Abstract
Diabetic foot ulcer (DFU) is a common and serious complication of diabetes mellitus, which influences patients’ quality of life. Recently, circRNA regulated the mRNA levels by functioning as miRNA sponge in various disease, including diabetes mellitus. Nevertheless, the circRNA-miRNA-mRNA regulatory network involved in DFU remains obscure. The aim of this study is to construct a competing endogenous RNA (ceRNA) network and screen biological indicators as diagnostic factors in DFU.
All the differentially expressed circRNAs, miRNAs and mRNAs were derived from Gene Expression Omnibus database. Furthermore, circRNAs identified by cytoHubba analysis and miRNAs obtained by human miRNA-disease database were used to construct DFU-specific ceRNA network with intersection of mRNAs. Functional enrichment analysis displayed the function and pathway of dysregulated mRNAs. Hub genes with high diagnostic value were screened by ClusterONE, GO semantic similarity and receiver operating characteristic (ROC) curve.
Here, the ceRNA network consisted of 8 circRNAs, 11 miRNAs and 91 mRNAs. Functional enrichment analysis demonstrated diabetic complications-related pathway including TGF-beta, FoxO and Wnt signaling pathway. GO semantic similarity and ROC curve analysis showed 6 hub genes with high diagnostic value (the area under the ROC curve ≥ 0.8) in patients with DFU, including BCL2, CCND1, IRAK4, SMAD4, SP1 and SUFU, which were identified as potential target genes for DFU diagnosis. In conclusion, the present study looked at a circRNA-miRNA-mRNA regulatory network with DFU and screened the potential function of mRNA, then identified novel diagnostic biomarkers and therapeutic targets for patients with DFU.
Keywords: Diabetic foot ulcer, ceRNA network, Diagnosis, Biomarkers
1. Introduction
Diabetic foot ulcer (DFU) is one of the most common diseases of lower extremity complications in diabetes mellitus, resulting in a heavy socioeconomic burden in families and societies. The patients with DFU usually increase costs and risk because of required long-term medical treatments and poor availability of tools for effective debridement [1]. The average medical cost of patients with diabetic foot infection in China was about $2685 per person in 2012, which is significantly higher than that in 2004 [2]. In addition, the enhancement of wound healing represents a major clinical challenge during the diabetic foot disease [3]. It has been showed that a large number of physiologic factors involved in the wound healing, including circular RNAs (circRNAs) [4,5].
CircRNA is an emerging class of noncoding RNA molecule of lacking 5′ caps and 3′ tails, which is consisted of a circular configuration through the 5′ to 3’ phosphodiester bond [6]. Early studies indicated that the circRNA may be a byproduct resulted from the process of mis-splicing [7]. With the development of next generation sequencing technology, accumulating evidence has emphasized the importance of the circRNA to discuss the roles in the competition of splicing machinery between linear RNA and circRNA [8]. Recent reports demonstrated that the hsa_circ_0067301 that competes for binding to miR-141–5p strongly suppressed miRNA activity and results in increased levels of Notch-1 during human epithelial-mesenchymal transition [9]. An increasing number of studies showed that circRNAs functioned as miRNA sponge in the competing endogenous RNA (ceRNA) network and played an important role in the disease process [10]. For instance, circRNA_010567 in ceRNA network has been shown to promote progression of myocardium and cardiac fibroblasts (CFs) cells by sponging miR-141 in diabetic mice [11]. Recent researches investigated the role of circRNA-associated ceRNA network in diabetes complications including diabetic nephropathy [12], diabetic cardiomyopathy [13] and DFU [4]. However, only one study that investigated the functional effects of hsa_circ_0084443 on DFU has been reported up to now [4]. The circRNA-mediated ceRNA regulatory network and biological function correlated to DFU remains obscure.
To investigate the effects of circRNAs on DFU, we collected the expression files of circRNA, miRNA and mRNA from Gene Expression Omnibus (GEO) datasets. Differentially expressed circRNAs, miRNAs and mRNAs were identified by “limma” package in R software. Next, the regulatory network of circRNA-miRNA-mRNA was constructed using bioinformatics approaches in accordance with the ceRNA theory. The main functional pathways of DFU were evaluated using gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The subnetwork of circRNA-miRNA-hub genes was established using the hub genes that were identified by protein–protein interaction (PPI) network and GO semantic similarity. The receiver operating characteristic (ROC) curve was used to identify the potential diagnostic biomarkers in the occurrence and development of DFU.
2. Materials and methods
2.1. Data collection
The microarray data were acquired from the Gene Expression Omnibus (GEO) database. The circRNA expression data was obtained from GSE114248 (5 DFU tissues and 5 non-DFU tissues). The miRNA data was derived from GSE84971 (3 DFU tissues and 3 non-DFU tissues). The mRNA data was obtained from GSE80178 (6 DFU tissues and 3 non-DFU tissues). Diabetes mellitus-related miRNAs were collected from the HMDD v3.0 database [14].
2.2. Differential expression analysis of circRNAs, miRNAs, and mRNAs
The analysis of differentially expressed of cirRNAs, miRNAs and mRNAs were performed using “limma” package in R software. The differentially expressed circRNAs (DECs), differentially expressed miRNAs (DEMs) and differentially expressed genes (DEGs) were screened with P-value < 0.05 and |logFC| > 1 as the threshold.
2.3. Construction of the circRNA-miRNA-mRNA regulatory network
The Information about circRNAs were available on the CircInteractome (http://circinteractome.nia.nih.gov) and circBase database (http://www.circbase.org/). In order to identify the key circRNAs participating in the ceRNA network, the closeness centrality, degree and betweenness centrality of cicrRNAs were calculated using cytoHubba plugin based on cytoscape software [15]. The target miRNAs of circRNAs were obtained using the Encyclopedia of RNA Interactomes (ENCORI, http://starbase.sysu.edu.cn/index.php). Furthermore, the intersections between the DEMs and the miRNAs that were collected using HMDD database were considered as candidate miRNAs to construct the ceRNA network. Then, the TargetScan [16], miRDB [17] and miRWalk [18] were used to predict the relationships among DEMs and their target DEGs. The intersecting miRNAs negatively regulated by circRNAs and mRNAs were used to construct the ceRNA network according to the ceRNA theory. The visualization of regulatory network was performed by cytoscape 3.7.2.
2.4. Functional enrichment analysis and construction PPI network
The miRNA KEGG pathway analysis were performed using DIANA-mirPath 3.0 [19]. Moreover, KEGG functional analysis was conducted using DEGs in the ceRNA network with “clusterProfiler” package in R. Gene Ontology (GO) analysis was conducted using KOBAS 2.0. The false discovery rate (FDR) of less 0.05 was set as the cut-off criterion. GO semantic similarity analysis was performed to compute semantic similarity among GO terms by “GOSemSim” package in R software.
The protein–protein interaction (PPI) network was conducted using the Search Tool for the Tetrieval of Interacting Genes (STRING) database (http://string-db.org). The ClusterONE algorithm was used to screen modules of hub genes from PPI network. The visualization was performed by cytoscape software. The ROC curve were performed by calculating the sensitivity and specificity of target expression level to screen the potential diagnostic target genes in patients with DFU.
3. Results
3.1. Convergence of gene expression signatures across different studies from GEO database
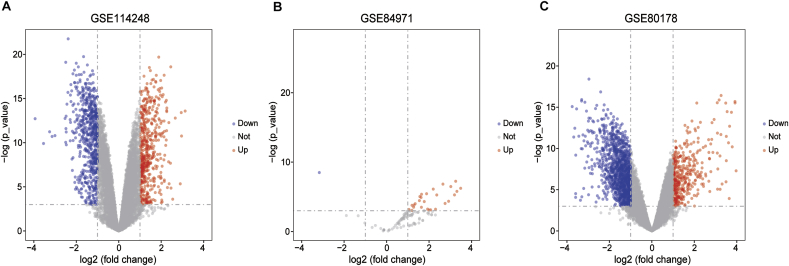
To investigate differentially expressed genes in each data sets, the “limma” package in R software were used for differentially expressed genes with P-value < 0.05 and |LogFC| > 1 as the threshold. The GSE114248 data set contained 945 DECs including 568 upregulated circRNAs and 645 downregulated circRNAs (Fig. 1A). The integrated analysis of GSE84971 dataset identified 30 DEMs, which included 29 upregulated miRNAs and 1 downregulated miRNAs (Fig. 1B). Furthermore, the DEGs were identified using the GSE80178 dataset. A total of 1715 DEGs were obtained, 430 of which were upregulated and 1285 of which were downregulated (Fig. 1C).
Fig. 1.
Volcano plots of differentially expressed circRNAs (A), miRNAs (B) and mRNAs (C) in GEO database. The red dots represent the upregulated genes with P-value < 0.05 and LogFC >1 as the threshold. The blue dots represent the downregulated genes with P-value < 0.05 and LogFC <1 as the cut-off criteria. The grey spots represent genes with no significant difference in expression. LogFC, Log2 (fold change); GEO, Gene Expression Omnibus.
3.2. Construction of the ceRNA network
The role of circRNAs as a miRNA sponge is the main mechanism of circRNA function in disease. Therefore, we explore the role of circRNA through establishing a ceRNA network in DFU. The circRNA-miRNA-mRNA ceRNA network were constructed using a combination of circRNA-miRNA pairs and miRNA-mRNA pairs. In order to screen the novel circRNAs of these DECs in DFU, the betweenness centrality, degree and closeness centrality of cicrRNAs were performed by using the cytoHubba plugin (Table 1). We predicted the target miRNAs of circRNAs using the ENCORI database, which has the advance that the results of the database could be filtered for experimentally-validated target miRNAs.
Table 1.
The betweenness centrality, degree and closeness centrality of cicrRNAs.
| CircRNAs | Betweenness | Degree | Closeness |
|---|---|---|---|
| hsa_circ_0089761 | 7280.369 | 11 | 63.200 |
| hsa_circ_0089762 | 68.769 | 2 | 42.643 |
| hsa_circ_0001222 | 632.959 | 2 | 42.943 |
| hsa_circ_0089763 | 1360.684 | 6 | 53.183 |
| hsa_circ_0000711 | 326.281 | 2 | 44.250 |
| hsa_circ_0001714 | 0 | 1 | 38.510 |
| hsa_circ_0000437 | 407.081 | 3 | 43.333 |
| hsa_circ_0000288 | 0 | 1 | 39.010 |
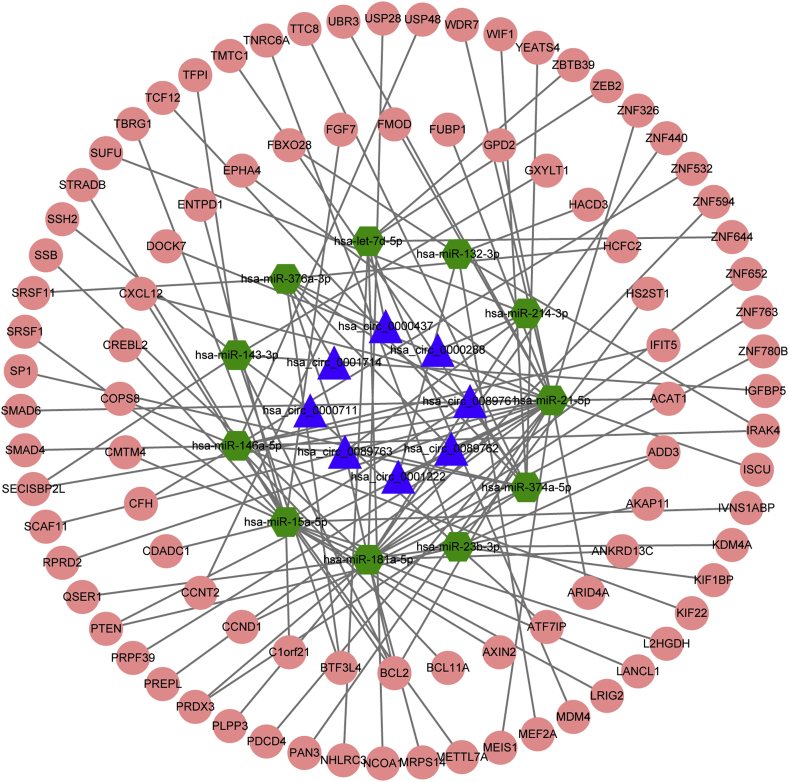
In addition, previous studies were performed using the open web-resource HMDD v3.0 to investigate the claim that miRNA are truly specific for DFU. The HMDD database collected miRNAs that are associated with the development of a broad range of diseases [20]. Among miRNAs included in the database, we selected the miRNAs associated with the spectrum of diabetes according to the categories: “diabetes mellitus”, “diabetes foot”. Finally, 19 diabetes-associated DEMs were selected to use for further analysis with intersections of 30 DEMs. For miRNA-mRNA pairs, the TargetScan, miRDB and miRWalk programs were used to predict the target mRNAs with intersections of 1715 DEGs. Taken together, we established the circRNA–miRNA–mRNA ceRNA network, including 8 DECs, 11 DEMs, 91 DEGs (Supplementary Table 1). The ceRNA network was plotted using cytoscape 3.7.2 software (Fig. 2).
Fig. 2.
The DFU-specific circRNA-miRNA-mRNA ceRNA network. The competing endogenous RNA network were constructed based on ceRNA theory. Blue triangle indicates circRNAs; green hexagon indicates miRNAs; pink ellipse indicates mRNAs.
3.3. Functional enrichment analysis
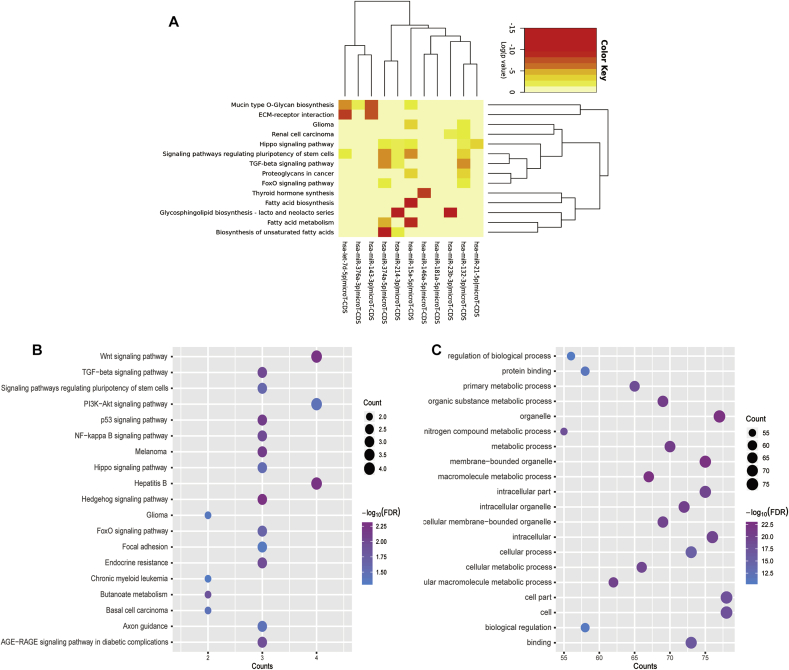
In order to explore the altered biological processes of ceRNA network, the basic characteristics for the 8 DECs were performed using circInteractome and circBase database, as described in Table 2. Three of DECs (hsa_circ_0089761, hsa_circ_0089762 and hsa_circ_0001222) were not found in these database. The role of circRNAs as a miRNA sponge is the molecular mechanism of circRNA function in diseases. Therefore, the combinatorial effect of miRNAs in pathways was investigated using DIANA-miRPath v3.0. The results demonstrated two novel pathway including TGF-beta and FoxO signaling pathway (Fig. 3A), which was verified in KEGG pathway analysis of DEGs.
Table 2.
Basic characteristics of the eight differentially expressed circRNAs in ceRNA network.
| CircRNA ID | Position | Genomic length (bp) | Stand | Best transcript | Gene symbol |
|---|---|---|---|---|---|
| hsa_circ_0089761 | chrM:7586-15888 | 8302 | – | None | None |
| hsa_circ_0089762 | chrM:7586-7982 | 396 | – | None | None |
| hsa_circ_0001222 | chr22:32874967–32881196 | 6229 | + | NM_012179 | FBXO7 |
| hsa_circ_0089763 | chrM:8366-14149 | 5783 | – | None | None |
| hsa_circ_0000711 | chr16:68155889–68160513 | 4624 | + | NM_173165 | NFATC3 |
| hsa_circ_0001714 | chr7:72861593-72884813 | 23220 | – | NM_032408 | BAZ1B |
| hsa_circ_0000437 | chr12:109046047–109048186 | 2139 | – | NM_014325 | CORO1C |
| hsa_circ_0000288 | chr11:34111725–34112225 | 500 | + | NM_005898 | CAPRIN1 |
Fig. 3.
Functional enrichment analysis of miRNAs and mRNAs in ceRNA network. A. DIANA mirPath analysis of 11 miRNAs in ceRNA network in patients with DFU. B. KEGG pathway enrichment analysis were performed using 91 differential target genes that were negatively associated with 11 miRNAs in ceRNA network. C. Gene Ontology (GO) analysis of differential expressed mRNAs in ceRNA network.
In addition, the functional enrichment analysis result of identified DEGs were performed by using KOBAS online database. The KEGG pathway analysis showed that the dysregulated mRNAs were enriched in diabetic complication-related pathways, including Wnt, TGF-beta, PI3K-Akt, p53, NF-kappa B and FoxO signaling pathway (Fig. 3B, Table 3). Furthermore, GO enrichment analyses were performed using KOBAS with P-value < 0.01 as the cutoff criteria. The top 20 terms were associated with multiple biological progress including cellular metabolic process, cell proliferation and protein metabolism (Fig. 3C, Table 4).
Table 3.
KEGG pathway analysis for the differentially expressed genes in ceRNA network (P < 0.01).
| KEGG pathway | KEGG entry | Count | Ratio | P-value |
|---|---|---|---|---|
| Wnt signaling pathway | hsa04310 | 4 | 0.027972 | 0.000340 |
| Hepatitis B | hsa05161 | 4 | 0.027397 | 0.000367 |
| PI3K-Akt signaling pathway | hsa04151 | 4 | 0.011696 | 0.007647 |
| Hedgehog signaling pathway | hsa04340 | 3 | 0.063830 | 0.000196 |
| p53 signaling pathway | hsa04115 | 3 | 0.043478 | 0.000575 |
| Melanoma | hsa05218 | 3 | 0.042254 | 0.000622 |
| TGF-beta signaling pathway | hsa04350 | 3 | 0.035714 | 0.000997 |
| NF-kappa B signaling pathway | hsa04064 | 3 | 0.032258 | 0.001324 |
| Endocrine resistance | hsa01522 | 3 | 0.030928 | 0.001489 |
| AGE-RAGE signaling pathway in diabetic complications | hsa04933 | 3 | 0.029703 | 0.001666 |
| FoxO signaling pathway | hsa04068 | 3 | 0.022388 | 0.003638 |
| Signaling pathways regulating pluripotency of stem cells | hsa04550 | 3 | 0.021127 | 0.004264 |
| Hippo signaling pathway | hsa04390 | 3 | 0.019481 | 0.005318 |
| Axon guidance | hsa04360 | 3 | 0.017045 | 0.007628 |
| Focal adhesion | hsa04510 | 3 | 0.014778 | 0.011163 |
| Butanoate metabolism | hsa00650 | 2 | 0.071429 | 0.002060 |
| Basal cell carcinoma | hsa05217 | 2 | 0.036364 | 0.007268 |
| Glioma | hsa05214 | 2 | 0.030769 | 0.009925 |
Table 4.
GO enrichment analysis for the differentially expressed genes in ceRNA network (top 20 and P-Value < 0.01).
| GO terms | GO entry | Count | Ratio | P-value |
|---|---|---|---|---|
| cell part | GO:0044464 | 78 | 0.004944 | 8.28E-21 |
| cell | GO:0005623 | 78 | 0.004936 | 9.30E-21 |
| organelle | GO:0043226 | 77 | 0.006067 | 1.90E-26 |
| intracellular | GO:0005622 | 76 | 0.005517 | 7.08E-23 |
| membrane-bounded organelle | GO:0043227 | 75 | 0.006353 | 1.94E-26 |
| intracellular part | GO:0044424 | 75 | 0.005585 | 1.29E-22 |
| binding | GO:0005488 | 73 | 0.005245 | 1.18E-19 |
| cellular process | GO:0009987 | 73 | 0.004984 | 3.09E-18 |
| intracellular organelle | GO:0043229 | 72 | 0.006164 | 1.28E-23 |
| metabolic process | GO:0008152 | 70 | 0.006578 | 3.30E-24 |
| organic substance metabolic process | GO:0071704 | 69 | 0.006714 | 3.67E-24 |
| Intracellular membrane-bounded organelle | GO:0043231 | 69 | 0.006464 | 3.81E-23 |
| macromolecule metabolic process | GO:0043170 | 67 | 0.007668 | 1.82E-26 |
| cellular metabolic process | GO:0044237 | 66 | 0.006703 | 2.13E-22 |
| primary metabolic process | GO:0044238 | 65 | 0.006616 | 1.57E-21 |
| cellular macromolecule metabolic process | GO:0044260 | 62 | 0.007653 | 1.95E-23 |
| protein binding | GO:0005515 | 58 | 0.005461 | 4.19E-14 |
| biological regulation | GO:0065007 | 58 | 0.005221 | 3.40E-13 |
| regulation of biological process | GO:0050789 | 56 | 0.005329 | 6.26E-13 |
| nitrogen compound metabolic process | GO:0006807 | 55 | 0.008227 | 2.86E-21 |
3.4. Construction of the PPI network and identification of the hub genes
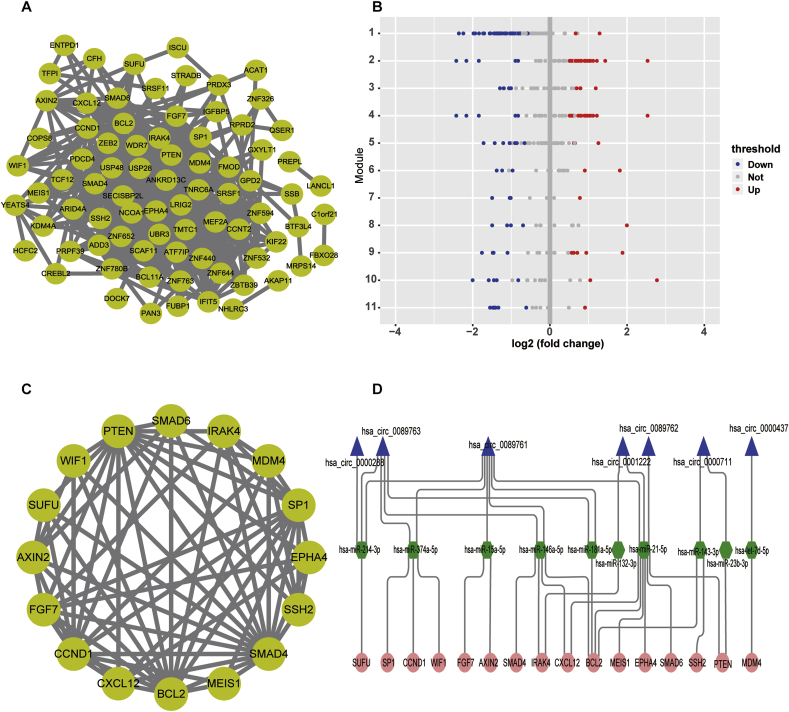
A total of 80 protein-protein interactions among 91 DEGs in ceRNA network were screened by applying STRING database (Fig. 4A). With the help of ClusterONE, we identified 10 PPI modules with P-value < 0.05 as the cutoff thresholds (Fig. 4B). Subsequently, in order to investigate the function of modules in PPI network, GO enrichment analysis results of module 8, 10 and 11 indicated enrichment in biosynthetic process. The results of module 6 and 9 suggested that these dysregulated genes were enriched in canonical Wnt signaling pathway and ephrin receptor signaling pathway, respectively. The enriched most significant function for the differentially expressed genes associated with the 10 modules were listed in Table 5.
Fig. 4.
Construction of protein‐protein interaction (PPI) network and ceRNA subnetworks. A. The PPI network was constructed for 80/91 differentially expressed mRNAs in ceRNA network. B. The modules were screened from the PPI network using “ClusterONE” algorithm containing 11 modules. Red dots represent upregulated mRNAs; blue dots represent downregulated mRNAs; grey dots represent mRNAs with no significant difference in expression. C. Interaction network of 16 hub genes. D. Sankey diagram for the circRNA-miRNA-hub gene subnetwork in diabetic foot ulcers, which was constructed using intersection between the top genes in the PPI network and potential genes that were associated with diabetic complication-related signaling pathways. Each rectangle indicates a gene. The size of the rectangle indicates the connection degree of each gene.
Table 5.
The enriched most significant function for the differentially expressed genes involved in 11 modules of PPI network.
| Module | Description | Term | Gene number | P-value |
|---|---|---|---|---|
| 1 | mRNA processing | GO:0006397 | 217 | 1.71E-37 |
| 2 | chemokine receptor activity | GO:0004950 | 125 | 5.13E-13 |
| 3 | Golgi vesicle transport | GO:0048193 | 79 | 2.83E-22 |
| 4 | G protein-coupled receptor activity | GO:0004930 | 108 | 1.75E-22 |
| 5 | peptide metabolic process | GO:0006518 | 90 | 4.37E-31 |
| 6 | canonical Wnt signaling pathway | GO:0060070 | 75 | 2.17E-27 |
| 7 | glycerophospholipid metabolic process | GO:0006650 | 51 | 8.98E-22 |
| 8 | nucleotide biosynthetic process | GO:0009165 | 38 | 5.50E-11 |
| 9 | ephrin receptor signaling pathway | GO:0048013 | 36 | 6.05E-17 |
| 10 | regulation of cellular macromolecule biosynthetic process | GO:2000112 | 43 | 5.09E-08 |
| 11 | aromatic compound biosynthetic process | GO:0019438 | 50 | 3.61E-08 |
The nodes with high degree are considered as important genes in network. In our study, the top genes were selected in the PPI network with degree >4 as the cutoff criteria. In addition, in order to investigate the function analysis of target genes. 16 dysregulated genes were enriched in diabetic complication-related signaling pathways. The intersection between the top genes in the network and genes that were associated with diabetic complication-related signaling pathways were AXIN2, BCL2, CCND1, CXCL12, EPHA4, FGF7, IRAK4, MDM4, MEIS1, PTEN, SMAD4, SMAD6, SP1, SSH2, SUFU and WIF1, which were considered as the hub genes of DFU (Fig. 4C). Furthermore, a circRNA-miRNA-hub gene subnetwork were construction according to the hub gene, as described in Fig. 4D.
3.5. ROC and GO semantic similarity analysis
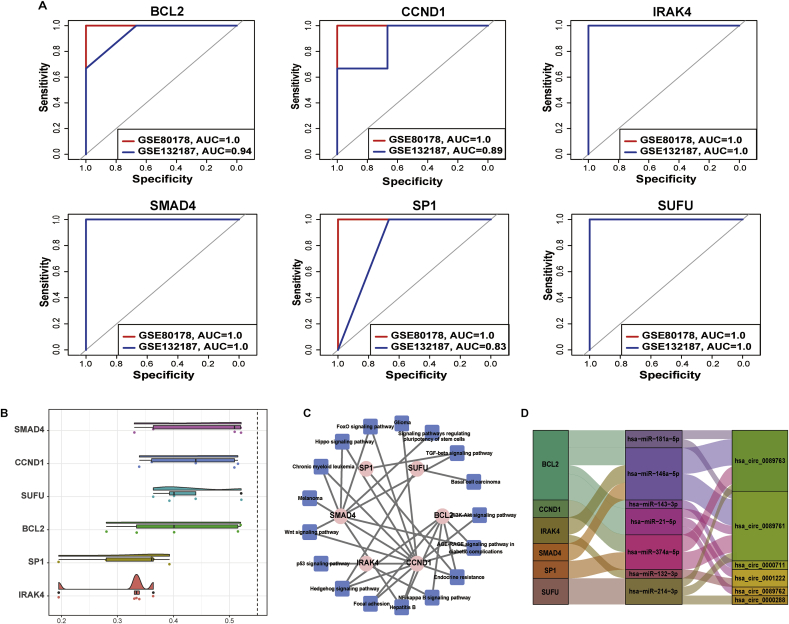
To explore specific genes and potential biomarker that might be capable of discriminating the patients with DFU from controls, the ROC analysis were performed for 16 hub genes in ceRNA subnetwork using the GSE80178 dataset as training set and GSE132187 dataset as test set. The best performance demonstrated to be the discriminating biomarkers with area under ROC curve (AUC) > 0.8 as the threshold, including BCL2, CCND1, IRAK4, SMAD4, SP1 and SUFU (Fig. 5A). SMAD4, SUFU and IRAK4 displayed the highest discriminatory power as a single marker, exhibiting a sensitivity of 100%, and specificity of 100%. Furthermore, in order to measure the strength of the relationship between each protein and its partners by considering functions and location of proteins, we ranked these 16 genes in the subnetwork with GO semantic similarity. The result showed that SMAD4 was considered as the most significantly strength the relationship between each protein and its partners (Fig. 5B). The intersection between ROC analysis and GO semantic similarity were SMAD4, which was regarded as the most potential and robust biomarker candidates for DFU. The KEGG analysis showed that SMAD4 was enriched in Wnt, FoxO, TGF-beta and AGE-RAGE signaling pathway (Fig. 5C). Finally, these findings provided evidence that SMAD4 that was identified by DFU-associated ceRNA might play an important role as potential diagnostic biomarker in patients with DFU.
Fig. 5.
Receiver operating characteristics (ROC) curve and GO semantic similarity analysis. A. ROC curve were performed in training (GSE80178) and test (GSE132187) dataset for hub genes with diagnostic values in DFU including BCL2, CCND1, IRAK4, SMAD4, SP1 and SUFU. B. Distributions of functional similarities between hub genes. The functional similarities were calculated using GO semantic similarity. C. KEGG enrichment analysis of hub genes with high diagnostic values in circRNA-miRNA-hub gene subnetwork.
4. Discussion
CircRNAs are ignored in classical transcriptome researches for a long time because of the lack of 5′ caps and 3′polyadenylated tails [21]. With the development of the technologies of next-generation sequencing (NGS) and the methods of biochemical and computational biology, a large number of circRNAs were studied in various tissues and cells [10]. An increasing number of studies have revealed the important role of circRNA-associated ceRNA network in screening diagnostic and prognostic biomarkers in a myriad of human diseases, such as cancer and diabetes mellitus [10,22]. In hepatocellular carcinoma, circMTO1 as a prognosis predictor suppressed hepatocellular carcinoma progression by acting as the sponge of oncogenic miR-9 to promote p21 expression [23]. Furthermore, study have been demonstrated the role of circHIPK3 in retinal vascular dysfunction induced by diabetes mellitus. The circHIPK3 expression was significantly upregulated in diabetic retinas and retinal endothelial cells following stressors related to diabetes mellitus [24]. Although several circRNAs have been screened as participating in the pathogenesis of complications in diabetes mellitus, the circRNA-associated ceRNA network and biological function corrected to the pathogenesis of extremity complications in diabetes mellitus including diabetic foot remain obscure. It is still necessary to conduct a circRNA-miRNA-mRNA regulatory network and advance our understanding of the molecular mechanism of DFU. In the present study, we constructed a ceRNA regulatory network in DFU consisting of 8 differentially expressed circRNAs (DECs), 11 differentially expressed miRNAs (DEMs) and 91 differentially expressed genes (DEGs).
In order to explore the interaction mechanism of ceRNA in patients with DFU, the miRNAs of circRNAs downstream were analyzed. Our results showed 11 DEMs were sponged by circRNAs involving in the ceRNA network. Among 11 DEMs, hsa-miR-146a-5p was identified as one of the highly up-regulated miRNA in the regulatory ceRNA network. In patients with type 1 diabetes mellitus, hsa-miR-146a-5p at 3 months correlated with residual beta cell function after diagnosis [25]. Furthermore, miR-146a as a potential molecular target contributed to inhibiting inflammation and apoptosis in the diabetic retina [26], suggesting miR-146a-5p might play an important role in pathogenesis and progression of diabetic complications. These findings were in consistence with our results that miR-146a-5p as a biological indicator was significantly increased in patients with DFU.
Previous studies showed that circRNA was considered for its active role as miRNA sponge to indirectly regulate the function of mRNA in ceRNA network [27]. Studies have showed that dysregulated mRNAs involved in diabetic complications [28]. In diabetic foot ulcer, pro-inflammatory S100A8 and IL-8 proteins contributed to impaired wound healing and caused persistent inflammation [29]. In our analysis, 91 differentially expressed mRNAs were involved in the ceRNA network. Furthermore, we performed numerous bioinformatics analysis to identify potential novel genes based functional enrichment, PPI network, GO semantic similarity and ROC analysis. The results indicated that BCL2, CCND1, IRAK4, SUFU, SP1 and SMAD4 were found to be the key differentially expressed genes. These dysregulated mRNAs might act as the essential molecules that mediated the progression of DFU.
BCL2 (BCL2 apoptosis regulator) encodes an integral outer mitochondrial membrane protein that blocks the apoptotic death of cells [30]. Recent study showed that Bcl-2 modified adipose-derived stem cells effectively contributed to the wound healing process in diabetic mice [31]. Similarity, our results showed the effects of BCL2 on DFU. Studies have showed that CCND1 (Cyclin D1) were diagnosed as the top hub genes in diabetes mellitus to insulin resistance [32]. IRAK4 (Interleukin 1 Receptor Associated Kinase 4) are serine-threonine kinases involving in essential regulators of IL-1R and Toll-like receptor-mediated signaling pathways [33]. It has been reported that inhibition of IRAK-4 activity contributed to attenuating the progression of diabetic nephropathy under diabetic conditions [34]. SUFU (Suppressor of Fused) are a negative regulator of the hedgehog signaling pathway [35]. Studies have showed that SUFU could inhibit the activation of the insulin promoter mediated by Glis3, which has been linked to both type I and type II diabetes [36]. SP1 (Specificity protein-1) is a zinc finger gene that binds to GC-rich motifs of many promoters and acts in many cellular processes, including cell differentiation, cell growth, apoptosis and immune responses [37]. Our result identified SP1 as a diagnostic gene in patients with DFU, which was in consistence with a recent report that Sp1 acted in diabetic wound healing by mediating matrix metalloproteinase-9 expression [38]. SMAD4 (SMAD family member 4) is a member of the Smad family of signal transduction proteins and is as an essential regulator of the transforming growth factor β (TGF-β) signaling pathway [39,40]. Previous researches showed that Smad4 was upregulated in human and mouse podocytes during diabetic nephropathy [41]. Depletion of Smad4 in podocytes protected mice from glomerulosclerosis in type 2 diabetes models [41]. Interestingly, GO semantic similarity analysis showed SMAD4 was the highest score among all of the hub genes in our results. ROC curve analysis showed that SMAD4 was a specific diagnostic biomarker in patients with DFU. These findings provided evidences that the candidate key genes played an important role in diagnosis and prognosis of diabetic complications. Our results identified these candidate key genes as diagnostic target indicators in patients with DFU. Although the molecular mechanism between some candidate key genes (CCND1, IRAK4, SUFU and SMAD4) and DFU have not been reported, further researches are needed to explore the molecular mechanism of novel target genes and implement combination therapy for DFU.
In order to investigate the potential causative roles of dysregulated genes in ceRNA network, the dysregulated biological pathways were identified by enrichment analysis. Our results showed that these dysregulated mRNA were enriched multiple signaling pathways, such as TGF-beta, FoxO and Wnt signaling pathway. Studies have found that the role of TGF-beta signaling pathway has been examined for the treatment of DFU [42]. TGF-β that was released from vicenin-2 film enhanced cell proliferation and wound contraction through the TGF-β signaling pathways [43]. In consistence with these findings, both functional enrichment analysis of dysregulated miRNAs and abnormal mRNAs in ceRNA network identified the novel TGF-β signaling pathways as an important regulator in patients with DFU in our results. Furthermore, Studies have shown that FOXO1 acted in the activation and regulation of key leukocytes needed to respond to bacterial challenge in oral mucosal tissues [44]. Although the effect of FoxO pathway on DFU has not been elucidated until now, the FoxO that are involved in diverse cellular function and clinical significance including immunity, wound healing and diabetic complications has been confirmed [45]. In addition, studies have showed that action of the Wnt/β-catenin pathway contributed to the inflammatory responses to diabetic ulcers and wound proliferation [46]. Another report demonstrated that inhibition of Wnt/β-catenin signaling was associated with impaired wound healing in patients with DFU [47]. Further study investigating strategies for molecular mechanism of wound healing in the DFU are needed. In our study, the comprehensive ceRNA regulatory network analysis of DFU was performed by using multiple bioinformatics analysis. However, there are some limitations to be acknowledged in our study. The biological experiments are still required to confirm the functions of the genes. Further studies warranted on the roles of hsa_circ_0089761, hsa_circ_0089763, hsa-miR-146a-5p and SMAD4 as potential diagnostic biomarkers and therapeutic targets for DFU.
In summary, the present study showed a comprehensive network analysis and key communication of circRNA, miRNA and mRNA using bioinformatics approach. Our study provided novel insights into circRNA-associated ceRNA network and identified potential diagnostic biomarkers in patients with DFU.
CRediT authorship contribution statement
Shuping Liao: Investigation, Writing - original draft. Xiaolan Lin: Data curation, Software, Validation. Changyu Mo: Conceptualization, Supervision, Project administration, Writing - review & editing.
Declaration of competing interest
The authors declared no conflicts of interest.
Acknowledgements
All the authors thank the public database for its open access.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2020.07.001.
Funding
Not applicable.
Availability of data and materials
The sequencing data have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE114248, GSE84971, GSE80178 and GSE132187.
CRediT authorship contribution statement
Shuping Liao designed experiments, collected data and helped analyze the data. Xiaolan Lin analyzed data and wrote the original draft. Weiqing Wu performed project administration and supervision. All authors read and approved the final manuscript.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Weng J.P., Bi Y. Epidemiological status of chronic diabetic complications in China. Chin. Med. J. 2015;128:3267–3269. doi: 10.4103/0366-6999.171350. http://10.4103/0366-6999.171350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z., Ran X. Diabetic foot care in China: challenges and strategy. Lancet Diabetes Endocrinol. 2016;4:297–298. doi: 10.1016/S2213-8587(16)00051-6. http://10.1016/S2213-8587(16)00051-6 [DOI] [PubMed] [Google Scholar]
- 3.Falanga V. Wound healing and its impairment in the diabetic foot. Lancet. 2005;366:1736–1743. doi: 10.1016/S0140-6736(05)67700-8. http://10.1016/S0140-6736(05)67700-8 [DOI] [PubMed] [Google Scholar]
- 4.Wang A., Toma M.A., Ma J. Circular RNA hsa_circ_0084443 is upregulated in diabetic foot ulcer and modulates keratinocyte migration and proliferation. Adv. Wound Care. 2020;9:145–160. doi: 10.1089/wound.2019.0956. http://10.1089/wound.2019.0956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brem H., Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J. Clin. Invest. 2007;117:1219–1222. doi: 10.1172/JCI32169. http://10.1172/JCI32169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu S., Yang X., Li X. Circular RNA: a new star of noncoding RNAs. Canc. Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. http://10.1016/j.canlet.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 7.Halbreich A., Pajot P., Foucher M. A pathway of cytochrome b mRNA processing in yeast mitochondria: specific splicing steps and an intron-derived circular DNA. Cell. 1980;19:321–329. doi: 10.1016/0092-8674(80)90506-1. http://10.1016/0092-8674(80)90506-1 [DOI] [PubMed] [Google Scholar]
- 8.Guan Y.J., Ma J.Y., Song W. Identification of circRNA-miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data. Canc. Cell Int. 2019;19:183. doi: 10.1186/s12935-019-0905-z. http://10.1186/s12935-019-0905-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang M., Wang S., Tang L. Downregulated circular RNA hsa_circ_0067301 regulates epithelial-mesenchymal transition in endometriosis via the miR-141/Notch signaling pathway. Biochem. Biophys. Res. Commun. 2019;514:71–77. doi: 10.1016/j.bbrc.2019.04.109. http://10.1016/j.bbrc.2019.04.109 [DOI] [PubMed] [Google Scholar]
- 10.Xiong D.D., Dang Y.W., Lin P. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J. Transl. Med. 2018;16:220. doi: 10.1186/s12967-018-1593-5. http://10.1186/s12967-018-1593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou B., Yu J.W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. http://10.1016/j.bbrc.2017.04.044 [DOI] [PubMed] [Google Scholar]
- 12.Hu W., Han Q., Zhao L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-beta1. J. Cell. Physiol. 2019;234:1469–1476. doi: 10.1002/jcp.26959. http://10.1002/jcp.26959 [DOI] [PubMed] [Google Scholar]
- 13.Yang F., Li A., Qin Y. A novel circular RNA mediates pyroptosis of diabetic cardiomyopathy by functioning as a competing endogenous RNA. Mol. Ther. Nucleic Acids. 2019;17:636–643. doi: 10.1016/j.omtn.2019.06.026. http://10.1016/j.omtn.2019.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Z., Shi J., Gao Y. HMDD v3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Res. 2019;47:D1013–D1017. doi: 10.1093/nar/gky1010. http://10.1093/nar/gky1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X., Chen Y. Identification of potentially functional CircRNA-miRNA-mRNA regulatory network in hepatocellular carcinoma by integrated microarray analysis. Med Sci Monit Basic Res. 2018;24:70–78. doi: 10.12659/MSMBR.909737. http://10.12659/MSMBR.909737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agarwal V., Bell G.W., Nam J.W. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. http://10.7554/eLife.05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32:1316–1322. doi: 10.1093/bioinformatics/btw002. http://10.1093/bioinformatics/btw002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dweep H., Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods. 2015;12:697. doi: 10.1038/nmeth.3485. http://10.1038/nmeth.3485 [DOI] [PubMed] [Google Scholar]
- 19.Vlachos I.S., Zagganas K., Paraskevopoulou M.D. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–466. doi: 10.1093/nar/gkv403. http://10.1093/nar/gkv403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimaldi A.M., Incoronato M. Clinical translatability of "identified" circulating miRNAs for diagnosing breast cancer: overview and update. Cancers. 2019;11 doi: 10.3390/cancers11070901. http://10.3390/cancers11070901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao K.Y., Sun H.S., Tsai S.J. Circular RNA - new member of noncoding RNA with novel functions. Exp. Biol. Med. 2017;242:1136–1141. doi: 10.1177/1535370217708978. http://10.1177/1535370217708978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J., Cui L., Yuan J. Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle cells proliferation and migration by sponging miR-124. Biochem. Biophys. Res. Commun. 2017;494:126–132. doi: 10.1016/j.bbrc.2017.10.068. http://10.1016/j.bbrc.2017.10.068 [DOI] [PubMed] [Google Scholar]
- 23.Han D., Li J., Wang H. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. http://10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 24.Shan K., Liu C., Liu B.H. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. http://10.1161/CIRCULATIONAHA.117.029004 [DOI] [PubMed] [Google Scholar]
- 25.Samandari N., Mirza A.H., Nielsen L.B. Circulating microRNA levels predict residual beta cell function and glycaemic control in children with type 1 diabetes mellitus. Diabetologia. 2017;60:354–363. doi: 10.1007/s00125-016-4156-4. http://10.1007/s00125-016-4156-4 [DOI] [PubMed] [Google Scholar]
- 26.Ye E.A., Steinle J.J. miR-146a suppresses STAT3/VEGF pathways and reduces apoptosis through IL-6 signaling in primary human retinal microvascular endothelial cells in high glucose conditions. Vis. Res. 2017;139:15–22. doi: 10.1016/j.visres.2017.03.009. http://10.1016/j.visres.2017.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin X., Feng C.Y., Xiang Z. CircRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of nonalcoholic steatohepatitis. Oncotarget. 2016;7:66455–66467. doi: 10.18632/oncotarget.12186. http://10.18632/oncotarget.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pichu S., Patel B.M., Apparsundaram S. Role of biomarkers in predicting diabetes complications with special reference to diabetic foot ulcers. Biomarkers Med. 2017;11:377–388. doi: 10.2217/bmm-2016-0205. http://10.2217/bmm-2016-0205 [DOI] [PubMed] [Google Scholar]
- 29.Singh K., Agrawal N.K., Gupta S.K. Increased expression of TLR9 associated with pro-inflammatory S100A8 and IL-8 in diabetic wounds could lead to unresolved inflammation in type 2 diabetes mellitus (T2DM) cases with impaired wound healing. J. Diabet. Complicat. 2016;30:99–108. doi: 10.1016/j.jdiacomp.2015.10.002. http://10.1016/j.jdiacomp.2015.10.002 [DOI] [PubMed] [Google Scholar]
- 30.Li J., Yanyan M., Mu L. The expression of Bcl-2 in adenomyosis and its effect on proliferation, migration, and apoptosis of endometrial stromal cells. Pathol. Res. Pract. 2019;215:152477. doi: 10.1016/j.prp.2019.152477. http://10.1016/j.prp.2019.152477 [DOI] [PubMed] [Google Scholar]
- 31.Ding S., Xu Y., Yan X. Effect of collagen scaffold with bcl-2-modified adipose-derived stem cells on diabetic mice wound healing. Int. J. Low. Extrem. Wounds. 2019 doi: 10.1177/1534734619880055. http://10.1177/1534734619880055 1534734619880055. [DOI] [PubMed] [Google Scholar]
- 32.Pujar M.K., Vastrad B., Vastrad C. Integrative analyses of genes associated with subcutaneous insulin resistance. Biomolecules. 2019;9 doi: 10.3390/biom9020037. http://10.3390/biom9020037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoarau C., Gerard B., Lescanne E. TLR9 activation induces normal neutrophil responses in a child with IRAK-4 deficiency: involvement of the direct PI3K pathway. J. Immunol. 2007;179:4754–4765. doi: 10.4049/jimmunol.179.7.4754. http://10.4049/jimmunol.179.7.4754 [DOI] [PubMed] [Google Scholar]
- 34.Kondo M., Tahara A., Hayashi K. Naunyn Schmiedebergs Arch Pharmacol; 2020. Therapeutic effects of interleukin-1 receptor-associated kinase 4 inhibitor AS2444697 on diabetic nephropathy in type 2 diabetic mice.http://10.1007/s00210-020-01816-2 [DOI] [PubMed] [Google Scholar]
- 35.Kogerman P., Grimm T., Kogerman L. Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1999;1:312–319. doi: 10.1038/13031. http://10.1038/13031 [DOI] [PubMed] [Google Scholar]
- 36.ZeRuth G.T., Yang X.P., Jetten A.M. Modulation of the transactivation function and stability of Kruppel-like zinc finger protein Gli-similar 3 (Glis3) by Suppressor of Fused. J. Biol. Chem. 2011;286:22077–22089. doi: 10.1074/jbc.M111.224964. http://10.1074/jbc.M111.224964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan N.Y., Midgley V.C., Kavurma M.M. Angiotensin II-inducible platelet-derived growth factor-D transcription requires specific Ser/Thr residues in the second zinc finger region of Sp1. Circ. Res. 2008;102:e38–51. doi: 10.1161/CIRCRESAHA.107.167395. http://10.1161/CIRCRESAHA.107.167395 [DOI] [PubMed] [Google Scholar]
- 38.Wang W., Yang C., Wang X.Y. MicroRNA-129 and -335 promote diabetic wound healing by inhibiting sp1-mediated MMP-9 expression. Diabetes. 2018;67:1627–1638. doi: 10.2337/db17-1238. http://10.2337/db17-1238 [DOI] [PubMed] [Google Scholar]
- 39.Wu L. Functional characteristics of a novel SMAD4 mutation from thoracic aortic aneurysms (TAA) Gene. 2017;628:129–133. doi: 10.1016/j.gene.2017.07.042. http://10.1016/j.gene.2017.07.042 [DOI] [PubMed] [Google Scholar]
- 40.Wan M., Cao X., Wu Y. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3:171–176. doi: 10.1093/embo-reports/kvf024. http://10.1093/embo-reports/kvf024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J., Sun Y.B.Y., Chen W. Smad4 promotes diabetic nephropathy by modulating glycolysis and OXPHOS. EMBO Rep. 2020;21 doi: 10.15252/embr.201948781. http://10.15252/embr.201948781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zubair M., Ahmad J. Role of growth factors and cytokines in diabetic foot ulcer healing: a detailed review. Rev. Endocr. Metab. Disord. 2019;20:207–217. doi: 10.1007/s11154-019-09492-1. http://10.1007/s11154-019-09492-1 [DOI] [PubMed] [Google Scholar]
- 43.Tan W.S., Arulselvan P., Ng S.F. Improvement of diabetic wound healing by topical application of Vicenin-2 hydrocolloid film on Sprague Dawley rats. BMC Compl. Alternative Med. 2019;19:20. doi: 10.1186/s12906-018-2427-y. http://10.1186/s12906-018-2427-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Graves D.T., Milovanova T.N. Mucosal immunity and the FOXO1 transcription factors. Front. Immunol. 2019;10:2530. doi: 10.3389/fimmu.2019.02530. http://10.3389/fimmu.2019.02530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tia N., Singh A.K., Pandey P. Role of Forkhead Box O (FOXO) transcription factor in aging and diseases. Gene. 2018;648:97–105. doi: 10.1016/j.gene.2018.01.051. http://10.1016/j.gene.2018.01.051 [DOI] [PubMed] [Google Scholar]
- 46.Zhang H., Nie X., Shi X. Regulatory mechanisms of the Wnt/beta-catenin pathway in diabetic cutaneous ulcers. Front. Pharmacol. 2018;9:1114. doi: 10.3389/fphar.2018.01114. http://10.3389/fphar.2018.01114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi W., Yang C., Dai Z. High levels of pigment epithelium-derived factor in diabetes impair wound healing through suppression of Wnt signaling. Diabetes. 2015;64:1407–1419. doi: 10.2337/db14-1111. http://10.2337/db14-1111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE114248, GSE84971, GSE80178 and GSE132187.