Highlights
-
•
Autosis is an autophagy-dependent, nonapoptotic, and non-necrotic form of cell death that is characterized by unique morphological and biochemical features, including the presence of ballooning of perinuclear space (PNS) and sensitivity to cardiac glycosides, respectively.
-
•
Autotic cell death may be initiated by excessive accumulation of autophagosomes rather than lysosomal degradation.
-
•
Autosis is stimulated during the late phase of reperfusion after a period of ischemia in the heart when up-regulation of rubicon in the presence of continuous autophagosome production induces massive accumulation of autophagosomes.
-
•
Suppression of autosis, which may reduce death of cardiomyocytes during the late phase of reperfusion, in combination with inhibition of apoptosis and necrosis targeting the early phase of injury, may enhance the effectiveness of treatment for I/R injury in the heart.
Key Words: autophagic cell death; autophagic flux; autosis; beclin 1; Na+,K+–adenosine triphosphatase; rubicon
Abbreviations and Acronyms: ATG, autophagy-related; ATPase, adenosine triphosphatase; ER, endoplasmic reticulum; HIV, human immunodeficiency virus; I/R, ischemia-reperfusion; LBR, lamin B receptor; PI3K, phosphatidylinositol 3 kinase; PNS, perinuclear space; Tat, transactivation of transcription
Summary
Excessive autophagy induces a defined form of cell death called autosis, which is characterized by unique morphological features, including ballooning of perinuclear space and biochemical features, including sensitivity to cardiac glycosides. Autosis is observed during the late phase of reperfusion after a period of ischemia and contributes to myocardial injury. This review discusses unique features of autosis, the involvement of autosis in myocardial injury, and the molecular mechanism of autosis. Because autosis promotes myocardial injury under some conditions, a better understanding of autosis may lead to development of novel interventions to protect the heart against myocardial stress.
Central Illustration
Macroautophagy (hereafter autophagy) is an essential process by which cells degrade intracellular organelles and cytosolic materials, characterized by the presence of double-membrane structures called autophagosomes that fuse with lysosomes. Traditionally, autophagy is believed to be a protective mechanism because it eliminates misfolded or dysfunctional proteins or organelles and supplies energy by recycling the amino acids and fatty acids obtained through degradation (1). However, emerging evidence suggests that the genetic machinery of autophagy is also essential for cell death under some conditions (2). Cell death mediated by autophagy, or autophagic cell death, was originally classified as a type II programmed cell death, defined by massive cytosolic vacuolization in dying cells. Autophagic cell death has been reported in cardiomyocytes during ischemia-reperfusion (I/R), pressure overload, and doxorubicin-induced cardiomyopathy (3, 4, 5). However, because of technical limitations that make it difficult to distinguish autophagic cell death from other forms of death accompanied by autophagy, the causative role of autophagy in cardiomyocyte cell death is largely unknown. Recently, a unique form of cell death induced by activation of autophagy was identified and termed autosis (6). Autosis is distinct from other forms of cell death, including necrosis and apoptosis, and is characterized by the presence of unique morphological and biochemical features. Increasing lines of evidence suggest that cell death through autosis occurs in various cell types and organs in response to some types of stress. We have also shown recently that cardiomyocytes die by autosis during reperfusion after a short period of ischemia in the heart (7,23). Here, we review the current understanding of autophagic cell death, with a focus on autosis, and we discuss the involvement of cardiomyocyte autosis in myocardial injury and the underlying molecular mechanisms.
Autophagy and Autophagic Cell Death in the Heart
To explain how autophagy can kill cells under some conditions, we will first describe the physiological function of autophagy and its molecular machinery. Autophagy is an essential catabolic process that is highly conserved in eukaryotes. The autophagy pathway primarily consists of evolutionarily conserved autophagy-related (ATG) proteins. These ATG proteins generate double-membrane vesicles, termed autophagosomes, which engulf cellular materials, generally in a nonselective manner but also in a selective manner under some conditions. The autophagy process is initiated by activation of the Unc-51-like kinase 1/Atg1 complex, which is regulated by mammalian target of rapamycin complex 1 and adenosine monophosphate–activated protein kinase (8). The Unc-51-like kinase 1/Atg1 complex activates the class III phosphatidylinositol 3 kinase (PI3K) complex, consisting of PI3K, beclin 1, vacuolar protein sorting 15 (VPS15), and/or additional partners, to initiate autophagosome nucleation. As a scaffold protein, beclin 1 recruits several proteins, such as Atg14l, autophagy and beclin regulator 1 (AMBRA1), ultraviolet resistance–associated gene (UVRAG), and rubicon, to the nucleation site to regulate the lipid kinase activity of the PI3K complex (9). Although most beclin 1 interactors enhance beclin 1 activity, rubicon negatively regulates both the autophagic and endocytic pathways by interacting with beclin 1 or Rab7, a member of the RAS oncogene family (10). Unlike in yeast, there is no pre-autophagosomal structure in mammalian cells. Instead, the endoplasmic reticulum (ER) exit sites, mitochondria, ER-mitochondria contact sites, the ER-Golgi intermediate compartment, the Golgi apparatus, and the plasma membrane have all been reported as sources of isolation membranes in mammalian cells (11). Whether dysregulated autophagosome formation leads to depletion of the aforementioned structures and contributes to malfunction of intracellular organelles is an open question. In subsequent steps, elongation and completion of autophagosomes are regulated by the ubiquitin-like conjugation system. First, the E1-like enzyme Atg7 binds to the ubiquitin-like protein Atg12, which promotes conjugation of Atg12 to Atg5 in the presence of an E2-like enzyme, Atg10, finally forming the Atg5-Atg12/Atg16 multimeric complex. The Atg5-Atg12/Atg16 complex localizes at expanding phagophores and acts as an E3-like ligase to conjugate another ubiquitin-like molecule, Atg8 (light chain 3-I), to phosphatidylethanolamine to form LC3 (MAP1LC3; a light chain of the microtubule-associated protein 1, ortholog of yeast Atg8), which is essential for targeting autophagosomes to lysosomes (12). Finally, autophagosomes fuse with lysosomes to degrade the sequestered materials. On completion of autophagosome formation, syntaxin 17, localized on the outer autophagosomal membrane, interacts with synaptosomal-associated protein 29 and lysosomal synaptosomal-associated protein receptor vesicle-associated membrane protein 8 to enable fusion with the lysosomes (13) (Figure 1). Within the lysosomal compartment, membrane components and proteins are degraded by phospholipases and proteolytic enzymes, respectively, and degraded phospholipids and amino acids are recycled (14). Because autophagy is executed through the coordinated actions of: 1) autophagosome formation; 2) fusion between autophagosomes and lysosomes; and 3) degradation of autophagosomes and its cargo in lysosomes, dysregulation of any step could potentially induce malfunction of autophagic degradation and disturb cellular homeostasis, thereby leading to cell death. For example, blocking autophagy at the level of lysosomal degradation could lead to accumulation of undigested autophagosomes and depletion of membrane sources that would otherwise be recycled to form new autophagosomes.
Figure 1.
Molecular Mechanism of Autophagy
The process of autophagy comprises multiple steps, including initiation, nucleation, elongation and completion, and fusion and degradation. In the initiation step, the Unc-51-like kinase 1 (ULK1)/autophagy-related protein 1 (Atg1) complex is regulated by mammalian target of rapamycin (mTOR) inhibition in response to autophagy-inducing conditions, such as starvation. Thereafter, the ULK1/Atg1 complex activates the phosphatidylinositol 3 kinase complex to nucleate autophagosomal membranes. In the elongation step, 2 ubiquitin-like conjugation systems, the Atg12 and light chain 3 (LC3) conjugation systems, expand autophagosomes. After completion of elongation, autophagosomes fuse with lysosomes to degrade cargo materials. Rubicon associated with the phosphatidylinositol 3 kinase complex negatively regulates the fusion process. Atg13 = autophagy-related protein 13; Atg16L = autophagy-related protein 16 like; Bcl2 = B-cell lymphoma 2; FIP200 = FAK family kinase-interacting protein of 200 kD; mLST8 = mammalian lethal with SEC13 protein 8; p = phosphorylation; PE = phosphatidylethanolamine; UVRAG = ultraviolet radiation resistance–associated gene; Vps15 = vacuolar protein sorting 15; Vps34 = vacuolar protein sorting 34.
Autophagic cell death was originally identified by a massive cytoplasmic vacuolization with autophagy activation (2). It is important to note that the presence of autophagic vacuoles in dying cells does not necessarily mean that autophagy promotes cell death. Because autophagy is often activated in response to stress as an adaptive mechanism, the presence of autophagy in dying cells could indicate that the last resort to prevent cell death may have failed. Unfortunately, because most of the currently available inhibitors are not 100% specific for autophagy, chemical inactivation of autophagy may not provide definitive evidence of autophagic cell death. Similarly, many molecules involved in autophagy, such as Atg, have autophagy-independent functions (15), which makes the use of genetically altered animal models for loss-of-function studies of autophagy challenging as well. For these reasons, autophagic cell death has been defined by the Nomenclature Committee on Cell Death as a cell death that is suppressed by at least 2-independent interventions to inhibit the autophagy pathway (16).
With these technical challenges in mind, under what conditions is autophagy involved in cell death? In many examples, autophagy is activated strongly, and molecules involved in autophagosome formation, including beclin 1, are up-regulated when autophagy-dependent cell death takes place. For example, these conditions are observed in the heart in response to some pathologically relevant stresses, including I/R, acute pressure overload, and doxorubicin-induced cardiotoxicity. It is noteworthy that several interventions to suppress the autophagic machinery, including down-regulation of beclin 1 and chemical inhibitors of autophagy such as 3-methyladenine, an inhibitor of class III PI3K activity and autophagosome formation, significantly suppress death of cardiomyocytes and myocardial injury. If we include other types of cell death that are suppressed when autophagy is inhibited, autophagy-dependent cell death may also include the following forms of cell death. First, some forms of autophagy selectively degrade proteins essential for cell survival. For example, in flies, autophagy selectively degrades Drosophila BIR repeat containing ubiquitin-conjugating enzyme (dBruce) dBruce, a caspase inhibitor, thereby activating caspase-dependent cell death, namely apoptosis (17). Autophagy may also degrade catalase, an antioxidant, which in turn disrupts the intracellular redox balance and induces cell death in mouse fibroblast cell lines (18). Second, activation of nonselective autophagy in a mouse model of diabetes reciprocally suppresses mitophagy, thereby inducing mitochondrial dysfunction and death in cardiomyocytes. Inhibition of autophagy by down-regulation of Becn1 or Atg16 improves diabetic cardiomyopathy by activating mitophagy through an unknown mechanism (19). Although autophagy is involved in these 2 types of cell death, the cell death itself is mediated through other forms of cell death, including apoptosis and necrosis. It has thus been recommended to use the terms “autophagy-associated cell death” or “autophagy-mediated cell death” unless the death is prevented only by multiple autophagy-inhibiting interventions but not by inhibition of other forms of cell death, such as apoptosis or necrosis. Along these lines, it is preferable to use the term “autophagy-dependent cell death” when the aforementioned criteria have been clearly proven (20).
New Form of Autophagic Cell Death, Autosis
Although autophagy is associated with multiple forms of cell death as discussed, an important question remains as to whether autophagy-dependent cell death has any morphological or biochemical features that distinguish it from other forms of cell death. Recently, Liu et al. (6) characterized a new form of autophagy-dependent cell death, which is triggered by high doses of autophagy-inducing peptides, starvation, and permanent brain ischemia, conditions in which autophagy is strongly activated. This form of cell death, termed autosis, is characterized by the presence of unique morphological and biochemical features and can be rescued by inhibitors of autophagy, but not by inhibitors of apoptosis or necrosis. Thus, autosis clearly fulfills the criteria of autophagy-dependent cell death. Although autosis may not be the sole form of autophagy-dependent cell death, it is certainly the most well-defined form of cell death in this category.
The unique morphological features of autotic cell death are induced in a time-dependent manner (Figure 2). In phase 1a, dilated and fragmented ER and an increased number of autophagosomes, autolysosomes, and empty vacuoles are observed. In phase 1b, a swollen perinuclear space (PNS) containing cytoplasmic materials and electron-dense mitochondria can be observed by transmission electron microscopy. In phase 2, the last step of autosis, cytoplasmic organelles are drastically decreased and focal nuclear concavity and focal ballooning of the PNS are observed (21). Cells that have died by autosis are generally more firmly attached to the culture dishes than are those that do not undergo autosis, and the increased adherence appears to be another morphological feature of autosis in vitro. To date, the best method for defining autosis is electron microscopy analysis. Alternatively, immunofluorescence assays can also help distinguish autotic cells by detecting fragmented ER or mitochondria with nuclear concavity.
Figure 2.
Morphological Features of Autosis
During the early phase of autosis, the numbers of autophagosomes (APs), autolysosomes (ALs), and empty vacuoles (EVs) are drastically increased and separation of the inner and outer nuclear membranes is observed. The later phase is characterized by focal nuclear concavity, focal ballooning of the perinuclear space (PNS), and disappearance of subcellular organelles. ER = endoplasmic reticulum; Nu = nucleus.
Although autosis can be inhibited by chemical inhibitors of autophagy, including 3-methyladenine, and down-regulation of the autophagic machinery, such as beclin 1 and Atg7, it cannot be inhibited by inhibitors of apoptosis, necrosis, or any other form of programmed cell death. It should be mentioned that suppression of lysosomes fails to inhibit autosis in either HeLa cells or cardiomyocytes. This suggests that autosis may be induced by excessive activation of autophagosome formation, but not by excessive degradation. We will discuss this issue in the section on the underlying mechanism of autosis.
Another important feature of autosis is its sensitivity to cardiac glycosides. A compound library screen revealed that inhibitors of Na+,K+–adenosine triphosphatase (ATPase) effectively suppress autosis (6,22). Consistent with this finding, cardiac glycoside, a chemical inhibitor of Na+,K+-ATPase, dramatically reduces Transactivator of transcription (Tat)–beclin 1–induced autosis in vitro and the autosis observed in neonatal rats subjected to brain ischemia in vivo (6). Moreover, injection of ouabain into cardiac glycoside–sensitive mice significantly reduces myocardial I/R injury in adult mice (23). We also noted that protein expression of Na+,K+-ATPase was up-regulated in the heart in response to I/R, the time course of which coincides with that of autosis. One caveat here is that Na+,K+-ATPase in rodent hearts is less sensitive to cardiac glycosides than that in human hearts is. Thus, it is difficult to test whether cardiomyocyte death can be rescued by cardiac glycosides in the murine heart. Humanized Na+,K+-ATPase α1 subunit knock-in mice can be used to overcome this issue (23). Alternatively, the α1 subunit of Na+,K+-ATPase can be down-regulated with short hairpin ribonucleic acid without obvious effects on contractility.
The features of autosis compared with other forms of programmed cell death are summarized in Table 1. Currently, no other criteria are available to distinguish autotic cell death from other forms of cell death. We expect, however, that progress in the field may provide additional criteria to help identify autosis more easily and precisely.
Table 1.
Comparison of Biochemical and Morphological Features Among Autosis, Apoptosis, and Necrosis
| Autosis | Apoptosis | Necrosis | |
|---|---|---|---|
| Biochemical features | |||
| Features | Inhibited by inhibitors of autophagy but not by inhibitors of apoptosis or necrosis | Caspase activation and internucleosomal DNA fragmentation | Rapid, extensive thiol oxidation and high level of intracellular Ca2+ |
| Regulators | ATG genes, rubicon, Na+,K+-ATPase | Caspases, PARP | RIP1, RIP3 |
| Morphological features | |||
| Nucleus | Focal ballooning of PNS, focal concavity of the nuclear surface, mild chromatin condensation | Nuclear chromatin condensation, fragmentation of DNA, irregularity of nucleus | Karyolysis and caspase-independent DNA fragmentation, lysis of nucleolus, dilation of nuclear membrane |
| Plasma membrane | Focal plasma membrane rupture | Intact, altered orientation of lipids | Disrupted |
| Cell size | Minor changes | Reduced (shrinkage) | Increased (swelling) |
| Mitochondria | Electron-dense mitochondria, abnormal internal structure, and swollen mitochondria | Release of cytochrome c, mitochondrial dysfunction | Failure of ATP production, collapse of mitochondrial membrane potential, and mPTP opening |
| Specific features | Ballooning of PNS under EM, increased adherence of cultured cells to culture dishes | Apoptotic bodies, pseudopod retraction | Swelling of ER and mitochondria, lysosome, and plasma membrane rupture |
ATG = autophagy-related; ATPase = adenosine triphosphatase; DNA = deoxyribonucleic acid; EM = electron microscopy; mPTP = mitochondrial permeability transition pore; PARP = poly adenosine diphosphate ribose polymerase; PNS = perinuclear space; RIP1 = receptor interacting serine/threonine kinase 1; RIP3 = receptor interacting serine/threonine kinase 3.
Autosis-Inducing Conditions
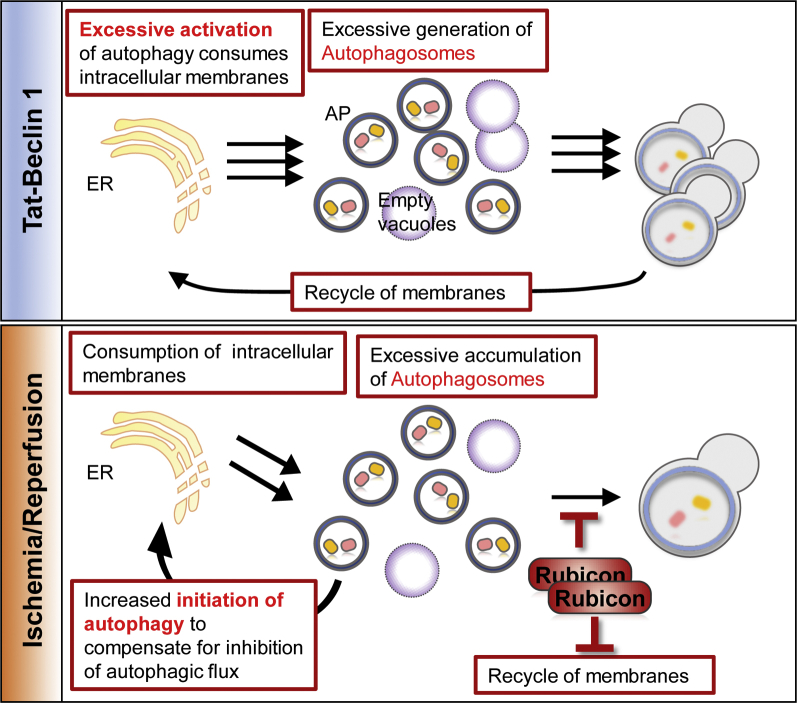
Although autosis was initially identified in HeLa cells when very high levels of autophagy were induced by Tat–beclin 1 in vitro (6), autosis has also been observed in vivo under various conditions (Central Illustration).
Central Illustration.
Biochemical Features of Autosis
Autosis is activated by the autophagy-inducing peptide, tyrosine aminotransferase (Tat)–beclin 1, hypoxia-ischemia in neonatal cerebral brain and ischemia-reperfusion in the heart. Autosis is regulated by the physical interaction between beclin 1 and Na+,K+–adenosine triphosphatase (ATPase), which is inhibited by cardiac glycosides, by up-regulation of rubicon, which attenuates fusion of autophagosome and lysosome, and by the core autophagy machinery. Atg5 = autophagy-related protein 5.
Hypoxia-ischemia and reperfusion appear to be common conditions in which strong activation of autophagy and autosis are observed in multiple organs. Cerebral hypoxia-ischemia in the brain of neonatal rats promotes autosis, and cardiac glycosides, such as neriifolin, digoxin, and digitoxigenin, rescue autotic cell death in vivo (6). Autosis is also induced in mouse kidneys subjected to I/R (22). More recently, autosis was also observed in the heart and the cardiomyocytes therein during the late phase of I/R. The fact that inhibition of autosis with cardiac glycosides reduces the size of infarct in all of these models suggests that autosis is widely involved in tissue injury in response to I/R (23).
Although human immunodeficiency virus (HIV) ultimately kills infected cells, HIV-infected macrophages escape cell death (24). Interestingly, autophagy-inducing peptides, including Tat–beclin 1 and Tat-viral FLICE inhibitory protein-α2, preferentially kill chronically HIV-infected human macrophages, whereas the cell death is inhibited by knockdown of ATG genes and Na+,K+-ATPase α1 (25). A major reservoir of HIV latent infection is resting central memory CD4+ T cells. Under antiretroviral treatment, HIV escapes clearance by staying inside CD4+ T cells (26). However, HIV-infected CD4+ T cells are also selectively killed by autophagy-inducing peptides through autosis (27). Thus, autosis selectively kills HIV-infected macrophages and CD4+ T cells. This is intriguing in that HIV utilizes autophagy proteins for replication (28) but down-regulates autophagy to avoid proteolytic degradation in activated CD4+ T cells (29). The molecular mechanisms through which HIV-infected macrophages and CD4+ T cells selectively undergo autosis remain to be elucidated.
Activation of autophagy in response to hypoxia has dichotomous functions in cancer cells (30,31). Autophagy inhibits cancer cell death 4 h after hypoxia but promotes cell death 72 h after hypoxia (21). Autophagic cell death at late time points of hypoxia showed morphological features of autosis and was inhibited by cardiac glycosides in A549 cell lines. During the initial stage of hypoxia, epidermal growth factor receptor (EGFR) is activated and interacts with beclin 1, which activates autophagy at physiological levels and contributes to survival of cancer cells. When hypoxia persists, however, EGFR is deactivated by autophagic degradation of caveolin 1 and dissociates from the beclin 1 complex, which in turn stimulates autophagic flux above physiological levels, thereby triggering autosis. The detailed mechanism through which protein-protein interaction between beclin 1 and EGFR regulates the level of autophagic flux and autosis is not well understood. Whether cancer cells become susceptible to autosis in response to prolonged hypoxia due to higher autophagic activity alone or whether additional modulatory mechanisms are needed to promote cell death remains to be elucidated. The fact that HIV-infected macrophages and/or T cells and cancer cells are more sensitive to autosis under some conditions is intriguing and suggests that induction of autosis may be utilized for selective elimination of certain cell populations.
Autosis has been reported in humans as well. Death of hepatocytes in patients with severe anorexia is accompanied by the presence of convoluted nuclei, electron-dense mitochondria, dilated ER, and numerous autophagic vacuoles and empty vacuoles, suggesting that these cells die by autosis (32). It would be interesting to test whether hepatic injury and decreased function in anorexia patients can be alleviated by cardiac glycosides. We recently showed that Tat–beclin 1 treatment induces autosis in human induced pluripotent stem cell–derived cardiomyocytes, suggesting that autosis can take place in human cells (23).
We expect that progress in the field will identify more examples of autosis in many other cell types under diverse conditions. It will be important to learn from the available examples when and how autosis is induced.
Induction of Autosis in the Heart in Response to I/R
Because I/R strongly induces autophagy in the heart, it is natural to speculate that autosis is induced, as in the case of I/R injury, in the brain and kidney. In fact, we have shown recently that autosis of cardiomyocytes is induced in the heart in vivo in response to I/R (23). Here, we describe the key findings of this study and discuss the underlying mechanism of autosis induced by I/R. Autophagy is up-regulated by a short period of ischemia and further stimulated during reperfusion in the heart, whereas myocardial injury is attenuated when autophagy is suppressed by down-regulation of beclin 1 or other interventions (3). Cardiomyocytes located in the ischemic area exhibit accumulation of autophagosomes and autolysosomes, fragmented ER, electron-dense mitochondria, and mild chromatin condensation in response to reperfusion in a time-dependent manner. Cardiomyocytes start to show a characteristic PNS during the late phase of reperfusion, namely 6 h after reperfusion and thereafter, which is consistent with the induction of autosis. These morphological findings, which are consistent with autosis, and myocardial injury, as evaluated with 2,3,5-triphenyl tetrazolium chloride staining, in response to I/R, were significantly suppressed in the presence of ouabain treatment in humanized Na+,K+-ATPase knock-in mice, suggesting that I/R induces autosis, which contributes significantly to the overall I/R injury in the heart.
Interestingly, autosis is increased in the late phase of reperfusion. Although the consensus has been that the process of cell death in the ischemic area should be completed within 1 to 2 h of reperfusion, our results suggest that cardiomyocytes die continuously throughout the late phase of reperfusion via mechanisms distinct from apoptosis or necrosis. This provides a strong rationale for extending interventions to prevent reperfusion injury through the late phase of reperfusion.
Molecular Mechanisms of Autosis
Here, we discuss why strong activation of autophagy leads to autotic cell death (Figure 3). An important consideration is the fact that induction of autosis cannot be alleviated by lysosome inhibitors. Thus, autosis is not mediated through lysosomal degradation. Increasing lines of evidence suggest that excessive accumulation of autophagosomes may mediate autotic cell death. Excessive accumulation of autophagosomes can take place when the balance between synthesis and degradation of autophagosomes is disrupted. We will discuss this issue, using the induction of autosis in response to myocardial I/R as an example.
Figure 3.
Cause of Autotic Cell Death
(Top) Excessive generation of autophagic vacuoles in response to tyrosine aminotransferase (Tat)–beclin 1 causes excessive consumption of intracellular membrane in cardiomyocytes in vitro. (Bottom) Accumulation of autophagic vacuoles induced by up-regulation of rubicon causes excessive consumption of intracellular membrane in cardiomyocytes in vivo. Abbreviations as in Figure 2.
Although autophagic flux is up-regulated during the initial hours of myocardial reperfusion, it gradually decreases thereafter (23). We found that autosis is observed 6 h after reperfusion, when autophagic flux is attenuated. This appears counterintuitive because, by definition, autosis should be stimulated by autophagy. Importantly, however, marked accumulation of autophagosomes is observed during the late phase of reperfusion due to a high level of autophagosome formation despite decreased lysosomal degradation. Interestingly, rubicon, a molecule known to inhibit fusion between autophagosomes and lysosomes, is up-regulated during the late phase of reperfusion, thereby inhibiting autophagic flux. When autophagic flux is slowed down by rubicon, cardiomyocytes appear to try compensating for the reduced level of autophagic flux by making more autophagosomes. Thus, we speculate that up-regulation of rubicon dramatically stimulates accumulation of autophagosomes during the late phase of reperfusion. Cardiac-specific heterozygous down-regulation of rubicon restores autophagic flux and inhibits accumulation of autophagosomes, suggesting that rubicon plays an important role in inhibiting autophagic flux and, in turn, inducing accumulation of autophagosomes. Loss of rubicon function attenuates autosis and reduces myocardial infarct size, similar to the results observed in ouabain-treated humanized Na+,K+-ATPase knock-in mice. These results suggest that endogenous rubicon plays an essential role in mediating autosis during myocardial reperfusion. Although down-regulation of rubicon may reduce myocardial injury solely by improving autophagic flux, we believe that the consequent prevention of excessive autophagosome accumulation is also important because treatment with Tat–beclin 1, which stimulates autophagy, exacerbates autosis if it is given during the late phase of reperfusion (23).
Why is excessive accumulation of autophagosomes detrimental for cells? Because endomembranes are used as a membrane source for autophagosomes (33), excessive formation of autophagosomes may occur at the expense of intracellular organelle membranes, including ER, mitochondria, and even plasma membrane. Consistent with this hypothesis, activation of autosis in the heart is accompanied by decreases in intracellular membranes and a consequent reduction in the function of intracellular organelles, including depolarization of mitochondrial membrane potentials. On the other hand, down-regulation of vesicle-associated membrane protein-associated proteins, which establish ER contact with autophagosome precursors, inhibits the consumption of ER membranes and even rescues Tat–beclin 1–induced autotic cell death (23). Importantly, whether decreases in intracellular membrane sources contribute to autotic cell death remains to be tested. To address this issue, it is necessary to investigate whether interventions to restore the endomembrane system level or intracellular organelles can rescue cells from autosis.
One of the most unique morphological features of autosis is the ballooning of the PNS. Previous reports showed a similar abnormal perinuclear morphology in human osteosarcoma U2OS cells, in the presence of overexpressed disease-associated mutant lamin B receptor (LBR) (34). LBR has sterol reductase activity, which is important for cholesterol biosynthesis (35). Expression of mutant LBRs and the related sterol reductases transmembrane 7 superfamily member 2 and 7-dehydrochlesterol reductase causes massive ER and PNS expansion, similar to the PNS in autosis. Because sterol content is not altered by overexpression of mutant LBRs, the sterol reductase activity may not be the direct trigger for the PNS-like structure. Alternatively, mutant LBRs or wild-type transmembrane 7 superfamily member 2 and 7-dehydrocholesterol reductase may change the membrane fluidity or permeability, thereby affecting osmolarity and signaling (34). Whether these mechanisms are involved in the pathogenesis of autosis remains to be elucidated.
Currently, the sensitivity to cardiac glycosides is among the most important clues to elucidating the molecular mechanism of autosis. Na+,K+-ATPase physically interacts with beclin 1 to stimulate autosis. Cardiac glycosides disrupt the interaction between Na+,K+-ATPase and beclin 1 by interacting with Na+,K+-ATPase (22). It is possible that this interaction alters ion pump activity or ion exchange–dependent effects of Na+,K+-ATPase. Na+,K+-ATPase modulates calcium signaling through interaction with its steroid agonist ouabain (36). However, treatment of cardiomyocytes with a calcium chelator, 1,2-Bis(2-aminophenoxy)ethane-N,N,N’, N’-tetraacetic acid tetrakis(acetoxymethyl ester), did not affect autosis (23), suggesting that calcium may not be involved in autosis. Interestingly, the interaction between Na+,K+-ATPase and beclin 1 takes place at many intracellular membranes, including ER, perinuclear membranes, mitochondria, and endosomes. Thus, the Na+,K+-ATPase/beclin 1 interaction may trigger autosis through its effect on membrane ion, osmolyte, and fluid homeostasis across intracellular membranes (37). Further investigation is required to elucidate the role of the Na+,K+-ATPase/beclin 1 interaction.
Another possibility is that Na+,K+-ATPase alters the function of beclin 1, thereby affecting autophagic activity or vesicle trafficking. Because rubicon also physically interacts with beclin 1 (38), modulation of beclin 1 may play a key role in mediating autosis. However, the interaction between Na+,K+-ATPase and beclin 1 alone may not affect the function of beclin 1 in such a way as to induce autophagy. Further investigation is required to clarify how the interaction between beclin 1 and either Na+,K+-ATPase or rubicon affects the function and modulates the activity of autosis.
Clinical Implications
Acute myocardial infarction is among the main causes of morbidity worldwide (39). Preventing death of cardiomyocytes is a major goal in the treatment of cardiac disease. Unfortunately, however, medical treatments targeting any single form of cell death in cardiomyocytes do not appear to be significantly effective for alleviating cardiac dysfunction in many clinical conditions (40). A possible reason for this could be that cardiomyocytes die by many mechanisms, with different time courses in response to different forms of stress. Thus, it is important to determine specifically how cardiomyocytes die in the presence of a given stress and clarify the time course of death. For example, the fact that autosis is observed during the late phase of I/R is unexpected in that it has generally been believed that death of cardiomyocytes takes place within the first few hours of reperfusion; thus, it has never been targeted. However, this might partially explain why a therapy focusing on either apoptosis or necrosis alone may not be able to reduce reperfusion injury. If autosis continuously kills cardiomyocytes during the subacute phase of I/R injury, it might prove useful to add a treatment targeting autosis to other forms of treatment targeting apoptosis and necrosis, which primarily take place during the early phase of I/R. Because autosis is induced by signaling mechanisms molecularly distinct from those of apoptosis and necrosis, treatment targeting autosis should show additive effects to those targeting other forms of cell death.
The human body produces endogenous cardiac glycoside–like substances. Inhibiting endogenous cardiac glycoside–like substances with DigiFab (BTG International Inc., West Conshohocken, Pennsylvania) exacerbates cardiac autosis during exercise (22), suggesting that endogenous cardiac glycoside–like substances have the ability to limit autosis during stress. Thus, if one could find an intrinsic mechanism to up-regulate endogenous cardiac glycosides, stimulation of this endogenous mechanism might prevent autosis and myocardial injury during I/R. Given that cardiac glycosides have the general side effect of inducing cardiac arrhythmia, however, it would be essential to develop a safer intervention to inhibit autosis.
Currently, other cardiac conditions besides I/R injury where autosis contributes to cardiomyocyte death and either myocardial injury or heart failure remain to be identified. For example, the level of autophagy is dramatically increased 3 to 4 days after permanent coronary ligation, during the acute phase of pressure overload or in response to doxorubicin-induced cardiomyopathy (4,41). In these conditions, suppression of autophagy effectively reduces myocardial cell death and improves either left ventricular function or the survival of the animals (4,42) (Table 2). Marked accumulation of autophagosomes in the heart is also observed in lysosomal storage disease (43). It would be important to investigate whether autosis is involved in any of these cardiac conditions and, if so, to test whether interventions to block autotic cell death can alleviate cardiac injury or cardiac dysfunction. It should be noted that the molecular mechanisms through which excessive autophagy induces cell death and, consequently, interventions to alleviate cell death induced by autophagy in these cardiac conditions may not be identical. For example, inhibition of rubicon may not be effective when a genetic defect responsible for a lysosomal disease causes an irreversible defect in lysosomal function.
Table 2.
Examples of Maladaptive Autophagy in the Heart
| Insult | Model | Autophagy Modulator | Autophagy Activity | Result | Ref. # |
|---|---|---|---|---|---|
| I/R | Adult mouse | Beclin 1+/− mouse | Autophagy is drastically activated during I/R in the heart | Reduced myocardial infarct size with reduced autophagy in beclin 1+/− mice | Matsui et al. (3) |
| Rubicon+/− mouse | Autophagic flux is increased during ischemic period but attenuated during reperfusion | Myocardial infarct size is reduced by restoration of autophagic flux in rubicon+/− mice during I/R | Nah et al. (23) | ||
| LncRNA CAIF | LncRNA CAIF directly binds to p53 and inhibits p53-mediated autophagy activation during I/R | Knockdown of myocardin inhibits autophagy and attenuates myocardial infarction during I/R | Liu et al. (44) | ||
| (ALDH2) | ALDH2 activates autophagy during ischemic period but inhibits autophagy during reperfusion | Overexpression of ALDH2 rescues myocardial injury by inhibiting autophagy during reperfusion | Ma et al. (45) | ||
| H/R | Neonatal rat CMs | Overexpression or knockdown of beclin 1 | Autophagosome clearance is impaired during I/R and H/R | Beclin 1 knockdown restores autophagosome processing and attenuates cell death by H/R | Ma et al. (46) |
| DOX-induced cardiomyopathy | Adult mouse | Beclin 1+/− mouse | DOX blocks autophagic flux in the heart | Beclin 1 haploinsufficiency protects against DOX cardiotoxicity | Li et al. (42) |
| Neonatal rat CMs | Transcription factor GATA4 | GATA4 inhibits DOX-induced autophagy | Overexpression of GATA4 reduces DOX-induced CM death by inhibiting autophagy | Kobayashi et al. (5) | |
| Cardiac hypertrophy | Adult mouse | HDAC inhibitors | HDAC inhibitors attenuate autophagy during TAC | HDAC inhibitors alleviate TAC-induced hypertrophy by inhibiting autophagy | Cao et al. (47) |
| miR-30 | miR-30 attenuates autophagy by targeting beclin 1 | Angiotensin II-induced cardiac hypertrophy is attenuated by miR-30 through down-regulation of beclin 1 | Pan et al. (48) | ||
| Diabetic cardiomyopathy | Adult mouse | Beclin 1+/− mouse | Autophagy activity is reduced in type 1 diabetic heart | Further reduction in autophagy by beclin 1+/− protects the heart against diabetic cardiomyopathy | Xu et al. (19) |
| Neonatal rat CMs | 3-MA, shAtg7, and shbeclin 1 | Hyperglycemia reduces autophagic flux | Additional suppression of autophagy by 3-MA or shAtg7 and shbeclin 1 attenuates high glucose-induced CM death | Kobayashi et al. (49) | |
| Arrhythmia | Rabbit | Lentivirus-mediated Atg7 knockdown or (CQ) | Autophagic flux is markedly activated in AF patient and rabbit model | Atg7 knockdown or CQ restored the shortened atrial effective refractory period and alleviated the AF vulnerability by inhibiting autophagy in rabbit | Yuan et al. (50) |
3-MA = 3-methyladenine; AF = atrial fibrillation; ALDH2 = aldehyde dehydrogenase 2; Atg7 = autophagy-related protein 7; CAIF = cardiac autophagy inhibiting factor; CM = cardiomyocyte; CQ = chloroquine; DOX = doxorubicin; GATA4 = GATA binding protein 4; HDAC = histone deacetylase; H/R = hypoxia-reoxygenation; I/R = ischemia-reperfusion; LncRNA = long noncoding ribonucleic acid; miR-30 = microribonucleic acid 30; sh = short hairpin; TAC = transverse aortic constriction.
If depletion of intracellular membranes is a key facilitator of autosis, molecular interventions to facilitate generation of the endomembrane system may be considered to prevent the loss of intracellular organelles during autosis. For example, testing whether facilitating the production of membrane phospholipid can restore cellular function and prevent cell death would be of great interest.
In summary, autosis is an attractive therapeutic target in cardiac disease because it is activated through unique signaling mechanisms with a distinct time course compared with other forms of cell death. Thus, one could expect additive effects when autosis inhibitors are applied in conjunction with existing modalities of treatment. Further investigation should be focused on identification of medical conditions in which autosis is activated and the elucidation of underlying signaling mechanisms.
Concluding Remarks
There is increasing evidence that cardiomyocytes can die through autosis under some conditions. This suggests that it may be possible to reduce the extent of cardiomyocyte death and myocardial injury in some conditions by targeting autosis. Even if autophagy is initially activated as an adaptive mechanism, it can become maladaptive in a time- and dose-dependent manner. Thus, if autosis is targeted for treatment of myocardial injury, it is important to correctly evaluate the extent of autophagy and identify the morphological and biochemical features of autosis to confirm that autophagy is no longer protective in a given condition. To date, several cardiac conditions have been shown to exhibit very strong autophagy and down-regulation of autophagy protects the heart in such conditions. It will be interesting to test whether autosis is observed in these conditions and, if so, whether cardiac glycosides can reduce cardiomyocyte death and improve cardiac function. If autotic cell death is identified in a given condition, it would be important to investigate how autophagy becomes dysregulated in that condition and how autosis kills cardiomyocytes. Apoptosis can be an orderly process and, thus, is physiological under some conditions. It remains unclear whether cardiomyocytes use autosis as a salutary mechanism. Because marked accumulation of autophagosomes is often accompanied by a mismatch between autophagosome formation and lysosomal degradation, how the mismatch takes place and amplifies the accumulation of autophagosomes should be clarified. Because beclin 1–interacting proteins, including Na+,K+-ATPase and cardiac glycosides, have been shown to be involved in autosis, it will be interesting to further investigate how beclin 1, rubicon, Na+,K+-ATPase, and possibly other interacting proteins induce cell death. By conducting a genome-wide small, interfering ribonucleic acid screen, Fernandez et al. (22) identified potential mediators of autosis besides Na+,K+-ATPase, including some involved in ion transport and cell-to-matrix adhesion. Further investigation to determine how those molecules participate in autosis would clarify the molecular mechanism of autosis. It would also be interesting to test whether chemical inhibitors against the molecules identified by the small, interfering ribonucleic acid screen can inhibit cardiac autosis in response to I/R. Because disappearance of intracellular organelles is an important feature of autosis and is likely to induce cellular malfunction, molecular mechanisms through which dysregulation of autophagosome formation leads to the disappearance of intracellular organelles should also be investigated. Elucidation of the underlying molecular mechanism of autosis should provide multiple options for identifying the occurrence more easily and conveniently and, eventually, should allow for the development of novel and selective interventions to treat cardiac disease. Because specific therapy for autosis has never been combined with therapies for other forms of cell death and because autosis occurs with a time course different from that of other forms of cell death, the development of a specific therapy for autosis could potentially represent a breakthrough in the treatment of myocardial I/R injury. Thus, further investigation regarding autosis in the heart is warranted.
Footnotes
This work was supported in part by the U.S. Public Health Service, the American Heart Association, and the Fondation Leducq. Dr. Nah has received support from the American Heart Association (postdoctoral fellowship 18POST34050036). Dr. Sadoshima has received research support from the U.S. Public Health Service (grant AG23039), American Heart Association (award 20 Merit35120374), and Fondation Leducq Transatlantic Network of Excellence (grant 15CBD04). Dr. Zablocki has reported that she has no relationships relevant to the contents of this paper to disclose.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the JACC: Basic to Translational Scienceauthor instructions page.
References
- 1.Pyo J.O., Nah J., Jung Y.K. Molecules and their functions in autophagy. Exp Mol Med. 2012;44:73–80. doi: 10.3858/emm.2012.44.2.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G., Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–1010. doi: 10.1038/nrm2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsui Y., Takagi H., Qu X. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and beclin 1 in mediating autophagy. Circ Res. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H., Tannous P., Johnstone J.L. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi S., Volden P., Timm D., Mao K., Xu X., Liang Q. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem. 2010;285:793–804. doi: 10.1074/jbc.M109.070037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y., Shoji-Kawata S., Sumpter R.M., Jr. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nah J., Fernandez A.F., Kitsis R.N., Levine B., Sadoshima J. Does autophagy mediate cardiac myocyte death during stress? Circ Res. 2016;119:893–895. doi: 10.1161/CIRCRESAHA.116.309765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zachari M., Ganley I.G. The mammalian ULK1 complex and autophagy initiation. Essays Biochem. 2017;61:585–596. doi: 10.1042/EBC20170021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang R., Zeh H.J., Lotze M.T., Tang D. The beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabata K., Matsunaga K., Sakane A., Sasaki T., Noda T., Yoshimori T. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol Biol Cell. 2010;21:4162–4172. doi: 10.1091/mbc.E10-06-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei Y., Liu M., Li X., Liu J., Li H. Origin of the autophagosome membrane in mammals. Biomed Res Int. 2018;2018:1012789. doi: 10.1155/2018/1012789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 13.Itakura E., Kishi-Itakura C., Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Schulze H., Kolter T., Sandhoff K. Principles of lysosomal membrane degradation: cellular topology and biochemistry of lysosomal lipid degradation. Biochim Biophys Acta. 2009;1793:674–683. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 15.Mishra P.K., Adameova A., Hill J.A. Guidelines for evaluating myocardial cell death. Am J Physiol Heart Circ Physiol. 2019;317:H891–H922. doi: 10.1152/ajpheart.00259.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galluzzi L., Vitale I., Abrams J.M. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nezis I.P., Shravage B.V., Sagona A.P. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu L., Wan F., Dutta S. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X., Kobayashi S., Chen K. Diminished autophagy limits cardiac injury in mouse models of type 1 diabetes. J Biol Chem. 2013;288:18077–18092. doi: 10.1074/jbc.M113.474650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky D.J., Abdelmohsen K., Abe A. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y., Henson E.S., Xiao W. Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy. 2016;12:1029–1046. doi: 10.1080/15548627.2016.1164357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez A.F., Liu Y., Ginet V. Interaction between the autophagy protein beclin 1 and Na+,K+-ATPase during starvation, exercise, and ischemia. JCI Insight. 2020;5:133282. doi: 10.1172/jci.insight.133282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nah J., Zhai P., Huan C.Y. Upregulation of rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest. 2020;130:2978–2991. doi: 10.1172/JCI132366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahabieh M.S., Battivelli E., Verdin E. Understanding HIV latency: the road to an HIV cure. Annu Rev Med. 2015;66:407–421. doi: 10.1146/annurev-med-092112-152941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang G., Luk B.T., Hamidy M., Zhang L., Spector S.A. Induction of a Na(+)/K(+)-ATPase-dependent form of autophagy triggers preferential cell death of human immunodeficiency virus type-1-infected macrophages. Autophagy. 2018;14:1359–1375. doi: 10.1080/15548627.2018.1476014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finzi D., Hermankova M., Pierson T. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 27.Zhang G., Luk B.T., Wei X. Selective cell death of latently HIV-infected CD4(+) T cells mediated by autosis inducing nanopeptides. Cell Death Dis. 2019;10:419. doi: 10.1038/s41419-019-1661-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyei G.B., Dinkins C., Davis A.S. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol. 2009;186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou D., Spector S.A. Human immunodeficiency virus type-1 infection inhibits autophagy. AIDS. 2008;22:695–699. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cui J., Hu Y.F., Feng X.M. EGFR inhibitors and autophagy in cancer treatment. Tumour Biol. 2014;35:11701–11709. doi: 10.1007/s13277-014-2660-z. [DOI] [PubMed] [Google Scholar]
- 31.Song J., Qu Z., Guo X. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy. 2009;5:1131–1144. doi: 10.4161/auto.5.8.9996. [DOI] [PubMed] [Google Scholar]
- 32.Kheloufi M., Boulanger C.M., Codogno P., Rautou P.E. Autosis occurs in the liver of patients with severe anorexia nervosa. Hepatology. 2015;62:657–658. doi: 10.1002/hep.27597. [DOI] [PubMed] [Google Scholar]
- 33.Gatica D., Chiong M., Lavandero S., Klionsky D.J. Molecular mechanisms of autophagy in the cardiovascular system. Circ Res. 2015;116:456–467. doi: 10.1161/CIRCRESAHA.114.303788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwerger M., Kolb T., Richter K., Karakesisoglou I., Herrmann H. Induction of a massive endoplasmic reticulum and perinuclear space expansion by expression of lamin B receptor mutants and the related sterol reductases TM7SF2 and DHCR7. Mol Biol Cell. 2010;21:354–368. doi: 10.1091/mbc.E09-08-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennati A.M., Schiavoni G., Franken S. Disruption of the gene encoding 3beta-hydroxysterol Delta-reductase (Tm7sf2) in mice does not impair cholesterol biosynthesis. FEBS J. 2008;275:5034–5047. doi: 10.1111/j.1742-4658.2008.06637.x. [DOI] [PubMed] [Google Scholar]
- 36.Tian J., Xie Z.J. The Na-K-ATPase and calcium-signaling microdomains. Physiology (Bethesda) 2008;23:205–211. doi: 10.1152/physiol.00008.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galva C., Artigas P., Gatto C. Nuclear Na+/K+-ATPase plays an active role in nucleoplasmic Ca2+ homeostasis. J Cell Sci. 2012;125:6137–6147. doi: 10.1242/jcs.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsunaga K., Saitoh T., Tabata K. Two beclin 1-binding proteins, Atg14L and rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 39.Roth G.A., Johnson C., Abajobir A. Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. J Am Coll Cardiol. 2017;70:1–25. doi: 10.1016/j.jacc.2017.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davidson S.M., Ferdinandy P., Andreadou I., for the CARDIOPROTECTION COST Action Investigators Multitarget strategies to reduce myocardial ischemia/reperfusion injury: JACC Review Topic of the Week. J Am Coll Cardiol. 2019;73:89–99. doi: 10.1016/j.jacc.2018.09.086. [DOI] [PubMed] [Google Scholar]
- 41.Wu X., He L., Chen F. Impaired autophagy contributes to adverse cardiac remodeling in acute myocardial infarction. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D.L., Wang Z.V., Ding G. Doxorubicin blocks cardiomyocyte autophagic flux by inhibiting lysosome acidification. Circulation. 2016;133:1668–1687. doi: 10.1161/CIRCULATIONAHA.115.017443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim Y.C., Park H.W., Sciarretta S. Rag GTPases are cardioprotective by regulating lysosomal function. Nat Commun. 2014;5:4241. doi: 10.1038/ncomms5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu C.Y., Zhang Y.H., Li R.B. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29. doi: 10.1038/s41467-017-02280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma H., Guo R., Yu L., Zhang Y., Ren J. Aldehyde dehydrogenase 2 (ALDH2) rescues myocardial ischaemia/reperfusion injury: role of autophagy paradox and toxic aldehyde. Eur Heart J. 2011;32:1025–1038. doi: 10.1093/eurheartj/ehq253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma X., Liu H., Foyil S.R. Impaired autophagosome clearance contributes to cardiomyocyte death in ischemia/reperfusion injury. Circulation. 2012;125:3170–3181. doi: 10.1161/CIRCULATIONAHA.111.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao D.J., Wang Z.V., Battiprolu P.K. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011;108:4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan W., Zhong Y., Cheng C. MiR-30-regulated autophagy mediates angiotensin II-induced myocardial hypertrophy. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi S., Xu X., Chen K., Liang Q. Suppression of autophagy is protective in high glucose-induced cardiomyocyte injury. Autophagy. 2012;8:577–592. doi: 10.4161/auto.18980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yuan Y., Zhao J., Gong Y. Autophagy exacerbates electrical remodeling in atrial fibrillation by ubiquitin-dependent degradation of L-type calcium channel. Cell Death Dis. 2018;9:873. doi: 10.1038/s41419-018-0860-y. [DOI] [PMC free article] [PubMed] [Google Scholar]