Abstract
The rhizosphere offers a quintessential habitat for the microbial communities and facilitates a variety of plant-microbe interactions. Members of the genus Bacillus constitute an important group of plant growth promoting rhizobacteria (PGPR), which improve growth and yield of crops. In a total of 60 bacterial isolates from the tomato rhizosphere, 7 isolates were selected based on distinct morphological characteristics and designated as tomato rhizosphere (TRS) isolates with a number suffixed viz., TRS-1, 2, 3, 4, 5, 7, and TRS-8. All the seven isolates were Gram positive, with in vitro plant growth promoting (PGP) traits like phosphate and zinc solubilization, and also produced indoleacetic acid (IAA), phytase, siderophore, hydrogen cyanide (HCN), and 1-aminocyclopropane-1-carboxylate (ACC) deaminase, besides being antagonistic to other microbes and formed biofilm. The seven isolates belonged to the genus Bacillus as per the 16S rDNA sequence analysis. Phylogenetic tree grouped the isolates into four groups, while BOX-PCR fingerprinting allowed further differentiation of the seven isolates. The PGP activity of the isolates was measured on tomato seedlings in plant tissue culture and greenhouse assays. A significant increase in root colonization was observed over 15 days with all the isolates. Greenhouse experiments with these isolates indicated an overall increase in the growth of tomato plants, over 60 days. Isolates TRS-7 and TRS-8 were best plant growth promoters among the seven isolates, with a potential as inoculants to increase tomato productivity.
Keywords: Microbiology, Agricultural soil science, Rhizosphere, Plant growth, Bacteria, Microorganism, PGPR, Tomato, Bacillus spp., BOX-PCR, Rhizobacteria
Microbiology; Agricultural Soil Science; Rhizosphere; Plant Growth; Bacteria; Microorganism; PGPR; tomato; Bacillus spp.; BOX-PCR
1. Introduction
The rhizosphere harbours a diverse group of plant beneficial microorganisms, possessing the inherent ability to improve plant growth and development, and soil health. Beneficial interactions between roots and microbes in the rhizosphere determine overall plant health and soil fertility (Parray et al., 2016; Kalam et al., 2017a). Such interactions play a vital role in regulating various biophysical and biogeochemical processes in the soil. A broad canopy of rhizosphere colonizing bacteria, referred to as plant growth promoting rhizobacteria (PGPR), produce growth promoting substances and elicit phytoprotective effects on plants through several direct and indirect mechanisms (Dutta and Podile, 2010). Inoculation with PGPR plays a significant role in facilitating plant growth and/or safeguarding crops against phytopathogens (biological control) (Parray et al., 2016; Qiao et al., 2017), thereby providing an eco-friendly alternative to chemical fertilizers and fungicides. Hence, exploring the diversity of potential PGPR strains suitable for different environmental conditions, including soil type, is relevant for sustainable agriculture.
The genus Bacillus represents one of the most abundant and phylogenetically diverse groups of easily cultivable PGPR (Orozco-Mosqueda et al., 2020). Bacilli, due to their avid rhizosphere colonization and PGP characteristics, offer considerable interest for improving crop productivity and yield (Zhou et al., 2016; Sansinenea, 2019). Bacillus spp. promote growth by increasing the bioavailability of minerals viz., phosphorus and zinc, fixing atmospheric nitrogen, sequestration of iron through siderophores, and also by the production of phytohormones. In addition, biosynthesis of ethylene catabolism related 1-aminocyclopropane-1-carboxylate (ACC) deaminase, antibiosis, lytic enzyme production, detoxification and degradation of pathogens’ virulence factors (Ahmad et al., 2008; Barea and Richardson, 2015) also contribute to the plant beneficial effects of Bacilli. Seed bacterization was often employed to study the effect of Bacilli or their formulations on plant growth (Kishore et al., 2005; Das et al., 2010). Beneficial Bacillus spp. have the potential to improve soil health and enhance crop yield as external inputs.
Tomato (Solanum lycopersicum L.) is one of the most commonly used vegetables all over the world. There is a need to adopt non-chemical alternatives to increase yield, safety and quality of tomato. Biofertilizers based on Bacillus PGPR have been widely documented to enhance tomato yield and fruit quality. Multiple Bacillus species such as B. licheniformis, B. subtilis, B. polymyxa, B. cereus, B. amyloliquefaciens, B. megaterium, and B. pumilus successfully colonize the tomato rhizosphere and contribute to better growth and yield (Chen et al., 2013; Vaikuntapu et al., 2014; Zhou et al., 2016).
BOX-PCR fingerprinting, along with 16S rRNA gene sequencing, is often employed for identification and molecular typing of bacterial species. It involves amplification of the BOX-elements (interspersed repetitive DNA sequences present in bacterial genomes) with BOX-A1R primer and demonstrates intraspecies diversity (Versalovic et al., 1994). Here, we report the characterization of multifarious PGPR from the tomato rhizosphere both in vitro and in planta. We also demonstrate the potential of BOX-PCR to distinguish different Bacillus spp.
2. Material and methods
2.1. Seed material
Tomato (Solanum lycopersicum L.) seeds (var. Arka Vikas) were procured from ICAR-Indian Institute of Horticulture Research, Bangalore, India.
2.2. Microbial cultures
A commercially available strain of Bacillus licheniformis (CBli) was procured from M/s Sri Biotech, Hyderabad, India. Fungal pathogens Curvularia sp. and Fusarium sp. were obtained from Osmania University, Hyderabad, India. Phytopathogenic bacterium Xanthomonas oryzae pv. oryzae (Xoo) strain BXO43 was obtained from CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India. Xanthomonas axonopodis pv. citri (Xac) strain was from our laboratory collection.
2.3. Rhizospheric soil sample collection
Healthy tomato plants were uprooted, and rhizospheric soil samples were collected from several tomato fields across different districts of Andhra Pradesh and Telangana, India. Sterile sample containers were used for sampling and storage of rhizospheric soil at 4 °C until further use.
2.4. Isolation of rhizobacteria
To isolate rhizobacteria, 1 g rhizospheric soil was added to 10 ml of PBS (phosphate buffer saline) [pH 7.0] followed by vortexing, serially diluting and finally plating the inoculum onto two different media viz., minimal medium-1 (M1) (gL−1)- KH2PO4, 0.2; NH4Cl, 0.25; KCl, 0.5; CaCl2.2H2O, 0.15; NaCl, 1.0; MgCl2.6H2O, 0.62; Na2SO4, 2.84; HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] (pH 6.8), 10 mM and minimal medium-2 (M2) (gL−1)- MgSO4, 0.5; KNO3, 0.5; K2HPO4, 1.3; Ca(NO3)2, 0.06; glucose, 0.06; casamino acids, 0.001; pH 7.5. The plates were incubated at 30 °C for 48 h. Phenotypically distinct isolates were subcultured in fresh medium and purified. The seven bacterial isolates used in the present study included TRS-1, 2, 3, 4, 5, 7 and 8, which were compared with CBli.
2.5. Selection of rhizobacterial isolates based on plant growth promotion traits
In vitro PGP traits of the rhizobacterial isolates were assessed using standard protocols for production of indoleacetic acid (IAA), ACC deaminase, chitinase, phytase, siderophore and HCN, solubilization of zinc and phosphate, and biofilm formation (Saravanan et al., 2004; Demirkan et al., 2014; Vaikuntapu et al., 2014). Antagonistic activity of the test isolates was determined against two soil-borne phytopathogenic fungi viz., Fusarium sp. and Curvularia sp. as described by Vaikuntapu et al. (2014) and also against two phytopathogenic Xanthomonas strains viz., Xac and Xoo, according to Sharma and Kaur (2010).
2.6. Characterization of rhizobacterial isolates
Physiological and biochemical characterization was done using standard protocols (Cappuccino and Sherman, 2014). Selected seven isolates were identified to the genus level using 16S rRNA gene sequencing. Further differentiation of the isolates to species and sub-species was done using BOX-PCR.
2.7. 16S rRNA gene-based molecular characterization of rhizobacteria
Rhizobacterial isolates were grown for 12 h in M1 and M2 broths at 30 °C with shaking at 160×g. Rhizobacterial genomic DNA was isolated, according to Sharma and Singh (2005). 16S rRNA gene was amplified using 100 ng of genomic DNA and employing the universal primers (Sigma-Aldrich, USA): 27F (5′-GTTTGATCCTGGCTCAG-3′) and 1494R (5′-ACGGCTACCTTGTTACGACTT-3′) as described earlier (Kalam et al., 2017a). The PCR products were electrophoresed in 1.5 % TAE-agarose gel, purified using Nucleospin® Extract II Kit (Macherey Nagel, Germany), and subjected to Sanger sequencing at First Base, Malaysia, using ABI PRISM 3730XI Genetic Analyzer (Applied Biosystems, USA). Resulting nucleotide sequences were analyzed using the BLAST® sequence analysis tool provided by the US National Center for Biotechnology Information (NCBI). Rhizobacterial isolates were identified based on the percentage of similarity with the top-hit taxon. The 16S rDNA sequences were deposited in GenBank, and accession numbers were obtained. All the sequences were aligned with MEGA6 (Molecular Evolutionary Genetics Analysis version 6.0) software (Tamura et al., 2013) for constructing a phylogenetic tree.
2.8. BOX-PCR analysis
For the genotypic fingerprinting of closely related Bacillus strains, BOX-PCR was performed using BOX-A1R primer (5′-CATACGGCAAGGCGACGCT-3′) as described by Versalovic et al. (1994). The PCR mixture contained 1μM of primer, 1X of PCR buffer with 1.5 mM MgCl2 (Sigma-Aldrich, USA), 10mM of each dNTP (Fermentas, USA) and 2 U of Taq DNA polymerase (Sigma-Aldrich, USA). The PCR was carried out for 30 cycles including an initial denaturation step for 5 min at 95 °C, denaturation for 1 min at 94 °C, annealing for 1 min at 50 °C, polymerization for 1 min at 72 °C and a final extension for 10 min at 72 °C. Amplicons were separated by electrophoresing on 2% TAE-agarose gel.
2.9. Plant growth in tissue culture
Surface sterilized tomato seeds (treated with 2 % sodium hypochlorite solution) were bacterized with culture suspensions of 1 × 108 colony forming units (CFU) mL−1 in 1 % sterile carboxymethyl cellulose (CMC). CBli and CMC were respectively used as positive and negative controls. The bacterized seeds were blot dried and transferred aseptically to plant tissue culture bottles containing half-strength Murashige and Skoog (MS) medium (Hi-media, India). The bottles were maintained in a plant growth chamber at 26 °C, 16h/8h photoperiod and 40 μmol m−2 s−1 light intensity for 15 days. The experiment was repeated four times with triplicates. After 15 days, three seedlings were randomly selected from each replication, and the root and shoot lengths were measured. The samples were dried to a constant weight in an oven to measure the dry weights.
For root colonization, 5-, 10- and 15-days old seedlings grown on MS medium were sampled. The roots were excised and serially diluted in 0.85% saline, and were grown onto M1 and M2 plates at 30 °C for 24–48 h followed by counting the colonies in the form of CFUs.
2.10. Plant growth in greenhouse
Pot experiments were conducted in the greenhouse with selected rhizobacterial isolates (TRS-2, 7, and 8) and controls (CBli and CMC). Before the start of the experiment, the physicochemical properties of the greenhouse soil were determined using Soil Test Kit (Hi-Media, India). Surface sterilized tomato seeds were bacterized and sown into plastic pots filled with greenhouse soil. The plants were maintained in a greenhouse (16 h/8 h photoperiod, 30 ± 2 °C, and 70 % relative humidity). The same volume of tap water was used to water the plants daily, without applying any other nutrients or PGPR inocula. The experiment was repeated three times with triplicates. Plant growth parameters (root length, shoot length and dry weight) were assessed at 20-, 40- and 60-days post-inoculation by randomly selecting and uprooting three plants from each treatment.
2.11. Statistical analysis
Data were analyzed using GraphPad Prism statistical software (Version 6.0) for significant mean differences via either one-way or two-way Analysis of Variance (ANOVA), respectively followed by Dunnett's or Bonferroni's post-hoc test for multiple mean comparisons, as per requirement. Depending on the comparisons made, Dunnett's test was used for multiple comparisons with the control mean, while Bonferroni's test was used for pairwise comparisons. Statistical significance was determined at the critical alpha level of 0.05.
3. Results
3.1. Isolation, selection, and characterization of rhizobacteria
A total of 60 distinct bacterial colonies were isolated from tomato rhizosphere on two minimal media using standard plating methods. Seven isolates were selected based on differential colony morphology, and designated as tomato rhizosphere (TRS) isolates with a number suffixed. Three isolates, TRS-1, TRS-3, and TRS-5, were isolated on M1, and four isolates, TRS-2, TRS-4, TRS-7, and TRS-8, were isolated on M2. Physiological and biochemical characteristics of the bacterial isolates are presented in Table 1.
Table 1.
Physiological and biochemical characterization of rhizobacterial isolates.
| Characteristics | TRS-1 | TRS-3 | TRS-5 | TRS-2 | TRS-4 | TRS-7 | TRS-8 |
|---|---|---|---|---|---|---|---|
| Gram stain | +ve | +ve | +ve | +ve | +ve | +ve | +ve |
| pH optimum (range) | 7.0 (6.5–8.0) | 7.0 (7.5–8.0) | 6.0 (7.0–8.5) | 7.0 (5.0–8.0) | 7.0 (6.0–8.0) | 7.0 (7.0–8.5) | 7.0 (7.0–8.5) |
| Temperature optimum (range) °C | 30 (30–40) | 30 (28–40) | 30 (30–40) | 37 (28–45) | 37 (28–45) | 28 (28–40) | 28 (28–40) |
| Motility | Motile | Motile | Motile | Motile | Motile | Non- motile | Non- motile |
| Nature |
Facultative Anaerobic |
Aerobic |
Aerobic |
Aerobic |
Aerobic |
Aerobic |
Aerobic |
|
Biochemical tests | |||||||
| Indole test | - | - | - | - | - | - | - |
| Methyl red test | + | - | - | - | - | - | - |
| Voges Proskauer test | + | + | + | + | + | + | + |
| Starch hydrolysis | + | + | - | - | - | + | + |
| H2S production test | + | - | - | - | - | - | - |
| Catalase test | + | + | + | + | + | + | + |
| Glucose utilization | + | + | + | + | + | + | + |
Physiological and biochemical characterization of rhizobacterial isolates was carried out under identical conditions. ‘+’, positive; ‘-’, negative result for the test.
3.2. PGP traits of rhizobacterial isolates
All the seven isolates viz., TRS-1, TRS-2, TRS-3, TRS-4, TRS-5, TRS-7, and TRS-8 were Gram positive and exhibited multiple PGP activities (Table 2). Phosphate solubilization was observed with the TRS-5 only. Out of seven isolates, TRS-1, TRS-3, and TRS-8 exhibited good zinc solubilization capability and phytase production. Siderophore, HCN, and IAA production was common for all the test isolates, while none of the isolates hydrolyzed chitin. Isolates TRS-1 and TRS-7 formed biofilm, while TRS-2, TRS-4, TRS-7, and TRS-8 produced ACC deaminase.
Table 2.
Characterization of rhizobacterial isolates for plant growth promoting activities.
| Isolate | PS | CP | SP | ZS | HP | IP | PP | BF | AD | Antibacterial |
Antifungal |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Xac | Xoo | C | F | ||||||||||
| TRS-1 | - | - | + | + | + | + | + | + | - | ++ | +++ | ++ | - |
| TRS-2 | - | - | ++ | - | ++ | + | - | - | + | - | - | - | - |
| TRS-3 | - | - | + | + | + | ++ | +++ | - | - | - | - | - | - |
| TRS-4 | - | - | +++ | - | + | ++ | - | - | + | - | - | - | - |
| TRS-5 | +++ | - | ++ | - | + | ++ | - | - | - | + | +++ | - | - |
| TRS-7 | - | - | +++ | - | ++ | +++ | - | + | + | - | - | - | - |
| TRS-8 | - | - | +++ | +++ | + | +++ | + | - | + | - | - | - | - |
PS- Phosphate solubilization; CP- Chitinase production; SP- Siderophore production; ZS- Zinc solubilization; HP- HCN production; IP- IAA production; PP- Phytase production; BF- Biofilm formation; AD- ACC deaminase activity. ‘+’, positive; ‘-’, negative result for the test. For phosphate and zinc solubilization, siderophore and chitinase production, antifungal and antibacterial assay: ‘+’, zone of clearance <0.2 mm; ‘++’, zone of clearance 0.2–0.4 mm; ‘+++’, zone of clearance >0.4 mm. For IAA production: ‘+’, absorbance <0.1; ‘++’, absorbance 0.1–0.3; ‘+++’, absorbance >0.3. Xac- Xanthomonas axonopodis pv. citri; Xoo- Xanthomonas oryzae pv. oryzae; C- Curvularia sp.; F- Fusarium sp.
All the seven rhizobacterial isolates were screened for their antagonistic ability against phytopathogenic fungi like Fusarium sp. and Curvularia sp., and phytopathogenic bacteria like Xanthomonas axonopodis pv. citri and X. oryzae pv. oryzae. None of the seven isolates was antagonistic to Fusarium sp. TRS-1 inhibited the growth of Curvularia sp., while TRS-1 and TRS-5 showed antibacterial activity against both the Xanthomonads (Xac and Xoo).
3.3. 16S rRNA gene-based molecular characterization
Amplicons of approximately 1500 bp were obtained after PCR amplification of the 16S rDNA. NCBI-BLAST analysis of the 16S rRNA gene sequences of all the test rhizobacterial isolates (GenBank accession nos. KJ572791, KJ572792, KJ572793, KJ631602, KJ631603, KJ631604, and KJ631605) indicated that all seven isolates are Bacillus spp., sharing 99–100% similarity with members of the genus Bacillus (Table 3).
Table 3.
Rhizobacterial isolates and their identity based on 16S rRNA gene sequence similarity.
| Isolate | Isolation medium | NCBI strain | Similarity (%) | GenBank Accession No. |
|---|---|---|---|---|
| TRS-1 | M1 | Bacillus licheniformis | 99 | KJ572792 |
| TRS-3 | M1 | Bacillus subtilis | 100 | KJ572793 |
| TRS-5 | M1 | Bacillus pumilus | 99 | KJ572791 |
| TRS-2 | M2 | Bacillus sp. | 99 | KJ631602 |
| TRS-4 | M2 | Bacillus sp. | 100 | KJ631603 |
| TRS-7 | M2 | Bacillus sp. | 99 | KJ631604 |
| TRS-8 | M2 | Bacillus sp. | 99 | KJ631605 |
Homology and phylogenetic identity of the rhizobacterial isolates were obtained by comparing the 16S rRNA gene sequence similarity with that of related strains available at the NCBI database.
3.4. BOX-PCR analysis
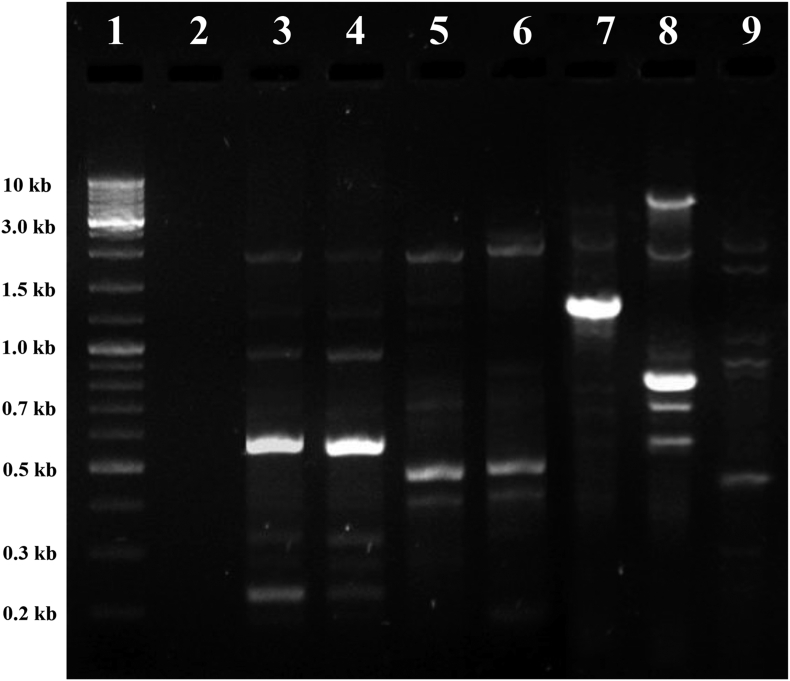
BOX-PCR amplification conditions were optimized to obtain fingerprints with distinct informative bands (Figure 1). BOX-PCR analysis allowed differentiation of individual strains, resulting in 6 different electrophoretic patterns or fingerprinting profiles for the seven isolates with isolates TRS-2 and TRS-4 sharing the same pattern. A large number of polymorphic bands of variable intensity were observed in the profiles, whose size ranged from 0.2–3.0 kb and remained consistent as the experiment was done in triplicate (n = 3) to confirm the reproducibility and stability. Although all the isolates showed a banding pattern typical of genus Bacillus, the intensity of a few bands was high in isolates TRS-1, TRS-2, TRS-3, and TRS-4.
Figure 1.
BOX-PCR patterns of the seven TRS isolates of Bacillus genus. (L–R) Lane 1, DNA molecular mass standard (Generuler 2-Log DNA Ladder, New England Biolabs, USA; size indicated in the left-hand margin); lane 2, no DNA control (sterile water); lanes 3–9, Bacillus spp. TRS-2, TRS-4, TRS-7, TRS-8, TRS-1, TRS-3 and TRS-5. Full, non-adjusted gel images are provided in Supplementary File 1.
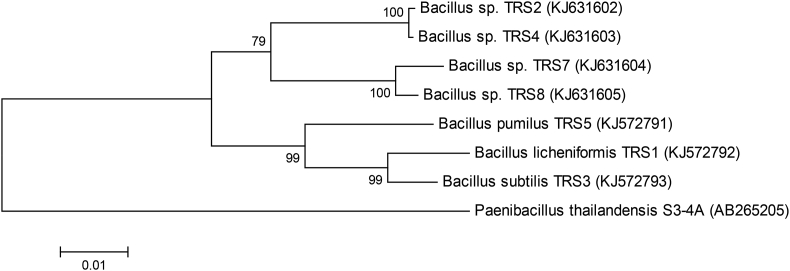
3.5. Phylogenetic tree
A phylogenetic tree, constructed based on the 16S rRNA gene sequences, indicated a considerable genetic homogeneity among the seven Bacillus isolates (Figure 2). The isolates could be divided into four groups, with the first, second, third, and fourth groups having 2, 2, 1, and 2 isolates, respectively.
Figure 2.
Phylogenetic tree of the seven TRS isolates based on 16S rRNA gene sequences. Genetic relatedness between the seven TRS isolates was inferred from Neighbor-Joining tree based evolutionary analyses with a Bootstrap value of 1000, using Paenibacillus thailandensis S3-4A (GenBank accession no. AB265205) as the outgroup. The phylogenetic tree was constructed using MEGA6 software. Numbers at nodes represent the percentage of replicate trees in which the associated taxa clustered together. The tree is drawn to scale, and the scale bar represents 0.01 substitutions per nucleotide position.
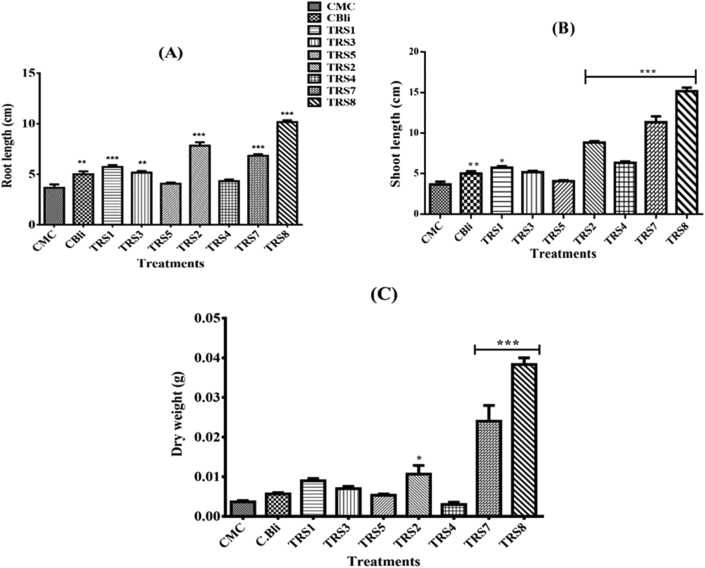
3.6. Plant growth promotion in tissue culture
The overall growth response of tomato seedlings to seed bacterization by the Bacillus isolates was assessed in MS medium (Figure 3). Rhizobacteria treated tomato plants exhibited variations in root length (Figure 3A). The response to seed bacterization with TRS-4 and TRS-5 did not vary significantly in comparison to both the controls. The remaining five treatments, along with the commercial isolate (CBli), significantly increased tomato root length in comparison to the CMC control. TRS-8 notably increased the root length more than the other isolates. Shoot length response to bacterial isolates differed considerably (Figure 3B). Isolates TRS-2, TRS-4, TRS-7, and TRS-8 improved shoot length more than the other isolates. Plants arising from TRS-8, followed by TRS-7, TRS-2, and TRS-4 bacterized seeds showed significant improvement in shoot length as compared to control (CMC) and CBli.
Figure 3.
Effect of seven rhizobacterial isolates, a commercial Bacillus licheniformis strain (CBli) and a CMC control on tomato plants in MS medium after 15 days of treatment: root length (A), shoot length (B), and dry weight (C). Values represent mean (n = 3), and the vertical lines represent ±standard error of the mean. For each of the growth response, i.e., root length, shoot length and dry weight, statistical analysis has been performed using one-way ANOVA followed by Dunnett's post-hoc test for multiple mean comparisons. ∗∗∗ highly significant, ∗∗ moderately significant, ∗ less significant compared to CMC control (p < 0.05).
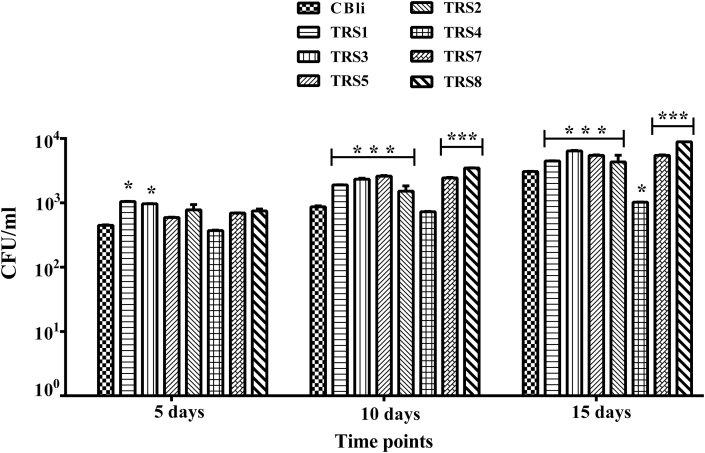
An increase in the dry weight due to seed bacterization varied significantly (Figure 3C) on treatment with TRS-2, TRS-7, and TRS-8. Isolate TRS-8, followed by TRS-7, significantly enhanced the plant dry weight in comparison to CMC control. A comparison of root colonization by the test rhizobacterial isolates was also done (Figure 4). All the isolates colonized tomato roots. There was a significant increase in root colonization from 5 days with the isolates TRS-1 and TRS-3. At 10 and 15 days, all the isolates colonized roots extensively except TRS-4. TRS-8 and TRS-7 were better colonizers of the root compared with CBli.
Figure 4.
Comparison of root colonization by test rhizobacterial isolates. Values represent mean (n = 3), and the vertical lines represent ±standard error of the mean. Statistical analysis was performed using two-way ANOVA for each of the test bacterial isolate against three different time points, followed by Bonferroni's post-hoc test for multiple mean comparisons. ∗∗∗ highly significant, ∗∗ moderately significant, ∗ less significant compared to the CBli positive control (p < 0.05).
3.7. Plant growth promotion in greenhouse
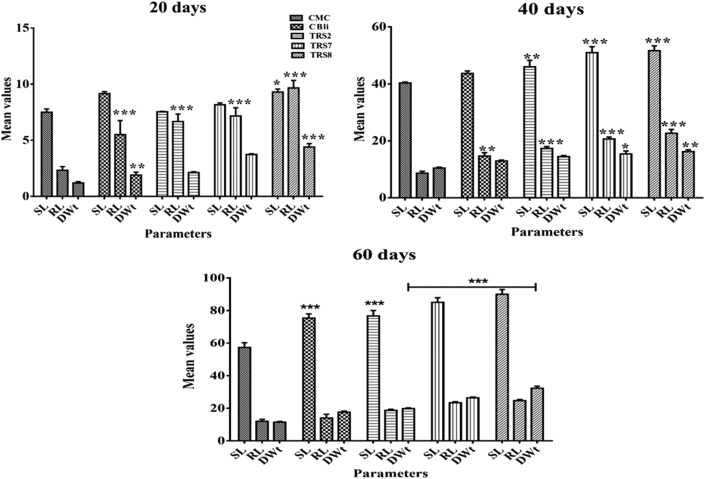
The greenhouse soil used in this study had the following physicochemical characteristics: red colour, sandy texture, 7.5 pH, 1–1.5% oxidizable organic carbon, 10–15 kg ha−1 ammoniacal nitrogen, 10–20 kg ha−1 nitrate nitrogen, 56–73 kg ha−1 available phosphorus, and 112–280 kg ha−1 available potassium. There was a gradual increase in shoot length, root length and dry weight of the bacterized tomato plants at 20-, 40- and 60-days post-inoculation (Figure 5). The isolates TRS-8 and TRS-7 produced significant responses on tomato plant growth parameters after 20, 40, or 60 days with respect to the CMC control.
Figure 5.
Influence of three rhizobacterial isolates (selected) along with a commercial strain (CBli) on tomato plant responses in greenhouse (20d, 40d and 60d): root length (RL in cm), shoot length (SL in cm), and dry weight (DWt in g). Values represent mean (n = 3), and the vertical lines represent ±standard error of the mean. For each of the growth response, i.e., root length, shoot length and dry weight with or without the isolates, statistical analysis has been performed using two-way ANOVA followed by Bonferroni's post-hoc test for multiple mean comparisons. ∗∗∗ highly significant, ∗∗ moderately significant, ∗ less significant compared to CMC control (p < 0.05).
4. Discussion
The growth and development of plants often depend on the type of plant-microbe interactions functioning in the rhizosphere. Several cultivation-dependent studies revealed the occurrence of multiple species of Bacillus in the soil and rhizosphere which were reported as PGPR (Kumar et al., 2012; Singh et al., 2014; Mumtaz et al., 2017; Akinrinlola et al., 2018) as they promoted plant growth and/or suppressed phytopathogens. Based on the 16S rRNA gene sequences, the seven tomato rhizobacterial isolates, in this study, had 99–100% similarity with the genus Bacillus. Identification of Bacillus species exclusively based on the 16S rRNA gene sequences was considered insufficient (Lima-Bittencourt et al., 2007). BOX-PCR fingerprinting is a well-documented and widely employed phylogenetic informative tool for molecular typing of various bacteria (Marques et al., 2008; Zhu et al., 2014). Köberl et al. (2011) showed BOX-PCR fingerprinting to be an effective tool to explore intraspecies diversity within Bacillus populations. In this study, BOX-PCR generated distinctive electrophoretic patterns among different Bacillus strains with clear and identifiable bands. The presence of similar or variable bands enabled differentiation of the Bacillus isolates.
To improve the screening approaches for selecting effective PGPR strains, identification of traits predicting PGP will be useful. Most of the seven Bacillus isolates exhibited multiple PGP characteristics. Phosphate solubilization by rhizobacteria promotes plant growth and yields (Lyngwi et al., 2016). Some species of bacteria, including Bacillus spp., possess the ability to mineralize and solubilize organic and inorganic phosphorus in the soil for quick access to the plant (Barea and Richardson, 2015). Microbial phytases, specially produced by Bacillus spp., were studied due to their PGP effects and diverse agrobiotechnological applications (Kumar et al., 2013; Sanguin et al., 2016). Besides solubilizing phytate phosphorous, extracellular phytases produced by Bacillus spp. release essential mineral nutrients like Ca2+, Zn2+, and Fe2+ from chelate-forming phytates (Sansinenea, 2019). Only one isolate (TRS-5) solubilized mineral phosphate effectively, and three of them (TRS-1, TRS-3, and TRS-8) produced phytases.
IAA and ethylene are growth regulators that regulate different stages of plant growth (Etesami et al., 2015). All seven isolates produced IAA. Highest IAA produced by TRS-7 and TRS-8 might play a role in enhancing the growth of tomato plants. The auxin IAA is known to strongly affect root growth and architecture. Exogenous IAA of rhizobacterial origin can increase root length and biomass, and enhance plant growth by regulating the expression of host genes related to auxin response, defense, hormone and cell wall synthesis (Ruzzi and Aroca, 2015; Backer et al., 2018). The isolates TRS-2, TRS-4, TRS-7 and TRS-8 produced ACC deaminase that breaks down the ethylene precursor ACC into ammonia and α-ketobutyrate, alleviating ethylene stress in plants and delaying senescence (Etesami et al., 2015). Bacteria can further metabolize these end products for their growth. Bacillus spp. exhibiting ACC deaminase activity are reported to be halotolerant and can promote plant growth under salinity stress conditions (Santoyo et al., 2019; Orozco-Mosqueda et al., 2020).
Bacillus spp. enhance plant growth and yield by solubilizing insoluble zinc compounds and increasing bioavailability of zinc in the soil (Mumtaz et al., 2017). TRS-1, TRS-3, and TRS-8 solubilized zinc with TRS-8 being the best zinc solubilizer. The presence of iron-chelating siderophore producing microorganisms in the rhizosphere makes iron available to the plant, aids in plant growth under iron-deficient conditions while limiting iron availability for phytopathogens (Saha et al., 2016; Sansinenea, 2019). Siderophore and HCN were produced by all the seven isolates. TRS-4, TRS-7, and TRS-8 produced more of siderophore, while TRS-2 and TRS-7 produced more of HCN. The PGPR that produce HCN suppress plant pathogens and reduce the severity of disease and also indirectly increase phosphorous availability through sequestration and metal chelation (Rijavec and Lapanje, 2016; Backer et al., 2018). All the seven isolates were positive for the Voges Proskauer test, indicating the production of acetylmethyl carbinol (acetoin) from glucose fermentation. Volatile organic compounds like acetoin produced by Bacillus strains were reported to increase leaf surface area and induce systemic resistance in Arabidopsis thaliana (Ryu et al., 2003).
Bacillus spp. exhibit remarkable antibacterial and/or antifungal activity against different phytopathogens (Kumar et al., 2012) that make them suitable biocontrol agents in agriculture. They are reported to be reservoirs of several biologically active molecules, including those with potential antifungal activity (Sansinenea, 2019). Rhizospheric Bacilli can aid in suppressing several soil-borne phytopathogens (Singh et al., 2014; Cao et al., 2018). In the present study, antagonistic activity was observed with TRS-1 and TRS-5 against both Xanthomonas axonopodis pv. citri and X. oryzae pv. oryzae. TRS-1 exhibited antifungal activity against Curvularia sp. The antagonistic effect of the test bacterial isolates could be useful for an effective biocontrol and other plant growth promotion activities.
Extensive root colonization is a prerequisite for the PGPR to establish successfully in the rhizosphere and rhizoplane (Zhou et al., 2016). The isolates TRS-8 and TRS-7 were better root colonizers in comparison to other test isolates. Root colonization by PGPR is often enhanced by the formation of biofilms on root surfaces that facilitates retention of moisture and protects plant roots from harmful microorganisms (Kalam et al., 2017b). Paenibacillus polymyxa and Bacillus subtilis colonize Arabidopsis roots by forming biofilms and render biocontrol activity (Timmusk et al., 2005; Beauregard et al., 2013; Vlamakis et al., 2013). Two isolates TRS-1 and TRS-7, from tomato rhizosphere formed biofilms. As active colonization of tomato roots by Bacillus subtilis PTS-394 (Qiao et al., 2017) promoted growth and suppressed soil pathogens with no durable impact on the tomato rhizosphere microbial community composition, a combination of factors may be essential for PGPR to be successful.
Seed bacterization was employed to monitor the effects of rhizobacterial strains on plant growth (Vaikuntapu et al., 2014; Kalam et al., 2017a). The response of tomato plants to seed bacterization in terms of shoot length, root length and dry weight varied significantly. PGPR enhance plant growth and yield by facilitating the uptake of mineral nutrients, synthesizing several phytohormones, and protecting plants from diverse phytopathogens. The genus Bacillus represents one of the most diverse Bacilli genera commonly used as bioinoculants to promote plant growth. Akinrinlola et al. (2018) identified 12 Bacillus strains promoting the growth of corn, wheat, and soybean. The strains exhibited multiple PGP traits, including phosphate solubilization, nitrogen fixation and IAA production. Similarly, wheat rhizobacteria were screened for multiple in vitro PGP attributes and were evaluated under controlled conditions in pot experiments (Rana et al., 2011).
The use of beneficial, environmentally safe microbial inoculants has been regarded as an alternative to synthetic agrochemicals. Bacillus species, present in the immediate vicinity of crop rhizospheres, are most widely used as bioinoculants. The present study selected TRS-8 and TRS-7 as potential PGPR isolates from tomato rhizosphere that can enhance plant growth and fertility. Further characterization of these isolates is required to assess their suitability as effective bioinoculants.
Declarations
Author contribution statement
Sadaf Kalam: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Anirban Basu: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Appa Rao Podile: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by the Department of Science and Technology (DST), Government of India, in the form of Women Scientist Scheme-A (WOS-A) project fellowship to SK [Grant no. SR/WOS-A/LS-294/2012(G)].
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors acknowledge University of Hyderabad – Department of Biotechnology, Centre for Research and Education in Biology and Biotechnology (UoH-DBT-CREBB), Department of Science and Technology, Government of India, Funds for Infrastructure in Science and Technology (DST-FIST), Department of Plant Sciences and Central Instrumentation Laboratory of University of Hyderabad for the infrastructural support. The authors gratefully acknowledge Prof. K. Satya Prasad, Department of Botany, Osmania University, Hyderabad, and Dr. Ramesh V. Sonti, CSIR- Centre for Cellular and Molecular Biology, Hyderabad, for kindly providing the phytopathogenic fungal and bacterial strains used in this study. SK and AB, respectively, acknowledge UoH-DBT-CREBB and University Grants Commission (UGC) for research fellowships.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary File 1
References
- Ahmad F., Ahmad I., Khan M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008;163:173–181. doi: 10.1016/j.micres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Akinrinlola R.J., Yuen G.Y., Drijber R.A., Adesemoye A.O. Evaluation of Bacillus strains for plant growth promotion and predictability of efficacy by in vitro physiological traits. Int. J. Microbiol. 2018;2018:1–11. doi: 10.1155/2018/5686874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer R., Rokem J.S., Ilangumaran G., Lamont J., Praslickova D., Ricci E., Subramanian S., Smith D.L. Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018;9:1473. doi: 10.3389/fpls.2018.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barea J.M., Richardson A.E. Phosphate mobilisation by soil microorganisms. In: Lugtenberg B., editor. Principles of Plant-Microbe Interactions. Springer International Publishing; Heidelberg, Switzerland: 2015. pp. 225–234. [Google Scholar]
- Beauregard P.B., Chai Y., Vlamakis H., Losick R., Kolter R. Bacillus subtilis biofilm induction by plant polysaccharides. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Pi H., Chandrangsu P., Li Y., Wang Y., Zhou H., Xiong H., Helmann J.D., Cai Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018;8:4360. doi: 10.1038/s41598-018-22782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappuccino J.C., Sherman N. tenth ed. Pearson; New York: 2014. Microbiology: a Laboratory Manual. [Google Scholar]
- Chen Y., Yan F., Chai Y., Liu H., Kolter R., Losick R., Guo J.H. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 2013;15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S.N., Dutta S., Kondreddy A., Chilukoti N., Pullabhotla S.V., Vadlamudi S., Podile A.R. Plant growth-promoting chitinolytic Paenibacillus elgii responds positively to tobacco root exudates. J. Plant Growth Regul. 2010;29(4):409–418. [Google Scholar]
- Demirkan E., Baygın E., Usta A. Screening of phytate hydrolysis Bacillus sp. isolated from soil and optimization of the certain nutritional and physical parameters on the production of phytase. Turk. J. Bioch. 2014;39(2):206–214. [Google Scholar]
- Dutta S., Podile A.R. Plant Growth Promoting Rhizobacteria (PGPR): the bugs to debug the root zone. Crit. Rev. Microbiol. 2010;36(3):232–244. doi: 10.3109/10408411003766806. [DOI] [PubMed] [Google Scholar]
- Etesami H., Alikhani H.A., Hosseini H.M. Indole-3-acetic acid and 1-aminocyclopropane-1-carboxylate deaminase: bacterial traits required in rhizosphere, rhizoplane and/or endophytic competence by beneficial bacteria. In: Maheshwari D.K., editor. Bacterial Metabolites in Sustainable Agroecosystem. Springer International; Switzerland: 2015. pp. 183–258. [Google Scholar]
- Kalam S., Basu A., Ankati S. Plant root-associated biofilms in bioremediation. In: Ahmad I., Husain F.M., editors. Biofilms in Plant and Soil Health. John Wiley & Sons; Hoboken, New Jersey, USA: 2017. pp. 337–355. [Google Scholar]
- Kalam S., Das S.N., Basu A., Podile A.R. Population densities of indigenous Acidobacteria change in the presence of plant growth promoting rhizobacteria (PGPR) in rhizosphere. J. Basic Microbiol. 2017;57(5):376–385. doi: 10.1002/jobm.201600588. [DOI] [PubMed] [Google Scholar]
- Kishore G.K., Pande S., Podile A.R. Phylloplane bacteria increase seedling emergence, growth and yield of field-grown groundnut (Arachis hypogaea L.) Lett. Appl. Microbiol. 2005;40:260–268. doi: 10.1111/j.1472-765X.2005.01664.x. [DOI] [PubMed] [Google Scholar]
- Köberl M., Müller H., Ramadan E.M., Berg G. Desert farming benefits from microbial potential in arid soils and promotes diversity and plant health. PloS One. 2011;6(9) doi: 10.1371/journal.pone.0024452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Dubey R.C., Maheshwari D.K. Bacillus strains isolated from rhizosphere showed plant growth promoting and antagonistic activity against phytopathogens. Microbiol. Res. 2012;167(8):493–499. doi: 10.1016/j.micres.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Kumar V., Singh P., Jorquera M.A., Sangwan P., Kumar P., Verma A.K. Isolation of phytase-producing bacteria from Himalayan soils and their effect on growth and phosphorus uptake of Indian mustard (Brassica juncea) World J. Microbiol. Biotechnol. 2013;29(8):1361–1369. doi: 10.1007/s11274-013-1299-z. [DOI] [PubMed] [Google Scholar]
- Lima-Bittencourt C.I., Astolfi-Filho S., Chartone-Souza E., Santos F.R., Nascimento A.M.A. Analysis of Chromobacterium sp. natural isolates from different Brazilian ecosystems. BMC Microbiol. 2007;7:58. doi: 10.1186/1471-2180-7-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyngwi N.A., Nongkhlaw M., Kalita D., Joshi S.R. Bioprospecting of plant growth promoting Bacilli and related genera prevalent in soils of pristine sacred groves: biochemical and molecular approach. PloS One. 2016;11(4) doi: 10.1371/journal.pone.0152951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A.S., Marchaison A., Gardan L., Samson R. BOX-PCR-based identification of bacterial species belonging to Pseudomonas syringae: P. viridiflava group. Genet. Mol. Biol. 2008;31(1):106–115. [Google Scholar]
- Mumtaz M.Z., Ahmad M., Jamil M., Hussain T. Zinc solubilizing Bacillus spp. potential candidates for biofortification in maize. Microbiol. Res. 2017;202:51–60. doi: 10.1016/j.micres.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Orozco-Mosqueda M.C., Glick B.R., Santoyo G. ACC deaminase in plant growth-promoting bacteria (PGPB): an efficient mechanism to counter salt stress in crops. Microbiol. Res. 2020;235:126439. doi: 10.1016/j.micres.2020.126439. [DOI] [PubMed] [Google Scholar]
- Parray J.A., Jan S., Kamili A.N., Qadri R.A., Egamberdieva D., Ahmad P. Current perspectives on plant growth-promoting rhizobacteria. J. Plant Growth Regul. 2016;35(3):877–902. [Google Scholar]
- Qiao J., Yu X., Liang X., Liu Y., Borriss R., Liu Y. Addition of plant-growth-promoting Bacillus subtilis PTS-394 on tomato rhizosphere has no durable impact on composition of root microbiome. BMC Microbiol. 2017;17:131. doi: 10.1186/s12866-017-1039-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana A., Saharan B., Joshi M., Prasanna R., Kumar K., Nain L. Identification of multi-trait PGPR isolates and evaluating their potential as inoculants for wheat. Ann. Microbiol. 2011;61:893–900. [Google Scholar]
- Rijavec T., Lapanje A. Hydrogen cyanide in the rhizosphere: not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 2016;7:1785. doi: 10.3389/fmicb.2016.01785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzzi M., Aroca R. Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 2015;196:124–134. [Google Scholar]
- Ryu C.M., Farag M.A., Hu C.H., Reddy M.S., Wei H.X., Paré P.W., Kloepper J.W. Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2003;100(8):4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha M., Sarkar S., Sarkar B., Sharma B.K., Bhattacharjee S., Tribedi P. Microbial siderophores and their potential applications: a review. Environ. Sci. Pollut. Res. 2016;23:3984–3999. doi: 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- Sanguin H., Wilson N.L., Kertesz M.A. Assessment of functional diversity and structure of phytate-hydrolysing bacterial community in Lolium perenne rhizosphere. Plant Soil. 2016;401:151–167. [Google Scholar]
- Sansinenea E. Bacillus spp.: as plant growth-promoting bacteria. In: Singh H.B., editor. Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms. Springer; Singapore: 2019. pp. 225–237. [Google Scholar]
- Santoyo G., Equihua A., Flores A., Sepulveda E., Valencia-Cantero E., Sanchez-Yañez J.M., Morales L.R., Govindappa M., de los Santos-Villalobos S. Plant growth promotion by ACC deaminase-producing Bacilli under salt stress conditions. In: Islam M.T., editor. Bacilli and Agrobiotechnology: Phytostimulation and Biocontrol. Springer; Cham: 2019. pp. 81–95. [Google Scholar]
- Saravanan V.S., Subramoniam S.R., Raj S.A. Assessing in vitro solubilization potential of different zinc solubilizing bacterial (ZSB) isolates. Braz. J. Microbiol. 2004;35:121–125. [Google Scholar]
- Sharma A.D., Singh J. A nonenzymatic method to isolate genomic DNA from bacteria and actinomycete. Anal. Biochem. 2005;337(2):354–356. doi: 10.1016/j.ab.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Sharma S., Kaur M. Antimicrobial activities of rhizobacterial strains of Pseudomonas and Bacillus strains isolated from rhizosphere soil of carnation (Dianthus caryophyllus cv. Sunrise) Indian J. Microbiol. 2010;50:229–232. doi: 10.1007/s12088-010-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.K., Kumar D.P., Singh P., Solanki M.K., Srivastava S., Kashyap P.L., Kumar S., Srivastava A.K., Singhal P.K., Arora D.K. Multifarious plant growth promoting characteristics of chickpea rhizosphere associated Bacilli help to suppress soil-borne pathogens. Plant Growth Regul. 2014;73:91–101. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S., Grantcharova N., Wagner E.G.H. Paenibacillus polymyxa invades plant roots and forms biofilms. Appl. Environ. Microbiol. 2005;71(11):7292–7300. doi: 10.1128/AEM.71.11.7292-7300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaikuntapu P.R., Dutta S., Samudrala R.B., Rao V.R.V.N., Kalam S., Podile A.R. Preferential promotion of Lycopersicon esculentum (tomato) growth by plant growth promoting bacteria associated with tomato. Indian J. Microbiol. 2014;54(4):403–412. doi: 10.1007/s12088-014-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic J., Schneider M., De Bruijn F.J., Lupski J.R. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell Biol. 1994;5(1):25–40. [Google Scholar]
- Vlamakis H., Chai Y., Beauregard P., Losick R., Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013;11(3):157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Huang X.F., Chaparro J.M., Badri D.V., Manter D.K., Vivanco J.M., Guo J. Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant Soil. 2016;401:259–272. [Google Scholar]
- Zhu L., Xu H., Zhang Y., Fu G., Wu P.Q., Li Y. BOX-PCR and PCR-DGGE analysis for bacterial diversity of a naturally fermented functional food (Enzyme®) Food Biosci. 2014;5:115–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1