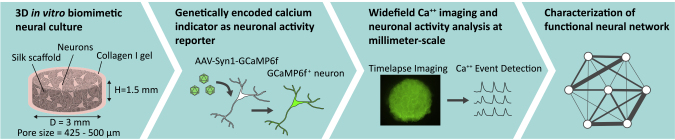
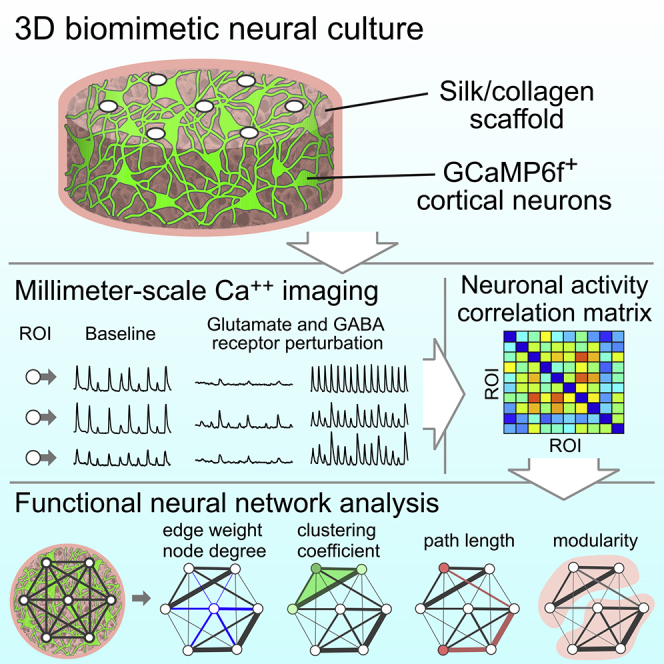
Summary
Three-dimensional (3D) in vitro cultures recapitulate key features of the brain including morphology, cell-cell and cell-extracellular matrix interactions, gradients of factors, and mechanical properties. However, there remains a need for experimental and computational tools to investigate network functions in these 3D models. To address this need, we present an experimental system based on 3D scaffold-based cortical neuron cultures in which we expressed the genetically encoded calcium indicator GCaMP6f to record neuronal activity at the millimeter-scale. Functional neural network descriptors were computed with graph-theory-based network analysis methods, showing the formation of functional networks at 3 weeks of culture. Changes to the functional network properties upon perturbations to glutamatergic neurotransmission or GABAergic neurotransmission were quantitatively characterized. The results illustrate the applicability of our 3D experimental system for the study of brain network development, function, and disruption in a biomimetic microenvironment.
Subject Areas: Optical Imaging, Techniques in Neuroscience, Biomedical Engineering
Graphical Abstract
Highlights
-
•
3D biomimetic in vitro cultures engineered to study functional neural networks
-
•
Millimeter-scale network analysis using GCaMP6f expression and widefield microscopy
-
•
Connected, unspecialized networks form in the developing 3D neural culture
-
•
Network properties change upon pharmacological perturbations of neurotransmission
Optical Imaging; Techniques in Neuroscience; Biomedical Engineering
Introduction
Three-dimensional (3D) neural cultures recapitulate the cell-cell and cell-extracellular matrix (ECM) interactions of the native brain tissue more accurately than conventional 2D cultures (Zhuang et al., 2018). Because they express in vivo-like gene, protein and biomarker expression patterns of development, diseases, and network firing, 3D tissue cultures are useful systems to study brain-like functions at the cellular and tissue level (Lovett et al., 2020). In the brain, adhesion contacts and axonal pathfinding are isotropically distributed in a 3D microenvironment, with neurons synapsing with their immediate neighbors as well as distant neurons in all three dimensions to form extensive neural networks. These interactions are not recapitulated in 2D cultures, where neuronal connections are restricted unnaturally to the x-y plane (Ulloa Severino et al., 2016).
Several 3D culture platforms have been developed to model brain tissues in the laboratory. These platforms generally utilize self-assembly and self-organization principles (e.g., spheroids and organoids) (Birey et al., 2017; Dingle et al., 2015; Lancaster et al., 2013; Quadrato et al., 2017), with or without biomaterial support (e.g., hydrogels or scaffolds) (Frega et al., 2014; Tang-Schomer et al., 2014; Ulloa Severino et al., 2016). One example is our versatile composite scaffold consisting of silk and ECM components for 3D in vitro culture. These systems accommodate the long-term growth and function of rodent primary neurons as well as human neurons derived from induced pluripotent stem cells (iPSCs), induced neural stem cells (iNSCs), or other cell types isolated from patient brain or tumor explants (Cairns et al., 2016; Cantley et al., 2018; Lovett et al., 2020; Rouleau et al., 2020; Sood et al., 2016, 2019; Tang-Schomer et al., 2014).
One of the key challenges with using 3D in vitro cultures is the functional analysis of the engineered brain-like circuitry. Among the neural network analysis technologies, multi-electrode arrays (MEA) and optical-based approaches such as calcium (Ca++) and voltage imaging provide investigators detailed information on the spatial-temporal aspects of brain network development and function of rodent and human neurons in vitro (Badura et al., 2014; Poli et al., 2015; Trujillo et al., 2019). Brain network architecture is a hierarchical entity at multiple spatial scales ranging from connections between synapses to connections between brain regions (Bassett et al., 2018; Betzel and Bassett, 2017). Although MEA offer excellent temporal resolution, cells needs to be in proximity to the electrodes to be detected, and the spatial resolution is physically fixed to the number and spacing of electrodes (typically in the tens to hundreds of μm) (Bourke et al., 2018; Obien et al., 2015; Smith et al., 2015; Tedesco et al., 2018). In contrast to MEA, optical-imaging-based approaches using dyes or genetically encoded reporter constructs have the spatial resolution as high as the microscope equipment allows (typically sub-μm) and have the potential to record contribution of all cells, enabling researchers to conduct analysis at different spatial scales. An optical approach can also be more widely used than MEA due to MEA's requirement of dedicated equipment. Optical analyses of neural networks generally focus on cellular activity patterns at a fraction of 3D space (100–500 μm field width at a single x-y plane) due to the technical limitations of imaging equipment (Bosi et al., 2015; Gu et al., 2016; Huang et al., 2014; Sakaguchi et al., 2019; Ulloa Severino et al., 2016), with few exceptions (Dana et al., 2014; Lu et al., 2020; Palazzolo et al., 2017; Wu et al., 2020). There is a demand for tools to evaluate functional networks at larger, millimeter-scale in 3D cultures.
The objective of the present study was to develop an experimental 3D in vitro system that addressed this need through the incorporation of bioengineering principles, genetically encoded calcium indicators (GECIs), widefield microscopy image acquisition, and computational analysis. The output of this experimental system is a set of mathematical parameters describing the functional neural networks that investigators can use to quantitatively compare networks formed under different conditions. Primary mouse cortical neurons were cultured in 3D scaffolds and virally infected with genetic constructs expressing the calcium reporter GCAMP6f (Chen et al., 2013) to monitor activity. Network analysis showed functionally connected neural networks in the 3D cortical cultures. We demonstrated the utility of this experimental system with pharmacological perturbation of glutamatergic excitatory inputs (treatment with D-2-amino-5-phosphonovalerate, AP5; or 2,3-Dioxo-6-nitro-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide, NBQX) or γ-aminobutyric acid (GABA) inhibitory inputs (treatment with bicuculline or picrotoxin) and presented quantitative outcomes in the network descriptors. Bicuculline and picrotoxin increased neuronal activity, NBQX greatly reduced neuronal activity and impaired the functional network, and AP5 reduced neuronal activity to an even greater degree than NBQX. Interestingly, between the two GABA receptor antagonists, only bicuculline resulted in changes in the functional network properties at the scale we analyzed. Collectively, these data demonstrate the applicability of our 3D in vitro system for understanding the links between neurotransmission and network connectivity and for further investigation of the function and disruption of neural networks in a pathophysiological context.
Results
3D In Vitro Biomimetic Cortical Culture System to Study Neural Network Structure and Function
To investigate neural network formation in 3D microenvironments, we adapted our previously published in vitro composite (silk and collagen) scaffold culture system that mimics the softness of native brain tissue and allows for growth and interaction of neurons and glial cells in three dimensions (Chwalek et al., 2015; Tang-Schomer et al., 2014). Additional elements incorporated into the system in this study were (1) expression of a GECI, GCaMP6f (Chen et al., 2013), as a reporter of neuronal activity, (2) widefield epifluorescence microscopy to record activity at micrometer- and millimeter-scales, and (3) computational tools adapted from published algorithms to investigate functional connectivity of the neural network (Muldoon et al., 2016; Newman, 2006; Onnela et al., 2005; Patel et al., 2015). The scaffolds were fabricated following established protocols, with dimensions optimized for high density cell seeding and for imaging the neural network in one single field of view (scaffold diameter = 3 mm, height = 1.5 mm, volume = 10.6 mm3 and pore size = 425–500 μm) (Figures 1 and S1). Two million cells were seeded per scaffold (Figure S1). The seeding efficiency was approximately 60%–70%, determined by amount of DNA in the 3D culture relative to a 2 x 106 cell pellet with a PicoGreen DNA quantification assay (Figure S2). The cell density in the 3D culture was approximately 1.2—1.3 x 106 cells/mm3.
Figure 1.
Schematic of the 3D Bioengineered Experimental Platform for Investigating Functional Neural Networks
Development of Neural Networks in the Biomimetic Cortical Culture
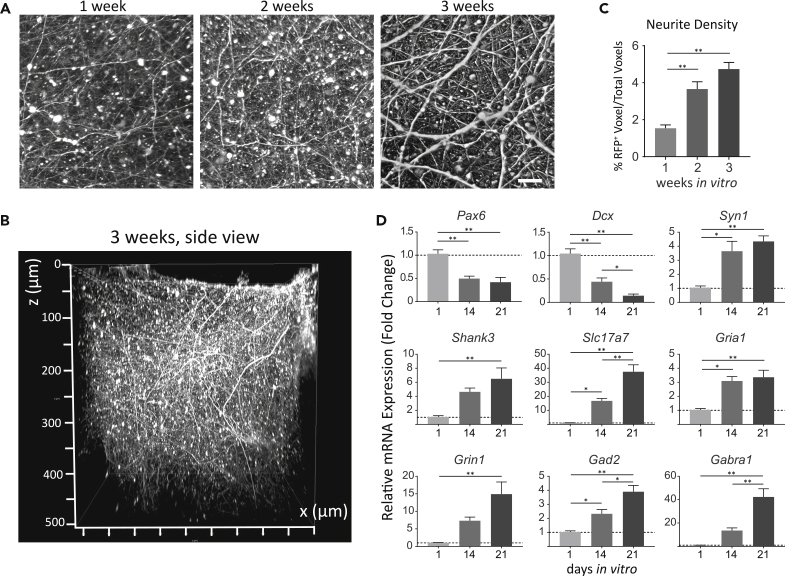
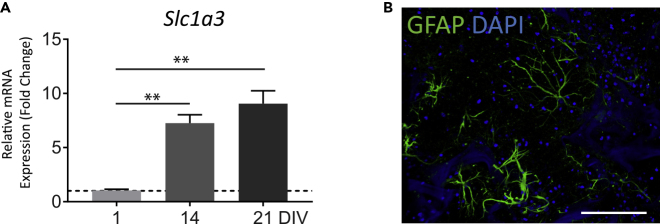
Confocal z stack images at 1, 2, and 3 weeks of culture showed that neurons formed extensive 3D structural networks (Figures 2A and 2B, and Video S1). Using a custom MATLAB code to quantify the 3D neurite density ((Liaudanskaya et al., 2020) and detailed in Transparent Methods), we observed an increase in neurite density over the course of development (Figure 2C). To provide further evidence of structural network formation, quantitative real-time PCR (qRT-PCR) was used to analyze the expression of genes known to change during neural development. As anticipated, we observed a downregulation in mRNA expression levels of the neural progenitor marker (Pax6) and the immature neuron marker (Dcx) (Figure 2D) (Francis et al., 1999; Manuel et al., 2015). We detected a significant upregulation of mRNA encoding proteins with roles in presynaptic vesicle trafficking (Syn1), postsynaptic scaffolding (Shank3), glutamatergic neurotransmission (Slc17a7, Gria1, Grin1), and GABAergic neurotransmission (GAD2, Gabra1) (Figure 2D). The increased gene expression of the Slc3a1 excitatory amino acid transporter together with immunofluorescence imaging of the astrocyte marker glial fibrillary acidic protein (GFAP) indicated the presence of astrocytes in the 3D cultures (Figure 3). Taken together, the data suggest neuronal maturation and synapse formation in the 3D culture model (Ben-Ari, 2001).
Figure 2.
Structural Neural Network Development of the 3D In Vitro Biomimetic Cortical Culture
(A) Representative 3D projections of confocal z stack images of RFP-expressing neurons at 1, 2, and 3 weeks. Scale bar, 20 μm.
(B) Representative 3D rendering of side (x-z) view of RFP-expressing neurons at 3 weeks.
(C) Neurite density in the 3D cortical culture (mean ± SEM). n = 6–8 samples from three independent experiments.
(D) mRNA expression over time (mean ± SEM) of proteins associated with neural progenitors (Pax6), immature neurons (Dcx, doublecortin), presynaptic vesicles (Syn1, synapsin 1), postsynaptic scaffolding protein (Shank3), glutamatergic transmission (Slc17a7, vesicular glutamate transporter 1; Gria1, AMPA receptor subunit 1;Grin1, NMDA receptor subunit 1), and GABAergic transmission (GAD2, glutamate decarboxylase 2; Gabra1, GABAA receptor subunit ɑ1). Expressions were normalized to housekeeping gene 18s. n = 7–11 samples from three to four independent experiments. For (C) and (D), ANOVA and post-hoc Tukey tests were used for statistical significance among groups. ∗p < 0.05; ∗∗p < 0.001. See also Video S1. See also Table S2.
Figure 3.
Astrocyte Markers in the 3D Cortical Culture
(A) Relative mRNA expressions at 1, 14, and 21 DIV of Slc1a3, excitatory amino acid transporter 1, marker of astrocytes. n = 7–11 samples from three to four independent experiments ANOVA with post-hoc Tukey test was used for statistical significance. ∗∗p < 0.001. Scale bar, 100 μm.
(B) Confocal image showing the presence of astrocytes identified by the expression of glial fibrillary acidic protein (GFAP). n = 3 samples from three independent experiments.
Cultures were infected with AAV1-hSyn1-TurboRFP at 1 day in vitro and confocal z-stack images were taken at 21 days in vitro.
3D Cultures Display Spontaneous Neuronal Activities Recorded with a Genetically Encoded Calcium Indicator
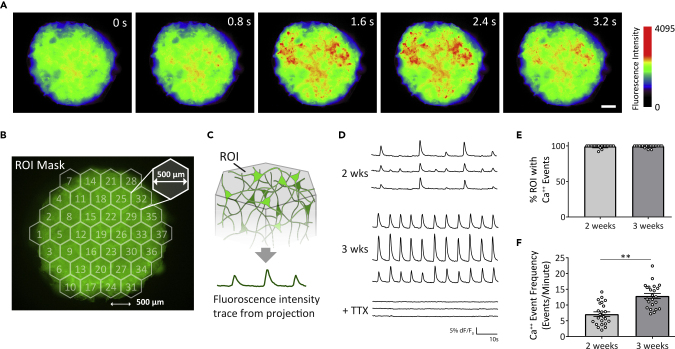
For functional analysis of network activity in our 3D biomimetic cortical culture, a GCaMP6f (Chen et al., 2013) genetic construct was expressed in neurons for recording temporal calcium (Ca++) concentration changes, called Ca++ transients. The cultures were infected at 1 day in vitro (DIV) with adeno-associated virus serotype 1 (AAV1) driving expression in neurons of GCaMP6f under the control of the synapsin 1 promoter (Figure S1). A standard widefield fluorescence microscope with a 4× objective (field size = 4.15× 3.51 mm2) was used to record the activity patterns from projection of the 3D culture (Figure 4A). Imaging was done for 1-min duration at 5 Hz, the temporal resolution that was sufficient to detect Ca++ events in a time-lapse series (Badura et al., 2014).
Figure 4.
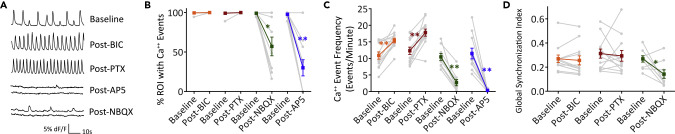
Spontaneous Neuronal Activity in the 3D In Vitro Biomimetic Cortical Culture
(A) Representative fluorescence time-lapse image stacks of 3-week-old 3D cultures expressing GCaMP6f. Images are pseudo-colored to highlight Ca++ transients. Scale bar, 500 μm.
(B) Mask with 37 hexagonal ROIs (dimensions: area = 0.22 mm2, side to side and center-to-center distances = 500 μm) is applied to the time-lapse image stack.
(C) Extraction of time-series trace of average fluorescence intensity from the projection of 3D culture in each ROI.
(D) Representative traces of spontaneous Ca++ transients from individual ROIs at 2 weeks, 3 weeks, and 3 weeks post-tetrodotoxin (TTX, 40 μM) treatment. Fluorescence changes are normalized to the basal fluorescence intensity (dF/F0).
(E) Percentage of all ROIs containing Ca++ event(s) (mean ± SEM). Each data point represents a unique 3D tissue culture sample.
(F) Ca++ event frequency at 2 and 3 weeks (mean ± SEM). Each data point represents average frequency of non-zero event ROIs of a sample. For (E) and (F), n = 23 samples from 3 to 4 matched experiments for 2-week and 3-week samples. An unpaired t test was used for statistical significance. ∗∗p < 0.001. See also Figure S5 and Video S2.
Neuronal activity was recorded at 2 and 3 weeks of culture, concurrent with the time frame in which rodent neurons grown in conventional 2D culture demonstrate increased activity (Chiappalone et al., 2006). At 2 and 3 weeks of culture, Ca++ transients were observed in individual neurons that synchronized within small, local clusters of neurons (size 30–100 μm) as well as larger clusters (>1 mm) (Figure 4A, Video S2). To perform quantitative analysis of the neural networks, the image areas were segmented into equal-size hexagonal regions of interest (ROIs) in a honeycomb configuration that covered the complete projection of the 3D culture sample (Figure 4B). We chose an ROI size of side-to-side distance of 500 μm for long-distance analysis based on previous studies, suggesting cortical neuron axon length in vitro (Virlogeux et al., 2018). It should be noted that the ROI size is user-defined, and computational analyses presented below were dependent on this initial ROI determination. Once an ROI mask was applied, fluorescence signal intensity from the projected image in each ROI was averaged to produce a time-series trace (Figure 4C). For each of the ROIs, Ca++ event detection was performed with the open-source software FluoroSNNAP (Patel et al., 2015). Spontaneous Ca++ events were present at both 2 and 3 weeks of culture in the majority (range 89–100%) of ROIs (Figures 4D, 4E, S3A, and S3B). The frequency of Ca++ events increased from 2 to 3 weeks (7.1 ± 0.7 events/min at 2 weeks, 12.9 ± 0.8 events/min at 3 weeks; mean ± SEM) (Figure 4F), correlating with the increased expression of markers of synaptic neurotransmission (Figure 2D). The Ca++ transients were abolished after tetrodotoxin (TTX) treatment, reflecting the action-potential-dependent nature of the neuronal activities (Figure 4D).
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm. This culture sample is the same one as in Video S3.
Pharmacological Perturbations in Excitatory and Inhibitory Neurotransmission Alters Neuronal Activity of 3D Cultures
To validate the responsiveness of our experimental system to network perturbations, a pharmacological strategy was employed to disrupt excitatory or inhibitory synaptic transmissions. The 3D cultures were treated with bicuculline (BIC, a competitive antagonist of inhibitory GABAA receptors), picrotoxin (PTX, a non-competitive antagonist of inhibitory GABAA and GABAρ receptors), NBQX (a competitive antagonist of excitatory AMPA receptors), or AP5 (a competitive antagonist of excitatory NMDA receptors). Experiments were conducted on 3-week cultures because of their higher baseline activity (Figure 4F).
Spontaneous neuronal activities of each 3D culture sample were recorded and analyzed pre- (“baseline”) and post-treatment (Figures 5 and S3, Videos S3, S4, and S5, and Table S1 for detailed numbers and p values). Following treatments with BIC or PTX, Ca++ events were present in 100% ROIs. Treatment with NBQX and AP5 significantly reduced the number of ROIs containing Ca++ events (post-NBQX, 57 ± 11.8%; post-AP5, 30 ± 10.2%; mean ± SEM). In the AP5-treatment group, 5 of 11 samples (n = 4 experiments) had complete loss of neuronal activity (Figure 5B). Treatment with equivalent concentrations of DMSO, used to dissolve BIC and PTX, had no measurable effects on all the parameters reported in this study (Figure S4).
Figure 5.
Drug Treatments Induced Neuronal Activity Changes in the 3D Cortical Culture
(A) Representative traces of Ca++ transients from individual ROIs in 3-weeks-old 3D cortical culture samples at baseline condition and post-treatments of GABAA receptor antagonist bicuculline (BIC, 10 μM), GABAA and GABAρ receptors antagonist picrotoxin (PTX, 50 μM), NMDA receptor antagonist AP5 (50 μM), or AMPA receptor antagonist NBQX (5 μM). Fluorescence changes were normalized to the basal fluorescence intensity (dF/F0).
(B) Percentage of ROIs in each culture sample containing at least one Ca++ event at baseline and post-treatments.
(C) Average Ca++ events of all ROIs per sample.
(D) Global synchronization index of Ca++ transients across all ROIs in a sample. For (B–D), gray data points and lines show baseline and post-treatment measurements of the same 3D tissue culture samples, and color lines show mean ± SEM. n = 9–12 per group from three to four independent experiments. Paired t tests were used for statistical significance comparing baseline and post-treatment of the same sample. ∗p < 0.05, ∗∗p < 0.001. See also Figure S3, Table S1, and Videos S2, S3, S4, and S5.
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm. This culture sample is the same one as in Video S2.
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm.
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm.
Both GABA receptor blockers resulted in an increase in Ca++ event frequency (BIC group: baseline 10.9 ± 1.1, post-treatment 15.4 ± 0.7; PTX group: baseline 12.3 ± 1.2, post-treatment 17.8 ± 1.1 events/min; mean ± SEM) (Figure 5C). AMPA receptor blocker NBQX reduced the neuronal activity (baseline 10.5 ± 1.0, post-NBQX 2.7 ± 0.9 events/min). Treatment with the NMDA blocker AP5 resulted in greater reduction than NBQX (baseline 11.5 ± 1.6, post-treatment 0.4 ± 0.1 events/min) (Figure 5C). Together, these data show the presence of inhibitory and excitatory transmissions in the 3D tissue cultures and the differential responses of neurons to selective antagonists.
Synchronization of neuronal activity in local clusters (5–50 neurons) has been observed in vivo during early development, with the level of synchronization changing in a brain-region-dependent manner as neural circuits become more complex (Garaschuk et al., 2000). For in vitro studies of networks, a numerical index is used to describe the degree of global synchronization. Here, the global synchronization index was computed by comparing the timing of events in all of the ROIs in each individual 3D culture using FluoroSNNAP software (Patel et al., 2015). An average index of 0.29 ± 0.02 (mean ± SEM) was observed for the 3-week culture baseline conditions. No changes were observed with BIC or PTX treatments (BIC, 0.26 ± 0.04; PTX, 0.29 ± 0.05). In contrast, application of NBQX reduced the global synchronization index (0.14 ± 0.04), suggesting a reduction of global connectivity (Figure 5D). Global synchronization index was not computed for the AP5 group due to lack of neuronal activity.
Functional Neural Network Characterization Using Graph Theory Models: Functional Connectivity, Clustering, Path Length, and Community Structure
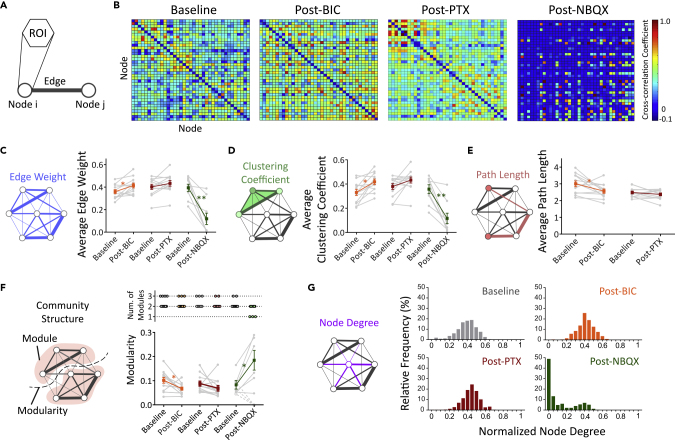
A fundamental goal of the field of network neuroscience is to understand brain networks as a complex web of interactions between functional units (Bassett et al., 2018; Lynn and Bassett, 2019). Graph theory is utilized extensively as a mathematical approach to analyze neural network functionality. To demonstrate the applicability of our 3D experimental system for evaluating functional neural networks, we applied previously published graph-theory-based network analyses to evaluate key attributes of the neural networks formed in our 3D cortical cultures at baseline and post-drug treatment conditions (Muldoon et al., 2016; Newman, 2006; Onnela et al., 2005). The basic principle of graph theory is to represent the network as a graph with each user-defined neural unit as a “node” and the connection between two nodes as an “edge” (Figure 6A). In this study, each of the 37 hexagonal ROIs is the equivalent of a node. The functional connectivity (edge) is defined as the statistical similarity of activity patterns of the two nodes, regardless of physical locations or the underlying structural connections.
Figure 6.
Graph Theory Model-Based Characterizations of Functional Neural Networks in 3D Biomimetic Cortical Cultures
(A) The ROI was designated as a node in the graph-theory-based network analysis.
(B) Representative matrices of cross-correlation coefficients of the 3-week-old 3D cortical culture samples at baseline and post-treatment of bicuculline (BIC; 10 μM), picrotoxin (PTX; 50 μM), or NBQX (5 μM). Self-correlations were removed. The cross-correlation coefficients were then used as edge weight.
(C–F) (C) Average edge weights, (D) average clustering coefficient, (E) average path length, and (F) community structure—number of modules (top) and modularity (bottom) at baseline and post-treatments, as indicated. Post-NBQX samples that lost community structure (1 module and modularity = 0, dotted lines) were excluded from the average modularity calculation.
(G) Frequency distribution of normalized weighted node degree. For (C–F), gray data points and lines represent baseline and post-treatment measurements of individual 3D culture sample. Colored lines show mean ± SEM. (n = 9–12 samples from three to four independent experiments per group). Paired t tests were used for statistical significance comparing baseline and post-treatment of the same sample. ∗p < 0.05, ∗∗p < 0.001. See also Figures S3 and S5, and Table S1.
The functional connectivity between node pairs in the 3D culture was assessed using cross-correlation analysis (Figure 6B). The cross-correlation coefficient of the paired nodes was used as the edge weight, a value to indicate the level of connectivity. These data were then used to compute average clustering coefficient, average path length, community structure, and node degree, which are each described in greater detail below (Figures 6C–6G and see Table S1 for detailed numbers and p values). Average edge weight of all node pairs at baseline and post-BIC, PTX, and NBQX treatments was calculated, but network analysis was not conducted for the AP5 group due to low level of activity. Average edge weight was significantly decreased by the AMPA receptor antagonist NBQX, suggesting a reduction in overall functional connectivity (Figure 6C). Interestingly, between the two GABA receptor antagonists, BIC treatment increased the average edge weight, whereas no significant changes were observed with PTX treatment. These results suggest that BIC, but not PTX, strengthened the overall functional connectivity of the 3D culture.
The average clustering coefficient, a measure of the intensity of functional connectivity among triplets of nodes (detailed in Transparent Methods), was evaluated for our cultures. A network with a high average clustering coefficient is thought to be better at local information integration and more robust to disruption (Bullmore and Sporns, 2009; Onnela et al., 2005). In our cultures, an increase in the average clustering coefficient was observed with BIC treatment, whereas no significant changes were observed with PTX treatment. NBQX treatment resulted in a reduction in the average clustering coefficient (Figure 6D). This suggests that acute modulation of GABA and NMDA neurotransmission may alter information integration in our 3D neural networks (Bullmore and Sporns, 2009).
We similarly investigated path length as a descriptor of neural network functionally in our 3D cultures. Inversely proportional to edge weight, the path length is the shortest distance to traverse from one node to another node in the network. In classical network studies, a short average path length suggests that information can be efficiently shared across the network (Muldoon et al., 2016). However, it is important to mention the disagreements with this interpretation in the field (Fornito et al., 2016), and here we use path length as an indication of statistical dependence and not necessarily to characterize information routes. Average path length was calculated from all node pairs and normalized to the number of active nodes (detailed in Transparent Methods). BIC treatment reduced the average path length, whereas PTX had no significant effect (Figure 6E). Because average path length becomes artificially short when only a small portion of nodes remain active (Figure 6B), this descriptor was not calculated for the NMDA-treated cultures.
Many complex networks have community structures in which the network divides naturally into communities, or modules, containing densely interconnected nodes (Newman, 2006). It is hypothesized that community structures play an important role in functional specialization. Examining the community structures can thus provide insights to the specialization of the network (Bullmore and Sporns, 2009). The measurement that describes the strength of segregation of different parts of network into modules is called the modularity (detailed in Transparent Methods). A higher modularity indicates that the network is more strongly divided and the nodes are more interconnected within modules than connected to nodes in other modules. At baseline conditions after 3 weeks of culture, two to three modules formed in the 3D neural network (Figure 6F, top panel) with low modularity (Figure 6F, bottom panel), suggesting a relatively weak community structure. BIC treatment reduced the modularity, whereas PTX had no significant effects. In the NBQX-treated 3D cultures, two different outcomes of community structures were observed. In cultures with fewer than 50% remaining active nodes (3 of 9 cultures), a single module with modularity of 0 was observed, suggesting that the network was deprived of a community structure after losing activity in the majority of nodes. In contrast, in cultures with 75%–100% active nodes, the modularity increased (6 of 9 cultures), suggesting that the network was more strongly divided (Figure 6F, bottom panel). Possible explanations to the variation among samples could be attributed to potential differences in expression level of AMPA receptors and relative level of AMPA receptor- and NMDA receptor-mediated glutamatergic neurotransmissions in each culture at this stage.
Another characteristic of complex networks is the presence of hubs, which facilitate efficient communication by shortening path lengths and bridging modules in a network (Barabási, 2009; Bullmore and Sporns, 2009). Hubs are nodes with a node degree (sum of edge weights touching the node) that greatly exceeds the average. The presence of hub nodes in a network can be inferred by the presence of a non-Gaussian distribution with a long right tail in a frequency distribution plot of node degrees (Schroeter et al., 2015). We computed the node degree for all of the individual nodes and graphed the frequency distribution. The absence of a long right tail in the frequency plots, combined with a normal distribution, indicate the absence of hub nodes under baseline conditions. Neither BIC nor PTX treatments resulted in the emergence of hub nodes. NBQX treatments resulted in a left shift of the node degree distribution because of the observed reduction in functional connectivity of the network, with 49% of the nodes having a node degree of zero (i.e. no remaining functional connectivity) (Figure 6G).
The above analyses were based on ROIs of side-to-side distances of 500 μm. To demonstrate that this model can be used at different spatial scales with other ROI sizes, we repeated the network analyses with smaller ROIs with side-to-side distance of 250 μm on BIC and PTX experiments (Figure S5). Our analyses showed that modifying ROI size from 500 μm to 250 μm changed the raw values of the network descriptors. It did not affect the pre- and post-treatment trends for synchronization, average edge weight, average path length, and modularity. At this scale of analysis, the increase of average clustering coefficient from pre- to post-PTX became statistically significant. This emphasizes that the network analysis is dependent on the initial determination of ROI, which should be chosen based on the question of interest.
In summary, the network analysis indicated that millimeter-sized, functional neural networks developed in the 3D biomimetic cortical cultures. The networks responded to pharmacological perturbations in neurotransmission with measurable changes in the network descriptors. The average edge weight and average clustering coefficient increased with BIC treatment and decreased with NBQX treatment. The average path length decreased with BIC treatment. The networks at 3 weeks of culture had a weak community structure (modularity) and were reduced by BIC treatment. This, combined with the lack of hubs, suggested that unspecialized networks developed in our 3D cultures. These results demonstrate the utility of this 3D culture system for the study of functional neural networks in vitro.
Discussion
Functional connection of a neural network is an important field of study for brain function and disorders. Conventional 2D culture with both rodent and human iPSC-derived neurons have been used to investigate neural networks using MEA and optical assays. For example, the correlation patterns of 2D rat cortical neuron cultures over the time frame of 7–35 DIV were evaluated with MEA of 60 electrodes with 200-μm spacing (Chiappalone et al., 2006). The emergency of hub structures in 2D mouse hippocampal neuron cultures were assessed with MEA of 64 electrodes with 200-μm spacing (Schroeter et al., 2015). In the past, network analysis has primarily been done with in-house, customized computer algorithms. FluoroSNNAP software, initially intended for 2D cellular scale analysis, was developed to facilitate the computational analysis in the field (Patel et al., 2015). Since that time, 3D in vitro cultures have become increasingly popular to investigate the development, functions, and disorders of brain networks as they better represent key features of the brain tissue (Lovett et al., 2020; Zhuang et al., 2018). One of the first studies demonstrating network activities in months-old human cortical organoids using MEA (200-μm spacing) and observing changes in synchronized network activities over time was reported (Trujillo et al., 2019). With the increasing popularity of 3D cultures, the field will benefit from tools designed for examining neural networks in greater detail in 3D.
Among the techniques for functional recordings and analysis, optical approaches have the advantages of contact-free, high and flexible spatial resolution, and cell-type-specific targeting (Badura et al., 2014). Here we present an in vitro experimental system that allows for interrogation of functional networks that form in a 3D biomimetic microenvironment, with graph-theory-based network analysis that had not been widely utilized by the 3D culture field. The use of a GECI driven by a neuron-specific promotor in combination with observation using widefield microscopy allows for imaging of spatiotemporal patterns in neuronal activity at the millimeter-scale (Figure 1). To describe the functional neural network characteristics and topological properties of our 3D biomimetic cortical culture, we computed several mathematical descriptors (edge weight, clustering coefficient, path length, number of modules, modularity, and node degree). Ca++ imaging has been widely used in tissues, both in vivo and ex vivo, and in other forms of 3D cultures, including organoids. With user-optimized ROI size and Ca++ event detection parameters according to the culture's neuronal activities, our network analysis methods can be readily adapted to a wide range of other culture models. We compiled MATLAB algorithms into a user-friendly code and made this available to researchers (https://github.com/yutingdingle/network-analysis) so that it can be applied to other in vitro culture models. Additional network descriptors (e.g., shortcuts, core-periphery) (Lynn and Bassett, 2019) can potentially be quantified by plugging in other available open source codes and software (Hassan et al., 2015; Rubinov and Sporns, 2010).
In our 3D experimental system, dense structural networks and spontaneous neuronal activities were detected within 2 weeks of culture. In the neonatal rodent brain, AMPA receptors are initially “silent,” and glutamatergic synaptic transmission is purely driven by NMDA receptors. In contrast, activation of AMPA receptor is required for the opening of NMDA receptors in the adult brain (Ben-Ari et al., 1997). At 3 weeks of our 3D culture, a large portion of NMDA receptor activity (blocked by AP5) was independent of AMPA receptors (blocked by NBQX). NMDA receptors were also observed to drive a higher percentage of excitatory activities than AMPA receptors. The glutamatergic neurotransmission of the 3D cultures thus appears to fall within the developmental stage of immature brains at the postnatal stage. In addition, we noticed variations among the 3D cultures' susceptibility to NMDA and AMPA receptors blockers, with one hypothesis being that differences in the expression levels of AMPA and NMDA receptors and the functionality of AMPA receptors among individual 3D samples during neural network formation is the underlying basis for this observation.
Functional networks were established by 3 weeks in the 3D cortical culture. The lack of hubs and weak community structures observed in the networks at this time point can be interpreted as an indication of an unspecialized network (Bullmore and Sporns, 2009; Lynn and Bassett, 2019; Schroeter et al., 2015). It is plausible that our 3D mouse cortical network may develop more specialized features with extended culture time and/or after application of external stimuli in the form of optogenetic actuators (such as channelrhodopsins) and/or biochemical stimuli, such as brain-derived neurotrophic factor (BDNF) (Afshar Saber et al., 2018; Mishchenko et al., 2019).
A functional, balanced neural network relies on the highly regulated integration of the flow of information from both excitatory and inhibitory inputs. We demonstrated that this experimental system can model disruptions in excitatory and inhibitory neurotransmissions and network dysfunctions. Blocking excitatory AMPA receptors with NBQX reduced neuronal activity and disrupted the functional neural network with reductions in edge weight, clustering coefficient, and community structure. Blocking inhibitory GABA receptors with antagonists BIC and PTX increased neuronal activity (indicated by the increase of Ca++ event frequency). Interestingly, only BIC treatment resulted in significant changes to functional network descriptors such as increased average edge weight, increased average clustering coefficient, decreased average path length, and decreased modularity, at our chosen spatial scale of analysis. In-depth examination of GABA receptor subtype expressions and distributions are needed to further address the mechanistic differences in network responses. However, possible explanations for these discrepancies include the differences in targets and mechanisms-of-action of BIC and PTX (i.e., competitive GABAA receptor antagonist versus non-competitive, allosteric antagonist of both GABAA and GABAρ, respectively). Considering that the brain undergoes critical changes of GABA, NMDA, and AMPA receptor isoforms and functions during development in vivo, our 3D culture system may provide a useful in vitro tool to further examine how changes in receptor expressions shape functional network formation.
The inherent flexibility of our bioengineering approach allows for expression of other genetically encoded constructs to induce (e.g., optogenetic tools) and record cell and network activities (e.g., voltage sensitive and glutamate reporters) (Afshar Saber et al., 2018; Mutoh et al., 2012). Furthermore, high-speed recording of large 3D networks with cellular resolution may be possible using light-sheet confocal microscopy. Another distinct advantage of our scaffold-based bioengineering approach is the possibility of tailoring the organization, compartmentalization, and mechanical properties of the 3D culture to better match that of the native brain. For instance, specific cell-types (e.g., astrocytes, microglia, oligodendrocytes, or other neural subtypes), as well as their ratio and density, can be customized in our 3D cultures to mimic white and gray matter structure (Tang-Schomer et al., 2014), to create specific cellular interactions (e.g., neural circuits), and/or to study interactions between cells and varied ECM compositions (Sood et al., 2016). We hypothesize that systematic testing of these input parameters will advance the bioengineering of 3D in vitro biomimetic models to more closely mimic the network density and functional connectivity of the brain.
The 3D in vitro biomimetic culture approach would be particularly useful to study the many human brain disorders that are thought to be caused by dysfunction in the structure and function of synapses and neural circuits (Busche and Konnerth, 2016; Cantley et al., 2018; Centeno et al., 2018; Watanabe and Rees, 2016). For instance, we recently observed Alzheimer-disease-related changes in our long-lived 3D human, iPSC-derived neuron-astrocyte co-cultures grown in silk scaffolds (Rouleau et al., 2020). This suggests that our 3D experimental system has utility as a model for long-term studies to the onset and progression of neurodegenerative and neurodevelopmental disorders. In summary, our interdisciplinary experimental system consisting of a 3D in vitro biomimetic neural culture, genetically encoded neuronal activity reporter, widefield imaging acquisition, and graph-theory-based network analysis adds a valuable new tool for investigating development, function, and disorders of brain networks.
Limitations of the Study
The brain is organized into circuits in which diverse types of neurons connect over multiple spatial scales in a 3D space, ranging from synaptic connections to connections between brain regions. In the present study, we utilized readily accessible standard widefield microscopy to conduct millimeter-scale analysis of 3D mouse cortical cultures at the expense of resolution and the analysis in the third dimension. In comparison to 2D cultures, where all the information flow within the field of view is captured, our platform integrated the 3D information onto a projection and information flow in the Z-dimension is lost. In order to capture Ca++ transients with high spatial resolution and at multilayers at the necessary speed for single-cell and/or 3D analysis, advanced microscopy technologies such as high-speed light-sheet confocal or two-photon microscopy will be crucial. These functional neural networks change dynamically during one's lifespan. Our functional network characterization was conducted in mouse cultures at a 3-week time point. Future studies could incorporate human iPSC-derived cultures to investigate time-dependent development of neural network functions and the biology of brain disease.
Resource Availability
Lead Contact
David Kaplan, Ph.D., david.kaplan@tufts.edu.
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The code generated during this study is available at https://github.com/yutingdingle/network-analysis.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
This work was supported by National Institutes of Health (NIH) research grants R01NS092847 and P41EB027062 to DLK, and NIH S10 OD021624 and National Science Foundation MRI 1531683 research infrastructure grants to Tufts University. The authors would like to thank Mattia Bonzanni, Ph.D., for guidance on network analysis models, constructing the computational framework and writing the MATLAB code for network analysis, and assistance with interpreting results; Martin Hunter for assistance with confocal microscopy; Nilay Vora for writing the preliminary Ca++ peak detection MATLAB code; Patrick Dingle for writing the MATLAB and Python codes for batch processing data; Allison Sweeney for data processing for neurite density imaging analysis; and Michael Lovett, Ph.D., for reading the manuscript.
Author Contributions
Conceptualization, Y.L.D., T.J.F.N., and D.L.K.; Methodology, Y.L.D., V.L., and T.J.F.N.; Software, Y.L.D, V.L., C.M., and I.G.; Formal Analysis, Y.L.D., V.L., and L.T.F.; Investigation, Y.L.D., V.L., L.T.F., and K.C.B.; Writing—Original Draft, Y.L.D. and T.J.F.N.; Writing—Review & Editing, I.G., T.J.F.N., and D.L.K; Visualization, Y.L.D.; Supervision, T.J.F.N. and D.L.K.; Funding Acquisition, D.L.K.
Declaration of Interests
The authors declare no competing interests.
Published: August 21, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101434.
Supplemental Information
References
- Afshar Saber W., Gasparoli F.M., Dirks M.G., Gunn-Moore F.J., Antkowiak M. All-optical assay to study biological neural networks. Front. Neurosci. 2018;12:451. doi: 10.3389/fnins.2018.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badura A., Sun X.R., Giovannucci A., Lynch L.A., Wang S.S. Fast calcium sensor proteins for monitoring neural activity. Neurophotonics. 2014;1:025008. doi: 10.1117/1.NPh.1.2.025008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A.-L. Scale-free networks: a decade and beyond. Science. 2009;325:412. doi: 10.1126/science.1173299. [DOI] [PubMed] [Google Scholar]
- Bassett D.S., Zurn P., Gold J.I. On the nature and use of models in network neuroscience. Nat. Rev. Neurosci. 2018;19:566–578. doi: 10.1038/s41583-018-0038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y. Developing networks play a similar melody. Trends Neurosci. 2001;24:353–360. doi: 10.1016/s0166-2236(00)01813-0. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y., Khazipov R., Leinekugel X., Caillard O., Gaiarsa J.-L. GABAA, NMDA and AMPA receptors: a developmentally regulated `ménage à trois. Trends Neurosci. 1997;20:523–529. doi: 10.1016/s0166-2236(97)01147-8. [DOI] [PubMed] [Google Scholar]
- Betzel R.F., Bassett D.S. Multi-scale brain networks. Neuroimage. 2017;160:73–83. doi: 10.1016/j.neuroimage.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosi S., Rauti R., Laishram J., Turco A., Lonardoni D., Nieus T., Prato M., Scaini D., Ballerini L. From 2D to 3D: novel nanostructured scaffolds to investigate signalling in reconstructed neuronal networks. Sci. Rep. 2015;5:9562. doi: 10.1038/srep09562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke J.L., Quigley A.F., Duchi S., O'Connell C.D., Crook J.M., Wallace G.G., Cook M.J., Kapsa R.M.I. Three-dimensional neural cultures produce networks that mimic native brain activity. J. Tissue Eng. Regen. Med. 2018;12:490–493. doi: 10.1002/term.2508. [DOI] [PubMed] [Google Scholar]
- Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Busche M.A., Konnerth A. Impairments of neural circuit function in Alzheimer's disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150429. doi: 10.1098/rstb.2015.0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns D.M., Chwalek K., Moore Y.E., Kelley M.R., Abbott R.D., Moss S., Kaplan D.L. Expandable and rapidly differentiating human induced neural stem cell lines for multiple tissue engineering applications. Stem Cell Reports. 2016;7:557–570. doi: 10.1016/j.stemcr.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley W.L., Du C., Lomoio S., DePalma T., Peirent E., Kleinknecht D., Hunter M., Tang-Schomer M.D., Tesco G., Kaplan D.L. Functional and sustainable 3D human neural network models from pluripotent stem cells. ACS Biomater. Sci. Eng. 2018;4:4278–4288. doi: 10.1021/acsbiomaterials.8b00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno E.G.Z., Cimarosti H., Bithell A. 2D versus 3D human induced pluripotent stem cell-derived cultures for neurodegenerative disease modelling. Mol. Neurodegener. 2018;13:27. doi: 10.1186/s13024-018-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.W., Wardill T.J., Sun Y., Pulver S.R., Renninger S.L., Baohan A., Schreiter E.R., Kerr R.A., Orger M.B., Jayaraman V. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappalone M., Bove M., Vato A., Tedesco M., Martinoia S. Dissociated cortical networks show spontaneously correlated activity patterns during in vitro development. Brain Res. 2006;1093:41–53. doi: 10.1016/j.brainres.2006.03.049. [DOI] [PubMed] [Google Scholar]
- Chwalek K., Tang-Schomer M.D., Omenetto F.G., Kaplan D.L. In vitro bioengineered model of cortical brain tissue. Nat. Protoc. 2015;10:1362–1373. doi: 10.1038/nprot.2015.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., Marom A., Paluch S., Dvorkin R., Brosh I., Shoham S. Hybrid multiphoton volumetric functional imaging of large-scale bioengineered neuronal networks. Nat. Commun. 2014;5:3997. doi: 10.1038/ncomms4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle Y.T., Boutin M.E., Chirila A.M., Livi L.L., Labriola N.R., Jakubek L.M., Morgan J.R., Darling E.M., Kauer J.A., Hoffman-Kim D. Three-dimensional neural spheroid culture: an in vitro model for cortical studies. Tissue Eng. Part C Methods. 2015;21:1274–1283. doi: 10.1089/ten.tec.2015.0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A., Zalesky A., Bullmore E. Academic Press; 2016. Fundamentals of Brain Network Analysis. [Google Scholar]
- Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M.-C., Friocourt G., McDonnell N., Reiner O., Kahn A. Doublecortin is a developmentally regulated, microtubule-associated protein expressed in migrating and differentiating neurons. Neuron. 1999;23:247–256. doi: 10.1016/s0896-6273(00)80777-1. [DOI] [PubMed] [Google Scholar]
- Frega M., Tedesco M., Massobrio P., Pesce M., Martinoia S. Network dynamics of 3D engineered neuronal cultures: a new experimental model for in-vitro electrophysiology. Sci. Rep. 2014;4:5489. doi: 10.1038/srep05489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaschuk O., Linn J., Eilers J., Konnerth A. Large-scale oscillatory calcium waves in the immature cortex. Nat. Neurosci. 2000;3:452–459. doi: 10.1038/74823. [DOI] [PubMed] [Google Scholar]
- Gu Q., Tomaskovic-Crook E., Lozano R., Chen Y., Kapsa R.M., Zhou Q., Wallace G.G., Crook J.M. Functional 3D neural mini-tissues from printed Gel-based bioink and human neural stem cells. Adv. Healthc. Mater. 2016;5:1429–1438. doi: 10.1002/adhm.201600095. [DOI] [PubMed] [Google Scholar]
- Hassan M., Shamas M., Khalil M., El Falou W., Wendling F. EEGNET: an open source tool for analyzing and visualizing M/EEG connectome. PLoS One. 2015;10:e0138297. doi: 10.1371/journal.pone.0138297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Sun Y., Liu W., Zhang W., Zheng W., Jiang X. Assembly of functional three-dimensional neuronal networks on a microchip. Small. 2014;10:2530–2536. doi: 10.1002/smll.201400513. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaudanskaya V., Chung J.Y., Mizzoni C., Rouleau N., Berk A.N., Wu L., Turner J.A., Georgakoudi I., Whalen M.J., Nieland T.J.F. Modeling controlled cortical impact injury in 3D brain-like tissue cultures. Adv. Healthc. Mater. 2020;9:2000122. doi: 10.1002/adhm.202000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett M.L., Nieland T.J.F., Dingle Y.-T.L., Kaplan D.L. Innovations in 3-dimensional tissue models of human brain physiology and diseases. Adv. Funct. Mater. 2020:1909146. doi: 10.1002/adfm.201909146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Liang Y., Meng G., Zhou P., Svoboda K., Paninski L., Ji N. Rapid mesoscale volumetric imaging of neural activity with synaptic resolution. Nat. Methods. 2020;17:291–294. doi: 10.1038/s41592-020-0760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn C.W., Bassett D.S. The physics of brain network structure, function and control. Nat. Rev. Phys. 2019;1:318–332. [Google Scholar]
- Manuel M.N., Mi D., Mason J.O., Price D.J. Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell. Neurosci. 2015;9:70. doi: 10.3389/fncel.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishchenko T.A., Mitroshina E.V., Usenko A.V., Voronova N.V., Astrakhanova T.A., Shirokova O.M., Kastalskiy I.A., Vedunova M.V. Features of neural network formation and their functions in primary hippocampal cultures in the context of chronic TrkB receptor system influence. Front. Physiol. 2019;9:1925. doi: 10.3389/fphys.2018.01925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muldoon S.F., Bridgeford E.W., Bassett D.S. Small-world propensity and weighted brain networks. Sci. Rep. 2016;6:22057. doi: 10.1038/srep22057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh H., Akemann W., Knopfel T. Genetically engineered fluorescent voltage reporters. ACS Chem. Neurosci. 2012;3:585–592. doi: 10.1021/cn300041b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.E. Modularity and community structure in networks. Proc. Natl. Acad. Sci. U S A. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obien M.E.J., Deligkaris K., Bullmann T., Bakkum D.J., Frey U. Revealing neuronal function through microelectrode array recordings. Front. Neurosci. 2015;8:423. doi: 10.3389/fnins.2014.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnela J.P., Saramaki J., Kertesz J., Kaski K. Intensity and coherence of motifs in weighted complex networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;71:065103. doi: 10.1103/PhysRevE.71.065103. [DOI] [PubMed] [Google Scholar]
- Palazzolo G., Moroni M., Soloperto A., Aletti G., Naldi G., Vassalli M., Nieus T., Difato F. Fast wide-volume functional imaging of engineered in vitro brain tissues. Sci. Rep. 2017;7:8499. doi: 10.1038/s41598-017-08979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel T.P., Man K., Firestein B.L., Meaney D.F. Automated quantification of neuronal networks and single-cell calcium dynamics using calcium imaging. J. Neurosci. Methods. 2015;243:26–38. doi: 10.1016/j.jneumeth.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli D., Pastore V.P., Massobrio P. Functional connectivity in in vitro neuronal assemblies. Front. Neural Circuits. 2015;9:57. doi: 10.3389/fncir.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau N., Cantley W.L., Liaudanskaya V., Berk A., Du C., Rusk W., Peirent E., Koester C., Nieland T.J.F., Kaplan D.L. A long-living bioengineered neural tissue platform to study neurodegeneration. Macromol. Biosci. 2020;20:e2000004. doi: 10.1002/mabi.202000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M., Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52:1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H., Ozaki Y., Ashida T., Matsubara T., Oishi N., Kihara S., Takahashi J. Self-organized synchronous calcium transients in a cultured human neural network derived from cerebral organoids. Stem Cell Reports. 2019;13:458–473. doi: 10.1016/j.stemcr.2019.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter M.S., Charlesworth P., Kitzbichler M.G., Paulsen O., Bullmore E.T. Emergence of rich-club topology and coordinated dynamics in development of hippocampal functional networks in vitro. J. Neurosci. 2015;35:5459–5470. doi: 10.1523/JNEUROSCI.4259-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I., Haag M., Ugbode C., Tams D., Rattray M., Przyborski S., Bithell A., Whalley B.J. Neuronal-glial populations form functional networks in a biocompatible 3D scaffold. Neurosci. Lett. 2015;609:198–202. doi: 10.1016/j.neulet.2015.10.044. [DOI] [PubMed] [Google Scholar]
- Sood D., Chwalek K., Stuntz E., Pouli D., Du C., Tang-Schomer M., Georgakoudi I., Black L.D., 3rd, Kaplan D.L. Fetal brain extracellular matrix boosts neuronal network formation in 3D bioengineered model of cortical brain tissue. ACS Biomater. Sci. Eng. 2016;2:131–140. doi: 10.1021/acsbiomaterials.5b00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood D., Tang-Schomer M., Pouli D., Mizzoni C., Raia N., Tai A., Arkun K., Wu J., Black L.D., 3rd, Scheffler B. 3D extracellular matrix microenvironment in bioengineered tissue models of primary pediatric and adult brain tumors. Nat. Commun. 2019;10:4529. doi: 10.1038/s41467-019-12420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang-Schomer M.D., White J.D., Tien L.W., Schmitt L.I., Valentin T.M., Graziano D.J., Hopkins A.M., Omenetto F.G., Haydon P.G., Kaplan D.L. Bioengineered functional brain-like cortical tissue. Proc. Natl. Acad. Sci. U S A. 2014;111:13811–13816. doi: 10.1073/pnas.1324214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedesco M.T., Di Lisa D., Massobrio P., Colistra N., Pesce M., Catelani T., Dellacasa E., Raiteri R., Martinoia S., Pastorino L. Soft chitosan microbeads scaffold for 3D functional neuronal networks. Biomaterials. 2018;156:159–171. doi: 10.1016/j.biomaterials.2017.11.043. [DOI] [PubMed] [Google Scholar]
- Trujillo C.A., Gao R., Negraes P.D., Gu J., Buchanan J., Preissl S., Wang A., Wu W., Haddad G.G., Chaim I.A. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25:558–569 e557. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa Severino F.P., Ban J., Song Q., Tang M., Bianconi G., Cheng G., Torre V. The role of dimensionality in neuronal network dynamics. Sci. Rep. 2016;6:29640. doi: 10.1038/srep29640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlogeux A., Moutaux E., Christaller W., Genoux A., Bruyere J., Fino E., Charlot B., Cazorla M., Saudou F. Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington's disease. Cell Rep. 2018;22:110–122. doi: 10.1016/j.celrep.2017.12.013. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Rees G. Anatomical imbalance between cortical networks in autism. Sci. Rep. 2016;6:31114. doi: 10.1038/srep31114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Liang Y., Chen S., Hsu C.-L., Chavarha M., Evans S.W., Shi D., Lin M.Z., Tsia K.K., Ji N. Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nat. Methods. 2020;17:287–290. doi: 10.1038/s41592-020-0762-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang P., Sun A.X., An J., Chua C.K., Chew S.Y. 3D neural tissue models: from spheroids to bioprinting. Biomaterials. 2018;154:113–133. doi: 10.1016/j.biomaterials.2017.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cultures were infected with AAV1-hSyn1-TurboRFP at 1 day in vitro and confocal z-stack images were taken at 21 days in vitro.
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm. This culture sample is the same one as in Video S3.
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm. This culture sample is the same one as in Video S2.
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm.
Cultures were infected with AAV-hSyn1-GCaMP6f at 1 day in vitro. Time-lapse images were taken at 5 Hz for 1-min duration. Video is displayed at 4X speed. Size of scaffold = 3 mm.
Data Availability Statement
The code generated during this study is available at https://github.com/yutingdingle/network-analysis.