Abstract
Background
PARP inhibitors are active in various tumour types beyond BRCA-mutant cancers, but their activity and molecular correlates in colorectal cancer (CRC) are not well studied.
Methods
Mutations and genome-wide mutational patterns associated with homologous recombination deficiency (HRD) were investigated in 255 primary CRCs with whole-exome sequencing and/or DNA copy number data. Efficacy of five PARP inhibitors and their molecular correlates were evaluated in 93 CRC cell lines partly annotated with mutational-, DNA copy number-, and/or gene expression profiles. Post-treatment gene expression profiling and specific protein expression analyses were performed in two pairs of PARP inhibitor sensitive and resistant cell lines.
Findings
A subset of microsatellite stable (MSS) CRCs had truncating mutations in homologous recombination-related genes, but these were not associated with genomic signatures of HRD. Eight CRC cell lines (9%) were sensitive to PARP inhibition, but sensitivity was not predicted by HRD-related genomic and transcriptomic signatures. In contrast, drug sensitivity in MSS cell lines was strongly associated with TP53 wild-type status (odds ratio 15.7, p = 0.023) and TP53-related expression signatures. Increased downstream TP53 activity was among the primary response mechanisms, and TP53 inhibition antagonized the effect of PARP inhibitors. Wild-type TP53-mediated suppression of RAD51 was identified as a possible mechanism of action for sensitivity to PARP inhibition.
Interpretation
PARP inhibitors are active in a subset of CRC cell lines and preserved TP53 function may increase the likelihood of response.
Keywords: Colorectal cancer, PARP inhibition, homologous recombination deficiency, TP53, RAD51, gene expression, mutational signatures
Research in context.
Evidence before this study
There are few targeted agents and companion diagnostics recommended for clinical use against colorectal cancers (CRCs), despite comprehensive molecular knowledge of this cancer type. PARP inhibitors have shown clinical efficacy in various tumour types, in particular for cancers with homologous recombination deficiency (HRD), but their activity and potential molecular correlates in CRC are not well studied.
Added value of this study
We show that PARP inhibitors are effective in a subset of CRC cell lines. Existing HRD-related biomarkers with proven predictive value of PARP inhibition in other cancer types was identified in a proportion of both primary CRCs and the cancer cell cultures, but did not predict in vitro sensitivity. In contrast, pharmacogenomic analyses showed a clear association between wild-type TP53 function and PARP inhibitor sensitivity. This was consistent at different molecular levels, and both prior to treatment exposure and in response to treatment. Wild-type TP53-mediated suppression of RAD51 was identified as a possible mechanism by which PARP inhibitors exhibit their activity in pre-clinical models of CRC.
Implications of all the available evidence
Emerging evidence suggests that the molecular underpinnings for efficacy of PARP inhibitors are context-dependent, and there is a need for disease-specific predictive biomarkers. We propose that wild-type TP53 activity can be involved with suppression of homologous recombination in CRC, and that TP53 wild-type CRCs represent a relevant stratum for clinical testing of PARP inhibitors.
Alt-text: Unlabelled box
1. Introduction
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer-related deaths [1]. The therapeutic repertoire for CRC remains limited, with few targeted agents and companion diagnostics recommended for clinical use [2]. PARP inhibitors have shown clinical efficacy in molecular subgroups of diverse cancer histologies [3], [4], [5], [6]. The poly(ADP-ribose) polymerase (PARP) enzymes are key components in repair of single-stranded DNA breaks and replication fork damage [7]. PARP inhibition causes accumulation of such lesions through catalytic inhibition and trapping of PARP, eventually resulting in double-stranded DNA breaks (DSBs). Homologous recombination is the optimal pathway for accurate repair of DSBs and restarting of replication forks [7]. Accordingly, PARP inhibitors were found to be selectively active in cells with homologous recombination deficiency (HRD) due to BRCA1 or BRCA2 mutations [8,9]. The demonstration of synthetic lethality between PARP inhibition and BRCA mutations led to clinical testing and eventually approval of olaparib as monotherapy in patients with germline BRCA-mutated metastatic ovarian or breast cancers [4, 10].
HRD also occurs in certain BRCA wild-type cancers, and further studies have identified BRCA wild-type tumours with sensitivity to PARP inhibitors based on three main approaches [11]. First, several DNA-based assays and signatures have been developed based on the assumption that HRD causes distinct genome-wide mutational patterns involving copy number aberrations, base substitutions and chromosomal rearrangements [12], [13], [14], [15], [16], [17]. Clinical relevance of this approach was shown by the ability of a composite HRD score [12] incorporating three different genomic instability signatures [15], [16], [17], to predict PARP inhibitor responses in ovarian and triple-negative breast cancer (TNBC) [3,12]. Second, inactivating mutations in additional genes involved in homologous recombination, such as ATM, ATR, CHEK1/2 and RAD51 have also been shown to confer sensitivity to PARP inhibitors [11]. Third, gene expression-based signatures indicative of HRD and PARP inhibitor sensitivity have been developed in patient-derived material and cell lines from different cancer types, but their predictive value have not been prospectively validated in clinical material [18], [19], [20].
The role of HRD and the efficacy of PARP inhibitors have been less studied in CRC. In one phase II trial including 33 patients with metastatic CRC refractory to standard therapies, olaparib did not demonstrate any meaningful anti-tumour activity as a single-agent, but the small sample size and lack of HRD-related molecular stratification of the patients prohibit any conclusions regarding the efficacy of PARP inhibitors in CRC [21]. Notably, ATM-deficient CRC cell lines have been shown to be sensitive to PARP inhibition [22]. ATM and other genes critical for homologous recombination, including BRCA1 and BRCA2, are somatically mutated in more than 20% of CRCs [11,23], and germline mutations in these genes have been detected in 3% of CRCs [24]. Furthermore, pan-cancer analyses of the HRD score identified a subset of CRCs with genomic scars associated with HRD [25], while a recent study discovered elevated HRD mutational signatures in brain metastases from CRC [26]. This suggests that a significant subset of CRCs harbour genetic alterations potentially conferring sensitivity to PARP inhibitors. Furthermore, PARP1 is the main molecular target of PARP inhibitors [7] and has been shown to be overexpressed and associated with disease progression in primary CRCs and transgenic mouse models [27].
Here, we investigated mutations and genome-wide mutational patterns associated with HRD in primary CRCs, including their clinicopathological and prognostic associations. We also evaluated the activity of PARP inhibitors in a panel of 93 CRC cell lines, followed by functional analyses to identify the molecular underpinnings of sensitivity to PARP inhibition in CRC.
2. Materials and Methods
2.1. Patient material
A total of 255 primary tumour samples from patients operated for CRC stages I–IV at Oslo University Hospital, Norway between 2005 and 2014 were analysed. Patients in this observational study were included from a larger population-based consecutive series based on availability of whole-exome sequencing (n = 176) and/or DNA copy number data (n = 199; Supplementary Table S1). The patients were treated with standard therapies in an adjuvant or palliative setting and no patients were treated with PARP inhibitors. All participants provided informed written consent. The research conformed to the Helsinki Declaration and was approved by the Regional Committee for Medical and Health Research Ethics (REC number 1.2005.1629). The research biobanks have been registered according to national legislation.
2.2. Exome sequencing and DNA copy number analysis in primary CRCs
Determination of MSI status and TP53 mutation analysis has previously been performed [28,29]. Matched tumour and normal colonic mucosa samples from 176 patients were subject to paired-end exome sequencing using the Agilent SureSelectXT Human All Exon V5 kit (Agilent, Santa Clara, CA, USA) and the Illumina HiSeq 2500 system (Illumina, San Diego, CA, USA) at the Oslo University Hospital Genomics Core Facility. Processing of raw sequencing reads (100 basepairs), as well as mutation calling, annotation and filtering were performed as previously described [30]. These data have partially been previously published (n = 33 MSI tumours) [30] or will be published elsewhere. In this study, whole-exome sequencing data were used to analyse mutations among 141 genes previously implicated in HRD [11], as described below.
DNA copy number profiles have been generated from 199 primary CRCs using Affymetrix SNP6.0 arrays (Affymetrix Inc., Santa Clara, CA, USA), and CEL files were pre-processed with PennCNV-Affy and ASCAT as previously described [31]. The fraction of the genome with aberrant copy number or loss of heterozygosity (LOH) was calculated as the percentage of aberrant bases out of the total number of bases with copy number/LOH estimates available. LOH was defined for segments in which either the A allele or B allele were zero and the remaining allele was non-zero. Complete losses were reported only for segments covered by at least 10 SNP or copy number probes.
2.3. Molecular profiles of CRC cell lines
Cell lines were obtained from American Type Culture Collection (ATCC), Cell Lines Service (CLS), DSMZ-German Collection of Microorganisms and Cell Cultures, NCI DCTD Tumour Repository, European Collection of Authenticated Cell Cultures (ECACC), JCRB Cell Bank, Korean Cell Line Bank (KCLB) and collaborators as indicated in Supplementary Table S2. Mutation data were available for 59 cell lines, either from in-house “kinome” sequencing [32] or publicly available datasets [[33], [34], [35]]. TP53 mutation status was retrieved from various publications for 14 additional cell lines (Supplementary Table S2). DNA copy number (n = 43 cell lines) and gene expression profiles (n = 75 cell lines) were taken from in-house data [32] (Gene Expression Omnibus accession number GSE97023) or publicly available datasets [34] (GSE59857 [36] and GSE57083) (Supplementary Table S2). MSI status and CMS classification of cell lines was determined as previously described [37]. Cell line authenticity has been verified by short tandem repeat profiling according to the AmpFℓSTR Identifiler PCR Amplification Kit (Thermo Fisher Scientific).
2.4. Drug sensitivity screening of CRC cell lines
Sensitivity to five PARP inhibitors (olaparib, niraparib, rucaparib, talazoparib and veliparib) was investigated in 93 unique CRC cell lines (Supplementary Table S2) as part of an in vitro drug screen using a high-throughput platform including 459–529 investigational or clinically approved drugs. These data have partially been published [37] and the rest will be published elsewhere. Drug sensitivity scores (DSS) were calculated for each drug in each cell line relative to negative (mean of twelve tests with 0.1% DMSO) and positive controls (mean of twelve tests with 100 µM benzethonium chloride) based on cell viability after 72 h drug treatment at five different concentrations over a 10,000-fold concentration range (Talazoparib 0.1–1000 nM, remaining 4 PARP inhibitors at 1–10,000 nM). All dose response models were visually inspected and for spurious curves, DSS was imputed by assessing the other PARP inhibitors in the same sample (n = 7 out 465 values including 6 NA's). The overall PARP inhibitor sensitivity index for each cell line was defined as the sum of the standardized DSS (z-score) for four highly correlated PARP-inhibitors (see Results). Cell viability was determined using the CellTiter-Glo luminescent assay (Promega, USA).
2.5. Genomic and transcriptomic profiles associated with HRD and PARP inhibitor sensitivity
Mutations were analysed in 141 genes implicated in homologous recombination and/or PARP inhibitor sensitivity (Supplementary Table S3) [11]. Due to varying coverage in the different sources of DNA sequencing data, not all cell lines had mutation data for all relevant genes (Supplementary Table S2). A total HRD score based on DNA copy numbers was calculated using an algorithm [25] that combines three genomic scar signatures associated with HRD; number of telomeric allelic imbalances (NtAI) [17], large scale transitions (LST) [16] and HRD score (HRD-LOH) [15]. The total HRD score was calculated as the unweighted numeric sum of the three individual signatures [12].
COSMIC base substitution signatures (https://cancer.sanger.ac.uk/cosmic/signatures_v2) were calculated from filtered somatic single nucleotide variants in the primary CRCs tumour sample using the R package SomaticSignatures (Supplementary Methods). A gene expression signature (PARP inhibitor sensitivity score) predictive of response to PARP inhibition in preclinical models of breast and ovarian cancer [20] were calculated for the CRC cell lines (Supplementary Methods). Gene set expression enrichment analyses comparing PARP inhibitor sensitive and resistant cell lines were performed using the R package GSA and gene sets from BioCarta (collected from MSigDB: https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp).
2.6. Drug treatment with Talazoparib, Niraparib and Pifithrin-β Hydrobromide
The two microsatellite stable (MSS) cell lines with strongest sensitivity to PARP inhibition in the high-throughput drug screen (SKCO1 and LS513) and two resistant cell lines (SW1222 and SNU61), matched based on potentially relevant molecular features (MSS, ATM mutation status, consensus molecular subtype [CMS]), were selected for additional analyses. Among the PARP inhibitors talazoparib was chosen because it showed the greatest potency in the drug screen, but selected experiments were also performed with niraparib for validation. 2.5–4 × 105 cells were seeded in 60 mm dishes (Falcon, #353002) 24 h prior to incubation with 100 and 1000 nM talazoparib, 10000 nM niraparib (MedChemExpress, #HY-16106), or 0.005% DMSO for control (Sigma, #D5879) for 48 h. After the drug incubation, cells were washed twice with 1xPBS (Gibco, #20010019) and collected after trypsinization.
Combination treatment with talazoparib and pifithrin-β hydrobromide (PFT-β) was performed for SKCO1 and LS513 in 384-well plates. Talazoparib and PFT-β as single agents or in combination were incubated for 48h and 72 h before measuring luminescence using the CellTiter-Glo luminescent assay. The effect of PFT-β on talazoparib activity was analysed with the response additivity method (additive combination drug effect predicted as the sum of the effects of single drugs), as well as combination indices using the CompuSyn 1.0 software.
2.7. Post-treatment gene expression profiling
Nucleic acids were isolated from cell pellets after treatment with talazoparib (100 nM and 1000 nM) or DMSO using the Allprep DNA/RNA/miRNA Universal Kit (Qiagen). Gene expression was analysed using the GeneChip Human Transcriptome Array 2.0 according to the manufacturer's instructions (Thermo Fisher Scientific, Waltham, MA, USA). Raw intensity data stored in CEL files were background corrected, quantile normalized and summarized at the gene-level according to the robust multi-array average (RMA) approach with modified Signal Space Transformation implemented in the Affymetrix Expression Console 1.1 software. The data (total n = 12 samples) have been deposited to GEO and can be accessed from GSE140258.
Differential gene expression analyses comparing treatment and control cultures of the two drug sensitive cell lines (SKCO1 and LS513) was performed using the R package limma (paired mode). Gene set enrichment analysis of the top differentially expressed genes (p < 0.001) was performed with the R package topGO (parameters: algorithm = classic; statistic = “fisher”), based on the Gene Ontology data base and the “biological process” category. A response signature was generated based on the top 5 differentially expressed genes from limma (upregulated), and sample-wise signature scores were calculated by single-sample gene set enrichment analysis (ssGSEA function in the R package GSVA).
2.8. Western blot analysis
Expression of TP53, P21, RAD51, β-actin; cleavage of PARP and phosphorylation of γH2A.X were analysed forty-eight hours after treatment with talazoparib, niraparib, idasanutlin or DMSO (Supplementary Methods).
2.9. Immunofluorescence and confocal microscopy
TP53 and RAD51 nuclear expression in cell lines was analysed after 48 h treatment with talazoparib and PFT-β as described in Supplementary Methods.
2.10. Statistical analyses
Statistical tests were performed with R (v 3.6.1) and SPSS 25.0 (SPSS Inc.) software packages. Survival curves were generated by the Kaplan-Meier method, while univariable and multivariable survival analyses were conducted according to the Cox proportional hazards model. In multivariable survival analyses all specified variables were entered into the models. The fraction of the genome with aberrant copy number or LOH, an indirect marker of chromosomal instability, was included as a continuous variable, while the remaining variables were categorical. Fisher's exact test was used to evaluate associations between categorical variables. The independent samples t-test or one-way ANOVA test were used, when appropriate, to compare continuous variables between groups. Correlation of drug sensitivity scores between PARP inhibitors was tested with the Pearson correlation coefficient (r), while the Spearman correlation coefficient was used to assess correlation between drug sensitivity and copy number- and gene expression-based signatures. Two-sided p values < 0.05 were considered significant.
3. Results
3.1. Truncating mutations in homologous recombination-related genes are mutually exclusive in MSS CRC
Exome sequencing of 176 primary CRCs showed that all MSI tumours (n = 33) had multiple mutations (range 4–34) in genes implicated in homologous recombination and/or PARP inhibitor sensitivity. However, this was likely related to their hypermutated phenotype, and truncating mutations resulting from the high frequency of frameshift mutations inherent to MSI accounted for almost half (46%) of the relevant mutations (Table 1, Supplementary Table S4). BRCA2 and ATM had truncating mutations in 24% and 15% of the tumours, respectively (Table 1), but previously published data from this cohort showed no signs of base substitution signatures associated with HRD [30] (signature 3; more or less equal representation of all base substitutions and their sequence context; [38]), as all MSI tumours had a base substitution signature dominated by defective DNA mismatch repair (signature 6).
Table 1.
Most frequently mutated homologous recombination-related in primary CRCs
| MSS tumours (n = 143) |
MSI tumours (n = 33) |
||||
|---|---|---|---|---|---|
| Gene | Frequency1 (%) | Truncating2 (%) | Gene | Frequency (%) | Truncating (%) |
| ATM | 10 (7) | 3 (2) | MSH3 | 20 (61) | 20 (61) |
| PTEN | 7 (5) | 1 (0.7) | ATR | 16 (48) | 13 (39) |
| BRCA2 | 6 (4) | 1 | BRCA2 | 11 (33) | 8 (24) |
| MSH3 | 4 (3) | 0 | G2E3 | 11 (33) | 10 (30) |
| SMARCA2 | 3 (2) | 1 | TOP3A | 11 (33) | 5 (15) |
| REV3L | 3 (2) | 1 | REV3L | 9 (27) | 8 (24) |
| SLX4 | 3 | 1 | SMARCA2 | 9 (27) | 1 (3) |
| DNMT3A | 3 | 0 | BLM | 9 (27) | 7 (21) |
| MUS81 | 3 | 0 | TOP2B | 9 (27) | 4 (12) |
| POLK | 3 | 0 | SHPRH | 8 (24) | 6 (18) |
| C1orf86 | 3 | 0 | ATM | 8 (24) | 5 (15) |
| DNASE1L2 | 3 | 0 | TP53BP1 | 7 (21) | 1 (3) |
Number of tumours with any non-silent mutation
Nonsense and frameshift mutations
Among the 143 MSS tumours, 74 (52%) harboured one or more mutations (range 1–5) in homologous recombination-related genes. A total of 111 mutations were detected, distributed across 59 (42%) of the 141 analysed genes. The large majority were missense mutations, while truncating (nonsense or frameshift) mutations accounted for 11% (Supplementary Table S4), were mutually exclusive, and detected in 8% of MSS tumours. ATM was the most frequently mutated gene (ten tumours, 7%, Table 1) and three ATM mutations were predicted to be truncating, although not likely to confer complete loss of function, due to mutant allele fractions in the range of 0.21 to 0.3. One tumour harboured a frameshift mutation in BRCA2 (p.Q1782fs), which is expected to lead to loss of BRCA2 protein function, but with a mutant allele fraction of only 0.34. None of the MSS tumours with truncating mutations in BRCA2 or ATM displayed base substitution signature 3.
3.2. Various mutational patterns indicative of HRD are non-overlapping in CRC
Subsequently, we investigated mutational patterns associated with HRD on a DNA copy number level by calculating a previously validated HRD score [25] in 199 primary CRCs. MSS tumours had significantly higher HRD scores than MSI tumours (mean MSS 17.8 versus MSI 5.6, p < 0.001, independent samples t-test, Fig. 1a, Supplementary Table S5a), and the HRD scores were associated with the fraction of the genome with aberrant copy number (r = 0.66, p < 0.001, Pearson's correlation). This is in line with previous results across cancer types [25] and supports that the frequent truncating mutations in homologous recombination-related genes in MSI tumours do not have a significant impact on the ongoing mutational processes.
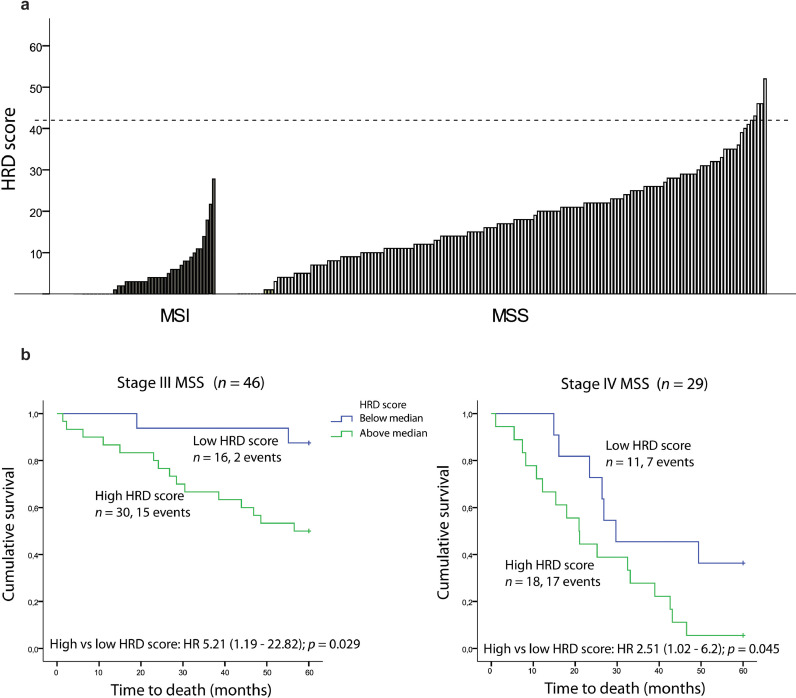
Fig. 1.
HRD scores and their prognostic impact in primary colorectal cancers. a DNA copy number-based HRD scores in 199 primary tumours, split according to MSI status. Dashed horizontal line marks HRD score ≥ 42 used as threshold for homologous recombination deficiency in previous non-CRC publications. b Kaplan-Meier survival curves showing five-year overall survival (OS) in patients with MSS tumours with high (above median) and low HRD scores (below median) in stage III (left panel) and stage IV (right panel).
High HRD scores were further associated with TP53 mutations, left sided tumour location, and stage III or IV tumours (Supplementary Table S5a). Univariable survival analysis encompassing all disease stages in MSS tumours showed that high HRD scores (above median) were associated with inferior five-year overall survival (hazard ratio (HR) 2.00; 1.16–3.44; p = 0.013, Supplementary Fig. 1a). Stage-specific analyses demonstrated negative prognostic impact of high HRD scores among stage III (HR 5.21; 1.19–22.82; p = 0.029, Fig. 1b) and IV MSS cancers separately (HR 2.51; 1.02–6.2; p = 0.045, Fig. 1b), while no association with survival was seen in stages I–II (Supplementary Fig. S1b). Stage-specific negative prognostic trends were seen also in multivariable analyses incorporating age, gender, primary tumour location, chemotherapy, TP53 mutation status and the extent of genomic copy number aberrations, although statistically significant only in stage IV (Stage III: HR 2.98; 0.56–16.02; p = 0.20, Stage IV: HR 3.04; 1.02–9.09; p = 0.046).
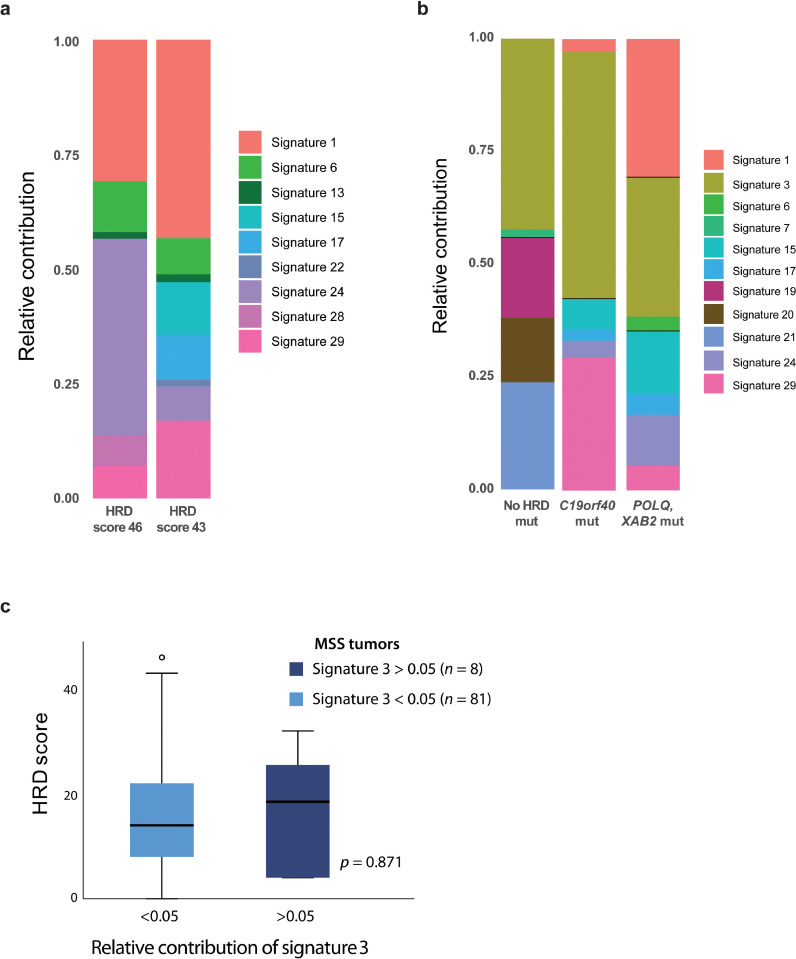
Five MSS tumours (2.5%) had HRD scores ≥ 42 (Supplementary Table S5b), which has been indicative of HRD and PARP inhibitor sensitivity in other cancer types [3,12]. Two of these five samples had whole-exome sequencing data available; one sample (HRD score 43) harboured three individual missense mutations in RAD23B, while the other (HRD score 46) had missense mutations in BRCA2 and FANCG. However, none of the two tumours had base substitution signatures associated with HRD (Fig. 2a). Signature 3 was the largest contributor to underlying mutational processes in three (2%) of the 143 MSS tumours (Fig. 2b). One of these harboured a frameshift mutation in POLQ, with a mutant allele fraction of 0.30. The other two did not have any truncating mutations in homologous recombination-related genes. Across all MSS tumours, there was no difference in HRD scores between tumours with a detectable contribution of signature 3 (at least 5% relative contribution) compared with tumours without contribution from this signature (p = 0.9, independent samples t-test, Fig. 2c). In summary, we identified gene-specific or genome-wide mutational patterns associated with HRD in a small subset of MSS tumours. However, there was no sample overlap between the features, and altogether non-consistent evidence of HRD in the genome of these primary CRCs.
Fig. 2.
Base substitution mutational signatures in primary colorectal cancers. a Relative contribution of the various base substitution mutational signatures (as designated in COSMIC and described by Alexandrov et al.) in the two samples with HRD scores ≥ 42 and available whole-exome sequencing data. b Relative contribution of mutational signatures in the three samples where signature 3 was most dominant. One sample harboured a frameshift mutation in POLQ and a missense mutation in XAB2 with mutant allele fractions of 0.30 and 0.66, respectively. Another sample had a missense mutation in C19orf40 with a mutant allele fraction of 0.21, while no mutations in homologous recombination-related genes were detected in the last sample. None of these three tumours had DNA copy number data and HRD scores available. The frequency of different types of base substitutions for Fig. 3a and b is shown in Supplementary Fig. S2a–b. c Comparison of HRD scores in MSS tumours according to relative contribution of signature 3. P value from independent samples t-test.
3.3. A small subset of CRC cell lines are sensitive to PARP inhibition
The in vitro drug screen of 93 CRC cell lines showed that PARP inhibition was active in a small subset of samples, based on the standardized and summed DSS (the PARP inhibitor sensitivity index) of four different PARP inhibitors (olaparib, niraparib, rucaparib and talazoparib; Fig. 3a). Among the five PARP inhibitors tested, talazoparib was the most potent in the 8 most sensitive cell lines (Fig. 3b). Veliparib was least potent and correlated weakly with the four others (Pearson's r < 0.3, Supplementary Table S6), possibly due to its previously reported lacking ability to trap PARP [7], and was therefore removed from the analyses.
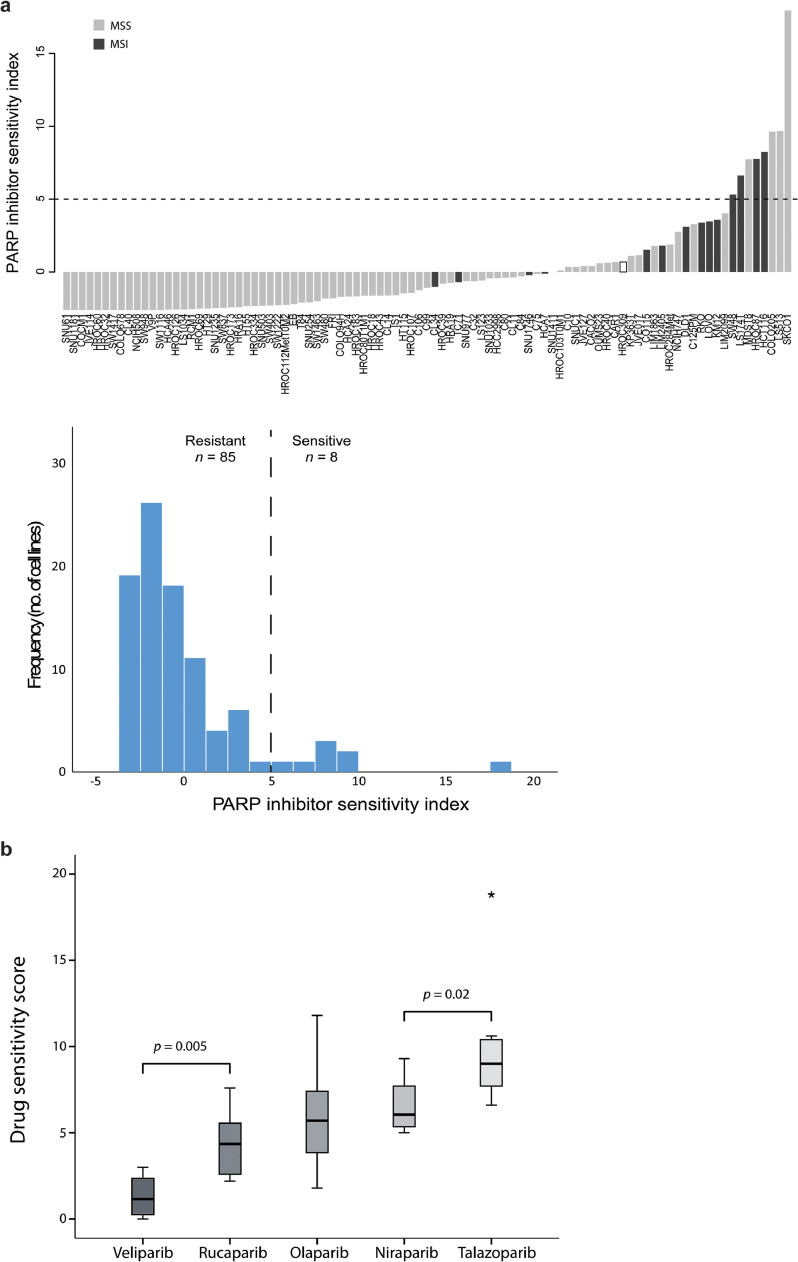
Fig. 3.
PARP inhibitor drug sensitivity scores per cell line. a Barplot with cell lines ordered according to a PARP inhibitor sensitivity index (the sum of the standardized drug sensitivity score for four PARP inhibitors with veliparib excluded due to low activity/low correlation). Dashed horizontal line indicates value used for dichotomy in downstream analyses. Samples are coloured according to MSI-status (dark grey: MSI; grey: MSS; white: unknown). Histogram shows distribution of PARP inhibitor drug sensitivity scores among 93 cell lines. b Comparison of potency of the five PARP inhibitors based on drug sensitivity scores in the eight cell lines classified as PARP inhibitor sensitive. P values are from paired-samples t-tests.
The target PARP1 was highly expressed in most cell lines, and correlated weakly with sensitivity (Pearson's r = 0.23; Supplementary Fig. S3a). MSI cell lines had in general a higher PARP inhibitor sensitivity index than MSS (p < 0.001, Wilcoxon rank sum test), but the three most sensitive cell lines were MSS. The cell lines were characterized as sensitive or resistant using a threshold value for the sensitivity index of 5 (defined by a discontinuity in the distribution of index values), classifying 8 of 93 cell lines (9%) as sensitive (Fig. 3a, Table 2). These included 4 MSS and 4 MSI cell lines. Although there was an enrichment with MSI among the sensitive cell lines (OR = 7.4; 95% confidence interval, 1.59 –34.35, p = 0.017, Fisher's exact test), the positive predictive value of MSI status for sensitivity to PARP inhibition was only 29%.
Table 2.
Molecular characteristics of PARP inhibitor sensitive cell lines
| MSI status | TP53 | CMS | HRD score | Biallelic aberration HR gene1 | No. of mutated HR genes2 | PARPi sensitivity score3 | |
|---|---|---|---|---|---|---|---|
| SKCO1 | MSS | wt | CMS3 | 50 | ATM | 1 | -0.37 |
| LS513 | MSS | wt | CMS3 | 9 | No | 4 | 0.06 |
| COLO205 | MSS | mut | n.a. | 42 | No | 1 | -0.16 |
| HCT116 | MSI | wt | CMS4 | 17 | MSH3 | 32 | 0.42 |
| HROC87 | MSI | n.a. | CMS1 | n.a. | n.a. | n.a. | n.a. |
| MDST8 | MSS | wt | CMS4 | 21 | PTEN, REV3L | 6 | 0.1 |
| LS174T | MSI | wt | CMS3 | 22 | No | 23 | 0.06 |
| SW48 | MSI | wt | n.a. | 7 | No | 23 | 0.45 |
Genes harbouring homozygous mutations, biallelic deletions or mutations in the single remaining allele included. 133 genes implicated in homologous recombination and/or PARP inhibitor sensitivity have been investigated.
See Supplementary Table S7 for full list of mutations in relevant genes
Gene expression-based signature of PARP inhibitor sensitivity
HR: homologous recombination, n.a.; not available, mut; mutated, PARPi; PARP inhibitor wt; wild-type
3.4. Existing HRD-related mutations and genomic signatures have limited predictive power for PARP inhibition in CRC cell lines
All the PARP inhibitor sensitive cell lines with available DNA sequencing data (n = 7) harboured mutations in at least one gene implicated in homologous recombination and/or PARP inhibitor sensitivity (range 1–32 mutated genes, Table 2, Supplementary Table S7), although biallelic alterations were detected in only 3 cell lines (2 MSS, 1 MSI, Table 2). SKCO1 (MSS) had a homozygous splice mutation in ATM, while HCT116 (MSI) and MDST8 (MSS) had homozygous mutations in MSH3 and PTEN, respectively. MDST8 also harboured a hemizygous mutation in REV3L. None of the sensitive samples with DNA copy number data had complete losses of any of the homologous recombination-related gene loci. Notably, 31% (16/52) of the PARP inhibitor resistant cell lines also had homozygous mutations in at least one of these genes, including truncating mutations in MSH3 (HCA7), PTEN (LIM2405) or ATM (SW1222). The positive predictive value of a homozygous mutation in any homologous recombination-related gene for PARP inhibitor sensitivity was only 16%, suggesting shortcomings in applying an HRD-related gene-specific strategy to predict PARP inhibitor sensitivity in CRC.
Next, we evaluated the predictive power of the DNA copy number based HRD score [12] and a gene expression-based signature of PARP inhibitor sensitivity [20]. Neither the genomic HRD scores (n = 43 cell lines; HRD scores: resistant 28.6 versus sensitive 24.0, p = 0.5, independent samples t-test) nor the transcriptomic sensitivity score (n = 75 cell lines; p= 0.46, independent samples t-test) were associated with efficacy of PARP inhibition in CRC cell lines. Both signatures lacked predictive value also when analysed separately in MSI and MSS cell lines (p > 0.6). These results indicate that predictive signatures of PARP inhibitor efficacy developed in other cancer types cannot be translated to CRC.
3.5. PARP inhibitor sensitivity is strongly associated with TP53 wild-type status in CRC cell lines
Further exploratory analyses of the molecular correlates of PARP inhibitor sensitivity were focused on the MSS subgroup to reduce the potential influence from DNA repair mechanisms not related to homologous recombination. Gene set expression enrichment analysis (GSEA) showed that DNA damage-, TP53-, and immune-related signatures were upregulated in sensitive compared to resistant cell lines (Fig. 4a). Moreover, a significant enrichment with TP53 wild-type status was seen among the PARP inhibitor sensitive samples (OR = 15.7; 95% confidence interval 1.5–168.1, p = 0.023, Fisher's exact test, Fig. 4b). Notably, this was also evident when including MSI cell lines (p = 0.001; Supplementary Table S8a and Fig. S3b), and multiple regression analysis incorporating TP53 and MSI status showed that only the former was significantly associated with PARP inhibitor sensitivity (p = 0.001 and p = 0.39, respectively, Supplementary Table S8b).
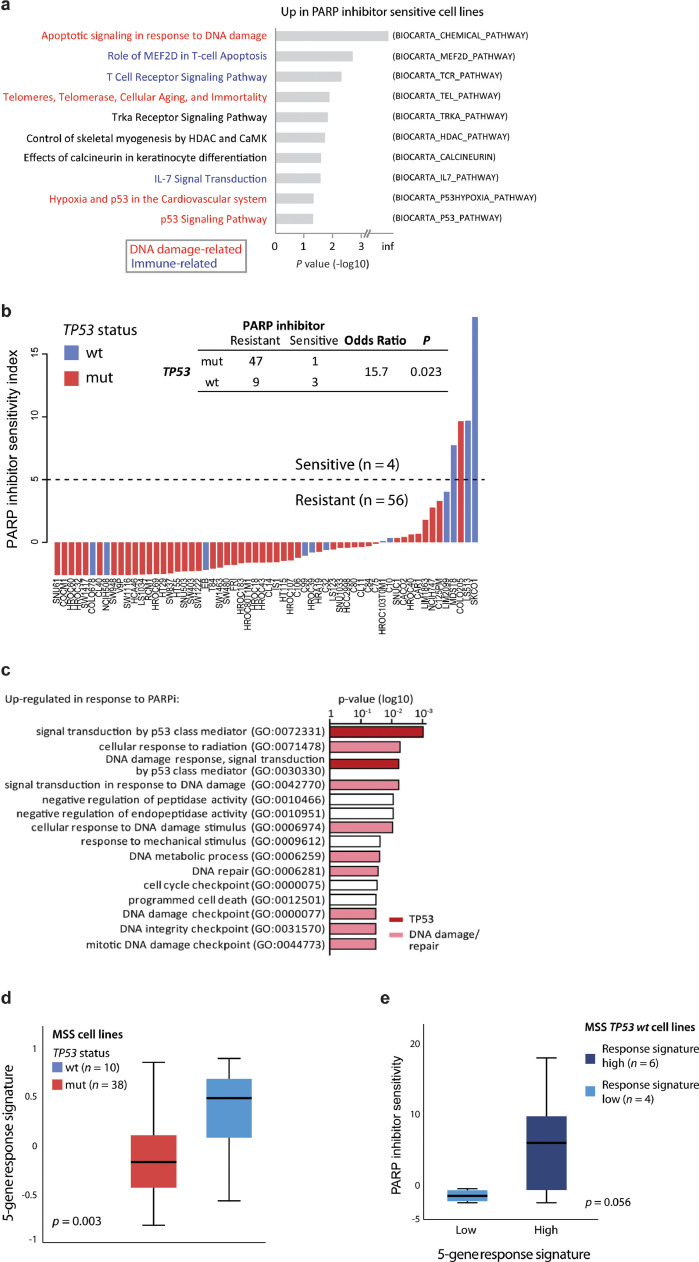
Fig. 4.
PARP inhibitor sensitivity is associated with TP53 activity. a Gene set enrichment analysis comparing PARP inhibitor resistant (n = 53) and sensitive (n = 4) colorectal cancer cell lines. Only microsatellite stable cell lines are included. Gene sets from BioCarta. The ten most upregulated gene sets in PARP inhibitor sensitive cell lines are shown, ranked according to p values (log10-scale). b Barplot shows summed standardized PARP inhibitor drug sensitivity scores per cell line, ranked according to drug sensitivity and coloured according to TP53 mutation status. Dashed horizontal line indicates value used for dichotomy in downstream analyses. Only MSS cell lines with known TP53 mutation status are included (n = 60). Similar plot including MSI cell lines is shown in Supplementary Fig. S3A. c Gene set enrichment analysis of genes associated with PARP inhibitor response. Analysis based on genes upregulated (paired limma-analysis) in PARP inhibitor sensitive cell lines (SKCO1 and LS513) after treatment with talazoparib compared to DMSO (control). d Five-gene response signature (GDF15, PLK2, MDM2, TP53INP1, RRM2B) according to TP53 mutation status in untreated MSS cell lines. e Comparison of PARP inhibitor sensitivity scores in MSS TP53 wild-type (wt) cell lines split according to the 5-gene response signature. Samples are dichotomized according to high or low gene response signature using a threshold of 0.3, reflecting the largest discontinuity in the distribution of response signature values. P value from Welch's t-test.
3.6. Transcriptional activity downstream of TP53 is a primary response to PARP inhibition in CRC cell lines
Response mechanisms of PARP inhibition were analysed by gene expression profiling of pairs of sensitive (SKCO1 and LS513; TP53 wild-type) and resistant (SW1222 and SNU61; TP53 mutated) MSS cell lines after treatment with talazoparib or DMSO for control (Supplementary Fig. S3c-d and Table S9). GSEA of the most upregulated genes (treatment versus control) in the two sensitive cell lines showed that DNA damage repair, and more specifically TP53 activity, was among the primary response mechanisms (Fig. 4c). Notably, the five most upregulated genes (GDF15, PLK2, MDM2, TP53INP1, RRM2B) were all known transcriptional targets of TP53, and were upregulated exclusively in the sensitive cell lines (Supplementary Fig. S3e). A treatment response signature based on these five genes was significantly upregulated in TP53 wild-type compared to mutated samples in the larger panel of MSS cell lines prior to treatment exposure (Fig. 4d). Furthermore, in the TP53 wild-type group, sensitivity to PARP inhibitors was indeed higher in cell lines with a high response signature (Fig. 4e), although the low number of cell lines within each subgroup precluded statistically significant associations (p = 0.056).
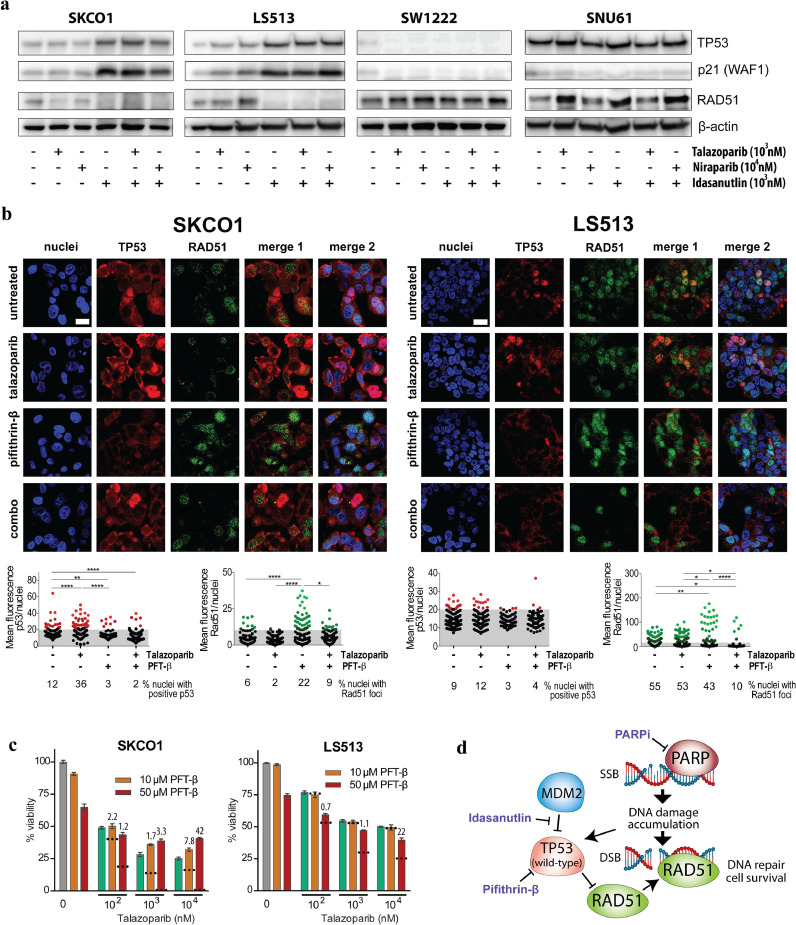
Increased transcriptional activity downstream of TP53 was further investigated at the protein level after treatment with two different PARP inhibitors (talazoparib and niraparib) and an MDM2-TP53 wild-type interaction inhibitor (idasanutlin) as a positive control (Fig. 5a and Supplementary Fig. S4 and S5). Idasanutlin increased the protein levels of both TP53 and its downstream cyclin-dependent kinase inhibitor P21 strongest in the PARP-inhibitor sensitive and TP53 wild-type cell lines. There was no further increase after combination with PARP inhibition, but single-agent treatment with either PARP inhibitor increased the expression levels of P21 exclusively in the sensitive cell lines. Collectively, these data suggest that PARP inhibitor sensitivity is associated with wild-type TP53 activity in CRC.
Fig. 5.
TP53 activation and regulation of RAD51 after PARP inhibition. a Western Blot analyses of TP53, p21 and RAD51 in PARP inhibitor sensitive (SKCO1 and LS513) and resistant cell lines (SW1222 and SNU61) after treatment with talazoparib, niraparib and idasanutlin for 48 h. b Representative images of TP53 expression and RAD51 foci analysed with fluorescence microscopy after 48 h treatment with 1 µM talazoparib, 50 µM pifithrin-β (PFT-β) or combination of the two. Scale bar = 20 µm. Scatter dot plots below depict analysed TP53- (red) and RAD51 (green) mean fluorescence per nuclei for the indicated treatment and cell lines, respectively. For statistical analyses, one-way ANOVA multiple comparison test with Tukey post correction was performed (****p < 0.0001, **p < 0.005, *p < 0.05). Each dot represents mean fluorescence in one nuclei and grey area indicates analysed nuclei (black) with no TP53 or RAD51 expression. c Viability of SKCO1 and LS513 cell lines after 72 h combination treatment with talazoparib and PFT-β. Values given are means ± standard error of the mean. Dotted lines indicate the predicted additive effect calculated as the sum of mean inhibitory effects from single drugs. Values above the bars for combination treatments indicate combination indexes (CI). CI < 1, CI = 1 or CI > 1 represent synergism, additive and antagonistic effects, respectively. d Schematic depiction of TP53 and RAD51 involvement after PARP inhibition in TP53 wild-type cell lines.
3.7. Wild-type TP53 inhibits RAD51 foci formation in response to PARP inhibition
To evaluate the involvement of homologous recombination in the association between wild-type TP53 and sensitivity to PARP inhibition, we analysed RAD51 expression and RAD51 foci formation, which are surrogate markers of homologous recombination functionality [39]. Consistent with a previous study reporting that wild-type TP53 can repress RAD51 transcription in response to double-stranded DNA breaks (40), we observed decreased RAD51 protein expression after talazoparib treatment in the two TP53 wild-type cell lines SKCO1 and LS513. Down-regulation of RAD51 was strongest in SKCO1, which was also the cell line most sensitive to PARP inhibition (Fig. 5a and Supplementary Fig. S6a and S6c). Similar results were found by immunofluorescence microscopy, showing that talazoparib significantly increased the number of TP53-positive nuclei (3 fold) and decreased the RAD51 foci (3 fold) in SKCO1, and to a lesser extent in LS513 (3% increase of TP53 expression and 2% decrease of RAD51 foci; Fig. 5b) Consequently, SKCO1 also exhibited the highest increase of the DNA double-strand break marker γH2A.X and the apoptotic marker cleaved PARP after talazoparib treatment (Supplementary Fig. S6a, d-e). In contrast to talazoparib, treatment with the TP53 inhibitor PFT-β had the opposite effect on TP53 and RAD51, with a significant decrease of TP53 expression and 4 fold decrease of expressing nuclei, as well as an increase of nuclei with RAD51 foci (4 fold) in SKCO1. A significant increase of RAD51 foci was also observed in the LS513 cell line after TP53 inhibition, despite the fact that fewer RAD51 expressing nuclei were counted compared to control (1.3 fold). Consequently, co-treatment with PFT-β and talazoparib in the two PARP inhibitor sensitive cell lines showed that TP53 inhibition antagonized the impact of PARP inhibition on cell viability after 48h and 72h, providing further support that wild-type TP53 is required for the activity of PARP inhibitors in CRC cell lines (Fig. 5c–d and Supplementary Fig. S7).
4. Discussion
The clinical potential of targeting low-frequency molecular aberrations in CRC has been demonstrated using several anticancer agents with different mechanisms of action [[41], [42], [43]]. Our results from high-throughput drug screening of a large panel of CRC cell lines suggest a potential for development of a PARP inhibitor strategy. However, only a small subset of the cell lines was sensitive to PARP inhibition, highlighting the need to identify accurate response prediction markers. Existing transcriptomic and genomic signatures related to HRD and with proven predictive value in other cancer types had poor predictive power in CRC. In contrast, a clear association was found between wild-type TP53 function and PARP inhibitor sensitivity. This was consistent at different molecular levels and in different analysis settings, including TP53 wild-type status, a TP53-related gene expression signature within the TP53 wild-type group, downstream TP53 activity as one of the main response mechanisms after treatment, and the antagonistic effect of TP53 inhibition. This is somewhat in contrast to the efficacy of PARP inhibitors seen in ovarian cancer, in which TP53 is almost invariably mutated [44]. However, recent publications have shown that synergy between PARP inhibition and either ionizing radiation or chemotherapy depends on functional TP53 [45,46]. Furthermore, reactivation of mutant TP53 in combination with olaparib showed highly effective tumour growth inhibition in pre-clinical models of TP53 mutant TNBC [47]. These findings indicate context-dependent molecular underpinnings for PARP inhibitor efficacy. While our study does not imply that TP53 mutation status has sufficient predictive value to be used as a solitary biomarker for PARP inhibition in CRC, it may point to the TP53 wild-type group as an overarching stratum more likely to contain responders. Our study is based on the in vitro activity of PARP inhibition, and additional pre-clinical analyses are needed to specify a strategy for clinical testing.
DNA damage-related mechanisms were indeed involved in responses to PARP inhibition also in CRC cell lines, but not necessarily related to the known genomic markers of HRD. In most genes implicated in homologous recombination and PARP inhibitor sensitivity, biallelic aberrations are required for oncogenic impact [11]. We identified biallelic aberrations in ATM, PTEN and MSH3 in three of the PARP inhibitor sensitive cell lines. Homozygous deletion of ATM in SKCO1 cells (TP53 wild-type) has been shown to induce sensitivity to olaparib [22], whereas PTEN deficiency has been shown to sensitize CRC cell lines to PARP inhibition [48]. However, the lack of PARP inhibitor sensitivity in other cells harbouring homozygous frameshift mutations in ATM (SW1222: TP53 mutated) and PTEN (LIM2405) in our study, demonstrates a more complex biomarker-drug association. Notably, while loss of BRCA1 or PTEN have been shown to cause HRD in diverse cancer cells, simultaneous inactivation of both genes reactivates homologous recombination and confers resistance to PARP inhibition [18]. Thus, each mutation must be interpreted in the context of other relevant co-occurring mutations to precisely characterize the effect on homologous recombination competence and PARP inhibitor sensitivity. Our results suggest that TP53 mutation status should be taken into account in CRC.
Analyses of response mechanisms to PARP inhibitors suggested that RAD51 may provide a link between wild-type TP53, HRD and treatment sensitivity in CRC. RAD51 is a crucial player in homologous recombination and assists in double-strand break repair by binding to DNA and mediating homologous pairing and strand exchange [49]. Up-regulation of RAD51 seems to reduce sensitivity to excessive DNA damage in cancer cells and to represent a compensatory pathway for other deficient repair mechanisms [50]. TP53 has been reported to down-regulate important homologous recombination proteins, including RAD51, and thus inhibit inappropriate DNA repair [40]. However, the frequent mutations of TP53 in tumour cells cause a tendency for RAD51 to be overexpressed and to increase the resistance to DNA damage. Notably, a recent study in patient-derived CRC models demonstrated that RAD51 foci formation after exposure to radiation was associated with resistance to olaparib, supporting the relevance of RAD51 as a marker of homologous recombination proficiency and sensitivity to PARP inhibitors [51]. These results are in line with our findings, which indicated that wild-type TP53-mediated suppression of RAD51 is a possible mechanism by which PARP inhibitors exhibit their activity.
The limited overlap of previously reported genomic and transcriptomic signatures of HRD among primary CRCs in our study supported the pre-clinical data and indicated that the molecular characteristics of HRD are different in CRC than other cancer types in which synthetic lethality with PARP inhibition is well described. The poor-prognostic association of a high DNA copy number-based HRD score in stage III-IV CRC is supported by analyses of CRCs in TCGA [52], but is in contrast to the positive prognostic impact of HRD in TNBC and ovarian cancer [15,17]. The latter analyses were performed in patient cohorts receiving the DNA damaging compounds cisplatin or carboplatin, to which HRD confers sensitivity through inefficient DNA repair, thereby contributing to the superior survival. Patients with stage III-IV CRC are commonly treated with oxaliplatin-containing chemotherapy, which in contrast to other platinum compounds does not kill cells through the DNA damage response [53]. Thus, HRD is not expected to be beneficial for treatment efficacy in this setting. However, these somewhat conflicting results reinforce the need for interpretation of genomic HRD scores according to cancer type. A recent study suggested that the phenotypic impact of BRCA mutations on mutational patterns and PARP inhibitor sensitivity is restricted to cancer types associated with BRCA-mediated heritable cancer risk (i.e. breast, ovary, pancreatic and prostate cancers) [54]. Thus, mutations in BRCA1/2 and other genes related to HRD found in CRC may be biologically neutral passenger mutations with limited impact on phenotype and drug sensitivity. However, the exact impact of each gene on homologous recombination and/or PARP inhibitor sensitivity is likely very diverse, both with respect to character and magnitude. The association between defects in ATM and abrogated homologous recombination and sensitivity to PARP inhibition is well-characterized in various tumour types [11]. Our finding of SKCO1 cells with deleted ATM and TP53 wild-type status to have clearly the strongest PARP inhibitor sensitivity and the highest HRD score among the sensitive cell lines provides multi-level support of ATM defects as a relevant predictive marker of PARP inhibition also in CRC.
In conclusion, we demonstrated activity of PARP inhibitors in a subset of CRC cell lines. The limited predictive value of existing HRD-related biomarkers suggested a need for development of novel stratified treatment strategies with respect to PARP inhibition in CRC, and this study points to wild-type TP53 function as a relevant starting point.
Acknowledgments
Acknowledgements
Not applicable.
Funding sources
This work was supported by the Norwegian Cancer Society (project numbers 6824048-2016 and 182759-2016); the foundation "Stiftelsen Kristian Gerhard Jebsen"; the South-Eastern Norway Regional Health Authority (project number 2017102), and the Research Council of Norway (FRIPRO Toppforsk, project number 250993). The funders had no role in study design, data acquisition, analysis, interpretation, writing or submission of the manuscript.
Declaration of Interests
The authors declare no competing interests.
Authors' contributions
RAL and AS conceived the study. RAL, AS, MGG, AN, JB and JS designed the study. AN, KK, KCGB, PWE, IAE and JB acquired data. KCGB, PWE, AS, RAL, KK, BJ and JS analysed and/or interpreted data. JS and AS drafted the manuscript. All authors revised and approved the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.ebiom.2020.102923.
Appendix. Supplementary materials
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 3.Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N Engl J Med. 2016;375(22):2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 4.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 5.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373(18):1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381(4):317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pommier Y, O'Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med. 2016;8(362) doi: 10.1126/scitranslmed.aaf9246. 362ps17. [DOI] [PubMed] [Google Scholar]
- 8.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 9.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 10.Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366(15):1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 11.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 12.Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC. Homologous Recombination Deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22(15):3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23(4):517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, Stratton MR. A mutational signature in gastric cancer suggests therapeutic strategies. Nat Commun. 2015;6:8683. doi: 10.1038/ncomms9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107(10):1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popova T, Manie E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72(21):5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 17.Birkbak NJ, Wang ZC, Kim JY, Eklund AC, Li Q, Tian R. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2(4):366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng G, Chun-Jen Lin C, Mo W, Dai H, Park YY, Kim SM. Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat Commun. 2014;5:3361. doi: 10.1038/ncomms4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010;28(22):3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGrail DJ, Lin CC, Garnett J, Liu Q, Mo W, Dai H. Improved prediction of PARP inhibitor response and identification of synergizing agents through use of a novel gene expression signature generation algorithm. NPJ Syst Biol Appl. 2017;3:8. doi: 10.1038/s41540-017-0011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leichman L, Groshen S, O'Neil BH, Messersmith W, Berlin J, Chan E. Phase II Study of Olaparib (AZD-2281) after standard systemic therapies for disseminated colorectal cancer. Oncologist. 2016;21(2):172–177. doi: 10.1634/theoncologist.2015-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, Jette N, Moussienko D, Bebb DG, Lees-Miller SP. ATM-deficient colorectal cancer cells are sensitive to the PARP inhibitor olaparib. Transl Oncol. 2017;10(2):190–196. doi: 10.1016/j.tranon.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol. 2017;35(10):1086–1095. doi: 10.1200/JCO.2016.71.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marquard AM, Eklund AC, Joshi T, Krzystanek M, Favero F, Wang ZC. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res. 2015;3:9. doi: 10.1186/s40364-015-0033-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun J, Wang C, Zhang Y, Xu L, Fang W, Zhu Y. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat Commun. 2019;10(1):3190. doi: 10.1038/s41467-019-10987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dörsam B, Seiwert N, Foersch S, Stroh S, Nagel G, Begaliew D. PARP-1 protects against colorectal tumor induction, but promotes inflammation-driven colorectal tumor progression. Proc Natl Acad Sci U S A. 2018;115(17):E4061–E4e70. doi: 10.1073/pnas.1712345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merok MA, Ahlquist T, Royrvik EC, Tufteland KF, Hektoen M, Sjo OH. Microsatellite instability has a positive prognostic impact on stage II colorectal cancer after complete resection: results from a large, consecutive Norwegian series. Ann Oncol. 2013;24(5):1274–1282. doi: 10.1093/annonc/mds614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smeby J, Sveen A, Eilertsen IA, Danielsen SA, Hoff AM, Eide PW. Transcriptional and functional consequences of TP53 splice mutations in colorectal cancer. Oncogenesis. 2019;8(6):35. doi: 10.1038/s41389-019-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sveen A, Johannessen B, Tengs T, Danielsen SA, Eilertsen IA, Lind GE. Multilevel genomics of colorectal cancers with microsatellite instability-clinical impact of JAK1 mutations and consensus molecular subtype 1. Genome Med. 2017;9(1):46. doi: 10.1186/s13073-017-0434-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg KCG, Sveen A, Holand M, Alagaratnam S, Berg M, Danielsen SA. Gene expression profiles of CMS2-epithelial/canonical colorectal cancers are largely driven by DNA copy number gains. Oncogene. 2019;38(33):6109–6122. doi: 10.1038/s41388-019-0868-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA. Multi-omics of 34 colorectal cancer cell lines - a resource for biomedical studies. Mol Cancer. 2017;16(1):116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74(12):3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 34.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45(D1):D777–DD83. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nature Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 37.Sveen A, Bruun J, Eide PW, Eilertsen IA, Ramirez L, Murumagi A. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res. 2018;24(4):794–806. doi: 10.1158/1078-0432.CCR-17-1234. [DOI] [PubMed] [Google Scholar]
- 38.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cruz C, Castroviejo-Bermejo M, Gutiérrez-Enríquez S, Llop-Guevara A, Ibrahim YH, Gris-Oliver A. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann Oncol. 2018;29(5):1203–1210. doi: 10.1093/annonc/mdy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arias-Lopez C, Lazaro-Trueba I, Kerr P, Lord CJ, Dexter T, Iravani M. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 2006;7(2):219–224. doi: 10.1038/sj.embor.7400587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738–746. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 43.Drilon A, Siena S, Ou S-HI, Patel M, Ahn MJ, Lee J. Safety and antitumor activity of the multi-targeted Pan-TRK, ROS1, and ALK Inhibitor Entrectinib (RXDX-101): combined results from two phase 1 trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017 doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cancer Genome Atlas Research N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474(7353):609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sizemore ST, Mohammad R, Sizemore GM, Nowsheen S, Yu H, Ostrowski MC. Synthetic lethality of PARP Inhibition and ionizing radiation is p53-dependent. Mol Cancer Res. 2018;16(7):1092–1102. doi: 10.1158/1541-7786.MCR-18-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Augustine T, Maitra R, Zhang J, Nayak J, Goel S. Sensitization of colorectal cancer to irinotecan therapy by PARP inhibitor rucaparib. Invest New Drugs. 2019;37(5):948–960. doi: 10.1007/s10637-018-00717-9. [DOI] [PubMed] [Google Scholar]
- 47.Na B, Yu X, Withers T, Gilleran J, Yao M, Foo TK. Therapeutic targeting of BRCA1 and TP53 mutant breast cancer through mutant p53 reactivation. NPJ breast cancer. 2019;5:14. doi: 10.1038/s41523-019-0110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mendes-Pereira AM, Martin SA, Brough R, McCarthy A, Taylor JR, Kim JS. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1(6-7):315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumann P, Benson FE, West SC. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87(4):757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 50.Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res. 2007;67(20):9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- 51.Arena S, Corti G, Durinikova E, Montone M, Reilly NM, Russo M. A Subset of Colorectal Cancers with Cross-Sensitivity to Olaparib and Oxaliplatin. Clin Cancer Res. 2020;26(6):1372–1384. doi: 10.1158/1078-0432.CCR-19-2409. [DOI] [PubMed] [Google Scholar]
- 52.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018;23(1) doi: 10.1016/j.celrep.2018.03.076. 239–54 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruno PM, Liu Y, Park GY, Murai J, Koch CE, Eisen TJ. A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress. Nat Med. 2017;23(4):461–471. doi: 10.1038/nm.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND. Tumour lineage shapes BRCA-mediated phenotypes. Nature. 2019;571(7766):576–579. doi: 10.1038/s41586-019-1382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.