Abstract
Both dengue virus (DENV) and hepatitis C virus (HCV) belong to the Flaviviridae family and could induce hepatitis. We aimed to investigate the interference between them. In total, 515 patients confirmed with dengue fever (DF) were enrolled. Thirty-two patients (6.21%) were seropositive for anti-HCV; 12 of 32 anti-HCV-positive patients had detectable HCV-RNA at presentation of DF. The proportion of dengue hemorrhagic fever was comparable between patients with or without anti-HCV and between those with or without HCV-RNA. Eleven of 32 patients received HCV-RNA testing during a median interval of 23 months after DF, which revealed significantly increased HCV-RNA levels (5.43 ± 0.77 vs 3.09 ± 1.24 log IU/mL, follow-up vs acute-DF phase; P = .003). Four of 11 patients with baseline HCV-RNA values before DF demonstrated a nadir viremia during acute DF. We also included age-, sex-, and follow-up duration–matched HCV-monoinfected patients as controls; higher delta HCV-RNA changes were demonstrated in patients with DF than in controls during the follow-up period (2.34 ± 1.15 vs –0.27 ± 0.76 log IU/mL; P < .001). Further in vitro experiments showed that HCV nonstructural protein 5A was downregulated in Con1 HCV replicon cells infected by DENV1. These clinical and experimental findings suggested possible viral interference in DENV/HCV. However, HCV viremia did not affect the disease outcomes of DF.
Keywords: dengue fever, DENV NS1, HCV NS5A, hepatitis C, viral interference
The global disease burden of dengue virus (DENV) infection is extremely high, reaching ~400 million new infections per year [1]. People in the tropical and subtropical regions, accounting for about half of the human population globally, are at risk for DENV infection. The clinical presentations vary among different age groups, ranging from asymptomatic disease to fever with generalized symptoms. The related complications include dengue fever (DF), dengue hemorrhagic fever (DHF), dengue shock syndrome, and severe DENV infection (SD) [2]. Because of convenient transportation and frequent communication, dengue has been prone to spread among countries in recent decades.
Hepatitis C virus (HCV), belonging to the genus Hepacivirus, is also a member of the Flaviviridae family. It affects >150 million people worldwide and is the primary etiology of liver diseases and liver-related morbidity or mortality [3]. HCV genotype 1 predominates globally and in Taiwan [4]. Besides, chronic hepatitis C (CHC) also leads to many extrahepatic manifestations, such as glucose or lipid abnormalities [5, 6]. Even after successful HCV eradication, the risk of the occurrence of hepatocellular carcinoma still exists [7, 8]. Lacking effective or safe vaccination for prevention, both DENV and HCV could lead to human hepatitis upon active replication in the cytoplasm [9]. However, the clinical characteristics of DENV/HCV dual infection, mostly CHC superimposed with DF, have rarely been investigated [10]. In addition, the phenomenon of viral interference mainly has mainly come from experimental studies. It has seldom been observed in a human study, especially between viruses of the same evolutional level. The interaction and/or viral interference, alteration of the natural course, and clinical outcomes between the 2 viruses of the same Flavirividae deserve to be elucidated.
Southern Taiwan is an HCV-endemic region with a prevalence of 8.6% among the general population [11, 12]. The most prevalent genotype is 1b in some townships in Kaohsiung [13]. Southern Taiwan is also a tropical geographic location, which facilitated many DF outbreaks in the past decade (dengue serotype 1 [DENV1] predominated in 2014 and DENV2 in 2015) [14, 15]. This unique epidemiological setting thus provides an excellent opportunity for exploring the viral interference between HCV and DENV. Consequently, we conducted the current study aiming to clarify the disease course of HCV infection and its viral kinetic changes in patients with acute DENV infection.
METHODS
Our study was conducted prospectively in 1 medical center and 2 regional core hospitals in Kaohsiung City, Taiwan, from July 2014 to December 2015. This research was performed in accordance with the guidelines of the International Conference on Harmonization for Good Clinical Practice and the Declaration of Helsinki. It was approved by the institutional review board and ethics committee of each hospital. All of the study participants signed informed consent forms before enrollment and serum processing.
Patients
DF is listed as a national important infectious disease. Affected patients are obligatorily reported to the Center for Disease Control (CDC), Taiwan. Serum samples are collected for further confirmative diagnosis within 1 week of the development of DF-associated clinical manifestations such as fever, headache, retro-orbital pain, bone pain, elevated transaminase levels, skin rash, and so forth. Eligible patients (1) were adults aged 18 years or older; (2) had DF confirmed by the CDC, Taiwan; (3) were seropositive for antibodies to HCV (anti-HCV) at presentation of DF. Patients with HIV infection, autoimmune hepatitis, primary biliary cirrhosis, sclerosing cholangitis, Wilson’s disease, and α1-antitrypsin deficiency were excluded from the study. We also excluded those patients who received ongoing antiviral therapy for HCV (interferon-based regimen or direct-acting agents). The baseline laboratory data included white blood cells, hemoglobin concentration, platelet counts, and levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), and creatinine. The patients who were positive for anti-HCV at presentation of DF underwent at least 1 subsequent HCV RNA test after the DF episode. Meanwhile, age-, sex-, and HCV genotype–matched chronic hepatitis C patients without DF were selected as controls with a ratio of 1:3.
Laboratory Tests
Dengue fever (DF) was confirmed by a record of body temperature ≥38.0°C and met 1 or more of the following diagnostic criteria: (1) positive Dengue rapid test by nonstructural protein 1 (NS1) Ag STRIP (Bio-Rad Laboratories, Marnes-la-Coquette, France) within 7 days after disease onset; (2) successful DENV isolation; (3) positive real-time polymerase chain reaction (PCR) testing; (4) seroconversion (or >4-fold elevation) of IgM or IgG antibodies to DENV from paired specimens collected in the acute phase and recovery phase of DF. Dengue hemorrhagic fever (DHF) was defined as fever, bleeding manifestations, thrombocytopenia (<100 000 cells/mm3), and, most importantly, plasma leakage (ascites or pleural effusions on image, hematocrit change >20%, or hypoalbuminemia <3g/dL). Severe dengue (SD) had to meet any of the following criteria: shock or respiratory distress induced by severe plasma leakage, severe bleeding evaluated by physicians, or severe organ injury (eg, AST or ALT >1000 IU/L). The above definition of DHF and SD was in line with the World Health Organization guidelines [16].
Anti-HCV was measured using a third-generation enzyme-linked immunoassay (Abbott Laboratories, North Chicago, IL, USA). HCV RNA levels and HCV genotypes were determined by real-time PCR assay (RealTime HCV; Abbott Molecular, Des Plaines, IL, USA; detection limit: 12 IU/mL) [17]. Hepatitis B surface antigen (HBsAg) was determined by means of a standard quantitative chemiluminescent microparticle immunoassay (ARCHITECT HBsAg, Abbott Diagnostics).
HCV Phylogenetic Analysis by Direct Sequencing
The sequenced target was from the 5’untranslated region (UTR) to the front end of the core gene. The primers for PCR amplification and DNA sequencing contained HCV 5’UTR-F (TTGTGGTACTGCCTGATAGGG) and HCV 5’UTR-R (GGATGTACCCCATGAGGTCG). First, whole nucleic acid was separated by the MagNA Pure Compact Nucleic Acid Isolation Kit on the MagNA Pure Compact System (Roche). Nucleic acid concentration was analyzed by Nanodrop ND-1000. Second, reverse transcription reaction was done using the High Capacity cDNA Reverse Transcription Kit based on the standardized protocol of the supplier (Applied Biosystems) for 120 minutes at 37°C. Third, the target regions were amplified using Platinum PCR SuperMix High Fidelity (Invitrogen) in a total reaction volume of 50 μL. The PCR amplicons were refined using the PCR Fragment Extraction Kit (Geneaid, Taiwan). Eventually, DNA sequencing was performed using the Sanger sequencing method with the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit, version 3.1 (Applied Biosystems), on the ABI PRISM 3730XL DNA Analyzer.
Con1 Cell Line Experiment
An in vitro study was performed to evaluate potential viral interaction by utilizing HCV Con1 cells (containing the subgenomic HCV Con1 replicon, genotype 1b) infected with DENV1 grown in Dulbecco’s modified minimal essential medium (DMEM, Gibco, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA, USA). All cells were incubated in 10-cm tissue culture dishes at 37°C and 5% CO2 and were subcultured every 3–4 days. The DENV1 was maintained and replicated in C6/36 cells, which grew in RPMI-1640 medium. For virus infection, Con1 cells were adsorbed with DENV1 at a multiplicity of infection (MOI) of 10 at 37°C for 2 hours, then they were washed 3 times with PBS and incubated at 37°C and 5% CO2 in culture medium for further processing. Each experiment was repeated in triplicate. The primary antibodies used for Western blotting included NS1 (Genetex, San Antonio, TX, USA), NS5A (Millipore, Billerica, MA, USA), and GAPDH (Abcam, Cambridge, UK).
All the viral proteins were prepared in RIPA cell lysis buffer (Biotools, Taiwan) supplemented with a cocktail of proteinase inhibitors (Sigma-Aldrich). Equal amounts of protein (20 µg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel and then transferred to PVDF membrane (Millipore). The membranes were blocked with PBST buffer containing 5% instant nonfat milk and incubated with the specified primary antibodies. The membranes were then incubated with HRP-conjugated secondary antibodies and were visualized using the ECL chemiluminescent method (Millipore). Finally, the membrane was exposed by BioSpectrum AC (UVP, Upland, CA, USA) and semiquantified by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Statistical Analysis
The independent t test (for normally distributed data) or Mann-Whitney U test (for non-normally distributed data) was used to assess the difference of quantitative variables between the groups. As for categorical variables, comparison of data was done by utilizing a chi-square test, with Yates correction, or the Fisher exact test. HCV RNA level was expressed as logarithmic values. The 2-sample Wilcoxon signed-rank test was applied for the comparison of HCV RNA levels during and after DF, as well as HCV RNA serial changes in the control group. All the statistical analyses were calculated by utilization of the SPSS 12.0 statistical package (SPSS, Chicago, IL, USA). Statistical significance was determined by 2-tailed hypothesis tests and a P value <.05.
RESULTS
Patient Characteristics, HCV Status, and Prognosis of DF
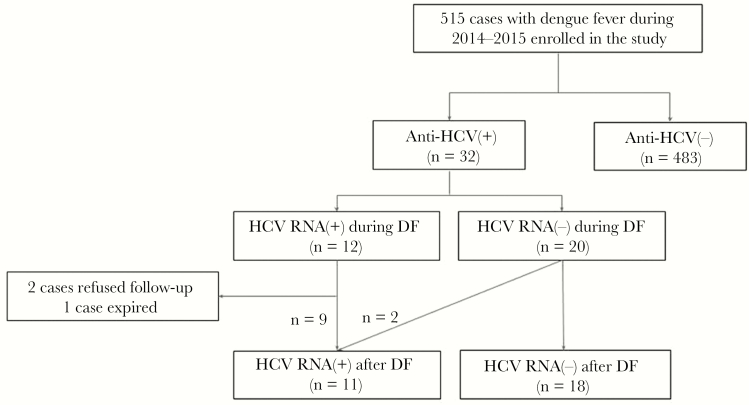
Figure 1 shows the study flow chart. Overall, a total of 1,192 patients were confirmed with DENV infection during 2014 and 2015. Of these, 515 participated in the current study. Their baseline characteristics and virological features are listed in Table 1. The mean age was 56.9 years, and 274 (53.2%) were male. Ninety-five patients (18.4%) were seropositive for HBsAg; 32 (6.21%) were seropositive for anti-HCV. At the presentation of DF, the clinical features showed significant hepatitis (mean AST and ALT levels, 242 and 133 IU/L, respectively), leukopenia (mean, 2900/uL) and thrombocytopenia (70.4 ×1000/μL). One hundred six (20.6%) patients experienced DHF; 34 (6.60%) developed severe dengue.
Figure 1. .
Study flow diagram. Abbreviations: anti-HCV: antibody to HCV; DF = dengue fever; HBV, hepatitis B virus; HCV, hepatitis C virus.
Table 1. .
Patient Characteristics and Virological Features at Presentation of Dengue Fever
| Anti-HCV(+) | |||||||
|---|---|---|---|---|---|---|---|
| Overall | Anti-HCV(-) | Anti-HCV(+) | HCV RNA(+) | HCV RNA(-) | |||
| n = 515 | n = 483 | n = 32 | P | n = 12 | n = 20 | P | |
| Male | 274 (53.2) | 258 (53.4) | 16 (50.0) | .708 | 5 (41.7) | 11 (55.0) | .465 |
| Age, y | 56.9 ± 15.8 | 56.6 ± 16.0 | 61.8 ± 11.3 | .018* | 61.1 ± 12.8 | 62.5 ± 10.5 | .641 |
| HCV RNA, log IU/mL | 3.51 ± 1.15 | ||||||
| HCV genotype 1b/2/UD | 7/4/1 | ||||||
| HCV tx-experienced | 10 (31.3) | 1 (8.30) | 9 (45.0) | <.05* | |||
| Anti-HCV titer, s/co | 10.4 ± 5.93 | 14.7 ± 3.86 | 7.75 ± 5.47 | <.001* | |||
| HBsAg (+) | 95 (18.4) | 88 (18.2) | 7 (21.9) | .606 | 1 (8.30) | 6 (30.0) | .212 |
| Liver cirrhosis | 3 (9.38) | 1 (7.14) | 2 (11.1) | 1.000 | |||
| The year of DF, 2014/2015 | 195/320 | 182/301 | 13/19 | .740 | 6/6 | 7/13 | .473 |
| Laboratory data during DF | |||||||
| AST, IU/L | 242 ± 1136 | 252 ± 1174 | 100 ± 81.6 | .468 | 138 ± 114 | 78.6 ± 44.4 | .102 |
| ALT, IU/L | 133 ± 282 | 137 ± 290 | 76.8 ± 94.7 | .240 | 68.8 ± 37.8 | 81.5 ± 117 | .723 |
| Bil(T), mg/dL | 0.97 ± 0.59 | 0.96 ± 0.61 | 1.03 ± 0.22 | .738 | 1.04 ± 0.23 | 1.02 ± 0.24 | .451 |
| Cr, mg/dL | 1.17 ± 1.24 | 1.18 ± 1.27 | 1.04 ± 0.62 | .561 | 1.18 ± 0.99 | 0.97 ± 0.29 | .930 |
| WBC, 1000/μL | 2.90 ± 1.28 | 2.89 ± 1.27 | 3.14 ± 1.43 | .283 | 3.01 ± 1.35 | 3.21 ± 1.50 | .706 |
| Hb, g/dL | 12.7 ± 2.00 | 12.7 ± 2.00 | 12.8 ± 2.02 | .823 | 12.3 ± 1.84 | 13.1 ± 2.02 | .164 |
| Plt, 1000/μL | 70.4 ± 53.1 | 70.3 ± 53.3 | 70.9 ± 50.0 | .954 | 68.9 ± 46.7 | 72.1 ± 52.8 | .858 |
| Prognosis of DF | |||||||
| Dengue hemorrhagic fever | 106 (20.6) | 97 (20.1) | 9 (28.1) | .276 | 3 (25.0) | 6 (30.0) | 1.000 |
| Severe dengue | 34 (6.60) | 32 (6.60) | 2 (6.30) | 1.000 | 1 (8.30) | 1 (5.00) | 1.000 |
Data are presented as No. (%) or mean ± SD. HCV RNA levels are presented as logarithmic values (base number 10).
Abbreviations: ALT, alanine aminotransferase; anti-HCV, antibody to HCV; AST, aspartate aminotransferase; Bil, total bilirubin; Cr, creatinine; DF, dengue fever; Hb, hemoglobin; HBsAg, hepatitis B virus surface antigen; HCV, hepatitis C virus; Plt, platelet; tx, treatment; UD, undetermined; WBC, white blood cells.
*P < .05.
Baseline characteristics were comparable between patients with and without anti-HCV seropositivity, except that anti-HCV-positive patients were older than those who were anti-HCV-negative (mean ± SD, 61.8 ± 11.3 vs 56.6 ± 16.0 years, respectively). The proportions of patients who experienced DHF and severe dengue were 20.1% (97/483) and 6.6% (32/483), respectively, in patients negative for anti-HCV; these rates were comparable to those of patients positive for anti-HCV: 28.1% (9/32) and 6.3% (2/32), respectively.
Of the 32 patients who were seropositive for anti-HCV, 12 (37.5%) were seropositive for HCV RNA at presentation of DF, with a mean (SD) HCV RNA level of 3.51 (1.15) log IU/mL, including 7 (58.3%) genotype 1b, 4 (33.3%) genotype 2, and 1 (8.3%) genotype unclassified. The baseline characteristics in anti-HCV-seropositive patients were similar between those with and without HCV RNA seropositivity, except that HCV-viremic patients had significantly higher anti-HCV s/co ratios (mean ± SD, 14.7 ± 3.86 vs 7.75 ± 5.4; P < .0001) and a lower proportion of prior interferon experience (8.30% [1/12] vs 45.0% [9/20]; P < .05) than HCV-nonviremic patients did. The proportions of anti-HCV-seropositive patients who experienced DHF and severe dengue were 25% (3/12) and 8.3% (1/12), respectively, in HCV-viremic patients; these rates were comparable to those of HCV-nonviremic patients: 30% (6/20) and 5% (1/20), respectively. There were 7 HBsAg-positive cases in the 32 anti-HCV-positive patients. Although the rate of HBsAg seropositivity was lower in HCV-viremic patients (8.3% [1/12]) than in HCV-nonviremic patients (30% [6/20]), the difference did not reach significance (P = .212) (Table1). For the 12 HCV-viremic patients, the mean HCV RNA levels were comparable between the only patient seropositive for HBsAg and the 11 patients seronegative for HBsAg (3.07 vs 3.55 ± 1.20 logs IU/mL; P = .71). All 7 HBsAg-positive patients had an inactive HBV status (HBV e antigen–negative and undetectable HBV DNA) or were on oral nucleos(t)ide therapy (Supplementary Table 2).
HCV Viral Kinetics at Presentation and After Recovery From DF
Of the 32 anti-HCV-seropositive patients, 2 patients were lost to follow-up; 1 patient died due to hepatocellular carcinoma after recovering from a DF episode. Therefore, HCV RNA testing was performed after recovery from acute DF (12–36 months after the presentation of DF) in 29 patients. All of the 9 patients who were HCV-viremic at presentation of DF remained HCV-viremic at follow-up, whereas 2 of the 20 patients without detectable HCV RNA at presentation of DF became HCV RNA–seropositive during the post-DF follow-up period.
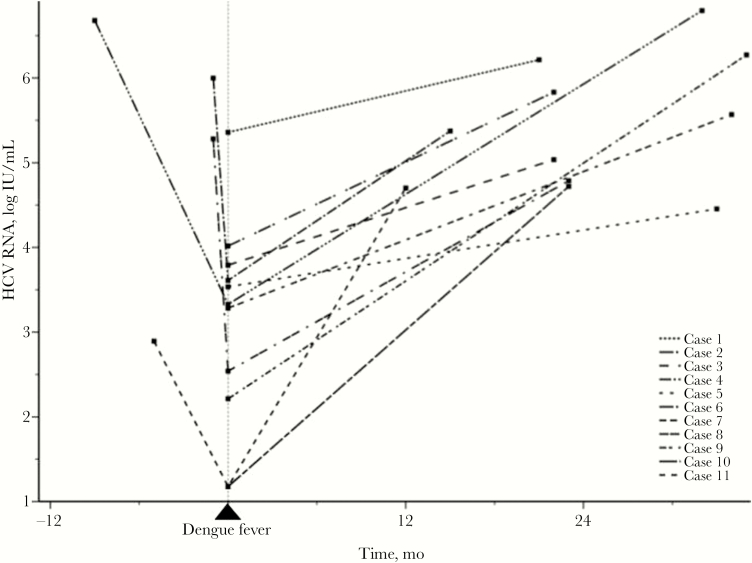
There was a significantly decreased HCV RNA level during the acute DF episode among the 4 patients who had documented HCV RNA levels before presentation of DF (2.64 ± 1.13 vs 5.21 ± 1.65 log IU/mL; P = .004). Viral loads significantly increased from the time of DF presentation to the time after DF recovery, with a mean follow-up interval (range) of 24.7 (12–35) months (3.09 ± 1.24 vs 5.43 ± 0.77 log IU/mL, respectively; P = .003) (Figure 2; Supplementary Table 1). All of these patients experienced a >0.5 log increase in viral load from the onset of DF through the follow-up period. The details of their viral loads and liver function tests are shown in Supplementary Table 1. None of the patients suffered from acute liver decompensation. Sequence alignment showed a 99.8% rate of HCV genetic homology between the samples collected before and after presentation of DF in a patient, indicating the same strain of HCV infection (case 6 in Supplementary Table 1 and Supplementary Data 1).
Figure 2. .
HCV RNA serial changes among the 11 HCV cases before, during, and after dengue fever. HCV RNA level was transformed into logarithmic values. Abbreviation: HCV, hepatitis C virus.
Comparison of HCV RNA Kinetics Between HCV Patients With and Without DF
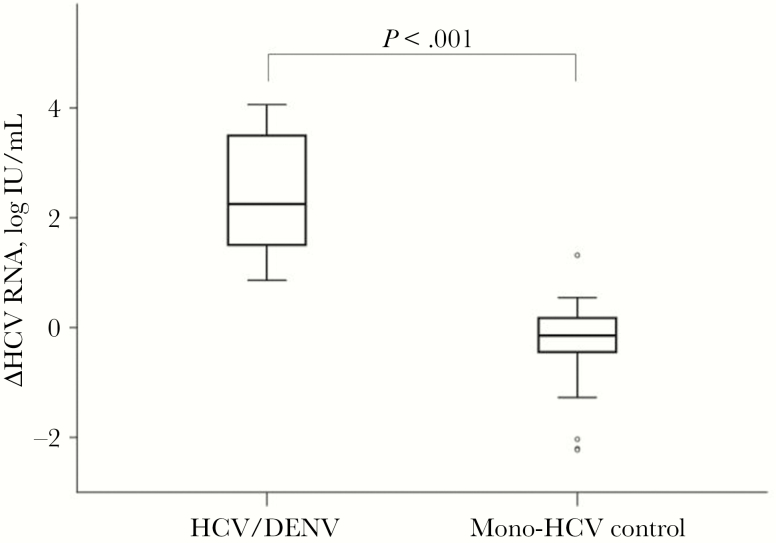
To elucidate whether the significant transient decrease in HCV viral load is relevant to patients with DF, we recruited more age-, sex-, genotype-, and follow-up interval–matched treatment-naïve CHC-monoinfected patients without DF as a control group, with a ratio of 1:3 (Table 2). In these mono-HCV controls, HCV levels changed from 5.91 ± 0.82 to 5.64 ± 1.07 log IU/mL, with a median interval of 26.5 months (Supplementary Data 2). The mean difference of HCV RNA changes among the 33 controls was –0.27 ± 0.76 log IU/mL (favoring fluctuation of HCV RNA in the natural course), which was significantly lower than that of the 11 patients with DF (2.34 ± 1.15 log IU/mL; P < .001) (Table 2, Figure 3). None of the 11 HCV patients with DF had an absolute HCV RNA change <0.5 log IU/mL, compared with 24 (72.7%) HCV patients without DF who did (P < .001). Interestingly, the mean HCV titers at DF presentation of DENV-HCV patients were significantly lower than those of mono-HCV controls at first measurement (3.09 ± 1.24 logs vs 5.91 ± 0.82 logs; P < .001). Later on, the mean HCV titers were comparable between DENV-HCV patients at post-DF follow-up and mono-HCV controls at second measurement (5.43 ± 0.77 vs 5.64 ± 1.07 log IU/mL, respectively; P = .557) (Supplementary Data 2). The data implicated reduced HCV levels in HCV patients during the acute period of DF.
Table 2. .
The Comparison of HCV Viral Loads Changes Between 11 HCV/DENV Cases and 33 HCV Controls
| HCV/DENV (n = 11) | HCV Control (n = 33) | P | |
|---|---|---|---|
| Age, y | 59.7 ± 14.2 | 60.2 ± 13.3 | .916 |
| Male | 6 (54.5) | 18 (54.5) | 1.000 |
| HCV genotype-1/2 | 7/4 | 22/11 | 1.000 |
| HCV RNA follow-up interval, mo | 23.0 (21.1–33.0) | 26.5 (18.3–42.0) | .283 |
| The changes of viral loads | |||
| ∆HCV RNA, log IU/mL | 2.34 ± 1.15 | –0.27 ± 0.76 | <.001 |
| ∆HCV RNA >0.5 log | 11 (100.0) | 2 (6.06) | <.001 |
| │∆HCV RNA│<0.5 log | 0 (0.00) | 24 (72.7) | <.001 |
| │∆HCV RNA│<1 log | 2 (18.2) | 28 (84.8) | <.001 |
Data are presented as No. (%), mean ± SD, or median (interquartile range). HCV RNA levels are presented as logarithmic values (base number 10). ∆HCV RNA was calculated by the individual difference of HCV RNA levels during the DF episode and the follow-up stage in the HCV/DENV dual-infected group or the individual difference of HCV RNA level between similar periods in HCV controls; │∆HCV RNA│ denotes the absolute value of ∆HCV RNA. HCV RNA follow-up interval is expressed as median (interquartile range).
Abbreviations: DENV, dengue virus; HCV, hepatitis C virus.
Figure 3. .
The comparison of HCV RNA changes between 11 HCV/DENV patients and 33 CHC controls. △HCV RNA was calculated by individual differences in HCV RNA levels during DF episodes and the follow-up stage in the HCV/DENV dual-infected group, or the individual difference of HCV RNA level between similar periods in the CHC group. Abbreviations: CHC, chronic hepatitis C; DENV, dengue virus; HCV, hepatitis C virus.
In Vitro Evidence of Interaction Between DENV and HCV
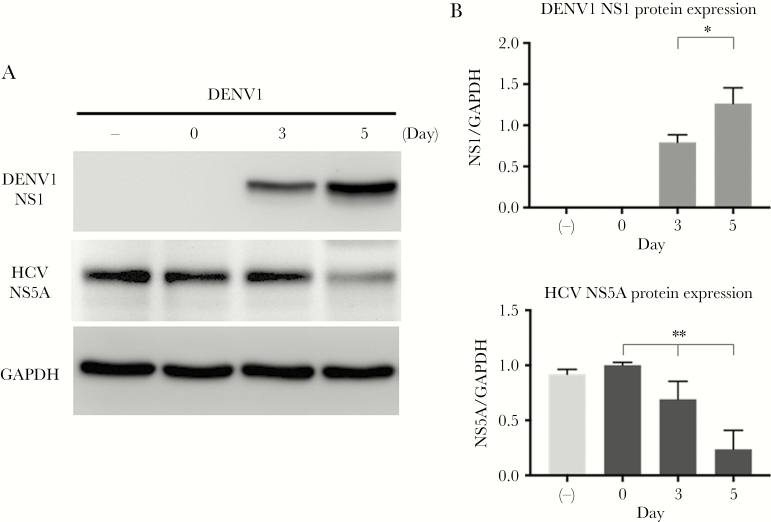
To elucidate the interplay between DENV and HCV viral replication, we performed an in vitro study by using Con1 HCV replicon cells infected with active DENV1 and heat-inactivated DENV1 (as a negative control) at the MOI of 10. The samples were collected at the third and fifth days post-infection (p.i.) to clarify whether DENV1 infection altered HCV replication in vitro. We demonstrated that DENV1 NS1 protein expression significantly increased over time, with significant concomitant suppression of HCV NS5A protein expression (linear trend P = .001) (Figure 4). Nevertheless, DENV1 infection at a lower MOI of 2 failed to result in a significant reduction trend for HCV NS5A (Supplementary Figure 3).
Figure 4. .
The reciprocal expression of DENV1-NS1 and HCV-NS5A after Con1 cells successfully infected by DENV serotype 1. Con1 cells were infected with either 10 MOI of DENV or inactive DENV1 as a negative control. A, One representative result of Western blots, which were detected with 3 specific antibodies, 1 binding up to the DENV NS1 protein, another to HCV NS5A, and the other to GAPDH. B, The bar graphs of protein expression of DENV1 NS1 and HCV NS5A. The protein levels were calculated by the quantification of bands of intensity of NS1 and NS5A after normalization of GAPDH. The data are demonstrated by the mean values of triplicate assays ± SD. *P = .019 (by independent t test); **P = .001 (by 1-way analysis of variance trend test). Abbreviations: DENV1, dengue virus serotype 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HCV, hepatitis C virus; NS1, nonstructural protein 1; NS5A, nonstructural protein 5A.
DISCUSSION
Viral interference is an intriguing phenomenon in nature. To our knowledge, viral interference between human viruses of the same family has rarely been investigated in a clinical setting. The recent outbreaks of DF in Southern Taiwan thus provided an excellent scope for a comprehensive observation. Our study demonstrated that HCV RNA levels of CHC patients were significantly suppressed during acute DENV infection. In 4 of our patients, a nadir change of HCV viral loads was observed at the time of acute DENV infection, and subsequently HCV viral loads were restored to the levels before the DF attack. Our data suggest a greatly transient suppression of HCV RNA (>2 log IU/mL) during an acute attack of DF when compared with the natural course of chronic HCV infection. In vitro experiments further confirmed the suppression of HCV replication by infection of DENV in Con1 HCV replicon cells. The current study provides both the clinical and experimental evidence of viral interference between 2 viruses of the same evolutionary level. Fortunately, dual DENV/HCV infection did not have an impact on the clinical outcome of acute DENV infection nor on the virological outcome of chronic HCV infection.
The pathogenic mechanisms involved in the viral interference between the 2 Flaviviridae viruses remain elusive. For one thing, the primary target cell of DENV is a monocyte, which is also able to be infected by HCV [18]. For another, both viruses could infect liver cells. Although HCV is hepatotropic and DENV is nonhepatotropic, the latter could result in hepatitis and/or liver dysfunction from organ hypoperfusion by dengue shock syndrome or through direct viral injury on hepatocytes [19]. At any given moment, DENV and HCV could potentially have a mutual interaction at the cellular level. As a consequence, 1 virus would have more opportunities to affect another because of the replication competition. Furthermore, DENV and HCV take control of many intracellular pathways and organelles during their infectious processes [20]. It is reasonable that both of them would have a competing relationship, owing to sharing many features in the replicative cycle. Cytokine storm, commonly triggered by acute DENV infection, is also an alternative explanation for HCV suppression. Cytokine storm not only changes the vascular endothelial morphology and contributes to plasma leakage, but also might be responsible for the inhibition of HCV replication. Accompanied with activation of T lymphocytes by DENV, robust production of many pro-inflammatory cytokines would be found in the plasma of patients with DF, such as interferon-α or -γ or -λ, tumor necrosis factor–α, interleukin-1β/2/6/8/10, etc. [21]. Among cytokines, interferon-α or -γ or -λ was considered to have antiviral activity in HCV replication [22, 23]. Interferon would further activate interferon-stimulated genes, which would result in HCV NS5A modulation [24, 25]. Taken collectively, all of the abovementioned could be the underlying mechanisms accounting for the potential interference between the 2 viruses.
Previous clinical studies addressing viral interference came mainly from the observations from HBV/HCV dual infections showing that HCV core protein could inhibit HBV gene expression and replication [26, 27]. Clinical observation discovered that HBV reactivation was not uncommon after successful anti-HCV treatment by interferon-based or direct-acting antiviral therapy [28–32]. By contrast, patients positive for HBsAg had a significantly higher chance of spontaneous clearance of HCV viremia [33, 34]. All of the evidence suggests that viral interference may exist between HCV and HBV through human innate or adaptive immune response [30]. Our study demonstrated that HCV RNA levels were much suppressed during acute DENV infection and then resumed after the acute episode. We further confirmed that the reduction of HCV viral load was significantly beyond the natural fluctuation of HCV RNA during its chronic infectious phase. The finding echoed previous studies indicating that the fluctuation of HCV viral loads was always <0.5 log in CHC patients [35]. All the HCV patients after DF in our study had a viral load elevation of >0.5 log IU/mL with a mean elevated level of 2.34 log IU/mL, which was considered a significant increase after the acute DF event compared with controls. Our results thus provided robust evidence showing the viral interference between the 2 viruses. The underlying immunological difference and mechanisms deserve to be clarified in future studies. We recently observed that a CHC patient achieved an SVR with an ultrashort course (<4 weeks) of antiviral treatment after an acute episode of DF during therapy [36]. However, whether DENV infection could enhance HCV elimination remains unclear.
Schaller et al. found that in coinfection of HCV Jc1 virus (MOI 10) and DENV2 (MOI 2) the expression of NS5A is not downregulated (detected by immunofluorescence analysis) in Huh7.5 cells for 24 hours [37]. Our cell model disclosed that there was a reciprocal change between DENV1-NS1 and HCV-NS5A in a higher titer of DENV1 infection. The inconsistency could be attributed to some different conditions (virus type, concentration, time, and cell line). Despite this, our clinical and experimental findings echoed each other. There should be more experiments to explore if the interplay exists between the 2 viruses.
Some limitations may exist in the present study. First, the limited case numbers may not make the observation a strong result. Hence, further large-scale studies would be needed to validate our findings. Second, we did not examine NS1 titers and DENV serotypes or viral loads to investigate whether these variables may lead to different outcomes. Third, immune profiles or cytokines were not checked for DF patients to test our hypothesis that HCV might be suppressed indirectly by DENV via the cytokine storm and immune response during DF. Lastly, for the anti-HCV-seropositive patients who were negative for HCV RNA at presentation and after recovery from DF, it is difficult to differentiate whether the episode was previously self-resolved HCV infection or DF-related HCV clearance.
In conclusion, the current study demonstrated a suppression of HCV during DENV infection both in vivo and in vitro, which implied probable interference between DENV and HCV. This transient viral load reduction did not change HCV virological outcomes after 1–2 years of follow-up. Additionally, HCV viremia did not influence disease outcomes of patients with acute DF.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
The authors especially thank the secretaries from the Taiwan Liver Research Foundation (TLRF) for their help.
Financial support. This study was partially supported by grants from The Ministry of Science and Technology, Taiwan (107-2314-B-037-082-MY3, 107-2314-B-037-083-MY3), Kaohsiung Medical University (MOST 107-2314-B-037-121, KMU-TC108B06), and Kaohsiung Medical University Hospital (S10806, KMUH107-7M04, KMUH107-7R08, MOHW 107-TDU-B-212-123006). The foundation did not influence how the study was conducted or the approval of the manuscript.
Disclaimer. The funder had no role in the study design, data acquisition or interpretation, manuscript writing, or decision to submit.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. Conception and design: Jee-Fu Huang, Ming-Lun Yeh, Ming-Lung Yu, Chia-Yen Dai, Chung-Feng Huang, Yen-Hsu Chen, Wan-Long Chuang. Acquisition of data: Po-Cheng Liang, Jee-Fu Huang, Chia-Yen Dai, Chung-Feng Huang, Shinn-Chern Chen, Ming-Lun Yeh, Ching-I Huang, Meng-Hsuan Hsieh. Data analysis and interpretation: Jee-Fu Huang, Chung-Feng Huang, Chia-Yen Dai, Wan-Long Chuang, Ming-Lung Yu, Ching-I Huang. Manuscript drafting and critical revision: Po-Cheng Liang, Jee-Fu Huang, Ming-Lung Yu, Wan-Long Chuang.
Prior presentation. Our data were partially presented as a poster at the International Liver Congress 2019 held by the European Association for the Study of the Liver (EASL).
References
- 1. Guzman MG, Harris E. Dengue. Lancet 2015; 385:453–65. [DOI] [PubMed] [Google Scholar]
- 2. Murray NE, Quam MB, Wilder-Smith A. Epidemiology of dengue: past, present and future prospects. Clin Epidemiol 2013; 5:299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thrift AP, El-Serag HB, Kanwal F. Global epidemiology and burden of HCV infection and HCV-related disease. Nat Rev Gastroenterol Hepatol 2017; 14:122–32. [DOI] [PubMed] [Google Scholar]
- 4. Chan CY, Lee SD, Hwang SJ, et al. . Quantitative branched DNA assay and genotyping for hepatitis C virus RNA in Chinese patients with acute and chronic hepatitis C. J Infect Dis 1995; 171:443–6. [DOI] [PubMed] [Google Scholar]
- 5. Huang JF, Yu ML, Dai CY, et al. . Reappraisal of the characteristics of glucose abnormalities in patients with chronic hepatitis C infection. Am J Gastroenterol 2008; 103:1933–40. [DOI] [PubMed] [Google Scholar]
- 6. Dai CY, Chuang WL, Ho CK, et al. . Associations between hepatitis C viremia and low serum triglyceride and cholesterol levels: a community-based study. J Hepatol 2008; 49:9–16. [DOI] [PubMed] [Google Scholar]
- 7. Huang CF, Yeh ML, Tsai PC, et al. . Baseline gamma-glutamyl transferase levels strongly correlate with hepatocellular carcinoma development in non-cirrhotic patients with successful hepatitis C virus eradication. J Hepatol 2014; 61:67–74. [DOI] [PubMed] [Google Scholar]
- 8. Wang JH, Yen YH, Yao CC, et al. . Liver stiffness-based score in hepatoma risk assessment for chronic hepatitis C patients after successful antiviral therapy. Liver Int 2016; 36:1793–9. [DOI] [PubMed] [Google Scholar]
- 9. Chatel-Chaix L, Bartenschlager R. Dengue virus- and hepatitis C virus-induced replication and assembly compartments: the enemy inside—caught in the web. J Virol 2014; 88:5907–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Machain-Williams C, Talavera-Aguilar L, Cetina-Trejo RC, et al. . Detection of hepatitis C virus coinfection in patients with dengue diagnosis. Biomed Res Int 2014; 2014:321286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang JF, Lu SN, Chue PY, et al. . Hepatitis C virus infection among teenagers in an endemic township in Taiwan: epidemiological and clinical follow-up studies. Epidemiol Infect 2001; 127:485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang JF, Lin CI, Huang JF, et al. . Viral hepatitis infections in Southern Taiwan: a multicenter community-based study. Kaohsiung J Med Sci 2010; 26:461–9. [DOI] [PubMed] [Google Scholar]
- 13. Yu ML, Chuang WL, Chen SC, et al. . Changing prevalence of hepatitis C virus genotypes: molecular epidemiology and clinical implications in the hepatitis C virus hyperendemic areas and a tertiary referral center in Taiwan. J Med Virol 2001; 65:58–65. [PubMed] [Google Scholar]
- 14. Wang SF, Chang K, Loh EW, et al. . Consecutive large dengue outbreaks in Taiwan in 2014-2015. Emerg Microbes Infect 2016; 5:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang K, Huang CH, Lee IK, et al. . Differences in mortality and clinical manifestations of dengue hemorrhagic fever in Taiwan in different years: a comparison for cases in 2014 and 2015 epidemics. Am J Trop Med Hyg 2017; 97:361–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dengue: Guidelines fo r Diagnosis, Treatment, Prevention and Control: New Edition. Geneva: World health organization (WHO ); 2009. [Google Scholar]
- 17. Vermehren J, Yu ML, Monto A, et al. . Multi-center evaluation of the Abbott RealTime HCV assay for monitoring patients undergoing antiviral therapy for chronic hepatitis C. J Clin Virol 2011; 52:133–7. [DOI] [PubMed] [Google Scholar]
- 18. Castillo I, Rodríguez-Iñigo E, Bartolomé J, et al. . Hepatitis C virus replicates in peripheral blood mononuclear cells of patients with occult hepatitis C virus infection. Gut 2005; 54:682–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seneviratne SL, Malavige GN, de Silva HJ. Pathogenesis of liver involvement during dengue viral infections. Trans R Soc Trop Med Hyg 2006; 100:608–14. [DOI] [PubMed] [Google Scholar]
- 20. Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol 2018; 16:125–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guabiraba R, Ryffel B. Dengue virus infection: current concepts in immune mechanisms and lessons from murine models. Immunology 2014; 141:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei X, Jia ZS, Lian JQ, et al. . Inhibition of hepatitis C virus infection by interferon-gamma through downregulating claudin-1. J Interferon Cytokine Res 2009; 29:171–8. [DOI] [PubMed] [Google Scholar]
- 23. Donnelly RP, Dickensheets H, O’Brien TR. Interferon-lambda and therapy for chronic hepatitis C virus infection. Trends Immunol 2011; 32:443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim N, Kim MJ, Sung PS, et al. . Interferon-inducible protein SCOTIN interferes with HCV replication through the autolysosomal degradation of NS5A. Nat Commun 2016; 7:10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim MJ, Yoo JY. Inhibition of hepatitis C virus replication by IFN-mediated ISGylation of HCV-NS5A. J Immunol 2010; 185:4311–8. [DOI] [PubMed] [Google Scholar]
- 26. Chen SY, Kao CF, Chen CM, et al. . Mechanisms for inhibition of hepatitis B virus gene expression and replication by hepatitis C virus core protein. J Biol Chem 2003; 278:591–607. [DOI] [PubMed] [Google Scholar]
- 27. Dai CY, Yu ML, Chuang WL, et al. . Influence of hepatitis C virus on the profiles of patients with chronic hepatitis B virus infection. J Gastroenterol Hepatol 2001; 16:636–40. [DOI] [PubMed] [Google Scholar]
- 28. Liu CJ, Chuang WL, Lee CM, et al. . Peginterferon alfa-2a plus ribavirin for the treatment of dual chronic infection with hepatitis B and C viruses. Gastroenterology 2009; 136:496–504.e3. [DOI] [PubMed] [Google Scholar]
- 29. Chuang WL, Dai CY, Chang WY, et al. . Viral interaction and responses in chronic hepatitis C and B coinfected patients with interferon-alpha plus ribavirin combination therapy. Antivir Ther 2005; 10:125–33. [PubMed] [Google Scholar]
- 30. Holmes JA, Yu ML, Chung RT. Hepatitis B reactivation during or after direct acting antiviral therapy - implication for susceptible individuals. Expert Opin Drug Saf 2017; 16:651–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mücke MM, Backus LI, Mücke VT, et al. . Hepatitis B virus reactivation during direct-acting antiviral therapy for hepatitis C: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol 2018; 3:172–80. [DOI] [PubMed] [Google Scholar]
- 32. Yeh ML, Huang CF, Huang CI, et al. . Hepatitis B-related outcomes following direct-acting antiviral therapy in Taiwanese patients with chronic HBV/HCV co-infection. J Hepatol 2020; 73(1):62–71. [DOI] [PubMed] [Google Scholar]
- 33. Yu ML, Dai CY, Huang CF, et al. ; FORMOSA-LIKE group High hepatitis B virus surface antigen levels and favorable interleukin 28B genotype predict spontaneous hepatitis C virus clearance in uremic patients. J Hepatol 2014; 60:253–9. [DOI] [PubMed] [Google Scholar]
- 34. Huang CF, Yeh ML, Lee JJ, et al. . Hepatitis C viremia interferes with serum hepatitis B virus surface antigen and DNA levels in hepatitis B uremics. Hepatol Int 2014; 8:224–32. [DOI] [PubMed] [Google Scholar]
- 35. Arase Y, Ikeda K, Chayama K, et al. . Fluctuation patterns of HCV-RNA serum level in patients with chronic hepatitis C. J Gastroenterol 2000; 35:221–5. [DOI] [PubMed] [Google Scholar]
- 36. Huang CF, Jang TY, Lu PL, Yu ML. Four weeks of paritaprevir/ritonavir/ombitasvir plus dasabuvir encountering dengue fever resulted in sustained virological response in an HCV patient: a case report. Medicine (Baltimore) 2016; 95:e5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schaller T, Appel N, Koutsoudakis G, et al. . Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J Virol 2007; 81:4591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.