Abstract
Cytosolic 80S ribosomes contain proteins of the mature cytosolic ribosome (r-proteins) as well as proteins with roles in ribosome biogenesis, protein folding or modification. Here, we refined the core r-protein composition in Arabidopsis thaliana by determining the abundance of different proteins during enrichment of ribosomes from cell cultures using peptide mass spectrometry. The turnover rates of 26 40S subunit r-proteins and 29 60S subunit r-proteins were also determined, showing that half of the ribosome population is replaced every 3–4 days. Three enriched proteins showed significantly shorter half-lives; a protein annotated as a ribosomal protein uL10 (RPP0D, At1g25260) with a half-life of 0.5 days and RACK1b and c with half-lives of 1–1.4 days. The At1g25260 protein is a homologue of the human Mrt4 protein, a trans-acting factor in the assembly of the pre-60S particle, while RACK1 has known regulatory roles in cell function beyond its role in the 40S subunit. Our experiments also identified 58 proteins that are not from r-protein families but co-purify with ribosomes and co-express with r-proteins; 26 were enriched more than 10-fold during ribosome enrichment. Some of these enriched proteins have known roles in translation, while others are newly proposed ribosome-associated factors in plants. This analysis provides an improved understanding of A. thaliana ribosome protein content, shows that most r-proteins turnover in unison in vivo, identifies a novel set of potential plant translatome components, and how protein turnover can help identify r-proteins involved in ribosome biogenesis or regulation in plants.
Keywords: Arabidopsis thaliana, proteomics, ribosomes
Introduction
The ribosome uses information encoded by messenger RNA (mRNA) to drive peptide-bond formation resulting in protein synthesis. All ribosomes consist of two subunits, a large and small, and each contain ribosomal RNAs (rRNAs) and ribosomal proteins (r-proteins). The 80S eukaryotic ribosome is larger and more complicated than the 70S prokaryotic ribosome due to the presence of additional r-proteins and rRNAs [1]. The composition, structure and organisation of r-proteins and their distribution between small and large subunits have been studied in detail in bacteria (Escherichia coli) [2] yeast (Saccharomyces cerevisiae) [3], Tetrahymena thermophila [4], humans (Homo sapiens) [5] and plants (Arabidopsis thaliana) [6–8].
Based on gene annotation, the 80S cytosolic ribosome in Arabidopsis thaliana (Arabidopsis) is understood to contain 81 different r-proteins and four rRNAs, including 33 proteins in the small subunit and 48 in the large subunit [6]. There is a high degree of conservation of ribosomal proteins between Arabidopsis and mammals with the exception of the acidic phosphoprotein (RPP) family, also known as acidic stalk P3 (RPP3), which only exists in plants [8]. Many of the Arabidopsis r-proteins are encoded by multiple genes. In a recent survey, 102 Arabidopsis genes were predicted to encode the 33 r-proteins of the small subunit and 146 genes to encode the 48 r-proteins of the large subunit; on average there are three predicted Arabidopsis genes for each r-protein type [8]. In comparison, yeast has 137 genes encoding 78 r-proteins [9], humans have 85 genes encoding 80 r-proteins [10], and chlamydomonas has 79 genes encoding 78 r-proteins [11]. However, in experimental assessments of r-protein content in isolated Arabidopsis ribosomes there are varying sets of r-proteins reported, indicating technical issues with different approaches or potentially variant ribosome contents from different Arabidopsis tissue sources [7,8,12–15]. Each of these reports also identified proteins in ribosome preparations that were not annotated as members of r-protein families. However, each study had limited experimental strategies to determine if such proteins were simply contaminants of ribosome preparations in the different plant tissues used or if they were real ribosome-associated proteins in vivo.
Here, we refined the core ribosome protein composition by determining the enrichment of r-proteins during the purification of ribosomes from Arabidopsis cell cultures. This protein list was further refined by determining the co-degradation and co-synthesis rates of r-proteins during cell growth. We identified many proteins that are not from r-protein families but clearly associate and co-purify with Arabidopsis cytosolic ribosomes. This analysis has provide a more robust understanding of Arabidopsis ribosome content, multiple lines of evidence to justify protein inclusion in the core cytosolic 80S r-protein set, a novel set of potential translatome components and evidence that some ribosomal proteins turnover much more rapidly that the ribosome as a whole, indicating potential roles for these proteins in ribosome biogenesis and/or regulation.
Materials and methods
Arabidopsis cell culture
Arabidopsis cell suspension (ecotype Landsberg erecta) was cultured in growth medium consisting of 1× Murashige and Skoog (MS) Modified Basal Salt Mixture (Phyto Technology M524) without vitamins, 3% w/v sucrose (Ajax Chemical A530), 0.5 mg/l naphthaleneacetic acid (Sigma–Aldrich 15165-79-4), 0.05 mg/l kinetin (Sigma–Aldrich K0753) and with a pH of 5.8. Cells were incubated at 22°C with orbital shaking at 100–120 rpm and under constant light (100 µmol m−2 s−1). Cultures were maintained in 250 ml Erlenmeyer flasks by the inoculation of 20 ml of 7 days old cells into 100 ml of fresh growth medium.
15N labelling of Arabidopsis cell culture
Seven-day-old Arabidopsis cell culture was transferred from non-labelled media (14N) to media without nitrogen, and the cells washed three times to eliminate 14N in the media. Washed cells were transferred to heavy media (15N) containing two nitrogen sources for optimal growth [16], namely 1.65 g/l 15NH415NO3 (Sigma–Aldrich 299278) and 1.9 g/l K15NO3 (Sigma–Aldrich 335134). To study protein turnover rate, the labelled Arabidopsis cell culture was collected by vacuum filtration after the first day, third day and fifth day following transfer into new media, then stored at −80°C until use.
Ribosome purification from cell culture
To isolate 80S cytosolic ribosomes from Arabidopsis cell culture, 10 g of 7-day-old cells were frozen in liquid N2 and ground in a pre-cooled mortar with a pestle. The powder was transferred into extraction buffer containing 0.45 M mannitol (Ajaxs Chemical A310), 30 mM HEPES (Sigma–Aldrich H3375), 100 mM KCl (Sigma–Aldrich P9333), 20 mM MgCl2·6H2O (Ajax Finechem A296), 0.5% (w/v) polyvinyl pyrrolidone-40 (Sigma–Aldrich 9003-39-8) and 0.5% (w/v) bovine serum albumin (Bovogen biologicals BSAS1.0) with the addition of 20 mM cysteine-L (Sigma–Aldrich A-9165) prior to grinding and pH adjusted to 7.5, followed by using an Ultra-Turrax for homogenisation of the mixture. The homogenised solution was filtered through two layers of miracloth (Merk Millipore, Darmstadt, Germany). The filtrate was transferred into a pre-cooled, sterilised, 25 ml Beckman Coulter centrifuge tubes and centrifuged at 30 000×g for 20 min at 4°C. The supernatant (low purity ribosome extract) was transferred into 1.5 M sucrose (Ajaxs chemical A530) dissolved in a mixture of 2× ribosome resuspension buffer consisting of 30 mM HEPES, 100 mM KCl and 20 mM MgCl2·6H2O, pH adjusted to 7.5 and centrifuged at 180 000 g for 90 min. After the first ultra-centrifugation, the pellet (partially purified ribosome extract) was resuspended in 2 ml of a solution that consisted of 5 mM DTT (Sigma–Aldrich D0632) and 1.5 M sucrose. The resuspended sample was mixed with 2× ribosome extraction buffer followed by second ultra-centrifugation at 180 000 g for 2.5 h. Finally, the purified ribosome pellet was resuspended in 1× ribosome extraction solution and stored in −80°C for later use. Ribosomal protein content was quantified using Amido black against a standard solution of bovine serum albumin.
Precipitation, denaturation and digestion of ribosomal proteins
Ribosomal extracts (50 µg protein) were precipitated in cold acetone and incubated for 1 h at −80°C and centrifuged for 20 min at 20 800×g at 4°C. The protein pellet was denatured, reduced and alkylated using 1.5 M urea, 10 mM DTT and 25 mM iodoacetamide. Denaturated protein was digested using trypsin (Invitrogen MSI0015) and incubated overnight at 37°C and the reaction terminated by acidification of the mixture with 1% formic acid. Peptides from digested protein was purified and concentrated before mass spectrometry analysis using micro spin column silica C18 in a reverse phase column.
LC–MS/MS and data analysis
Mass spectrometry analysis was performed with an Thermo Scientific Orbitrap Fusion mass spectrometer. The raw data files (. raw) files were converted to .mzML files by using open source MS convert software. Then the .mzML files were converted to .mgf files via the Convert mZ[X]ML tool in the trans-proteomic pipeline v. 4.8 (TPP). The Mascot search algorithm (Matrix Science) was used to search tandem mass spectra against proteins from the TAIR 10 release of Arabidopsis (https://www.arabidopsis.org/) database to convert the .mgf files to both .dat and .csv files. Search parameters were: variable modifications: carbamidomethyl (C) and oxidation (M), monoisotopic, trypsin selected as digesting enzyme, and maximum missed cleavages set to one. Moreover, the peptide charges chosen as (+2, +3 and +4), 13C error set to one and peptide tolerance selected as ±50 ppm. Finally, MS/MS ion searches were activated, and MS/MS tolerance selected as ±0.6 Da. Mascot results were exported as a .dat file, and then converted to pep.xml using the PepXML tool in TPP for determination of peptide and protein probabilities (protein FDR <1%). Pep.XML files were then further converted to interact.pep.XML by using the peptide prophet tool in TPP. Labelled peptide fractions (LPFs) were estimated from the interact.pep.xml and the mzML files by an in-house solution as described previously [17], rewritten in R and including an extra filter. The heavy population, calculated by substracting the natural abundance envelope from the observed isotopes as described in Nelson et al. [17], was also filtered by calculating the correlation coefficient between the estimated heavy distribution as per NNLS result and the observed data. We found this correlation filter to more accurately remove data with poor signal-to-noise or contamination by other peptides than the fit of a Gaussian distribution. The minimum correlation to pass the filter was set to 0.5. Fifteen seconds was chosen as retention time tolerance, m/z tolerance was 10 ppm, and 95% selected as maximum label enrichment. For 15N labelled protein studies nitrogen (N) was chosen as the labelled element and 15 as the isotope number. For each sample that was analysed by this pipeline a .tsv file was generated. Protein level LPFs were obtained from these .tsv files in conjunction with TPP .prot.xml files by taking the median of peptide LPFs for each protein. The mass spectrometry proteomics data for identification analyses and protein labelling and turnover analyses have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012839. MS1 quantiation was performed using the Skyline MS1 filtering workflow. Peak integrations were manually inspected and areas of monoisotopic peaks exported for statistical analysis (t-test). Raw data files were searched against araport11 peptide sequence database with Comet under the Crux toolkit v3.2 [18]. Results were post processed by Percolator and normalised spectral abundance factors (NSAF) was calculated by the ‘spectral-counts’ function of Crux. The percentage of total fraction NSAF associated with r-proteins was calculated using the r-protein list in Supplemental Table S2.
Bioinformatics analysis
To check if the non-core ribosomal proteins identified in this study were related with core ribosomal proteins in previous studies we investigated seven score summaries from Arabidopsis String-db (https://string-db.org/cgi/download.pl) [19]. This data scores the interaction between each pair of genes as summarised from transcript studies, protein : protein interaction studies, etc. We calculated interaction scores for each of the genes whose protein product was identified in the high enrichment samples by calculating an average of its interaction scores with a core set of bonafide ribosomal proteins as judged by their inclusion in previous Arabidopsis ribosomal studies.
Statistical analysis
The analysis of data including one-way and two-way variance analysis (ANOVA) and the Kolmogorov–Smirnov test for comparison of samples using XLSTAT version 2018.5 (<0.05 was considered significant). For ANOVAs, 0.0001 was set as tolerance and a confidence interval of 95% was chosen. For model selection, we chose the best model based on adjusted R2, and number 2 was selected as minimum and maximum variables. For the Kolmogorov–Smirnov (K-S) test, the criteria were selected as follows: Alternative hypothesis F1(x) ≠ F2(x), 5% significant level, hypothesised difference (D) = 0 and an asymptotic P-value.
Results
Enrichment proteomics of 80S cytosolic r-proteins
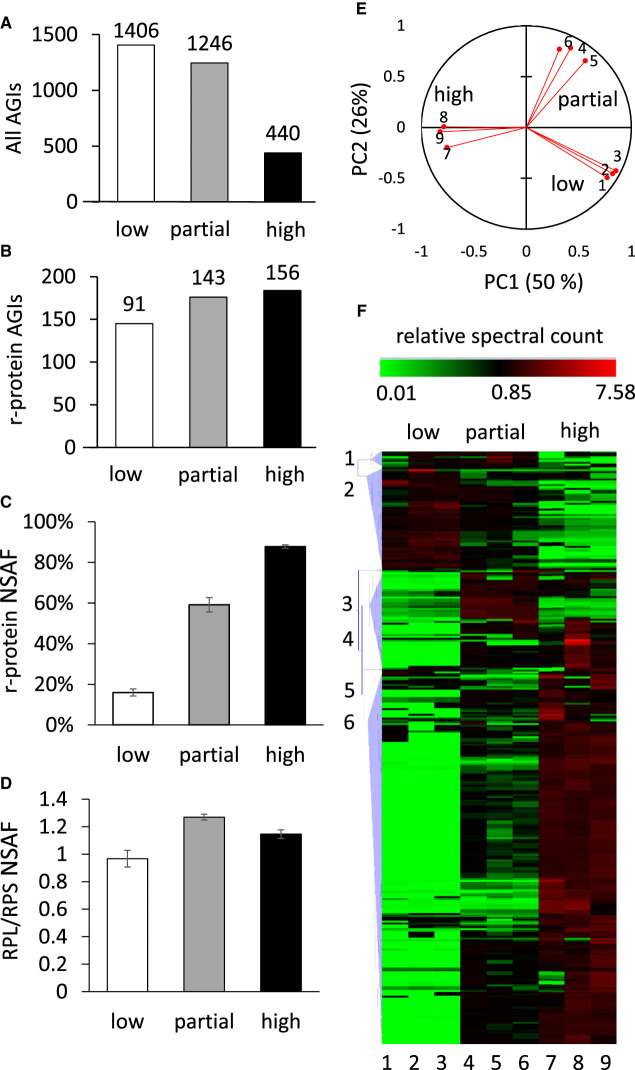
To quantify the enrichment of proteins during the process of ribosome purification, ribosomal particles from 7-day-old Arabidopsis cell culture were extracted, subjected to a three-step enrichment process and analysed in triplicate. Low purity, partially enriched and highly enriched ribosome samples were obtained by differential centrifugation in normal and high-density solutions as outlined in methods. We previously used this method to purify 80S ribosomes from this cell culture and the resultant ribosomes were shown to be intact using salt denaturation and fractionation of sucrose density gradients to quantify the subsequent separation of 60S and 40S subunits [8]. Proteins in each sample were digested with trypsin and subjected to data-dependent (shotgun) analysis by reversed-phase LC peptide mass spectrometery. The total number of non-redundant proteins identified in all three replicates decreased 3-fold as the level of enrichment increased (Figure 1A), while the number of proteins annotated as 80S cytosolic r-proteins increased step by step through the purification (Figure 1B). Analysis of the NSAF for all proteins in each sample allowed a percentage of all protein that was associated with r-proteins in each sample to be determined (Figure 1C). This showed that while r-proteins represented only 15% of the summed NSAF in low purity samples, they represented 88% of summed NSAF in high purity samples. Average NSAF values for r-proteins were typically 100 times higher than the NSAF for non r-proteins in high purity ribosome samples. SDS–PAGE analysis of the high purity samples revealed a typical r-protein pattern (Supplemental Figure S1), similar to other published gel-based analyses of the Arabidopsis 80S ribosome [8,13]. To verify the ratio of 60S to 40S subunits, the ratio of summed NSAF for 60S (RBL) and 40S (RBS) r-proteins was calculated and shown to be 0.96 in slow purity ribosome samples and 1.15 in high purity ribosome samples (Figure 1D). This indicates that nearly equal proportions of 60S and 40S subunits are present in the highly purified samples. Details of the identified proteins are provided in Supplemental Table S1 and comparisons to other reports in the literature are shown in Supplemental Table S2.
Figure 1. Purification and composition of the 80S cytosolic ribosome in Arabidopsis cell culture.
(A) Number of identified Arabidopsis proteins (AGIs) found in all of three replicates following differential centrifugation steps for low, partial and high enrichment of ribosomes. (B) The number of Arabidopsis 80S r-proteins isoforms identified across the three replicates. (C) Percentage of normalised spectral abundance factors (NSAF) in each ribosome enrichment that are r-proteins. (D) The ratio of NSAF for 60S large subunit r-proteins/40S small subunit r-proteins (RPL/RPS). (E) Principal component analysis of spectral counts assigned to the 319 proteins found by the differential centrifugation process across the three replicates and three purification steps (1–9). (F) Hierarchical clustering analysis of spectral counts for the 319 proteins identified in the purification process. The relative number of normalised spectral counts is shown in green for numbers <1 and in red for numbers >1. The vertical numbers (1–6) indicate cluster number based on Pearson correlation. Cluster 6 contains 193 proteins, including 135 80S cytosolic ribosome r-proteins. The horizontal numbers (1–9) indicate replicate number as in (E).
The intensity information for each peptide was extracted from the raw MS data files to estimate their relative abundance across the three ribosome enrichment steps. A total of 6084 peptides detected in all nine samples (three enrichment steps, three biological replicates) were included in the analysis (Supplemental Table S3). Where more than one peptide per protein was available, a protein intensity value was then calculated by averaging the peptide values on a protein basis per replicate. This analysis resulted in quantitation of enrichment for 319 non-redundant proteins derived from ribosome preparations. PCA analysis of the peptide quantification data showed that two principal components explained more than 75% of the variation (Figure 1C). The clustering of samples in the PCA demonstrated that the samples analysed were distinct between enrichment steps but consistent within biological replicates.
The protein intensity values were analysed by hierarchical clustering and Pearson correlation to assess the similarity of the enrichment profiles of known r-proteins with non-ribosomal proteins in the set of 319 proteins. Six clusters based on a minimum similarity of 0.70 [20] were chosen for further analysis. Cluster 6 was the largest cluster and contained 193 proteins (Figure 1D), including 135 cytosolic r-proteins. This cluster was significantly enriched in cytosolic r-proteins (P ≤ 0.00001 Fisher's exact) when compared with the whole proteome. Functional biological annotation of these 193 proteins showed that more than 70% (141 proteins) were annotated as components of protein synthesis and 135 of the 141 were annotated as located in the cytosol (based on SUBACon [21]; http://suba.live/); the remaining proteins were annotated to be located in the plastid or mitochondrion (Supplemental Figure S2). These results indicated the methodology employed successfully enriched 80S cytosolic r-proteins. A total of 11 putative r-protein isoforms were identified that have not been previously reported in Arabidopsis ribosome purifications (Table 1, Supplemental Table S2 for details). We did not find new evidence of methylation, acetylation or phosphorylation modifications of r-proteins beyond those we have already reported for Arabidopsis r-proteins [8].
Table 1. Newly identified and re-annotated Arabidopsis cytosolic ribosomal protein genes.
| Family | AGIs | Arabidopsis protein name | Unique peptides |
|---|---|---|---|
| eL20 | At1g29965 | RPL18aD | 9 |
| eL24 | At2g36620 | RPL24A | 7 |
| At2g44860 | RPL24C | 3 | |
| eL36 | At2g37600 | RPL36A | 5 |
| eL37 | At1g15250 | RPL37A | 2 |
| bL12 | At3g44590 | RPP2D | 3 |
| At5g40040 | RPP2E | 9 | |
| uS19 | At5g43640 | RPS15E | 2 |
| eS21 | At5g27700 | RPS21C | 2 |
| eS31 | At1g23410 | RPS27aA | 5 |
| At2g47110 | RPS27aB | 5 |
Enrichment of ribosome-associated proteins
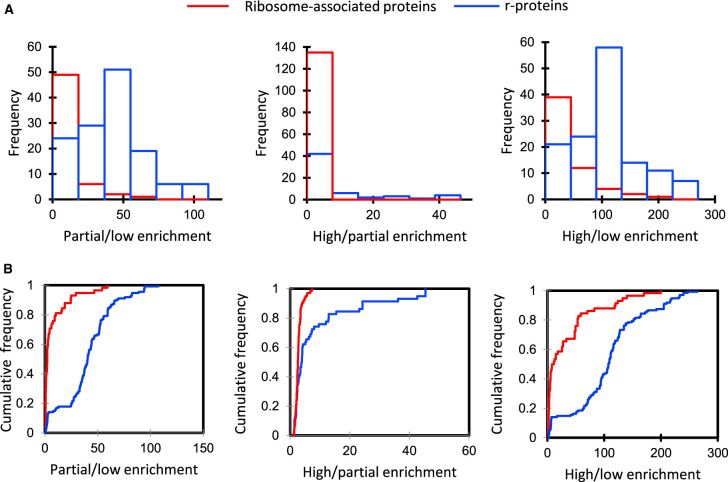
To determine if the r-protein set and the other ribosome-associated protein set in cluster 6 differed significantly in their enrichment, or if they came from the same distribution, the ratio of abundances (partial/low enrichment), (high/partial enrichment), and (high/low enrichment) were calculated for each protein in both groups. The results showed a non-normal distribution between these two sets with the value of the D statistic being 0.75, 0.40, and 0.66, respectively, for each enrichment comparison (Figure 2A). The corresponding P-value using the Kolmogorov–Smirnov test suggests a significant difference between the enrichment characteristics of the r-protein and ribosome-associated protein sets (P < 0.0001). The distributions of these enrichments as histograms of the two protein sets are shown in Figure 2B. Even though the enrichment characteristics were different, it is possible that the 58 ribosome-associated proteins are part of a less tightly bound translatome complex that partially dissociates during purification. As a further stringency filter the high/low enrichment of non-ribosomal proteins was set at a minimum of 10-fold with a P-value ≤ 0.05. Twenty-six of the 58 proteins met this enrichment criteria and are shown in Table 2 as significantly enriched ribosome-associated proteins.
Figure 2. Frequency distribution graphs of the ratio of spectral counts for r-proteins and ribosome-associated proteins in pairs of ribosomal enrichments.
(A) Frequency histogram of the ratio of spectral counts in samples of different enrichment for r-proteins and ribosome-associated proteins; the blue bars indicate the distribution of ratios for r-proteins and red bars indicate the distribution of rations for ribosome-associated proteins. (B) Cumulative relative frequency distribution graphs of (A). Statistical significance was estimated by the Kolmogorov–Smirnov test for 193 proteins including 135 r-proteins (blue) and 58 ribosome-associated proteins (red) (P < 0.0001) from cluster 6 (Figure 1E).
Table 2. Non-ribosomal proteins found to be ribosome-associated proteins in Arabidopsis cell culture.
| AGIs | Fold enrichment (high/low) | P-value | Protein annotation |
|---|---|---|---|
| AT5G53460.1 | 140 | 0.049 | NADH-dependent glutamate synthase 1 |
| AT5G20010.1 | 124 | 0.000 | RAS-related nuclear protein-1 |
| AT1G62850.2 | 120 | 0.041 | Class I peptide chain release factor |
| AT5G55730.1 | 118 | 0.001 | Fasciclin-like arabinogalactan 1 |
| AT4G13780.1 | 83 | 0.045 | Methionyl-tRNA synthetase |
| AT3G09840.1 | 63 | 0.043 | Cell division cycle 48 |
| AT3G53230.1 | 63 | 0.050 | ATPase, AAA-type, CDC48 protein |
| AT3G27850.1 | 59 | 0.001 | 50S ribosomal protein L12-C |
| AT1G48920.1 | 55 | 0.020 | Nucleolin like 1 |
| AT2G27030.3 | 53 | 0.041 | Calmodulin 5 |
| AT2G01140.1 | 50 | 0.042 | Aldolase superfamily protein |
| AT2G21330.1 | 50 | 0.049 | Fructose-bisphosphate aldolase 1 |
| AT1G22840.1 | 48 | 0.000 | Cytochrome C-1;encodes cytochrome c |
| AT3G09820.1 | 48 | 0.010 | Adenosine kinase 1 |
| AT5G03300.1 | 48 | 0.010 | Adenosine kinase 2 |
| AT4G05400.1 | 33 | 0.000 | Copper ion binding |
| AT1G08580.1 | 31 | 0.004 | Not assigned |
| AT4G35460.1 | 28 | 0.006 | NADPH-dependent thioredoxin reductase B |
| AT2G17420.1 | 28 | 0.026 | NADPH-dependent thioredoxin reductase A |
| AT2G21870.1 | 27 | 0.049 | Copper ion binding;cobalt ion binding;zinc ion binding |
| AT3G51160.1 | 26 | 0.001 | NAD(P)-binding Rossmann-fold superfamily protein |
| AT3G58610.1 | 26 | 0.048 | Ketol-acid reductoisomerase |
| AT3G55620.1 | 15 | 0.038 | Translation initiation factor IF6 |
| AT1G15270.1 | 15 | 0.004 | Translation-machinery associated TMA7 |
| AT3G55410.1 | 13 | 0.017 | 2-oxoglutarate dehydrogenase |
| AT5G08570.1 | 10 | 0.039 | Pyruvate kinase family protein |
Non-ribosomal proteins were selected based of their >10-fold enrichment in the third centrifugation (high purity) compared with the first centrifugation (low purity) (P ≤ 0.05). Columns show the Arabidopsis gene index number (AGI), the significant (P < 0.05) fold enrichment (high/low) for each protein and their functional annotation.
Protein turnover rates of ribosomal proteins
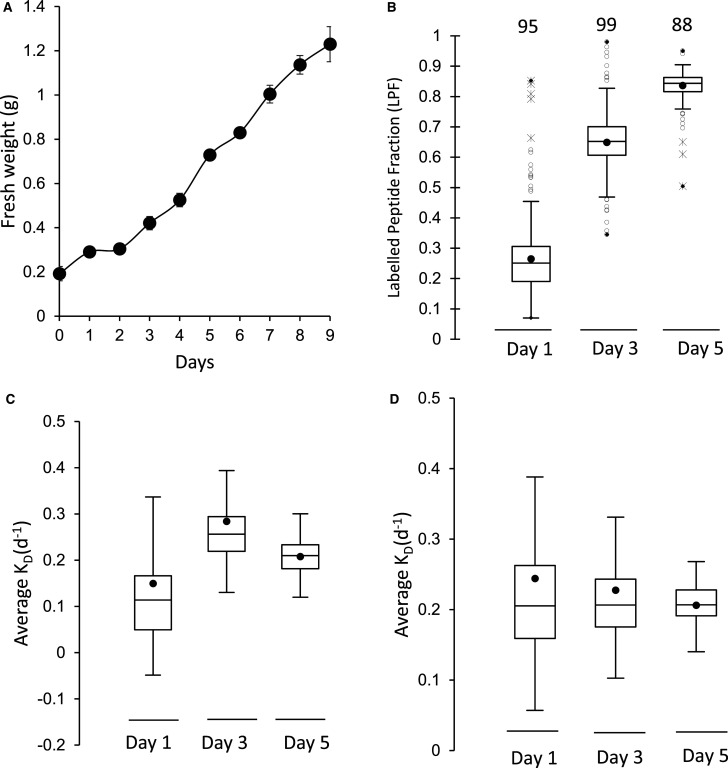
As a further criteria to verify if proteins were part of a ribosomal complex in vivo, we sought to calculate the protein turnover rate of proteins in ribosomal extracts in Arabidopsis cell culture to determine which proteins turned over as a similar rate. We had previously shown the distribution of turnover rates in Arabidopsis cell culture varied more than 100-fold [16] but that members of the same complex tend to turnover at a similar rate [22]. Cell cultures were labelled by changing the growth medium from a solution that contained inorganic 14N to one that only contained inorganic 15N. As proteins degrade over time, newly synthesised proteins increased in the amount of 15N they contained as the amino acid pool was progressively labelled. Calculation of the fraction of the protein labelled with 15N (labelled peptide fraction or LPF) could be achieved by a binary differentiation between the natural abundance population before labelling and the newly synthesised labelled population of peptides by a non-linear least-squares analysis [23]. Determining the proportional degradation rate (KD) and a normalised synthesis rate (Ks/A) for each protein requires the LPFs, the growth rate of the cell culture calculated by determining the fresh weight (g) of the cell culture from day 1 to day 5 (Figure 3A), and a series of calculations as previously reported [23].
Figure 3. Protein turnover of ribosomes in Arabidopsis cell culture.
(A) Growth rate of Arabidopsis cell culture over 9 days after transferred into new media (FW, n = 4, standard error). (B) Box plot of the average 15N enrichment of r-protein peptides after 1, 3 and 5 days of growth in 98% 15N media. The number of r-proteins included for each time point is shown above the box plot (n = 4). (C,D) box plots representing the average degradation rate (Kd) of r-proteins after day 1, 3 and 5 through the course of the growth experiment. (C) Kd for each time point using measured FCP values. (D) KD using the median polish strategy for FCP calculation.
LPFs of ribosomal protein in Arabidopsis cell culture were calculated from three times points during the growth of the cell culture (day 1, 3 and 5) using four biological replicates. Highly enriched ribosomal protein fractions from the cytosol was isolated as indicated above, digested by trypsin and the peptides analysed by LC–MS/MS. The 15N enrichment of the labelled peptide fractions were calculated at each time point, showing 15N enrichment in new peptides was ∼60% after 1 day and rose to 83% by day 5 (Supplemental Figure S3). Calculated LPF showed a clear increase from 25% at 1 day to more than 85% by day 5 (Figure 3B). From LPF data the degradation rate for protein isoforms were calculated at each timepoint with the criteria that peptides for a protein were found in three or more biological replicates and the protein calculation was supported by at least five independently quantified peptides. The measured KD of these proteins over day 1, day 3 and 5 showed variability based on fluctuations in FCP value coming from measuring the fresh weight of Arabidopsis cell culture (Figure 3C). To gain a more precise degradation rate in each replicate a median polish strategy [24] was used for experimental normalisation and to calculate FCP (Figure 3D). The overall correlation between the measured FCP and calculated FCP was very high (R = 0.96), and the median polish approach gave a more precise estimate of growth of the cell culture in each labelled replicate, lowering KD variation.
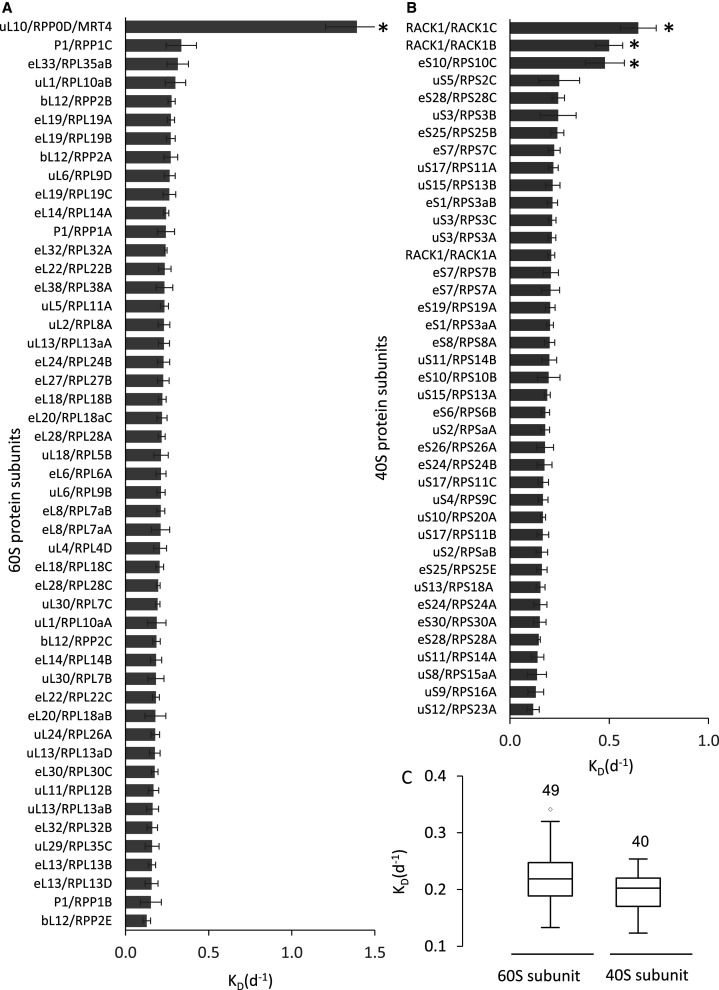
Of the proteins in the cytosolic ribosome of Arabidopsis, the degradation rate (KD) and relative synthesis rate (Ks/A) from 54 protein families encoded by 89 genes were calculated. These proteins were all r-proteins rather than the ribosomal-associated proteins; presumably due to the lower abundance of the latter. The turnover rate set included proteins from 29 r-protein families from the large subunit (Figure 4A) and from 25 r-protein families from the small subunit (Figure 4B) which were encoded by 49 and 40 genes, respectively.
Figure 4. Turnover rate of specific r-proteins in Arabidopsis cell culture.
(A) Degradation rate of core 60S ribosomal subunit proteins and the putative 60S-transacting factor Mrt4/RPP0D. (B) Degradation rate of 40S ribosomal subunit proteins. In both (A) and (B) the mean degradation rate value for each protein was calculated using data from ≥ 3 biological replicates and included data from ≥ 5 peptide spectra. Error bars: standard error. * indicates significant difference from the mean of the ribosome (based on t-tests P < 0.05). (C) Box plots of KD values for r-proteins of the 60S large and 40S small subunits.
More than 90% of the r-protein degradation rates fell within a narrow range of 0.18–0.25 d−1 (half-lives of 3–5 days) (Supplemental Table S4). There was no significant difference in degradation rate between 60S ribosomal subunits and 40S subunits with averages of 0.25 and 0.22 d−1, respectively (Figure 4C). Relative synthesis rate (Ks/A) data mirrored the degradation rate data for each r-protein isoform (Supplemental Table S4). However, there were several r-proteins that showed degradation and synthesis rate characteristics that were significantly different from those of the other r-proteins in each ribosome subunit. In the 60S ribosomal subunit, an uL10 r-protein annotated as RPP0D in Arabidopsis (60S acidic ribosomal protein family/ribosomal protein L10 family) was very unstable with a KD of 1.39 d−1 and Ks/A of 1.7 d−1 and thus a half-life of 0.5 d. In the 40S ribosomal subunit, two isoforms of RACK1, namely RACK1b and RACK1c, had KD of 0.5–0.65 d−1 and Ks/A of 0.74–0.87 d−1 and half-lives of 1–1.4 days. Indepth visual inspection the LPF data supporting these exceptional degradation and synthesis rates (e.g. Supplemental Figure S4) confirmed the KD and Ks/A calculations. The eS10 isoform RPS10C also had a significantly higher KD than most RPS subunits and other eS10 isoforms (Figure 4B).
Discussion
Core 80S ribosomal r-protein composition in plants defined by quantitative enrichment, co-synthesis and co-degradation
This analysis has provided the most robust foundation yet for the Arabidopsis r-protein content of 80S ribosomes by providing multiple lines of evidence to justify protein inclusion in the core r-protein set and aligning Arabidopsis r-proteins with the unifying nomenclature of r-proteins in other organisms [10] (Supplemental Table S4). Typical plant ribosomal enrichment protocols [7,8,14] do not yield ribosomal preparations devoid of all contaminants, hence the quantitative enrichment and other information provided here help to clarify which proteins are part of the ribosome complex (Supplemental Table S4). We now have information on the degree of enrichment during ribosome purification and the in vivo turnover rate of putative r-proteins in Arabidopsis to support this effort to full characterized the plant 80S ribosome (Supplemental Table S3, summarised in Supplemental Table S4). A full numerical comparison showing the improvement in coverage over previous research [7,8,13,14] is shown graphically in Supplemental Figure S5A,B. This analysis was only conducted in cell culture. Given the slight apparent variation in the reported ribosome composition from different tissues of origin in plants (Supplemental Figure S5A,B) it would be valuable to conduct studies similar to this one in whole plants or specific plant tissues to determine if various ribosome compositions might be differentially expressed in plant cell types.
Five of the r-proteins first identified here (Table 1) are proposed by homology to be located in the 60S subunit and six are proposed to be in the 40S subunit [7,8,13,14]. In total at least one isoform of 76 out of 81 ribosomal protein families predicted in Arabidopsis were identified in this analysis. This included 108 protein isoforms in the 60S large subunit and 76 protein isoforms in the 40S small subunit (Supplemental Table S4).
Comparing the r-proteins observed here and in previous reports with a systematic assessment of r-protein families and groups from other kingdoms of life [10], did not reveal any significant difference between the turnover rate or fold-enrichment during ribosome purification for eukaryotic specific subunits versus those present across all kingdoms (Supplementary Table S4). The eukaryotic protein families predicted in plants but not found experimentally to date in Arabidopsis are the large subunit r-proteins eL29 (RPL29A and RPL29B), eL39 (RPL39A, RPL39B and RPL39C), eL40 (RPL40A and RPL40B) and eL41 (RPL41A-G). uS14 (RPS29) was found in previous studies in Arabidopsis (Supplementary Table S2) but we were unable to confirm this identification. This is likely because their short amino acid sequences (25–128 AAs) and many Arg and Lys residues yield short and few tryptic peptides which are not easily detected by mass spectrometry [7].
Turnover of ribosomal proteins
In whole tissue studies in yeast [25], Barley [17] and Arabidopsis [23] it had been shown that turnover rates of r-proteins fall within a relatively narrow range and are among the longest-lived protein complexes in cells. We have confirmed this is also the case for isolated Arabidopsis ribosomes and that degradation rates and synthesis rates of proteins in both 60S and 40S subunits were very similar (Figure 4C, Supplemental Table S4). However, we did observe three proteins annotated as Arabidopsis r-proteins with unusually rapid degradation and synthesis rates, namely RACK1b, RACK1c and a protein annotated as RPP0D.
RACK1b and RACK1c have been previously found in ribosomes [7,8] and their function as r-protein subunits has been investigated [26,27]. RACK1 is required for efficient translation of mRNA during the translation initiation process within the 40S ribosomal subunit [28]. RACK1 also acts as a scaffold protein between ribosomal machinery and many signalling pathways in the cell [29]. In Arabidopsis, knockout of RACK1A results in developmental defects [30] and hypersensitivity to hormonal treatment [26]. Knockout of RACK1B or RACK1C alone did not lead to significant phenotypes in the presence of RACK1A, but their loss exacerbated the rack1a phenotype in double mutants leading to the suggestion they are functionally redundant with RACK1a [27]. A more specialised role for RACK1B,C could be infered from the combination of these prior data with our evidence of more rapid turnover rates for RACK1B and RACK1C than RACK1A.
A protein annotated in Arabidopsis as RPP0D (an isoform of uL10) was previously reported as a ribosomal subunit isolated from Arabidopsis cell culture [8], however, it was not reported in ribosomal purifications from Arabidopsis leaves or whole seedlings [7,13,14]. Examination of whole Arabidopsis proteome datasets from multiple tissues show this protein is present in cell culture, flowers and siliques, but not in vegetative tissues [31]. Carroll et al. (2008) named this protein RPP0D based on its presence in ribosomes and its homology with other uL10/P0 isoforms. But the homology with other uL10/P0 sequences is only in one region of the protein and the overall sequence identity is only 20%. Re-analysis by sequence homology searches show that RPP0D shares more than 60% sequence identity with a yeast and human protein called Mrt4. Mrt4 is not a uL10 isoform, it is a pre-60S trans-acting factor [32–34]. Mrt4 shares sequence similarity to uL10/P0 proteins in yeast and humans, and together with P1, bL12 and uL11, it forms the ribosomal stalk responsible for translation factor-dependent GTP hydrolysis [35]. Mrt4 binds to pre-60S during its shuttling between the nucleus and the cytoplasm at the same site in the ribosome as uL10 but is displaced by uL10 in the cytosol during ribosome maturation [36]. Our data suggest that the very rapid degradation rate of uL10/RPP0 in Arabidopsis and its homology with Mrt4 warrants the renaming of At1g25260 as Mrt4 and its further investigation in ribosome biogenesis in plants as a pre-60S trans-acting factor.
Analysis of r-protein turnover rates in mouse liver has shown that r-proteins with roles in peptide-bond formation or that reside at the interface of the 60S and 40S subunits are replaced during the lifespan of ribosomes [37]. Direct comparison between homologous Arabidopsis and mouse r-protein turnover rates (Supplemental Figure S6) shows that both mammals and plants share this characteristic of high turnover rate for the same set of r-proteins, namely eL24, eL33, eL19, eL38 and bL12. Two of these (eL19, eL24) are reported to act structurally to secure the 40S to the 60S subunit, while bL12 is reported to be localised at the interface between the subunits [37]. This conservation of higher than average turnover for the same subunits across kingdoms provides further confidence that it is a feature of the eukaryotic ribosome and its maintenance.
Ribosome-associated proteins as potential translatome components in Arabidopsis
Identification and further characterisation of ribosome-associated proteins that are not in r-protein families are essential in order to obtain a better understanding of the physical organisation of the process of translation and post-translational folding and modification of nascent polypeptides in plants. An earlier study in yeast showed that proteins that are associated with the ribosome are enriched in proteins needed for the translational process; so called translation-machinery-associated (TMA) proteins [38]. The deletion of one of these proteins (TMA7) resulted in an alteration in both protein synthesis rate and translation-related processes in yeast [38].
Here, we identified a significant number of ribosome-associated proteins in the purification process of ribosomes in Arabidopsis. By using the open source STRING database version 10.5 (https://string-db.org/) [19] the protein interaction score was compared between known r-proteins in Arabidopsis and these ribosome-associated proteins. Most of the proteins that are considered ribosome-associated proteins in this study showed a significant interaction score with r-proteins based on a combination of co-expression and protein–protein interaction data, confirming their likely associations with ribosomes (Supplemental Table S3). By imposing a strict definition of enrichment on our data of ≥10-fold enrichment during co-purification, 26 putative ribosome-associated proteins are proposed for Arabidopsis (Table 2).
Eight of these proteins have been previously found to be involved in translation and ribosomal function in different organisms, including TMA7 [38], eIF6 [39], cytochrome c [40], Class I peptide chain release factor [41], methionyl-tRNA synthetase [42], nucleolin like 1 or PARL1 [43], fructose-bisphosphate aldolase 1[44] and AAA-ATPases [45]. AT1G15270, which is annotated as Arabidopsis gene translation-machinery associated TMA7, gained this annotation based on homology to the yeast protein found in a similar enrichment study [38]. However, to our knowledge this is the first direct experimental evidence of its association with ribosomes in a plant species. No further information on its function in yeast has yet been revealed since the Flesicher et al. [38] report. AT3G55620, annotated as a translation initiation factor IF6, is also shown to be ribosome-associated in our study. This protein has been found in Arabidopsis to physically interact with the small ribosomal RACK1 protein [39] as well as to be involved in the formation of the 60S large subunit [46]. eIF6 in yeast (S. cerevisiae) is required for pre-rRNA processing and the biogenesis of 60S ribosome subunit, and the depletion of this protein resulted in imbalance of the 60S to 40S subunit ratio and reduction in protein synthesis and eventually reduction in cell growth [47]. Unexpectedly, cytochrome c, a mitochondrial protein involved in electron transport, was found in the ribosomal enrichment list (Table 2). In mammals, this protein has been reported to be a tRNA-associated protein that is essential for disabling the formation of an apoptosis complex in the cytosol [40]. It is still unclear the functionality of other proteins that interacts with the 80S cytosolic ribosome and how they participate, if at all, in protein synthesis in plant cells. Further study is required to reveal the role of ribosome-associated proteins during the translational process in plant cells.
Data Availability
PRIDE Project Name: Ribosomal protein turnover under normal conditions
Project accession: PXD012839
Acknowledgements
This work used the services of the WA Proteomics Facility supported by Bioplatforms Australia as a component of NCRIS and the Government of Western Australia.
Abbreviations
- LPFs
labelled peptide fraction
- mRNA
messenger RNA
- MS
Murashige and Skoog
- NSAF
normalised spectral abundance factors
- TMA
translation-machinery-associated
- TPP
trans-proteomic pipeline
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
K.J.S. was a recipient of a PhD Scholarship from the Higher Committee for Educational Development in Iraq (HCED). This work was supported through funding by the Australian Research Council [CE140100008, DP180104136] to A.H.M.
Open access
Open access for this article was enabled by the participation of The University of Western Australia in an all-inclusive Read & Publish pilot with Portland Press and the Biochemical Society under a transformative agreement with CAUL.
Author Contribution
K.J.S., O.D., L.L. and A.H.M. designed the research; K.J.S. performed cell culture and biochemical experiments with supervision from O.D., L.L. and A.H.M. Mass spectrometry and anaylsis was performed by K.J.S. and O.D., J.T. provided bioinformatics analysis support. All authors contributing to the writing and revision of the article.
Supplementary Material
References
- 1.Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G. and Yusupov M. (2011) The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 2.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M. et al. (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310, 827–834 10.1126/science.1117230 [DOI] [PubMed] [Google Scholar]
- 3.Jenner L., Melnikov S., Garreau de Loubresse N., Ben-Shem A., Iskakova M., Urzhumtsev A. et al. (2012) Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 22, 759–767 10.1016/j.sbi.2012.07.013 [DOI] [PubMed] [Google Scholar]
- 4.Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S. and Ban N. (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948 10.1126/science.1211204 [DOI] [PubMed] [Google Scholar]
- 5.Khatter H., Myasnikov A.G., Natchiar S.K. and Klaholz B.P. (2015) Structure of the human 80S ribosome. Nature 520, 640–645 10.1038/nature14427 [DOI] [PubMed] [Google Scholar]
- 6.Barakat A., Szick-Miranda K., Chang I.F., Guyot R., Blanc G., Cooke R. et al. (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 127, 398–415 10.1104/pp.010265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hummel M., Dobrenel T., Cordewener J.J., Davanture M., Meyer C., Smeekens S.J. et al. (2015) Proteomic LC-MS analysis of Arabidopsis cytosolic ribosomes: identification of ribosomal protein paralogs and re-annotation of the ribosomal protein genes. J. Proteom. 128, 436–449 10.1016/j.jprot.2015.07.004 [DOI] [PubMed] [Google Scholar]
- 8.Carroll A.J., Heazlewood J.L., Ito J. and Millar A.H. (2008) Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteom. 7, 347–369 10.1074/mcp.M700052-MCP200 [DOI] [PubMed] [Google Scholar]
- 9.Planta R.J. and Mager W.H. (1998) The list of cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Yeast 14, 471–477 [DOI] [PubMed] [Google Scholar]
- 10.Ban N., Beckmann R., Cate J.H., Dinman J.D., Dragon F., Ellis S.R. et al. (2014) A new system for naming ribosomal proteins. Curr. Opin. Struct. Biol. 24, 165–169 10.1016/j.sbi.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manuell A.L., Yamaguchi K., Haynes P.A., Milligan R.A. and Mayfield S.P. (2005) Composition and structure of the 80S ribosome from the green alga Chlamydomonas reinhardtii: 80S ribosomes are conserved in plants and animals. J. Mol. Biol. 351, 266–279 10.1016/j.jmb.2005.06.022 [DOI] [PubMed] [Google Scholar]
- 12.Carroll A.J. (2013) The Arabidopsis cytosolic ribosomal proteome: from form to function. Front. Plant. Sci. 4, 32 10.3389/fpls.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang I.F., Szick-Miranda K., Pan S. and Bailey-Serres J. (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol. 137, 848–862 10.1104/pp.104.053637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giavalisco P., Wilson D., Kreitler T., Lehrach H., Klose J., Gobom J. et al. (2005) High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant. Mol. Biol. 57, 577–591 10.1007/s11103-005-0699-3 [DOI] [PubMed] [Google Scholar]
- 15.Salih K.J., Duncan O., Li L., O'Leary B., Fenske R., Trosch J. et al. (2020) Impact of oxidative stress on the function, abundance, and turnover of the Arabidopsis 80S cytosolic ribosome. Plant J. 103, 128–139 10.1111/tpj.14713 [DOI] [PubMed] [Google Scholar]
- 16.Li L., Nelson C.J., Solheim C., Whelan J. and Millar A.H. (2012) Determining degradation and synthesis rates of Arabidopsis proteins using the kinetics of progressive 15N labeling of two-dimensional gel-separated protein spots. Mol. Cell. Proteom. 11, M111.010025 10.1074/mcp.M111.010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson C.J., Alexova R., Jacoby R.P. and Millar A.H. (2014) Proteins with high turnover rate in barley leaves estimated by proteome analysis combined with in planta isotope labeling. Plant Physiol. 166, 91–108 10.1104/pp.114.243014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park C.Y., Klammer A.A., Kall L., MacCoss M.J. and Noble W.S. (2008) Rapid and accurate peptide identification from tandem mass spectra. J. Proteome Res. 7, 3022–3027 10.1021/pr800127y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J. et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R. et al. (2012) The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210 10.1093/nar/gkr1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper C.M., Castleden I.R., Tanz S.K., Aryamanesh N. and Millar A.H. (2017) SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 45, D1064–D1074 10.1093/nar/gkw1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li L., Nelson C.J., Carrie C., Gawryluk R.M., Solheim C., Gray M.W. et al. (2013) Subcomplexes of ancestral respiratory complex I subunits rapidly turn over in vivo as productive assembly intermediates in Arabidopsis. J. Biol. Chem. 288, 5707–5717 10.1074/jbc.M112.432070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L., Nelson C.J., Trosch J., Castleden I., Huang S. and Millar A.H. (2017) Protein degradation rate in Arabidopsis thaliana leaf growth and development. Plant Cell. 29, 207–228 10.1105/tpc.16.00768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L., Nelson C., Fenske R., Trosch J., Pruzinska A., Millar A.H. et al. (2017) Changes in specific protein degradation rates in Arabidopsis thaliana reveal multiple roles of Lon1 in mitochondrial protein homeostasis. Plant J. 89, 458–471 10.1111/tpj.13392 [DOI] [PubMed] [Google Scholar]
- 25.Christiano R., Nagaraj N., Frohlich F. and Walther T.C. (2014) Global proteome turnover analyses of the yeasts S. cerevisiae and S. pombe. Cell Rep. 9, 1959–1965 10.1016/j.celrep.2014.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo J., Wang J., Xi L., Huang W.D., Liang J. and Chen J.G. (2009) RACK1 is a negative regulator of ABA responses in Arabidopsis. J. Exp. Bot. 60, 3819–3833 10.1093/jxb/erp221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo J. and Chen J.G. (2008) RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant. Biol. 8, 108 10.1186/1471-2229-8-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson M.K., Rojas-Duran M.F., Gangaramani P. and Gilbert W.V. (2016) The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. eLife 5, e11154 10.7554/eLife.11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Islas-Flores T., Rahman A., Ullah H. and Villanueva M.A. (2015) The receptor for activated C kinase in plant signaling: tale of a promiscuous little molecule. Front. Plant. Sci. 6, 1090 10.3389/fpls.2015.01090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J.G., Ullah H., Temple B., Liang J., Guo J., Alonso J.M. et al. (2006) RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J. Exp. Bot. 57, 2697–2708 10.1093/jxb/erl035 [DOI] [PubMed] [Google Scholar]
- 31.Baerenfaller K., Grossmann J., Grobei M.A., Hull R., Hirsch-Hoffmann M., Yalovsky S. et al. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 10.1126/science.1157956 [DOI] [PubMed] [Google Scholar]
- 32.Kemmler S., Occhipinti L., Veisu M. and Panse V.G. (2009) Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 186, 863–880 10.1083/jcb.200904111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo K.Y., Li Z., Wang F., Marcotte E.M. and Johnson A.W. (2009) Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol. 186, 849–862 10.1083/jcb.200904110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Mateos M., Garcia-Gomez J.J., Francisco-Velilla R., Remacha M., de la Cruz J. and Ballesta J.P. (2009) Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 7519–7532 10.1093/nar/gkp806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michalec B., Krokowski D., Grela P., Wawiorka L., Sawa-Makarska J., Grankowski N. et al. (2010) Subcellular localization of ribosomal P0-like protein MRT4 is determined by its N-terminal domain. Int. J. Biochem. Cell Biol. 42, 736–748 10.1016/j.biocel.2010.01.011 [DOI] [PubMed] [Google Scholar]
- 36.Michalec-Wawiorka B., Wawiorka L., Derylo K., Krokowski D., Boguszewska A., Molestak E. et al. (2015) Molecular behavior of human Mrt4 protein, MRTO4, in stress conditions is regulated by its C-terminal region. Int. J. Biochem. Cell Biol. 69, 233–240 10.1016/j.biocel.2015.10.018 [DOI] [PubMed] [Google Scholar]
- 37.Mathis A.D., Naylor B.C., Carson R.H., Evans E., Harwell J., Knecht J. et al. (2017) Mechanisms of in vivo ribosome maintenance change in response to nutrient signals. Mol. Cell. Proteom. 16, 243–254 10.1074/mcp.M116.063255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleischer T.C., Weaver C.M., McAfee K.J., Jennings J.L. and Link A.J. (2006) Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 20, 1294–1307 10.1101/gad.1422006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J., Wang S., Valerius O., Hall H., Zeng Q., Li J.F. et al. (2011) Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 155, 370–383 10.1104/pp.110.160663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mei Y., Yong J., Stonestrom A. and Yang X. (2010) tRNA and cytochrome c in cell death and beyond. Cell Cycle 9, 2936–2939 10.4161/cc.9.15.12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petropoulos A.D., McDonald M.E., Green R. and Zaher H.S. (2014) Distinct roles for release factor 1 and release factor 2 in translational quality control. J. Biol. Chem. 289, 17589–17596 10.1074/jbc.M114.564989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon N.H., Kang T., Lee J.Y., Kim H.H., Kim H.R., Hong J. et al. (2011) Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc. Natl. Acad. Sci. U.S.A. 108, 19635–19640 10.1073/pnas.1103922108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petricka J.J. and Nelson T.M. (2007) Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 144, 173–186 10.1104/pp.106.093575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziveri J., Tros F., Guerrera I.C., Chhuon C., Audry M., Dupuis M. et al. (2017) The metabolic enzyme fructose-1,6-bisphosphate aldolase acts as a transcriptional regulator in pathogenic Francisella. Nat. Commun. 8, 853 10.1038/s41467-017-00889-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bassler J., Kallas M., Pertschy B., Ulbrich C., Thoms M. and Hurt E. (2010) The AAA-ATPase rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol. Cell 38, 712–721 10.1016/j.molcel.2010.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brina D., Miluzio A., Ricciardi S. and Biffo S. (2015) eIF6 anti-association activity is required for ribosome biogenesis, translational control and tumor progression. Biochim. Biophys. Acta 1849, 830–835 10.1016/j.bbagrm.2014.09.010 [DOI] [PubMed] [Google Scholar]
- 47.Basu U., Si K., Warner J.R. and Maitra U. (2001) The Saccharomyces cerevisiae TIF6 gene encoding translation initiation factor 6 is required for 60S ribosomal subunit biogenesis. Mol. Cell. Biol. 21, 1453–1462 10.1128/MCB.21.5.1453-1462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PRIDE Project Name: Ribosomal protein turnover under normal conditions
Project accession: PXD012839