Abstract
Preservation of denatured dermis exerts promotive functions in wound healing and improves the appearance and function of skin. Angiogenesis is crucial for wound healing during burn injury. However, the potential molecular mechanism of angiogenesis in the recovery after burn injury remains to be elucidated. Herein, RNA chromatin immunoprecipitation (ChIP) sequencing analysis revealed upregulation of long intergenic non-coding RNA 00174 (linc00174) in the post-burn tissues. linc00174 overexpression promoted angiogenic activities of human umbilical vein endothelial cells (HUVECs) in the heat-denatured cell model, characterized by the promotion of cell proliferation, migration, and tube formation. Mechanistically, linc00174 directly bound to enhancer of zeste homolog 2 (EZH2), thus stimulating the protein level of trimethylation at lysine 27 of histone H3 (H3K27me3). Moreover, inhibition of EZH2 resulted in downregulation of ZNF24 and Runx1, as well as a decline of vascular endothelial growth factor A (VEGFA). Furthermore, EZH2 modulated epigenetic repression of ZNF24 and Runx1 through the promoter of H3K27me3. Additionally, ZNF24 and Runx1 both functioned as transcriptional inhibitors of VEGFA. Taken together, these findings uncover that linc00174 epigenetically inhibits ZNF24 and Runx1 expression through binding to EZH2, thus attenuating the suppression of VEGFA, contributing to the facilitation of angiogenesis during the recovery of heat-denatured endothelial cells.
Keywords: linc00174, EZH2, ZNF24, Runx1, VEGFA, angiogenesis, heat denature, wound healing
Graphical Abstract
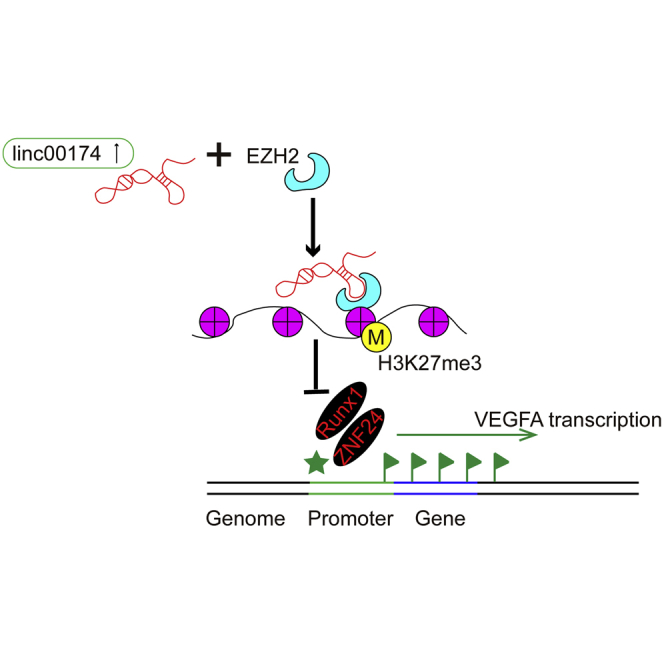
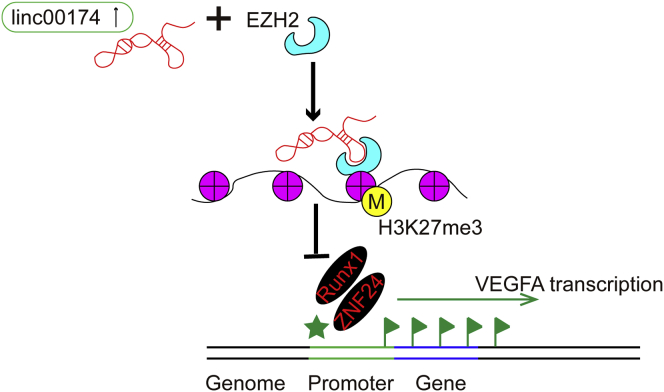
In the present study, we discovered that upregulation of linc00174 promotes endothelial cell-mediated angiogenesis by a mechanism of the linc00174-EZH2-ZNF24/Runx1-VEGFA regulatory axis, and it facilitates post-burn wound healing. This discovery provides a new regulation of post-burn wound healing, and we elicited a new role of linc00174 in the regulation of angiogenesis.
Introduction
Severe burn injury is considered as a global public health concern since it brings a high burden to patients.1 According to data from the World Health Organization (WHO), fire-related burn injuries led to more than 300,000 death and the loss of 10 million disability-adjusted life years (DALYs) each year.2 The post-burn wound healing process is mainly composed of three stages, including inflammation, proliferation, and remodeling.3 It has been proven that neutrophils4 and macrophages5 contribute to the inflammation process of wound healing. Keratinocytes and fibroblasts are the major cell types for the proliferative stage.3 For the remodeling stage, fibroblasts produce collagens and other matrices for the maturation of new-born tissues and the promotion of scarring.6 However, the molecular mechanism of post-burn wound healing remains to be elucidated.
According to the previously established theory, denatured dermis plays a critical role in post-burn skin healing. Denatured dermis appears in burned skin, and it guides morphological changes, cell metabolism disturbance, and function impairments; importantly, denatured dermis has the ability to restore normal skin morphology and promote post-burn wound healing by improving their surrounding microenvironment.7 Two cell types were found in the denatured dermis, including vascular endothelial cells (VECs) and fibroblasts.8 Fibroblasts synthesize extracellular matrix (ECM) and play a critical role in wound healing.9 Vascular endothelial cells are of great importance in angiogenic neogenesis of the vascular system, which is a major activity in post-burn skin wound healing.
Angiogenesis plays a major role in the regulation of wound healing.10 During the process of wound healing, angiogenesis results in the generation of neonatal capillaries, which will bring oxygen and nutrients to the growing tissues and remove catabolic wastes. In this way, angiogenesis assists the repairing of post-burn wound tissues.11 Angiogenesis is tightly regulated by a variety of factors, including pro-angiogenic and anti-angiogenic factors.12,13 Pro-angiogenic factors include vascular endothelial growth factor (VEGF),14,15 platelet-derived growth factor (PDGF),15,16 and transforming growth factor β (TGF-β),16,17 among others. VEGF is recognized as playing the vital role in the regulation of angiogenesis by stimulating the proliferation, migration, and tube formation of endothelial cells.14 Therefore, VEGF has been considered as a critical therapeutic target for the regulation of angiogenesis.
Long non-coding RNAs (lncRNAs) are the kind of non-protein-coding transcripts that have more than 200 nt.18 Recently, the functions of lncRNAs in regulating various pathophysiological events have been studied intensively.12,19, 20, 21, 22 lncRNAs are crucial mediators in modulating cell proliferation, migration, and invasion, especially for cancer cells.20,23,24 Additionally, lncRNAs are also involved in the regulation of the proliferation, migration, and tube formation of endothelial cells, subsequently contributing to angiogenesis.25 For example, lncRNA HULC was identified to regulate endothelial cells to stimulate angiogenesis by modulating the activity of miR-107 and SPHK1 signaling.26 lncRNA PVT1 promotes angiogenesis by activating the signal transducer and activator of transcription 3 (STAT3)/VEGFA axis.27 lncRNA ATB promotes viability, migration, and angiogenesis in human microvascular endothelial cells by sponging miR-195.28 However, the potential roles and underlying mechanisms of lncRNAs in post-burn wound healing remain largely unknown.29
Previous studies have confirmed that lncRNAs regulate downstream gene expression through epigenetic modification via direct or indirect interaction with their target genes.30 Further studies demonstrate that lncRNAs exert epigenetic regulation by maternal effect, DNA methylation, and histone modification. One example is lncRNA GClnc1, which acts as a molecular scaffold to recruit WDR2 and KAT2A complexes for the modification of specific histones.31 Herein, EZH2 (enhancer of zeste homolog 2), a histone H3 lysine 27 (H3K27) methyltransferase, is considered to be involved in the epigenetic regulation by a mass of lncRNAs.32 EZH2 is the catalytic subunit of polycomb-repressive complex 2 (PRC2), and promotes transcriptional silencing by modification of histones and regulation of chromatin structure.32 For example, lncRNA XIST was found to modulate the suppression of KLF2 transcription via directly binding with EZH2, thus inducing a stimulation of lung cancer cell proliferation.33 Therefore, the interaction of lncRNAs and EZH2 is a critical mechanism for epigenetic regulations.
In the present study, we identified a novel lncRNA, long intergenic non-coding RNA 00174 (linc00174), in the regulation of angiogenesis during the recovery of heat-denatured endothelial cells, which might provide important insights for post-burn wound healing therapy.
Results
Upregulation of linc00174 in Post-burn Tissues
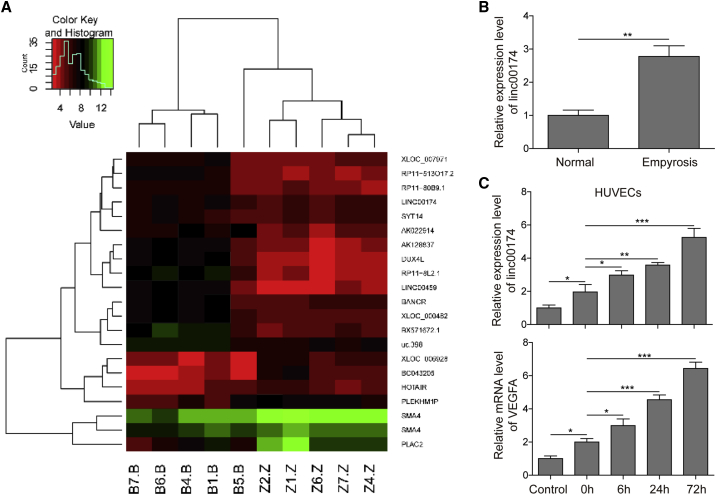
With the aim to study the expression profile of lncRNAs in post-burn wound tissues, clinical burned tissues (n = 5) were selected for microarray assays. Figure 1A shows the top 20 upregulated genes in burned wound tissues, compared to the normal tissues. According to the microarray data, the expression of linc00174 was greatly promoted in the post-burn wound tissues. This result was confirmed by a qRT-PCR assay, as shown in Figure 1B, suggesting that linc00174 may be involved in the regulation of post-burn wound healing. Since the activity of angiogenesis is initiated after injury, we speculate that linc00174 may be associated with angiogenesis. Therefore, we sought to study the expression of linc00174 in a post-burn angiogenic cell model (endothelial cells), which can simulate the activation of angiogenesis after burn wounding. In this cell model, human umbilical vein endothelial cells (HUVECs) were pre-heated by incubation at 52°C for 35 s. 72 h after heating, the expression of linc00174 was found to be notably stimulated in the pre-heated HUVECs, compared to the negative control, as indicated by qRT-PCR results (Figure 1C). Interestingly, the expression of VEGFA was found to be stimulated in pre-heated HUVECs (72 h after heating) as well, suggesting the enhanced angiogenic activity of HUVECs. Moreover, the coordinated expression profiles between linc00174 and VEGFA implied their putative correlation.
Figure 1.
Expression of linc00174 and VEGFA in Post-burn Skin Tissues and the Heat-Denatured In Vitro Model
(A) High-throughput sequencing of RNAs of the five clinical post-burn skin tissues. The top 20 upregulated or downregulated genes are shown in the heatmap. (B) Relative gene expression of linc00174 in post-wound skin tissues compared with that of the normal control measured by qRT-PCR. (C) Establishment of the heat-denatured cell model, by a 52°C, 35-s heating of HUVECs. qRT-PCR was performed to measure the expression levels of linc00174 and VEGFA in HUVECs after heating stimulation. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
linc00174 Promoted Angiogenic Activities of HUVECs in the Heat-Denatured Cell Model
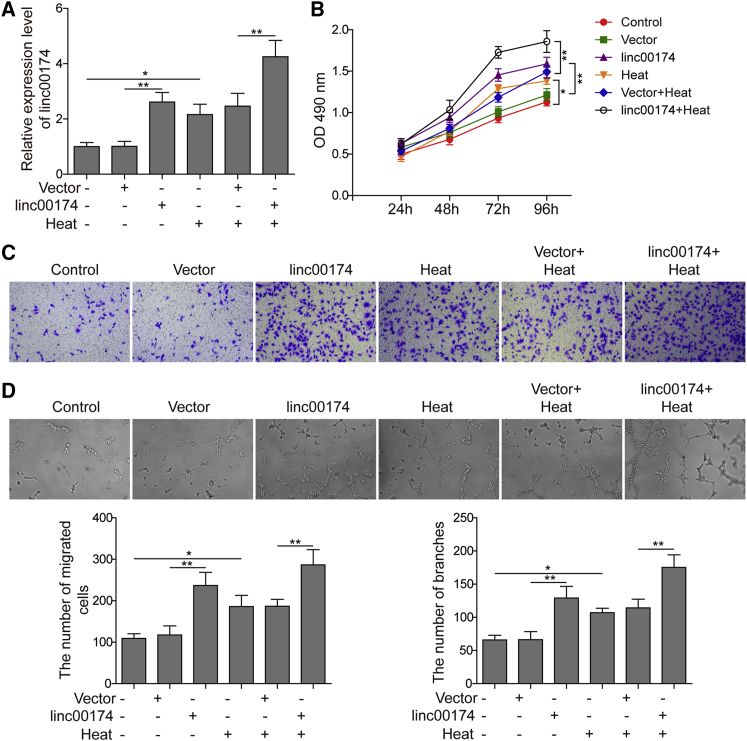
To investigate the potential role of linc00174 in the regulation of angiogenesis, a linc00174-overexpressing plasmid was constructed and transfected to the endothelial cell line HUVECs. The transfection efficiency of linc00174-overexpressing plasmids was confirmed by qRT-PCR analysis. As indicated in Figure 2A, the level of linc00174 was significantly increased in the overexpression group, compared to the negative control. Additionally, consistent with the data shown above, the pre-heated group of HUVECs also presented an increasing level of linc00174. Moreover, the linc00174-overexpressing and pre-heated HUVECs showed the highest linc00174 level. These results demonstrated that overexpression of linc00174 was successfully established in HUVECs. As shown in Figure 2B, the data of an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay indicated that HUVECs with linc00174 overexpression presented the stronger proliferative ability, compared to the negative control. Additionally, the pre-heated HUVECs with linc00174 overexpression presented a higher proliferation rate than did HUVECs with single pre-heated treatment. Additionally, in Figure 2C, the number of migratory HUVECs with linc00174 overexpression was significantly increased, according to the results of a cell migration assay in a transwell system. Additionally, the cell migratory rate of HUVECs that were treated with both pre-heating and linc00174 overexpression was found to be significantly higher than that for HUVECs treated singly with pre-heating. Moreover, the capacity of angiogenesis was also stimulated by linc00174 overexpression. As shown in Figure 2D, HUVECs with linc00174 overexpression displayed better angiogenesis compared to the negative control; additionally, HUVECs treated with both pre-heating and linc00174 overexpression presented significant higher tube formation rate than did the one with single pre-heating treatment.
Figure 2.
Effects of linc00174 on Angiogenic Activities of HUVECs
(A) A qRT-PCR assay was subjected to detect the expression of linc00174. (B) An MTT assay was used to assess the cell viability. (C) The capacity of cell migration was measured using a transwell assay. (D) The capacity of angiogenesis was detected using a tube formation assay. The data are presented as the mean ± SD obtained from at least three independent experiments. Significance was determined by a Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
linc00174 Regulated the Expression of VEGFA via Binding to EZH2
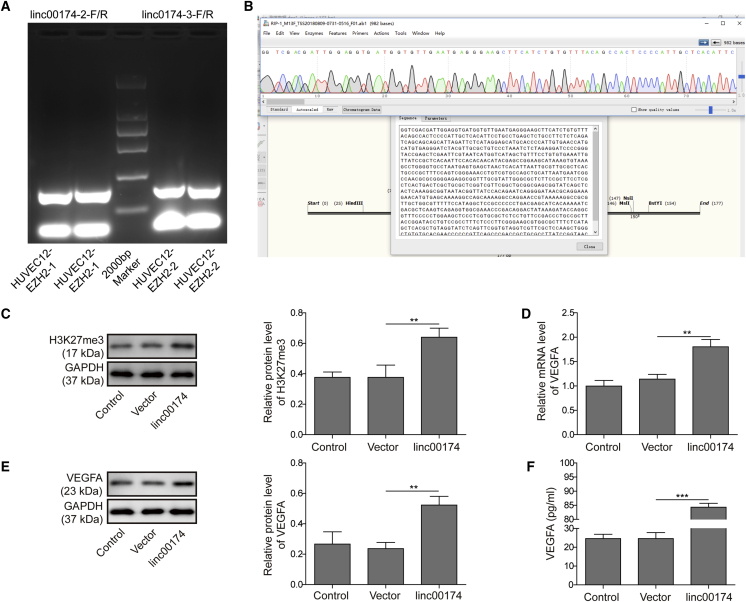
According to the prediction from bioinformatics studies, there are complementary sequences between linc00174 and EZH2, suggesting their direct interaction. To further verify our hypothesis, an RNA immunoprecipitation (RIP) experiment was performed. As shown in Figure 3A, the PCR amplifications with linc00174 primers and anti-EZH2 mediated immunoprecipitations resulted in strong positive DNA bands, suggesting that there was a direct binding relationship of linc00174 to EZH2. Subsequently, the sequences of the precipitated RNA were confirmed to be linc00174, as shown in Figure 3B. To ascertain the putative regulation of histone modifications by linc00174/EZH2 complex, we detected the protein level of trimethylation at lysine 27 of histone H3 (H3K27me3) via western blot analysis. As shown in Figure 3C, knockdown of linc00174 resulted in a decrease of H3K27me3 in HUVECs; oppositely, overexpression of linc00174 led to a promotion of H3K27me3. Therefore, this result suggested that linc00174 may promote the methylation of histone H3 at the amino acid lysine 27 (K27). To clarify whether linc00174 regulates the expression of VEGFA, we performed knockdown and overexpression of linc00174 in HUVECs and examined the expression of VEGFA in mRNA and protein levels. As shown in Figures 3D and 3E, knockdown of linc00174 induced the inhibition of VEGFA both in mRNA and protein levels, while overexpression of linc00174 led to a notable upregulation of VEGFA expression. Meanwhile, the secreted form of VEGFA was promoted by linc00174 overexpression and suppressed by linc00174 knockdown (Figure 3F). These findings indicated that linc00174 stimulates the expression of VEGFA.
Figure 3.
linc00174 Regulated the Expression of VEGFA via EZH2
(A) Results of a RIP assay verified that the EZH2-binding RNA can be amplified by linc00174 primers. (B) The bioinformatics prediction presented the binding site between linc00174 and EZH2. (C) Western blot analysis was performed to detect the level of H3K27me3. (D) qRT-PCR assay was performed to determine the expression of linc00174, and the results indicated that overexpression of linc00174 stimulated the mRNA level of VEGFA. (E) Western blot analysis was performed to detect the level of VEGFA. (F) The concentration of VEGFA was detected using an ELISA assay. The data are presented as the mean ± SD obtained from at least three independent experiments. Significance was determined by a Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
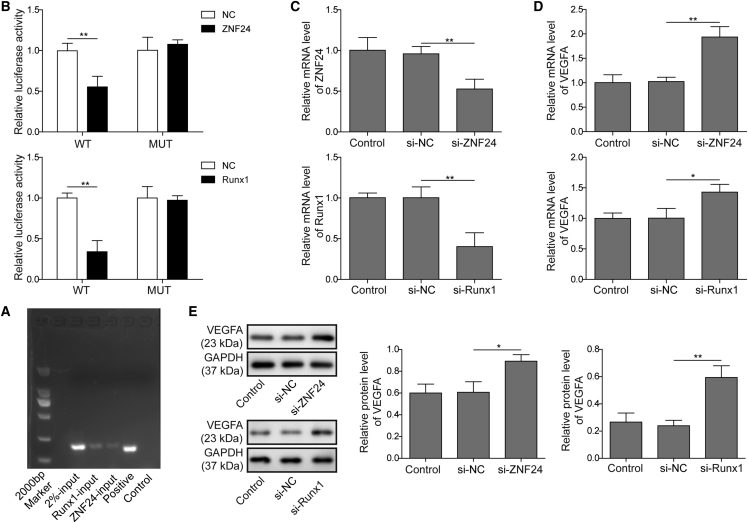
EZH2 Negatively Regulated the Expression of ZNF24 and Runx1 through Recruiting H3K27me3 to the Promoter
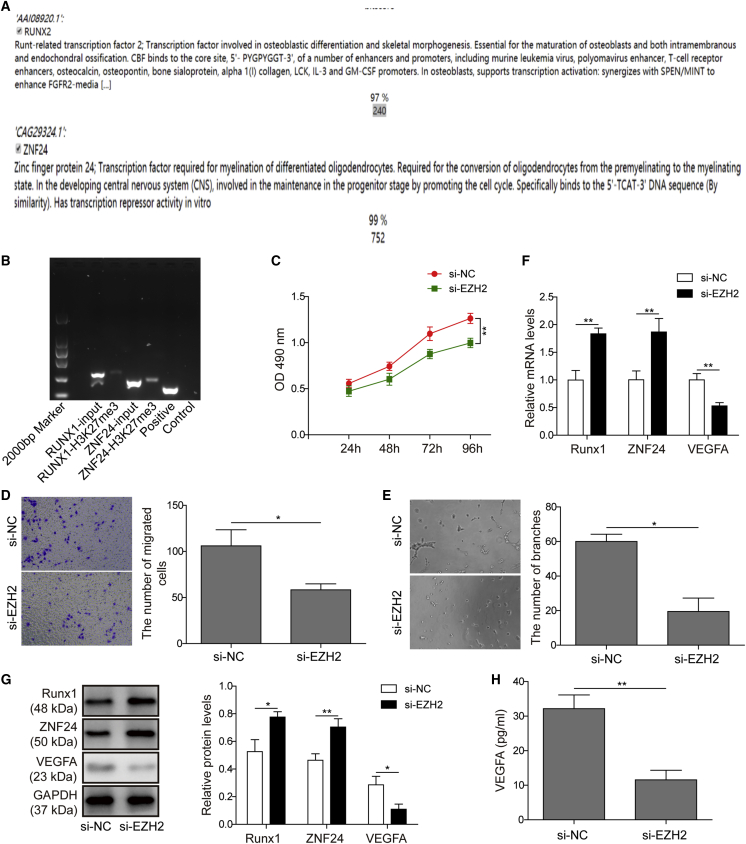
According to previous reports, EZH2 and ZNF24 have been proven to be transcriptional repressors for the gene expression of VEGFA.34 Thus, we hypothesized that linc00174/EZH2 may regulate the expression of VEGFA viaZNF24 and Runx1. Bioinformatics analysis was performed to predict the putative connections between EZH2 and ZNF24/Runx1 promoters (Figure 4A). Because the histone methyltransferase EZH2 mediates the methylation of H3 histone protein on the K27 position, a chromatin immunoprecipitation (ChIP) experiment using anti-H3K27me3 antibody for chromatin co-precipitation may reveal whether the H3K27me3 protein is located in the promoter of ZNF24 or Runx1. ChIP results indicated positive PCR amplifications with ZNF24 and Runx1 primers, suggesting that H3K27me3 protein bound to the promoter region of these two genes (Figure 4B). Next, we sought to clarify whether EZH2 affects the angiogenic activity of HUVECs. As shown in Figures 4C–4E, silencing EZH2 in HUVECs significantly inhibited the capacity of cell proliferation, migration, and angiogenesis, suggesting that EZH2 might positively modulate the angiogenic activity of HUVECs. To explore the regulatory mechanism of EZH2 on angiogenesis, the expression levels of VEGFA and its suppressors ZNF24/Runx1 were evaluated. As shown in Figures 4F and 4G, the mRNA and protein levels of ZNF24 and Runx1 were significantly promoted by EZH2 inhibition, while VEGFA expression was obviously reduced. This is consistent with the level of secreted VEGFA, as indicated by the ELISA results in Figure 4H.
Figure 4.
EZH2 Regulated the Expression of ZNF24 and Runx1
(A) Bioinformatics predictions indicated that EZH2 binds to the promoter of ZNF24 and Runx1. (B) A ChIP assay certified that H3K27me3 binds to the promoter of ZNF24 and Runx1. (C) An MTT assay was performed and the data showed that EZH2 knockdown resulted in the suppression of cell proliferation in HUVECs. (D) Transwell assay of cell migration indicated that EZH2 knockdown caused the inhibition of HUVEC migration. (E) EZH2 knockdown caused the inhibition of tube formation in HUVECs. (F and G) qRT-PCR (F) and western blot analysis (G) were performed, and the data revealed that EZH2 knockdown increased the mRNA and protein levels of ZNF24 and Runx1, but reduced VEGFA expression. (H) An ELISA assay was subjected to assess the level of VEGFA, and the results showed that EZH2 knockdown caused a decreasing level of secreted VEGFA. The data are presented as the mean ± SD obtained from at least three independent experiments. Significance was determined by a Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
VEGFA Transcriptional Activity Was Regulated by ZNF24 and Runx1
To confirm the direct regulation of VEGFA by ZNF24 and Runx1, ChIP and luciferase reporter assays were performed. As shown in Figure 5A, ChIP results indicated that the DNA fragments of the VEGFA promoter can be detected from the anti-ZNF24 and anti-Runx1 antibody-mediated immunoprecipitations, suggesting that ZNF24 and Runx1 directly bound to the promoter of VEGFA in HUVECs. Next, luciferase reporter assays indicated that VEGFA promoter-mediated luciferase activities were suppressed by overexpression of ZNF24 and Runx1 (Figure 5B). These results demonstrated that ZNF24 and Runx1 bind to the promoter of VEGFA directly to suppress its gene expression. To examine the regulation of VEGFA expression by ZNF24 and Runx1, gene knockdown cell models were established by transfecting ZNF24 or Runx1 short hairpin RNA (shRNA) to HUVECs, and the expression levels of these two genes were downregulated successfully, as confirmed by qRT-PCR (Figure 5C). As shown in Figure 5D, the mRNA level of VEGFA was found to be sharply stimulated by knockdown of ZNF24 or Runx1. This is consistent with the promotion of VEGFA protein by ZNF24 or Runx1 knockdown, as indicated by the results of Figure 5E.
Figure 5.
VEGFA Expression Was Regulated by ZNF24 and Runx1
(A) Results of a ChIP assay indicated that ZNF24 and Runx1 are binding to the promoter of VEGFA. (B) Results of a luciferase reporter assay indicated that ZNF24 and Runx1 promoted the wild-type form of VEGFA promoter-mediated luciferase activity, but there was no significant effect for the mutant one. (C) qRT-PCR results indicated significant reduced of ZNF24 and Runx1 mRNA level in the respective gene knockdown HUVECs. (D and E) qRT-PCR (D) and western blot analysis (E) were performed, and the data revealed that knockdown of ZNF24 or Runx1 upregulated the mRNA and protein levels of VEGFA in HUVECs. The data are presented as the mean ± SD obtained from at least three independent experiments. Significance was determined by a Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
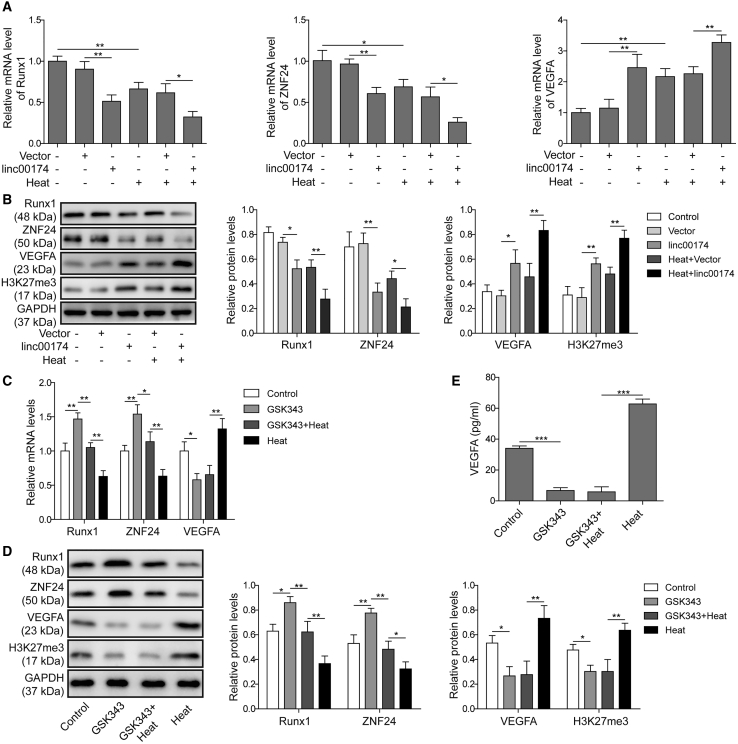
linc00174/EZH2 Regulated VEGFA Expression through ZNF24 and Runx1 in the Heat-Denatured Cell Model
To ascertain the exact roles of the factors mentioned above in the regulation of post-burn wound healing, we detected the expression levels of each gene. As shown in Figure 6A, compared to negative control, in pre-heated HUVECs, the expression levels of ZNF24 and Runx1 were suppressed; however, the mRNA level of VEGFA was stimulated. In the group of HUVECs with linc00174 overexpression, the mRNA levels of ZNF24 and Runx1 were dramatically suppressed; however, the mRNA level of VEGFA was promoted. Moreover, as shown in Figure 6B, the protein levels of ZNF24, Runx1, and VEGFA using western blot analysis also displayed the corresponding trend with their mRNAs. Additionally, the level of H3K27me3 exerted the similar results with VEGFA (Figure 6B). To investigate the role of EZH2 on the regulation of VEGFA, the EZH2 inhibitor GSK343 was applied for pre-treated HUVECs. As shown in Figures 6C and 6D, the pre-heated treatment of HUVECs caused the suppression of ZNF24 and Runx1 expression, while notably increasing VEGFA and H3K27me3 expression levels, and this was opposite to the effects mediated by GSK343. In addition, GSK343 partly reversed the effects induced by the pre-heating treatment. Similar results were also observed in Figure 6E.
Figure 6.
linc00174/EZH2 Regulated VEGFA Expression through ZNF24 and Runx1 in the Heat-Denatured Cell Model
(A and B) The expression levels of Runx1, ZNF24, VEGFA, and H3K27me3 were detected using qRT-PCR (A) and western blot analysis (B). (C and D) Effects of the EZH2 inhibitor GSK343 in the heat-denatured cell model were determined by qRT-PCR (C) and western blot analysis (D). (E) The concentration of VEGFA in the supernatant was detected by ELISA. The data are presented as the mean ± SD obtained from at least three independent experiments. Significance was determined by a Student’s t test. ∗p < 0.05, ∗∗p < 0.01.
Discussion
lncRNAs are found to be critical in the regulation of various pathophysiology events, especially tumors.35 Recently, the regulations of gene expression, cell activity, and pathophysiology by lncRNAs have been studied intensively.12,35, 36, 37, 38, 39 lncRNAs have been identified to be involved in the regulation of wound healing.40 For example, lncRNA TINCR regulates the differentiation of keratinocyte to affect wound healing.41 However, few of those studies focused on the role of lncRNAs in regulating post-burn wound healing. Previous studies demonstrated that denatured dermis is important for post-burn skin healing.20,42,43 However, the investigations on lncRNA-mediated regulation of denatured dermis are still rare to date. In the present study, we analyzed the expression profile of lncRNAs in post-burn tissues. Compared to the control, the upregulated and downregulated lncRNAs in post-burn tissues were identified. Among them, linc00174 expression was found to be promoted significantly, and this was confirmed by qRT-PCR, suggesting a potential regulatory role of this gene in post-burn wound healing. The stimulation of linc00174 expression was found in a post-burn angiogenic endothelial cell model (HUVECs), suggesting that linc00174 may function in the angiogenic activity of endothelial cells. Additionally, the expression profile of linc00174 was found to be correlated with VEGFA, suggesting that there may be a correlation of gene expression between the two genes. linc00174 is a lncRNA that is almost unexplored to date. The only publication about linc00174 to date includes the discovery that linc00174 facilitates colorectal carcinoma progression by regulating TAZ expression.44 In the present study, our results suggested that linc00174 may play a critical role in the regulation of endothelial cell-mediated post-burn wound healing.
To validate the regulation of endothelial cell activities by linc00174 in the post-burn condition, an overexpression of linc00174 in HUVECs was performed. Our results indicated that overexpression of linc00174 facilitated cell proliferation, migration, and tube formation rate of endothelial cells in a post-burn model. It is well accepted that the promotion of angiogenesis contributes greatly to post-burn wound healing.45 There is abundant evidence demonstrating that the stimulation of angiogenesis results in a promotion of wound healing. For example, miR-135a-3p was recently identified to regulate endothelial cell-mediated angiogenesis and promote wound healing by targeting the p38 signaling pathway.23 Our results demonstrated that high expression of linc00174 promoted the angiogenic activity of endothelial cells, and most importantly it stimulated the angiogenesis in a post-burn endothelial cell model; therefore, it is reasonable to conclude that linc00174 may positively contribute to post-burn wound healing by promoting angiogenesis. In our previous studies, we proved that the denatured dermis, which is the part of dermis in burned skin, exhibits the function of promoting skin wound healing.8, 9, 10 In the present study, we provide evidence to show that the angiogenic activity of denatured dermis vascular endothelial cells can be regulated by linc00174. The upregulation of linc00174 in denatured dermis plays a critical role in promoting angiogenesis and wound healing. We propose that the artificial upregulation of linc00174 in the clinical wound skin may facilitate skin wound healing by promoting angiogenesis, and thus the upregulation of linc00174 may be a novel type of gene therapy for skin wound healing. However, there are still challenges for RNA therapeutics, such as off-target effects, gene delivery, and others.46 Therefore, more investigations on this topic are necessary. In future studies, we will critically examine the function of lin00174 skin wound healing in an animal model to validate its role in vivo, so as to confirm whether lin00174 can be a therapeutic target for burned skin wound healing.
Typically, the angiogenic activity of endothelial cells is determined by the ratio of pro-angiogenic factors and anti-angiogenic factors.47 VEGFA is probably one of the most important pro-angiogenic factors, and it plays a crucial role for angiogenesis.14 Thus, the expression and secretion of VEGFA in endothelial cells may primarily determine the angiogenic activity. For a deeper insight into the molecular mechanism behind linc00174-mediated promotion of the angiogenic activity of endothelial cells, we focused on investigating the regulation of VEGFA gene expression by linc00174. Our results indicated that overexpression of linc00174 in endothelial cells resulted in a promotion of VEGFA expression, including VEGFA mRNA and protein level, and also the secreted form of VEGFA. In contrast, knockdown of linc00174 in endothelial cells resulted in reduction of VEGFA production. These results demonstrated that linc00174 positively regulated the expression of VEGFA in endothelial cells. Due to the critical role of VEGFA in the regulation of angiogenesis, our results lead to the conclusion that linc00174 promotes the angiogenic activity of endothelial cells by stimulating the gene expression of VEGFA.
To understand more molecular details of linc00174-mediated regulation of VEGFA gene expression, a combination of bioinformatics predictions and experimental validations were performed. According to the bioinformatics analysis, linc00174 can bind to the protein EZH2, which is a catalytic subunit of polycomb-repressive complex 2.48 This binding activity was subsequently confirmed by a RIP experiment. Typically, the protein EZH2 exerts a function for catalyzing the methylation reaction at the amino acid H3K27.12 Generally, EZH2-mediated histone methylation of H3K27 results in a repression of target gene expression.49 EZH2-mediated epigenetic regulations of gene expression have been proven to be crucial for various pathophysiologic mechanisms. For example, recent studies have demonstrated that EZH2 plays a critical role in the regulation of tumorigenesis.50 As a transcriptional repressor, the gain-of-function mutations of EZH2 result in abnormal gene transcriptions for a variety of cancer-related genes and subsequently induce an abnormal level of various oncogenes and tumor suppressor genes. In the present study, we proved that EZH2 was modulated by linc00174 via direct binding.
Next, we sought to discover the link between the linc00174/EZH2 complex and VEGFA expression. Again, bioinformatics analysis indicated that the target genes regulated by EZH2 included ZNF24 and Runx1, both of which have been proven to be suppressors of VEGFA expression.34. This analysis led to a hypothesis that the linc00174/EZH2 complex affects VEGFA gene expression by suppressing the VEGFA suppressors ZNF24 and Runx1. Subsequently, this hypothesis was proven by a series of experiments. ChIP results indicated that H3K27me3, the methylated form of histone H3 that was mediated by EZH2, binds to the promoter of ZNF24 and Runx1 directly. Therefore, this result demonstrated that EZH2 regulated the expression of ZNF24 and Runx1 by catalyzing the methylation of H3K27. Furthermore, the inhibition of EZH2 by its chemical inhibitor resulted in a promotion of ZNF24 and Runx1 expression, and simultaneously a reduction of VEGFA expression, demonstrating that EZH2 exerted a suppressive function of ZNF24 and Runx1 expression, but probably an indirect promotion of VEGFA expression. Additionally, the inhibition of EZH2 led to a suppression of cell proliferation and migration of HUVECs, suggesting that EZH2 positively regulates the angiogenic activity of endothelial cells. This is consistent with the regulatory function of VEGFA for angiogenesis. Further experiments were performed to prove the regulation of VEGFA by EZH2. First, ChIP experiments indicated that ZNF24 and Runx1 directly bound to the promoter of VEGFA. Second, luciferase assays indicated that knockdown of ZNF24 or Runx1 inhibited the VEGFA promoter-derived luciferase activity. Third, knockdown of ZNF24 and Runx1 resulted in a promotion of VEGFA expression, both in mRNA and protein levels. Thus, these results demonstrated that ZNF24 and Runx1 suppressed the expression of VEGFA by a direct binding to its promoter.
In the final step, we provided more evidence to thoroughly prove the regulation of post-burn wound healing by the linc00174-EZH2-ZNF24/Runx1-VEGFA regulatory axis. In this section of experiments, linc00174 was overexpressed in a post-burn endothelial cell model, and the expression levels of ZNF24, Runx1, VEGFA, and H3K27me3 were monitored. Results indicated that post-burn cells exhibited a suppression of ZNF24 and Runx1, but a promotion of VEGFA and H3K27me3 levels, compared to the negative control; meanwhile, overexpression of linc00174 resulted in a suppression of ZNF24 and Runx1 expression, but a promotion of VEGFA and H3K27me3 levels as well. Overexpression of linc00174 in the heat-denatured cell model resulted in the strongest suppression of ZNF4 and Runx1, and also the strongest promotion of VEGFA expression and the H3K27me3 level. This evidence proved that, in the post-burn condition, linc00174 stimulated VEGFA expression in endothelial cells by inhibiting ZNF24 and Runx1 expression and promoting the methylation of H3K27. Our further experiments indicated that inhibition of EZH2 resulted in an opposite effect, compared to linc00174 overexpression, in the heat-denatured endothelial cell model. These results included a promotion of ZNF24 and Runx1 expression, alongside with a suppression of VEGFA expression and the H3K27me3 level. Similarly, these results demonstrated that, in the post-burn condition, EZH2 stimulated VEGFA expression in endothelial cells by inhibiting ZNF24 and Runx1 expression and promoting the methylation of H3K27. Based on the above conclusion that linc00174 binds to EZH2 and stimulated its activity, we further conclude that linc00174 binds to EZH2 directly and activates its function of catalyzing the H3K27 methylation in the chromatin region of ZNF24 and Runx1 promoters. Subsequently, ZNF24 and Runx1 expression levels are suppressed, and VEGFA expression is promoted indirectly. Considering that in post-burn wound healing the expression of linc00174 is upregulated, we conclude that the linc00174-EZH2-ZNF24/Runx1-VEGFA regulatory pathway plays a crucial role in endothelial cell-mediated angiogenesis.
Collectively, in the present study, we investigated the function of linc00174, a novel lncRNA with the regulatory functions principally unexplored, in the regulation of endothelial cell-mediated angiogenesis and post-burn wound healing. Our findings lead to the conclusion that, in the heat-denatured cell model, upregulation of linc00174 promotes endothelial cell-mediated angiogenesis by a mechanism of the linc00174-EZH2-ZNF24/Runx1-VEGFA regulatory axis, and finally contributes greatly to post-burn wound healing (Figure 7). This discovery provides a new regulation of post-burn wound healing, and, for the first time, we delineated a new role for linc00174 in the regulation of angiogenesis. We suggest that linc00174 will be a new therapeutic target for developing agents to accelerate post-burn wound healing.
Figure 7.
Theoretical Diagram Illustrating the Molecular Mechanisms of linc00174 in Post-burn Wound Healing
Materials and Methods
Collection of Tissue Specimens and ChIP Sequencing of lncRNAs
All experiments conducted in this study were approved by the Ethics Committee of Xiangya Hospital of Central South University. Written informed consent was obtained from all patients. A total of five tissue samples were collected from five males (37.2 ± 5.6 years) with second- and third-degree burns admitted <24 h after the time of burn injury.44 The samples were collected during tangential excision of eschar and large sheets of split thickness autograft with preservation of dermis at day 4 after burn. Normal skin was remnant donor skin taken from the trunk, while the dermis was from the extremities. Denatured dermis was distinguished as follows: the eschar and yellow necrotic tissue were debrided, and then a layer of white dermis with a small amount of scattered tiny bleeding points was seen, and this white dermis was confirmed as denatured dermis and used for further experiments. All of the samples were collected at day 4 after burn. Total cellular RNA was isolated from the fresh tissues using an RNeasy mini kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol, and then quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The purified total RNA was used for a lncRNA ChIP sequencing experiment.
Cell Culture and Treatment
HUVECs (Lonza, Walkerville, MD, USA) were cultured in endothelial cell basal medium bullet kits (Lonza) containing 20% fetal bovine serum (FBS; Gibco, Auckland, New Zealand) supplemented with l-glutamine and antibiotics (1% penicillin/streptomycin) on 100-mm cell-culturing plates. The plates were coated with type I rat tail collagen (Millipore/Upstate, Temecula, CA, USA) at 10 μg/cm2. For the heat-denatured cell model, HUVECs were incubated at 52°C for 35 s, then shifted to 37°C for incubation. For the inhibition of EZH2, the specific inhibitor of EZH2 GSK343 was used, with the concentration 5 μM and treatment for 72 h.
Construction of Plasmids
For linc00174 overexpression, cells were transfected with the plasmid porcine cytomegalovirus (pCMV)-linc00174. A DNA fragment of linc00174 was chemically synthesized (GenScript, NanJing, China) and inserted into pCMV plasmid to generate pCMV-linc00174. Similarly, the plasmids pCMV-ZNF24 and pCMV-Runx1 were constructed by inserting the chemically synthesized DNA fragments ZNF24 and Runx1, respectively. shRNA-ZNF24 and shRNA-Runx1 were constructed by GenePharma (Shanghai, China).
For luciferase assays, the plasmid pGL3-VEGFA promoter wild-type (PRWT) was constructed by inserting the chemically synthesized VEGFA promoter wild-type DNA fragment into pGL3-basic empty vector (Promega, USA). Similarly, the plasmid pGL3-VEGFA promoter mutant (PRMT) was constructed by inserting the chemically synthesized VEGFA promoter mutant DNA fragment into pGL3-basic empty vector.
Plasmid Transfection
For the transfection of the plasmids, cells were used at a confluence of 50%–60% in a 24-well plate. The transfection was mediated by Lipofectamine 3000 (Thermo Fisher Scientific, USA). Cells were added with 100 μL of mixed serum-free Opti-MEM I medium with 0.8 μg of DNA and 3 μL of Lipofectamine 3000 and incubated at 37°C for 4 h. Afterward, 500 μL of endothelial cell basal medium (Lonza) was added and cells were incubated at 37°C for normal culturing.
Quantitative Real-Time PCR
Cells were harvested at indicated times, and total RNA was extracted by TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. 2–5 μg of RNA was used for reversed transcription, with 200 U of SuperScript II (Invitrogen) and synthetic oligo(dT). The resultant cDNA was used for PCR reaction with SYBR Green Premix Ex Taq II (Takara, Dalian, China), according to the manufacturer’s instructions. GAPDH served as an endogenous control. PCR was carried out in an ABI 7500 thermocycler with fluorescence detection (Applied Biosystems). Data were quantified using the 2−ΔΔCt method.
Primers are as follows: VEGFA, forward, 5′-CTGCCGTCCGATTGAGACC-3′, reverse, 5′-CCCCTCCTTGTACCACTGTC-3′; linc00174, forward, 5′-GTGGTTTGATCTTGGCTCAC-3′, reverse, 5′-CCAGGGGCTTCTTGTTGCAT-3′; GAPDH, forward, 5′-AGGTCGGTGTGAACGGATTTG-3′, reverse, 5′-GGGGTCGTTGATGGCAACA-3′; ZNF24, forward, 5′-CTGATGGCGAAGAGGGATCAA-3′, reverse, 5′-CCAGCACTACCAGCTCCAAG-3′; Runx1, forward, 5′-CTGCCCATCGCTTTCAAGGT-3′, reverse, 5′-GCCGAGTAGTTTTCATCATTGCC-3′.
RIP Assays
Rabbit polyclonal anti-EZH2 antibody (Abcam) was used for the RIP assay. The RIP assay was performed by a Magna RIP RNA-binding protein immunoprecipitation kit (Merck Millipore, USA). The cell pellet from 5 to 7 × 107 HUVECs was suspended in 100 μL of RIP lysis buffer. The resulting cell lysate was incubated with either 10 μg of rabbit polyclonal anti-EZH2 antibody or 10 μg of normal mouse immunoglobulin G (IgG). Antibodies were attached to magnetic beads in RIP immunoprecipitation buffer for 6 h at 4°C. After washing five times, the magnetic beads were incubated with proteinase K for 30 min at 55°C. After centrifugation, total RNA from the supernatant was extracted by phenol/chloroform/isoamyl alcohol followed by ethanol precipitation. Total RNA was used for reverse transcription, with 200 U of SuperScript II (Invitrogen) and synthetic oligo(dT) (GenScript, Nanjing, China). PCR was performed by using rTaq DNA polymerase (Takara, Dalian, China).
ChIP Assays
ChIP assays were performed via a ChIP kit (Millipore, Temecula, CA, USA). Cells were first cross-linked by formaldehyde at a final concentration of 1% for 10 min. Then, cells were washed twice with cold PBS (with protease inhibitors). Afterward, cells were collected and resuspended in 200 μL of SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]) and incubated for 10 min on ice. The lysates were then sonicated five times. Next, the samples were centrifuged, and the supernatants were collected and diluted 10-fold in ChIP dilution buffer. Cross-linked chromatin was incubated overnight with 10 μg of anti-histone H3K27me3 antibody or control IgG (all purchased from Abcam) in a total volume of 1 mL at 4°C. Antibody-protein-DNA complexes were immunoprecipitated by 60 μL of salmon sperm DNA/protein A. Pellets were washed and eluted by elution buffer (1% SDS, 0.1 M NaHCO3). 5 M NaCl was used for the reverse of cross-linking. Samples were purified through a QIAquick PCR purification kit (QIAGEN, Chatsworth, CA, USA). PCR amplifications were performed using the purified DNA.
Luciferase Reporter Assay
Cells were transiently co-transfected with pGL3-VEGFA PR and pCMV-ZNF24, or co-transfected with pGL3-VEGFA PR and pCMV-Runx1. Luciferase assay was performed by a Dual-Luciferase reporter assay system (Promega), following the instructions of the manufacturer.
Western Blot
Total proteins of the cells were extracted by Golden lysis buffer (Tris-HCl [pH 8.0], 400 mM NaCl, 5 mM EDTA, 1 mM EGTA, 1 mM Na pyrophosphate, 1% Triton X-100, 10% glycerol), with a supplement of protease inhibitors (Roche, Indianapolis, IN, USA). The concentration of total protein was measured with a bicinchoninic acid (BCA) kit (Pierce, Rockford, IL, USA). 20 mg of protein was loaded onto 10% SDS-PAGE gel and was transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Membranes were then incubated with primary and secondary antibodies. Protein signals were detected via the enhanced chemiluminescence (ECL) method. The primary antibodies used in this study include anti-VEGFA (1:1,000, Abcam), anti-ZNF24 (1:1,000, Abcam), anti-GAPDH (1:2,000, Abcam), anti-Runx1 (1:1,000, Abcam), and anti-H3K27me3 (1:1,000, Abcam). All primary antibodies were incubated overnight at 4°C. The horseradish peroxidase (HRP)-conjugated secondary antibody (1:5,000, Sigma-Aldrich) was incubated for 2 h at room temperature.
MTT Assay for Cell Viability
Cells were seeded in a 96-well plate at 15,000 cells/well and incubated for the indicated times. Each well of the cells was added with 20 μL of MTT (5 mg/mL) and incubated for 3 h. Next, the culture medium containing MTT solution was removed and the formazan crystals were dissolved in 100 μL of DMSO. Absorbance was read with a microplate reader (Thermo Fisher Scientific) at 490 nm.
Tube Formation Assay
12-well plates containing Matrigel (BD Biosciences, Bedford, MA, USA) were used. Cells in low serum medium (M199 containing 5% FBS) were seeded on the top of the polymerized Matrigel. After incubation for 2, 6, and 12 h, tube formation was imaged under a light microscope. To calculate the tube formation rate, the length of the capillary-like network and number of tubules were measured.
Transwell Assay
2 × 105 cells were seeded on the upper chamber of a transwell and fed with HUVEC growth medium. For the lower chamber, HUVEC growth medium was filled. After a 24-h incubation, the filters were stained with Harris hematoxylin solution (Sigma, St. Louis, MO, USA) and peeled off after washing and mounted on the slides. The migrated cells were counted under a light microscope.
ELISA
The concentrations of VEGFA in culturing supernatants of cells were measured using VEGFA ELISA kits (Thermo Fisher Scientific), following the protocols provided by the manufacturer.
Statistical Analysis
Data are expressed as mean ± standard deviation (SD) values. Comparisons between more than two groups were analyzed by one-way ANOVA followed by Tukey’s post hoc tests. Comparisons between two groups were made with two-tailed t tests. Statistical analysis was performed using SPSS 10.0 for Windows. p <0.05 was considered statistically significant.
Author Contributions
M.H., X.H., B.J., P.Z., and P.L. contributed to the conception and design of the entire study. M.H., X.H.. and L.G. participated in literature research. L.R., M.Z., and J.Z. performed the clinical studies. M.H., X.H., and S.Z performed the experimental studies. X.C. and B.J. performed the statistical analysis. M.H., X.H., and P.L. contributed to drafting the article. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (nos. 81974287, 81971820, and 81770306) and by the Key R&D Project in Hunan Province (no. 2018SK2089).
References
- 1.Jeschke M.G., Pinto R., Costford S.R., Amini-Nik S. Threshold age and burn size associated with poor outcomes in the elderly after burn injury. Burns. 2016;42:276–281. doi: 10.1016/j.burns.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadiq A., Shah A., Jeschke M.G., Belo C., Qasim Hayat M., Murad S., Amini-Nik S. The role of serotonin during skin healing in post-thermal injury. Int. J. Mol. Sci. 2018;19:1034. doi: 10.3390/ijms19041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landén N.X., Li D., Ståhle M. Transition from inflammation to proliferation: a critical step during wound healing. Cell. Mol. Life Sci. 2016;73:3861–3885. doi: 10.1007/s00018-016-2268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovtun A., Messerer D.A.C., Scharffetter-Kochanek K., Huber-Lang M., Ignatius A. Neutrophils in tissue trauma of the skin, bone, and lung: two sides of the same coin. J. Immunol. Res. 2018;2018:8173983. doi: 10.1155/2018/8173983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh T.J., DiPietro L.A. Inflammation and wound healing: the role of the macrophage. Expert Rev. Mol. Med. 2011;13:e23. doi: 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.desJardins-Park H.E., Foster D.S., Longaker M.T. Fibroblasts and wound healing: an update. Regen. Med. 2018;13:491–495. doi: 10.2217/rme-2018-0073. [DOI] [PubMed] [Google Scholar]
- 7.Zhou S., Zhang P., Liang P., Huang X. The expression of miR-125b regulates angiogenesis during the recovery of heat-denatured HUVECs. Burns. 2015;41:803–811. doi: 10.1016/j.burns.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Liang P., Jiang B., Lv C., Huang X., Sun L., Zhang P., Huang X. The expression and proangiogenic effect of nucleolin during the recovery of heat-denatured HUVECs. Biochim. Biophys. Acta. 2013;1830:4500–4512. doi: 10.1016/j.bbagen.2013.05.027. [DOI] [PubMed] [Google Scholar]
- 9.Jiang B., Li Y., Liang P., Liu Y., Huang X., Tong Z., Zhang P., Huang X., Liu Y., Liu Z. Nucleolin enhances the proliferation and migration of heat-denatured human dermal fibroblasts. Wound Repair Regen. 2015;23:807–818. doi: 10.1111/wrr.12339. [DOI] [PubMed] [Google Scholar]
- 10.Zhou J., Zhang X., Liang P., Ren L., Zeng J., Zhang M., Zhang P., Huang X. Protective role of microRNA-29a in denatured dermis and skin fibroblast cells after thermal injury. Biol. Open. 2016;5:211–219. doi: 10.1242/bio.014910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra A., Belinha J., Jorge R.N. Modelling skin wound healing angiogenesis: a review. J. Theor. Biol. 2018;459:1–17. doi: 10.1016/j.jtbi.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Pan M.R., Hsu M.C., Chen L.T., Hung W.C. Orchestration of H3K27 methylation: mechanisms and therapeutic implication. Cell. Mol. Life Sci. 2018;75:209–223. doi: 10.1007/s00018-017-2596-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C., Wang Q., Gao W., Zhang Z., Lou Y., Jin H., Chen X., Lei B., Xu H., Mao C. Highly efficient local delivery of endothelial progenitor cells significantly potentiates angiogenesis and full-thickness wound healing. Acta Biomater. 2018;69:156–169. doi: 10.1016/j.actbio.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 14.Potente M., Gerhardt H., Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 15.Johnson K.E., Wilgus T.A. Vascular endothelial growth factor and angiogenesis in the regulation of cutaneous wound repair. Adv. Wound Care (New Rochelle) 2014;3:647–661. doi: 10.1089/wound.2013.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai Y., Bai L., Zhou J., Chen H., Zhang L. Sequential delivery of VEGF, FGF-2 and PDGF from the polymeric system enhance HUVECs angiogenesis in vitro and CAM angiogenesis. Cell. Immunol. 2018;323:19–32. doi: 10.1016/j.cellimm.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Lin S., Xie J., Gong T., Shi S., Zhang T., Fu N., Ye L., Wang M., Lin Y. TGFβ signalling pathway regulates angiogenesis by endothelial cells, in an adipose-derived stromal cell/endothelial cell co-culture 3D gel model. Cell Prolif. 2015;48:729–737. doi: 10.1111/cpr.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cen X., Huang X.Q., Sun W.T., Liu Q., Liu J. Long noncoding RNAs: a new regulatory code in osteoarthritis. Am. J. Transl. Res. 2017;9:4747–4755. [PMC free article] [PubMed] [Google Scholar]
- 19.Peng L., Yuan X.Q., Zhang C.Y., Peng J.Y., Zhang Y.Q., Pan X., Li G.C. The emergence of long non-coding RNAs in hepatocellular carcinoma: an update. J. Cancer. 2018;9:2549–2558. doi: 10.7150/jca.24560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bo H., Fan L., Li J., Liu Z., Zhang S., Shi L., Guo C., Li X., Liao Q., Zhang W. High expression of lncRNA AFAP1-AS1 promotes the progression of colon cancer and predicts poor prognosis. J. Cancer. 2018;9:4677–4683. doi: 10.7150/jca.26461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian B., Wang X., Mao C., Jiang Y., Shi Y., Chen L., Liu S., Wang B., Pan S., Tao Y., Shi H. Long non-coding RNA linc01433 promotes migration and invasion in non-small cell lung cancer. Thorac. Cancer. 2018;9:589–597. doi: 10.1111/1759-7714.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu J., Li M., Zhong W., Hu C. Competing endogenous RNA in cancer: a new pattern of gene expression regulation. Int. J. Clin. Exp. Med. 2015;8:17110–17116. [PMC free article] [PubMed] [Google Scholar]
- 23.Icli B., Wu W., Ozdemir D., Li H., Haemmig S., Liu X., Giatsidis G., Cheng H.S., Avci S.N., Kurt M. MicroRNA-135a-3p regulates angiogenesis and tissue repair by targeting p38 signaling in endothelial cells. FASEB J. 2019;33:5599–5614. doi: 10.1096/fj.201802063RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Chang Y., Lu S., Xiang Y.Y. Downregulation of long noncoding RNA DGCR5 contributes to the proliferation, migration, and invasion of cervical cancer by activating Wnt signaling pathway. J. Cell. Physiol. 2019;234:11662–11669. doi: 10.1002/jcp.27825. [DOI] [PubMed] [Google Scholar]
- 25.Yu B., Wang S. Angio-LncRs: lncRNAs that regulate angiogenesis and vascular disease. Theranostics. 2018;8:3654–3675. doi: 10.7150/thno.26024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin D., Li Y., Fu C., Feng Y. Pro-angiogenic role of lncRNA HULC in microvascular endothelial cells via sequestrating miR-124. Cell. Physiol. Biochem. 2018;50:2188–2202. doi: 10.1159/000495060. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J., Du P., Cui P., Qin Y., Hu C., Wu J., Zhou Z., Zhang W., Qin L., Huang G. lncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094–4109. doi: 10.1038/s41388-018-0250-z. [DOI] [PubMed] [Google Scholar]
- 28.Zhu A.D., Sun Y.Y., Ma Q.J., Xu F. lncRNA-ATB promotes viability, migration, and angiogenesis in human microvascular endothelial cells by sponging microRNA-195. J. Cell. Biochem. 2019;120:14360–14371. doi: 10.1002/jcb.28692. [DOI] [PubMed] [Google Scholar]
- 29.Luan A., Hu M.S., Leavitt T., Brett E.A., Wang K.C., Longaker M.T., Wan D.C. Noncoding RNAs in wound healing: a new and vast frontier. Adv. Wound Care (New Rochelle) 2018;7:19–27. doi: 10.1089/wound.2017.0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C., Wang L., Ding Y., Lu X., Zhang G., Yang J., Zheng H., Wang H., Jiang Y., Xu L. lncRNA structural characteristics in epigenetic regulation. Int. J. Mol. Sci. 2017;18:2659. doi: 10.3390/ijms18122659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun T.T., He J., Liang Q., Ren L.L., Yan T.T., Yu T.C., Tang J.Y., Bao Y.J., Hu Y., Lin Y. lncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6:784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 32.Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang J., Sun C.C., Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem. Biophys. Res. Commun. 2016;478:811–817. doi: 10.1016/j.bbrc.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Jia D., Hasso S.M., Chan J., Filingeri D., D’Amore P.A., Rice L., Pampo C., Siemann D.W., Zurakowski D., Rodig S.J., Moses M.A. Transcriptional repression of VEGF by ZNF24: mechanistic studies and vascular consequences in vivo. Blood. 2013;121:707–715. doi: 10.1182/blood-2012-05-433045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie Y., Dang W., Zhang S., Yue W., Yang L., Zhai X., Yan Q., Lu J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer. 2019;18:37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang M., Wang M., Li X., Xie Y., Xia X., Tian J., Zhang K., Tang A. The role of lncRNAs in signaling pathway implicated in CC. J. Cell. Biochem. 2019;120:2703–2712. doi: 10.1002/jcb.26835. [DOI] [PubMed] [Google Scholar]
- 37.Fan C., Tang Y., Wang J., Xiong F., Guo C., Wang Y., Zhang S., Gong Z., Wei F., Yang L. Role of long non-coding RNAs in glucose metabolism in cancer. Mol. Cancer. 2017;16:130. doi: 10.1186/s12943-017-0699-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L., Peng F., Tao Y., Fan X., Li N. Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 2016;223:131–139. doi: 10.1016/j.virusres.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 39.Marchese F.P., Raimondi I., Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18:206. doi: 10.1186/s13059-017-1348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herter E.K., Xu Landén N. Non-coding RNAs: new players in skin wound healing. Adv. Wound Care (New Rochelle) 2017;6:93–107. doi: 10.1089/wound.2016.0711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kretz M., Siprashvili Z., Chu C., Webster D.E., Zehnder A., Qu K., Lee C.S., Flockhart R.J., Groff A.F., Chow J. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo L., Huang X., Liang P., Zhang P., Zhang M., Ren L., Zeng J., Cui X., Huang X. Role of XIST/miR-29a/LIN28A pathway in denatured dermis and human skin fibroblasts (HSFs) after thermal injury. J. Cell. Biochem. 2018;119:1463–1474. doi: 10.1002/jcb.26307. [DOI] [PubMed] [Google Scholar]
- 43.Liang P., Jiang B., Li Y., Liu Z., Zhang P., Zhang M., Huang X., Xiao X. Autophagy promotes angiogenesis via AMPK/Akt/mTOR signaling during the recovery of heat-denatured endothelial cells. Cell Death Dis. 2018;9:1152. doi: 10.1038/s41419-018-1194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen Y., Gao X., Tan W., Xu T. STAT1-mediated upregulation of lncRNA LINC00174 functions a ceRNA for miR-1910-3p to facilitate colorectal carcinoma progression through regulation of TAZ. Gene. 2018;666:64–71. doi: 10.1016/j.gene.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 45.DiPietro L.A. Angiogenesis and wound repair: when enough is enough. J. Leukoc. Biol. 2016;100:979–984. doi: 10.1189/jlb.4MR0316-102R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonçalves G.A.R., Paiva R.M.A. Gene therapy: advances, challenges and perspectives. Einstein (Sao Paulo) 2017;15:369–375. doi: 10.1590/S1679-45082017RB4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bodnar R.J. Chemokine regulation of angiogenesis during wound healing. Adv. Wound Care (New Rochelle) 2015;4:641–650. doi: 10.1089/wound.2014.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moritz L.E., Trievel R.C. Structure, mechanism, and regulation of polycomb-repressive complex 2. J. Biol. Chem. 2018;293:13805–13814. doi: 10.1074/jbc.R117.800367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao S.B., Zheng Q.F., Xu B., Pan C.B., Li K.L., Zhao Y., Zheng Q.L., Lin X., Xue L.X., Jin G.H. EZH2 represses target genes through H3K27-dependent and H3K27-independent mechanisms in hepatocellular carcinoma. Mol. Cancer Res. 2014;12:1388–1397. doi: 10.1158/1541-7786.MCR-14-0034. [DOI] [PubMed] [Google Scholar]
- 50.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat. Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]