Summary
Latent factor modeling applied to single-cell RNA sequencing (scRNA-seq) data is a useful approach to discover gene signatures. However, it is often unclear what methods are best suited for specific tasks and how latent factors should be interpreted.
Here, we compare four state-of-the-art methods and propose an approach to assign derived latent factors to pathway activities and specific cell subsets. By applying this framework to scRNA-seq datasets from biopsies of patients with rheumatoid arthritis and systemic lupus erythematosus, we discover disease-relevant gene signatures in specific cellular subsets. In rheumatoid arthritis, we identify an inflammatory OSMR signaling signature active in a subset of synovial fibroblasts and an efferocytic signature in a subset of synovial monocytes.
Overall, we provide insights into latent factors models for the analysis of scRNA-seq data, develop a framework to identify cell subtypes in a phenotype-driven way, and use it to identify novel pathways dysregulated in rheumatoid arthritis.
Subject Areas: Immunology, Bioinformatics, Transcriptomics
Graphical Abstract

Highlights
-
•
Benchmarking four latent factor models for analysis of scRNA-seq data
-
•
Map biological pathways associated with latent factors to specific cell subsets
-
•
OSMR pathway dysregulated in a subset of RA synovial fibroblasts
-
•
MERTK-expressing monocytes with efferocytic function depleted in RA
Immunology; Bioinformatics; Transcriptomics
Introduction
Single-cell RNA sequencing (scRNA-seq) is a powerful technique that enables gene expression measurements in thousands of individual cells. Resolving cellular heterogeneity by scRNA-seq has enabled groundbreaking discovery in the biomedical domain, such as finding key disease drivers in cancer (Patel et al., 2014; Puram et al., 2017; Tirosh et al., 2016), neurodegeneration (Keren-Shaul et al., 2017; Mathys et al., 2019), and immune-mediated diseases (Der et al., 2019; Smillie et al., 2019; The Accelerating Medicines Partnership in SLE Network et al., 2019; Zhang et al., 2019). From a data analysis standpoint, a crucial step in a standard scRNA-seq pipeline is clustering (Luecken and Theis, 2019), where discrete cell populations sharing a common transcriptional profile are defined. These cell clusters are used in a variety of downstream analyses, such as differential expression (Crowell et al., 2019; Ma et al., 2019; Soneson and Robinson, 2018), compositional analysis (Fonseka et al., 2018), and cellular interaction analysis (Vento-Tormo et al., 2018; Yuan et al., 2019; Zhou et al., 2017). Phenotypic identification of clusters is usually performed by means of a hybrid approach that entails prior knowledge of the biological system and gene set enrichment analysis on cluster markers. An alternative, cluster-free approach to phenotypic identification of cellular states is trajectory analysis (Saelens et al., 2019), which aims to derive differentiation processes by using a pseudo-temporal ordering of single cells. However, in addition to identity- and differentiation-specific activities, transcriptional programs encompass a variety of cellular processes such as metabolism, growth, stress, and cell signaling, which are not necessarily captured by these approaches. Nevertheless, such expression programs are of great interest in a disease setting, where several communicating cell populations might act within the same dysregulated pathway. Thus, an in-depth characterization of such pathogenic signaling cascades at single-cell resolution is of great interest from a disease understanding perspective.
Latent factor models aim to decompose the global expression profile in its underlying transcriptional programs (Stein-O’Brien et al., 2018). These models project both genes and cells in a low-dimensional space, with latent dimensions approximating cells' transcriptional programs and summarizing the contributions of several genes. Standard matrix factorization approaches, such as principal component analysis (PCA), non-negative matrix factorization (NMF), and independent component analysis (ICA), have been widely applied to scRNA-seq data (Kotliar et al., 2019). Nevertheless, novel methods have been developed that account for the specificities of single-cell data, using meaningful prior distributions and enforcing sparsity (Bielecki et al., 2018; Buettner et al., 2015; Levitin et al., 2019; Lopez et al., 2018; Pierson and Yau, 2015; Stein-O’Brien et al., 2019). A key parameter choice of these methods that is left to the analyst is the number of latent dimensions to use. Despite a few heuristics having been proposed based on stability analysis or model selection (Bielecki et al., 2018; Kotliar et al., 2019; Stein-O’Brien et al., 2019; Way et al., 2020), it is unclear whether these strategies could be applied effectively to datasets with different characteristics and whether such heuristics are appropriate for different downstream tasks. For example, it has been shown that different biological processes are captured at different dimensionalities of the latent space (Way et al., 2020), suggesting that approaches considering a varying number of latent dimensions could be more robust in fully recapitulating the underlying biology of the dataset under consideration.
To explore the potential of this family of methods to uncover previously unidentified pathway activities, we perform a systematic comparison of four recent latent factor models that specifically account for the sparsity of scRNA-seq data: scCoGAPS (Stein-O’Brien et al., 2019), LDA (Bielecki et al., 2018; Dey et al., 2017), scHPF (Levitin et al., 2019), and scVI (Lopez et al., 2018; Svensson et al., 2020). The first three methods are built on probabilistic approaches to matrix factorization and have been successfully used to extract gene signatures from scRNA-seq data (Clark et al., 2019; Svensson et al., 2020; Xu et al., 2019; Zhao et al., 2020), whereas the last one is based on a deep variational autoencoder with a linear decoder, making the inferred gene weights interpretable (Svensson et al., 2020). We test these on two scRNA-seq datasets from patients with autoimmune diseases. The first dataset consists of single cells isolated from synovial biopsies of patients with rheumatoid arthritis (RA) and sorted into four main cell subsets: monocytes, B cells, T cells, and fibroblasts (referred to as the RA dataset) (Zhang et al., 2019). The second dataset consists of single cells isolated from the kidney of patients with systemic lupus erythematosus (SLE) with lupus nephritis (LN) and enriched for the leukocyte component (referred to as the SLE dataset) (The Accelerating Medicines Partnership in SLE Network et al., 2019). We evaluate the stability over iterations of the four methods across the dimensionality of the latent space by using three different metrics and highlight the predictive power of these methods to discriminate cells isolated from patients or controls. Then, we assess the methods' ability to recover gene signatures by evaluating the coverage across 13 different gene set collections. Reasoning that latent factors can be used as surrogates of pathway activities, we devise a simple method to assign gene signatures to cell clusters, thus enabling the identification of cell subsets from a functional perspective. We then extend this analytical framework to integrate ligand – receptor interactions across cell subsets. Finally, we explore the reported gene signatures and discover two previously unidentified pathways in the RA dataset: the OSMR signaling pathway in a subpopulation of fibroblasts and the MERTK signaling pathway in a monocyte subset. We show that these signatures are potentially disease associated, thus highlighting the power of latent factor modeling to inform the discovery of novel pathogenic pathways.
Results
Evaluation of Latent Factor Models Show Differences in Performance across Tasks and Latent Dimensions
It has been shown that factorization solutions are not strictly convex, thus resulting in different outputs for different iterations of the algorithms (Kotliar et al., 2019; Nordhausen, 2009). A common heuristics to select an appropriate latent dimension is to calculate the algorithm's stability across iterations and select the number of dimensions with results that are more consistent across iterations (Kotliar et al., 2019; Wu et al., 2016). For each method, we performed 10 iterations across 13 dimensionalities of the latent space (from k = 16 to k = 40, with step 2) and computed three stability metrics: Amari distance, silhouette score on the k-medoids-defined clusters, and the singular value canonical correlation analysis (SVCCA) score (Raghu et al., 2017) (see Methods for details). We performed this evaluation for both the RA and the SLE datasets (Figures 1A and 1B). scCoGAPS and LDA emerge as having better stability properties, across latent dimensions as well as across the three chosen metrics. In contrast, scVI shows poor stability, showing better performance than scHPF only for the SVCCA score. Overall, all methods report a lower performance as the number of latent dimension increases, consistent with the increased model complexity. End-of-training values for the objective functions of the four methods, at different values of k, were also investigated (Figures S1A and S1B).
Figure 1.
Evaluation of Latent Factor Models Show Differences in Performance across Tasks and Latent Dimensions
(A and B) Stability metrics in (A) the RA dataset and (B) the SLE dataset. Y axis reports the mean value of the metric across 10 iterations. X axis reports k, the number of latent dimensions.
(C) Cross-validation AUPR curve in a disease-control classification task using latent variables as predictors, in the RA dataset and the SLE dataset. Y axis reports cross-validation AUPR value across 10 iterations. X axis reports k, the number of latent dimensions.
(D) Mean gene set collection coverage across latent dimensions, in the RA dataset and the SLE dataset. Y axis reports mean collection coverage value, averaged across 13 gene set collections. X axis reports k, the number of latent dimensions.
To assess whether these latent factors retained information on the disease state of the samples, we used them as predictors of an elastic net regression model with the task of classifying disease and control cells (Figures 1C, S1C, and S1D). Interestingly, we found the performance to vary considerably between datasets, but not methods. In the RA dataset, almost all methods fail to reach an AUPR >0.5 regardless of the dimensionality of the latent space. In contrast, for the SLE dataset, all methods see a consistent increase in predictive power as the dimensionality increases, with scCoGAPS and LDA showing the best performance at low (16–24) and high (26–40) dimensionality of the latent space, respectively. Taken together, these results suggest that the dimensionality of the latent space is critical for extracting biological features related to the disease state of the cell.
The ability of latent factor models to recover biological signal is a key feature in their application to discover cellular phenotypes. Gene set enrichment analysis is a widely used approach for this task, as it allows mapping each latent variable to a specific pathway or biological process. To evaluate the methods' ability to recapitulate biologically meaningful gene signatures in a systematic manner, we used an enrichment approach based on heterogeneous network (Himmelstein et al., 2017; Way et al., 2020). Briefly, at each dimensionality of the latent space, we compute the gene set coverage score (number of unique gene sets significantly associated with each latent variable divided by the total number of gene set in the collection) for the gene set collection of interest (see Methods for details). We considered thirteen gene set collections, covering most of the known pathways and biological processes, as well as several other gene signatures (Figure 1D). As expected, for all methods we could observe an increase in the gene set coverage as the dimensionality of the latent space increases. By comparing the gene set coverage on the latent variables with the standard enrichment on clusters' marker genes (Figure S1E), we showed that the number of significant gene sets is an order of magnitude higher for the factorization methods, pointing to a higher sensitivity in the discovery of pathway activities. Interestingly, scHPF clearly outperformed the other methods in the majority of the gene set collections in both datasets (Figures S1F and S1G). This suggests that scHPF can decompose the expression matrix in a latent space that retains the highest degree of biological signal, which prompted us to use this method for all downstream analyses.
Systematic Assignment of Latent Variables to Cell Clusters Allows Identification of Cell Types Based on Their Phenotype or Function
An open challenge in single-cell transcriptomics is the phenotypic identification of cell populations after clustering. Usually, this is performed by means of a combination of prior knowledge of cell-specific markers and gene set enrichment analysis performed on the marker genes list for each cell subset (Luecken and Theis, 2019). However, as latent variables provide a surrogate of pathway activities across cells, we devised a simple framework to assign each pathway to cell clusters (Figure 2A). This approach directly allows the identification of cell subsets in a function- or phenotype-driven way. Briefly, we start by employing a standard clustering procedure to identify cell subsets (see Methods). For each gene set, we collapse redundant assignments to multiple latent variables in unique pathway activities, by means of an iterative clustering approach. Then, we regress pathway activity weights (i.e., the numerical results of the factorization) using the cell cluster labels as predictors. The coefficient of each cluster represents an indicator of how important that cell subset is to explain the pathway activity, therefore linking the activity of the pathway to the cell cluster, which can then be functionally interpreted. The heatmap in Figure 2B shows the coefficients for the most significant KEGG gene sets mapped to the latent variables obtained from the RA dataset, across the different cell clusters. Interestingly, broadly defined cell populations cluster together, showing that consistent activities across different biological processes recapitulate cell lineages. Importantly, this approach allows discovery of pathway activities that are unique to specific cell types or that are shared across different cell subsets in an unsupervised way. For instance, “B cell signaling” and “NK cell cytotoxicity” pathways (Figures 2C and 2D) show a distinct activity in the expected cell populations (see Figure S1H and S1I for an overview of the identified clusters). This strategy can also be used to annotate known, yet unidentified, cell types, such as plasmacytoid dendritic cells (Figure 2E). Finally, in the SLE dataset, we could identify a type I interferon signature specifically active in a distinct subset of B cells and T cells, as previously reported (The Accelerating Medicines Partnership in SLE Network et al., 2019) (Figure 2F). Overall, this framework to define pathway activities is a powerful approach to assign cell identity and cell states to clusters based on their function or phenotype. Complete results for the RA and SLE datasets are reported in Figures S2 and S3 and Tables S1 and S2, respectively.
Figure 2.
Systematic Assignment of Latent Variables to Cell Clusters Allows Identification of Cell Types Based on Their Phenotype or Function
(A) Schematic of the framework to derive pathway activities and assign them to cell subsets.
(B–F) (B) Heatmap of the KEGG collection's gene sets assigned to cell clusters in RA. The reported value represents the coefficient of the regression model and the “# loadings” color scale represents the number of latent variables that were found significant for that specific gene set. Factor weights for NK cell cytotoxicity gene set from the KEGG collection (C), B cell receptor signaling gene set from the KEGG collection (D), plasmacytoid dendritic cell signature from the C7 immunological signature collection (E), and type I interferon signaling gene set from the REACTOME collection (F).
OSMR Signaling Is Active in Specific Subsets of Rheumatoid Arthritis Fibroblasts that Share a Similar Inflammatory Profile to Stromal Cells from Patients with Inflammatory Bowel Disease
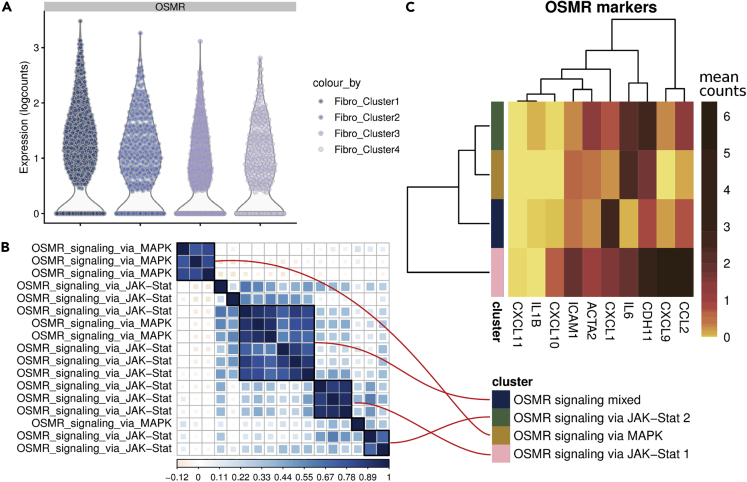
By using latent variables as surrogates of pathway activities, we sought to discover novel pathways potentially involved in RA. We focused on Oncostatin M (OSM) receptor (OSMR) signaling, whose expression level was found to be low yet widespread across fibroblast subsets (Figure 3A). However, we found 17 latent factors enriched for OSMR signaling-related gene sets. We collapsed these redundant gene sets in four pathway activities (Figure 3B), which showed a distinct distribution and composition of fibroblast subsets (Figures S4A and S4B). OSMR has been recently discovered to be a driver of increased inflammatory state of stromal cells in inflammatory bowel disease (IBD) (Oxford IBD Cohort Investigators et al., 2017). To investigate whether the RA fibroblast populations with high OSMR pathway activity also exhibited a similar inflammatory phenotype, we retrieved the gene signature associated with OSMR-high expression (Oxford IBD Cohort Investigators et al., 2017) and visualized the mean gene expression in the OSMR-signaling pathway activity (Figure 3C). Interestingly, two of the OSMR-related pathway activities showed a higher expression for the previously identified gene signature. Furthermore, these pathway activities seem to be mostly constituted by cells belonging to fibroblast clusters 1 and 2, which exhibit sublining markers (Figures S4C and S4D). These results indicate that OSMR signaling is active in synovial sublining fibroblasts in RA; as these cells share an inflammatory gene signature with OSMR-high stromal cells from patients with IBD, they suggest that OSMR could be a potential driver of inflammation also in RA.
Figure 3.
OSMR Signaling Is Active in Specific Subsets of Rheumatoid Arthritis Fibroblasts that Share a Similar Inflammatory Profile to Stromal Cells from Patients with Inflammatory Bowel Disease
(A) Expression levels of OSMR across fibroblast clusters.
(B) Correlation matrix of latent variables that maps to OSMR signaling pathways, as annotated by the METABASE gene set collection. Black frames enclose the correlation clusters that were selected to be representative of specific pathways activity.
(C) Mean expression level of genes found to be associated with OSMR-high-stromal cells in IBD.
Integration of Ligand-Receptor Interactions Reveals MERTK-Driven Apoptotic Cell Clearance by a Monocyte Subset in Rheumatoid Arthritis
To further explore the potential of pathway activities to uncover novel gene signatures, we sought to integrate this information with ligand-receptor interaction analysis (see Methods). In short, the expression level of interacting ligands and receptors was correlated with, and filtered for, latent variables with a significant enrichment for pathways where either protein was present (Figure 4A). Among the filtered cellular interactions, we found GAS6-MERTK. MERTK has a distinct expression across monocyte subsets (Figure 4B), and both monocyte clusters 1 and 3 showed interactions with GAS6 in fibroblasts and B cells subsets via MERTK (Figures S5A and S5B). We found MERTK-associated pathways to cluster in two main groups (Figure 4C): one with gene sets related to cell motility and cell signaling, the other one related to endocytosis and phagocytosis. As MERTK is a known marker for endocytic and phagocytic activity (particularly in the context of apoptotic cell clearance [Graham et al., 2014]), we set up to assess whether cells that were showing an endocytic-related activity showed an efferocytosis signature (Roberts et al., 2017; Waterborg et al., 2018). We observed that cells characterized by endocytic activity indeed recapitulated this gene signature to a higher degree as compared with the other activity clusters (Figure 4D). This cell subset, which is mostly constituted of monocytes from cluster 3 (Figure S5C, see Figure S5D for the signature enrichment across monocyte clusters), was found to be depleted in RA when compared with controls (Figure 4E), suggesting that reduced apoptotic cell clearance by MERTK-signaling monocytes could be a pathogenic mechanism in RA.
Figure 4.
Integration of Ligand-Receptor Interactions Reveals MERTK-Driven Apoptotic Cell Clearance by a Monocyte Subset in Rheumatoid Arthritis
(A) Ligand-receptor interaction network as computed by CellPhoneDB and filtered as described in the main text.
(B) Expression levels of MERTK across monocyte clusters.
(C) Correlation matrix of latent factors that are associated with MERTK expression. Black frames enclose the correlation clusters that were selected to be representative of specific pathways activities.
(D) Mean expression levels of genes found to be associated with infiltrating macrophages showing an efferocytic activity.
(E) Proportion of cell types from disease (RA) or control (OA) across monocytes clusters.
Discussion
Latent factor models are a flexible approach to uncover transcriptional programs in an unsupervised fashion, thus allowing a functional annotation of cells in an unbiased way.
Here, we conducted an evaluation of four state-of-the-art latent factor models specifically developed for scRNA-seq data (scCoGAPS [Stein-O’Brien et al., 2019], LDA [Bielecki et al., 2018; Dey et al., 2017], scHPF [Levitin et al., 2019], and scVI [Lopez et al., 2018; Svensson et al., 2020]), assessing stability, predictive power, and gene set coverage of latent variables across the dimensionality of the latent space. Although the four methods represent considerably different approaches to the latent factor modeling paradigm, our evaluation highlighted some of the strengths and weaknesses of these methods across different tasks and can be used as a starting point for selecting the method of choice depending on the user needs.
We devised a novel framework to collapse redundant gene sets into pathway activities and assign these to cell clusters. We show that such an approach is able to retrieve known cellular phenotypes and that it can be used to identify cell subpopulations based on existing cell identity signatures, without having to rely on marker genes. Although we focused on two autoimmune disease cohorts, the described framework is generally applicable to any scRNA-seq datasets and provides an intuitive way to directly define cell subpopulations based on their function or phenotype.
Importantly, we also show that our framework can be used to discover previously unidentified pathways active in specific cell subsets. Among the pathways activities we retrieved that were not previously reported by the authors of the original publication (Zhang et al., 2019), we noticed the OSM signaling pathway. Interestingly, OSM has been recently reported to be a key driver of intestinal inflammation in IBD and to be associated with response to anti-TNF therapy (Oxford IBD Cohort Investigators et al., 2017). We showed that RA fibroblasts with an OSMR pathway activity also express higher levels of a gene signature associated with high expression of OSMR in IBD stromal cells. This points to the potential involvement of the OSM/OSMR axis in establishing the inflammatory microenvironment in patients with RA and suggests the pathway might be similarly dysregulated in RA and IBD. Of note, cells with increased OSMR signaling activity mainly belong to sublining fibroblast subsets. Since sublining fibroblasts have also been recently associated with a specific inflammatory phenotype (Croft et al., 2019) (as opposed to a more cartilage degradation phenotype of the lining fibroblasts), we can speculate that the OSM pathway is one of the drivers of inflammation not only in IBD but also in RA.
Through integration of ligand-receptor interaction analysis with our approach, we recovered a link between GAS6 on B cells and fibroblasts and MERTK in monocytes. As one of the pathway activities correlated to MERTK expression related to endocytosis, and because MERTK is known to be involved in efferocytosis, we hypothesized that MERTK-expressing monocytes could be involved in apoptotic cell clearance. To test this, we evaluated the expression of genes part of an efferocytosis signature (Roberts et al., 2017; Waterborg et al., 2018) and were able to show that this monocyte subset does indeed show expression of this signature and is depleted in disease. Interestingly, a recent report (Alivernini et al., 2020) confirmed the presence of MERTK+ synovial macrophages driving remission in RA, thus substantiating our findings.
Limitations of the Study
We acknowledge an important limitation of our study is the relatively small selection of the latent factor models we pick for our initial evaluation. Our selection was guided by previous evidence that they could extract relevant transcriptional signatures from scRNA-seq data, as well as by the availability of implementations at the time of the analysis. Inclusion of other matrix factorization models such as f-scLVM (Buettner et al., 2017, 2015) and consensus NMF (Kotliar et al., 2019), or other deep learning approaches such as DCA (Eraslan et al., 2019), would certainly make any benchmarking efforts more comprehensive.
Another limitation is that both scRNA-seq datasets we analyzed were generated with the CEL-Seq2 technology and had a relatively low number of cells compared with more recently generated datasets (Martin et al., 2019; Schafflick et al., 2020; Smillie et al., 2019). The inclusion of larger datasets generated with different technologies would benefit a more comprehensive benchmarking effort and could potentially lead to different conclusions regarding the performance of the latent factor methods across different tasks.
Finally, as our approach to map latent factors to pathway activities and assign these to cell subsets relies heavily on gene sets annotation, the quality of the resulting pathway activities is inevitably tied to the quality of the original gene sets in the collection. As such, some of the identified pathway activities might be false positives. One example is the olfactory transduction gene set from the KEGG collection, which was found to be significantly associated with 64 latent factors, further collapsed in 19 pathway activities. Therefore, we suggest that hypothesis-driven explorations of the pathways activities assignments are needed to draw meaningful interpretations.
Resource Availability
Lead Contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Enrico Ferrero (enrico.ferrero@novartis.com).
Materials Availability
No materials were generated or used as part of this study.
Data and Code Availability
No new data was generated as part of this study. All code of the analysis is available at https://github.com/giovp/latent_factors_autoimmune.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to thank Jonas Zierer, Christine Huppertz, Grigory Ryzhakov, Dominik Hartl, Stephan Spiegel, James Rush, and Richard Siegel for helpful discussions and comments on the manuscript and Patrick Dunn for helping with data access.
Author Contributions
Conceptualization: E.F.; Methodology: E.F., G.P.; Formal Analysis: G.P.; Writing – Original Draft: G.P.; Writing – Review and Editing: E.F., G.P.; Visualization: G.P.; Supervision: E.F.
Declaration of Interests
E.F. is a Novartis employee and shareholder.
Published: September 25, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101451.
Supplemental Information
References
- Alivernini S., MacDonald L., Elmesmari A., Finlay S., Tolusso B., Gigante M.R., Petricca L., Di Mario C., Bui L., Perniola S. Distinct synovial tissue macrophage subsets regulate inflammation and remission in rheumatoid arthritis. Nat. Med. 2020;26:1295–1306. doi: 10.1038/s41591-020-0939-8. [DOI] [PubMed] [Google Scholar]
- Bielecki P., Riesenfeld S.J., Kowalczyk M.S., Vesely M.C.A., Kroehling L., Yaghoubi P., Dionne D., Jarret A., Steach H.R., McGee H.M. Skin inflammation driven by differentiation of quiescent tissue-resident ILCs into a spectrum of pathogenic effectors. bioRxiv. 2018 [Google Scholar]
- Buettner F., Natarajan K.N., Casale F.P., Proserpio V., Scialdone A., Theis F.J., Teichmann S.A., Marioni J.C., Stegle O. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- Buettner F., Pratanwanich N., McCarthy D.J., Marioni J.C., Stegle O. f-scLVM: scalable and versatile factor analysis for single-cell RNA-seq. Genome Biol. 2017;18:212. doi: 10.1186/s13059-017-1334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark B.S., Stein-O’Brien G.L., Shiau F., Cannon G.H., Davis-Marcisak E., Sherman T., Santiago C.P., Hoang T.V., Rajaii F., James-Esposito R.E. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron. 2019;102:1111–1126.e5. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft A.P., Campos J., Jansen K., Turner J.D., Marshall J., Attar M., Savary L., Wehmeyer C., Naylor A.J., Kemble S. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570:246–251. doi: 10.1038/s41586-019-1263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell H.L., Soneson C., Germain P.-L., Calini D., Collin L., Raposo C., Malhotra D., Robinson M.D. On the discovery of subpopulation-specific state transitions from multi-sample multi-condition single-cell RNA sequencing data. bioRxiv. 2019 doi: 10.1038/s41467-020-19894-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der E., Suryawanshi H., Morozov P., Kustagi M., Goilav B., Ranabothu S., Izmirly P., Clancy R., Belmont H.M., Koenigsberg M. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat. Immunol. 2019;20:915–927. doi: 10.1038/s41590-019-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey K.K., Hsiao C.J., Stephens M. Visualizing the structure of RNA-seq expression data using grade of membership models. PLoS Genet. 2017;13:e1006599. doi: 10.1371/journal.pgen.1006599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraslan G., Simon L.M., Mircea M., Mueller N.S., Theis F.J. Single-cell RNA-seq denoising using a deep count autoencoder. Nat. Commun. 2019;10:1–14. doi: 10.1038/s41467-018-07931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseka C.Y., Rao D.A., Teslovich N.C., Korsunsky I., Hannes S.K., Slowikowski K., Gurish M.F., Donlin L.T., Lederer J.A., Weinblatt M.E. Mixed-effects association of single cells identifies an expanded effector CD4$\mathplus$T cell subset in rheumatoid arthritis. Sci. Transl. Med. 2018;10:eaaq0305. doi: 10.1126/scitranslmed.aaq0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D.K., DeRyckere D., Davies K.D., Earp H.S. The TAM family: phosphatidylserine-sensing receptor tyrosine kinases gone awry in cancer. Nat. Rev. Cancer. 2014;14:769–785. doi: 10.1038/nrc3847. [DOI] [PubMed] [Google Scholar]
- Himmelstein D.S., Lizee A., Hessler C., Brueggeman L., Chen S.L., Hadley D., Green A., Khankhanian P., Baranzini S.E. Systematic integration of biomedical knowledge prioritizes drugs for repurposing. Elife. 2017;6:e26726. doi: 10.7554/eLife.26726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B. A unique microglia type Associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Kotliar D., Veres A., Nagy M.A., Tabrizi S., Hodis E., Melton D.A., Sabeti P.C. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. Elife. 2019;8:e43803. doi: 10.7554/eLife.43803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin H.M., Yuan J., Cheng Y.L., Ruiz F.J., Bush E.C., Bruce J.N., Canoll P., Iavarone A., Lasorella A., Blei D.M., Sims P.A. De novo gene signature identification from single-cell RNA -seq with hierarchical Poisson factorization. Mol. Syst. Biol. 2019;15:e8557. doi: 10.15252/msb.20188557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez R., Regier J., Cole M.B., Jordan M.I., Yosef N. Deep generative modeling for single-cell transcriptomics. Nat. Methods. 2018;15:1053–1058. doi: 10.1038/s41592-018-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15:e8746. doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B.X., Korthauer K., Kendziorski C., Newton M.A. A compositional model to assess expression changes from single-cell Rna-seq data. bioRxiv. 2019 doi: 10.1214/20-aoas1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J.C., Chang C., Boschetti G., Ungaro R., Giri M., Grout J.A., Gettler K., Chuang L., Nayar S., Greenstein A.J. Single-cell analysis of crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. 2019;178:1493–1508.e20. doi: 10.1016/j.cell.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., Menon M., He L., Abdurrob F., Jiang X. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–337. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordhausen K. The elements of statistical learning: data mining, inference, and prediction, second edition by Trevor Hastie, robert Tibshirani, Jerome Friedman. Int. Stat. Rev. 2009;77:482. [Google Scholar]
- Oxford IBD Cohort Investigators. West N.R., Hegazy A.N., Owens B.M.J., Bullers S.J., Linggi B., Buonocore S., Coccia M., Görtz D., This S., Stockenhuber K. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017;23:579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson E., Yau C. ZIFA: dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol. 2015;16:241. doi: 10.1186/s13059-015-0805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu M., Gilmer J., Yosinski J., Sohl-Dickstein J. SVCCA: singular vector canonical correlation analysis for deep learning dynamics and interpretability. arxiv. 2017 [Google Scholar]
- Roberts A.W., Lee B.L., Deguine J., John S., Shlomchik M.J., Barton G.M. Tissue-Resident macrophages are locally programmed for silent clearance of apoptotic cells. Immunity. 2017;47:913–927.e6. doi: 10.1016/j.immuni.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saelens W., Cannoodt R., Todorov H., Saeys Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019;37:547–554. doi: 10.1038/s41587-019-0071-9. [DOI] [PubMed] [Google Scholar]
- Schafflick D., Xu C.A., Hartlehnert M., Cole M., Schulte-Mecklenbeck A., Lautwein T., Wolbert J., Heming M., Meuth S.G., Kuhlmann T. Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun. 2020;11:247. doi: 10.1038/s41467-019-14118-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C.S., Biton M., Ordovas-Montanes J., Sullivan K.M., Burgin G., Graham D.B., Herbst R.H., Rogel N., Slyper M., Waldman J. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell. 2019;178:714–730.e22. doi: 10.1016/j.cell.2019.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Robinson M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- Stein-O’Brien G.L., Arora R., Culhane A.C., Favorov A.V., Garmire L.X., Greene C.S., Goff L.A., Li Y., Ngom A., Ochs M.F. Enter the matrix: factorization uncovers knowledge from omics. Trends Genet. 2018;34:790–805. doi: 10.1016/j.tig.2018.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein-O’Brien G.L., Clark B.S., Sherman T., Zibetti C., Hu Q., Sealfon R., Liu S., Qian J., Colantuoni C., Blackshaw S. Decomposing cell identity for transfer learning across cellular measurements, platforms, tissues, and species. Cell Syst. 2019;8:395–411.e8. doi: 10.1016/j.cels.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Accelerating Medicines Partnership in SLE Network. Arazi A., Rao D.A., Berthier C.C., Davidson A., Liu Y., Hoover P.J., Chicoine A., Eisenhaure T.M., Jonsson A.H., Li S. The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 2019;20:902–914. doi: 10.1038/s41590-019-0398-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson V., Gayoso A., Yosef N., Pachter L. Interpretable factor models of single-cell RNA-seq via variational autoencoders. Bioinformatics. 2020;36:3418–3421. doi: 10.1093/bioinformatics/btaa169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.-E., Stephenson E., Polański K., Goncalves A. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg C.E.J., Beermann S., Broeren M.G.A., Bennink M.B., Koenders M.I., van Lent P.L.E.M., van den Berg W.B., van der Kraan P.M., van de Loo F.A.J. Protective role of the MER tyrosine kinase via efferocytosis in rheumatoid arthritis models. Front. Immunol. 2018;9:742. doi: 10.3389/fimmu.2018.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way G.P., Zietz M., Rubinetti V., Himmelstein D.S., Greene C.S. Compressing gene expression data using multiple latent space dimensionalities learns complementary biological representations. Genome Biol. 2020;21:109. doi: 10.1186/s13059-020-02021-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Joseph A., Hammonds A.S., Celniker S.E., Yu B., Frise E. Stability-driven nonnegative matrix factorization to interpret spatial gene expression and build local gene networks. Proc. Natl. Acad. Sci. U S A. 2016;113:4290–4295. doi: 10.1073/pnas.1521171113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Ding J., Porter C.B.M., Wallrapp A., Tabaka M., Ma S., Fu S., Guo X., Riesenfeld S.J., Su C. Transcriptional Atlas of intestinal immune cells reveals that neuropeptide α-CGRP modulates group 2 innate lymphoid cell responses. Immunity. 2019;51:696–708.e9. doi: 10.1016/j.immuni.2019.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D., Tao Y., Chen G., Shi T. Systematic expression analysis of ligand-receptor pairs reveals important cell-to-cell interactions inside glioma. Cell Commun. Signal. 2019;17:48. doi: 10.1186/s12964-019-0363-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Wei K., Slowikowski K., Fonseka C.Y., Rao D.A., Kelly S., Goodman S.M., Tabechian D., Hughes L.B., Salomon-Escoto K. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019;20:928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Dovas A., Spinazzi E.F., Levitin H.M., Upadhyayula P., Sudhakar T., Marie T., Otten M.L., Sisti M., Bruce J.N. Deconvolution of cell type-specific drug responses in human tumor tissue with single-cell RNA-seq. bioRxiv. 2020 doi: 10.1186/s13073-021-00894-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.X., Taramelli R., Pedrini E., Knijnenburg T., Huang S. Extracting intercellular signaling network of cancer tissues using ligand-receptor expression patterns from whole-tumor and single-cell transcriptomes. Sci. Rep. 2017;7:8815. doi: 10.1038/s41598-017-09307-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data was generated as part of this study. All code of the analysis is available at https://github.com/giovp/latent_factors_autoimmune.