Abstract
Dengue virus (DENV) infection is a major global public health concern, and there is no effective vaccine for it. In this study, we describe the design and characterization of three nucleotide-modified mRNA vaccines (prME-mRNA, E80-mRNA, and NS1-mRNA) for DENV-2. Our results showed that vaccination with E80-mRNA alone or a combination of E80-mRNA and NS1-mRNA can induce high levels of neutralizing antibodies and antigen-specific T cell responses; furthermore, these vaccines confer complete protection against DENV-2 challenge in immunocompetent mice. These data provide foundations for further development of a tetravalent DENV vaccine based on nucleotide-modified mRNA.
Graphical Abstract
Dengue virus (DENV) infection is a major public health concern for which the current vaccine is not satisfactory. Zhang et al. developed new mRNA vaccines for DENV-2 that confer complete protection against viral challenge in mice. These data provide foundations for further development of a tetravalent DENV mRNA vaccine.
Introduction
Dengue virus (DENV) infection is the most widely transmitted arboviral disease in tropical and subtropical regions, affecting 390 million people1 per year, of whom 96 million have clinical manifestations.2 DENV is divided into four serotypes (DENV-1, DENV-2, DENV-3, and DENV-4) that can cause life-threatening dengue hemorrhagic fever (DHF) and dengue shock syndrome (DSS).3 The current effort for the development of vaccine against DENV is thought to be hampered by antibody-dependent enhancement (ADE), a phenomenon that seems to exacerbate some DENV infections.4 Although primary infection with one DENV serotype can elicit lifelong protective immunity, a secondary infection by a different DENV serotype could aggravate the disease. Immune complexes formed by non-neutralizing or sub-neutralizing antibodies and DENV can lead to the aggravation of DENV infection in immune cells bearing the Fcγ receptor.5,6 ADE may be related to antibody titer, immunoglobulin G (IgG) subclass, IgG glycosylation, and Fcγ receptor polymorphism.7 Therefore, the quality and quantity of neutralizing antibody are crucial for protection against DENV, and these parameters need to be carefully measured in vaccine studies.
DENV belongs to the Flavivirus genus of the Flaviviridae family. Its genome encodes three structural (C, prM, and E) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins.8,9 The E protein mediates virus binding to putative cell-surface receptors10 and contains multiple epitopes for neutralizing antibodies. The N-terminal ectodomain, comprising 80% of the E protein and, thus, termed E80 protein, has often been tested as a vaccine antigen.11 prM is the precursor of the M protein, which helps the correct folding of the E protein; prME can form viral-like particles (VLPs) that are released from infected or transfected cells.12 The NS1 protein, which is involved in virus replication and immune evasion, has also been reported to activate antibody Fc-mediated effector functions and to provide partial protection against flavivirus.13, 14, 15, 16
Despite many vaccine candidates having been tested,11,12,17, 18, 19, 20, 21 CYD-TDV (Dengvaxia) of Sanofi Pasteur is the only licensed dengue vaccine that provides protection for people who have already been infected with DENV; however, the vaccine appears to have increased the incidence of severe dengue disease in those who were naive to DENV infection.22 This has been observed in the Philippines, where 19 children who had been vaccinated with Dengvaxia died of a subsequent DENV infection.21,23 Recent reports indicate that Takeda’s chimeric live-attenuated dengue vaccine DENVax has 73.7%, 97.7%, and 62.6% effectiveness rates against DENV-1, DENV-2, and DENV-3, respectively.24 However, the protective effect of the vaccine against DENV-4 infection could not be determined, because there were not enough cases of DENV-4 infection detected in the regions where the vaccine was tested. Additionally, whether the Takeda vaccine can provide long-term protection against DENV infections in the immunized population is currently unknown. Despite other vaccines being in clinical trials, the general consensus is that the development of a safe and effective dengue vaccine is still needed.
With the rapid development of research on RNA biology, the stability of mRNA and the efficiency of its delivery have been greatly improved.25 The mRNA vaccine technology has been applied to the development of vaccines for many infectious diseases, including influenza virus,26, 27, 28 HIV,29,30 Zika virus (ZIKV),31,32 and Ebola virus.33 Compared to DNA vaccines, mRNA vaccines cannot integrate into host genome, thus avoiding the risk of insertional mutagenesis and potential oncogenesis.34,35 The mRNA vaccine generally includes a 5′ cap, a 5′ UTR, a gene encoding one antigen or more, a 3′ UTR, and a poly(A) tail; it expresses proteins of different kinds, such as transmembrane, secretory, or intracellular.25 Importantly, modified mRNA not only can stimulate innate immunity through Toll-like receptors and RIG-I-like receptors, but also can be used directly without the need of any additional adjuvant.36,37
In this study, we developed a DENV mRNA vaccine based on two structural proteins (prME and E80) and one non-structural protein (NS1) from DENV-2 using mRNA encapsulated by lipid nanoparticles (LNPs). These mRNA vaccine candidates induced high levels of DENV-2-specific neutralizing antibodies and T cell immune responses and provided sterilizing immunity against DENV-2 challenge in immunocompetent BALB/c mice.
Results
Design of mRNA Vaccines for DENV-2
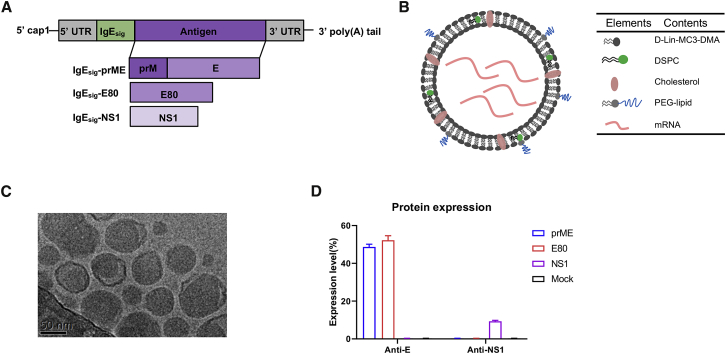
To apply the mRNA vaccine platform in the development of DENV vaccine, we designed three modified mRNA vaccines encoding the prME protein, E80 protein, and NS1 protein of DENV-2 strain 16681; a Cap1 (N7mGpppAm) sequence and a signal peptide sequence from human IgE were added to each of these proteins (Figure 1A).
Figure 1.
Design and Characterization of DENV-2 mRNA LNP Vaccines
(A) Schematic illustration of DENV-2 mRNA comprising a 5′ cap, a 5′ UTR, a signal peptide, an antigen (either prME, E80, or NS1), a 3′ UTR, and a 3′ poly(A) tail. (B) Schematic representation of the LNP-encapsulated mRNA. (C) Cryo-electron microscopy image of a preparation of mRNA LNP. (D) Protein expression levels in mRNA-transfected HEK293T cells. Protein expression was analyzed by flow cytometry with an antibody to DENV envelope (4G2) or an antibody to the DENV NS1 protein using staining of mock-transfected HEK293T cells as negative control.
The modified mRNA containing the modified nucleoside 1-methylpseudouridine-5′-triphosphate (1 mΨ) was chemically synthesized and packed into LNPs. The LNP consisted of the four lipids D-Lin-MC3-DMA, DSPC, cholesterol, and PEG-lipid, mixed at a molar ratio of 50:10:38.5:1.5 (Figure 1B); the resultant nanoparticle appeared to have an approximate diameter of 80 nm on examination with cryo-electron microscopy (Figure 1C). DENV-2 prME-mRNA- and E80-mRNA-transfected HEK293T cells were, on average, 48% and 52% positive for E protein expression, respectively, while the NS1-mRNA-transfected HEK293T cells were, on average, 9% positive for NS1 protein (Figure 1D).
All Three Modified mRNA-LNP Vaccines Induce Antigen-Specific Immune Responses
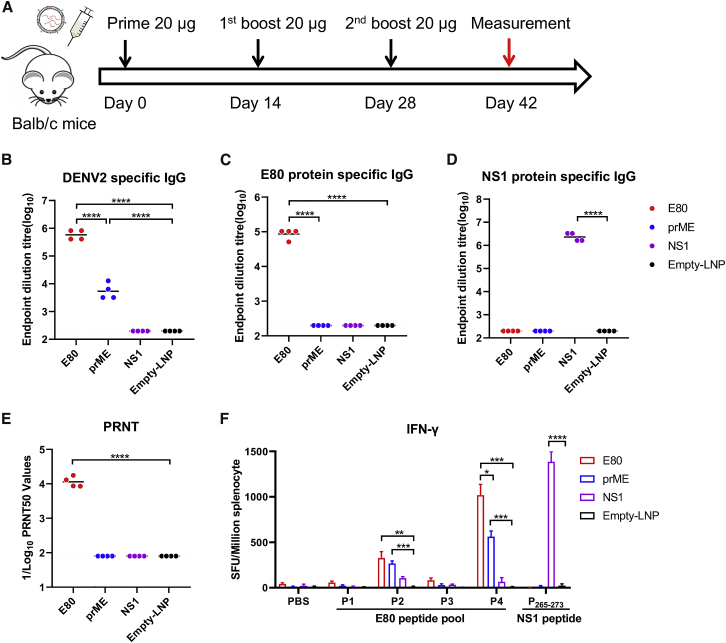
We first assessed the immunogenicity of prME-mRNA, E80-mRNA, and NS1-mRNA vaccines. Eight-week-old female BALB/c mice were divided into four groups receiving 20 μg each of the three mRNA-LNPs or the same amount of empty LNP (RNA-free LNP) as a negative control via intramuscular (i.m.) inoculation at days 0, 14, and 28 (Figure 2A). At day 42 (2 weeks after the last immunization), the mice were sacrificed, and serum samples and spleen cells were harvested for the measurement of antibody and T cell responses, respectively. Both E80-mRNA and prME-mRNA vaccines elicited virion-binding antibodies. The average endpoint titer of DENV-2 virion-specific IgG was 1,158,000 in the E80-mRNA immune sera group but only 5,300 in the prME-mRNA group, indicating a more than 200-fold difference (Figure 2B). E80-mRNA also elicited a high level of E-specific IgG, with an average endpoint titer of 86,000, whereas prME-mRNA stimulated an E80-specific IgG titer of less than 200, which is comparable to its level in the negative controls (Figure 2C), suggesting that most antibodies elicited by prME were prM targeting. The NS1-mRNA group showed high levels of antibody (endpoint titer of 106) against the DENV-2 NS1 protein (Figure 2D). Importantly, mice that received E80-mRNA had the strongest neutralizing antibody response against DENV-2, with an average 50% plaque reduction neutralization titer (PRNT50) of 11,000 (Figure 2E). Antigen-specific T cell responses were also induced by prME-mRNA, E80-mRNA, and NS1-mRNA immunization. In E80-mRNA or prME-mRNA immunized mice, 300 to 1,000 interferon gamma (IFN-γ)-producing splenocytes per 106 input cells were detected upon stimulation with DENV-2 E80 peptide pools (P2 and P4), but not with P1 and P3, indicating antigen specificity; in the NS1-mRNA group, 1,300 antigen-specific splenocytes per 106 input cells produced IFN-γ upon stimulation with a known immunodominant NS1 peptide epitope (P265–P273)38 (Figure 2F).
Figure 2.
Immunization with Modified DENV-2 mRNA-LNPs Elicits Potent Antigen-Specific T Cell and Neutralizing Antibody Responses
(A) Female BALB/c mice were immunized by i.m. injection with three doses of 20 μg nucleoside-modified prME-mRNA LNP, E80-mRNA LNP, NS1-mRNA LNP, or empty LNP at weeks 0, 2, and 4. Sera were collected at week 6. (B–D) Endpoint dilution titers were measured by ELISA plates coated with DENV-2 16681 virion (B), DENV-2 E80 protein (C), or NS1 protein (D). Sera neutralization against DENV-2 16681 was determined by PRNT50 assay (E). Each point represents an individual mouse, while horizontal lines indicate the mean. (F) At week 6, splenocytes were harvested and used for measuring antigen-specific IFNγ-producing T cells using the ELISPOT assay. Four peptide pools (P1, P2, P3, and P4) spanning the entire DENV-2 E80 protein and a single immunodominant epitope peptide from DENV2 NS1 (P265–273) were used for stimulation, while PBS was used as negative control. Four mice were used in each group. Statistical differences were determined using Student’s t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
E80-mRNA and NS1-mRNA Vaccines Elicit Antigen-Specific Immune Responses in a Dose-Dependent Manner
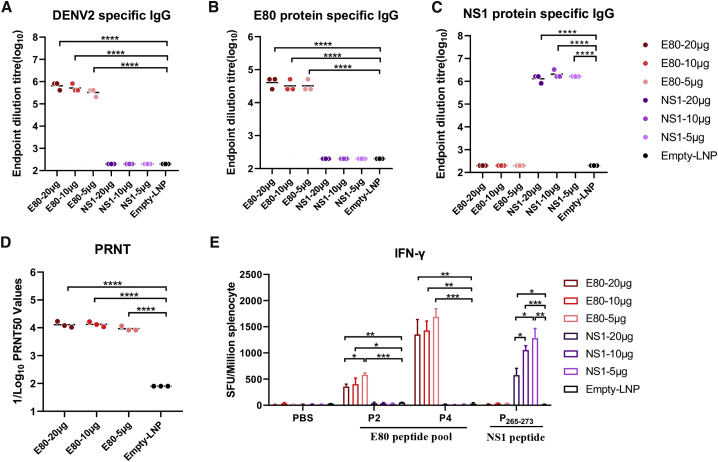
To further optimize the vaccination doses with these mRNAs, female BALB/c mice were immunized via i.m. inoculation with 20 μg, 10 μg, or 5 μg E80-mRNA or NS1-mRNA vaccine. IgG antibodies specific for both DENV-2 virion and E80 protein were efficiently induced by all three different doses of E80-mRNA vaccines, with an average virion-binding antibody titer of 650,000 (Figure 3A) and without statistically significant differences among these doses (Figures 3A and 3B). Similarly, vaccination with NS1-mRNA induced antibodies specific for the NS1 protein without significant differences among different dosages (Figure 3C). Notably, the average titer of neutralizing antibodies was similarly high, at a PRNT50 around 12,000, regardless of the dose group (Figure 3D). As expected, antigen-specific T cells were also activated. An inverse trend was observed, wherein E-protein-specific IFN-γ producing cells were activated in splenocytes stimulated with E80 peptide pools (P2 and P4) or the NS1 peptide in a dose-dependent manner, and the lowest dose of 5 μg mRNA instead of the highest dose of 20 μg mRNA appeared to be optimal (Figure 3E).
Figure 3.
Evaluation of Immunogenicity of Various Doses of E80-mRNA or NS1-mRNA Vaccine
Groups of female BALB/c mice were vaccinated via i.m. inoculation with 20 μg, 10 μg, or 5 μg E80-mRNA and NS1-mRNA, or empty LNP as negative control at weeks 0, 2, and 4, and sera were collected at week 6 for analysis. (A–C) Endpoint dilution titers were measured by ELISA plates coated with DENV-2 16681 virion (A), E80 protein (B), or NS1 protein (C). (D) Sera neutralization against DENV2 16681 was determined by PRNT50 assay. Each point represents an individual mouse, while horizontal lines indicate the mean. (E) Splenocytes were used to quantify antigen-specific IFNγ-producing T cells using ELISPOT assay with P2, P4, P265–273, or PBS control as stimuli. Three mice were used in each group. Statistical differences were determined using Student’s t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
mRNA-LNP Vaccines Confer Complete Protection against DENV-2 in BALB/c Mice
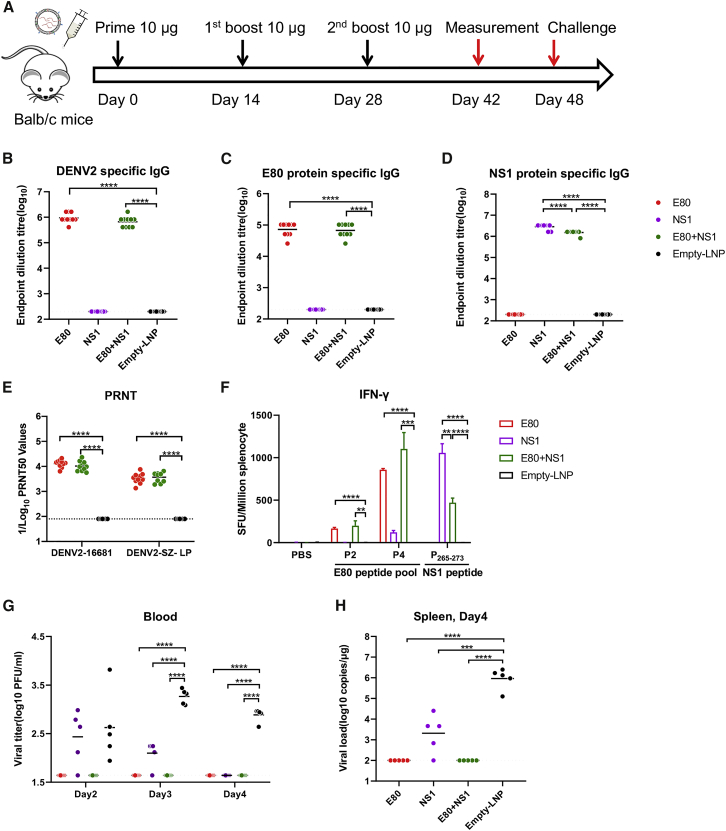
Having demonstrated that each of the mRNA-LNP vaccines elicited desirable immune responses, we went on to determine the protective effects of these vaccines, either alone or in combination (E80-mRNA, NS1-mRNA, E80-mRNA+NS1-mRNA, or empty LNP) with three immunizations using the intermediate dose of 10 μg per injection (Figure 4A). Briefly, mice were immunized via i.m. inoculation with mRNA LNPs, bled at 2 weeks after the third vaccination for measuring antigen-specific antibody, and then challenged with DENV-2. Results showed that E80-mRNA and E80-mRNA+NS1-mRNA vaccines elicited high levels of serum IgG against DENV-2 virions (Figure 4B) and E80 protein (Figure 4C); in addition, NS1-mRNA and E80-mRNA+NS1 mRNA vaccines stimulated strong antibody responses against the NS1 protein (Figure 4D). Notably, all three vaccination groups elicited neutralizing antibody and antigen-specific T cell responses (Figure 4F).
Figure 4.
mRNA LNP Vaccines Confer Protection against Viral Challenge in BALB/c Mice
(A) Female BALB/c mice (n = 10) were vaccinated via i.m. inoculation with 10 μg E80-mRNA, NS1-mRNA, or empty LNP at weeks 0, 2, and 4, and sera were collected at week 6 for analysis. At week 7, vaccinated BALB/c mice (n = 5) were administered 1 mg anti-INFα/βR blocking antibody and challenged with 5 × 106 PFUs of mouse-adapted DENV-2-GZ-LP 1 day later. (B–D) At days 2, 3, and 4 after viral challenge, binding antibody endpoint dilution titers were measured by ELISA with plates coated with DENV-2 16681 virion (B), E80 protein (C), or NS1 protein (D). (E) Sera neutralization against DENV-2 16681 was determined by PRNT50 assay. (F) Splenocytes were used to measure antigen-specific IFN-γ-producing T cells using the ELISPOT assay (n = 5). E protein peptide pools (P2 and P4), NS1 peptide (P265–273), or PBS was used for stimulation. (G and H) Viral loads in serum (G) or spleen (H) were measured by PRNT50 assay (G) or by qRT-PCR (H). Each point represents an individual mouse, while horizontal lines indicate the mean. Statistical differences were determined using Student’s t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
To create a DENV challenge model in immunocompetent mice, we passively transferred 1 mg of an anti-IFNAR1 blocking antibody 1 day prior to infection, then challenged the mice with 5 × 106 plaque-forming units (PFUs) of a mouse-adapted DENV-2-GZ-LP viral strain as described previously,39 and assessed viremia between 2 and 4 days post-infection. This model was used to examine the protective efficacy of the aforementioned characterized E80-mRNA, E80-mRNA+NS1-mRNA, and NS1-mRNA vaccines. Mouse sera were collected and used for analyzing neutralization activity against DENV-2-16681 and DENV-2-GZ-LP strains before the challenge. Immune sera in the E80-mRNA group showed potent neutralizing antibody responses against DENV-2-16681 and DENV-2-GZ-LP with PRNT50 values of 13,000 and 3,300, respectively; in the E80-mRNA+NS1-mRNA group, these values were 10,000 and 3,600, respectively (Figure 4E). As expected, DENV-2 immune sera showed cross-neutralization with the other three DENV serotypes, albeit with 10- to 100-fold lower PRNT50 values; specifically, these PRNT50 values were 1,000, 1,400, and 130 for DENV-1, DENV-3, and DENV-4, respectively, in the E80-mRNA group and 955, 1,100, and 120 for DENV-1, DENV-3, and DENV-4, respectively, in the E80-mRNA+NS1-mRNA group (Figure S1). Consistent with the high neutralizing titers and potent antigen-specific T cell responses, mice immunized with E80-mRNA and E80-mRNA+NS1-mRNA had no measurable viremia at days 2, 3, and 4 after infection with DENV-2-GZ-LP; in comparison, the control empty-LNP group had a detectable viremia in all mice at days 2, 3, and 4 post-infection (Figure 4G). Mice were euthanized at day 4 after infection, and their spleens were harvested for measurement of the viral RNA levels. No viral RNA was detected in the spleens from the E80-mRNA and E80-mRNA+NS1-mRNA-vaccinated groups, whereas up to 105–106 viral RNA copies per microgram of RNA were detected in spleens from the controls (Figure 4H).
NS1-mRNA Vaccine Alone Provides Partial Protection against DENV-2 Challenge
Because the NS1 protein has multiple functions in DENV viral infections,16 we also examined whether NS1-mRNA LNP alone could induce protection against DENV-2. Sera from NS1-mRNA-vaccinated mice had high levels of DENV-2 NS1 protein-specific binding antibody after immunization (Figure 4D) but no DENV-2 neutralizing antibodies (Figure 2E). At day 2 after DENV-2 challenge, mice vaccinated with NS1-mRNA had a viremia level similar to those inoculated with empty LNP; however, the viremia was not detected 4 days after the challenge, when viremia was still detectable in the controls (Figure 4G). Moreover, mice in the NS1-mRNA group had a significantly lower viral load in spleen (p < 0.001) than the control group (empty LNP) at day 4 post-challenge (Figure 4H). These results indicate that NS1 alone has some capacity to protect mice against DENV-2 challenge and that the protection is not mediated by virus neutralization.
NS1-mRNA and E80-mRNA Vaccines Act Together to Alter the ADE Response and IgG Subclass
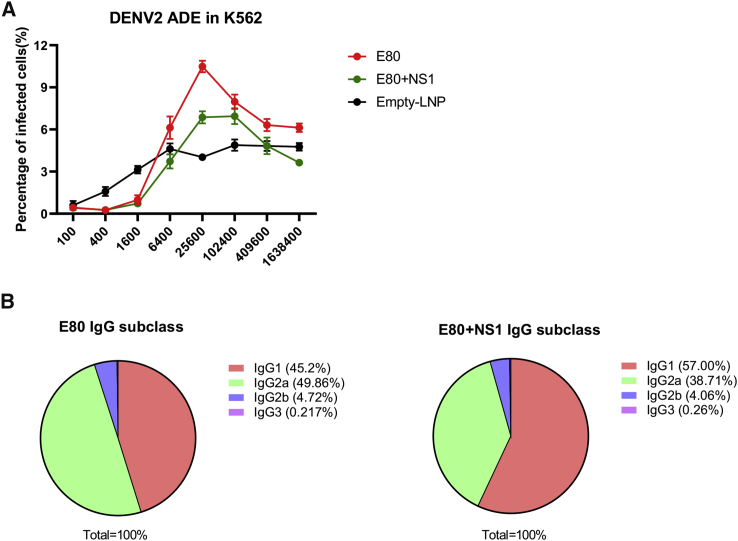
Finally, we assessed whether the addition of a NS1 component to the E-based vaccine could minimize ADE response and whether the IgG subclass distribution was altered as a consequence. We first compared the abilities of the sera from the E80-mRNA and E80-mRNA+NS1-mRNA groups to induce ADE using a standard K562 cell infection assay as described previously.11,40 As shown with a representative result in Figure 5A, the average percentage of DENV-2 infected cells reached a peak of 10% at a 1:25,600 dilution of serum samples from E80-mRNA-immunized mice; in comparison, the sera from E80-mRNA+NS1-mRNA-immunized mice had a lower level of infection (6%) at dilutions between 1:25,600 and 1:102,400. These data indicate that the presence of NS1-specific immune responses induced by the NS1-mRNA vaccine reduced the ADE activity of DENV2 infection via the action of E80-specific antibodies elicited by the E80-mRNA vaccine. As expected, serum from mice vaccinated with either E80-mRNA or the combination of E80-mRNA+NS1-mRNA also mediated ADE activity for DENV-1, DENV-3, and DENV-4 infections in K562 cells, although the peak enhancement levels for these three viruses were reached at dilutions from 1:400 to 1:3,200, which were 10- to 100-fold higher concentrations than those for DENV-2 (Figure S2). Together, these data reinforced the concept that neutralization titers are inversely correlated with ADE capacities.
Figure 5.
Characterization of ADE Activities and IgG Subclass Using Immune Sera Obtained from Mice Immunized with mRNA Vaccines
(A) Assessing antibody-dependent enhancement of DENV-2 infection of immune sera in K562 cells. Serial dilutions of heat-inactivated mouse sera were pre-incubated with DENV-2, followed by the addition of K562 cells. Cells were harvested 48 h after infection with immune-complexes and stained with a primary antibody against E protein (4G2 antibody) and then with a secondary anti-mouse antibody labeled with Alexa Fluor 488. The percentages of infection were determined by fluorescence-activated cell sorting (FACS). (B) Sera from E80-mRNA vaccine (left) and E80-mRNA+NS1-mRNA vaccine (right) were analyzed for IgG1, IgG2a, IgG2b, and IgG3 ratio by isotype ELISA using coating antigen DENV-2 16681 virion.
Because we and others have found that neutralization activity is associated with antibody subclasses,41,42 we also assessed the subclass distribution in immune sera. Half of IgGs elicited by E80-mRNA vaccine were IgG2as (Figure 5B, left), which are indicative of an underlying Th1 immune response; in comparison, sera from E80-mRNA+NS1-mRNA-immunized mice had a dominant IgG1 response (Figure 5B, right), indicating that Th2 immune response was induced.
Discussion
DENV infection remains a major public health concern; there are no specific antiviral drugs to treat DENV infection, and the currently available vaccine is not satisfactory.21,23,43 Here, we developed a highly efficacious mRNA-LNP vaccine platform for DENV, which is based on the expression of prME, E80, or NS1 antigens of DENV-2. We showed that vaccines based on E80-mRNA alone or in combination with NS1-mRNA induced both strong neutralizing antibodies and T cell immune response; more importantly, these vaccines provided full protection against DENV-2 challenge. Interestingly, vaccination with only NS1-mRNA also elicited antigen-specific T cell responses and binding antibodies, conferring partial protection against DENV-2 viral challenge in immunocompetent BALB/c mice. This is the first demonstration showing that mRNA-based DENV vaccines are not only immunogenic but also protective in an animal model.
The high efficacy of protection may be related to the high titers of antibodies induced by these vaccines: E80-mRNA induced high titers of E80 protein binding antibodies with an average endpoint dilution titer of 86,000 and DENV virions with an average endpoint dilution titer of 1,158,000, as well as high levels of neutralizing antibodies with an average PRNT50 of 13,000. Additionally, NS1-mRNA elicited high levels of NS1 protein binding antibodies with an average endpoint dilution titer of 2,800,000. In comparison, previous studies using other forms of DENV vaccines could only induce PRNTs50 of up to 500–10,000 in murine models.11,44,45
Another interesting but not fully explained finding is that doses as low as 5 μg of the three mRNA LNPs can elicit immune responses just as potent as those from the 20-μg dose, possibly due to a limitation of antigen uptake or antigen processing in vaccinated mice when the amount of antigen is in excess. Similar results have been shown in a previous study, wherein 2 μg ZIKV prME-mRNA could induce neutralizing-antibody titers with a PRNT50 of approximately 10,000, comparable to those induced by a 10 μg dose.32 It is also possible that different formulations of mRNA vaccines may have different optimal dosages and that the underlying molecular mechanisms still need to be investigated.
The observed protective effect of NS1-mRNA, either alone or in combination with E-mRNA, is also interesting. Among reported dengue vaccine studies, E protein is regarded as the major target antigen with multiple neutralizing antibody sites, while the NS1 protein, which has no neutralizing antibody sites, is often neglected. However, some experimental studies have shown that ZIKV and DENV prototype NS1 vaccines can prevent the development of lethal infections in experimental conditions,20,46,47 possibly due to FcR-mediated complement activation or antibody-dependent, cell-mediated cytotoxicity (ADCC) induced by NS1-specific antibodies elicited by NS1 protein vaccines.48,49 We observed that E80-mRNA+NS1-mRNA elicited a lower titer of NS1-specific antibody than NS1-mRNA did, probably due to antigenic competition between E80 and NS1 proteins, as reported in some studies.50,51 Although the incorporation of NS1-mRNA did not significantly enhance the protective efficacy of the DENV-2 E80-mRNA vaccine in our study, it apparently reduced ADE response to DENV-2 and altered IgG subclass distribution. This is consistent with previous reports that the incorporation of NS1 into E-protein-based vaccine enhanced its protective efficacy against experimental DENV and ZIKV infections.19,50,51 In these models, the specific relationships between immune responses and the NS1 protein in the overall protection are yet to be understood. We reason that there are several ways in which NS1 specific immune responses play a beneficial role. First, it was reported that the NS1 protein circulating in mammalian host blood can increase flavivirus infectivity in mosquitoes in vivo52 and that NS1 antibodies may reduce viral infectivity in vivo in experimentally infected mice. Second, DENV NS1 protein has been reported to trigger endothelial permeability and vascular leakage;53 thus, anti-NS1 antibodies can antagonize the pathological activity exerted by the NS1 protein and thereby prevent endothelial permeability and vascular leakage.
Reducing ADE effect is an important goal in DENV vaccine design. NS1 protein is not expressed on the surface of the DENV virion; therefore, it cannot elicit ADE, as the E80 protein is expressed on the virion surface.16 We found that NS1-mRNA vaccination can confer partial protection against viremia in immunocompetent BALB/c mice challenged with DENV-2 without the induction of neutralizing antibodies. This is consistent with a previous report that mRNA vaccine with selected NS peptides provided partial protection in transgenic mice against experimental DENV infection.54
Current efforts in subunit vaccines have focused mostly on prME or E80 proteins that can theoretically induce potent neutralizing antibodies. However, these antibodies can also induce ADE against DENV and other closely related flaviviruses that share antigenic structures.55 NS1 vaccine induces NS1-specific antibodies but does not cause an ADE response and can, therefore, be modified and included as a component in future vaccines. The fact that DENV-2 immune sera showed not only cross-neutralization but also ADE effect against DENV-1, DENV-3, and DENV-4 emphasizes the importance of developing tetravalent vaccines to provide full-range protection against dengue viruses.
In summary, with the mRNA LNP system, we have developed candidate DENV vaccines expressing prME, E80, and NS1 proteins and further showed that all of these three vaccines are immunogenic. Notably, we demonstrated that both E80-mRNA vaccine alone and its combination with NS1-mRNA conferred sterilizing immunity against DENV-2 challenge in vaccinated immunocompetent mice. Additionally, NS1-mRNA also elicited strong immune response and conferred partial protection against DENV-2. These results form the foundation for further development of a tetravalent DENV vaccine based on mRNA technology.
Materials and Methods
Animals
Female BALB/c mice (6–8 weeks old) purchased from Shanghai Laboratory Animal ( Shanghai, China) were used for the assessment of immunogenicity. All procedures were performed according to protocols approved by the Institut Pasteur of Shanghai Animal Experimentation Committee (No. A2018016).
mRNA and LNP Production
Based on methods essentially as previously described,32 mRNA was produced using T7 RNA polymerase on linearized plasmids (puc57-5′ UTR-ORF-3′ UTR) encoding codon-optimized E80, prME, or NS1 genes from DENV-2 strain 16681. The UTP (Uridine triphosphate) was fully substituted with 1 mΨ (1-methylpseudouridine-5′-triphosphate) (TriLink BioTechnologies, San Diego, CA, USA) (5′ UTR: 5′-AAATAAGAGAGAAAAGAAGAGTAAGAAGAAATATAAGAGCCACC-3′; 3′ UTR: 5′-TGATAATAGGCTGGAGCCTCGGTGGCCATGCTTCTTGCCCCTTGGGCCTCCCCCCAGCCCCTCCTCCCCTTCCTGCACCCGTACCCCCGTGGTCTTTGAATAAAGTCTGA-3′). Cap1 was added to the synthesized RNA by using the Vaccinia Capping System, and an mRNA Cap 2′-O-methyltransferase and poly(A) tail were generated by E. coli Poly(A) Polymerase (New England Biolabs, Ipswich, MA, USA). The mRNA was stored frozen at −80°C.
LNP-mRNA formulations were made according to a described method.56 Briefly, D-Lin-MC3-DMA (MedChemExpress), DSPC (Avanti Polar Lipids), cholesterol (Sigma), and PEG-lipid (Avanti Polar Lipids) were solubilized in ethanol at a molar ratio of 50:10:38.5:1.5. The lipid mixture was added to an aqueous buffer (50 mM citrate buffer [pH 4.0]), resulting in the final ethanol and lipid concentration of 30% (v/v) and 6.1 mg/mL. After equilibrating at room temperature for 2 min, the lipid mixture was added into a lipid extruder (LF-1; Avestin, Ottawa, ON, Canada) and then extruded at room temperature through two stacked 80-nm pore-sized filters (Nuclepore: Whatman, GE Healthcare, Chicago, IL, USA) to form vesicles of about 80 nm in diameter. The mRNA solubilized in 50 mM citrate buffer was mixed with lipid at a ratio of 3:1. LNP-encapsulated mRNA samples were dialyzed against PBS (pH 7.4) in Slide-A-Lyzer G2 Dialysis Cassettes (10 K MWCO; Thermo Fisher Scientific) for 24 h and then concentrated using Amicon Ultra Centrifugal Filters (Millipore, Burlington, MA, USA), passed through 0.45-μm filters, and stored at 4°C until use. The encapsulation efficiency was measured with the Quant-iT RiboGreen RNA Assay Kit (Life Technologies, Carlsbad, CA, USA) using a microplate reader.
Viruses and Cell Lines
DENV-1 strain 16007, DENV-2 strain 16681, DENV-3 strain 16562, and DENV-4 strain 1036 were kindly provided by Dr. Claire Huang (Centers for Disease Control and Prevention [CDC], Ft. Collins, CO, USA). The virus was propagated in C6/36 cells, and viral titers were determined in Vero cells through plaque assay. HEK293T cells (ATCC) were cultured at 37°C in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Carlsbad, CA, USA) and 10% fetal bovine serum ([qualified, New Zealand origin] FBS; GIBCO Gaithersburg, MD, USA). C6/36 cells were maintained in modified Eagle’s medium (MEM; GIBCO, Gaithersburg, MD, USA) supplemented with nonessential amino acids (GIBCO, Gaithersburg, MD, USA) and 10% FBS at 28°C with 5% CO2.
mRNA Transfection
Transfection of HEK293T cells was performed with Lipofectamine 2000 Reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions: 2.5 μg mRNA in 125 μL OPTI-MEM (GIBCO, Gaithersburg, MD, USA) was incubated with 5 μL Lipofectamine 2000 in 130 μL OPTI-MEM for 30 min; then, the Lipofectamine 2000-mRNA complex mix was immediately added to the 6-well plates with 1.0 × 106 cells per well. Supernatant was collected, and cells were lysed for 30 min on ice in RIPA buffer (Beyotime Biotechnology, Shanghai, China) at 24 h after transfection.
Flow Cytometry Analyses of mRNA-Transfected 293T Cells
The transfected HEK293T cells (1.0 × 106) were fixed in 4% paraformaldehyde (PFA) at room temperature for 10 min, followed by permeabilization (eBioscience, San Diego, CA, USA). Cells were then incubated at 4°C for 30 min with flavivirus-specific antibody 4G2, or a DENV-2 NS1 specific antibody (Sigma, Germany), followed by staining with Alexa Fluor 488-conjugated goat anti-mouse IgG or Alexa Fluor 488-conjugated goat anti-rabbit IgG (Life Technologies, Carlsbad, CA, USA) for 30 min on ice. The samples were analyzed using a BD LSR II flow cytometer. A minimum of 10,000 events for each sample were recorded and analyzed with FlowJo software.
Animal Experiments
mRNA-LNP was diluted in PBS and administered via i.m. inoculation with 40 μL each by syringe (BD Biosciences). Groups of 6- to 8-week-old BALB/c mice were immunized at weeks 0, 2, and 4. For DENV2 challenge studies, 1 mg anti-IFNα/β Receptor blocking antibody (MAR1-5A3) was administered via intraperitoneal injection at 24 h prior to viral infection. Mice were infected with 5 × 106 PFUs of mouse-adapted DENV-2-GZ-LP39 at 7 weeks after the initial vaccination. Blood or spleen samples were obtained at days 2, 3, and 4 after challenge. Before viral challenge, blood samples from each group of mice were collected 2 weeks after the third immunization. Serum samples were collected by centrifugation and kept at −80°C until use.
IFN-γ Enzyme-Linked Immunosorbent Spot (ELISPOT) Assay
ELISPOT assays were performed according to the manufacturer’s protocol (Mabtech, Nacka Strand, Sweden) to detect IFN-γ-producing T cells in the splenocytes. Briefly, 96-well ELISPOT plates (Millipore, Burlington, MA, USA) were pre-coated with anti-mouse IFN-γ antibody (AN18, Mabtech) at 4°C overnight. After removing the coating antibody, wells were blocked with RPMI 1640 medium containing 10% FBS for 1 h. The splenocytes isolated from immunized mice were added to ELISPOT plates and then stimulated with four peptide pools spanning DENV-2 E80 protein (P1, P2, P3, and P4), a single immunodominant peptide (P265–273) from DENV-2 NS1, or PBS as negative control at 37°C for 48 h. After incubation, the wells were washed five times with PBS and then incubated with a biotinylated anti-mouse IFN-γ detection antibody (R4-6A2-biotin; Mabtech) diluted in PBS containing 0.5% FBS at 0.2 μg per well for 2 h at room temperature, followed by the addition of diluted alkaline-phosphatase-conjugated streptavidin for 1 h. Immune spots were developed using TMB substrate and counted with the ImmunoSpot Analyzer (CellularTechnology, Kennesaw, GA, USA).
ELISA
DENV-2-specific IgG antigen in sera was measured by ELISA using 96-well flat-bottom plates (Corning, Corning, NY, USA) that were coated with 1.0 × 105 PFUs of UV-inactivated DENV-2 virions, E80 protein, or NS1 protein overnight at 4°C. The plates were blocked for 2 h with non-fat 5% milk in PBS containing 0.05% Tween 20 (PBST) and washed five times with PBST. Inactivated mouse serum underwent 2-fold serial dilution in 1% milk/PBST and incubation for 1 h, followed by washing for five times. Secondary horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Santa Cruz Biotechnology, Dallas, TX, USA) was diluted in 1:10,000 in 1% milk/PBST and incubated for 1 h. TMB substrate (Life Technology, Carlsbad, CA, USA) was applied to the plates, and the reaction was stopped with 2 M HCl. The absorbance was measured at 450 nm using a microplate reader. The endpoint dilution of the antibody titer was defined as the highest serum dilution that gave an optical density 450 (OD450) value above two times of that of control sera. To measure antibody isotypes, ELISA was performed according to the manufacturer’s protocol (SouthernBiotech, Birmingham, AL, USA) with inactivated DENV-2 strain 16681 as capture antigen. The concentration of the IgG subtype was calculated based on the standard curve and the relative percentages of IgG1, IgG2a, IgG2b, and IgG3 were normalized to total IgG.
Virus Neutralization Assay
100 PFUs of DENV-2 strain 16681 were incubated with serially diluted serum in serum-free DMEM for 1 h at 37°C. The virus-serum mixture (200 μL) was added into Vero cells pre-seeded in 48-well culture plates for 1 h at 37°C. Then, a 700 μL medium overlay containing 50% (v/v) DMEM, 1.5% (v/v) FBS, 0.45% (w/v) NaCl, and 1.5% carboxylmethylcellulose was added to each well, and the plates were incubated for 48–96 h at 37°C. The plates were washed with warmed PBS and then fixed with 4% PFA. The fixed cells were stained with the flavivirus-specific antibody 4G2 for 3 h at room temperature, followed by a secondary anti-mouse IgG antibody (Promega, Madison, WI, USA) for 3 h. Finally, foci of infected cells were visualized with a BCIP/NBT reaction provided in a commercial kit. The PRNT50 was defined by non-linear regression analysis to determine the dilution of sera required to inhibit 50% of infection. All neutralization data were normalized with foci number of infected cells without pre-incubation with mouse sera.
Quantification of Virus Load
At different time points after the DENV-2 challenge, blood samples were collected, and spleens were harvested. Spleen was homogenized using a bead-beater apparatus (MagNA Lyser, Roche), and serum was prepared from blood after centrifugation. Total RNA was extracted from spleen and serum samples with TRIzol (Life Technologies, Carlsbad, CA, USA). DENV infection levels in serum were determined by titration assay, and DENV RNA levels from spleen were determined by a TaqMan one-step quantitative reverse transcriptase PCR (qRT-PCR) kit on a Light Cycler 96 Real-Time PCR system (Roche Diagnostics, Mannheim, Germany). Viral load was expressed on a log10 scale as viral RNA equivalents per microgram for spleen or per milliliter for serum. For DENV, the following primer sets were used: forward: 5′-AAGGACTAGAGTTAGAGGAGAC-3′; reverse: 5′-GGCGTTCTGTGCCTGGAATGAT-3′; and probe: 5′-FAM/CCAGAGATCCTGCTGTCTC/MGB-3′.
For viral titration assay, mouse sera were serially diluted and added into Vero cells with serial dilution and incubated for 96 h at 37°C. The plates were washed with warmed PBS and then fixed with 4% PFA. The fixed cells were stained with 4G2 antibody for 3 h at room temperature, followed by a secondary anti-mouse IgG antibody (Promega, Madison, WI, USA) for 3 h. Finally, foci of infected cells were visualized by commercial test kit BCIP/NBT reaction.
ADE Assay
Serial dilutions of heat-inactivated mouse sera were incubated with DENV for 1 h at 37°C, followed by the addition of K562 cells expressing the Fcγ receptor CD32A. Two days later, the cells were harvested and fixed with 4% PFA, permeabilized, and then stained with DENV-specific primary antibody D1-11 or 4G2, followed by the addition of Alexa Fluor 488-conjugated goat anti-mouse IgG. The percentage of infected cells was analyzed by flow cytometry.
Statistical Analysis
All data were analyzed with GraphPad Prism 6 software, and the statistical differences between vaccination groups were determined by Student’s t test. PRNT50 titers were obtained through Probit regression analysis (SPSS). Statistical significance was reported as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001.
Author Contributions
X.J. and M.Z. designed the study. M.Z. carried out the experiments with technical assistance from M.L. and J.S. X.J. and M.Z. analyzed data. M.Z. and X.J. wrote the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported in part by grants from the National Key R&D Program of China (2016YFC1201000 to X.J.) and the National Science Foundation of China (31670941 and 31870916 to X.J.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2020.07.013.
Supplemental Information
References
- 1.Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beatty M.E., Beutels P., Meltzer M.I., Shepard D.S., Hombach J., Hutubessy R., Dessis D., Coudeville L., Dervaux B., Wichmann O. Health economics of dengue: a systematic literature review and expert panel’s assessment. Am. J. Trop. Med. Hyg. 2011;84:473–488. doi: 10.4269/ajtmh.2011.10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kularatne S.A. Dengue fever. BMJ. 2015;351:h4661. doi: 10.1136/bmj.h4661. [DOI] [PubMed] [Google Scholar]
- 4.Guzman M.G., Halstead S.B., Artsob H., Buchy P., Farrar J., Gubler D.J., Hunsperger E., Kroeger A., Margolis H.S., Martínez E. Dengue: a continuing global threat. Nat. Rev. Microbiol. 2010;8(12, Suppl):S7–S16. doi: 10.1038/nrmicro2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halstead S.B., Nimmannitya S., Cohen S.N. Observations related to pathogenesis of dengue hemorrhagic fever. IV. Relation of disease severity to antibody response and virus recovered. Yale J. Biol. Med. 1970;42:311–328. [PMC free article] [PubMed] [Google Scholar]
- 6.Halstead S.B., O’Rourke E.J. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J. Exp. Med. 1977;146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilder-Smith A., Ooi E.E., Horstick O., Wills B. Dengue. Lancet. 2019;393:350–363. doi: 10.1016/S0140-6736(18)32560-1. [DOI] [PubMed] [Google Scholar]
- 8.Simmons C.P., Farrar J.J., Nguyen V., Wills B. Dengue. N. Engl. J. Med. 2012;366:1423–1432. doi: 10.1056/NEJMra1110265. [DOI] [PubMed] [Google Scholar]
- 9.Lindenbach B.D., Rice C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 10.Kuhn R.J., Zhang W., Rossmann M.G., Pletnev S.V., Corver J., Lenches E., Jones C.T., Mukhopadhyay S., Chipman P.R., Strauss E.G. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J., Li M., Wang Y., Hao P., Jin X. Elaboration of tetravalent antibody responses against dengue viruses using a subunit vaccine comprised of a single consensus dengue envelope sequence. Vaccine. 2017;35:6308–6320. doi: 10.1016/j.vaccine.2017.09.063. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz I.C., Allison S.L., Heinz F.X., Helenius A. Folding and dimerization of tick-borne encephalitis virus envelope proteins prM and E in the endoplasmic reticulum. J. Virol. 2002;76:5480–5491. doi: 10.1128/JVI.76.11.5480-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackenzie J.M., Jones M.K., Young P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 14.Chung K.M., Nybakken G.E., Thompson B.S., Engle M.J., Marri A., Fremont D.H., Diamond M.S. Antibodies against West Nile Virus nonstructural protein NS1 prevent lethal infection through Fc gamma receptor-dependent and -independent mechanisms. J. Virol. 2006;80:1340–1351. doi: 10.1128/JVI.80.3.1340-1351.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung K.M., Thompson B.S., Fremont D.H., Diamond M.S. Antibody recognition of cell surface-associated NS1 triggers Fc-gamma receptor-mediated phagocytosis and clearance of West Nile Virus-infected cells. J. Virol. 2007;81:9551–9555. doi: 10.1128/JVI.00879-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glasner D.R., Puerta-Guardo H., Beatty P.R., Harris E. The Good, the Bad, and the Shocking: The Multiple Roles of Dengue Virus Nonstructural Protein 1 in Protection and Pathogenesis. Annu. Rev. Virol. 2018;5:227–253. doi: 10.1146/annurev-virology-101416-041848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guy B., Barrere B., Malinowski C., Saville M., Teyssou R., Lang J. From research to phase III: preclinical, industrial and clinical development of the Sanofi Pasteur tetravalent dengue vaccine. Vaccine. 2011;29:7229–7241. doi: 10.1016/j.vaccine.2011.06.094. [DOI] [PubMed] [Google Scholar]
- 18.Coller B.A., Clements D.E., Bett A.J., Sagar S.L., Ter Meulen J.H. The development of recombinant subunit envelope-based vaccines to protect against dengue virus induced disease. Vaccine. 2011;29:7267–7275. doi: 10.1016/j.vaccine.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu H., Xu X.F., Gao N., Fan D.Y., Wang J., An J. Preliminary evaluation of DNA vaccine candidates encoding dengue-2 prM/E and NS1: their immunity and protective efficacy in mice. Mol. Immunol. 2013;54:109–114. doi: 10.1016/j.molimm.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Pinto P.B.A., Assis M.L., Vallochi A.L., Pacheco A.R., Lima L.M., Quaresma K.R.L., Pereira B.A.S., Costa S.M., Alves A.M.B. T Cell Responses Induced by DNA Vaccines Based on the DENV2 E and NS1 Proteins in Mice: Importance in Protection and Immunodominant Epitope Identification. Front. Immunol. 2019;10:1522. doi: 10.3389/fimmu.2019.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilder-Smith A., Hombach J., Ferguson N., Selgelid M., O’Brien K., Vannice K., Barrett A., Ferdinand E., Flasche S., Guzman M. Deliberations of the Strategic Advisory Group of Experts on Immunization on the use of CYD-TDV dengue vaccine. Lancet Infect. Dis. 2019;19:e31–e38. doi: 10.1016/S1473-3099(18)30494-8. [DOI] [PubMed] [Google Scholar]
- 22.Villar L., Dayan G.H., Arredondo-García J.L., Rivera D.M., Cunha R., Deseda C., Reynales H., Costa M.S., Morales-Ramírez J.O., Carrasquilla G., CYD15 Study Group Efficacy of a tetravalent dengue vaccine in children in Latin America. N. Engl. J. Med. 2015;372:113–123. doi: 10.1056/NEJMoa1411037. [DOI] [PubMed] [Google Scholar]
- 23.Arkin F. Dengue researcher faces charges in vaccine fiasco. Science. 2019;364:320. doi: 10.1126/science.364.6438.320. [DOI] [PubMed] [Google Scholar]
- 24.Biswal S., Reynales H., Saez-Llorens X., Lopez P., Borja-Tabora C., Kosalaraksa P., Sirivichayakul C., Watanaveeradej V., Rivera L., Espinoza F., TIDES Study Group Efficacy of a Tetravalent Dengue Vaccine in Healthy Children and Adolescents. N. Engl. J. Med. 2019;381:2009–2019. doi: 10.1056/NEJMoa1903869. [DOI] [PubMed] [Google Scholar]
- 25.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines – a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogel A.B., Lambert L., Kinnear E., Busse D., Erbar S., Reuter K.C., Wicke L., Perkovic M., Beissert T., Haas H. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018;26:446–455. doi: 10.1016/j.ymthe.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pardi N., Parkhouse K., Kirkpatrick E., McMahon M., Zost S.J., Mui B.L., Tam Y.K., Karikó K., Barbosa C.J., Madden T.D. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018;9:3361. doi: 10.1038/s41467-018-05482-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J. Preclinical and Clinical Demonstration of Immunogenicity by mRNA Vaccines against H10N8 and H7N9 Influenza Viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guardo A.C., Joe P.T., Miralles L., Bargalló M.E., Mothe B., Krasniqi A., Heirman C., García F., Thielemans K., Brander C., iHIVARNA consortium Preclinical evaluation of an mRNA HIV vaccine combining rationally selected antigenic sequences and adjuvant signals (HTI-TriMix) AIDS. 2017;31:321–332. doi: 10.1097/QAD.0000000000001276. [DOI] [PubMed] [Google Scholar]
- 30.Pardi N., LaBranche C.C., Ferrari G., Cain D.W., Tombácz I., Parks R.J., Muramatsu H., Mui B.L., Tam Y.K., Karikó K. Characterization of HIV-1 Nucleoside-Modified mRNA Vaccines in Rabbits and Rhesus Macaques. Mol. Ther. Nucleic Acids. 2019;15:36–47. doi: 10.1016/j.omtn.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C. Modified mRNA Vaccines Protect against Zika Virus Infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer M., Huang E., Yuzhakov O., Ramanathan P., Ciaramella G., Bukreyev A. Modified mRNA-Based Vaccines Elicit Robust Immune Responses and Protect Guinea Pigs From Ebola Virus Disease. J. Infect. Dis. 2018;217:451–455. doi: 10.1093/infdis/jix592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 35.Pardi N., Weissman D. Nucleoside Modified mRNA Vaccines for Infectious Diseases. Methods Mol. Biol. 2017;1499:109–121. doi: 10.1007/978-1-4939-6481-9_6. [DOI] [PubMed] [Google Scholar]
- 36.Desmet C.J., Ishii K.J. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012;12:479–491. doi: 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- 37.Edwards D.K., Jasny E., Yoon H., Horscroft N., Schanen B., Geter T., Fotin-Mleczek M., Petsch B., Wittman V. Adjuvant effects of a sequence-engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J. Transl. Med. 2017;15:1. doi: 10.1186/s12967-016-1111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao G., Wang Q., Dai Z., Calcedo R., Sun X., Li G., Wilson J.M. Adenovirus-based vaccines generate cytotoxic T lymphocytes to epitopes of NS1 from dengue virus that are present in all major serotypes. Hum. Gene Ther. 2008;19:927–936. doi: 10.1089/hum.2008.011. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Z., Li M., Liu Z., Jin X., Sun J. Establishment of murine infection models with biological clones of dengue viruses derived from a single clinical viral isolate. Virol. Sin. 2020;25:1–11. doi: 10.1007/s12250-020-00229-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li M., Wang X., Wang Q., Yu L., Wang L., Yan J., Zhang F., Zhang L., Gao G.F., Jin X. Both structure and function of human monoclonal antibodies contribute to enhancement of Zika virus infectivity in vitro. Sci. China Life Sci. 2017;60:1396–1398. doi: 10.1007/s11427-017-9192-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodrigo W.W., Block O.K., Lane C., Sukupolvi-Petty S., Goncalvez A.P., Johnson S., Diamond M.S., Lai C.J., Rose R.C., Jin X., Schlesinger J.J. Dengue virus neutralization is modulated by IgG antibody subclass and Fcgamma receptor subtype. Virology. 2009;394:175–182. doi: 10.1016/j.virol.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang H., Tang J., Liu Z., Liu Y., Huang Y., Xu Y., Hao P., Yin Z., Zhong J., Ye L. ZIKV infection induces robust Th1-like Tfh cell and long-term protective antibody responses in immunocompetent mice. Nat. Commun. 2019;10:3859. doi: 10.1038/s41467-019-11754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Halstead S.B. Licensed Dengue Vaccine: Public Health Conundrum and Scientific Challenge. Am. J. Trop. Med. Hyg. 2016;95:741–745. doi: 10.4269/ajtmh.16-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urakami A., Ngwe Tun M.M., Moi M.L., Sakurai A., Ishikawa M., Kuno S., Ueno R., Morita K., Akahata W. An Envelope-Modified Tetravalent Dengue Virus-Like-Particle Vaccine Has Implications for Flavivirus Vaccine Design. J. Virol. 2017;91:e01181-17. doi: 10.1128/JVI.01181-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khalil S.M., Tonkin D.R., Mattocks M.D., Snead A.T., Johnston R.E., White L.J. A tetravalent alphavirus-vector based dengue vaccine provides effective immunity in an early life mouse model. Vaccine. 2014;32:4068–4074. doi: 10.1016/j.vaccine.2014.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costa S.M., Freire M.S., Alves A.M. DNA vaccine against the non-structural 1 protein (NS1) of dengue 2 virus. Vaccine. 2006;24:4562–4564. doi: 10.1016/j.vaccine.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 47.Bailey M.J., Broecker F., Duehr J., Arumemi F., Krammer F., Palese P., Tan G.S. Antibodies Elicited by an NS1-Based Vaccine Protect Mice against Zika Virus. mBio. 2019;10:e02861-18. doi: 10.1128/mBio.02861-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai Y.C., Chuang Y.C., Liu C.C., Ho T.S., Lin Y.S., Anderson R., Yeh T.M. Antibodies Against Modified NS1 Wing Domain Peptide Protect Against Dengue Virus Infection. Sci. Rep. 2017;7:6975. doi: 10.1038/s41598-017-07308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S.S., Li I.H., Hong P.D., Yeh M.K. Evaluation of protective efficacy using a nonstructural protein NS1 in DNA vaccine-loaded microspheres against dengue 2 virus. Int. J. Nanomedicine. 2013;8:3161–3169. doi: 10.2147/IJN.S49972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X., Qu L., Ye X., Yi C., Zheng X., Hao M., Su W., Yao Z., Chen P., Zhang S. Incorporation of NS1 and prM/M are important to confer effective protection of adenovirus-vectored Zika virus vaccine carrying E protein. NPJ Vaccines. 2018;3:29. doi: 10.1038/s41541-018-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li A., Yu J., Lu M., Ma Y., Attia Z., Shan C., Xue M., Liang X., Craig K., Makadiya N. A Zika virus vaccine expressing premembrane-envelope-NS1 polyprotein. Nat. Commun. 2018;9:3067. doi: 10.1038/s41467-018-05276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu J., Liu Y., Nie K., Du S., Qiu J., Pang X., Wang P., Cheng G. Flavivirus NS1 protein in infected host sera enhances viral acquisition by mosquitoes. Nat. Microbiol. 2016;1:16087. doi: 10.1038/nmicrobiol.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beatty P.R., Puerta-Guardo H., Killingbeck S.S., Glasner D.R., Hopkins K., Harris E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci. Transl. Med. 2015;7:304ra141. doi: 10.1126/scitranslmed.aaa3787. [DOI] [PubMed] [Google Scholar]
- 54.Roth C., Cantaert T., Colas C., Prot M., Casadémont I., Levillayer L., Thalmensi J., Langlade-Demoyen P., Gerke C., Bahl K. A Modified mRNA Vaccine Targeting Immunodominant NS Epitopes Protects Against Dengue Virus Infection in HLA Class I Transgenic Mice. Front. Immunol. 2019;10:1424. doi: 10.3389/fimmu.2019.01424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halstead S.B. Which Dengue Vaccine Approach Is the Most Promising, and Should We Be Concerned about Enhanced Disease after Vaccination? There Is Only One True Winner. Cold Spring Harb. Perspect. Biol. 2018;10:a030700. doi: 10.1101/cshperspect.a030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Jun Q. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.