Abstract
Increasing evidence indicates that lymphocyte cytosolic protein 1 (LCP1) overexpression contributes to tumor progression; however, its role in osteosarcoma (OS) remains unclear. We aimed to investigate the potential effect of LCP1 in OS and the underlying mechanisms. We first demonstrated that LCP1 is upregulated in OS cell lines and tissues. Then, we found that aberrant expression of LCP1 could induce the proliferation and metastasis of OS cells in vitro and in vivo by destabilizing neuregulin receptor degradation protein-1 (Nrdp1) and subsequently activating the JAK2/STAT3 signaling pathway. When coculturing OS cells with bone marrow-derived mesenchymal stem cells (BMSCs) in vitro, we validated that oncogenic LCP1 in OS was transferred from BMSCs via exosomes. Moreover, microRNA (miR)-135a-5p, a tumor suppressor, was found to interact upstream of LCP1 to counteract the pro-tumorigenesis effects of LCP1 in OS. In conclusion, BMSC-derived exosomal LCP1 promotes OS proliferation and metastasis via the JAK2/STAT3 pathway. Targeting the miR-135a-5p/LCP1 axis may have potential in treating OS.
Keywords: osteosarcoma, exosomal LCP1, BMSCs, miR-135a-5p, JAK2/STAT3 pathway
Graphical Abstract

Ge et al. elucidate that the oncogenic role of lymphocyte cytosolic protein 1 (LCP1) in osteosarcoma and exosomal LCP1 derived from human BMSCs plays a crucial role in malignance of osteosarcoma by inducing degradation of Nrdp1 and subsequent activation of the JAK2/STAT3 signaling pathway.
Introduction
Osteosarcoma (OS) is the most common primary malignant tumor of bone in children and adolescents (one to three cases annually per million worldwide), accounting for approximately 20% of bone neoplasia and 2% of pediatric tumors.1,2 OS typically arises from the metaphysis of long bones, and distant metastasis to the lungs is confirmed in 20% of patients at the time of diagnosis.3 Currently, the standard treatment strategies for OS are surgery combined with chemotherapy, and this has remained unchanged for the past 30 years.4,5 Despite the effort to improve therapy, the 5-year overall survival rate of patients with distant metastasis is still at a low level.6 Therefore, it is urgent to develop more potential strategies for OS.
The actin-binding protein lymphocyte cytosolic protein 1 (LCP1) was initially found to be expressed only in hematopoietic cells.7 After the discovery that LCP1 occurs in many nonhematopoietic malignancies, its role as a tumor biomarker was widely investigated. It was reported that LCP1 was positively correlated with lymph node metastasis in prostate cancer.8 In addition, the overexpression of LCP1 could induce proliferation and metastasis in colorectal and oral cancers.9, 10, 11 Although studies have confirmed that LCP1 plays important roles in several cancers, the impact of LCP1 on OS development is still unknown.
Bone marrow-derived mesenchymal stem cells (BMSCs) are reported to have self-renewing capability and multipotential, which have been exploited as therapies for many human diseases;12, 13, 14 however, in the treatment of cancer, their effects are debatable. Some studies have shown that BMSCs inhibit the development of bladder and pancreatic cancers,15,16 whereas others have indicated that BMSCs functioned as a tumor promoter in lung and hepatocellular cancers.17, 18, 19 In the tumor microenvironment, locally adjacent to OS tissues, BMSCs definitely play important roles in OS growth. Previous studies have suggested that BMSCs could promote progression of OS;20,21 however, the underlying mechanisms are still unclear.
Exosomes are small extracellular vesicles, 50–150 nm in diameter, that are released by mammalian cells through the membranes of multivesicular bodies. They were discovered in 1983, and since then, their roles in the tumor microenvironment have been widely investigated.22 Exosomes are rich in genetic material, proteins, and lipids, and they mediate interactions between cancer cells and other cells in the tumor microenvironment via a paracrine effect.23 Accumulating evidence has shown that exosomes can realize the cell–cell communication between BMSCs and tumor cells by packaging protein and RNA, thus exerting the effects of BMSCs on tumor progression.24,25 In addition, previous studies have revealed that exosomes could mediate the crosstalk between BMSCs and OS cells, and BMSC-derived exosome additions could promote OS progression.26 However, the underlying mechanisms of BMSC-derived exosomes aggravating OS still need further investigation.
Post-transcriptional regulation plays a key role in protein expression. MicroRNAs (miRNAs) are small noncoding RNAs, and their dysregulation is involved in oncogenesis.27, 28, 29 By binding to the 3′ untranslated region (UTR) of target mRNA, miRNAs negatively regulate the translation of mRNA.30 Levels of microRNA (miR)-135a-5p, a tumor suppressor, are reported to be decreased in several malignant cancers, and its aberrant expression is correlated with worse clinical outcomes.31, 32, 33, 34 However, the role of miR-135a-5p in OS remains unclear.
In this study, we explored the role of LCP1 in OS and uncovered the potential underlying mechanism in vitro and in vivo. We found that aberrant LCP1 transferred from BMSCs via exosomes plays pro-tumorigenesis and pro-metastasis effects on OS through destabilization of neuregulin receptor degradation protein-1 (Nrdp1) and activation of the JAK2/STAT3 pathway, whereas miR-135a-5p can inhibit these effects. Our findings may provide a potential therapeutic target for OS.
Results
LCP1 Is Upregulated in OS Specimens and Cells
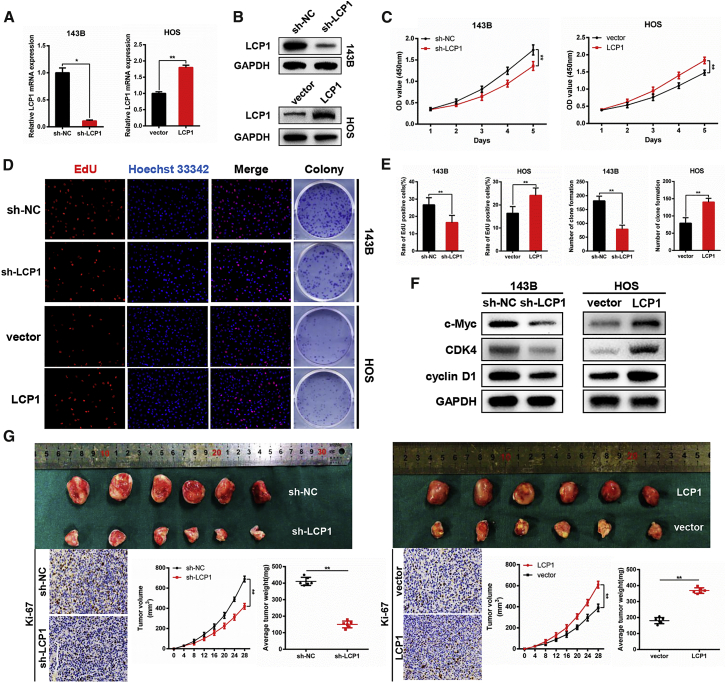
We first analyzed three OS-related datasets (GEO: GSE12865, GSE14359, and GSE33382), and all of the three public datasets showed that the expression level of LCP1 was higher in OS tissues (Figures 1A–1C). The mRNA expression level of LCP1 in human fetal osteoblastic (hFOB) cell line 1.19, human OS (HOS), U-2 OS, Saos-2, 143B, and clinical samples was measured via qRT-PCR. As shown in Figure 1D, the expression level of LCP1 was significantly upregulated in OS cell lines (p = 0.000) as well as OS tumor tissues (p = 0.000). Western blot analysis demonstrated similar results with qRT-PCR (Figure 1E). In an immunohistochemistry assay, LCP1 and Ki-67 expression was markedly higher in OS tissues than in adjacent tissues (Figure 1F), which was consistent with the above results. Furthermore, we explored the relationship between LCP1 expression and clinicopathological characteristics in 40 OS patients (Table 1), and we found that the expression level of LCP1 was positively correlated with tumor node metastasis (TNM) stage, tumor size, and metastasis.
Figure 1.
LCP1 Is Upregulated in Osteosarcoma (OS) Cell Lines and Tissues
(A) The hub genes upregulated in OS from the GEO: GSE12865, GSE14359, and GSE33382 databases. (B) Log2 fold-change values of LCP1 in three databases. (C) The volcano map of differentially expressed genes in GEO: GSE12865. (D) The mRNA expression level of LCP1 in hFOB 1.19 and five OS cell lines and clinical samples. (E) The protein level of LCP1 in hFOB 1.19 and five OS cell lines and clinical samples. (F) Immunohistochemistry staining was used to determine the protein levels of LCP1 and Ki-67 between OS tissues and adjacent tissues.
Table 1.
Expression of LCP1 and miR-135a-5p According to Patients’ Clinical Features
| Characteristics | Number | LCP1 Expression |
p Value | miR-135a-5p Expression |
p Value | ||
|---|---|---|---|---|---|---|---|
| High Group | Low Group | High Group | Low Group | ||||
| Age (Years) | |||||||
| <18 | 25 | 10 | 15 | 0.41 | 11 | 14 | 0.80 |
| ≥18 | 15 | 8 | 7 | 6 | 9 | ||
| Gender | |||||||
| Female | 22 | 12 | 10 | 0.95 | 9 | 13 | 0.57 |
| Male | 18 | 10 | 8 | 9 | 9 | ||
| Location | |||||||
| Femur/tibia | 33 | 16 | 17 | 0.79 | 14 | 19 | 0.48 |
| Elsewhere | 7 | 3 | 4 | 4 | 3 | ||
| TNM Stage | |||||||
| I | 19 | 8 | 11 | 0.028a | 13 | 6 | 0.027a |
| II/III | 21 | 16 | 5 | 7 | 14 | ||
| Tumor Size (cm) | |||||||
| <5 | 18 | 7 | 11 | 0.031a | 12 | 6 | 0.028a |
| ≥5 | 22 | 16 | 6 | 7 | 15 | ||
| Metastasis | |||||||
| Yes | 16 | 12 | 4 | 0.02a | 5 | 11 | 0.028a |
| No | 24 | 9 | 15 | 16 | 8 | ||
Expression levels of LCP1 and miR-135a-5p were correlated with tumor TNM stage and metastasis, according to the clinical features of 40 patients.
p < 0.05 (chi-square test).
LCP1 Promotes the Proliferation of OS Cells In Vitro and In Vivo
To investigate the effect of LCP1 on OS cells in vitro, HOS and 143B cells were selected for transfection with the LCP1-coding sequence and short hairpin (sh)RNA-targeting LCP1 (LCP1 and sh-LCP1), respectively. Transfection efficiency is shown in Figure 2A (p = 0.020 and p = 0.000 for knockdown and overexpression assays) and Figure 2B. The results of a cell-counting assay showed that cell proliferation was significantly inhibited after silencing LCP1 (p = 0.000); conversely, LCP1 overexpression promoted the proliferation of OS cells (p = 0.000; Figure 2C). Moreover, the clonality of OS cells was suppressed by LCP1 knockdown (p = 0.001) but increased by LCP1 upregulation (p = 0.006; Figures 2D and 2E). Furthermore, a 5-ethynyl-2′-deoxyuridine (EdU) proliferation assay indicated that LCP1 plays a key role in cell proliferation, as the percentage of mitotic cells was decreased by LCP1 downregulation (p = 0.004) and vice versa (p = 0.004; Figures 2D and 2E). Results of western blotting demonstrated that LCP1 depletion significantly attenuated the levels of G1/S checkpoint proteins, including c-Myc, cyclin-dependent kinase-4 (CDK4), and cyclin D1, whereas LCP1 overexpression conversely enhanced their expression (Figure 2F), which suggested that LCP1 promotes OS cell proliferation by accelerating cell cycle from G1 phase to S phase.
Figure 2.
LCP1 Promotes OS Proliferation In Vitro and In Vivo
(A and B) Transfection efficiency was examined by qRT-PCR (A) and western blot analysis (B). (C–E) The effect of LCP1 on proliferation in vitro was detected using CCK-8 (C), EdU (D [left] and E [left]), and colony-formation assays (D [right] and E [right]). (F) Western blot analysis of cell-cycle-related proteins following LCP1 depletion and overexpression. (G) Knockdown of LCP1 inhibited tumor growth in vivo, whereas the upregulation of LCP1 promoted tumor growth. Tumor volume was measured every 4 days after injection. Tumor weight and Ki-67 expression were determined.
In a subcutaneous tumorigenesis assay, stably transfected cells were subcutaneously injected into nude mice. The weight (p = 0.000) and size (p = 0.000) of tumors were significantly decreased after LCP1 knockdown. An immunohistochemistry assay showed that Ki-67 expression levels in the tumor nodes were attenuated in a sh-LCP1 group, whereas the opposite result was observed in an LCP1 overexpression group (p = 0.000; Figure 2G).
LCP1 Promotes Metastasis of OS Cells In Vitro and In Vivo
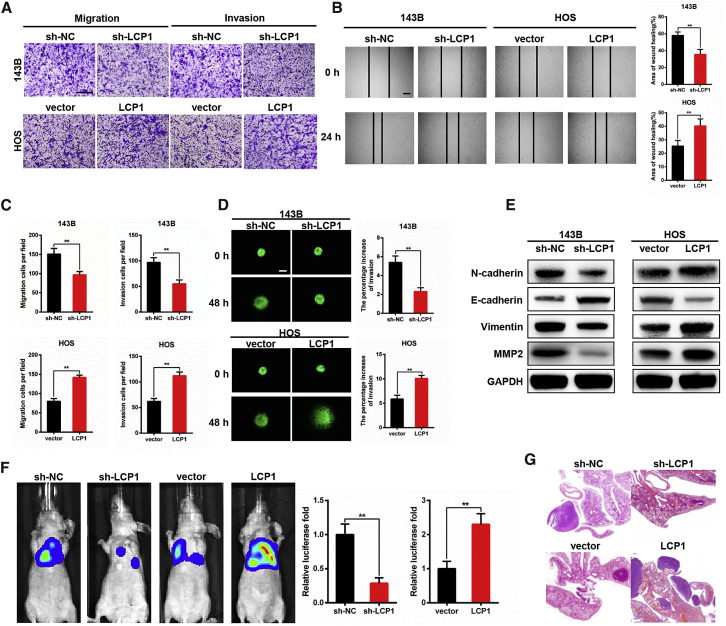
To explore whether LCP1 plays a crucial role in OS cell metastasis, Transwell and wound-healing assays were conducted. In the Transwell assay, the number of migrated cells was significantly decreased after LCP1 downregulation (p = 0.000) but increased by LCP1 overexpression (p = 0.000). Similarly, silencing LCP1 reduced cell invasion (p = 0.000), whereas upregulating LCP1 expression enhances the invasive ability of cells (p = 0.000; Figures 3A and 3C). A wound-healing assay showed that the migration rate was restrained in the LCP1 knockdown group (p = 0.006); however, the opposite result was observed in the LCP1 overexpression group (p = 0.002; Figure 3B). Results of the 3D tumor spheroid invasion assay also demonstrated that a higher expression level of LCP1 led to the stronger invasive ability of OS cells (p = 0.002 and p = 0.002 for knockdown and overexpression assays; Figure 3D).
Figure 3.
LCP1 Promotes OS Metastasis In Vitro and In Vivo
(A–C) The effect of LCP1 on cell migration and invasion was evaluated by Transwell (A and C) and wound-healing assays (B). (D) 3D tumor spheroid invasion assay was performed to determine the effect of LCP1 on invasion. (E) Western blot analysis showed that LCP1 promoted epithelial-mesenchymal transition (EMT). (F) Representative photographs of metastatic nodules were taken by an IVIS imaging system. Quantification of the luciferase is shown. (G) H&E staining was used to characterize the lung metastatic nodules. Scale bar, 200 μm.
We next analyzed the relationship between LCP1 and proteins involved in epithelial-mesenchymal transition (EMT) through western blotting. The expression levels of N-cadherin, vimentin, as well as matrix metalloproteinase-2 (MMP-2) were attenuated following LCP1 knockdown, whereas E-cadherin was upregulated (Figure 3E), and the opposite result was observed when LCP1 was overexpressed, which revealed that LCP1 promotes tumor metastasis via activating the EMT process.
To generate a pulmonary metastasis model, cells stably transfected with luciferase were injected into the tail vein of nude mice to determine the role of LCP1 on tumor metastasis in OS. As expected, LCP1 knockdown greatly suppressed the invasion of OS cells and alleviated pulmonary metastasis compared with the control group (p = 0.002), whereas LCP1 overexpression could reverse these effects (p = 0.004; Figures 3F and 3G). Taken together, in vitro and in vivo experiments further confirmed that LCP1 accelerated OS metastasis via activating the EMT process.
LCP1 Promotes Metastasis via Degradation of Nrdp1 and Activation of the JAK2/STAT3 Signaling Pathway
To uncover the downstream signaling pathways of LCP1, a single-gene gene-set enrichment analysis (GSEA) was performed using the correlation coefficient of each gene with LCP1 in three public databases. Results indicated that the JAK2/STAT3 signaling pathway was enriched after LCP1 overexpression (p = 0.007; Figures 4A and 4B). As expected, a western blot assay further confirmed that LCP1 knockdown markedly decreased the phosphorylation of JAK2 and STAT3. Moreover, the expression level of phosphorylated STAT3 in the nuclei also decreased (Figure 4C). To investigate whether the JAK2/STAT3 signaling pathway participated in LCP1-induced metastasis and EMT activation, an agonist (coumermycin A1 [C-A1]) and inhibitor (fedratinib) of the JAK2/STAT3 pathway were used, and the expression levels of EMT-related proteins were examined. As shown in Figure 4C, expression levels of N-cadherin, vimentin, and MMP-2 were downregulated following LCP1 knockdown, whereas E-cadherin was upregulated. The C-A1 could partly reverse these effects. The opposite result was observed in the LCP1 overexpression group. In addition, an immunofluorescence assay revealed that knockdown of LCP1 significantly suppressed the transport of phosphorylated STAT3 from the cytoplasm into the nuclei, whereas upregulation of LCP1 reversed this effect (Figure 4D).
Figure 4.
LCP1 Participates in the JAK2/STAT3 Signaling Pathway
(A and B) Single-gene GSEA analysis showed top activated gene sets ordered by gene ratio (A) and the JAK2/STAT3 signaling pathway was enriched after LCP1 overexpression (B). (C) Western blot analysis indicated that LCP1 promoted the phosphorylation of JAK2 and STAT3 and EMT. (D) Immunofluorescence showed that LCP1 promoted the transport of phosphorylated STAT3 from the cytoplasm to the nucleus in OS cells. Scale bar, 50 μm.
To explore underlying mechanisms of how LCP1 activates the JAK2/STAT3 pathway, we predicted the potential proteins interacting with LCP1 online (Uniprot; https://www.uniprot.org/) and selected those that are involved in the JAK2/STAT3 pathway, including CXC receptor 2, Nrdp1, and transcription factor AP-1 for the coimmunoprecipitation (coIP) assay. We found that only Ndrp1, which was reported to negatively regulate JAK2 activation,35,36 could interact with LCP1 (Figure 5A). The fluorescence colocation assay further demonstrated that LCP1 and Nrdp1 are colocated in the cytoplasm (Figure 5B). Interestingly, we identified no significant differences of Nrdp1 mRNA expression after LCP1 overexpression in OS cells (p = 0.693 for 143B and p = 0.657 for HOS; Figure 5C), whereas the Nrdp1 protein level was attenuated in LCP1-overexpressed OS cells (Figure 5D), so we asked whether the post-translational modification of LCP1 on Nrdp1 is due to ubiquitination (Ub). Addition of 10 μM proteasome inhibitor MG132 reversed the downregulation of the Nrdp1 level following LCP1 overexpression (Figure 5D). Meanwhile, endogenous Nrdp1 was degraded more rapidly in LCP1-overexpressed cells compared with control cells in the presence of cycloheximide (100 μg/mL), an inhibitor of protein translation (p = 0.000; Figure 5E). The ubiquitylation assay was then performed to determine whether LCP1 regulates Nrdp1 destabilization via proteasomal degradation. We found that overexpression of LCP1 significantly increased the Nrdp1 polyubiquitination in 143B and HOS cell lines (Figure 5F), which suggested that LCP1 destabilizes Nrdp1 by mediating its polyubiquitination and proteasomal degradation. Taken together, the above results confirmed that LCP1 promoted EMT via degradation of Nrdp1 and activation of the JAK2/STAT3 signaling pathway.
Figure 5.
Overexpression of LCP1 Increased Nrdp1 Polyubiquitination and Proteasomal Degradation
(A) Coimmunoprecipitation analysis demonstrated that LCP1 could interact with Nrdp1. (B) Fluorescence colocation analysis indicated that LCP1 is colocated with Nrdp1 in the cytoplasm. (C) Overexpression of LCP1 did not affect the Nrdp1 mRNA level in 143B and HOS cells. (D) Western blot analysis of LCP1 and Nrdp1 in 143B and HOS cells, which were overexpressed LCP1 in the presence or absence of 10 μM MG132. (E) 143B and HOS cells transfected with the LCP1-coding sequence were treated with cycloheximide (100 μg/mL) and collected for indicated times for western blot analysis. Quantification of Nrdp1 expression is shown. (F) 143B and HOS cells transfected with HA-Ub and the LCP1-coding sequence were pretreated with MG132 for 8 h before harvest. Nrdp1 was immunoprecipitated with anti-Nrdp1 antibody and then immunoblotted with anti-HA antibody.
Coculturing with BMSCs Promotes OS Progression In Vitro
Previous studies revealed that BMSCs functioned as a regulator of several tumors via intercellular communication,18,19,37,38 but its role in the OS tumor microenvironment is still unclear. To explore further the effect of BMSCs on OS, we cocultured BMSCs with OS cells; a sketch map of the coculture is shown in Figure 6A. Surprisingly, coculture with BMSCs increased mRNA and protein expression of LCP1 in OS cells (p = 0.000; Figures 6E and 6F). In addition, we found that OS cell proliferation was significantly increased, as shown by the results of the cell-counting assay (p = 0.000; Figure 6B), colony-formation assay (p = 0.000; Figure 6C), and EdU assay (p = 0.000; Figure 6D). Correspondingly, levels of c-Myc, CDK4, and cyclin D1 were upregulated (Figure 6F). Given that exosomes could realize cell–cell communication through a paracrine effect in the tumor microenvironment and to investigate further whether BMSCs promote OS progression through exosomes, we first extracted and identified the exosomes of BMSCs (Figures S1A–S1C), and we identified that BMSC-derived exosomes could be taken up by OS cells (Figure S1D). We then pretreated BMSCs with GW4869, an inhibitor of exosomal secretion, before coculturing with OS cells. The results demonstrated that addition of GW4869 downregulated expression of LCP1 in OS cells when coculturing with BMSCs (p = 0.000; Figures 6E and 6F). Also, the tumor-promoting effect of BMSCs was attenuated by GW4869 (Figures 6B–6D and 6F). Moreover, similar results were observed in the metastatic ability of OS cells (Figures 6G–6J), and the expression levels of proteins related to EMT process were consistent with the above results (Figure 6K). Taken together, BMSCs promoted OS proliferation and metastasis in vitro, and these effects could be reversed by GW4869. It is well known that exosomes contain genetic material, which is taken up by other cells where they exert their functions.23 Thus, we hypothesized that LCP1 was delivered by exosomes from BMSCs to act as a tumor promotor to accelerate OS progression.
Figure 6.
OS Cell Proliferation, Migration, and Invasion Abilities Were Enhanced When Cocultured with BMSCs In Vitro
(A) Diagrammatic sketch of BMSCs and OS cell coculture. (B) The CCK-8 assay indicated that coculturing with BMSCs promoted tumor proliferation, which was reversed by the exosomal inhibitor GW4869. (C and D) Results of the colony-formation assay (C) and EdU assay (D) were in accordance with the above results. (E) The LCP1 mRNA expression in 143B and HOS cells cocultured with BMSCs with or without treatment of GW4869. (F) Western blot analysis of c-Myc, CDK4, cyclin D1, and LCP1 in 143B and HOS cells cocultured with BMSCs with or without treatment of GW4869. (G–J) The Transwell (G and J [top]), wound-healing (H and J [lower left]), and 3D tumor spheroid assays (I and J [lower right]) showed that coculture with BMSCs promoted tumor migration and invasion, whereas GW4869 could partly reverse this effect. (K) EMT in OS cells was activated when cocultured with BMSCs, whereas the administration of GW4869 could attenuate this effect. Scale bars, 200 μm.
LCP1 Transferred from BMSCs via Exosomes Promotes OS Cell Proliferation and Metastasis In Vitro
To confirm our hypothesis, we silenced LCP1 in BMSCs using shRNA and then examined the expression level of exosomal LCP1 via qRT-PCR (p = 0.015; Figure 7A). The mRNA and protein levels of LCP1 in OS cells cocultured with indicated BMSCs are shown in Figure 7B (p = 0.015 for 143B and p = 0.004 for HOS) and Figure 7F. Compared to the control group, we found that OS cell proliferation was significantly inhibited after coculture with LCP1-knockdown-BMSCs (Figures 7C–7E), and the expression levels of c-Myc, CDK4, and cyclin D1 were significantly attenuated in the LCP1-knockdown-BMSC group as well (Figure 7F). In addition, Transwell, wound-healing, and 3D tumor spheroid invasion assays all indicated that cell-invasive ability was also attenuated after coculture with LCP1-knockdown-BMSCs compared to BMSCs (Figures 7G–7J). As expected, EMT was suppressed in the sh-LCP1 group compared with the control group (Figure 7K).
Figure 7.
OS Cell Proliferation, Migration, and Invasion Abilities Were Reversed When Cocultured with LCP1-Knockdown-BMSCs Compared to BMSCs
(A) Efficiency of LCP1 knockdown in BMSC-derived exosomes. (B) Relative LCP1 mRNA expression of 143B and HOS cells cocultured with LCP1-knockdown-BMSCs compared to BMSCs. (C) The CCK-8 assay showed that coculturing with LCP1-knockdown-BMSCs inhibited OS cell proliferation compared to BMSCs. (D and E) The colony-formation assay (D) and EdU assay (E) showed similar results with CCK-8 assay. (F) Western blot analysis of c-Myc, CDK4, cyclin D1, and LCP1 in 143B and HOS cells cocultured with LCP1-knockdown-BMSCs compared to BMSCs. (G–J) The Transwell assay (G and J [top]), wound-healing assay (H and J [lower left]), and 3D tumor spheroid assay (I and J [lower right]) indicated that coculturing with LCP1-knockdown-BMSCs significantly attenuated OS cell migrative and invasive abilities compared to BMSCs. (K) EMT in OS cells was inhibited by coculturing with LCP1-knockdown-BMSCs compared to BMSCs. Scale bars, 200 μm.
To confirm further our hypothesis, we examined the expression level of exosomal LCP1 in the serum of healthy volunteers and OS patients. Transmission electron micrographs of exosomes from healthy volunteers and OS patients are shown in Figure 8A. It is worth mentioning that the expression level of exosomal LCP1 derived from the blood serum of healthy volunteers was significantly lower than that of OS patients (p = 0.014; Figure 8B). All of the above results demonstrate that the exosomal transfer of LCP1 from BMSCs promotes OS development.
Figure 8.
Identification of Exosomes in the Serum of Patients and Graphical Abstract of the Potential Underlying Mechanism of Exosomal LCP1 in Promoting OS Progression
(A) Transmission electron micrographs of exosomes derived from healthy volunteers and OS patients. (B) The exosomal LCP1 level in serum of volunteers and OS patients was determined by qRT-PCR. (C) The potential underlying mechanism of exosomal LCP1 derived from BMSCs in promoting OS progression. Scale bars, 100 nm.
LCP1 Is Negatively Regulated by miR-135a-5p
We next investigated miRNAs that may regulate LCP1 expression through a bioinformatics website (http://starbase.sysu.edu.cn/starbase2/index.php). Four hub miRNAs were predicted as potential regulators (Figure S2A). To determine if LCP1 was regulated by the predicted miRNAs, the mRNA level of LCP1 was examined after upregulating the miRNAs, and only miRNA-135a-5p had a significant inhibitory effect on LCP1 expression (p = 0.000; Figure S2B). qRT-PCR demonstrated that miR-135a-5p was downregulated in OS cell lines and tumor tissues (p = 0.000; Figures S2C and S2D), and it was negatively correlated with the expression level of LCP1 (p = 0.002; Figure S2E). Furthermore, the expression level of miR-135a-5p was found to be negatively correlated with TNM stage, tumor size, and metastasis (Table 1). The results of a luciferase reporter assay indicated that ectopic expression of miR-135a-5p significantly reduced the luciferase activity of wild type (WT)-LCP1-3′ UTR but had no effect on mutant (MUT)-LCP1-3′ UTR in 143B (p = 0.002) and HOS cell lines (p = 0.011; Figure S2G), which confirmed the binding between miR-135a-5p and LCP1. Moreover, a western blot assay showed that miR-135a-5p downregulated the protein level of LCP1, whereas its inhibitor could counteract this effect (Figure S2H). This further confirmed that miR-135a-5p, a tumor-suppressing miRNA, could bind to and directly regulates LCP1 expression.
miR-135a-5p Inhibits Tumor Proliferation and Metastasis by Suppressing LCP1 in OS Cells
To verify that LCP1 was a downstream target of miR-135a-5p, a series of rescue experiments were performed. LCP1 was overexpressed in 143B cells following transfection with miR-135a-5p mimics, whereas it was silenced in HOS cells following transfection with the miR-135a-5p inhibitor. The cell counting (Figure S3A), colony-formation (Figure S3B), EdU (Figure S3C), Transwell (Figure S4A), wound-healing (Figure S4B), and 3D tumor spheroid invasion assays (Figure S4C) suggested that cotransfection of LCP1 in miR-135a-5p-upregulated 143B cells partly reversed the inhibitory role of miR-135a-5p in OS cell proliferation and metastasis. However, silencing LCP1 in HOS cells with miR-135a-5p downregulated abolished the effect of miR-135a-5p inhibition on OS cell proliferation and metastasis. These findings revealed that miR-135a-5p suppressed tumor proliferation and metastasis by suppressing LCP1 in OS cells.
miR-135a-5p Inhibits the JAK2/STAT3 Signaling Pathway and EMT Process by Targeting LCP1
Given that LCP1 knockdown inhibits the phosphorylation of the JAK2/STAT3 signaling pathway and EMT process, western blot analysis was conducted to determine whether miR-135a-5p could target LCP1 to regulate these effects. As expected, miR-135a-5p reduced the phosphorylation level of the JAK2/STAT3 signaling pathway and suppressed the EMT process. Furthermore, this inhibitory effect was partly rescued by LCP1 re-expression. An opposite result was observed in the miR-135a-5p downregulated cells (Figure S5).
Discussion
LCP1, an actin-binding protein, is upregulated in several cancer types, and its tumor-promoting effects have been widely investigated.7 For instance, redox-modified LCP1 is known to inhibit the actin-based functions of tumor cells,39 and an in vivo analysis indicated that the knockdown of LCP1 blocked the migration of chronic lymphocytic leukemia to bone marrow.40 It is worth mentioning that phosphorylated LCP1 is recognized as the form that promotes tumor progression in breast and prostate cancers.41,42 Moreover, LCP1 could be transferred by breast cancer cell-derived exosomes to facilitate metastatic bone osteolysis,43 which provides an insight into the transfer of LCP1 between cells in the tumor microenvironment. Based on these studies, we examined the expression level of LCP1 in OS and verified that LCP1 was overexpressed in OS cell lines and tumor tissues, which was consistent with the findings from bioinformatics analysis. Furthermore, we identified endogenous LCP1 as a tumor-promoting factor in OS proliferation and metastasis.
BMSCs are mesenchymal stem cells, isolated from bone marrow, that play crucial roles in tumor progression. For OS, studies have suggested that OS may be originated from BMSCs,44, 45, 46, 47 and it is well established that the bone microenvironment is critical for OS developed from BMSCs. Baglio et al.48 reported that tumor-educated BMSCs promote tumor growth and metastasis in vivo by increasing interleukin (IL)-6 production. Another study reported that once in contact with OS cells, BMSCs would transdifferentiate into cancer-associated fibroblasts, increasing cytokines in the tumor microenvironment and further increasing aggressiveness of OS cells.49 Coincidentally, the STAT3 pathway was previously shown to participate in the crosstalk between BMSCs and OS cells.50,51 To investigate further the relationship between BMSCs and OS cells, we cocultured the two cell types and found that BMSCs could transfer LCP1 to OS cells via delivering exosomes and further aggravate OS malignancy. The contribution of endogenous and exosomal LCP1 in OS development was then balanced through silencing LCP1 in OS cells and coculturing them with BMSCs; we identified that LCP1 expression was upregulated in OS cells cocultured with BMSCs compared with those without coculture, which means that exosomal LCP1 could promote OS progression even if endogenous LCP1 was knockdown. It is well established that BMSC-derived exosomes could transfer genetic material to other cells and exert biofunctions similar to those of BMSCs.52,53 Huang et al.54 reported that exosomes derived from BMSCs promote OS development via activating oncogenic autophagy in vivo. Another study demonstrated that BMSC-derived exosomal miR-208a promotes the progression of OS by downregulating expression of programmed cell death factor 4 (PDCD4) and activating the extracellular signal-regulated kinase (ERK)1/2 pathway.55 There are a great number of similar studies, which further demonstrate that BMSC-derived exosomes play a crucial role in OS development.
The EMT process has been identified as a driver of tumor metastasis by degrading extracellular matrix proteins and triggering the dissociation of cancer cells from the primary tumor.56 Mediated by multiple transcription factors, the EMT process is characterized by modifying adhesion molecules expressed by cells, including the downregulation of E-cadherin and the upregulation of N-cadherin, vimentin, and MMPs.57 Studies have shown that LCP1 expression is correlated with cell motility and could induce E-cadherin downregulation in colorectal carcinoma cells.9 In the current study, we found that LCP1 accelerates the metastasis of OS and regulates the expression of EMT-related proteins, and these effects could be partly rescued by a JAK2 inhibitor, which further illustrates that LCP1 regulates the EMT process via the JAK2/STAT3 pathway. However, when we blocked the JAK2/STAT3 pathway in LCP1-overexpressed OS cells with fedratinib, we found the proliferation was not reversed, which demonstrated that the JAK2/STAT3 pathway is not involved in the proliferative effects of LCP1 on OS cells, and there must be some other underlying mechanism leading to LCP1-induced proliferation. In the 1990s, JAKs and STATs were recognized as being associated with malignancy, and since then, the JAK2/STAT3 pathway has been widely investigated in cancers.58,59 In response to cytokines, JAK2 becomes activated and thereby phosphorylates STAT3. Then, phosphorylated STAT3 dimerizes and moves into the nucleus to activate the transcription of cytokine-responsive genes.60 STAT3-induced EMT was also reported in lung cancer pretreated with hypoxic BMSC-derived exosomes.17 Moreover, Wu et al.61 revealed that aberrant STAT3 leads to hepatocellular carcinoma aggressiveness through the induction of EMT. The chromatin immunoprecipitation (ChIP) assay performed by Lin et al.62 further confirmed the direct binding between STAT3 and Slug, which is an EMT-related protein. These studies revealed that the JAK2/STAT3 signaling pathway is an inductor of the EMT process and regulates tumor metastasis.
Clearly, ubiquitination is a post-translational modification controlling the proteasomal degradation of proteins. Starting by attachment with Ub, substrates will ultimately be degraded by the 26S proteasome.63 In our study, we found that LCP1 could interact with and ubiquitinate Nrdp1, leading to subsequent degradation of Nrdp1 and activation of the JAK2/STAT3 pathway. Previous studies reported that Nrdp1 regulated degradation and shedding of JAK2-associated cytokine receptors to inactivate the STAT signaling pathway,36 which is induced by interaction between Nrdp1 and Ub-specific protease 8 (USP8)35. Moreover, Wu et al.64 confirmed that rhodanese and catalytic domains of USP8 could bind to Nrdp1 and enhance its stability. The effect of LCP1 on Nrdp1 was probably due to its competitive combination with USP8, and this needs further investigation.
Bioinformatics analysis and luciferase reporter assay indicated that miR-135a-5p correlates with LCP1. Previous studies have demonstrated that miR-135a-5p plays a tumor-suppressing role in different cancer types. By targeting HOXA10, it inhibited proliferation and enhanced apoptosis in head and neck squamous cell carcinoma;31 assisted by miR-124-3p, miR-135a-5p significantly interfered in the malignancy of glioblastoma cells;32 and Zheng et al.34 showed that the tumor-suppressing effects of miR-135a-5p could be regulated by long noncoding RNA (lncRNA) differentiation antagonizing nonprotein coding RNA (DANCR) in tongue squamous cell carcinoma cells. Our findings indicate that miR-135a-5p suppresses OS progression by negatively regulating the LCP1/JAK2/STAT3/EMT axis.
Taken together, our findings demonstrate that exosomal LCP1 derived from BMSCs promotes OS progression via degradation of Nrdp1 and activation the JAK2/STST3 signaling pathway and is negatively regulated by miR-135a-5p (Figure 8C). Our findings may enhance the understanding of OS development and provide a potential therapeutic target for OS.
Materials and Methods
Ethics Approval and Consent to Participate
This study was approved by the Institutional Review Board and The First Affiliated Hospital of Nanjing Medical University Ethics Committee, and informed consent was obtained from all patients and volunteers. Forty pairs of OS tissues and matched adjacent tissues were collected for experiments during surgery from the Department of Orthopedics, The First Affiliated Hospital of Nanjing Medical University, and the blood serum of OS patients and healthy volunteers were obtained for exosome extraction. The clinical features of the patients are shown in Table 1.
Reagents and Antibodies
The exosomal secretion inhibitor GW4869 was obtained from Sigma-Aldrich (St. Louis, MO, USA). C-A1 was bought from Santa Cruz Biotechnology (Santa Cruz, CA, USA), and fedratinib was obtained from MedChemExpress (Monmouth Junction, NJ, USA). The primary antibodies used for western blot and immunofluorescence assay in our study included anti-LCP1 (Cell Signaling Technology [CST], USA; Proteintech, China), anti-Nrdp1 (Santa Cruz Biotechnology), anti-hemagglutinin (HA; CST), anti-Ki-67 (CST), anti-c-Myc (CST), anti-CDK4 (CST), anti-cyclin D1 (CST), anti-N-cadherin (CST), anti-E-cadherin (CST), anti-vimentin (CST), anti-MMP-2 (CST), anti-JAK2 (CST), anti-phospho-JAK2 (CST), anti-STAT3 (CST), anti-phospho-STAT3 (CST), anti-CD63 (Abcam, UK), anti-CD9 (CST), anti-calnexin (CST), anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH; CST), and anti-Lamin B1 (CST). The horseradish peroxidase (HRP)- and fluor-conjugated secondary antibodies (Jackson ImmunoResearch, USA) were utilized for western blot and immunofluorescence assay, respectively.
Microarray Data
Three gene-expression profiles (GEO: GSE12865, GSE14359, and GSE33382) of OS were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Before analyzing differentially expressed genes (DEGs), unrelated samples were excluded. Following data preprocessing and the identification of DEGs, single-gene GSEA analysis was performed to further investigate functions of LCP1 in OS.
Cell Culture
All human OS cell lines (143B, HOS, Saos-2, and U-2 OS), human osteoblast cells (hFOB 1.19), and BMSCs were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). OS cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin and incubated with 5% CO2 at 37°C. The hFOB 1.19 cell line was cultured in DMEM/F-12 containing 10% FBS, 1% penicillin-streptomycin, and 0.3 mg/mL of G418 and incubated with 5% CO2 at 33.5°C. BMSCs were cultured in alpha-modified eagle medium (αMEM) containing 5% FBS, 1% mesenchymal stem cell growth supplement, and 1% penicillin-streptomycin and incubated with 5% CO2 at 37°C.
Cell Transfection and Lentivirus Construction
Lentivirus vectors containing LCP1- and shRNA-coding sequences were constructed by GenePharma (Shanghai, China) to up- or downregulate LCP1 in OS cells. When cells were at 40%–50% confluency, they were transfected with LCP1, vector, sh-LCP1, and sh-negative control (NC). Stably transfected cell lines were filtered by puromycin (Sigma-Aldrich) for 1 week. The transfection efficiency was examined by qRT-PCR and western blot analysis. miR-135a-5p mimics and miR-135a-5p inhibitor were purchased from RiboBio (Guangzhou, China).
Cell Coculture
A six-well culture plate insert with 0.4 μm pores (Millipore, USA) was used to investigate the effects of BMSCs on OS cells. BMSCs were seeded onto the upper chamber, whereas OS cells were planted in the lower dish. The cellular secretion of BMSCs was allowed to pass through the membrane to stimulate OS cells. Following coincubation for 48 h, cell proliferation and metastatic ability were assessed.
Exosome Extraction
Exosomes were extracted from the culture supernatant of stably transfected BMSCs, according to our previous study.65 Exosomes in the blood serum of patients with OS and healthy volunteers were isolated using the method mentioned above. The identification of exosomes was performed via transmission electron microscopy, nanoparticle tracking analysis, and western blotting.
Exosome Uptake by OS Cells
Dil solution (4 mg/mL; Molecular Probes, USA) was incubated with exosomes containing PBS solution for fluorescent labeling. Excessive dye was removed through centrifugation at 100,000 × g at 4°C, and labeled exosomes were washed three times by resuspending in PBS. These Dil-labeled exosomes were cocultured with OS cells for 24 h, and the cells were then washed with PBS and fixed in 4% paraformaldehyde. Uptake of exosomes by OS cells was visualized by laser confocal microscopy.
Cell Viability Assay
Cell viability was accessed by using a Cell Counting Kit-8 (CCK-8; Dojindo, Japan). The cells were seeded in 96-well plates (1,000 cells per well) and cultured in DMEM with 10% FBS. After different periods of incubation, 10 μL of CCK-8 reagent was added to each well to determine cell viability using a microplate spectrophotometer at 450 nm.
Colony-Formation Assay
The OS cells were seeded into six-well plates (500 cells per well) and cultured in DMEM containing 10% FBS for 2 weeks. Clones were fixed with methanol and stained with 0.1% crystal violet when they were visible to the naked eye. All colonies were counted manually and photographed under a scanner (Microtek, China).
EdU Assay
The cells were plated in 96-well plates (6,000 cells per well) with DMEM containing 10% FBS for 24 h. Following incubation in 50 μM EdU reagent for 2 h, cells were fixed with 4% paraformaldehyde, permeated by 0.5% Triton X-100, and stained with 1 × Apollo reagent for 30 min. Nuclei were stained with 1 × Hoechst 33342, and the cells were visualized under a fluorescence microscope (Carl Zeiss Microscopy, Germany).
Cell Migration and Invasion Assay
A Transwell insert with 8 μm pores (Millipore) was used to access the migratory and invasive abilities of OS cells. A total of 2 × 104 cells was cultured in the upper chamber with 200 μL of serum-free DMEM, and 600 μL of medium containing 10% FBS was added to the lower chamber. Following 24 h incubation, the migrated cells were fixed with 4% paraformaldehyde and stained by 0.1% crystal violet for 20 min. For the invasion assay, Transwell chambers were pretreated with 80 μL of Matrigel (BD Biosciences, USA), and the remaining steps were the same as with the migration assay. Ultimately, cells were visualized and photographed under a microscope (Nikon, Japan), and the number of cells per field was calculated.
Wound-Healing Assay
Cell motility was accessed by a wound-healing assay. The OS cells were seeded in six-well plates and allowed to grow until 100% confluency. A scratch was made by a P200 pipette tip and migration distance was measured at 0 and 24 h after scratching.
3D Tumor Spheroid Invasion Assay
A 3D tumor spheroid invasion assay was conducted to better simulate the invasion environment of tumor cells. 20 μL of cell suspension, containing approximately 1,000 cells, was added dropwise to the lid of a dish; then the lid was inverted over dishes with 10 mL PBS. After incubation for 48 h, cellular aggregates were collected and seeded into 3D collagen I gels (Purecol, USA). Following polymerization at 37°C, the gel was overlaid with 400 μL DMEM with 10% FBS for 48 h. The motion of cells was observed under fluorescence microscopy (Carl Zeiss Microscopy).
RNA Extraction and Quantitative Real-Time PCR
The total RNA of cells or tissues was extracted using RNAiso Plus reagent (Takara, Japan). Exosomal RNA was isolated using an exosomal RNA and protein extraction kit (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. miRNA and mRNA were subsequently reverse transcribed by a Hairpin-it miRNA qPCR Quantitation Kit (GenePharma, China) and a PrimeScript RT Reagent Kit (Takara), respectively. qRT-PCR was performed using a TB Green Premix Ex Taq Kit (Takara). The relative expression levels of mRNA and miRNA were normalized to GAPDH and U6, respectively. All groups were repeated in triplicate, and relative expression was calculated using the 2−ΔΔCT method. The primer sequences used in this study are as follows: LCP1 forward, 5′-GGGGTTCCTGGTCATACACC-3′, and LCP1 reverse, 5′-CAAGCAAGCAGCCTTGAACA-3′; Nrdp1 forward, 5′-GAGCCAGTACAGGCACCTC-3′, and Nrdp1 reverse, 5′-GACCGTCACAACACTACGGT-3′; GAPDH forward, 5′-AATGGGCAGCCGTTAGGAAA-3′, and GAPDH reverse, 5′-GCGCCCAATACGACCAAATC-3′; miR-135a-5p forward, 5′-CCGGCGTATGGCTTTTTATTCC-3′, and miR-135a-5p reverse, 5′-CAGTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′, and U6 reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Western Blot Analysis
The total protein of cells or tissues was extracted by radioimmunoprecipitation assay buffer (RIPA) lysis buffer (KeyGEN BioTECH, China), and nuclear protein was extracted using a Nuclear Protein Extraction Kit (Thermo Fisher Scientific). Exosomal protein was isolated by using an exosomal RNA and protein extraction kit (Thermo Fisher Scientific) following the manufacturer’s instructions. The protein was separated through SDS-PAGE gel after denaturation and transferred to a polyvinylidene fluoride (PVDF) membrane. Following blocking with QuickBlock Blocking Buffer (Beyotime Biotechnology, China) and incubation with primary antibodies at 4°C overnight and secondary antibodies at room temperature for 2 h, protein bands were ultimately visualized with the enhanced chemiluminescence (ECL) Detection Kit (Millipore). The relative expression levels were normalized to GAPDH.
CoIP Assay
The coIP assay was performed with the Co-IP Kit (Pierce, Thermo Fisher Scientific), following the protocol. Cell lysates of 143B and HOS cells were prepared in lysis buffer and were incubated with agarose bead-conjugated antibodies (20 μg antibody added into 1 mg cell lysates) overnight at 4°C with gentle rotation. Following washed with lysis buffer, proteins bound to beads were eluted by elution buffer for subsequent western blot detection.
Immunohistochemistry
After antigen retrieval and blocking, tissue sections were incubated with primary antibodies against LCP1 and Ki-67 overnight at 4°C and secondary antibodies for 2 h at room temperature. Then, the sections were treated with diaminobenzidine (Beyotime Biotechnology) for staining. To quantify the protein level of LCP1, the sections were observed and photographed with a microscope (Nikon).
Immunofluorescence Assay
Cells were fixed by 4% paraformaldehyde and permeated by 0.5% Triton X-100. Following blocking with 5% bovine serum albumin (Invitrogen, USA), cells were incubated with primary antibodies overnight. Fluorescence-labeled secondary antibodies and 4′,6-diamidino-2-phenylindole
(DAPI; Invitrogen) were then used for staining. Ultimately, cells were observed and photographed under fluorescence microscopy (Carl Zeiss Microscopy).
Luciferase Reporter Analysis
A potential interaction site between LCP1 and miR-135a-5p was predicted by using the TargetScan database (http://www.targetscan.org/vert_72/). WT LCP1 (WT-LCP1-3′ UTR) and MUT LCP1 (MUT-LCP1-3′ UTR) were designed and synthesized by GenePharma (Shanghai, China). Cells were cotransfected with miR-135a-5p mimics and MUT-LCP1-3′ UTR or their NC. After 48 h of incubation, luciferase activity was detected by using a Dual-Luciferase Assay Kit (Solarbio, China).
Animal Experiments
The animal experiments were approved by the Committee on the Ethics of Animal Experiments of Nanjing Medical University. For the subcutaneous tumorigenesis assay, 5-week-old male nude mice were randomly divided into four groups (143B-NC, 143B-shRNA, HOS-NC, and HOS-LCP1; n = 6 per group) and subcutaneously injected with 2 × 106 stably transfected cells in 100 μL PBS. The tumor volume was measured every 4 days until 28 days and calculated by the following formula: volume = length × width2 × 0.5. Tumor weight was also measured. The immunohistochemistry of tumors was performed following the above method. For the metastasis model, a total of 2 × 106 cells (143B-NC, 143B-shRNA, HOS-vector, and HOS-LCP1) with luciferase were injected into the tail vein of mice (n = 6 per group). Lung metastasis was detected by using an IVIS imaging system (Caliper Life Sciences, USA), and lung tissues were subjected to H&E staining.
Statistical Analysis
All data are presented as mean ± SD of at least three independent experiments. All statistical analyses were performed with GraphPad Prism 6.0 and data comparison between two groups was analyzed by the Student’s t test. One-way ANOVA test or two-way ANOVA test was used for multivariate analysis. Correlation analysis was conducted by the Spearman method. p <0.05 was considered significant.
Author Contributions
W.C., J.B., and Y.W. participated in the conception and design of this study. X.G. and Wei Liu performed the experiments and cowrote the manuscript. A.D., J.F., S.F., and W.Z. provided suggestions. K.S., Wanshun Liu, Z.X., X.L., and J.Z. helped to perform the analysis and interpretation. D.J., Y.R., F.G., J.W., and C.J. provided insightful discussions and comments on the manuscript. All authors read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (grant no. 81974335), China; Natural Science Foundation of Jiangsu Province (grant no. BK20181490), China; Postgraduate Research & Practice Innovation Program of Jiangsu Province (grant no. SJCX20_0475), China; Six Talent Peaks Project in Jiangsu Province (grant no. TD-SWYY-010), China; and Wu Jieping Medical Foundation (grant no. 320-2745-16-117), China.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.07.025.
Contributor Information
Yongzhong Wei, Email: weiyongzhong@jsph.org.cn.
Jianling Bai, Email: baijianling@njmu.edu.cn.
Weihua Cai, Email: caiwhspine@sina.com.
Supplemental Information
References
- 1.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 2.Rickel K., Fang F., Tao J. Molecular genetics of osteosarcoma. Bone. 2017;102:69–79. doi: 10.1016/j.bone.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cortini M., Avnet S., Baldini N. Mesenchymal stroma: Role in osteosarcoma progression. Cancer Lett. 2017;405:90–99. doi: 10.1016/j.canlet.2017.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Isakoff M.S., Bielack S.S., Meltzer P., Gorlick R. Osteosarcoma: Current Treatment and a Collaborative Pathway to Success. J. Clin. Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayles L.C., Breese M.R., Koehne A.L., Leung S.G., Lee A.G., Liu H.Y., Spillinger A., Shah A.T., Tanasa B., Straessler K. Genome-Informed Targeted Therapy for Osteosarcoma. Cancer Discov. 2019;9:46–63. doi: 10.1158/2159-8290.CD-17-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saraf A.J., Fenger J.M., Roberts R.D. Osteosarcoma: Accelerating Progress Makes for a Hopeful Future. Front. Oncol. 2018;8:4. doi: 10.3389/fonc.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C.S., Park T., Chen Z.P., Leavitt J. Human plastin genes. Comparative gene structure, chromosome location, and differential expression in normal and neoplastic cells. J. Biol. Chem. 1993;268:2781–2792. [PubMed] [Google Scholar]
- 8.Chen C., Cai Q., He W., Lam T.B., Lin J., Zhao Y., Chen X., Gu P., Huang H., Xue M. AP4 modulated by the PI3K/AKT pathway promotes prostate cancer proliferation and metastasis of prostate cancer via upregulating L-plastin. Cell Death Dis. 2017;8:e3060. doi: 10.1038/cddis.2017.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foran E., McWilliam P., Kelleher D., Croke D.T., Long A. The leukocyte protein L-plastin induces proliferation, invasion and loss of E-cadherin expression in colon cancer cells. Int. J. Cancer. 2006;118:2098–2104. doi: 10.1002/ijc.21593. [DOI] [PubMed] [Google Scholar]
- 10.Koide N., Kasamatsu A., Endo-Sakamoto Y., Ishida S., Shimizu T., Kimura Y., Miyamoto I., Yoshimura S., Shiiba M., Tanzawa H., Uzawa K. Evidence for Critical Role of Lymphocyte Cytosolic Protein 1 in Oral Cancer. Sci. Rep. 2017;7:43379. doi: 10.1038/srep43379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otsuka M., Kato M., Yoshikawa T., Chen H., Brown E.J., Masuho Y., Omata M., Seki N. Differential expression of the L-plastin gene in human colorectal cancer progression and metastasis. Biochem. Biophys. Res. Commun. 2001;289:876–881. doi: 10.1006/bbrc.2001.6047. [DOI] [PubMed] [Google Scholar]
- 12.Alfaifi M., Eom Y.W., Newsome P.N., Baik S.K. Mesenchymal stromal cell therapy for liver diseases. J. Hepatol. 2018;68:1272–1285. doi: 10.1016/j.jhep.2018.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Karantalis V., Hare J.M. Use of mesenchymal stem cells for therapy of cardiac disease. Circ. Res. 2015;116:1413–1430. doi: 10.1161/CIRCRESAHA.116.303614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger T., Hart M., Patarroyo M., Rolauffs B., Aicher W.K., Klein G. Mesenchymal Stromal Cells for Sphincter Regeneration: Role of Laminin Isoforms upon Myogenic Differentiation. PLoS ONE. 2015;10:e0137419. doi: 10.1371/journal.pone.0137419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai H., Yang X., Gao Y., Xu Z., Yu B., Xu T., Li X., Xu W., Wang X., Hua L. Exosomal MicroRNA-9-3p Secreted from BMSCs Downregulates ESM1 to Suppress the Development of Bladder Cancer. Mol. Ther. Nucleic Acids. 2019;18:787–800. doi: 10.1016/j.omtn.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu D.M., Wen X., Han X.R., Wang S., Wang Y.J., Shen M., Fan S.H., Zhang Z.F., Shan Q., Li M.Q. Bone Marrow Mesenchymal Stem Cell-Derived Exosomal MicroRNA-126-3p Inhibits Pancreatic Cancer Development by Targeting ADAM9. Mol. Ther. Nucleic Acids. 2019;16:229–245. doi: 10.1016/j.omtn.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Zhang X., Sai B., Wang F., Wang L., Wang Y., Zheng L., Li G., Tang J., Xiang J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer. 2019;18:40. doi: 10.1186/s12943-019-0959-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mi F., Gong L. Secretion of interleukin-6 by bone marrow mesenchymal stem cells promotes metastasis in hepatocellular carcinoma. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170181. BSR20170181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F., Zhang L.Y., Sai B.Q., Wang L.J., Zhang X.N., Zheng L.L., Tang J., Li G., Xiang J. BMSC-derived leptin and IGFBP2 promote erlotinib resistance in lung adenocarcinoma cells through IGF-1R activation in hypoxic environment. Cancer Biol. Ther. 2020;21:61–71. doi: 10.1080/15384047.2019.1665952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu B., Du L., Fan Q.M., Tang Z., Tang T.T. STAT3 activation by IL-6 from mesenchymal stem cells promotes the proliferation and metastasis of osteosarcoma. Cancer Lett. 2012;325:80–88. doi: 10.1016/j.canlet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Zhang P., Dong L., Yan K., Long H., Yang T.T., Dong M.Q., Zhou Y., Fan Q.Y., Ma B.A. CXCR4-mediated osteosarcoma growth and pulmonary metastasis is promoted by mesenchymal stem cells through VEGF. Oncol. Rep. 2013;30:1753–1761. doi: 10.3892/or.2013.2619. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L., Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim. Biophys. Acta Rev. Cancer. 2019;1871:455–468. doi: 10.1016/j.bbcan.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li I., Nabet B.Y. Exosomes in the tumor microenvironment as mediators of cancer therapy resistance. Mol. Cancer. 2019;18:32. doi: 10.1186/s12943-019-0975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Che Y., Shi X., Shi Y., Jiang X., Ai Q., Shi Y., Gong F., Jiang W. Exosomes Derived from miR-143-Overexpressing MSCs Inhibit Cell Migration and Invasion in Human Prostate Cancer by Downregulating TFF3. Mol. Ther. Nucleic Acids. 2019;18:232–244. doi: 10.1016/j.omtn.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Lang F.M., Hossain A., Gumin J., Momin E.N., Shimizu Y., Ledbetter D., Shahar T., Yamashita S., Parker Kerrigan B., Fueyo J. Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro-oncol. 2018;20:380–390. doi: 10.1093/neuonc/nox152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin S., Zhu B., Huang G., Zeng Q., Wang C. Microvesicles derived from human bone marrow mesenchymal stem cells promote U2OS cell growth under hypoxia: the role of PI3K/AKT and HIF-1α. Hum. Cell. 2019;32:64–74. doi: 10.1007/s13577-018-0224-z. [DOI] [PubMed] [Google Scholar]
- 27.Rupaimoole R., Calin G.A., Lopez-Berestein G., Sood A.K. miRNA Deregulation in Cancer Cells and the Tumor Microenvironment. Cancer Discov. 2016;6:235–246. doi: 10.1158/2159-8290.CD-15-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 29.Liu W., Jiang D., Gong F., Huang Y., Luo Y., Rong Y., Wang J., Ge X., Ji C., Fan J., Cai W. miR-210-5p promotes epithelial-mesenchymal transition by inhibiting PIK3R5 thereby activating oncogenic autophagy in osteosarcoma cells. Cell Death Dis. 2020;11:93. doi: 10.1038/s41419-020-2270-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L.M., Ding G.F., Xu W., Ge H., Jiang Y., Chen X.J., Lu Y. MiR-135a-5p represses proliferation of HNSCC by targeting HOXA10. Cancer Biol. Ther. 2018;19:973–983. doi: 10.1080/15384047.2018.1450112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J., Wen X., Zhang X., Sun X., Yunzhi L., Peng R., Zhu M., Wang M., Zhang Y., Luo W. miR-135a-5p and miR-124-3p Inhibit Malignancy of Glioblastoma by Downregulation of Syndecan Binding Protein. J. Biomed. Nanotechnol. 2018;14:1317–1329. doi: 10.1166/jbn.2018.2579. [DOI] [PubMed] [Google Scholar]
- 33.Luo W., Sun C., Zhou J., Wang Q., Yu L., Bian X.W., Zhou X., Hua D., Wang R., Rao C. miR-135a-5p Functions as a Glioma Proliferation Suppressor by Targeting Tumor Necrosis Factor Receptor-Associated Factor 5 and Predicts Patients’ Prognosis. Am. J. Pathol. 2019;189:162–176. doi: 10.1016/j.ajpath.2018.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Y., Zheng B., Meng X., Yan Y., He J., Liu Y. LncRNA DANCR promotes the proliferation, migration, and invasion of tongue squamous cell carcinoma cells through miR-135a-5p/KLF8 axis. Cancer Cell Int. 2019;19:302. doi: 10.1186/s12935-019-1016-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Ceuninck L., Wauman J., Masschaele D., Peelman F., Tavernier J. Reciprocal cross-regulation between RNF41 and USP8 controls cytokine receptor sorting and processing. J. Cell Sci. 2013;126:3770–3781. doi: 10.1242/jcs.131250. [DOI] [PubMed] [Google Scholar]
- 36.Wauman J., De Ceuninck L., Vanderroost N., Lievens S., Tavernier J. RNF41 (Nrdp1) controls type 1 cytokine receptor degradation and ectodomain shedding. J. Cell Sci. 2011;124:921–932. doi: 10.1242/jcs.078055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khakoo A.Y., Pati S., Anderson S.A., Reid W., Elshal M.F., Rovira I.I., Nguyen A.T., Malide D., Combs C.A., Hall G. Human mesenchymal stem cells exert potent antitumorigenic effects in a model of Kaposi’s sarcoma. J. Exp. Med. 2006;203:1235–1247. doi: 10.1084/jem.20051921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Hendrix A., Hernot S., Lemaire M., De Bruyne E., Van Valckenborgh E., Lahoutte T., De Wever O., Vanderkerken K., Menu E. Bone marrow stromal cell-derived exosomes as communicators in drug resistance in multiple myeloma cells. Blood. 2014;124:555–566. doi: 10.1182/blood-2014-03-562439. [DOI] [PubMed] [Google Scholar]
- 39.Balta E., Hardt R., Liang J., Kirchgessner H., Orlik C., Jahraus B., Hillmer S., Meuer S., Hübner K., Wabnitz G.H., Samstag Y. Spatial oxidation of L-plastin downmodulates actin-based functions of tumor cells. Nat. Commun. 2019;10:4073. doi: 10.1038/s41467-019-11909-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dubovsky J.A., Chappell D.L., Harrington B.K., Agrawal K., Andritsos L.A., Flynn J.M., Jones J.A., Paulaitis M.E., Bolon B., Johnson A.J. Lymphocyte cytosolic protein 1 is a chronic lymphocytic leukemia membrane-associated antigen critical to niche homing. Blood. 2013;122:3308–3316. doi: 10.1182/blood-2013-05-504597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lommel M.J., Trairatphisan P., Gäbler K., Laurini C., Muller A., Kaoma T., Vallar L., Sauter T., Schaffner-Reckinger E. L-plastin Ser5 phosphorylation in breast cancer cells and in vitro is mediated by RSK downstream of the ERK/MAPK pathway. FASEB J. 2016;30:1218–1233. doi: 10.1096/fj.15-276311. [DOI] [PubMed] [Google Scholar]
- 42.Riplinger S.M., Wabnitz G.H., Kirchgessner H., Jahraus B., Lasitschka F., Schulte B., van der Pluijm G., van der Horst G., Hämmerling G.J., Nakchbandi I., Samstag Y. Metastasis of prostate cancer and melanoma cells in a preclinical in vivo mouse model is enhanced by L-plastin expression and phosphorylation. Mol. Cancer. 2014;13:10. doi: 10.1186/1476-4598-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiedemann K., Sadvakassova G., Mikolajewicz N., Juhas M., Sabirova Z., Tabariès S., Gettemans J., Siegel P.M., Komarova S.V. Exosomal Release of L-Plastin by Breast Cancer Cells Facilitates Metastatic Bone Osteolysis. Transl. Oncol. 2019;12:462–474. doi: 10.1016/j.tranon.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y., Wang G., Chen R., Hua Y., Cai Z. Mesenchymal stem cells in the osteosarcoma microenvironment: their biological properties, influence on tumor growth, and therapeutic implications. Stem Cell Res. Ther. 2018;9:22. doi: 10.1186/s13287-018-0780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubio R., Abarrategi A., Garcia-Castro J., Martinez-Cruzado L., Suarez C., Tornin J., Santos L., Astudillo A., Colmenero I., Mulero F. Bone environment is essential for osteosarcoma development from transformed mesenchymal stem cells. Stem Cells. 2014;32:1136–1148. doi: 10.1002/stem.1647. [DOI] [PubMed] [Google Scholar]
- 46.Rubio R., Gutierrez-Aranda I., Sáez-Castillo A.I., Labarga A., Rosu-Myles M., Gonzalez-Garcia S., Toribio M.L., Menendez P., Rodriguez R. The differentiation stage of p53-Rb-deficient bone marrow mesenchymal stem cells imposes the phenotype of in vivo sarcoma development. Oncogene. 2013;32:4970–4980. doi: 10.1038/onc.2012.507. [DOI] [PubMed] [Google Scholar]
- 47.Velletri T., Xie N., Wang Y., Huang Y., Yang Q., Chen X., Chen Q., Shou P., Gan Y., Cao G. P53 functional abnormality in mesenchymal stem cells promotes osteosarcoma development. Cell Death Dis. 2016;7:e2015. doi: 10.1038/cddis.2015.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baglio S.R., Lagerweij T., Pérez-Lanzón M., Ho X.D., Léveillé N., Melo S.A., Cleton-Jansen A.M., Jordanova E.S., Roncuzzi L., Greco M. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017;23:3721–3733. doi: 10.1158/1078-0432.CCR-16-2726. [DOI] [PubMed] [Google Scholar]
- 49.Pietrovito L., Leo A., Gori V., Lulli M., Parri M., Becherucci V., Piccini L., Bambi F., Taddei M.L., Chiarugi P. Bone marrow-derived mesenchymal stem cells promote invasiveness and transendothelial migration of osteosarcoma cells via a mesenchymal to amoeboid transition. Mol. Oncol. 2018;12:659–676. doi: 10.1002/1878-0261.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu B., Zhu J., Liu S., Wang L., Fan Q., Hao Y., Fan C., Tang T.T. Mesenchymal stem cells promote osteosarcoma cell survival and drug resistance through activation of STAT3. Oncotarget. 2016;7:48296–48308. doi: 10.18632/oncotarget.10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Chu Y., Yue B., Ma X., Zhang G., Xiang H., Liu Y., Wang T., Wu X., Chen B. Adipose-derived mesenchymal stem cells promote osteosarcoma proliferation and metastasis by activating the STAT3 pathway. Oncotarget. 2017;8:23803–23816. doi: 10.18632/oncotarget.15866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu W., Li L., Rong Y., Qian D., Chen J., Zhou Z., Luo Y., Jiang D., Cheng L., Zhao S. Hypoxic mesenchymal stem cell-derived exosomes promote bone fracture healing by the transfer of miR-126. Acta Biomater. 2020;103:196–212. doi: 10.1016/j.actbio.2019.12.020. [DOI] [PubMed] [Google Scholar]
- 53.Liu W., Rong Y., Wang J., Zhou Z., Ge X., Ji C., Jiang D., Gong F., Li L., Chen J. Exosome-shuttled miR-216a-5p from hypoxic preconditioned mesenchymal stem cells repair traumatic spinal cord injury by shifting microglial M1/M2 polarization. J. Neuroinflammation. 2020;17:47. doi: 10.1186/s12974-020-1726-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Y., Liu W., He B., Wang L., Zhang F., Shu H., Sun L. Exosomes derived from bone marrow mesenchymal stem cells promote osteosarcoma development by activating oncogenic autophagy. J. Bone Oncol. 2020;21:100280. doi: 10.1016/j.jbo.2020.100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin F., Tang H., Zhang Y., Zhang Z., Huang P., Zhu J. Bone marrow-derived mesenchymal stem cell-derived exosomal microRNA-208a promotes osteosarcoma cell proliferation, migration, and invasion. J. Cell. Physiol. 2020;235:4734–4745. doi: 10.1002/jcp.29351. [DOI] [PubMed] [Google Scholar]
- 56.Nieto M.A., Huang R.Y.J., Jackson R.A., Thiery J.P. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 57.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leonard W.J., O’Shea J.J. Jaks and STATs: biological implications. Annu. Rev. Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 59.Johnson D.E., O’Keefe R.A., Grandis J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Shea J.J., Schwartz D.M., Villarino A.V., Gadina M., McInnes I.B., Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu. Rev. Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu J., Zhang J., Shen B., Yin K., Xu J.W., Gao W.C., Zhang L. Long noncoding RNA lncTCF7, induced by IL-6/STAT3 transactivation, promotes hepatocellular carcinoma aggressiveness through epithelial-mesenchymal transition. J. Exp. Clin. Cancer Res. 2015;34:116. doi: 10.1186/s13046-015-0229-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin J.C., Tsai J.T., Chao T.Y., Ma H.I., Liu W.H. The STAT3/Slug Axis Enhances Radiation-Induced Tumor Invasion and Cancer Stem-like Properties in Radioresistant Glioblastoma. Cancers (Basel) 2018;10:512. doi: 10.3390/cancers10120512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swatek K.N., Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu X., Yen L., Irwin L., Sweeney C., Carraway K.L., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 2004;24:7748–7757. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu W., Wang Y., Gong F., Rong Y., Luo Y., Tang P., Zhou Z., Zhou Z., Xu T., Jiang T. Exosomes Derived from Bone Mesenchymal Stem Cells Repair Traumatic Spinal Cord Injury by Suppressing the Activation of A1 Neurotoxic Reactive Astrocytes. J. Neurotrauma. 2019;36:469–484. doi: 10.1089/neu.2018.5835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.