Abstract
Background
Tumour Necrosis Factor (TNF) family members play important roles in mounting anti-tumour immune responses, and clinical trials targeting these molecules are ongoing. However, the expression patterns and clinical significance of TNF members in lung adenocarcinoma (LUAD) remain unrevealed. This study aimed to explore the gene expression profiles of TNF family members in LUAD and constructed a TNF family-based prognosis signature.
Methods
In total, 1300 LUAD cases from seven different cohorts were collected. Samples from The Cancer Genome Atlas (TCGA) were used as the training set, and the RNA data from five Gene Expression Omnibus (GEO) datasets and qPCR data from 102 samples were used for validation. The immune profiles and potential immunotherapy response prediction value of the signature were also explored.
Findings
After univariate Cox proportional hazards regression and stepwise multivariable Cox analysis, a TNF family-based signature was constructed in the TCGA dataset that significantly stratified cases into high- and low-risk groups in terms of OS. This signature remained an independent prognostic factor in multivariate analyses. Moreover, the clinical significance of the signature was well validated in different clinical subgroups and independent validation cohorts. Further analysis revealed that signature high-risk patients were characterized by distinctive immune cell proportions and immune-suppressive states. Additionally, signature scores were positively related to multiple immunotherapy biomarkers.
Interpretation
This was the first TNF family-based model for predicting outcomes and immune landscapes for patients with LUAD. The capability of this signature for predicting immunotherapy response needs further validation.
Keywords: Lung cancer, Immunotherapy, Immune checkpoint, TNF, TMB
Research in context.
Evidence before this study
Tumour Necrosis Factor (TNF) family members comprise key communication systems that regulate the body's anti-tumour immune responses, in addition to immune checkpoint molecules from the B7-CD28 family. Modulating the interactions between the TNFSF/TNFRSF families holds great potential as a novel cancer therapy. To date, there is no relevant study concerning the expression landscape of TNF family members in lung adenocarcinoma.
Added value of this study
Our study conducted a first systematic investigation of the expression details and clinical significance of TNF family members in lung adenocarcinoma. Additionally, we developed and validated a TNF family-based prognostic model using 1300 LUAD cases from seven different cohorts. Importantly, the novel signature was closely associated with distinct immune profiles and biomarkers of the immunotherapy response.
Implications of all the available evidence
Our findings provide a better understanding of the TNF family profile and the clinical and immunological characteristics of lung adenocarcinoma. The TNF family-based novel signature could be a clinically useful tool for prognostic management and the determination of targeted immunotherapies for patients with lung adenocarcinoma.
Alt-text: Unlabelled box
1. Introduction
According to the latest worldwide cancer statistics, lung cancer remains the leading cause of cancer-related incidence and mortality, and represented almost 20% of cancer deaths predicted in 2018 [1]. Lung cancer mainly consists of two subtypes: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC accounts for almost 80% of lung cancer cases and is made up of two major types, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma [2]. LUAD is the predominant histology, and the rates are still increasing [3]. Despite the clinical improvements in, and applications of, a combination of therapeutic strategies and individualized therapies—which primarily consist of tyrosine kinase inhibitors (TKIs)—the 5-year overall survival (OS) rate for LUAD remains only about 16% [4,5]. Therefore, continued efforts to discover specific prognostic methods for patient-specific survival are still needed so that the most suitable therapeutic and management schemes can be designed for distinct subsets of patients with LUAD. With the advancements in multi-omics profiling, numerous studies, which used different expression profiles and bioinformatic methods, have provided additional prognostic evaluation for patients with LUAD [3,6,7]. However, most of the enrolled parameters in these studies were derived from the entire genome or transcriptome with no consideration of biological processes. Consequently, these signatures possessed a natural bias in that they were simply mathematic models that did not reflect the intrinsic character of the cancer itself.
Nowadays, cancer immunotherapy by immune checkpoint blockade (ICB) is becoming a pillar of lung cancer treatment, alongside surgery, chemoradiotherapy, and TKIs [8]. The immune checkpoints inhibitors targeting programmed cell death protein 1 (PD-1) and programmed cell death 1 ligand 1 (PD-L1), which belongs to the B7-CD28 family [9], are the best-described immunotherapy targets and are becoming first-line therapies of choice for treating advanced NSCLCs [10]. However, a significant limitation of these immune checkpoint inhibitors is that more than half of the patients do not respond to PD-1/PD-L1 immunotherapy [11], indicating the existence of another co-stimulatory signal in the LUAD tumour microenvironment.
Recently, studies have revealed that promoting T-cell responsiveness through engaging co-stimulatory receptors from the tumour necrosis factor (TNF) family is another potential treatment, in addition to blocking co-inhibitory immune checkpoints from the B7-CD28 family [12]. The TNF family, which consists of a 19 TNF ligands superfamily (TNFSF) and a 29 TNF receptor superfamily (TNFRSF), mediates signalling which controls the survival, proliferation, differentiation, and effector functions for both immune and non-immune cells [13]. While members of the TNF family generally show proinflammatory functions through activating the NF-κB pathway, the activation of the TNFSF/TNFRSF family may also trigger apoptosis or other forms of cell death, leading the activation or suppression of the immune response in the tumour microenvironment [14]. Therefore, modulating the interactions between the TNFSF/TNFRSF families holds great potential as a cancer therapy. Actually, many therapeutic approaches that target TNF family members—including CD40, OX40, 4–1BB, GITR, and CD27—are now under active investigation in clinical trials for various cancers, including lung cancer [13,15,16]. Nevertheless, the expression patterns and clinical significance of TNF members in LUAD remain unrevealed.
This was a systematic investigation of the expression details and clinical significance of TNF family members in LUAD. Additionally, we developed and validated a TNF family-based prognostic model using 1300 LUAD cases from seven different cohorts. Considering the specific role of the TNFSF/TNFRSF family in the interaction between tumour-infiltrating lymphocytes and tumour cells, and the potential role of this family in controlling the body's response to immunotherapy, we further explored the relationship between the signature and the landscape of immune-related profiles and the association with immunotherapy responses. A better understanding of the TNF family profile and the clinical and immunological characteristics of the TNF family-based signature in LUAD may help optimize cancer immunotherapies.
2. Materials and methods
2.1. Public mRNA expression datasets
We enrolled 1198 LUAD cases from six public datasets in this study. The Cancer Genome Atlas (TCGA) level three RNA-seq data of 502 LUAD samples (Illumina HiSeq 2000) with clinical annotations and overall survival information, acquired from Cancer Genomics Browser of University of California Santa Cruz (UCSC) (https://genomecancer. ucsc.edu), was used as the training set. All the corresponding clinical data and mutation data of these 502 patients with LUAD were also collected from UCSC. Several reprehensive Gene Expression Omnibus (GEO) datasets (http://www.ncbi.nlm.nih.gov/geo) that contained relatively large populations of patients with LUAD (n>80) with clinical annotations and overall survival information were enrolled—including 90 cases from GSE11969 [17], 117 cases from GSE13213 [18], 83 cases from GSE30219 [19], 226 cases from GSE31210 [20] and 180 cases from GSE41271 [21]—as the public validation sets. The mRNA expression from GEO microarray data was first log2 transformed and quantile normalized, and the mean expression was selected as the expression of genes with more than one probe. Several reprehensive GEO datasets containing a relatively large population of LUAD patients (n>80) with clinical annotation and overall survival information were enrolled.
2.2. Samples and quantitative real-time polymerase chain reaction (qRT-PCR) analysis
From May 2013 to September 2014, we gathered a total of 102 frozen surgically resected lung adenocarcinoma tissues from patients at the First Affiliated Hospital of Zhengzhou University. Based on the standard protocols, total RNA was extracted from these collected samples by RNAiso Plus reagent (Takara, #9109). Then, the Prime Script™ RT reagent kit (Takara, #RR047A) was used for the total RNA to reverse single-stranded cDNA total RNA. All cDNA samples were prepared for qRT-PCR. In the qRT-PCR analysis, the enrolled five genes in this TNF family-based signature were detected. SYBR Premix Ex Taq II (Takara, #RR820A) was used in the qRT-PCR, and Agilent Mx3005P was used for analyzing the data. The expression value of the target genes was first normalized to GAPDH, and then log2 transformed for further analysis. The primer sequences of the included five genes and GAPDH are shown in Table S1. This study was approved by the Institutional Review Boards of The First Affiliated Hospital of Zhengzhou University. All patients and, where appropriate, their families provided written informed consent for participation.
2.3. Biological process and pathway enrichment analysis
The Gene Ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analyses of the TNF family-based signature related genes from TCGA were performed using DAVID 6.8 (http://david.abcc.ncifcrf.gov).
2.4. Immune cell infiltration analysis
The abundance of immune cell infiltration between different groups in this study was estimated by CIBERSORT [22]. CIBERSORT is a novel method widely used for characterizing the cell composition of complex tissues through the gene expression values in solid tumours [23,24]. These characterizations are highly consistent with ground-truth estimations in different cancers [22]. When we used CIBERSORT, the LM22 signature algorithm was applied. LM22 is a specific gene signature containing 547 genes that distinguish 22 immune cell subtypes downloaded from the CIBERSORT web portal (http://cibersort.stanford.edu/). The fractions of 22 immune cell types of patients with LUAD from TCGA, including different T cell types, B cell types, plasma cells, natural killer (NK) cells, and different myeloid subsets, were calculated using CIBERSORT with LM22.
2.5. Mutation, neoantigen and PD-L1 protein analysis
The mutation burden, number of neoantigens, number of clonal neoantigens, and number of subclonal neoantigens in patients with LUAD, obtained from the TCGA dataset, were obtained through The Cancer Immunome Atlas (TCIA) (https://tcia.at/home) [25]. The protein expression of PD-L1 was based on the reverse-phase protein array (RPPA) analysis from the TCGA dataset, which was retrieved from cbioPortal (http://www.cbioportal.org).
2.6. Tumour Immune Dysfunction and Exclusion (TIDE) analysis
TIDE is a computational framework construct by Jiang et al. [26] to predict immune checkpoint blockade response. TIDE integrates the expression of two primary mechanisms of tumour immune evasion: T cell dysfunction and T cell exclusion, to model tumour immune evasion. The TIDE signature was validated and outperformed known immunotherapy biomarkers that could predict immunotherapy response in melanoma and lung cancer, especially in patients treated with anti-CTLA4 and anti-PD-1/PDL1 [26]. The TIDE score, T cell dysfunction score, and T cell exclusion score of patients with LUAD from the TCGA dataset were downloaded from the TIDE web (http://tide.dfci.harvard.edu) after uploading the transcriptome profiles.
2.7. Prognostic meta-analysis
To comprehensively assess the prognostic significance of the TNF family-based signature in different public cohorts, STATA software (version 12.0) was used to conduct the prognostic meta-analysis. The pooled HR value was then calculated using the random-effects model.
2.8. Signature construction and statistical analysis
A univariate Cox proportional hazards regression analysis was used to evaluate the relationship between the gene expression value of TNF family members and the OS of patients with LUAD. Then, a stepwise Cox proportional hazards regression model was used to shrink the variables and screen the most predictive markers among the survival-related genes. The selected genes formed a risk formula that was determined by a linear combination of the gene expression levels and weighted with the corresponding regression coefficients from the stepwise Cox proportional hazards regression model. The proportional hazards assumption was checked by Schoenfeld's partial residuals and none of the variables revealed any indication of violation of this assumption. By ranking the risk formula score, patients were classified into high- and low-risk groups. The Kaplan–Meier method was used to assess the OS scores in the high- and low-risk groups, and a log-rank test was used to calculate the difference in OS between the two groups. The Mann–Whitney U-test was used to analyse the distribution of estimate immune cell-type fractions, the TMB load, number of neoantigens, number of clonal neoantigens, number of subclonal neoantigens, PD-L1 protein expression, TIDE score, T cell dysfunction score, and T cell exclusion score between high- and low-risk groups. In patients with advanced stage disease, the correlations between TMB and the cut-off score and signature were analysed using the chi-square test. Independent prognostic factors were calculated by Cox proportional hazards regression model. P < 0.05 was set as a significant difference in all statistical methods. R software version 3.5.1 (https://www.r-project.org) was used for data analysis and generation of figures.
3. Results
3.1. The landscape and prognostic significance of the TNF family genes in LUAD
We enrolled 47 well-defined TNF family genes in this study, including 18 TNFSF members and 29 TNFRSF members. Firstly, we used 502 patients with TCGA LUAD with complementary prognostic information to explore the TNF family gene expression profile. The demographics of this cohort are listed in Table 1. The expression correlation between the TNF family members is shown in Figure S1. Most of the members showed a strong positive correlation with each other. The univariate Cox proportional hazards regression analysis was used to evaluate the relationship between the gene expression value of TNF family members and the OS of patients with LUAD. Here, we found that 17 genes were significantly associated with OS (P < 0.05, Table 2), including six TNFSF members and 11 TNFRSF members. Among the 17 genes, three genes (TNFRSF1A, LTBR, and TNFRSF6B) were identified as “high-risk” factors, with hazard ratios (HR) greater than 1. There were 14 genes (CD27, TNFRSF10C, TNFRSF17, TNFRSF13B, TNFRSF13C, TNFRSF14, EDA2R, TNFRSF19, TNFSF12, LTB, LTA, CD40LG, TNFSF8, and TNFSF13) that were identified as protective factors because they demonstrated HRs less than 1.
Table 1.
Clinical characteristics of the patients from multiple institutions.
| Characteristics | TCGA n=502 | GSE11969n=90 | GSE13213 n=117 | GSE30219 n=83 | GSE31210 n=226 | GSE41271 n=180 | Independentn=102 |
|---|---|---|---|---|---|---|---|
| Age, year | |||||||
| Median (IQR) | 66 (13.8) | 62.0 (12.0) | 61.0 (13.0) | 60.0 (14.0) | 61.0 (10.0) | 63.9 (13.8) | 60.0 (9.0) |
| Gender | |||||||
| Male | 231 (46.0%) | 47 (52.2%) | 60 (51.3%) | 65 (78.3%) | 105 (46.5%) | 91 (50.6%) | 56 (54.9%) |
| Female | 271 (54.0%) | 43 (47.8%) | 57 (48.7%) | 18 (21.7%) | 121 (53.5%) | 89 (49.4%) | 46 (45.1%) |
| Smoking history | |||||||
| Yes | 416 (82.9%) | 45 (50.0%) | 61 (52.1%) | – | 111 (49.1%) | – | 61 (59.8%) |
| No | 72 (14.3%) | 45 (50.0%) | 56 (47.9%) | – | 115 (50.9%) | – | 41 (40.2%) |
| NA | 14 (2.9%) | 0 (0.0%) | 0 (0.0%) | – | 0 (0.0%) | – | 0 (0.0%) |
| TNM stage | |||||||
| I and II | 388 (77.3%) | 65 (72.2%) | 92 (78.6%) | 83 (100.0%) | 226 (100.0%) | 129 (71.7%) | 80 (78.4%) |
| III and Ⅳ | 105 (20.9%) | 25 (27.8%) | 25 (21.4%) | 0 (0.0%) | 0 (0.0%) | 51 (28.3%) | 22 (21.6%) |
| NA | 9 (1.8%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| OS state | |||||||
| Alive | 320 (63.7%) | 50 (55.6%) | 68 (58.1%) | 40 (48.2%) | 191 (84.5%) | 111 (61.7%) | 78 (76.5%) |
| Death | 182 (36.3%) | 40 (44.4%) | 49 (41.9%) | 43 (51.8%) | 35 (15.5%) | 69 (38.3%) | 24 (23.5%) |
Data are n (%). IQR, interquartile range; NA, not available; OS, overall survival.
Table 2.
Univariate Cox analysis of TNF family genes in TCGA Cohort.
| Official symbol | Aliases | Family | HR | 95%CI | P value |
|---|---|---|---|---|---|
| CD27 | TNFRSF7 | TNFRSF | 0.8682 | 0.7858-0.9592 | 0.0055 |
| CD40 | TNFRSF5 | TNFRSF | 0.9089 | 0.8063-1.0246 | 0.1182 |
| CD40LG | TNFSF5, CD154 | TNFSF | 0.8194 | 0.7421-0.9049 | 0.0000 |
| CD70 | TNFSF7, CD27L | TNFSF | 1.0688 | 0.9757-1.1708 | 0.1522 |
| EDA | EDA-A1, EDA-A2 | TNFSF | 0.9561 | 0.8727-1.0474 | 0.3345 |
| EDA2R | TNFRSF27, XEDAR | TNFRSF | 0.9107 | 0.8387-0.9888 | 0.0259 |
| EDAR | EDA-A1R | TNFRSF | 1.0425 | 0.9744-1.1154 | 0.2275 |
| FAS | TNFRSF6, CD95 | TNFRSF | 0.9501 | 0.8396-1.0752 | 0.4176 |
| FASLG | TNFSF6, CD95-L | TNFSF | 0.9083 | 0.8204-1.0056 | 0.0641 |
| LTA | TNFSF1 | TNFSF | 0.8918 | 0.8002-0.9939 | 0.0385 |
| LTB | TNFSF3 | TNFSF | 0.8771 | 0.7949-0.9679 | 0.0091 |
| LTBR | TNFRSF3 | TNFRSF | 1.3077 | 1.0591-1.6147 | 0.0126 |
| NGFR | TNFRSF16, CD271 | TNFRSF | 1.0011 | 0.9123-1.0986 | 0.9808 |
| RELT | TNFRSF19L | TNFRSF | 0.9259 | 0.7884-1.0873 | 0.3477 |
| TNF | TNFSF2, TNFA | TNFSF | 0.9098 | 0.8257-1.0024 | 0.0560 |
| TNFRSF10A | TRAILR1, CD261 | TNFRSF | 1.0582 | 0.9025-1.2408 | 0.4858 |
| TNFRSF10B | TRAILR2, CD262 | TNFRSF | 1.0336 | 0.8482-1.2596 | 0.7428 |
| TNFRSF10C | TRAILR3, CD263 | TNFRSF | 0.8628 | 0.7727-0.9634 | 0.0087 |
| TNFRSF10D | TRAILR4, CD264 | TNFRSF | 1.0799 | 0.9334-1.2495 | 0.3015 |
| TNFRSF11A | RANK, CD265 | TNFRSF | 1.1316 | 0.9925-1.2902 | 0.0647 |
| TNFRSF11B | OPG | TNFRSF | 1.0088 | 0.9139-1.1135 | 0.8619 |
| TNFRSF12A | FN14, TWEAKR, CD266 | TNFRSF | 1.1040 | 0.9768-1.2477 | 0.1131 |
| TNFRSF13B | TACI, TNFRSF14B, CD267 | TNFRSF | 0.8682 | 0.7987-0.9438 | 0.0009 |
| TNFRSF13C | BAFFR, CD268 | TNFRSF | 0.8788 | 0.7977-0.9682 | 0.0090 |
| TNFRSF14 | LIGHTR, HVEM, CD270 | TNFRSF | 0.8253 | 0.7112-0.9577 | 0.0114 |
| TNFRSF17 | BCMA, TNFRSF13A, CD269 | TNFRSF | 0.9023 | 0.8370-0.9727 | 0.0073 |
| TNFRSF18 | GITR, AITR, CD357 | TNFRSF | 0.9919 | 0.9060-1.0860 | 0.8607 |
| TNFRSF19 | TROY, TAJ | TNFRSF | 0.8596 | 0.7735-0.9553 | 0.0050 |
| TNFRSF1A | TNFR1, CD120A | TNFRSF | 1.3583 | 1.0542-1.7500 | 0.0179 |
| TNFRSF1B | TNFR2, CD120B | TNFRSF | 0.8640 | 0.7360-1.0142 | 0.0739 |
| TNFRSF21 | DR6, CD358 | TNFRSF | 1.1041 | 0.9425-1.2935 | 0.2201 |
| TNFRSF25 | DR3, TNFRSF12 | TNFRSF | 0.9262 | 0.8376-1.0241 | 0.1349 |
| TNFRSF4 | OX40, CD134 | TNFRSF | 0.9510 | 0.8323-1.0867 | 0.4607 |
| TNFRSF6B | DCR3 | TNFRSF | 1.1399 | 1.0481-1.2397 | 0.0022 |
| TNFRSF8 | CD30 | TNFRSF | 0.9513 | 0.8343-1.0847 | 0.4560 |
| TNFRSF9 | 4-1BB, CD137, ILA | TNFRSF | 0.9837 | 0.9007-1.0744 | 0.7156 |
| TNFSF10 | TRAIL, CD253 | TNFSF | 0.9741 | 0.8621-1.1007 | 0.6740 |
| TNFSF11 | RANKL, CD254 | TNFSF | 1.0685 | 0.9942-1.1484 | 0.0717 |
| TNFSF12 | TWEAK | TNFSF | 0.7643 | 0.6331-0.9226 | 0.0051 |
| TNFSF13 | APRIL, CD256 | TNFSF | 0.7978 | 0.6847-0.9296 | 0.0038 |
| TNFSF13B | BAFF, CD257 | TNFSF | 0.9889 | 0.8846-1.1056 | 0.8448 |
| TNFSF14 | LIGHT, HVEML, CD258 | TNFSF | 1.0094 | 0.9061-1.1246 | 0.8648 |
| TNFSF15 | TL1A | TNFSF | 0.9212 | 0.8391-1.0113 | 0.0848 |
| TNFSF18 | GITRL | TNFSF | 0.8992 | 0.7838-1.0316 | 0.1296 |
| TNFSF4 | OX-40L, CD134L, CD252 | TNFSF | 1.0323 | 0.9090-1.1723 | 0.6246 |
| TNFSF8 | CD30L, CD153 | TNFSF | 0.8864 | 0.7979-0.9848 | 0.0247 |
| TNFSF9 | 4-1BB-L, CD137L | TNFSF | 1.0888 | 0.9787-1.2112 | 0.1177 |
TCGA, The Cancer Genome Atlas; HR, hazard ratio; CI, confidence interval.
3.2. Construction of the TNF family based-signature with LUAD in the TCGA cohort
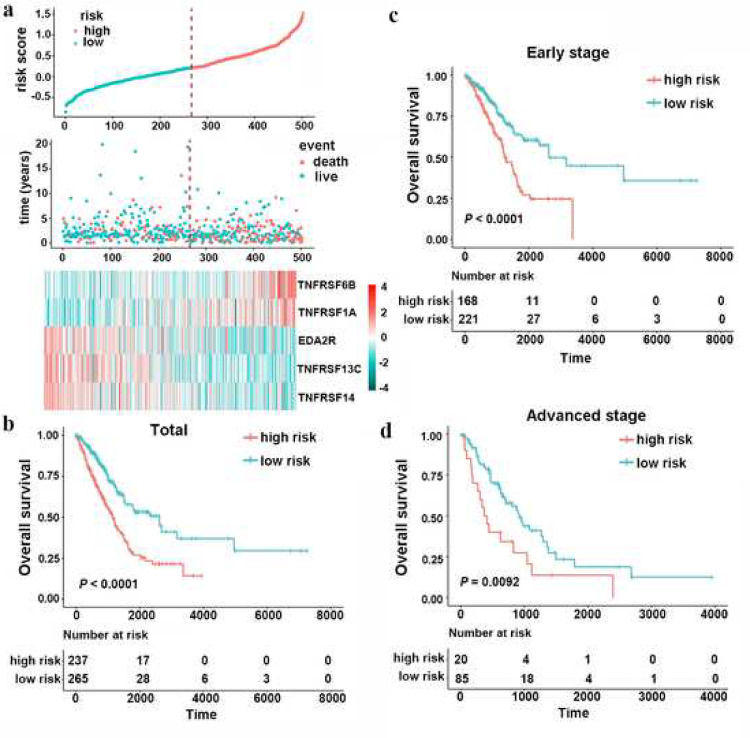
After filtered out the genes without prognostic significance, leaving 17 genes for further analysis. Then, a stepwise Cox proportional hazards regression model was used to shrink the variables and optimize the model by screening out five genes: TNFRSF6B, TNFRSF13C, TNFRSF14, TNFRSF1A and EDA2R (Table S2). Then, a risk formula was constructed with the expression levels of five genes and the corresponding regression coefficients: risk score = 0.1633 × TNFRSF6B – 0.1153 × TNFRSF13C – 0.2234 × TNFRSF14 + 0.1992 × TNFRSF1A – 0.1042 × EDA2R. The expression details of the five enrolled genes and the corresponding risk scores are shown in Fig. 1(a). This allowed patients to be divided into high-risk (n=237, score value ≥ 0.2085) and low-risk (n=265, score value < 0.2085) groups based on the optimal cut-off point. Compared with the low-risk group, patients in the high-risk group showed significantly worse OS (Fig. 1(b), HR 2.4163, 95% confidence interval (CI) 1.5903–2.8976, P < 0.0001). Because patients with early- (clinical stage I and II) and advanced-stage diseases (clinical stage III and IV) require different therapy strategies and hold different prognoses [27], we further applied the TNF family-based signature in patients with a clinical stage in the TCGA training set. Similarly, a high-risk score was associated with significantly worse OS regardless if the patient exhibited early- (Fig. 1(c), HR 2.2004, 95% CI 1.5246–3.1756, log-rank test P < 0.0001) or advanced-stage LUAD (Fig. 1(d), HR 2.0769, 95% CI 1.1834–3.6453, log-rank test P = 0.0092).
Fig. 1.
Construction of the TNF family-based signature in the TCGA dataset. (a), the distribution of risk score, survival status and gene expression panel. (b), Kaplan–Meier curves of OS in total LUAD (n=502) are stratified by TNF family-based signature in high and low risk groups based on the risk score. (c), Kaplan–Meier curves of OS in early-stage (stage I and II) LUAD (n=389) are stratified by TNF family-based signature in high- and low-risk groups based on the risk score. (d), Kaplan–Meier curves of OS in advanced-stage (stage III and IV) LUAD (n=105) are stratified by TNF family-based signature in high and low risk groups based on the risk score.
3.3. Validation of the TNF family based-signature in independent cohorts
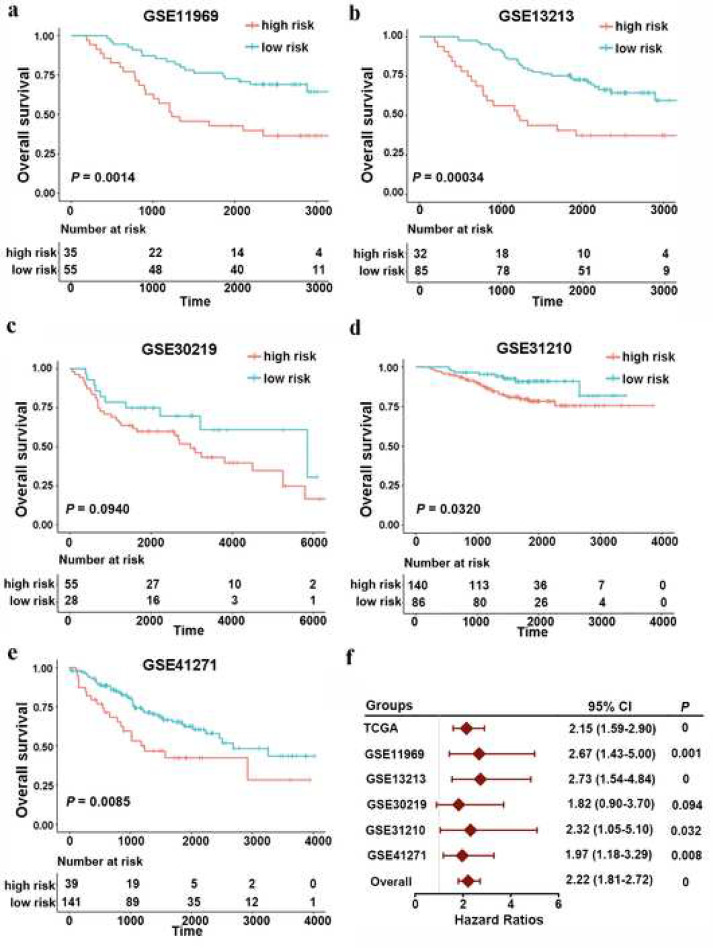
To validate the reproducibility of the TNF family based-signature in patients with LUAD, we first calculated the risk value for each patient in five independent GEO datasets using the same formula. All the demographics of these public GEO datasets are listed in Table 1. Patients in different cohorts were also classified into high- and low-risk groups based on the optimal risk value cut-off point. As expected, the Kaplan–Meier analysis revealed that patients in the high-risk group showed an increased risk of mortality compared to those in the low-risk group, either in GSE11969 (Fig. 2(a), cut-off value = −0.0852, HR 2.6731, 95% CI 1.4300–4.9970, log-rank test P = 0.0014), GSE12313 (Fig. 2(b), cut-off value = −0.5631, HR 2.7285, 95% CI 1.5396–4.8354, log-rank test P = 0.0003), GSE30219 (Fig. 2(c), cut-off value = −0.5066, HR 1.8202, 95% CI 0.8957–3.6988, log-rank test P = 0.0937), GSE31210 (Fig. 2(d), cut-off value = −0.6048, HR 2.3162, 95% CI 1.0518–5.1005, log-rank test P = 0.0318), or GSE41271 (Fig. 2(e), cut-off value = −0.0869, HR 1.9686, 95% CI 1.1774–3.2915, log-rank test P = 0.0085). Moreover, we determined the prognostic significance of TNF family-based signature in these public cohorts from TCGA and GEO datasets by conducting a prognostic meta-analysis based on these six groups (n=1198). Our results confirmed that the TNF family based-signature was a risk factor for patients with LUAD (combined HR= 2.22, 95% CI= 1.81–2.72, meta-analysis P < 0.001) (Fig. 2(f)).
Fig. 2.
Validation of TNF family-based signature in LUAD from different GEO datasets. Kaplan–Meier curves of OS in different GEO datasets, (a) GSE11969 (n=90); (b) GSE13213 (n=117); (c) GSE30219 (n=83); (d) GSE31210 (n=226); (e) GSE41217 (n=180). (f), a meta-analysis was performed using the prognostic results of TCGA and GEO datasets. The overall P value was calculated by meta-analysis.
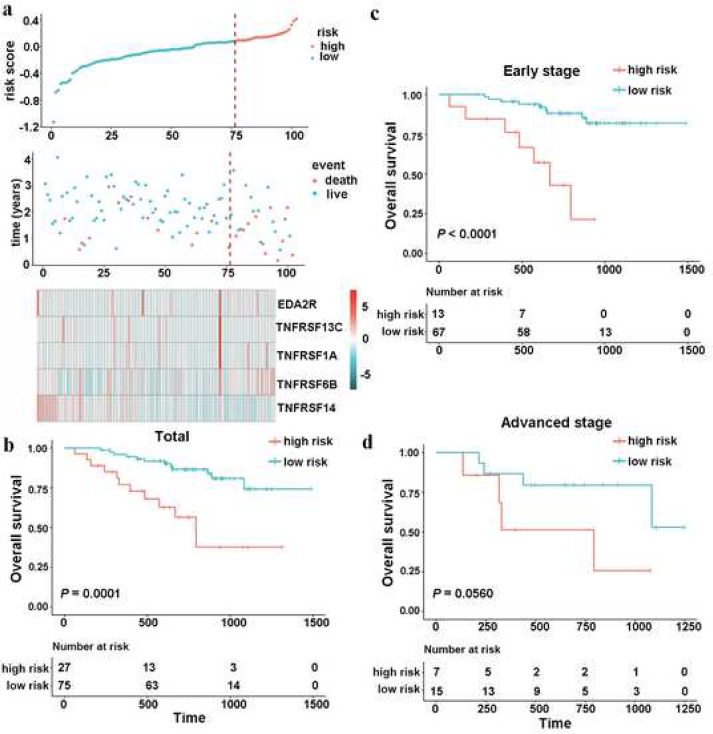
To evaluate the robustness of this five-gene signature for predicting OS for patients with LUAD in clinical practice, we further validated the TNF family-based signature in an independent cohort that included 102 LUAD frozen tissue samples using qRT-PCR. Using the same formula, patients were divided into two groups according to the optimal cut-off point (Fig. 3(a)). As a result, significantly different mortality was found between the high-risk group and low-risk group (Fig. 3(b), cut-off value = 0.0830, HR 4.2524, 95% CI 1.8936–9.5491, log-rank test P = 0.0001). When carried out a stratified analysis in patients with early- and advanced-stage disease with LUAD to evaluate the prognostic significance of the signature. The results confirmed that the signature could classify the patients into high- and low-risk groups with significantly different OS results (Fig. 3(c) and (d)).
Fig. 3.
Validation of the prognostic performance of TNF family-based signature in an independent cohort based on 102 frozen tissues. (a), the distribution of risk score, survival status and gene expression panel. (b), Kaplan–Meier curves of OS in total LUAD (n=102) are stratified by the signature in high- and low-risk groups based on risk score. (c), Kaplan–Meier curves of OS in early-stage (stage I and II) LUAD (n=80) are stratified by the signature in high- and low-risk groups based on the risk score. (d), Kaplan–Meier curves of OS in advanced-stage (stage III and IV) LUAD (n=22) are stratified by the signature in high and low risk groups based on the risk score.
3.4. Validation of the TNF family based-signature in different clinical subgroups
To further verify the stability of the signature in different clinical subgroups, we first evaluated the predictive ability of the signature in patients of different sexes, ages, and smoking histories. All patients were ranked by the formula score and divided into high- and low-risk groups. Kaplan–Meier survival analyses were then used to estimate the difference in OS probabilities between high- and low-risk groups. Our results indicated that the OS in the high-risk group was significantly shorter than that in the low-risk group, across all subgroups (Supplementary Fig. 2, log-rank test P < 0.05). Next, we further explored the performance of the signature in patient subsets with different mutations. Likewise, we found that no matter in EGFR wild-type (WT), EGFR mutation (MUT), KRAS WT, KRAS MUT, or EGFR/KRAS WT group, the TNF family-based signature risk group provided statistically significant OS stratification (Supplementary Fig. 3, log-rank test P < 0.05). Simultaneously, we tested the signature in three expression subtypes in LUAD: magnoid, squamiod and bronchioid [28]. The TNF family-based signature also statistically significantly stratified the patients in different subtypes, through the optimal cut-off point (Supplementary Fig. 4, log-rank test P < 0.05).
3.5. The TNF family-based signature is an independent risk factor for patients with LUAD
To explore whether the prognostic value of the TNF family-based signature performed independently of other clinicopathological factors, univariate and multivariate Cox regression analyses were first conducted on the TCGA set. Our results revealed that the risk score was independently correlated with OS in patients with LUAD (Table 3). Consistently, the TNF family-based signature was validated as an independent factor after Cox regression analyses in 102 patients with LUAD with qPCR data (Table S3). All the number of events in each category of each variable in TGCA and independent cohorts were shown in Table S4.
Table 3.
Univariable and multivariable Cox regression analysis of the TNF family-based signature and survival in TCGA dataset.
| Univariable analysis |
Multivariable analysis |
|||||
|---|---|---|---|---|---|---|
| Variable | HR | 95%CI | P value | HR | 95%CI | P value |
| Age | ||||||
| ≥60 or <60 | 1.1575 | 0.7957–1.6838 | 0.4445 | |||
| Gender | ||||||
| Male or Female | 1.1568 | 0.8401–1.5928 | 0.3722 | |||
| Smoking history | ||||||
| Yes or No | 1.0374 | 0.6532–1.6476 | 0.8763 | |||
| T stage | ||||||
| 1, 2, 3 or 4 | 1.5458 | 1.2602–1.8961 | <0.0001 | 1.2142 | 0.9556–1.5428 | 0.1122 |
| Lymphatic metastasis | ||||||
| Yes or No | 2.4053 | 1.7466–3.3124 | <0.0001 | 1.6476 | 1.0696–2.5381 | 0.0235 |
| TNM stage | ||||||
| I, II, III or Ⅳ | 1.5587 | 1.3381–1.8156 | <0.0001 | 1.2344 | 0.9670–1.5757 | 0.0908 |
| ERFR status | ||||||
| MUT or WT | 1.4658 | 0.9584–2.2418 | 0.0777 | |||
| KRAS status | ||||||
| MUT or WT | 1.2159 | 0.8598–1.7195 | 0.2689 | |||
| Risk score | ||||||
| High or low | 2.0558 | 1.4802–2.8554 | <0.0001 | 2.0003 | 1.4146–2.8285 | 0.0001 |
Abbreviations: HR, hazard ratio; CI, confidence interval; WT, wild-type; MUT, mutation.
3.6. Identification of the TNF family-based signature related biological pathways
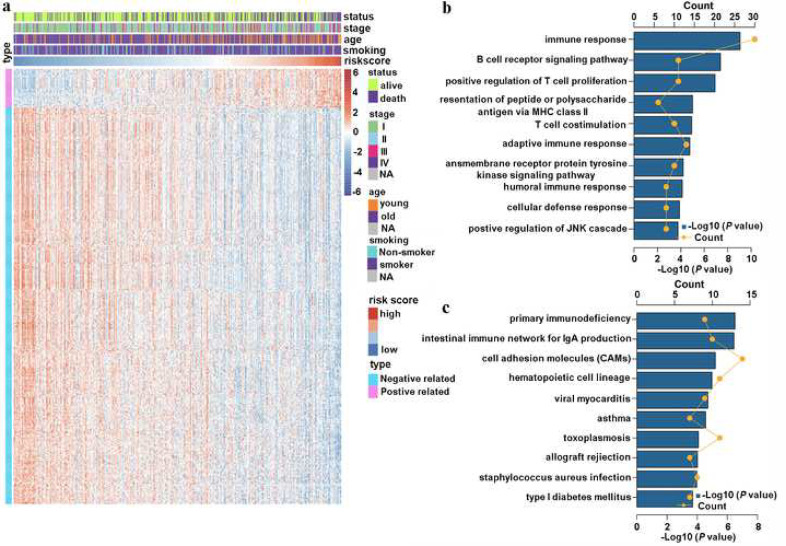
The strong stratification power of the TNF family-based signature in predicting the OS of patients with LUAD led us to explore signature-related biological pathways. We first sought to determine the genes that strongly correlated with the risk score (Pearson |R| > 0.4). As shown in Fig. 4(a), there were 38 positively related genes, and 392 negatively related genes that were screened out. Then these significantly related genes were chosen for GO and KEGG analysis DAVID (http://david.abcc.nci fcrf.gov). We found that these genes were more involved in the biological processes of the immune response—T cell proliferation and costimulation—and other immune-specific processes (Fig. 4(b)). Meanwhile, the KEGG analysis revealed that these genes were more relevant to primary immunodeficiency and other immune-related pathways (Fig. 4(c)).
Fig. 4.
TNF family-based signature-related biological pathways. (a), the most related genes of TNF family-based signature in LUAD (Pearson |R| > 0.4). (b) and (c), GO and KEGG analysis of the identified genes.
3.7. Immune cell infiltration and inflammatory profiles of the TNF family-based signature
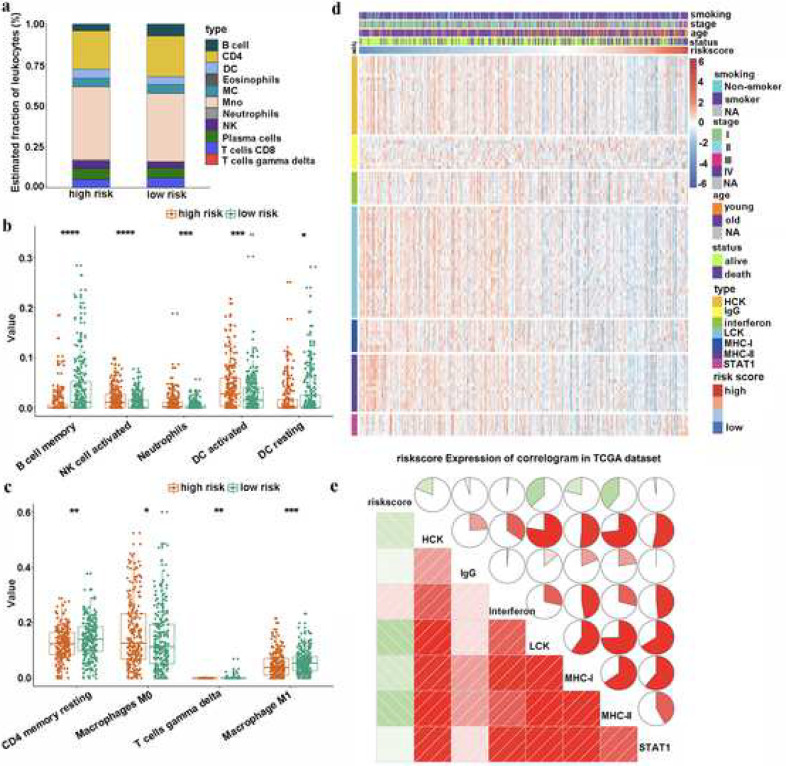
Due to the close relationship between the TNF family-based signature and immune-related biological pathways, we decided to drive deeper into the risk score as it related to immune cell infiltration and inflammatory profiles in patients with LUAD. First, CIBERSORT with LM22, a signature matrix containing 547 genes that distinguish 22 immune cell subtypes [22], was used in each sample in the TCGA cohort to estimate the immune cell infiltration. The distributions of different immune cells between high- and low-risk groups are shown in Fig. 5(a). More specifically, the high-risk group had a higher population of activated NK cells, neutrophils, activated dendritic cells (DCs), and macrophages M0 (Fig. 5(b) and (c)). In contrast, the low-risk group had a higher population of memory B cells, resting DCs, resting CD4 memory T cells, gamma delta T cells and macrophages M1 (Fig. 5(b) and (c)). The close relationship between risk score and immune cell infiltration reminded us of another immune profile-related prognostic marker—FOXM1—in LUAD [23]. Therefore, we compared the prognostic value of our signature with FOXM1. Our results indicated that our signature performed better than FOXM1 for predicting OS in patients with LUAD (Fig. S5).
Fig. 5.
The immune landscape of TNF family-based signature in LUAD. (a) Estimated immune cell expression proportion in high- and low-risk groups. (b) and (c) The details of different expression immune cells in high- and low-risk groups. (d) The relationship between risk score and inflammatory activities in patients with LUAD. (e) Correlogram was generated based on Pearson R-value between risk score and metagenes. *, **, ***, and **** represent P < 0.05, P < 0.01, P < 0.001 and P < 0.0001, respectively.
Next, to better understand risk score-related inflammatory activities, we examined the relationships among the seven clusters of metagenes [29]. These differences represent different inflammatory and immune responses. The TNF family-based signature was explored and the details of these metagenes are shown in Fig. 5(d). To validate what we found in the expression detail, a Gene Sets Variation Analysis (GSVA) was used to emulate the results of corresponding clusters of seven metagenes [26,30]. Our results showed that the risk score was negatively related to HCK, LCK, MHC_I, and MHC_II (Fig. 5(e)).
3.8. Relationship between the TNF family-based signature and immunotherapy response
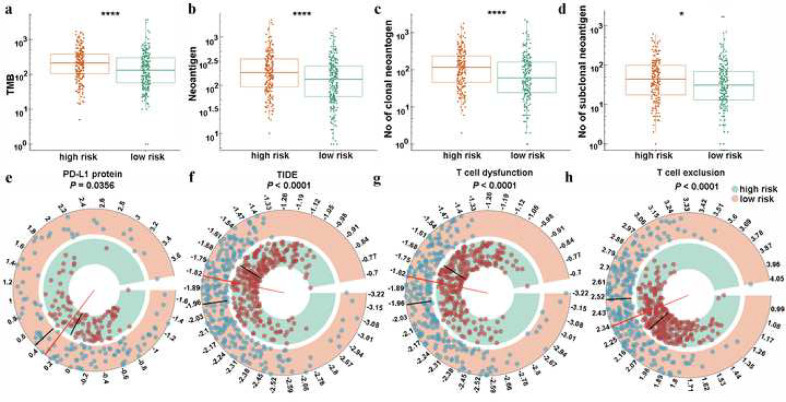
Immunotherapy targets immune checkpoints and is now the first-line treatment in lung cancer. The TNF family members are potential candidate molecules for this therapeutic regimen. We explored the relationship between the TNF family-based signature and the immunotherapy response by analysing the correlation of risk score and a series of widely used biomarkers [26,31]. First, the tumour mutation burden (TMB), number of neoantigens, number of clonal neoantigens, and number of subclonal neoantigens were calculated for the high- and low-risk groups. Our results showed that high-risk patients showed a significant mutation load and neoantigens (Fig. 6(a)–(c), Mann–Whitney U-test P < 0.05). It is widely accepted that patients with a high TMB (TMB≥10 mut/Mb) will achieve a higher objective response rate to immunotherapy [32]. Given the TMB cut-off point in many retrospective studies was derived from patients with advanced lung cancers who underwent immunotherapy, we further analysed the signature with the TMB cut-off point in patients with advanced stage (stage III and IV) disease. Using the same risk score cut-off point (0.2085), our results confirmed that a high risk score was significantly associated with a high TMB (Figure S6, chi-square test P = 0.0368). Next, the protein level of PD-L1, another well-known biomarker of the immunotherapy response in these two groups, was also investigated. We found that higher PD-L1 protein levels were observed in high-risk patients (Fig. 6(d), Mann–Whitney U-test P < 0.05). Finally, the TIDE score—a more accurate biomarker than the T cell dysfunction score and T cell exclusion score [26]—was introduced into our analysis. As expected, high-risk patients were characterized by a significantly lower TIDE scores, and higher T cell dysfunction and exclusion scores (Fig. 6(e)–(g), Mann–Whitney U-test P < 0.0001).
Fig. 6.
Distribution of immunotherapy response markers in high- and low-risk groups. (a), (b), (c), and (d), the distribution of TMB, number of neoantigens, number of clonal neoantigens, and number of subclonal neoantigens in high- and low-risk groups. (e), the distribution of protein level of PD-L1 in high- and low-risk groups. (f), (g), and (h) The distribution of TIDE score, T cell dysfunction score, and T cell exclusion score in high- and low-risk groups. * and **** represent P < 0.05 and P < 0.0001, respectively.
Besides, two subtypes of LUAD—EGFR MUT patients and STK11/LKB1 and KRAS co-MUT patients—are reported to have a low response to immune checkpoint inhibitors [33,34]. Therefore, we also explored the risk score distribution in different mutation subgroups. As shown in Fig. S7, although there was no statistical difference between STK11/LKB1 and KRAS co-mutation groups, and between the STK11/LKB1-WT and KRAS-MUT groups, a distinct difference was found between the EGFR-WT group and the EGFR-MUT group. These results suggested that the TNF family-based signature identified high-risk patients who may be appropriate candidates for immunotherapy, especially ICB.
4. Discussion
With advancements in high-throughput sequencing technology over past decades, increasing numbers of prognostic markers and therapeutic targets have been continually identified. This has increased our understanding of cancer. However, reliable biomarkers—based on the intrinsic microenvironment of tumorigenesis for the immunotherapy response and prognosis in LUAD—are still very rare. To bolster clinical tools and the understanding of co-stimulatory signalling in LUAD, we present the first TNF family-based gene prognostic signature. This was the first systematic analysis of the relationship and prognostic value of the TNF signature based on the gene expression levels of 47 well-defined TNF family members from the TCGA dataset. Using a Univariate Cox proportional hazards regression analysis and the stepwise Cox proportional hazards regression model, a five-gene based prognostic signature was constructed. The robust of this signature was well validated in five independent public cohorts and 102 cases from frozen tissues by qRT-PCR, which was confirmed by meta-analysis. The TNF family-based signature was identified as an independent risk factor for patients with LUAD and was significantly associated with the OS in different clinical and mutation subgroups. After applying a series of immune profile relevant analytical methods, the TNF family-based signature was found significantly related to different tumour-infiltrating immune cells and inflammatory activities. Interestingly, we found that the TNF family-based signature score was positively related to different immunotherapy biomarkers. This indicates that high-risk patients may be more suitable for immune checkpoint inhibitor-based immunotherapies. As far as we are aware, this study is the first and most comprehensive research to demonstrate the prognostic accuracy of the TNF family-based signature in patients with LUAD. With further validation, our signature might provide a more complete understanding and facilitate precise application of immunotherapy in patients with LUAD.
TNF family members are promising candidates for immune checkpoint blockade after B7-CD28 family members [14]. To identify genes that were able to predict prognosis in the TNF family, we systematically analysed a panorama of TNFSF/TNFRSF members with LUAD from the TCGA cohort. We found that most of the significant genes were protective factors, which is consistent with the predominant co-stimulatory function of TNF family members in the tumour microenvironment [13]. Finally, five genes, including TNFRSF6B, TNFRSF13C, TNFRSF14, TNFRSF1A, and EDA2R, were filtered out to establish the signature. TNFRSF6B, also known as Decoy receptor 3 (DcR3), is a soluble decoy receptor that belongs to TNFRSF, which can inhibit apoptosis and enhance angiogenesis by neutralizing three members of TNFSF: Fas ligand (FasL), LIGHT, and TL1A [[35], [36], [37], [38]]. A recent study revealed that switching off DcR3 expression in the tumour microenvironment enhanced the efficacy of cancer therapy [39]. TNFRSF13C, also known as B cell-activating factor receptor (BAFFR), is a receptor of BAFF, which belongs to the TNFRSF family and is involved in B lymphocyte development and maturation [40]. A recent study revealed that antibodies that targeted BAFFR showed greater potential for translation into clinical use [41]. TNFRSF14, also known as the Herpes Virus Entry Mediator (HVEM), is a member of the TNFRSF family and can bind LIGHT to enhance T cell proliferation and cytokine production, or engage with BTLA triggered the inhibitory signals of T cells [42,43]. Moreover, HVEM had a broader expression than PD-L1 and acted as a negative prognostic marker for cancer [44]. TNFRSF1A, also known as tumour necrosis factor receptor type 1 (TNFR1) or CD120A, is a member of the TNFRSF family, and can mediate signals for either cell survival or cell death after TNFα stimulation [45]. Blocking of the TNFα-TNFR1 axis can overcome resistance to anti-PD-1 in experimental melanoma [46], indicating a close relationship between the TNFSF/TNFRSF family and immune checkpoint-based immunotherapy. EDA2R, also known as X-linked Ectodermal Dysplasia Receptor (XEDAR), binds to the related legend EDA2, while the function of the EDA2-XEDAR pair remains enigmatic [13,47]. With the exception of TNFRSF14 and TNFRSF1A [48,49], the expression details and functions of all the other three molecules in this signature in LUAD are not clear, and further investigations are needed.
The performance of the TNF family-based signature was validated in several independent cohorts and different clinical subgroups. Despite the fact that the P value in the GSE30219 cohort do not reach statistically significant, our meta-analysis confirmed the robust performance of the signature in these multiple cohorts. Additionally, the borderline significance of the signature in the patients with advanced stage disease, sampled from the independent cohort, might be attributable to the small size of this cohort. Overall, the signature was well-validated in different cohorts, which led us to explore the potential underlying mechanism. Through correlation analysis, we found that signature-related genes were more involved in immune-specific biological processes and pathways. This indicated that an immune heterogeneity between the signature high- and low-risk groups may be the main cause of the difference in OS. Then, seven immune-related clusters and immune cell infiltration analyses were used to provide additional insight into the inflammatory landscape that exists between these two groups. Our results confirmed that high-risk patients were under an immunosuppressive state with a low level of B cell function (HCK) and reduced antigen-presenting capacity (MHC_I and MHC_II). Low-risk patients featured high infiltration of gamma delta T cells and macrophages M1, indicating that these patients may be in a relatively active anti-tumour immune response state. However, all these findings were estimated by bioinformatics and further experimental verification are need.
The interesting finding in our study was the relationship between the TNF signature and the well-studied immunotherapy predictive biomarkers. Although the clinical use of monoclonal antibodies targeting PD-1 and PD-L1 has yielded significant benefits for patients with NSCLC through inhibiting immune checkpoint activity, predictors of the response to these immunotherapy regimens remain incompletely characterized. To preliminarily assess the predictive ability of the TNF family-based signature, three different well-validated immunotherapy biomarkers were used. The protein levels of PD-L1 and TMB are two known classic biomarkers of the response to anti-PD-1/PD-L1 therapies [50,51]. The TIDE score was created to serve as a more accurate biomarker for the immune checkpoint blockade response than traditional biomarkers [26]. We found that signature high-risk patients had significantly higher levels of PD-L1 protein expression, tumour mutation burden, T cell dysfunction, and exclusion scores. These findings were consistent with the results of seven clusters of metagenes, indicating that high-risk patients were under an immunosuppressive state. These novel findings suggest the potential usage of the TNF family-based signature as a predictive biomarker for immunotherapy response. However, based on the signature, high-risk patients were characterized by their immune suppressive states. It is possible that these high-risk patients may not respond to immune checkpoint inhibitors. Therefore, further studies are needed to verify the ability of this signature to predict immunotherapy response.
Although the TNF family-based signature could act as an effective independent determiner of prognoses and may predict immunotherapy responses for patients with LUAD, there are still some limitations that should be acknowledged. Firstly, all the cases in our study were retrospective samples, and validation of prospective samples is still needed. Secondly, the candidate genes enrolled in this study were restricted to the TNF family members and the immune tumour microenvironment has high spatial heterogeneity. Hence, the prognosis predictive power of the signature was limited. However, the signature provides more information about the immune microenvironment profile and immunotherapy response. Thirdly, patients treated with immunotherapy were not examined in this study, so the predictive ability of the signature for immunotherapy response was evaluated indirectly. Further well-powered prospective studies are still needed.
In conclusion, this was the first and most comprehensive investigation of the expression profiles and clinical significance of TNF family members in patients with LUAD. We also developed and tested the first TNF family-based prognostic model in patients with LUAD. Moreover, these findings may provide a clinically useful tool for better prognostic management and optimize the associated immunotherapy for patients with LUAD.
Funding sources
This work was supported by the CAMS Innovation Fund for Medical Sciences (2017-I2M-1-005, 2016-I2M-1-001), the National Key R&D Program of China (2016YFC1303201), the National Natural Science Foundation of China (81802299, 81502514), the Fundamental Research Funds for the Central Universities (3332018070), the National Key Basic Research Development Plan (2018YFC1312105). These funding sources had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declarations of Competing Interests
The authors declare that they have no conflicts of interest. The authors have a patent pending to the State Intellectual Property Office of P. R. China (application number: 202010565226.1), which is relevant to this work.
Author contributions
JH and QX supervised the project, designed, edited, and led out the experiments of this study. CQZ and ZZ organized the public data and prepared all the figures and tables. ZZ and YC conducted the experiments and data analysis. CQZ, GCZ and NS drafted the manuscript. ZZH and YJL revised the manuscript. All the authors reviewed and approved the final manuscript.
Acknowledgements
All authors would like to thank the specimen donors and research groups for the TCGA, GSE11969, GSE13213, GSE30219, GSE31210, and GSE41271 which provided data for this collection.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102959.
Contributor Information
Qi Xue, Email: xueqi@cicams.ac.cn.
Jie He, Email: prof.jiehe@gmail.com.
Appendix. Supplementary materials
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T. Non-small-cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. doi: 10.1038/nrdp.2015.9. [DOI] [PubMed] [Google Scholar]
- 3.Shukla S, Evans JR, Malik R, Feng FY, Dhanasekaran SM, Cao X. Development of a RNA-Seq based prognostic signature in lung adenocarcinoma. J Natl Cancer Inst. 2017;109(1) doi: 10.1093/jnci/djw200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12(2):175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]
- 5.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 6.Liu XX, Yang YE, Liu X, Zhang MY, Li R, Yin YH. A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J Transl Med. 2019;17(1):50. doi: 10.1186/s12967-019-1800-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng F, Wang R, Zhang Y, Zhao Z, Zhou W, Chang Z. Differential expression analysis at the individual level reveals a lncRNA prognostic signature for lung adenocarcinoma. Mol Cancer. 2017;16(1):98. doi: 10.1186/s12943-017-0666-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burugu S, Dancsok AR, Nielsen TO. Emerging targets in cancer immunotherapy. Semin Cancer Biol. 2018;52(Pt 2):39–52. doi: 10.1016/j.semcancer.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Zhang C, Zhang Z, Li F, Shen Z, Qiao Y, Li L. Large-scale analysis reveals the specific clinical and immune features of B7-H3 in glioma. Oncoimmunology. 2018;7(11) doi: 10.1080/2162402X.2018.1461304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reck M, Rodriguez-Abreu D, Robinson AG, Hui R, Csoszi T, Fulop A. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016;375(19):1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 11.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017;168(4):707–723. doi: 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Ven K, Borst J. Targeting the T-cell co-stimulatory CD27/CD70 pathway in cancer immunotherapy: rationale and potential. Immunotherapy. 2015;7(6):655–667. doi: 10.2217/imt.15.32. [DOI] [PubMed] [Google Scholar]
- 13.Dostert C, Grusdat M, Letellier E, Brenner D. The TNF family of ligands and receptors: communication modules in the immune system and beyond. Physiol Rev. 2019;99(1):115–160. doi: 10.1152/physrev.00045.2017. [DOI] [PubMed] [Google Scholar]
- 14.Croft M, Benedict CA, Ware CF. Clinical targeting of the TNF and TNFR superfamilies. Nat Rev Drug Discov. 2013;12(2):147–168. doi: 10.1038/nrd3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowak AK, Cook AM, McDonnell AM, Millward MJ, Creaney J, Francis RJ. A phase 1b clinical trial of the CD40-activating antibody CP-870,893 in combination with cisplatin and pemetrexed in malignant pleural mesothelioma. Ann Oncol. 2015;26(12):2483–2490. doi: 10.1093/annonc/mdv387. [DOI] [PubMed] [Google Scholar]
- 16.Polesso F, Sarker M, Weinberg AD, Murray SE, Moran AE. OX40 agonist tumor immunotherapy does not impact regulatory T cell suppressive function. J Immunol. 2019;203(7):2011–2019. doi: 10.4049/jimmunol.1900696. [DOI] [PubMed] [Google Scholar]
- 17.Takeuchi T, Tomida S, Yatabe Y, Kosaka T, Osada H, Yanagisawa K. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol. 2006;24(11):1679–1688. doi: 10.1200/JCO.2005.03.8224. [DOI] [PubMed] [Google Scholar]
- 18.Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27(17):2793–2799. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 19.Rousseaux S, Debernardi A, Jacquiau B, Vitte AL, Vesin A, Nagy-Mignotte H. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5(186) doi: 10.1126/scitranslmed.3005723. 186ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 21.Sato M, Larsen JE, Lee W, Sun H, Shames DS, Dalvi MP. Human lung epithelial cells progressed to malignancy through specific oncogenic manipulations. Mol Cancer Res. 2013;11(6):638–650. doi: 10.1158/1541-7786.MCR-12-0634-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Cui Y, Diehn M, Li R. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non-small cell lung cancer. JAMA Oncol. 2017;3(11):1529–1537. doi: 10.1001/jamaoncol.2017.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes DN, Monti S, Parmigiani G, Gilks CB, Naoki K, Bhattacharjee A. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24(31):5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 29.Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hastings K, Yu HA, Wei W, Sanchez-Vega F, DeVeaux M, Choi J. EGFR mutation subtypes and response to immune checkpoint blockade treatment in non-small-cell lung cancer. Ann Oncol. 2019;30(8):1311–1320. doi: 10.1093/annonc/mdz141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8(7):822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin WW, Hsieh SL. Decoy receptor 3: a pleiotropic immunomodulator and biomarker for inflammatory diseases, autoimmune diseases and cancer. Biochem Pharmacol. 2011;81(7):838–847. doi: 10.1016/j.bcp.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P. Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature. 1998;396(6712):699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]
- 37.Yu KY, Kwon B, Ni J, Zhai Y, Ebner R, Kwon BS. A newly identified member of tumor necrosis factor receptor superfamily (TR6) suppresses LIGHT-mediated apoptosis. J Biol Chem. 1999;274(20):13733–13736. doi: 10.1074/jbc.274.20.13733. [DOI] [PubMed] [Google Scholar]
- 38.Migone TS, Zhang J, Luo X, Zhuang L, Chen C, Hu B. TL1A is a TNF-like ligand for DR3 and TR6/DcR3 and functions as a T cell costimulator. Immunity. 2002;16(3):479–492. doi: 10.1016/s1074-7613(02)00283-2. [DOI] [PubMed] [Google Scholar]
- 39.Hsieh SL, Lin WW. Decoy receptor 3: an endogenous immunomodulator in cancer growth and inflammatory reactions. J Biomed Sci. 2017;24(1):39. doi: 10.1186/s12929-017-0347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gross JA, Dillon SR, Mudri S, Johnston J, Littau A, Roque R. TACI-Ig neutralizes molecules critical for B cell development and autoimmune disease. impaired B cell maturation in mice lacking BLyS. Immunity. 2001;15(2):289–302. doi: 10.1016/s1074-7613(01)00183-2. [DOI] [PubMed] [Google Scholar]
- 41.Qin H, Wei G, Sakamaki I, Dong Z, Cheng WA, Smith DL. Novel BAFF-receptor antibody to natively folded recombinant protein eliminates drug-resistant human B-cell malignancies in vivo. Clin Cancer Res. 2018;24(5):1114–1123. doi: 10.1158/1078-0432.CCR-17-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duhen T, Pasero C, Mallet F, Barbarat B, Olive D, Costello RT. LIGHT costimulates CD40 triggering and induces immunoglobulin secretion; a novel key partner in T cell-dependent B cell terminal differentiation. Eur J Immunol. 2004;34(12):3534–3541. doi: 10.1002/eji.200425598. [DOI] [PubMed] [Google Scholar]
- 43.Croft M. The evolving crosstalk between co-stimulatory and co-inhibitory receptors: HVEM-BTLA. Trends Immunol. 2005;26(6):292–294. doi: 10.1016/j.it.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Malissen N, Macagno N, Granjeaud S, Granier C, Moutardier V, Gaudy-Marqueste C. HVEM has a broader expression than PD-L1 and constitutes a negative prognostic marker and potential treatment target for melanoma. Oncoimmunology. 2019;8(12) doi: 10.1080/2162402X.2019.1665976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nat Rev Immunol. 2015;15(6):362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 46.Bertrand F, Montfort A, Marcheteau E, Imbert C, Gilhodes J, Filleron T. TNFalpha blockade overcomes resistance to anti-PD-1 in experimental melanoma. Nat Commun. 2017;8(1):2256. doi: 10.1038/s41467-017-02358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan M, Wang LC, Hymowitz SG, Schilbach S, Lee J, Goddard A. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science. 2000;290(5491):523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 48.Ren S, Tian Q, Amar N, Yu H, Rivard CJ, Caldwell C. The immune checkpoint, HVEM may contribute to immune escape in non-small cell lung cancer lacking PD-L1 expression. Lung Cancer. 2018;125:115–120. doi: 10.1016/j.lungcan.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 49.Miller MA, Sullivan RJ, Lauffenburger DA. Molecular pathways: receptor ectodomain shedding in treatment, resistance, and monitoring of cancer. Clin Cancer Res. 2017;23(3):623–629. doi: 10.1158/1078-0432.CCR-16-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.