Abstract
Background & Aims
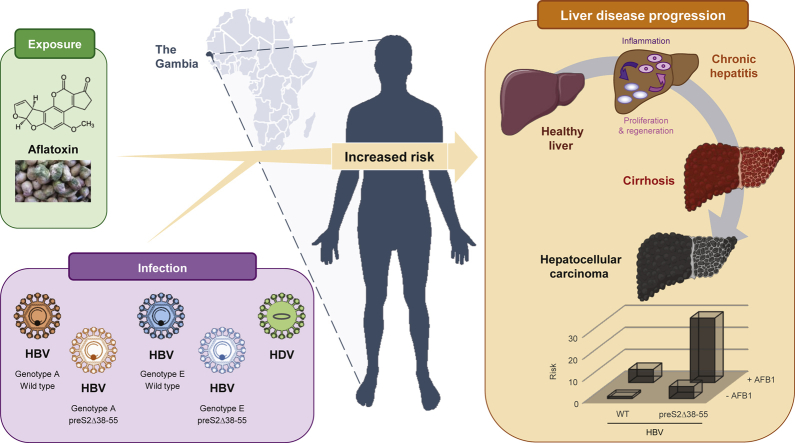
Although HBV is a major cause of death in Africa, its genetic variability has been poorly documented. This study aimed to address whether HBV genotype and surface gene variants are associated with HBV-related liver disease in The Gambia.
Methods
We conducted a case-control study nested in the Prevention of Liver Fibrosis and Cancer in Africa programme. Consecutive treatment-naive patients with chronic HBV infection and detectable viral load were recruited: 211 controls with no significant liver disease and 91 cases (56 cirrhosis and 35 HCC cases). HBV genotypes and surface gene variants were determined by Sanger sequencing or next-generation sequencing (NGS) in serum DNA. Aflatoxin B1 (AFB1)-specific codon 249 TP53 mutation was determined by NGS in circulating cell-free plasma DNA.
Results
In phylogenetic analysis, 85% of individuals carried HBV genotype E, 14% genotype A, and 1% A/E recombinant viruses. Surface gene variants were more frequently observed in cases (43% and 57% in cirrhosis and HCC cases, respectively) than controls (25%; p <0.001), with preS2 deletions between nucleotides 38–55 (preS2Δ38–55) being the main genetic variant detected. In multivariable analysis, HBeAg seropositivity, low HBsAg levels, and HDV seropositivity were significantly associated with cirrhosis and HCC, whilst older age, higher viral load, genotype A, preS2Δ38–55, and AFB1 exposure were only associated with HCC. There was a multiplicative joint effect of preS2Δ38–55 variants with HBeAg seropositivity (odds ratio [OR] 43.1 [10.4–177.7]), high viral load >2,000 IU/ml (OR 22.7 [8.0–64.9]), HBsAg levels <10,000 IU/ml (OR 19.0 [5.5–65.3]), and AFB1 exposure (OR 29.3 [3.7–230.4]) on HCC risk.
Conclusions
This study identified a hotspot for HBV preS2 deletions as a strong independent factor for HCC in The Gambia, with HBV genotypes and AFB1 exposure contributing to the high liver cancer risk.
Lay summary
Although HBV-related liver disease is highly prevalent in sub-Saharan Africa, the associated virological characteristics are poorly studied. Using clinical data from African patients chronically infected with HBV, an assessment of the virological variability (genotypes and mutations) and exposure to AFB1, a toxin often contaminating food, was carried out. Our results show that HBV genotypes, the presence of a highly prevalent mutant form of HBV, and AFB1 exposure contribute to the high liver cancer risk in this population.
Keywords: Aflatoxin B1, Africa, Carcinogenesis, Cirrhosis, Genotype, Hepatitis B virus, Hepatocellular carcinoma, PreS deletion
Abbreviations: AFB1, aflatoxin B1; AFP, alpha-fetoprotein; ER, endoplasmic reticulum; LSM, liver stiffness measurement; NBS1, Nijmegen breakage syndrome 1; NGS, next-generation sequencing; OR, odds ratio; PROLIFICA, Prevention of Liver Fibrosis and Cancer in Africa; ROC, receiver operating characteristic; SSA, sub-Saharan Africa; WT, wild type
Graphical abstract
Highlights
-
•
The rarer HBV genotype A (vs. E) is associated with a higher risk of HCC (odds ratio 5.2).
-
•
A genotype-independent deletion hotspot was found in the preS2 domain (preS2Δ38–55).
-
•
PreS2Δ38–55 was significantly associated with HCC and is an independent risk factor.
Introduction
Worldwide, 248 million individuals are positive for HBsAg, with the highest prevalence (>8%) found in sub-Saharan Africa (SSA).1 Chronic infection with HBV is a major cause of cirrhosis and accounts for more than a half of the global 770,000 HCC cases.2
The natural history of chronic HBV infection involves complex viral-host interactions, and the development of HBV-related HCC might be influenced by HBV genotypes and genetic variants that emerge during chronic infection. Ten genotypes (from A to J) exist based on a 7.5% nucleotide divergence that cluster geographically and can be distinguished by surface gene sequencing.3,4 For instance, genotype A is found in North America, Europe, South Africa, and India, whereas genotype E is predominant in West Africa. Links have been suggested between the genotypes, transmission routes, and impacts on clinical outcome.5 However, these associations have not been completely elucidated worldwide, particularly for the rarer HBV genotypes A and E circulating in SSA.
HBV genetic variants have been reported in the 3.2 kb HBV genome that encodes the viral polymerase, surface (preS/S), precore/core, and X proteins.6 This includes base substitutions, deletions, or insertions, which can emerge naturally during chronic infection caused by error-prone viral replication. They can persist as a dominant population during liver disease progression, modifying disease severity and cancer risk. For instance, preS deletions reported in longitudinal and cross-sectional studies conducted in Europe and Asia7 are generally associated with liver disease progression and could promote immune escape.8
HBV-associated HCC risk can also be modified by aflatoxin B1 (AFB1) exposure. This food contaminant is a class 1 human carcinogen found in SSA and China, and may multiplicatively amplify the effect of chronic HBV infection on HCC risk.9 The molecular hallmark of AFB1-induced liver carcinogenesis is the TP53 tumour suppressor gene codon 249 AGG>AGT mutation (p53R249S), with its presence in circulating cell-free (cf)DNA isolated from blood components considered as a surrogate biomarker of AFB1 exposure.10
In The Gambia, West Africa, previous studies found that the risk of HBV-related HCC is elevated in chronic HBV carriers with high viral load,11 positive HBeAg,11 coinfected with HCV12 or HDV,13 and previous exposure to AFB1.14 However, the impact of HBV genotypes and variants has not been elucidated yet. Therefore, to address this research gap, we conducted a case-control study nested in the Prevention of Liver Fibrosis and Cancer in Africa (PROLIFICA; www.prolifica.africa) programme15 in The Gambia.
Materials and methods
Study population
We investigated 302 treatment-naive HBsAg-positive patients with detectable viral load consecutively enrolled in the PROLIFICA programme from December 2012 to June 2015 (Fig. S1). They were identified as HBsAg positive through either community-based HBV screening15 or referral to the PROLIFICA liver clinic for a suspected chronic liver disease.16
All patients had a standardised comprehensive liver assessment, including physical examination, fasting liver stiffness measurement (LSM) using FibroScan® (Echosens, Paris, France), abdominal ultrasound with Doppler, liver function tests, and virological investigations (including HDV, HCV, and HIV serologies), as described previously.15 Cirrhosis was diagnosed using an LSM cut-off (9.5 kPa) previously validated in this population,17 or histologically or clinically in case of decompensated cirrhosis. HCC cases were either identified histopathologically or clinically if they fulfilled at least 2 of the following criteria: focal liver lesion ≥2 cm consistent with HCC by ultrasound; alpha-fetoprotein (AFP) levels ≥200 ng/ml; and cirrhosis.16
Controls were defined as HBsAg-positive patients without cirrhosis or HCC. The study was approved by The Gambia Government/MRC Joint Ethics Committee (SCC 1266) and conducted according to the guidelines of the Declaration of Helsinki. Ethical approval was granted by the institutional ethics committees, and all participants gave informed consent.
Viral DNA extraction, PCR, and sequencing
Viral DNA extracted from patient sera (200 μl) using the QIAamp® DNA Mini Kit (QIAGEN, Hilden, Germany) was used to PCR amplify the preS1/preS2/S gene before bidirectional Sanger sequencing (GenBank submission ID 2208379).18,19 First-round PCR was performed in 25 μl containing 5 μl HBV DNA and the F8-5′-ATTTGCATACTCTTTGGAAGGC-3′ and R5-5′-AAAGCCCAAAAGACCCACAAT-3′ primers. Then, 1 μl of this product was used in a nested PCR (primers F3-5ʹ-CGCCTCATTTTGTGGGTCAC-3ʹ and R8 5ʹ-CTTTGACAAACTTTCCAATCAAT-3ʹ) resulting in 1.4 kb (approximate) fragment. PCR products were checked on 1% agarose gels and purified using QIAquick® PCR Purification Kit (QIAGEN). The SeqScape™ Software version 3.0 (Thermo Fisher Scientific, Waltham, MA, USA) was used to join contigs with respect to the reference HBV genotype C AB014360. The Multiple Alignment using Fast Fourier Transform programme20 was used for alignments and CLC Sequence Viewer 8 (QIAGEN Bioinformatics) for mutation observation. Sequence logos were generated using an online application.21
Genotyping and phylogenetic tree
Genotypes and recombinants were determined using the jumping profile hidden Markov model.22 A/E recombinants were cut at the break point and analysed separately (Fig. S2). Three sequences were discarded from the data set (N-terminal of EG7151, EG7194, and EG7190) because of their very short length (<50 nt) leading to a set of 304 HBV S gene sequences from 302 individuals. Four S gene sequences of HBV genotype C (AB014360), B (AB010289), D (AB048701), and F (AB036905) have been added as external references.6 The sequences have been aligned using MACSE version 2.03 (default parameters),23 and the resulting alignment was trimmed using BMGE version 1.024 (option -t CODON). The phylogenetic tree was inferred with PhyML version 3.125 using the GTR+G4 evolutionary model proposed by ModelFinder,26 according to the Bayesian information criterion. The branch robustness was measured using the non-parametric bootstrap procedure implemented in PhyML (100 replicates of the original alignment). The inferred tree was rooted by HBV genotype F sequence and visualised using Interactive Tree Of Life.27
AFB1 assessment by isolation of circulating cfDNA and p53R249S fragment amplification
Circulating cfDNA was isolated from plasma (600 μl) of 117 patients, using the QIAamp® Circulating Nucleic Acid Kit (QIAGEN), quantified using the Quant-iT™ PicoGreen® dsDNA Assay (Molecular Probes/Invitrogen), and used to PCR amplify a TP53 fragment covering codon 249 (Supplementary Methods and materials).
Next-generation sequencing analysis by Ion Torrent technology
The NEBNext® Fast DNA Library Prep Set for Ion Torrent® kit (New England Biolabs) was used for TP53 mutation analysis (Supplementary Methods and materials). The same protocol was used to analyse the preS1/preS2/S amplicon. Purified barcoded libraries were sequenced on Ion Torrent Proton™ systems (Thermo Fisher Scientific). The accuracy and detection threshold and read error rate for the p53R249S mutation were determined using genomic DNA from p53R249S homozygous PLCPRF5 cells diluted into human wild-type (WT) DNA. For HBV sequences, the detection limits for variant sequences were estimated at 2% with a coverage of 10,000 reads.
Next-generation sequencing bioinformatics and statistical analyses
For the HBV genome, signal processing and base calling were performed with Torrent Suite™ Software version 5.0.2. Variant calling was obtained using Torrent Variant Caller 5.8 and applying somatic variant frequency, including down-sampling to coverage of 50,000 using the HBV EG0270 genome (genotype E) as reference. Next-generation sequencing (NGS) reads are available at the NCBI Sequence Read Archive (accession number PRJNA527947). HBV variants were filtered and highlighted using either IGV_2.3.55 or a Python local script.
For TP53, variant calling was performed using the Needlestack algorithm in extra-robust regression mode (Supplementary Methods and materials). A threshold of Phred scale q values QVAL >30 was used to call variants (QVAL = −10 log10 [q value]).
Statistical analysis
Data were analysed using Stata 13.0 (StataCorp, USA). Characteristics were compared between the controls and cases using chi-square test or Fisher's exact test for categorical variables, and Kruskal-Wallis test for continuous variables. Logistic regression was computed to identify factors associated with cirrhosis or HCC. All the variables significantly associated with liver disease in a crude analysis (p <0.05) were further included in a multivariable model to obtain adjusted odds ratio (OR). The interactions between preS2Δ38–55 and HBeAg seropositivity, HBV DNA, HBsAg levels, HBV genotype, or AFB1 exposure were examined using a likelihood ratio test by adding interaction terms in the logistic regression model.
The capability of the allelic fraction to correctly discriminate HBV-related HCC cases from people with chronic HBV infection without HCC was evaluated using the receiver operating characteristic (ROC) curve. Its optimal cut-off was selected to maximise the sum of sensitivity and specificity.
Results
Study population
In this study, 302 patients were enrolled: 211 controls and 91 cases (56 individuals with cirrhosis and 35 HCC cases). The demographic and clinical characteristics are presented in Table 1. The majority were men: 74% in controls, 84% in cirrhosis cases, and 71% in HCC cases. Median viral load (inter-quartile range [IQR]; log10 IU/ml) and HBeAg positivity were 2.7 (2.2–3.3) and 8.5% in the control group, 4.0 (2.3–5.9) and 31% in cirrhosis cases, and 5.2 (3.2–6.3) and 40% in HCC cases, respectively. Age, median LSM, median liver function test levels, and serum AFP levels were higher in cases than in the control group (p <0.001).
Table 1.
Characteristics of the HBsAg-positive controls, cirrhosis cases, and HCC cases (n = 302).
| Controls (n = 211) | Cirrhosis cases (n = 56) | HCC cases (n = 35) | p value | |
|---|---|---|---|---|
| Male | 156 (74) | 47 (84) | 25 (71) | 0.3 |
| Median age (years) | 33 (29–39) | 36 (27–42) | 40 (32–48) | 0.03 |
| Born in rural area | 146 (69) | 37 (66) | 23 (66) | 0.9 |
| Family history of liver cancer | 11 (5) | 1 (2) | 2 (6) | 0.5 |
| Ever drunk alcohol | 11 (5) | 6 (11) | 3 (9) | 0.3 |
| Obesity | 15 (7) | 2 (4) | 0 | 0.2 |
| Median LSM (kPa) | 5.3 (4.4–6.7) | 15.9 (10.3–27.7) | 75 (46.4–75) | <0.001 |
| Median ALT (IU/L) | 27 (21–33) | 39 (26–61) | 65 (47–107) | <0.001 |
| Median AST (IU/L) | 30 (26–38) | 52 (33–102) | 351 (195–429) | <0.001 |
| Median GGT (IU/L) | 27 (21–39) | 58 (36–100) | 363 (201–624) | <0.001 |
| Median ALP (IU/L) | 84 (69–106) | 129 (85–182) | 365 (261–452) | <0.001 |
| Median total bilirubin (mg/dl) | 0.6 (0.5–0.9) | 0.8 (0.6–2.1) | 2.0 (1.2–5.5) | <0.001 |
| Median albumin (g/ml) | 4.3 (4.0–4.5) | 4.0 (2.9–4.3) | 3.1 (2.8–3.3) | <0.001 |
| Median creatinine (mg/dl) | 0.89 (0.76–1.03) | 0.86 (0.71–0.98) | 0.71 (0.60–1.11) | 0.1 |
| Median platelets (109 cells/L) | 199 (156–251) | 118 (81–174) | 208 (143–329) | <0.001 |
| Median AFP (ng/ml) | 4.2 (2.5–6.5) | 13.4 (7.8–58.0) | 1,559.0 (167.9–6,172.0) | <0.001 |
| HBeAg seropositivity | 18 (8.5) | 17 (30.9) | 14 (40.0) | <0.001 |
| Median HBV DNA (log10 IU/ml) | 2.7 (2.2–3.3) | 4.0 (2.3–5.9) | 5.2 (3.2–6.3) | <0.001 |
| Median HBsAg (log10 IU/ml) | 4.1 (3.8–4.4) | 3.7 (3.3–3.9) | 3.6 (3.2–4.0) | <0.001 |
| Genotype | <0.001 | |||
| E | 188 (89) | 48 (86) | 20 (57) | |
| A | 23 (11) | 7 (12) | 12 (34) | |
| A and E | 0 | 1 (2) | 3 (9) | |
| HBV mutations | <0.001 | |||
| Wild type | 159 (75) | 32 (57) | 15 (43) | |
| Other mutation | 11 (5) | 2 (4) | 1 (3) | |
| PreS2Δ38–55 | 41 (20) | 22 (39) | 19 (54) | |
| Positive anti-HCV | 0 | 2 (4) | 3 (9) | 0.001 |
| Positive anti-HDV | 1 (1) | 4 (7) | 5 (17) | <0.001 |
| Positive anti-HIV= | 4 (2) | 2 (4) | 0 | 0.5 |
| AFB1 exposure, n (%)a | ||||
| No | 60 (81) | 22 (73) | 6 (46) | 0.03 |
| Yes | 14 (19) | 8 (27) | 7 (54) | |
| Median allelic fraction in those positive for AFB1 (IQR) | 0.009 (0.003–0.224) | 0.039 (0.002–0.342) | 0.332 (0.064–0.419) | <0.001 |
Data are presented as n (%) for categorical variables and median (IQR) for continuous variables. Obesity was defined as a body mass index ≥30 kg/m2. Characteristics were compared between the controls and cases using chi-squared test or Fisher’s exact test for categorical variables, and Kruskal-Wallis test for continuous variables. Logistic regression was computed to identify factors associated with cirrhosis or HCC. All the variables significantly associated with liver disease in a crude analysis (p < 0.05) were further included in a multivariable model to obtain adjusted OR. The interactions between PreS2Δ38-55 and HBeAg-seropositivity, HBV DNA, HBsAg levels, HBV genotype, or AFB1 exposure were examined using a likelihood ratio test by adding interaction terms in the logistic regression model. The capability of the allelic fraction to correctly discriminate HBV-related HCC cases from people with chronic HBV infection without HCC was evaluated using the ROC curve. Its optimal cut-off was selected to maximize the sum of sensitivity and specificity.
AFB1, aflatoxin B1; AFP, alpha-fetoprotein; ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate aminotransferase; GGT, gamma-glutamyltransferase; LSM, liver stiffness measurement.
AFB1 exposure was assessed by next-generation sequencing using the presence of TP53 R249S mutation in cell-free DNA extracted from plasma as a surrogate biomarker.
Risk factors of liver disease
HBeAg seropositivity was significantly associated with liver disease severity; adjusted OR 3.2 (95% CI 1.0–10.5; p = 0.04) for cirrhosis and 5.4 (95% CI 1.1–26.8; p = 0.04) for HCC (Table 2). Similarly, after adjusting for confounding factors, high viral loads were significantly associated with HCC: compared with patients with low viral load (<2,000 IU/ml), the ORs were 6.3 (95% CI 1.5–26.4) for 2,000–106 IU/ml and 15.9 (95% CI 2.6–97.3) for ≥106 IU/ml. However, no association was observed between viral load level and cirrhosis in an adjusted analysis (Table 2). Controls had higher median HBsAg levels (4.1 log10 IU/ml; IQR 3.8–4.4) than cirrhosis (3.7; IQR 3.3–3.9) or HCC cases (3.6; IQR 3.2–4.0) (p <0.001) (Table 1). HBsAg levels <4.0 log10 IU/ml were associated with an increased risk of cirrhosis and HCC (adjusted p <0.001 and p = 0.01, respectively; Table 2).
Table 2.
Factors associated with HBV-related cirrhosis and/or HCC.
| Variables | Cirrhosis |
HCC |
||||||
|---|---|---|---|---|---|---|---|---|
| Crude ORs |
Adjusted ORsa |
Crude ORs |
Adjusted ORsb |
|||||
| OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | OR (95% CI) | p value | |
| Gender | ||||||||
| Women | 1.0 | 0.1 | 1.0 | 0.8 | ||||
| Men | 1.8 (0.8–4.0) | 0.9 (0.4–1.9) | ||||||
| Age (OR per unit increase) | 1.0 (1.0–1.0) | 0.7 | 1.1 (1.0–1.1) | 0.01 | 1.0 (0.9–1.1) | 0.2 | ||
| HBeAg | ||||||||
| Negative | 1.0 | <0.001 | 1.0 | 0.04 | 1.0 | <0.001 | 1.0 | 0.04 |
| Positive | 4.8 (2.3–10.1) | 3.2 (1.0–10.5) | 7.1 (3.1–16.4) | 5.4 (1.1–26.8) | ||||
| HBV DNA (IU/ml)c | ||||||||
| <2,000 | 1.0 | <0.001 | 1.0 | 0.1 | 1.0 | <0.001 | 1.0 | 0.002 |
| 2,000–106 | 3.2 (1.6–6.4) | 1.5 (0.6–3.4) | 4.9 (2.1–11.7) | 6.3 (1.5–26.4) | ||||
| ≥106 | 4.8 (2.0–11.5) | 2.9 (0.8–11.5) | 8.8 (3.2–23.9) | 15.9 (2.6–97.3) | ||||
| HBsAg (IU/ml) | ||||||||
| <10,000 | 1.0 | <0.001 | 1.0 | <0.001 | 1.0 | <0.001 | 1.0 | 0.01 |
| ≥10,000 | 0.2 (0.1–0.3) | 0.1 (0.1–0.3) | 0.2 (0.1–0.5) | 0.1 (0.1–0.6) | ||||
| Genotype | ||||||||
| E | 1.0 | 0.7 | 1.0 | <0.001 | 1.0 | 0.02 | ||
| A | 1.2 (0.5–2.9) | 4.9 (2.1–11.3) | 5.2 (1.4–19.7) | |||||
| PreS2Δ38–55 | ||||||||
| Negative | 1.0 | 0.002 | 1.0 | 0.09 | 1.0 | <0.001 | 1.0 | 0.03 |
| Positive | 2.7 (1.4–5.1) | 1.9 (0.9–4.2) | 4.9 (2.3–10.4) | 3.3 (1.1–9.7) | ||||
| Anti-HDV | ||||||||
| Negative | 1.0 | 0.01 | 1.0 | 0.04 | 1.0 | 0.001 | 1.0 | 0.002 |
| Positive | 16.4 (1.8–149.8) | 15.5 (1.1–219.7) | 41.8 (4.7-372.3) | 135.7 (5.9–3,124.3) | ||||
| Anti-HIV | ||||||||
| Negative | 1.0 | 0.5 | 1.0 | n.a. | ||||
| Positive | 1.9 (0.3–10.7) | n.a. | ||||||
| AFB1 exposured | ||||||||
| No | 1.0 | 0.4 | 1.0 | 0.01 | ||||
| Yes | 1.6 (0.6–4.2) | 5.0 (1.5–17.2) | ||||||
AFB1, aflatoxin B1; n.a., not applicable; OR, odds ratio.
Adjusted for HBeAg, HBV DNA levels, HBsAg levels, preS2Δ38–55, and anti-HDV.
Adjusted for age, HBeAg, HBV DNA levels, HBsAg levels, HBV genotype, preS2Δ38–55, and anti-HDV.
p values are based on a test for trend.
AFB1 exposure was evaluated in a subset of study participants and is not included in the multivariable analysis.
The association of other known risk factors with liver disease severity was examined. Alcohol consumption and coinfection with HIV were relatively rare in this population (Table 1), and no association was observed probably owing to the lack of statistical power. HCV coinfection was more frequent in liver disease cases: 0% in control group, 4% in cirrhosis cases, and 9% in HCC cases (p = 0.001) (Table 1). Nevertheless, logistic regression was not performed to assess the effect of HCV because of the small number of coinfected patients. Additionally, in this HBsAg-positive population, coinfection with HDV, although infrequent (10/302), was significantly associated with an increased risk of both cirrhosis (adjusted OR 15.5; 95% CI 1.1–219.7) and HCC (adjusted OR 135.7; 95% CI 5.9–3,124.3) (Table 2).
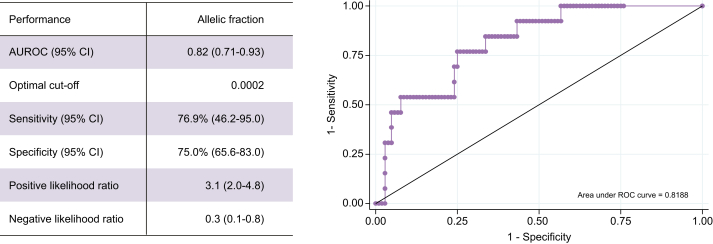
AFB1 exposure was evaluated using a surrogate biomarker in a subset of participants (74 controls, 30 cirrhosis cases, and 13 HCC cases) and found in 54% of HCC cases, 27% of cirrhosis cases, and 19% of HBsAg-positive controls (p = 0.03) (Table 1). AFB1 exposure was significantly associated with HCC risk (crude OR 5.0; 95% CI 1.5–17.2) (Table 2). Of note, no significant difference was observed in the distribution of age, sex, alanine transaminase, and viral load levels between those with or without AFB1 assessment (data not shown). Interestingly, the median allelic fraction in those positive for this AFB1-associated TP53 mutation was significantly higher in HCC cases than controls (Table 1). We assessed the performance of the allelic fraction to discriminate between those with HCC and without (HBsAg-positive controls and cirrhosis cases), and found an area under the ROC curve of 0.82 (95% CI 0.71–0.93) with a sensitivity of 76.9% and specificity of 75.0% at a cut-off level of 0.0002 (Fig. 1).
Fig. 1.
Performance of allelic fraction to discriminate HCC cases from non-HCC cases.
ROC curve for the allelic fraction to discriminate between HCC cases (n = 13) and those without HCC (n = 104, including 74 controls and 30 cirrhosis cases) and performance assessment. AUROC, area under the receiver operating characteristic curve; ROC, receiver operating characteristic.
HBV genetic variability
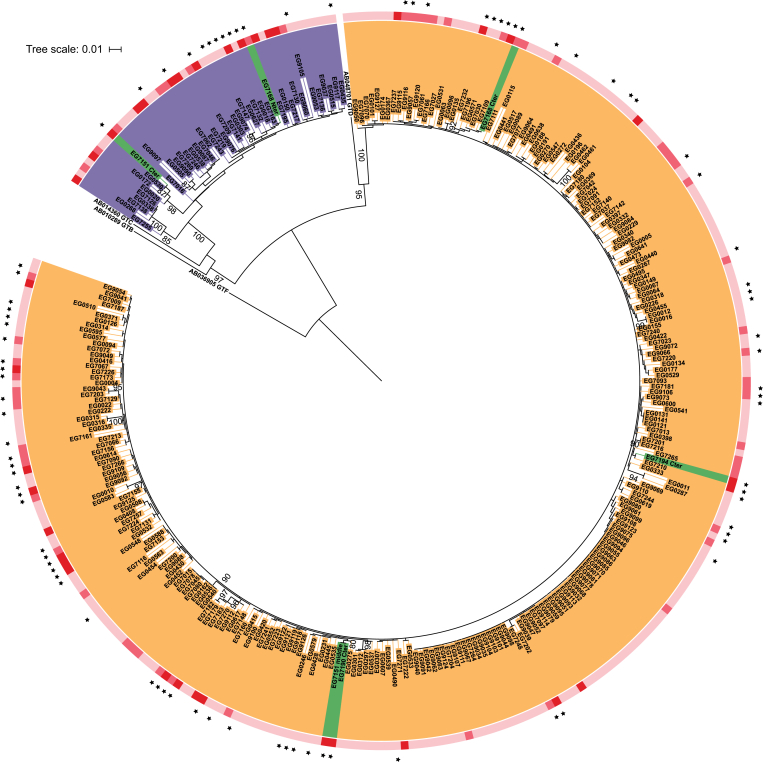
Two HBV genotypes, as assessed by S region sequencing, were detected in the study population: 85% (256/302) were genotype E and 14% (42/302) were genotype A with, in addition, 1% (4/302) A/E recombinants. Three recombinants had an A genotype N-terminal and an E genotype C-terminal, and one N- and C-terminals corresponding to the E genotype and a section between corresponding to the A genotype (see Fig. S1; Fig. 2). The prevalence of genotype A was 34% in HCC cases, 12% in cirrhosis cases, and 11% in controls (Table 1). A significant association was observed between genotype A (vs. E) and HCC (adjusted OR 5.2; 95% CI 1.4–19.7), but not with cirrhosis (Table 2). The distribution of covariates by each genotype in the control group is presented in Table 3. Genotype A was associated with a family history of liver cancer (p = 0.005) and low HBsAg levels (p <0.001), but no association was observed with other covariates.
Fig. 2.
Phylogenetic tree of HBV preS2 region.
Phylogenetic tree based on the HBV S gene sequences shows that genotype E was the most frequent (red), followed by genotype A (blue), and that 4 recombinants were detected (green). Recombinant sequences were cut at the break point and analysed (Fig. S2). The sequences containing preS2Δ38–55 (dots) are spread amongst the tree. The clinical status of patients is indicated by the outer striped ring: light pink, chronic carriers; pink, liver cirrhosis; red, HCC. Numbers at branches correspond to bootstrap values higher than 80%. The scale represents the average number of substitutions per site.
Table 3.
Characteristics of HBsAg-positive controls by the presence of preS2Δ38–55 deletions and different HBV genotypes (n = 211).
| Variables | PreS2Δ38–55 |
HBV genotypes |
||||
|---|---|---|---|---|---|---|
| Absent (n = 170) | Present (n = 41) | p value | E (n = 188) | A (n = 23) | p value | |
| Demographic factors | ||||||
| Gender | ||||||
| Women | 47 (85) | 8 (15) | 0.3 | 49 (89) | 6 (11) | 0.9 |
| Men | 123 (79) | 33 (21) | 139 (89) | 17 (11) | ||
| Age (years) | ||||||
| <40 | 134 (82) | 30 (18) | 0.4 | 145 (88) | 19 (12) | 0.6 |
| ≥40 | 36 (77) | 11 (23) | 43 (91) | 4 (9) | ||
| Birth place | ||||||
| Rural | 114 (78) | 32 (22) | 0.2 | 131 (90) | 15 (10) | 0.7 |
| Urban | 56 (86) | 9 (14) | 57 (88) | 8 (12) | ||
| Family history of liver cancer | ||||||
| No | 161 (81) | 39 (19) | 0.9 | 181 (90) | 19 (10) | 0.005 |
| Yes | 9 (82) | 2 (18) | 7 (64) | 4 (36) | ||
| Lifestyle factors | ||||||
| Alcohol intake | ||||||
| Never | 162 (81) | 38 (19) | 0.5 | 180 (90) | 20 (10) | 0.07 |
| Ever | 8 (73) | 3 (27) | 8 (73) | 3 (27) | ||
| AFB1 exposure | ||||||
| No | 44 (73) | 16 (27) | 0.7 | 54 (90) | 6 (10) | 0.2 |
| Yes | 11 (79) | 3 (21) | 14 (100) | 0 | ||
| Virological factors | ||||||
| HBeAg seropositivity | ||||||
| Negative | 155 (80) | 38 (20) | 0.8 | 171 (89) | 22 (11) | 0.4 |
| Positive | 15 (83) | 3 (17) | 17 (94) | 1 (6) | ||
| HBV DNA (IU/ml) | ||||||
| <2,000 | 125 (81) | 30 (19) | 0.9 | 137 (88) | 18 (12) | 0.8 |
| 2,000–106 | 32 (80) | 8 (20) | 36 (90) | 4 (10) | ||
| ≥106 | 13 (81) | 3 (19) | 15 (94) | 1 (6) | ||
| HBsAg levels (IU/ml) | ||||||
| <10,000 | 46 (74) | 16 (26) | 0.3 | 48 (77) | 14 (23) | <0.001 |
| ≥10,000 | 87 (81) | 21 (19) | 103 (95) | 5 (5) | ||
| Genotype | ||||||
| E | 154 (82) | 34 (18) | 0.2 | n.a. | n.a. | |
| A | 16 (70) | 7 (30) | n.a. | n.a. | ||
| Anti-HCV | ||||||
| Negative | 170 (81) | 41 (19) | n.a. | 188 (89) | 23 (11) | n.a. |
| Positive | 0 | 0 | 0 | 0 | ||
| Anti-HDV | ||||||
| Negative | 168 (80) | 41 (20) | 0.6 | 187 (89) | 22 (11) | 0.7 |
| Positive | 1 (100) | 0 | 1 (100) | 0 | ||
| Anti-HIV | ||||||
| Negative | 165 (80) | 41 (20) | 0.3 | 184 (89) | 22 (11) | 0.4 |
| Positive | 4 (100) | 0 | 3 (75) | 1 (25) | ||
Data are presented as n (%).
AFB1, aflatoxin B1; n.a., not applicable.
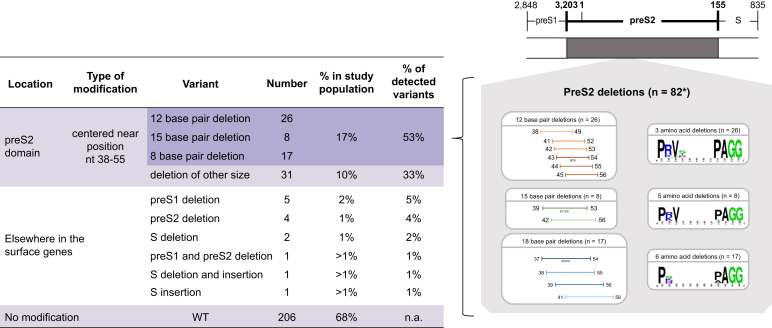
Multiple sequence alignments revealed a higher frequency of preS1/preS2/S gene deletions in HCC cases (57%) than in cirrhosis cases (43%) or control groups (25%) (p <0.001) (Table 1). The vast majority were located in the preS2 domain (84.4%; n = 82/96) (Fig. 3). These genotype-independent deletions were predominantly in-frame leading to 12, 15, or 18 bp (53%; n = 51/96). Losses clustered around nucleotides 38–55 (Δ38–55), resulting in the absence of up to 6 amino acids, but, as seen in the sequence logo, the amino acid sequence 3ʹ to the deletion is conserved (Fig. 3).
Fig. 3.
Variants detected in HBV surface (S) genes, their positions, and prevalence.
As shown, 96/302 S gene sequences had mutations localised for the vast majority (82/96) in the preS2 domain at positions 38–55 (EcoRI nomenclature). Of these, 53% lost 12, 15, or 18 bp (shades of orange, green, and blue, respectively). Corresponding amino acid changes are shown using a sequence logo. The thickness of the bar reflects the frequency of the deletion. The other mutations (14/96) were localised elsewhere in the S genes. n.a., not applicable; WT, wild type.
This mutation profile, taken together with its high atypical prevalence in controls (n = 41/211; 20%; Table 1), led to further phylogenetic analyses that showed that preS2Δ38–55 was spread amongst the inferred tree (Fig. 2). This would suggest that its carriage was not a consequence of infection by a unique HBV strain, but more likely arises independently and multiple times during or after the initial infection of different hosts. Of the 4 A/E recombinant genotypes detected, 3 were from HCC cases and 1 from a cirrhosis case, and all carried preS2Δ38–55 (Table 1). Despite the small sample size, shared recombination break points were observed: twice between nucleotides 146 and 193.5, at nucleotide 2,891, and between nucleotides 2,897 and 3,035 (Fig. S2).
The degree of coexistence of variant and WT populations and the quasi-species was assessed using NGS in 9 randomly selected samples identified as either WT or preS2Δ38–55 by Sanger sequencing. As shown for 3 representative examples (Fig. S3), the WT samples had a constant level of reads in the preS2 domain; by contrast, all those defined as preS2Δ38–55 showed the coexistence of mutant and WT sequences.
Factors associated with the presence of preS2Δ38–55
The presence of the preS2Δ38–55 hotspot mutation was not associated with any of the demographic, lifestyle, or virological factors in the control group (Table 3). However, it was marginally associated with an increased risk of cirrhosis (adjusted OR 1.9; 95% CI 0.9–4.2) and significantly with HCC risk (adjusted OR 3.3; 95% CI 1.1–9.7) (Table 2). AFB1 exposure was not associated with preS2Δ38–55 carriage or HBV genotype in the HBsAg-positive controls (Table 3; Fig. S4A).
Joint effects between preS2Δ38–55 and the other risk factors
The interaction was examined between the effects of preS2Δ38–55 and each of the following HCC risk factors: HBeAg seropositivity, viral load, HBsAg, HBV genotype, and AFB1 exposure (Table 4). HDV was not assessed because of the small number of coinfected patients. There was no evidence of interaction between the effects of preS2Δ38–55 and HBsAg levels (p = 0.6) or preS2Δ38–55 and AFB1 exposure (p = 0.9) on HCC risk, supporting that these effects combine multiplicatively. Although the test for heterogeneity was not statistically significant, the joint effect on HCC was more than multiplicative between the preS2Δ38–55 carriage and HBeAg seropositivity (p = 0.1) and the preS2Δ38–55 carriage and viral load levels (p = 0.1). By contrast, there was strong evidence of the interaction between the effects of preS2Δ38–55 and viral genotype on HCC (p <0.01). Compared with the reference group with the lowest HCC risk (genotype E/deletion[−]), the ORs were 8.4 (95% CI 3.1–22.7) in genotype E/deletion(+), 12.4 (95% CI 4.1–37.7) in genotype A/deletion(−), and 9.4 (95% CI 2.0–44.4) in genotype A/deletion(+). These dissimilarities suggest that preS2Δ38–55 is associated with an increased HCC risk in patients infected with genotype E, but not in those infected with genotype A. However, because of the low frequency of genotype A carriers, this differential effect needs to be confirmed in a larger cohort. In addition, a multiplicative effect on HCC risk was seen, albeit with a wide CI interval, between the preS2Δ38–55 carriage and AFB1 exposure with a 29-fold (95% CI 3.7–230.4) increased risk observed compared with either the deletion (5.5-fold) or AFB1 (6.0-fold) exposure alone (Table 4).
Table 4.
Joint effect of preS2Δ38–55 and HBeAg seropositivity, viral load, HBsAg levels, HBV genotype, and AFB1 exposure, on HCC risk.
| Variables | Controls (n = 211) | HCC cases (n = 35) | OR (95% CI) | p value for heterogeneity |
|---|---|---|---|---|
| HBeAg/preS2Δ38–55 | 0.1 | |||
| (−)/(−) | 155 (74) | 12 (34) | 1.0 | |
| (−)/(+) | 38 (18) | 9 (26) | 3.1 (1.2–7.8) | |
| (+)/(−) | 15 (7) | 4 (11) | 3.4 (1.0–12.0) | |
| (+)/(+) | 3 (1) | 10 (29) | 43.1 (10.4–177.7) | |
| HBV DNA/preS2Δ38–55 | 0.1 | |||
| <2,000/(−) | 125 (59) | 8 (23) | 1.0 | |
| <2,000/(+) | 30 (14) | 3 (9) | 1.6 (0.4–6.2) | |
| ≥2,000/(−) | 45 (22) | 8 (23) | 2.8 (1.0–7.8) | |
| ≥2,000/(+) | 11 (5) | 16 (45) | 22.7 (8.0–64.9) | |
| HBsAg/preS2Δ38–55 | 0.6 | |||
| ≥10,000/(−) | 87 (51) | 4 (12) | 1.0 | |
| ≥10,000/(+) | 21 (12) | 5 (14) | 5.2 (1.3–21.0) | |
| <10,000/(−) | 46 (27) | 12 (34) | 5.7 (1.7–18.6) | |
| <10,000/(+) | 16 (10) | 14 (40) | 19.0 (5.5–65.3) | |
| Genotype/preS2Δ38–55 | <0.01 | |||
| E/(−) | 154 (73) | 7 (22) | 1.0 | |
| E/(+) | 34 (16) | 13 (41) | 8.4 (3.1–22.7) | |
| A/(−) | 16 (8) | 9 (28) | 12.4 (4.1–37.7) | |
| A/(+) | 7 (3) | 9 (9) | 9.4 (2.0–44.4) | |
| AFB1/preS2Δ38–55 | ||||
| (−)/(−) | 44 (59) | 2 (15) | 1.0 | 0.9 |
| (−)/(+) | 16 (22) | 4 (31) | 5.5 (0.9–33.0) | |
| (+)/(−) | 11 (15) | 3 (23) | 6.0 (0.9–40.4) | |
| (+)/(+) | 3 (4) | 4 (31) | 29.3 (3.7–230.4) |
AFB1, aflatoxin B1; OR, odds ratio.
Discussion
This study has, for the first time, investigated the association between HBV genetic diversity and liver disease severity in a West African population exposed to the African environment.
In terms of virological factors, HBV genotype A, a minor genotype in West Africa, was significantly associated with HCC risk in The Gambia (OR 5.2 [1.3–21.0]). This is in line with our previous longitudinal study in The Gambia that showed an association between genotype A and significant liver fibrosis.28 In southern Africa, a study estimated that patients infected with HBV genotype A had a significantly higher HCC risk (risk ratio 4.5), which occurred 6.5 years earlier as compared to non-genotype A-infected patients.29
Our study also found that the preS2Δ38–55 variants are associated with an increased HCC risk in those infected with genotype E, despite genotype E carriers being at lower risk of HCC than genotype A patients. PreS deletions have been extensively described in Asian cohorts,[30], [31], [32] mainly in HBV genotypes B and C. A systematic literature review and meta-analysis performed on 109 such studies and including 1,511 patients has also reported an association between preS2 variants and HCC with an overall OR of 3.0 (2.3–3.9).33 However, a clear distinction between the studies is the location of the preS deletion. In these Asian cohorts, they are scattered throughout the preS region,[30], [31], [32], [33] whereas in the PROLIFICA cohort a deletion hotspot was found specifically in the preS2 domain. The phylogenetic tree based on sequences from the study population suggests that the mutated viruses arise independently many times and are not a unique viral strain. This aspect has not been investigated in other populations and clearly needs to be addressed in future studies.
The finding of the preS2Δ38–55 deletion hotspot raises a number of questions:
-
•
its origin and whether it is a consequence of the presence or processing of DNA adducts on the viral genome caused by exposure to an environmental mutagen;
-
•
if the deleted virus has a selective advantage in the Gambian environment; and
-
•
the cellular consequences of the circulating mutant virus.
AFB1 is one candidate mutagen that has been shown to interact with HBV DNA.34 Based on the p53R249S mutation analysis in cfDNA from individuals for whom adequate plasma samples were available, a high proportion of the study population (25%) has been exposed to AFB1, and this exposure was significantly associated with HCC risk. In addition, we noted that the median allelic fraction in those positive for this AFB1-associated TP53 mutation was significantly higher in HCC cases than in controls. The p53R249S allelic fraction is an integrated marker of both past AFB1 exposure and the presence of a tumour carrying this mutation, and indicative of HCC. The sensitivity and specificity of the technique used in this study to detect mutant p53 alleles are comparable with those obtained using another highly sensitive technology, the droplet digital PCR,35 and highlight their potential in assessing this biomarker in liquid biopsy studies for identifying and confirming HCC status as reviewed in 1 study.36 Moreover, AFB1 exposure could contribute to the low viral load observed in chronic HBV carriers in The Gambia as in vitro AFB1 exposure has been reported to reduce HBV replication.34 In addition, we report for the first time a potential multiplicative effect of AFB1 exposure and HBV preS2Δ38–55 carriage on HCC risk, clearly highlighting the impact of this environmental carcinogen.
In terms of possible impacts, the preS2Δ38–55 variant did not affect viral replication as similar viral loads, median HBsAg levels, and HBeAg prevalence were found in individuals infected with the preS2Δ38–55 or WT virus (Fig. S4B–D). This is likely caused by a cohabitation of the WT HBV with the preS2Δ38–55 form within hepatocytes. Under such a coexistence, the WT viral DNA could compensate the variants with full-length surface proteins, which in turn allows the release of variant surface protein encoding virions, as described previously.37 Our NGS data lend support to this situation and raise an important caveat: using Sanger sequencing, the prevalence and proportion of preS2Δ38–55 in populations are likely underestimated (Fig. S3).
Finally, previous literature suggests that preS2 deletions modulate cellular processes with a potential impact on liver disease. In addition to reducing envelope protein levels and favouring their retention in the endoplasmic reticulum (ER) resulting in ER stress,38 similar preS2 mutant proteins interact with importin α1, a key nuclear transportation factor. This results in reduced nuclear Nijmegen breakage syndrome 1 (NBS1) protein levels and defects in NBS1-mediated homologous recombination that could contribute to genetic instability.39 Other reports identified preS2 variants as associated with ground glass hepatocytes, which might be associated with preneoplastic lesions in chronic HBV infection.[38], [39], [40], [41] Further in vitro studies are needed to fully decipher the molecular mechanisms generating this mutation, its cellular consequences, and whether this quasi-species context of preS2 deletions and WT species could explain the persistence of the variant that would otherwise be negatively selected owing to its altered surface proteins.
Our study has limitations. Firstly, it was conducted in a cross-sectional manner, and therefore care must be taken to interpret the association between virological factors and liver disease severity. A longitudinal cohort study is underway to confirm these associations observed here. Secondly, the number of study participants, particularly those with HCC, was limited, and numerous variables were assessed for their potential associations with HCC. This might have led to a lack of statistical power to identify associations of some of the variables, and also might have resulted in generating ‘chance findings’. It was indeed difficult to recruit and obtain blood samples from patients with end-stage liver disease. Nevertheless, the strength of association observed in several analyses in this study supports the robustness of the study findings. Thirdly, because of limited access to liver biopsy in this resource-limited setting, we were unable to distinguish between HCC cases with and without cirrhosis, or to report their degree of fibrosis. In addition, as patients with HCC in SSA present at advanced stages, we were unable to recruit individuals with early small tumours, and all patients had tumours >5 cm (data not shown). Finally, whilst our partial HBV sequences (surface genes) are sufficient for genotyping,3,4 they are not for sub-genotyping as numerous misclassifications will arise,[42], [43], [44] and this issue needs to be resolved in future studies.
In conclusion, this study, which provides novel findings on HBV quasi-species and its impact on liver disease severity in The Gambia, identified the HBV preS2Δ38–55 variant as a strong independent risk factor and a biomarker for HCC in West African patients. Future studies need to use highly sensitive sequencing technologies to be able to fully validate this risk factor and address the mechanistic impact of its carriage on liver disease. This could help improve our understanding and the significance of the full virological profile of patients with chronic HBV infection to establish appropriate screening criteria and treatment strategies in such high-risk communities.
Financial support
European Commission (FP7); Agence Nationale de Recherches sur le Sida et les Hepatites Virales (12357); the ARC Santé from the Region Auvergne Rhône-Alpes; NIHR Imperial Biomedical Research Centre.
Authors' contributions
DC, SG, YS, ML, JH, and IC drafted the paper, and all the authors reviewed and approved it. IC, ML, MT, MM, and FZ designed the study. ML, YS, NR, and GN recruited the patients. DC, SG, AD, CG, and NK-ND carried out laboratory analyses. YS carried out statistical analysis. PSG and CB-A carried out phylogenetic analyses. SA, GD, and CV carried out next-generation sequencing (NGS) analyses. FLC-K designed NGS TP53 assay and supervised these analyses. JH, IC, MM, NR, and GN supported the conduct of the study.
Conflicts of interest
The authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
The authors thank Emilie Guérin and Rémi Moulinas from the genomic platform GenoLim at Limoges University Hospital for next-generation sequencing and analysis, and Christophe Combet for his expertise on HBV genotypes.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2020.100144.
Supplementary data
References
- 1.Schweitzer A., Horn J., Mikolajczyk R.T., Krause G., Ott J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 2.Maucort-Boulch D., de Martel C., Franceschi S., Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide: liver cancer attributable to hepatitis viruses worldwide. Int J Cancer. 2018;142:2471–2477. doi: 10.1002/ijc.31280. [DOI] [PubMed] [Google Scholar]
- 3.Norder H., Couroucé A.-M., Coursaget P., Echevarria J.M., Lee S.-D., Mushahwar I.K. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 4.Hussain M., Chu C.-J., Sablon E., Lok A.S.F. Rapid and sensitive assays for determination of hepatitis B virus (HBV) genotypes and detection of HBV precore and core promoter variants. J Clin Microbiol. 2003;41:3699–3705. doi: 10.1128/JCM.41.8.3699-3705.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajoriya N., Combet C., Zoulim F., Janssen H.L.A. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J Hepatol. 2017;67:1281–1297. doi: 10.1016/j.jhep.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Hayer J., Jadeau F., Deléage G., Kay A., Zoulim F., Combet C. HBVdb: a knowledge database for hepatitis B virus. Nucleic Acids Res. 2013;41:D566–D570. doi: 10.1093/nar/gks1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malve B., Eschlimann M., Galgey S., Fenaux H., Zoulim F., Goehringer F. Impact of deletions and mutations in hepatitis B virus envelope proteins on serological profile and clinical evolution. Virus Res. 2017;238:141–147. doi: 10.1016/j.virusres.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Pollicino T., Cacciola I., Saffioti F., Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol. 2014;61:408–417. doi: 10.1016/j.jhep.2014.04.041. [DOI] [PubMed] [Google Scholar]
- 9.Wild C.P., Montesano R. A model of interaction: aflatoxins and hepatitis viruses in liver cancer aetiology and prevention. Cancer Lett. 2009;286:22–28. doi: 10.1016/j.canlet.2009.02.053. [DOI] [PubMed] [Google Scholar]
- 10.Gouas D., Shi H., Hainaut P. The aflatoxin-induced TP53 mutation at codon 249 (R249S): biomarker of exposure, early detection and target for therapy. Cancer Lett. 2009;286:29–37. doi: 10.1016/j.canlet.2009.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Mendy M.E., Welzel T., Lesi O.A., Hainaut P., Hall A.J., Kuniholm M.H. Hepatitis B viral load and risk for liver cirrhosis and hepatocellular carcinoma in the Gambia, West Africa. J Viral Hepat. 2010;17:115–122. doi: 10.1111/j.1365-2893.2009.01168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirk G.D., Lesi O.A., Mendy M., Akano A.O., Sam O., Goedert J.J. The Gambia liver cancer study: infection with hepatitis B and C and the risk of hepatocellular carcinoma in West Africa. Hepatology. 2004;39:211–219. doi: 10.1002/hep.20027. [DOI] [PubMed] [Google Scholar]
- 13.Mahale P., Aka P., Chen X., Pfeiffer R.M., Liu P., Groover S. Hepatitis D virus infection, cirrhosis and hepatocellular carcinoma in the Gambia. J Viral Hepat. 2019;26:738–749. doi: 10.1111/jvh.13065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirk G.D., Lesi O.A., Mendy M., Szymañska K., Whittle H., Goedert J.J. 249(ser) TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene. 2005;24:5858–5867. doi: 10.1038/sj.onc.1208732. [DOI] [PubMed] [Google Scholar]
- 15.Lemoine M., Shimakawa Y., Njie R., Taal M., Ndow G., Chemin I. Acceptability and feasibility of a screen-and-treat programme for hepatitis B virus infection in the Gambia: the Prevention of Liver Fibrosis and Cancer in Africa (PROLIFICA) study. Lancet Glob Health. 2016;4:e559–e567. doi: 10.1016/S2214-109X(16)30130-9. [DOI] [PubMed] [Google Scholar]
- 16.Shimakawa Y., Lemoine M., Bottomley C., Njai H.F., Ndow G., Jatta A. Birth order and risk of hepatocellular carcinoma in chronic carriers of hepatitis B virus: a case-control study in the Gambia. Liver Int. 2015;35:2318–2326. doi: 10.1111/liv.12814. [DOI] [PubMed] [Google Scholar]
- 17.Lemoine M., Shimakawa Y., Nayagam S., Khalil M., Suso P., Lloyd J. The gamma-glutamyl transpeptidase to platelet ratio (GPR) predicts significant liver fibrosis and cirrhosis in patients with chronic HBV infection in West Africa. Gut. 2016;65:1369–1376. doi: 10.1136/gutjnl-2015-309260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh S., Banerjee P., RoyChoudhury A., Sarkar S., Ghosh A., Santra A. Unique hepatitis B virus subgenotype in a primitive tribal community in eastern India. J Clin Microbiol. 2010;48:4063–4071. doi: 10.1128/JCM.01174-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh S., Banerjee P., Deny P., Mondal R.K., Nandi M., RoyChoudhury A. New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in eastern India. J Viral Hepat. 2013;20:209–218. doi: 10.1111/j.1365-2893.2012.01655.x. [DOI] [PubMed] [Google Scholar]
- 20.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schultz A.-K., Bulla I., Abdou-Chekaraou M., Gordien E., Morgenstern B., Zoulim F. jpHMM: recombination analysis in viruses with circular genomes such as the hepatitis B virus. Nucleic Acids Res. 2012;40:W193–W198. doi: 10.1093/nar/gks414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranwez V., Harispe S., Delsuc F., Douzery E.J.P. MACSE: multiple Alignment of Coding SEquences accounting for frameshifts and stop codons. PLoS One. 2011;6:e22594. doi: 10.1371/journal.pone.0022594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Criscuolo A., Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 26.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letunic I., Bork P. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimakawa Y., Lemoine M., Njai H.F., Bottomley C., Ndow G., Goldin R.D. Natural history of chronic HBV infection in West Africa: a longitudinal population-based study from the Gambia. Gut. 2016;65:2007–2016. doi: 10.1136/gutjnl-2015-309892. [DOI] [PubMed] [Google Scholar]
- 29.Kew M.C., Kramvis A., Yu M.C., Arakawa K., Hodkinson J. Increased hepatocarcinogenic potential of hepatitis B virus genotype A in Bantu-speaking sub-Saharan Africans. J Med Virol. 2005;75:513–521. doi: 10.1002/jmv.20311. [DOI] [PubMed] [Google Scholar]
- 30.Chen B., Liu C., Jow G., Chen P., Kao J., Chen D. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130:1153–1168. doi: 10.1053/j.gastro.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Yeung P., Wong D.K.-H., Lai C.-L., Fung J., Seto W.-K., Yuen M.-F. Association of hepatitis B virus pre-S deletions with the development of hepatocellular carcinoma in chronic hepatitis B. J Infect Dis. 2011;203:646–654. doi: 10.1093/infdis/jiq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B.-F. Hepatitis B virus pre-S/S variants in liver diseases. World J Gastroenterol. 2018;24:1507–1520. doi: 10.3748/wjg.v24.i14.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W.-C., Wu I.-C., Lee Y.-C., Lin C.-P., Cheng J.-H., Lin Y.-J. Hepatocellular carcinoma-associated single-nucleotide variants and deletions identified by the use of genome-wide high-throughput analysis of hepatitis B virus. J Pathol. 2017;243:176–192. doi: 10.1002/path.4938. [DOI] [PubMed] [Google Scholar]
- 34.Lereau M., Gouas D., Villar S., Besaratinia A., Hautefeuille A., Berthillon P. Interactions between hepatitis B virus and aflatoxin B1: effects on p53 induction in HepaRG cells. J Gen Virol. 2012;93:640–650. doi: 10.1099/vir.0.032482-0. [DOI] [PubMed] [Google Scholar]
- 35.Marchio A., Amougou Atsama M., Béré A., Komas N.-P., Noah Noah D., Atangana P.J.A. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin Exp Med. 2018;18:421–431. doi: 10.1007/s10238-018-0502-9. [DOI] [PubMed] [Google Scholar]
- 36.Su Y.-H., Kim A.K., Jain S. Liquid biopsies for hepatocellular carcinoma. Transl Res. 2018;201:84–97. doi: 10.1016/j.trsl.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerken G., Kremsdorf D., Capel F., Petit M.A., Dauguet C., Manns M.P. Hepatitis B defective virus with rearrangements in the PreS gene during chronic HBV infection. Virology. 1991;183:555–565. doi: 10.1016/0042-6822(91)90984-j. [DOI] [PubMed] [Google Scholar]
- 38.Hsieh Y.-H., Su I.-J., Wang H.-C., Chang W.-W., Lei H.-Y., Lai M.-D. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023–2032. doi: 10.1093/carcin/bgh207. [DOI] [PubMed] [Google Scholar]
- 39.Hsieh Y.-H., Chang Y.-Y., Su I.-J., Yen C.-J., Liu Y.-R., Liu R.-J. Hepatitis B virus pre-S2 mutant large surface protein inhibits DNA double-strand break repair and leads to genome instability in hepatocarcinogenesis. J Pathol. 2015;236:337–347. doi: 10.1002/path.4531. [DOI] [PubMed] [Google Scholar]
- 40.Su I.-J., Wang H.-C., Wu H.-C., Huang W.-Y. Ground glass hepatocytes contain pre-S mutants and represent preneoplastic lesions in chronic hepatitis B virus infection. J Gastroenterol Hepatol. 2008;23:1169–1174. doi: 10.1111/j.1440-1746.2008.05348.x. [DOI] [PubMed] [Google Scholar]
- 41.Su I.-J., Wang L.H.-C., Hsieh W.-C., Wu H.-C., Teng C.-F., Tsai H.-W. The emerging role of hepatitis B virus pre-S2 deletion mutant proteins in HBV tumorigenesis. J Biomed Sci. 2014;21:98. doi: 10.1186/s12929-014-0098-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia L., Hu F., Li H., Li L., Tang X., Liu Y. Characterization of small genomic regions of the hepatitis B virus should be performed with more caution. Virol J. 2018;15:188. doi: 10.1186/s12985-018-1100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pourkarim M.R., Amini-Bavil-Olyaee S., Lemey P., Maes P., Van Ranst M. Are hepatitis B virus “subgenotypes” defined accurately? J Clin Virol. 2010;47:356–360. doi: 10.1016/j.jcv.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 44.McNaughton A.L., Revill P.A., Littlejohn M., Matthews P.C., Ansari M.A. Analysis of genomic-length HBV sequences to determine genotype and subgenotype reference sequences. J Gen Virol. 2020;101:271–283. doi: 10.1099/jgv.0.001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.