox homeostasis. As expected, compared with monolayer cells, spheroid MCF-7 and A549 cells were characterized by elevated ROS levels, accompanied by upregulated ME1 expression (Fig. 3A). ME1 knockdown significantly decreased the size of cancer cell spheroids (Supplementary Fig. 3A and B). In suspension culture, cells with control knockdown showed resistance to anoikis, while cells with ME1 knockdown showed significantly increased apoptosis (Fig. 3B), indicating that ME1 is important for the anchorage-independent survival of cancer cells. Moreover, the Transwell assay results showed that the fraction of ME1 knockdown cells passing through the membrane was reduced compared with the fraction of control cells (Fig. 3C). In addition, the wound healing assay results also revealed that ME1 knockdown cells migrated slower than control knockdown cells to close the gap of the scratch wound, while the impaired migratory ability was partially rescued by NADPH supplementation (Fig. 3D), indicating that reduced NADPH level induced by ME1 knockdown impacts on cancer cell migration. Altogether, these results confirm that ME1 is vital in supporting the anchorage-independent survival and migration of cancer cells and thereby cancer metastasis.
Fig. 3.
ME1 regulates cancer cell migration and anchorage-independent survival.
A. Intracellular ROS and ME1 protein levels in monolayer and spheroid MCF-7 and A549 cells (n = 3).
B. Apoptosis rate of cells with control transfection or ME1 knockdown cultured in suspension for 48 h (n = 3).
C. Transwell migration assay of MCF-7 and A549 cells with control transfection or ME1 knockdown (n = 3). Scale bar, 100 μm.
D. Wound healing assay of MCF-7 (n = 12) and A549 cells (n = 12) with control transfection, ME1 knockdown or ME1 knockdown with NADPH supplementation (200 μM for 24 h). Scale bar, 50 μm.
Data are represented as mean ± SD. The data represent at least two independent experiments. Statistical significance was determined by Student's t-test. **p < 0.01, ***p < 0.001. See also Supplementary Fig. 3.
3.4. ME1 interference adaptively upregulates IDH2 as a compensatory mechanism
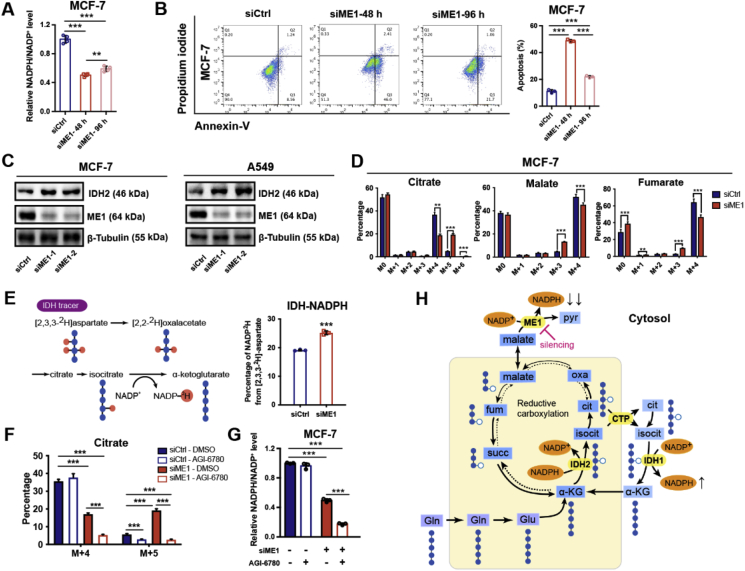
Considering that cancer cells have evolved powerful adaptability and that at least four pathways and dozens of enzymes contribute to NADPH production, we sought to determine whether cancer cells could recover from ME1 interference via adaptive activation of other enzymes for NADPH replenishment and thereby causes acquired resistance. To validate this assumption, we extended the cell culture time to 96 h. When ME1 was silenced in MCF-7 cells, the NADPH/NADP+ ratio gradually increased at 96 h compared with that detected at 48 h (Fig. 4A). In addition, detachment-induced anoikis decreased in cells with ME1 knockdown after 96 h compared with that detected after 48 h (Fig. 4B). These results indicate that cancer cells have adaptively developed a compensatory survival strategy to cope with the NADPH deficiency stress induced by ME1 knockdown, possibly by upregulating or activating other enzymes involved in NADPH production. We thus screened the expression of NADPH-producing enzymes after ME1 knockdown (Supplementary Fig. 4A) and found that IDH2 levels were markedly increased (Fig. 4C and Supplementary Fig. 4B and C). Previous studies have shown that reductive glutamine metabolism mediated by cytosolic IDH1 is an important metabolic reprogramming mechanism in replenishing mitochondrial NADPH pool to combat oxidative stress [24]. Therefore, we proposed that the adaptive upregulation of mitochondrial IDH2 may lead to increased reductive glutamine metabolism to compensate for the cytosolic NADPH loss induced by ME1 knockdown. To test this hypothesis, we used [U-13C] glutamine in stable isotope tracing experiments to analyze the flux of reductive glutamine metabolism in MCF-7 cells (Supplementary Fig. 4D). We noted that, after ME1 knockdown, the m+4 fraction of citrate produced from oxidative glutamine metabolism was reduced, while the m+5 fraction contributed by reductive carboxylation of glutamine was increased. Citrate m+5 produces acetyl-CoA m+2 and oxaloacetate m+3, which further produces fumarate m+3 and malate m+3. Consistently, the malate m+3 and fumarate m+3 fractions generated from reductive glutamine metabolism were also increased in ME1 knockdown cells (Fig. 4D). In addition to MCF-7 cells, we also found a decreased citrate m+4 and increased citrate m+5 fraction after ME1 knockdown in A549 cells (Supplementary Fig. 4E).
Fig. 4.
ME1 interference adaptively upregulates IDH2-mediated NADPH production as a compensatory mechanism.
A. Effect of ME1 knockdown on the relative NADPH/NADP+ ratio in MCF-7 cells cultured for prolonged durations (n = 4).
B. Apoptosis rate of ME1 knockdown cells cultured in suspension at the indicated times (n = 3).
C. Immunoblot analysis of ME1 and IDH2 expression in MCF-7 and A549 cells under the indicated treatments. β-Tubulin was used as the loading control.
D. Analysis of citrate, fumarate and malate mass isotopologues in MCF-7 cells cultured in medium containing 2 mM [U-13C] glutamine for 24 h (n = 5).
E. Percentage of NADP2H labeling from [2,3,3-2H] aspartate (2 mM, 24 h) in MCF-7 cells with control transfection or ME1 knockdown (n = 3).
F. Citrate m+4 and m+5 isotopologues in MCF-7 cells treated as indicated and cultured in medium containing 2 mM [U-13C] glutamine for 24 h (n = 3 for the siME1 DMSO group and n = 5 for others). The concentration of AGI-6780 used in this study is 2.5 μM.
G. Effect of combinatorial targeting ME1 and IDH2 (2.5 μM AGI-6780 for 24 h) on relative NADPH/NADP+ ratio in MCF-7 cells (n = 3).
H. Mitochondrial reductive carboxylation and subsequent isocitrate/citrate transport is adaptively upregulated by ME1 knockdown and functions as a compensatory route to replenish the cytosolic NADPH pool.
Data are represented as mean ± SD. The experiments described in A and F were conducted once, and all other experiments were repeated at least twice. Statistical significance was determined by Student's t-test. *p < 0.05, **p < 0.01, ***p < 0.001. See also Supplementary Fig. 4.
Together with the above evidences of an adaptive increase in reductive carboxylation after ME1 knockdown, flux analysis with the tracer [2,3,3-2H] aspartate confirmed that cells produced more NADPH via the IDH-mediated route than the control knockdown cells (Fig. 4E), implying that IDH2 upregulation is responsible for mitigating the ME1-mediated NADPH stress. To establish the causal link, we inhibited IDH2 using AGI-6780 [34], and found the inhibition significantly diminished the adaptive increase of the citrate m+5, malate m+3 and fumarate m+3 fractions in ME1-knockdown MCF-7 cells (Fig. 4F and Supplementary Fig. 4F). Expectedly, IDH2 inhibition in ME1 knockdown cells led to an almost depleted NADPH pool, reiterating the pivotal role of mitochondria-cytosol citrate/isocitrate shuttle in replenishing the cytosolic NADPH pool upon ME1 interference (Fig. 4G and Supplementary Fig. 4G). Silencing both ME1 and IDH2 phenocopied the pronounced NADPH/NADP+ reduction in MCF-7 and A549 cells with ME1 knockdown and IDH2 inhibition (Supplementary Fig. 4H). Moreover, since the IDH-dependent NADPH compensation also requires the transport of citrate between the cytosol and mitochondria [35,36], we silenced the citrate transporter protein (CTP, SLC25A1) and indeed found the knockdown blocked the IDH-mediated NADPH supplementary route (Supplementary Fig. 4I), which ultimately resulted in marked decrease of NADPH/NADP+ level (Supplementary Fig. 4J). Together, these results revealed that cancer cells with cytosolic ME1 knockdown developed a compensatory mechanism by adaptively upregulating IDH2 and subsequently increasing isocitrate/citrate produced reductively in mitochondria that then enters the cytosol and participates in NADPH generation to sustain the cytosolic NADPH supply and suppress ROS. This compensatory change underpins the acquired resistance to ME1 interference (Fig. 4H).
3.5. The AMPK-FoxO1 axis underlies adaptive IDH2 upregulation
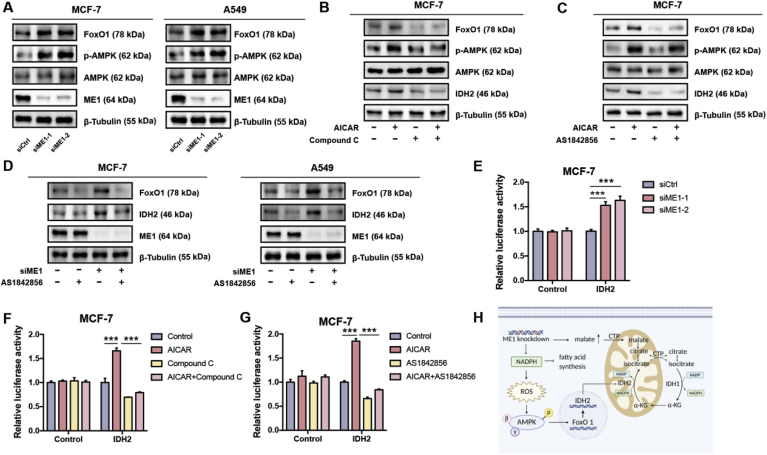
Since both the mRNA and protein levels of IDH2 were adaptively enhanced upon ME1 knockdown, we speculated that this change is transcriptionally regulated. We searched for potential transcription factors with IDH2-binding capacity in the GenoMatix database and found FoxO1 response elements within the IDH2 promoter region (Supplementary Fig. 5A). Interestingly, both the mRNA and protein levels of FoxO1 were elevated after ME1 knockdown (Fig. 5A and Supplementary Fig. 5B). We next investigated the mechanistic link between ME1 knockdown and FoxO1 upregulation. Previous studies demonstrated that increased ROS levels can lead to AMPK activation [37,38], an established indicator of metabolic stress. Activated AMPK further promote FoxO1 translocation to the nucleus for transcriptional activity [[39], [40], [41]]. We thus investigated whether AMPK activation is involved in ME1 knockdown-induced FoxO1 upregulation. As expected, ME1 knockdown led to drastic AMPK activation (Fig. 5A), while N-acetylcysteine (NAC) treatment eliminated ROS and largely abolished the regulation of downstream effectors, as evidenced by the decreased phosphorylated AMPK, FoxO1 and IDH2 levels (Supplementary Fig. 5C). Consistent with these results, administration of AICAR, an AMPK activator, increased the expression of FoxO1 and IDH2 at both the mRNA level and protein levels and promoted nuclear translocation of FoxO1 (Fig. 5B and Supplementary Fig. 5D and E). In contrast, when compound C, an AMPK inhibitor, was coadministered with AICAR, the effect of AICAR was largely abrogated (Fig. 5B and Supplementary Fig. 5D and E). In addition, treatment with the FoxO1 inhibitor AS1842856 abrogated AICAR- and ME1 knockdown-induced adaptive upregulation of IDH2 (Fig. 5C and D and Supplementary Fig. 5F) and diminished AICAR-induced nuclear translocation of FoxO1 (Supplementary Fig. 5G), suggesting that acquired resistance to ME1 silencing was driven by activation of the AMPK-FoxO1-IDH2 pathway. In further support of the transcriptional regulation of IDH2 by FoxO1 following AMPK activation, we found that, apart from IDH2, two known target genes of FoxO1, MnSOD and p21 [39,42], were increased at mRNA level after ME1 knockdown as well (Supplementary Fig. 5B). Moreover, such upregulation can also be induced by AICAR treatment, and further blocked by either coadministering compound C or AS1842856 with AICAR, respectively (Supplementary Fig. 5D and 5F). To further validate this transcriptional mechanism, we cloned 0.8 kb of genomic DNA upstream of the transcription start site of the IDH2 gene into a luciferase reporter plasmid and performed a luciferase reporter assay. ME1 knockdown significantly activated the luciferase activity of the IDH2 promoter (Fig. 5E). In addition, AICAR increased the luciferase activity of the IDH2 promoter, whereas this increase was revoked by compound C and AS1842856 (Fig. 5F and G). Moreover, treatment with AS1842856 pronouncedly reduced NADPH/NADP+ in ME1-deficient MCF-7 and A549 cells, phenocopying the effect induced by ME1 knockdown and IDH2 inhibition (Supplementary Fig. 5H). Collectively, these results indicate that IDH2 is transcriptionally upregulated upon ME1 knockdown through the AMPK-FoxO1 signaling pathway (Fig. 5H).
Fig. 5.
The AMPK-FoxO1 axis underlies adaptive IDH2 upregulation.
A. Immunoblot analysis of FoxO1, p-AMPK and AMPK expression in MCF-7 and A549 cells with control transfection or ME1 knockdown. β-Tubulin was used as the loading control.
B. Immunoblot analysis of p-AMPK, AMPK, FoxO1 and IDH2 in MCF-7 cells treated with AICAR (1 mM), compound C (5 μM) or both for 24 h. β-Tubulin was used as the loading control.
C. Immunoblot analysis of p-AMPK, AMPK, FoxO1 and IDH2 in MCF-7 cells treated with AICAR (1 mM), AS1842856 (0.2 μM) or both for 24 h. β-Tubulin was used as the loading control.
D. Immunoblot analysis of FoxO1 and IDH2 in MCF-7 cells with control transfection or ME1 knockdown and treated with or without AS1842856 (0.2 μM) for 24 h. β-Tubulin was used as the loading control.
E. Dual-luciferase reporter assay in MCF-7 cells with control transfection or ME1 knockdown (n = 3).
F. Dual-luciferase reporter assay in MCF-7 cells treated with AICAR (1 mM), compound C (5 μM) or both for 24 h (n = 3).
G. Dual-luciferase reporter assay in MCF-7 cells treated with AICAR (1 mM), AS1842856 (0.2 μM) or both for 24 h (n = 3).
H. The pathway through which IDH2 is activated by ME1 knockdown in a manner dependent on AMPK-FoxO1 can contribute to the replenishment of the cytosolic NADPH pool to reduce redox stress and support lipogenesis.
Data are represented as mean ± SD. The data represent at least two independent experiments. Statistical significance was determined by Student's t-test. ***p < 0.001. See also Supplementary Fig. 5.
3.6. Combined interference of ME1 and IDH2 synergistically induce cancer cell apoptosis
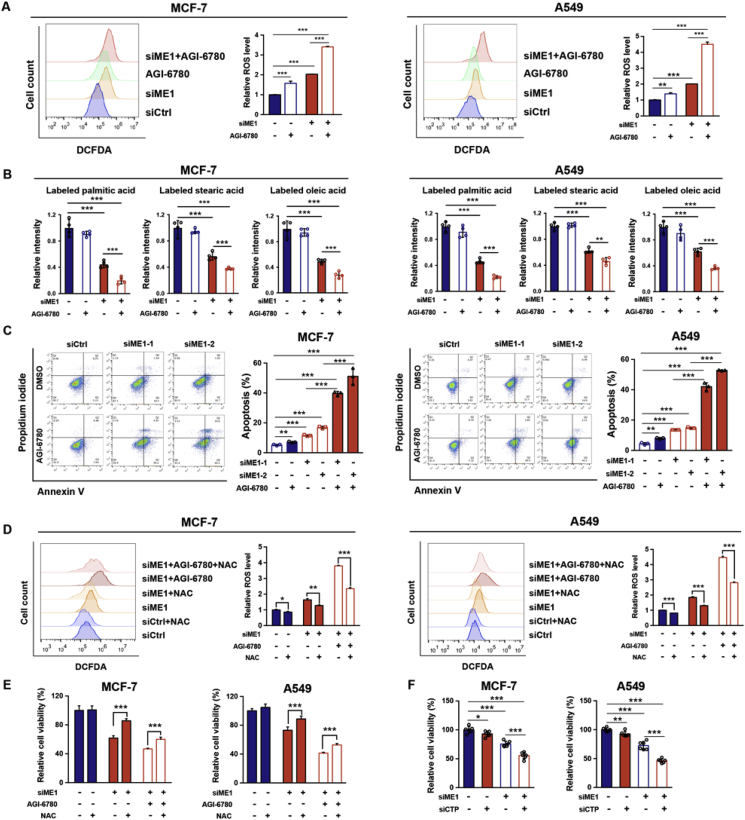
Although the level of G6PD, in addition to that of IDH2, was adaptively increased after ME1 knockdown (Supplementary Fig. 4A), previous studies and our data indicated that G6PD is essential for maintaining NADPH/NADP+ homeostasis in both the assayed normal cells and cancer cells, while IDH2 knockdown has little influence in the examined normal cells (Fig. 1B–D). Consistently, treatment with RRx-001, a G6PD inhibitor, exhibited non-specific toxicity to the examined normal and cancer cells (Supplementary Fig. 6A). Hence, we proposed a rationale of combining IDH2 rather than G6PD inhibition with ME1 ablation for targeting NADPH and thereby triggering apoptosis in cancer cells. Notably, ME1 knockdown alone increased intracellular ROS levels despite no significant change in mitochondrial ROS (Supplementary Fig. 6B and Fig. 2A). To further determine whether the compensatory upregulation of mitochondrial IDH2 mitigates the cytosolic ROS, ME1-knockdown cells were treated with IDH2 inhibitor, AGI-6780. This cotreatment significantly increased both the mitochondrial and total intracellular ROS levels compared to those in ME1 knockdown cells (Fig. 6A and Supplementary Fig. 6B). Mitochondrial outer membrane permeability was also pronouncedly enhanced when compared to cells with interference of single enzyme (ME1/IDH2) as indicated by JC-1 staining (Supplementary Fig. 6C). Accordingly, a marked disruption in fatty acid synthesis was observed after cotreatment (Fig. 6B). Consequently, combining IDH2 inhibitor treatment with ME1 knockdown drastically increased the apoptosis of both MCF-7 and A549 cells due to NADPH depletion-induced ROS accumulation and lipogenesis disruption (Fig. 6C). Consistently, knockdown of both ME1 and IDH2 also substantially decreased cell viability (Supplementary Fig. 6D). The increases of ROS level and apoptotic rate were partially rescued by pretreating the cells with NAC, a ROS scavenger (Fig. 6D and E). Apart from targeting IDH2, knockdown of CTP phenocopied the effects of IDH2 inhibition in synergizing with ME1 knockdown to increase cytotoxicity in both MCF-7 and A549 cells (Fig. 6F). In addition, inhibition of IDH1, another essential enzyme involved in mitochondrial-cytosol citrate/isocitrate shuttle, with GSK864 in ME1 knockdown cells also decreased the cell viability (Supplementary Fig. 6E). Importantly, co-interference of ME1 and IDH2 did not affect the viability of MCF-10A and MRC-5 cells (Supplementary Fig. 6F). These results indicate that combinatorial targeting of ME1 and IDH2 can induce apoptosis in certain cancer cells while sparing the normal cells that do not primarily rely on ME1-mediated NADPH production.
Fig. 6.
ME1 and IDH2 synergistically induce cancer cell apoptosis.
A. Effect of IDH2 inhibition by 2.5 μM AGI-6780 for 24 h, ME1 knockdown and the combined treatment on intracellular ROS levels in MCF-7 and A549 cells (n = 3).
B. Effect of IDH2 inhibition by 2.5 μM AGI-6780 for 24 h, ME1 knockdown and the combined treatment on labeled fatty acid abundance levels in MCF-7 and A549 cells cultured in medium containing 10 mM [U-13C] glucose for 72 h (n = 4).
C. Effect of IDH2 inhibition by 2.5 μM AGI-6780 for 24 h, ME1 knockdown and the combined treatment on apoptosis rate in MCF-7 and A549 cells (n = 3).
D. Intracellular ROS levels in MCF-7 and A549 cells after ME1 knockdown and dual targeting of ME1 and IDH2 in culture with or without 10 mM NAC treatment for 24 h (n = 3).
E. Relative viability of MCF-7 and A549 cells after ME1 knockdown and dual targeting of ME1 and IDH2 in culture with or without 10 mM NAC treatment for 24 h (n = 5).
F. Effect of combinatorial targeting ME1 and CTP on relative cell viability of MCF-7 and A549 cells (n = 6).
Data are represented as mean ± SD. The experiments in B were conducted once, and all the other experiments were repeated at least twice. Statistical significance was determined by Student's t-test. **p < 0.01, ***p < 0.001. See also Supplementary Fig. 6.
3.7. Dual targeting of ME1 and IDH2 inhibits tumor growth and metastasis
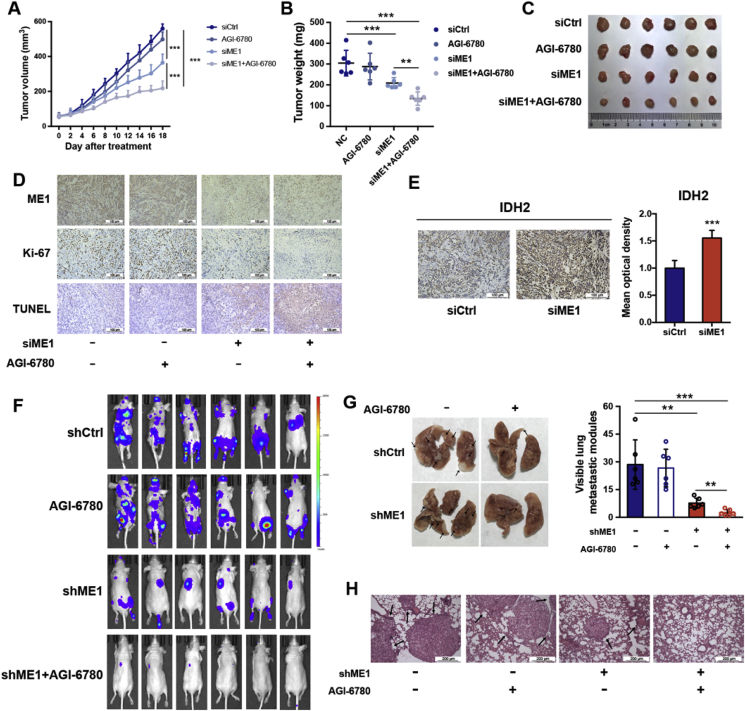
We next sought to determine whether the dual targeting strategy is also effective in vivo. ME1 siRNA was directly injected into A549 xenografts every other day in combination with daily intragastric administration of the IDH2 inhibitor AGI-6780 (60 mg/kg) for combinatorial targeting of ME1 and IDH2. Remarkably, this dual targeting strategy significantly slowed tumor growth (Fig. 7A–C) as compared with targeting single enzyme alone. The intratumorally-injected ME1 siRNA successfully diminished ME1 protein level in tumors indicated by IHC staining (Fig. 7D). The in vivo antitumoral effect by dual interference with ME1 and IDH2 was further confirmed by Ki-67 staining and TUNEL assays that show repressed cell proliferation and increased apoptosis (Fig. 7D). Moreover, consistent with the results in cultured cells, the protein level of IDH2 in A549 tumor xenografts was drastically increased in response to ME1 knockdown, demonstrating the physiological relevance of ME1-IDH2 pathways in replenishing NADPH (Fig. 7E).
Fig. 7.
ME1 and IDH2 synergistically inhibit cancer growth and metastasis.
A. Tumor volume progression in mice subcutaneously xenografted with A549 cells treated as indicated (n = 6). siME1s (5nmol/animal) were injected intratumorally every other day and AGI-6780 (60 mg/kg) was administrated orally everyday.
B. Tumor weights in mice subcutaneously xenografted with A549 cells treated as indicated (n = 6).
C. Photograph and size comparison of excised tumors (n = 6).
D. Representative ME1, Ki-67 immunostaining and TUNEL in xenografted tumors (n = 6). Scale bar, 100 μm.
E. Representative immunohistochemical staining of IDH2 in xenograft tumors with in vivo ME1 knockdown (n = 6). Scale bar, 100 μm.
F. Bioluminescence images of mice treated as indicated were obtained on day 60. AGI-6780 (60 mg/kg) was administrated orally everyday.
G. Left, representative images of metastatic nodules on the surface of lungs in nude mice. Right, metastatic nodules were counted and recorded with or without a microscope (n = 6).
H. Representative images of H&E-stained lung sections (n = 6). Scale bar, 200 μm.
Data are represented as mean ± SD. Statistical significance was determined by Student's t-test. **p < 0.01, ***p < 0.001.
Since ME1 interference alone resulted in resistance to anchorage-independent apoptosis, we proposed that dual targeting of ME1 and IDH2 would lead to a synergistic effect in impeding tumor metastasis. To test this hypothesis, A549-shCtrl or A549-shME1 cells were injected into the tail vein of nude mice, and these two groups of mice were further separated into groups treated either with or without AGI-6780. The resulting four groups of mice were sacrificed at 60 days post injection to examine the metastatic nodules formed on the lung surfaces. Despite significant inhibition of metastasis, shME1 mice still exhibited metastatic nodules, whereas dual targeting of ME1 and IDH2 inhibited tumor metastasis (Fig. 7F and G). Hematoxylin and eosin (H&E) staining further validated this synergistic effect in vivo (Fig. 7H). Collectively, our results strongly indicate that dual targeting of ME1 and IDH2 is a promising therapeutic strategy to combat tumor growth and metastasis.
4. Discussion
Metabolic reprogramming that favors uncontrolled proliferation and metastasis is a prominent hallmark of tumor development [6,43]. Substantial efforts have thus been made to uncover potential targetable nodes responsible for the metabolic adaptation of cancer cells [44,45]. NADPH production is one of these important metabolic nodes and represents a central hub connecting oncogene activation, metabolic reprogramming, and tumor growth and metastasis [46,47]. Notably, a global landscape of cancer cell line metabolism unveiled NADPH increase in relation to genetic events such as KEAP1 mutation, implying the essentiality of NADPH's buffering capacity against redox stress [11]. Thus, enzymes involved in the generation of NADPH including ME, IDH, G6PD, 6PGD and MTHFD have been exploited for their potential as druggable therapeutic targets and holds promise for cancer therapy [10,[13], [14], [15], [16]]. However, the mechanism by which cancer cells and normal cells differentially exploit NADPH-producing pathways and the optimal approaches to prevent the highly possible resistance to therapeutic interventions targeting a single metabolic enzyme are unclear, considering that cancer cells are well recognized for their powerful adaptability and metabolic plasticity. Here, we show that compared with the assayed normal cells, cancer cells preferentially depend on ME1-mediated production of NADPH to support uncontrolled growth and metastasis. ME1 interference slows tumor growth and inhibits metastasis while sparing normal cells; however, it results in resistance upon prolonged cell culture by adaptively increasing mitochondrial IDH2-mediated NADPH production as a compensatory mechanism in an AMPK-FoxO1-dependent manner. Combinatorial interference with ME1 and IDH2 thus leads to synergistic inhibition of NADPH production and thereby greatly impedes tumor growth and metastasis.
Selectively and specifically targeting cancer cells while sparing normal cells is an important requirement for exploiting metabolic vulnerabilities for anticancer therapy [48]. At least four metabolic pathways and dozens of enzymes are involved in NADPH production. A critical question is thus how tumor cells and normal cells differentially exploit these pathways and enzymes. Based on siRNA screening, metabolic tracing and functional validation, we identified a preferential usage of ME1-mediated NADPH production route for cancer cells using three established human cancer cell lines and three human non-transformed cell lines. This identification allows ME1 to be harnessed as a viable target that can selectively kill cancer cells while sparing normal cells. Our results agree with those of previous studies indicating that repression of ME1 leads to oxidative stress, suppressed cell growth, attenuated migratory capacity and remodeled cellular metabolism in several types of cancer cells, such as nasopharyngeal, colorectal and bladder cancer cells [20,[49], [50], [51], [52], [53]]. In addition, increased ME1 expression and activity are associated with poor prognosis in breast cancer and hepatocellular carcinoma and with radiation resistance in lung cancer [[54], [55], [56]]. Taken together, our results and previous findings support ME1 as a promising target specific for slowing tumor growth while sparing normal cells.
Consistent with previous quantitative flux analysis results [9], we noted substantial contribution from both G6PD and ME1 to NADPH production in cancer cell lines, including MCF-7, A549 and HCT116 cells. It is thus not surprising that previous findings indicated that disrupting G6PD also reduced NADPH and GSH levels, increased ROS levels [10], and ultimately promoted chemotherapy-induced apoptosis in cell-based xenografts and PDX models in colorectal cancer [13]. However, a critical concern raised by our findings is that G6PD, unlike ME1, is also indispensable for maintaining redox homeostasis in the assayed normal cells. Its physiological importance has been demonstrated by embryonic lethality after homozygous deletion of G6PD [57], and reiterated by a recent study showing pronounced defects in folate metabolism upon G6PD interference [10]. Although we cannot exclude the possibility that ME1 is also important for the function of other normal cells, our data indicate that these subtypes of cancer cells rely on ME1 preferentially to G6PD for NADPH production to a much higher extent than the examined normal cells. In addition to its effect on NADPH generation to maintain redox homeostasis and lipid synthesis, ME1 can also negatively regulate the key tumor suppressor p53 by promoting its MDM2-dependent degradation [20]. Thus, it is reasonable to expect that targeting ME1 may confer another advantage by activating p53 to inhibit tumor growth.
Although previous results together with these findings indicate that ME1 interference can slow cancer cell proliferation [50,53], resistance to ME1 silencing was observed in cancer cells cultured for extended durations, suggesting that cancer cells may adaptively upregulate or activate other enzymes involved in NADPH production. As expected, screening for changes in enzyme expression levels showed that cytosolic G6PD and mitochondrial IDH2 levels were significantly enhanced. This compensatory mechanism by increasing NADPH production may thus result in resistance to ROS-triggered cell death and may partially restore the proliferative capacity of cancer cells. We thus investigated whether combinatorial targeting of ME1 and IDH2 could lead to a synergistic effect on reducing tumor growth and metastasis. Although the influence of IDH2 inhibition alone was negligible, ME1 silencing combined with IDH2 inhibition dramatically slowed tumor growth, triggered apoptotic cell death, and completely inhibited metastasis, indicating that combinatorial targeting of ME1 and IDH2 is a promising therapeutic strategy for cancer types that are sensitive to NADPH interference.
Cytosolic IDH1 and mitochondrial IDH2 function in the bidirectional conversion of α-ketoglutarate and isocitrate [58]. Therefore, these enzymes play important roles in balancing the transfer of NADPH between the cytoplasm and mitochondria, considering that NADPH per se cannot be transported between these two compartments and relies on the isocitrate/citrate transport. Consistent with previous findings [24], we found that IDH2 minimally affected the growth and survival of cells in monolayer culture. Thus, IDH2 likely functions as a sophisticated backup mechanism that is activated only under stress conditions, such as those encountered after ME1 silencing or during detachment. Silencing of cytosolic ME1 resulted in an increase in cytosolic but not mitochondrial ROS levels. These results can be explained by the adaptive upregulation of mitochondrial IDH2, which may help cancer cells to withstand the ME1 knockdown-induced redox challenge, since IDH2 inhibition drastically increased mitochondrial ROS levels in ME1-silenced cells. Notably, previous studies have also shown that nicotinamide nucleotide transhydrogenase (NNT) transfers hydride from NADH to NADP+ to supply the mitochondrial NADPH required by IDH2 for reductive carboxylation [59,60]. It is thus important to determine in future studies whether the NNT-mediated NADPH formation is also adaptive to cytosolic ME1 knockdown, which is necessary in supporting increased NADPH consumption by the upregulated IDH2 and maintaining the redox balance in mitochondria. This explains the constant mitochondrial ROS levels in MCF-7 and A549 cells following the ME1 knockdown as shown in Supplementary Fig. 6B.
Although most previous reports focused on the gain-of-function of mutated IDH2 in producing oncometabolite 2-HG [34,61], our study highlights that the wild type IDH2, integrated with cytosolic ME1, is essential for tumor growth and metastasis. In further support of the compensatory role of IDH2-mediated NADPH transport between the cytosol and mitochondria, CTP knockdown largely phenocopied the effect of IDH2 inhibition and silencing. Previous findings indicated that the increased mitochondrial ROS induced by cell detachment or complex I inhibition activates the cytosolic reductive carboxylation pathway, soliciting the transport of isocitrate/citrate into mitochondria and thereby supplementing the mitochondrial NADPH pool to mitigate mitochondrial redox stress [24,62,63]. In contrast, our results indicate that in conditions of ME1 interference, the accumulated malate and the facilitated conversion of α-KG to isocitrate mediated by the upregulated IDH2 may contribute to replenish the cytosolic NADPH pool via the CTP-mediated citrate/isocitrate shuttle. In addition to the adaptive upregulation of other NADPH producers, the decreased NADPH pool from the deletion of NADPH-producing enzymes may be also restored by increasing the metabolic flux of other compensatory pathways [10]. Collectively, these results strongly indicate that targeting one enzyme may not be sufficient for limiting NADPH in cancer cells. Moreover, our results together with previous findings [24] support that the allocation and transport of NADPH between mitochondria and cytosol are the key to sustaining the proliferation and anchorage-independent survival of certain NADPH-sensitive cancer cells and that approaches for disrupting this transport can be exploited for cancer therapy.
Mechanistically, the increased cytosolic ROS level due to ME1 knockdown can activate AMPK [37], which promotes nuclear translocation of FoxO1 [39,41,64]. The transcriptionally active FoxO1 then upregulates the expression of IDH2, a newly identified target gene of FoxO1. Thus, the AMPK-FoxO1 pathway underscores why increased cytosolic ROS level adaptively upregulates a mitochondrial enzyme for NADPH compensation. Previously, AMPK activation in response to glucose deprivation or G6PD knockdown has been demonstrated to be crucial for NADPH maintenance by inhibiting acetyl-CoA carboxylases to decrease NADPH consumption involved in fatty acid synthesis [65]. These two signaling pathways both ramified from AMPK activation for NADPH compensation represent redundant metabolic adaptation machinery equipped by cancer cells when they encounter energy stress [66,67].
Because of the close connection of mitochondrial to cytosolic production of NADPH in supporting tumor growth and metastasis, we suppose that combined interference of cytosolic together with mitochondrial enzymes, as the case for ME1 and IDH2, would represent promising NADPH targeting approach to cancer therapy. Nevertheless, since cancer cells have evolved powerful adaptability, disrupting the function of a single gene/protein may be insufficient, and resistance to almost all kinds of targeted therapies has been observed [68,69]. Recent evidence indicates that targeting metabolic nodes also tends to cause acquired metabolic resistance [70,71]. For instance, PDAC cells treated with glutaminase (GLS) inhibitors were found to switch to GLS-independent glutamate production to fuel glutamine metabolism [72]. We believe that this risk also applies to targeting the NADPH metabolic network, particularly considering that at least four pathways and dozens of enzymes are involved in NADPH production. Here, we propose that combinatorial targeting of cytosolic ME1 and mitochondrial IDH2 is promising as a widely applicable NADPH-targeting strategy for certain cancer types. Nevertheless, a global study on cell type, tissue and environmental stimulus-dependent NADPH pathway usage is important for evaluating the translational value of the ME1-IDH2 targeting strategy. In addition, an IDH2 inhibitor is already available; despite being designed to inhibit mutated IDH2, it is also effective against wild-type IDH2. Therefore, the initial discovery of an inhibitor that dually targets ME1 and IDH2, currently underway in the authors’ laboratory, will ultimately allow clinical translation of the present findings.
Data availability
The raw data for immunoblotting can be accessed via https://doi.org/10.17632/d2gb5wz88r.1 on Mendeley Data.
Author contributions
C.S., H.Y., G.W. and H.H. conceived all the experiments. Y.D. and W.Y. carried out the human studies. C.S, Q.B., W.L. and W.Y. carried out the animal studies. C.S., W.L., H.Y., T.Y. and G.W. performed the experiments and interpreted the data. C.S., H.Y. and H.H. wrote the manuscript. H.Y., G.W. and H.H. supervised the project.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the National Key R&D Program of China (2018YFD0901101), National Natural Science Foundation of China (grants 81930109, 81720108032 to H.H., grants 81872838 to H.Y.), the Natural Science Foundation of Jiangsu Province (BK20180079), the Project for Major New Drug Innovation and Development (grants 2018ZX09711001-002-003 and 2018ZX09711002-001-004), Overseas Expertise Introduction Project for Discipline Innovation (grant G20582017001 to H.H.), Double-First Class Initiative Project (grants CPU2018GF09 to H.H., CPU2018GY09 to H.Y.), Sanming Project of Medicine in Shenzhen (SZSM201801060) and International Industrial Technology Research Collaboration of Nanjing (201911008).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101685.
Contributor Information
Hui Ye, Email: cpuyehui@cpu.edu.cn.
Haiping Hao, Email: haipinghao@cpu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Frisch S.M., Screaton R.A. Anoikis mechanisms. Curr. Opin. Cell Biol. 2001;13(5):555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 2.Simpson C.D., Anyiwe K., Schimmer A.D. Anoikis resistance and tumor metastasis. Canc. Lett. 2008;272(2):177–185. doi: 10.1016/j.canlet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- 3.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg Effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishikawa M. Reactive oxygen species in tumor metastasis. Canc. Lett. 2008;266(1):53–59. doi: 10.1016/j.canlet.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 5.Chio I.I.C., Tuveson D.A. ROS in cancer: the burning question. Trends Mol. Med. 2017;23(5):411–429. doi: 10.1016/j.molmed.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pavlova Natalya N., Thompson Craig B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23(1):27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulze A., Harris A.L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491(7424):364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 8.Mohammadi F., Soltani A., Ghahremanloo A., Javid H., Hashemy S.I. The thioredoxin system and cancer therapy: a review. Canc. Chemother. Pharmacol. 2019;84:925–935. doi: 10.1007/s00280-019-03912-4. [DOI] [PubMed] [Google Scholar]
- 9.Fan J., Ye J., Kamphorst J.J., Shlomi T., Thompson C.B., Rabinowitz J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510(7504):298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L., Zhang Z., Hoshino A., Zheng H.D., Morley M., Arany Z., Rabinowitz J.D. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1(3):404–415. [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Ning S., Ghandi M., Kryukov G.V., Gopal S., Deik A., Souza A., Pierce K., Keskula P., Hernandez D., Ann J., Shkoza D., Apfel V., Zou Y., Vazquez F., Barretina J., Pagliarini R.A., Galli G.G., Root D.E., Hahn W.C., Tsherniak A., Giannakis M., Schreiber S.L., Clish C.B., Garraway L.A., Sellers W.R. The landscape of cancer cell line metabolism. Nat. Med. 2019;25(5):850–860. doi: 10.1038/s41591-019-0404-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Olszewski K., Zhang Y., Lim E.W., Shi J., Zhang X., Zhang J., Lee H., Koppula P., Lei G., Zhuang L., You M.J., Fang B., Li W., Metallo C.M., Poyurovsky M.V., Gan B. Cystine transporter regulation of pentose phosphate pathway dependency and disulfide stress exposes a targetable metabolic vulnerability in cancer. Nat. Cell Biol. 2020;22(4):476–486. doi: 10.1038/s41556-020-0496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ju H.Q., Lu Y.X., Wu Q.N., Liu J., Zeng Z.L., Mo H.Y., Chen Y., Tian T., Wang Y., Kang T.B., Xie D., Zeng M.S., Huang P., Xu R.H. Disrupting G6PD-mediated redox homeostasis enhances chemosensitivity in colorectal cancer. Oncogene. 2017;36(45):6282–6292. doi: 10.1038/onc.2017.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calvert A.E., Chalastanis A., Wu Y., Hurley L.A., Kouri F.M., Bi Y., Kachman M., May J.L., Bartom E., Hua Y., Mishra R.K., Schiltz G.E., Dubrovskyi O., Mazar A.P., Peter M.E., Zheng H., James C.D., Burant C.F., Chandel N.S., Davuluri R.V., Horbinski C., Stegh A.H. Cancer-associated IDH1 promotes growth and resistance to targeted therapies in the absence of mutation. Cell Rep. 2017;19(9):1858–1873. doi: 10.1016/j.celrep.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ju H., Lu Y., Chen D., Zuo Z., Liu Z., Wu Q., Mo H., Wang Z., Wang D., Pu H., Zeng Z., Li B., Xie D., Huang P., Hung M., Chiao P.J., Xu R. Modulation of redox homeostasis by inhibition of MTHFD2 in colorectal cancer: mechanisms and therapeutic implications. J. Natl. Cancer Inst. 2019;111(6):584–596. doi: 10.1093/jnci/djy160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey P., Baddour J., Muller F., Wu C.C., Wang H., Liao W.-T., Lan Z., Chen A., Gutschner T., Kang Y., Fleming J., Satani N., Zhao D., Achreja A., Yang L., Lee J., Chang E., Genovese G., Viale A., Ying H., Draetta G., Maitra A., Wang Y.A., Nagrath D., DePinho R.A. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature. 2017;542(7639):119–123. doi: 10.1038/nature21052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Son J., Lyssiotis C.A., Ying H., Wang X., Hua S., Ligorio M., Perera R.M., Ferrone C.R., Mullarky E., Shyh-Chang N., Kang Y., Fleming J.B., Bardeesy N., Asara J.M., Haigis M.C., DePinho R.A., Cantley L.C., Kimmelman A.C. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496(7443):101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine A.J., Puzio-Kuter A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 19.Audet-Walsh É., Papadopoli D.J., Gravel S.-P., Yee T., Bridon G., Caron M., Bourque G., Giguère V., St-Pierre J. The PGC-1α/ERRα axis represses one-carbon metabolism and promotes sensitivity to anti-folate therapy in breast cancer. Cell Rep. 2016;14(4):920–931. doi: 10.1016/j.celrep.2015.12.086. [DOI] [PubMed] [Google Scholar]
- 20.Jiang P., Du W., Mancuso A., Wellen K.E., Yang X. Reciprocal regulation of p53 and malic enzymes modulates metabolism and senescence. Nature. 2013;493(7434):689–693. doi: 10.1038/nature11776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang P., Du W., Wang X., Mancuso A., Gao X., Wu M., Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 2011;13(3):310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno-Sánchez R., Gallardo-Pérez J.C., Rodríguez-Enríquez S., Saavedra E., Marín-Hernández Á. Control of the NADPH supply for oxidative stress handling in cancer cells. Free Radical Biol. Med. 2017;112:149–161. doi: 10.1016/j.freeradbiomed.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Liu L., Shah S., Fan J., Park J.O., Wellen K.E., Rabinowitz J.D. Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nat. Chem. Biol. 2016;12(5):345–352. doi: 10.1038/nchembio.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang L., Shestov A.A., Swain P., Yang C., Parker S.J., Wang Q.A., Terada L.S., Adams N.D., McCabe M.T., Pietrak B., Schmidt S., Metallo C.M., Dranka B.P., Schwartz B., DeBerardinis R.J. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532(7598):255–258. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nat. Rev. Canc. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 26.Lewis C.A., Parker S.J., Fiske B.P., McCloskey D., Gui D.Y., Green C.R., Vokes N.I., Feist A.M., Vander Heiden M.G., Metallo C.M. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell. 2014;55(2):253–263. doi: 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Chen L., Liu L., Su X., Rabinowitz J.D. Chemical basis for deuterium labeling of fat and NADPH. J. Am. Chem. Soc. 2017;139(41):14368–14371. doi: 10.1021/jacs.7b08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Díaz M.E., González L., Miquet J.G., Martínez C.S., Sotelo A.I., Bartke A., Turyn D. Growth hormone modulation of EGF-induced PI3K-Akt pathway in mice liver. Cell. Signal. 2012;24(2):514–523. doi: 10.1016/j.cellsig.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sethi N., Kang Y. Unravelling the complexity of metastasis — molecular understanding and targeted therapies. Nat. Rev. Canc. 2011;11(10):735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen D.X., Massague J. Genetic determinants of cancer metastasis. Nat. Rev. Genet. 2007;8(5):341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 31.Schafer Z.T., Grassian A.R., Song L., Jiang Z., Gerhart-Hines Z., Irie H.Y., Gao S., Puigserver P., Brugge J.S. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461(7260):109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taddei M.L., Giannoni E., Fiaschi T., Chiarugi P. Anoikis: an emerging hallmark in health and diseases. J. Pathol. 2012;226(2):380–393. doi: 10.1002/path.3000. [DOI] [PubMed] [Google Scholar]
- 33.Hawk M.A., Schafer Z.T. Mechanisms of redox metabolism and cancer cell survival during extracellular matrix detachment. J. Biol. Chem. 2018;293(20):7531–7537. doi: 10.1074/jbc.TM117.000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang F., Travins J., DeLaBarre B., Penard-Lacronique V., Schalm S., Hansen E., Straley K., Kernytsky A., Liu W., Gliser C., Yang H., Gross S., Artin E., Saada V., Mylonas E., Quivoron C., Popovici-Muller J., Saunders J.O., Salituro F.G., Yan S., Murray S., Wei W., Gao Y., Dang L., Dorsch M., Agresta S., Schenkein D.P., Biller S.A., Su S.M., Botton Sd, Yen K.E. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan R.S., Morris H.P., Coleman P.S. Kinetic characteristics of citrate influx and efflux with mitochondria from Morris Hepatomas 3924A and 16. Canc. Res. 1982;42:4399–4407. [PubMed] [Google Scholar]
- 36.Palmieri F., Monne M. Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim. Biophys. Acta. 2016;1863(10):2362–2378. doi: 10.1016/j.bbamcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Alexander A., Cai S.L., Kim J., Nanez A., Sahin M., MacLean K.H., Inoki K., Guan K.L., Shen J., Person M.D., Kusewitt D., Mills G.B., Kastan M.B., Walker C.L. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. U. S. A. 2010;107(9):4153–4158. doi: 10.1073/pnas.0913860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang A.B., Ryu E.-A., Artan M., Chang H.-W., Kabir M.H., Nam H.-J., Lee D., Yang J.-S., Kim S., Mair W.B., Lee C., Lee S.S., Lee S.-J. Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 2014;111(42):E4458–E4467. doi: 10.1073/pnas.1411199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yun H., Park S., Kim M.J., Yang W.K., Im D.U., Yang K.R., Hong J., Choe W., Kang I., Kim S.S., Ha J. AMP-activated protein kinase mediates the antioxidant effects of resveratrol through regulation of the transcription factor FoxO1. FEBS J. 2014;281(19):4421–4438. doi: 10.1111/febs.12949. [DOI] [PubMed] [Google Scholar]
- 40.Canto C., Gerhart-Hines Z., Feige J.N., Lagouge M., Noriega L., Milne J.C., Elliott P.J., Puigserver P., Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frescas D., Valent L., Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 2005;280(21):20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 42.Chae Y.C., Kim K.B., Kang J.Y., Kim S.R., Jung H.S., Seo S.B. Inhibition of FoxO1 acetylation by INHAT subunit SET/TAF-Ibeta induces p21 transcription. FEBS Lett. 2014;588(17):2867–2873. doi: 10.1016/j.febslet.2014.06.053. [DOI] [PubMed] [Google Scholar]
- 43.Boroughs L.K., DeBerardinis R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015;17(4):351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y., Butler E.B., Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hay N. Reprogramming glucose metabolism in cancer: can it be exploited for cancer therapy? Nat. Rev. Canc. 2016;16(10):635–649. doi: 10.1038/nrc.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piskounova E., Agathocleous M., Murphy M.M., Hu Z., Huddlestun S.E., Zhao Z., Leitch A.M., Johnson T.M., DeBerardinis R.J., Morrison S.J. Oxidative stress inhibits distant metastasis by human melanoma cells. Nature. 2015;527(7577):186–191. doi: 10.1038/nature15726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heiden M.G.V., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolpaw A.J., Dang C.V. Exploiting metabolic vulnerabilities of cancer with precision and accuracy. Trends Cell Biol. 2018;28(3):201–212. doi: 10.1016/j.tcb.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng F., Ye H., Wu M., Lian Y., Qian C., Zeng Y. Repressing malic enzyme 1 redirects glucose metabolism, unbalance the redox state, and attenuates migratory and invasive abilities in nasopharyngeal carcinoma cell lines. Chin. J. Canc. 2012;31(11):519–531. doi: 10.5732/cjc.012.10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murai S., Ando A., Ebara S., Hirayama M., Satomi Y., Hara T. Inhibition of malic enzyme 1 disrupts cellular metabolism and leads to vulnerability in cancer cells in glucose-restricted conditions. Oncogenesis. 2017;6(5):e329. doi: 10.1038/oncsis.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen H., Xing C., Cui K., Li Y., Zhang J., Du R., Zhang X., Li Y. MicroRNA-30a attenuates mutant KRAS-driven colorectal tumorigenesis via direct suppression of ME1. Cell Death Differ. 2017;24(7):1253–1262. doi: 10.1038/cdd.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu M., Chen Y., Huang B., Mao S., Cai K., Wang L., Yao X. Tumor-suppressing effects of microRNA-612 in bladder cancer cells by targeting malic enzyme 1 expression. Int. J. Oncol. 2018;52:1923–1933. doi: 10.3892/ijo.2018.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y., Ju H., Liu Z., Chen D., Wang Y., Zhao Q., Wu Q., Zeng Z., Qiu H., Hu P., Wang Z., Zhang D., Wang F., Xu R. ME1 regulates NADPH homeostasis to promote gastric cancer growth and metastasis. Canc. Res. 2018;78(8):1972–1985. doi: 10.1158/0008-5472.CAN-17-3155. [DOI] [PubMed] [Google Scholar]
- 54.Liao R., Ren G., Liu H., Chen X., Cao Q., Wu X., Li J., Dong C. ME1 promotes basal-like breast cancer progression and associates with poor prognosis. Sci. Rep. 2018;8(1) doi: 10.1038/s41598-018-35106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen D., Liu D., Tang J., Dong L., Liu Y., Tao Z., Wan J., Gao D., Wang L., Sun H., Fan J., Wu W. Malic enzyme 1 induces epithelial-mesenchymal transition and indicates poor prognosis in hepatocellular carcinoma. Tumor Biol. 2015;36(8):6211–6221. doi: 10.1007/s13277-015-3306-5. [DOI] [PubMed] [Google Scholar]
- 56.Chakrabarti G. Mutant KRAS associated malic enzyme 1 expression is a predictive marker for radiation therapy response in non-small cell lung cancer. Radiat. Oncol. 2015;10:145–151. doi: 10.1186/s13014-015-0457-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nicol C.J., Zielenski J., Tsui L-c, Wells P.G. An embryoprotective role for glucose-6-phosphate dehydrogenase in developmental oxidative stress and chemical teratogenesis. Faseb. J. 2000;14(1):111–127. doi: 10.1096/fasebj.14.1.111. [DOI] [PubMed] [Google Scholar]
- 58.Gnoni G.V., Priore P., Geelen M.J., Siculella L. The mitochondrial citrate carrier: metabolic role and regulation of its activity and expression. IUBMB Life. 2009;61(10):987–994. doi: 10.1002/iub.249. [DOI] [PubMed] [Google Scholar]
- 59.Gameiro P.A., Laviolette L.A., Kelleher J.K., Iliopoulos O., Stephanopoulos G. Cofactor balance by nicotinamide nucleotide transhydrogenase (NNT) coordinates reductive carboxylation and glucose catabolism in the tricarboxylic acid (TCA) cycle. J. Biol. Chem. 2013;288(18):12967–12977. doi: 10.1074/jbc.M112.396796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mullen A.R., Hu Z., Shi X., Jiang L., Boroughs L.K., Kovacs Z., Boriack R., Rakheja D., Sullivan L.B., Linehan W.M., Chandel N.S., DeBerardinis R.J. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7(5):1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dang L., Jin S., Su S.M. IDH mutations in glioma and acute myeloid leukemia. Trends Mol. Med. 2010;16(9):387–397. doi: 10.1016/j.molmed.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.-H., Sullivan L.B., Cheng T., Yang Y., Linehan W.M., Chandel N.S., DeBerardinis R.J. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.To T.-L., Cuadros A.M., Shah H., Hung W.H.W., Li Y., Kim S.H., Rubin D.H.F., Boe R.H., Rath S., Eaton J.K., Piccioni F., Goodale A., Kalani Z., Doench J.G., Root D.E., Schreiber S.L., Vafai S.B., Mootha V.K. A compendium of genetic modifiers of mitochondrial dysfunction reveals intra-organelle buffering. Cell. 2019;179(5):1222–1238. doi: 10.1016/j.cell.2019.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blattler S.M., Rencurel F., Kaufmann M.R., Meyer U.A. In the regulation of cytochrome P450 genes, phenobarbital targets LKB1 for necessary activation of AMP-activated protein kinase. Proc. Natl. Acad. Sci. U. S. A. 2007;104(3):1045–1050. doi: 10.1073/pnas.0610216104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jeon S.-M., Chandel N.S., Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485(7400):661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lavi O. Redundancy: a critical obstacle to improving cancer therapy. Canc. Res. 2015;75(5):808–812. doi: 10.1158/0008-5472.CAN-14-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia D., Lu M., Jung K.H., Park J.H., Yu L., Onuchic J.N., Kaipparettu B.A., Levine H. Elucidating cancer metabolic plasticity by coupling gene regulation with metabolic pathways. Proc. Natl. Acad. Sci. U. S. A. 2019;116(9):3909–3918. doi: 10.1073/pnas.1816391116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watson P.A., Arora V.K., Sawyers C.L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Canc. 2015;15(12):701–711. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kolch W., Fey D., Ryan C.J., Harrison Peter T., Huang Paul H. Exploiting vulnerabilities in cancer signalling networks to combat targeted therapy resistance. Essays Biochem. 2018;62(4):583–593. doi: 10.1042/EBC20180016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boudreau A., purkey H.E., Hitz A., Robarge K., Peterson D., Labadie S., Kwong M., Hong R., Gao M., Nagro C.D., Pusapati R., Ma S., Salphati L., Pang J., Zhou A., Lai T., Li Y., Chen Z., Wei B., Yen I., Sideris S., McCleland M., Firestein R., Corson L., Vanderbilt A., Williams S., Daemen A., Belvin M., Eigenbrot C., Jackson P.K., Malek S., Hatzivassiliou G., sampath D., Evangelista M., O'Brien T. Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat. Chem. Biol. 2016;12(10):779–786. doi: 10.1038/nchembio.2143. [DOI] [PubMed] [Google Scholar]
- 71.Vriens K., Christen S., Parik S., Broekaert D., Yoshinaga K., talebi A., Dehairs J., Escalona-Noguero C., Schmieder R., Cornfield T., Charlton C., Romero-Pérez L., Rossi M., Rinaldi G., Orth M.F., Boon R., Kerstens A., Kwan S.Y., Faubert B., Méndez-Lucas As, Kopitz C.C., Chen T., Fernandez-Garcia J., Duarte JoAG., Schmitz A.A., Steigemann P., Najimi M., Hägebarth A., Ginderachter J.A.V., Sokal E., Gotoh N., Wong K.-K., Verfaillie C., Derua R., Munck S., Yuneva M., Beretta L., DeBerardinis R.J., Swinnen J.V., Hodson L., Cassiman D., Verslype C., Christian S., Grünewald S., Grünewald T.G.P., Fendt S.-M. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566(7744):403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biancur D.E., Paulo J.A., Małachowska B., Rey M.Q.D., Sousa CoM., Wang X., Sohn A.S.W., Chu G.C., Gygi S.P., Harper J.W., Fendler W., Mancias J.D., Kimmelman A.C. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat. Commun. 2016;8 doi: 10.1038/ncomms15965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data for immunoblotting can be accessed via https://doi.org/10.17632/d2gb5wz88r.1 on Mendeley Data.