Abstract
Background
Metastasis is the leading cause of death in patients with osteosarcoma. Some of these patients fail to respond to chemotherapy and die of metastasis within a short period. Therefore, it is important to identify novel biomarkers to improve the diagnosis and treatment of osteosarcoma. TRIM7 is a member of the tripartite motif (TRIM) family protein that is involved in various pathological conditions including cancer; however, its role in osteosarcoma remains elusive.
Methods
Cell proliferation, invasion and migration were measured by CCK-8 and Transwell. Immunoprecipitation and mass spectrometry analysis were used to identify candidate proteins associated with TRIM7. Immunoprecipitation, immunofluorescence, pull down and ubiquitination assay were performed to examine the regulation between TRIM7 and its candidate protein. m6A modification of TRIM7 was measured by RNA immunoprecipitation.
Findings
TRIM7 expression was upregulated in osteosarcoma tissues and was an independent risk factor in predicting poor prognosis. TRIM7 regulates osteosarcoma cell migration and invasion through ubiquitination of breast cancer metastasis suppressor 1 (BRMS1). Moreover, chemoresistance was readily observed in osteosarcoma cells and in patient-derived xenograft (PDX) mice with higher TRIM7 levels. Loss of TRIM7 m6A modification was observed in osteosarcoma tissues. METTL3 and YTHDF2 were the main factors involved in the aberrant m6A modification of TRIM7.
Interpretation
Overall, our findings show that TRIM7 plays a key role in regulating metastasis and chemoresistance in osteosarcoma through ubiquitination of BRMS1.
Funding
This work was financially supported by grants of NSFC (81001192, 81672658 and 81972521) and National Key Research Project of Science and Technology Ministry (2016YFC0106204).
Keywords: Osteosarcoma, TRIM7, BRMS1, Metastasis, Ubiquitination, Chemoresistance, m6A methylation
1. Introduction
Osteosarcoma is the most common type of bone cancer and the second leading cause of cancer-related deaths in children and young adults [1]. For several decades, the treatment of osteosarcoma with multidrug chemotherapy has increased the 5-year event-free survival rate for local high-grade osteosarcomas from less than 20% to about 60%. However, few evidence-based treatment options have been shown to improve survival since then [2]. Furthermore, if metastasis occurs, which occurs in the lung in more than 85% of those cases, the survival rate goes down to 10–20% [3]. In addition, a number of osteosarcoma patients die of recurrence and metastasis within a short period due to failed chemotherapy. The fundamental causes of the recurrence and metastasis in osteosarcoma are tumour cell migration, invasion, and chemoresistance. Adriamycin (ADR) and methotrexate (MTX) are commonly used to treat osteosarcoma, and patients, who are resistant to these chemotherapy drugs, have a poor prognosis [4]. Therefore, understanding the molecular mechanism of metastasis and chemoresistance in osteosarcoma is of great importance for the prevention and treatment of osteosarcoma.
Tripartite motif-containing protein 7 (TRIM7) is a member of the tripartite motif (TRIM) family that contains a RING finger, one or two B-box, and a coiled-coil region. However, TRIM7 lacks the coiled-coil domain and localizes to both the nucleus and the cytoplasm. The RING finger region is characterized by E3 ubiquitin ligase activity and this region mediates the ubiquitination of different substrates by interacting with related E2 ubiquitin-binding enzymes [5]. The B-box and coiled-coil regions, on the other hand, promote protein-protein interactions and homo-oligomerization [6]. A growing body of evidence now suggests that the TRIM family of proteins can act as tumour suppressors or oncogenes and are involved in osteosarcoma development [7], [8], [9]. Analyses from cDNA microarrays have shown that TRIM7 is up-regulated in mucinous colorectal carcinomas compared with non-mucinous colorectal carcinomas [10]. TRIM7 is also overexpressed in human lung adenocarcinomas, and its depletion suppresses cell growth of primary murine lung adenocarcinoma as well as significantly decreases tumour growth in xenograft mice after doxycycline treatment [11]. Meanwhile, TRIM7 suppresses hepatocellular carcinoma progression by directly targeting Src protein [12] and regulates hepatocellular carcinoma cell proliferation via the DUSP6/p38 pathway [13]. TRIM7 has also been found to be positively regulated by CD26, a protein closely linked to cell cycle progress, apoptosis, and chemotherapy resistance in malignant pleural mesothelioma [14]. Taken together, these results suggest that TRIM7 plays an important role in cancer development; however, its function in regulating invasion, migration, and chemoresistance in osteosarcoma, as well as the underlying molecular mechanism, remains unexplored.
Our previous study showed that breast cancer metastasis suppressor 1 (BRMS1), a tumour suppressor in osteosarcoma, was associated with TRIM7 [15]. Since TRIM7 also promoted the ubiquitination of BRMS1, these studies suggest that TRIM7 and BRMS1 may closely interact with each other in osteosarcoma cells. BRMS1 was first found in breast cancer, and its anti-metastasis functions were also confirmed in other cancers such as osteosarcoma, prostate, gastric and lung cancer [15], [16], [17], [18], where lower expression of BRMS1 was associated with increased cancer cell metastasis. Additionally, knockdown of BRMS1 displays mesenchymal characteristics and enhances cell invasion and migration, along with decreased E-cadherin and increased Vimentin and Twist1 [19,20]. Given the important roles of the E-cadherin, Vimentin, and Twist1 in regulating cell invasion and migration [21], we hypothesized that BRMS1 may regulate cell metastasis by targeting these proteins.
Our results show that in osteosarcoma tissues, compared with the expression levels in normal tissues, the expression of TRIM7 is higher while the expression of BRMS1 is lower. The higher expression of TRIM7 was modulated by RNA m6A methylation. TRIM7 knockdown inhibits osteosarcoma cell invasion and migration both in vitro and in vivo; conversely, BRMS1 overexpression inhibited TRIM7 overexpression-induced increase in cell invasion and migration. Moreover, both in vitro and in vivo studies show that higher TRIM7 levels were associated with chemoresistance. Finally, TRIM7 ubiquitinates BRMS1 at the K184 site, which indicates a close relationship between TRIM7 and BRMS1 in osteosarcoma development. Our work offers a novel approach for understanding the metastasis and chemoresistance in osteosarcoma and may lead to prevention and control strategies in osteosarcoma treatment in the future.
2. Materials and methods
2.1. Patients and tissue samples
The study was approved by the Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital. A total of 100 high-grade extremity conventional osteosarcoma patients at Enneking's stage IIA and IIB were recruited for this study. For control, adjacent nontumorous tissues from 20 patients were resected when the patients underwent surgery. All the patients signed an informed consent form. The patients’ clinical characteristics, including age, sex, tumour size, local recurrence, Enneking's stage, AJCC stage, anatomic location, lung metastasis (latent metastasis after the operation), and prognosis were collected for statistical analysis. Fresh or formalin-fixed, paraffin-embedded osteosarcoma specimens were collected for the detection of TRIM7 expression levels. The expression levels were detected by quantitative real-time PCR, or immunohistochemistry (IHC) analysis using anti-TRIM7 antibody (Novus Biologicals, LLC.; NBP1-89751) as previously described [22]. The proportion of tumour cells with positive staining was determined. All the patients that had more than 25% of positive cells were classified as high expression group; in contrast, those that had positive cells below 25% were classified as low expression group.
2.2. Cell culture
Human osteosarcoma cell lines, HOS, SAOS2, U2-OS, and MG63, as well as human osteoblastic cell line hFOB1.19, were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cell lines were validated at the start of the project by short tandem repeat profiling, carried out by American Type Culture Collection. The cells were maintained at 37 °C with 5% CO2. HOS and MG63 cells were grown in minimum Eagle's medium (MEM; Hyclone, Logan, UT, USA), while U2-OS and SAOS2 cells were maintained in Dulbecco's Modified Eagle's medium (DMEM; Hyclone). After fixing, permeation, and blocking, HOS and MG63 cells were collected for the detection of TRIM7 and BRMS1 protein expression by immunofluorescence staining using anti-TRIM7 antibody (Novus Biologicals, LLC., Centennial, CO, USA; NBP1-89751), anti-BRMS1 antibody (Santa Cruz Biotech., Santa Cruz, CA, USA; sc-101219) and Alexa Fluor-labeled IgG (H+L) antibody (Beyotime Biotechnology, Shanghai, China; A0428 and A0453) as previously described [23].
2.3. Plasmids construction and lentivirus production
TRIM7 and BRMS1 overexpression plasmids were constructed by cloning full-length human TRIM7 and BRMS1 into the lentiviral expression vector pLVX-Puro (Addgen, Cambridge, MA, USA), produced in 293T cells as previously described [19], and these plasmids were used to transduce SAOS2 cells. The RNAi (RNA interference) sequences (Table 1) targeting the human TRIM7, BRMS1, METTL3, METTL14, FTO, ALKBH4, or YTHDF2 gene were cloned into the lentiviral vector pLKO.1 (Addgen), transfected into 293T cells as previously described [19], and used to transduce HOS and MG63 cells. Cells transduced with pLKO.1-scramble shRNA (shNC) or blank lentivirus pLVX-Puro (Vector) were used as negative controls.
Table 1.
shRNA sequences used in this study.
| Gene (position)-number | Sequences (5′-3′) |
|---|---|
| TRIM7 (584-602)-1 | GCGAACCCTTCAAGCTCTA |
| TRIM7 (726-744)-2 | GGGTCTTGAAGAAGGAACT |
| TRIM7 (728-746)-3 | GGGTCTTGAAGAAGGAACT |
| BRMS1 (450-468)-1 | GGAGCCTCAAGATTCGCAT |
| BRMS1 (479-497)-2 | GGGATCTACAAGGGCTTCT |
| BRMS1 (750-768)-3 | GCCCATACATCGTGTACAT |
| METTL3 (1005-1023)-1 | GCTGCACTTCAGACGAATT |
| METTL3 (1086-1104)-2 | GGATACCTGCAAGTATGTT |
| METTL14 (232-250)-1 | GCATTGGTGCCGTGTTAAA |
| METTL14 (561-579)-2 | GCTGACAGATTTGAAGAAT |
| FTO (206-224)-1 | GGAGCTCCATAAAGAGGTT |
| FTO (353-371)-2 | CCTGAACACCAGGCTCTTT |
| ALKBH4 (197-215)-1 | GCACAGAGGAGTCTGACTT |
| ALKBH4 (353-371)-2 | GATGCTGATCGAGGACTTT |
| YTHDF2 (768-786)-1 | GCACAGAAGTTGCAAGCAA |
| YTHDF2 (1482-371)-2 | TTGCTTGCAACTTCTGTGT |
Full-length or mutant BRMS1 cDNA was cloned and inserted into a pCMV-Tag 2B vector, and the generated plasmids were designated as BRMS1 (WT), BRMS1 (K8R), BRMS1 (K69R), and BRMS1 (K184R). Mutations were introduced into BRMS1 with the QuikChange II Site-directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). Myc-tagged TRIM7 sequence was purchased from GENEWIZ, lnc. (Suzhou, China) and cloned into a p-DONR221 vector to express myc-TRIM7. For his-ubiquitin (Ub), human Ub was cloned into a pcDNA-DEST40 vector with a His tag. All constructs and mutants were confirmed by sequencing.
2.4. Cell proliferation analysis
MG63 cells were transduced with pLKO.1-shTRIM7 and/or pLKO.1-shBRMS1 lentivirus in the presence or absence of adriamycin (ADR; Wyeth Lederle, Latina, Italy; 5 μg/ml) or methotrexate (MTX; Sandoz, Varese, Italy; 2 ng/ml); SAOS2 cells were transduced with lentivirus expressing TRIM7 and/or BRMS1 in the presence of ADR (5 μg/ml) or MTX (2 ng/ml). After treatment, cells were incubated with 10% Cell Counting Kit-8 (CCK-8) solution (SAB Biotech., College Park, MD, USA) for 1 h at 37 °C. The optical density (OD) value of each well was recorded at 450 nm and detected by a microplate reader (Tecan, Grödig, Austria).
2.5. Transwell analysis
HOS and MG63 cells were transduced with pLKO.1-shTRIM7 and/or pLKO.1-shBRMS1 lentivirus; SAOS2 cells were transduced with lentivirus expressing TRIM7 and/or BRMS1. After treatment, the cell migration and invasion were measured in Transwell inserts (Corning Costar, NY, USA) containing polycarbonate filters with 8-μm pores coated with or without Matrigel (Becton Dickinson, Bedford, MA, USA) as detailed previously [20].
2.6. Liquid chromatography/mass spectrometry (LC/MS) analysis
Full-length TRIM7 was constructed and cloned into a pCMV-Flag vector. 293T cells stably expressing Flag-TRIM7 were constructed and lysed in pre-cooled RIPA lysis buffer (20 mM Tris pH7.5, 150 mM NaCl, 1% Triton X-100). Cell lysates were then incubated with anti-Flag magnetic beads (Sigma-Aldrich) overnight at 4 °C. The bound proteins were eluted with Flag peptide by incubating 1 h at 4 °C. Protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The eluted proteins were detected by LC/MS as previously described [24].
2.7. Co-IP and ubiquitination analysis
To confirm the interaction between TRIM7 and BRMS1, cell lysates were prepared from HOS and MG63 cells with RIPA buffer, reacted with anti-TRIM7 (Biorbyt, Cambridge, UK; orb1892), anti-BRMS1 (Abcam, Cambridge, MA, USA; ab134968) or control IgG (Santa Cruz Biotech.; sc-2027) overnight at 4 °C and then with protein A/G Plus agarose (Santa Cruz Biotech.) for 2 h at 4 °C. The immunoprecipitated complexes were washed three times in lysis buffer and analyzed by Western blot. SAOS2 cells transduced with pLVX-Puro or pLVX-Puro-TRIM7 vector were lysed with RIPA buffer. The lysates were incubated with anti-BRMS1 or control IgG antibody, respectively. The immunoprecipitated complexes were subjected to western blot analysis using anti-Ub (Abcam; ab7780).
2.8. His-ubiquitin pull-down assay
293T cells were co-transfected with the BRMS1 (WT) or mutant BRMS1 constructs along with myc-TRIM7 and His-Ub constructs. After 48 h of transfection, cell lysates were incubated with Ni2+-NTA agarose beads (QIAGEN Sciences, Germantown, MD, USA). The washed complexes were eluted by boiling in SDS sample buffer and separated by SDS-PAGE and the interactions were analyzed by western blot using anti-Flag antibody (Abcam; ab205606) or anti-Ub antibody (Abcam; ab7780) and secondary antibody (Beyotime Biotechnology; A0208).
2.9. Quantitative real-time PCR analysis
Total RNA from the osteosarcoma tissues and cell lines was extracted using RNAiso (TaKaRa, Dalian, China). cDNA was synthesized with DNase-treated RNA by using the cDNA First-Strand Synthesis kit (Fermentas, Hanover, MD, USA). Quantitative real-time PCR (qRT-PCR) was performed with the Fast SYBR Green PCR kit (Applied Biosystems, Foster City, CA, USA) on ABI 7300 system (Applied Biosystem). The PCR primers were listed in Table 2. Data were analyzed with the 2−ΔΔCt method. The expression of target genes was normalized to those of GADPH.
Table 2.
Primes sequences used in this study.
| Gene | Sequences (5′-3′) |
|---|---|
| TRIM4-forward | AATGCTAAAGCGATTCCAAGTG |
| TRIM4-reverse | CCAGTAATGTTTCCCTGAGGTG |
| TRIM6-forward | AGGTGACCTGCCCTATCTGC |
| TRIM6-reverse | CTCTGAGCCGCCTCACTATG |
| TRIM7-forward | CTATGGAACCGCAGAATC |
| TRIM7-reverse | GGACAGCTCCAGTTTAAG |
| TRIM10-forward | CCTGCCTTACCCGCTACTG |
| TRIM10-reverse | CACGCACAACTGCATCTCATC |
| TRIM17-forward | CGCATTGTGCTGGAGTTTG |
| TRIM17-reverse | ACAGTCCTGGGTCTGGTTGG |
| TRIM22-forward | TGCCAGCACGCTCATCTC |
| TRIM22-reverse | GATTCAGCATCACGTCCACC |
| TRIM25-forward | GTCCAGGGCTCGCCATAC |
| TRIM25-reverse | ACACCAAGCACGTCTTCACG |
| TRIM27-forward | GGACCTGCCTGACAACCCC |
| TRIM27-reverse | TCCAAGAAAATCCCCACCC |
| BRMS1-forward | CCGAAGTCAAAGCCACATC |
| BRMS1-reverse | CAAGAACTGCCCTGAGAAC |
| METTL3-forward | CCTTTGCCAGTTCGTTAGTC |
| METTL3-reverse | TCCTCCTTGGTTCCATAGTC |
| METTL14-forward | CTGGGAATGAAGTCAGGATAG |
| METTL14-reverse | CCAGGGTATGGAACGTAATAG |
| FTO-forward | ACCTCCAGCATTAGATTC |
| FTO -reverse | GAAACTACCGCATTTACC |
| ALKBH4-forward | ATCACTTGATGCCAGGAGTTC |
| ALKBH4-reverse | AAGCAATTCTCCCACCTTAGC |
| YTHDF2-forward | ACACATTCGCCTAGAGAAC |
| YTHDF2-reverse | ACACATTCGCCTAGAGAAC |
| GAPDH-forward | AATCCCATCACCATCTTC |
| GAPDH-reverse | AGGCTGTTGTCATACTTC |
2.10. Western blotting
Western blotting was conducted as previously described [25]. Proteins were analyzed with the following antibodies: rabbit anti-TRIM7 (Proteintech Group, Inc., Rosemont, IL, USA; 26285-1-AP), mouse anti-BRMS1 (Abcam; ab172430), rabbit anti-Twist1 (Cell Signaling Technology, Danvers, MA, USA; 46702), mouse anti-E-cadherin (Cell Signaling Technology; 14472), rabbit anti-Vimentin (Cell Signaling Technology; 5741), rabbit anti-METTL3 (Abcam; ab195352), mouse anti-METTL14 (Abcam; ab220030), rabbit anti-FTO (Abcam; ab124892), rabbit anti-ALKBH4 (Abcam; ab195379), rabbit anti-YTHDF2 (Abcam; ab220163), and rabbit anti-GAPDH (Cell Signaling Technology; 5174). The primary antibodies and the secondary antibodies (Beyotime Biotechnology; A0208 and A0216) were used at a dilution of 1:1,000.
2.11. Animal model
Animal experiments were approved by the Animal Care and Use Committee of Shanghai Jiao Tong University Affiliated Sixth People's Hospital. Male BALB/c nude mice (4–6 weeks old) were kept in specific pathogen-free conditions. MG63 cells transduced with lentivirus expressing shTRIM7 or shNC, and SAOS2 cells transduced with lentivirus expressing TRIM7, BRMS1, TRIM7 plus BRMS1 or control vector, were injected via the tail vein into the nude mice (1 × 106 cells/mouse) (n = 11 per group). At 21 days after injection, mice were sacrificed and the lung tissues were collected and stained in hematoxylin and eosin (H&E) as previously described [19]. Otherwise, mice were injected via the tail vein with MG63 or SAOS2 cells and intraperitoneal injection of 1 mg/kg ADR every 2 days or 0.2 mg/kg MTX every 7 days (n = 20 per group), and then, lung metastasis and survival time were measured. To establish the patient-derived xenograft (PDX) model, tumour tissues from osteosarcoma patients, divided into two groups according to the TRIM7 expression (high vs low according to IHC staining), were collected at the time of surgery at Shanghai Jiao Tong University Affiliated Sixth People's Hospital. These tissues (about 5 × 5 × 3 mm3) were subcutaneously transplanted into 6–8-week-old nude mice within 1 h of removal of tissues. ADR (1 mg/kg; every 2 days) and MTX (0.2 mg/kg; every 7 days) chemotherapy were initiated when tumour volumes reached around 700 mm3 (n = 5 per group). On day 21 days following the injection, mice were sacrificed and tumour size was measured.
2.12. m6A content analysis
The content of m6A in total RNA was analyzed with the m6A RNA Methylation Assay Kit (Abcam, ab185912).
2.13. RNA immunoprecipitation assays
RNA immunoprecipitation (RIP) assays were performed using the Magna RIP RNA-Binding Protein Immunoprecipitation kit (Millipore). Total RNA (input control) and isotype control (IgG) for each antibody were assayed simultaneously. Precipitation was performed using an anti-m6A antibody (Synaptic Systems, 202003), which was bound to magnetic Dynabeads in RIP immunoprecipitation buffer (Magna RIP Kit, Millipore) and incubated with fragmented RNAs. RNAs were extracted, reverse transcribed, and subjected to qRT-PCR using the primers for TRIM7 (F: 5’-TCGCAGTGTAACCTGTAGCC-3’ and R: 5’-TCCTTCTTTTCCGTGGACC-3’).
2.14. mRNA stability measurements
Briefly, osteosarcoma cell lines were incubated with 0.2 mM actinomycin D for 30 min and collected as 0 h samples. The 3 and 6 h samples were collected and subjected to total RNA extraction. cDNAs were generated with SuperScript IV reverse transcriptase (Thermo Scientific) using the oligo d(T) primer. mRNA levels were quantified by qRT-PCR.
2.15. Statistical analysis
The experiments were performed in triplicates and repeated at least three times. All values are expressed as mean ± standard deviation (SD). Statistical analysis was conducted using GraphPad Prism 8.0.2 (GraphPad Software, La Jolla, CA, USA) and Mann–Whitney U test, unpaired Student's t-test or ANOVA followed by Bonferroni post-tests were carried out. The level of statistical significance was set at P < 0.05. Overall survival (OS) and disease-free survival (DFS) of patients or mice were analyzed by the Kaplan–Meier survival curves and log-rank test.
2.16. Availability of data
The Proteomics data have been deposited in the PeptideAtlas (http://www.peptideatlas.org/) under accession number PASS01605. All relevant data supporting the key findings are available from the corresponding author upon reasonable request.
3. Results
3.1. TRIM7 expression is upregulated, and the upregulation is correlated with poor prognosis in osteosarcoma
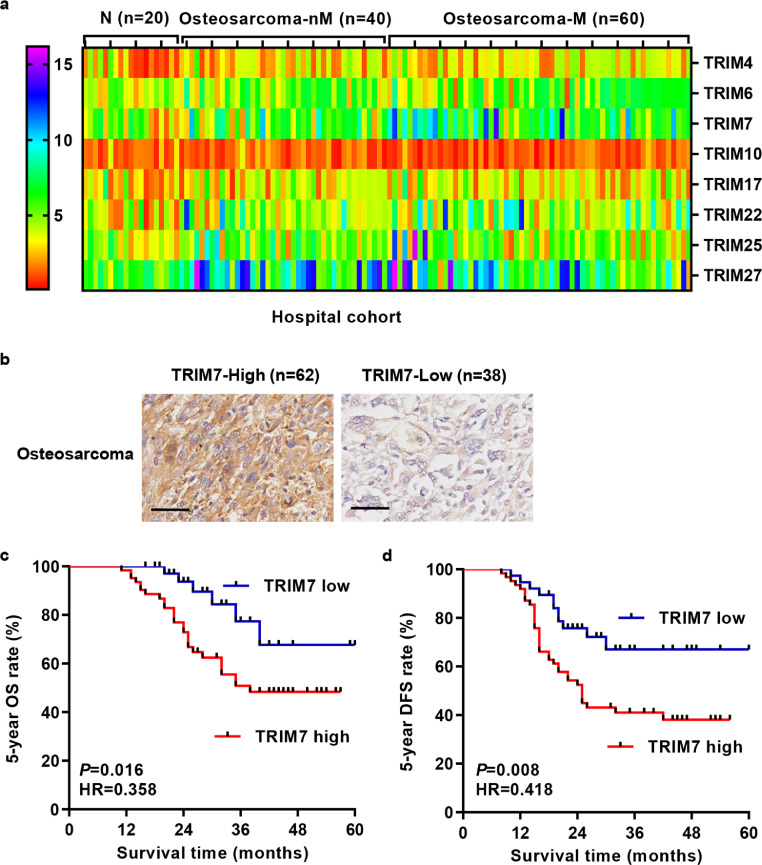
Given that osteosarcoma is the most common, primary solid malignancy of the bone and its high mortality usually correlates with early metastasis, the mRNA expression of several TRIM proteins, including TRIM4, 6, 7, 10, 17, 22, 25, and 27, was measured in tumour tissues of osteosarcoma patients with (n = 60) or without lung metastasis (n = 40). These proteins share the same structural domain [26] and are rarely reported in osteosarcoma. Among them, TRIM4 (P = 0.0011), TRIM6 (P = 0.0034), TRIM7 (P = 0.0001), TRIM17 (P = 0.0350), TRIM22 (P = 0.0024), TRIM25 (P = 0.0493), and TRIM27 (P = 0.0137) expression was significantly increased in osteosarcoma tissues compared with adjacent nontumorous tissues and only TRIM7 expression (P = 0.0044) was much more higher in metastatic osteosarcoma tissues compared with non-metastatic osteosarcoma tissues (Fig. 1(a); Mann–Whitney U test). Therefore, to further investigate the clinical significance of TRIM7 in osteosarcoma, IHC stainings were performed on the paraffin-embedded osteosarcoma samples (n = 100). Our data showed that 62% of osteosarcoma tissues expressed high levels of TRIM7 (Fig. 1(b)). Kaplan–Meier analysis and log-rank test showed that the OS and DFS rate of patients with lower TRIM7 expression were markedly higher than those of patients with higher TRIM7 expression (Fig. 1(c) and (d)). In addition to osteosarcoma, higher TRIM7 expression was also associated with poor survival of patients with breast cancer, esophageal adenocarcinoma, hepatocellular carcinoma, pancreatic ductal adenocarcinoma, sarcoma and uterine corpus endometrial carcinoma (Fig. S1), which indicates that higher TRIM7 expression usually occurs in most of the cancer patients. Chi-square test indicated that the expression of TRIM7 was correlated with Enneking's stage, AJCC stage, tumour size, and lung metastasis, but not with age (median value as cut off), sex, local recurrence and anatomic location (Table 3). In addition, a Cox regression model was established to analyze the effect of each variable on overall survival. In terms of overall survival, univariate analysis revealed that TRIM7 expression, local recurrence, lung metastasis, AJCC stage, and Enneking's stage were prognostic factors. The results of the multivariate analysis showed that AJCC stage and TRIM7 expression were independently prognostically relevant in terms of overall survival (Table 4). These results indicate that TRIM7 may play important roles in osteosarcoma's malignant transformation.
Fig. 1.
TRIM7 expression and survival analysis in patients with osteosarcoma. (a) qRT-PCR and (b) immunohistochemical staining showing the expression of TRIM7 in tumour tissues of osteosarcoma patients. Scale bar: 100 μm. (c, d) The 5-year overall survival (OS) and disease-free survival (DFS) rate of patients with osteosarcoma. The statistical analysis was performed using Log-rank (Mantel-Cox) test. All the experiments were repeated at least three times, and data are represented as mean ± SD. N, adjacent nontumorous tissues; nM, non-lung metastasis; M, lung metastasis.
Table 3.
Correlation of TRIM7 expression with clinicopathological features of osteosarcoma
| Clinicopathological parameter | TRIM7 protein expression |
P value | |
|---|---|---|---|
| Low (n = 38) | High (n = 62) | ||
| Age | 0.4628 | ||
| <14 | 15 | 20 | |
| ≥14 | 23 | 42 | |
| Sex | 0.2658 | ||
| Female | 28 | 39 | |
| Male | 10 | 23 | |
| Tumour size (cm) | 0.0284 | ||
| <8 | 22 | 22 | |
| ≥8 | 16 | 40 | |
| Local recurrence | 0.4297 | ||
| Yes | 2 | 6 | |
| No | 36 | 56 | |
| Enneking's stage | 0.0146 | ||
| IIA | 5 | 22 | |
| IIB | 33 | 40 | |
| AJCC stage | 0.0025 | ||
| IIA | 24 | 20 | |
| IIB | 14 | 42 | |
| Anatomic location | 0.3267 | ||
| Femur | 19 | 23 | |
| Tibia | 10 | 16 | |
| Humerus | 6 | 10 | |
| Others | 3 | 13 | |
| Lung metastasis | 0.0147 | ||
| Yes | 17 | 43 | |
| No | 21 | 19 | |
Differences between groups were done by the Chi-square test.
Table 4.
Univariate and multivariate analysis of overall survival in patients with osteosarcoma.
| Variables | Univariate analysis |
Multivariate analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age (≥14 vs. <14) | 1.040 (0.612–1.693) | 0.880 | ||
| Sex (female vs. male) | 1.108 (0.668–1.944) | 0.704 | ||
| Tumour size (≥8 cm vs. <8 cm) | 0.838 (0.617–1.102) | 0.207 | ||
| Local recurrence (yes vs. no) | 0.230 (0.041–0.887) | 0.023 | ||
| Enneking's stage (IIB vs. IIA) | 1.768 (1.129–2.666) | 0.014 | ||
| AJCC stage (IIB vs. IIA) | 0.650 (0.463–0.867) | 0.003 | 0.740 (0.536–0.977) | 0.034 |
| Anatomic location (femur, tibia, humerus vs. others) | 0.647 (0.359–1.377) | 0.245 | ||
| Lung metastasis (yes vs. no) | 0.571 (0.351–0.928) | 0.024 | ||
| TRIM7 expression (high vs. low) | 1.810 (1.243–2.635) | 0.002 | 1.622 (1.104–2.384) | 0.014 |
3.2. TRIM7 knockdown inhibits the migration and invasion of osteosarcoma cells
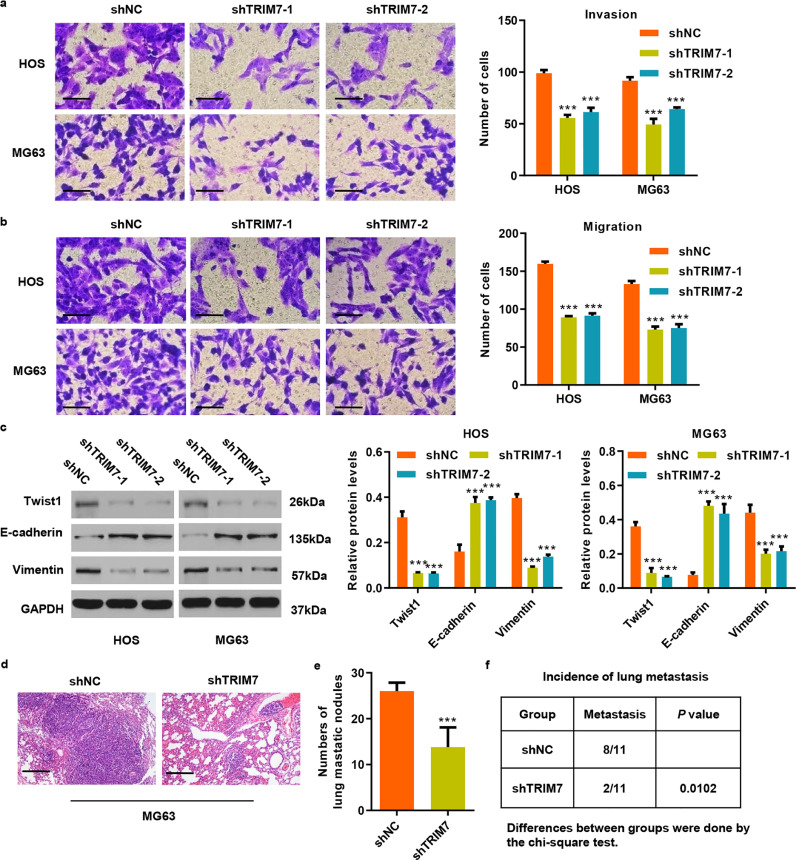
To examine the potential biological function of TRIM7 in osteosarcoma, we first measured TRIM7 expression in osteosarcoma cell lines. Our results show that TRIM7 expression was significantly increased in four osteosarcoma cell lines than that in the human osteoblastic cell line hFOB1.19, with the highest expression detected in HOS and MG63 cell lines (Fig. S2(a)). These two cell lines were therefore transduced with pLKO.1-shTRIM7 lentivirus (Fig. S2(a) and (c)). Since TRIM7 expression was associated with the metastasis of OS patients, the invasion and migration of HOS and MG63 cells were measured by Transwell analysis in vitro. As shown in Fig. 2(a), shTRIM7-1 significantly inhibited cell invasion by 43.8% and 46.2% in HOS and MG63 cells, respectively, compared with the shNC group. Moreover, shTRIM7-1 also significantly inhibited cell migration, by 44.1% and 45.1% in HOS and MG63 cells compared with the shNC group, respectively (Fig. 2(b)). Similarly, the decreased cell invasion and migration were also found in HOS and MG63 cells transduced with shTRIM7-2. Given the important roles of the epithelial-to-mesenchymal transition (EMT) in cancer pathogenesis and progression, we further measured the expression levels of epithelial gene (E-cadherin) and mesenchymal genes (vimentin and Twist1) in HOS and MG63 cells after transduction with shTRIM7-1 or shTRIM7-2. Interestingly, we found that TRIM7 knockdown reduced Twist1 and Vimentin expressions and increased E-cadherin expression in HOS and MG63 cells compared with the shNC group (Fig. 2(c)). These results suggest that the downregulation of TRIM7 may inhibit OS metastasis by inducing mesenchymal to epithelial transition (MET). To explore the effect of TRIM7 on lung metastasis of osteosarcoma cells in vivo, we injected MG63 cells expressing either shTRIM7 or shNC via the tail vein into nude mice. As shown in Fig. 2(d–f), the number of lung metastasis nodules and the instances of metastasis in mice were significantly decreased by TRIM7 knockdown. Taken together, these results suggest that TRIM7 is an important regulator of cancer cell invasion and migration in osteosarcoma.
Fig. 2.
TRIM7 silencing inhibits cell invasion and migration in osteosarcoma cell lines. (a, b) Transwell and (c) western blot analysis showing the effect of pLKO.1-shTRIM7 lentivirus transduction on the invasion, migration, and expression of Twist1, E-cadherin and Vimentin in HOS and MG63 cells. Scale bar: 50 μm. (d) After 21 days of intravenous injection with MG63 cells that have been transduced with pLKO.1-shTRIM7 lentivirus, a histological inspection was measured by H&E. Scale bar: 200 μm. (e) Quantification of lung microscopic nodules and (f) incidence of metastasis in mice of each group (n = 11). All the experiments were repeated at least three times, and data are represented as mean ± SD. (a–c) ***P < 0.001 (two-way ANOVA followed by Bonferroni post-tests) compared with the shNC group. (e) ***P < 0.001 (unpaired Student's t-test) compared with the shNC group.
3.3. TRIM7 regulates the response of osteosarcoma cells to chemotherapy
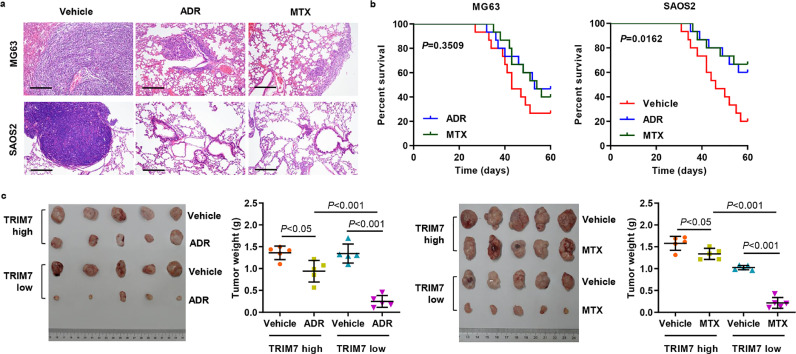
Recent studies have shown that EMT is associated with drug resistance; hence, we examined the role of TRIM7 in osteosarcoma response to chemotherapy [27]. MG63 and SAOS2 osteosarcoma cells with different TRIM7 expression were transplanted into mice, and these mice were inoculated with the chemotherapy agent as well. We measured lung metastasis foci and survival outcomes in these mice. As shown in Fig. 3(a) and (b), the lung metastasis was significantly inhibited in mice injected with SAOS2 or MG63 cells, and survival outcomes were significantly improved in the mice injected with SAOS2 cells compared with the mice injected with MG63 cells. Moreover, PDX mice following ADR or MTX chemotherapy with high-TRIM7-expression demonstrated larger tumour size than those with low-TRIM7-expression (Fig. 3(c)). These data reveal that TRIM7 may play an important role in regulating chemosensitivity in osteosarcoma.
Fig. 3.
TRIM7 regulates osteosarcoma cells’ response to chemotherapy. (a) After 21 days of intravenous injection with MG63 or SAOS2 cells as well as intraperitoneal injection with 1 mg/kg ADR, 0.2 mg/kg MTX, or vehicle (n = 5), a histological inspection was measured by H&E. Scale bar: 200 μm. (b) The survival period of the mice from each group is shown (n = 15). The statistical analysis was performed using Log-rank (Mantel-Cox) test. (c) Tumour weight was measured in mice with the patient-derived xenograft (PDX) following ADR or MTX chemotherapy (n = 5). P < 0.05, P < 0.01, P < 0.001 (one-way ANOVA followed by Bonferroni post-tests). All the experiments were repeated at least three times, and data are represented as mean ± SD.
3.4. TRIM7 interacts with and mediates the ubiquitination of BRMS1
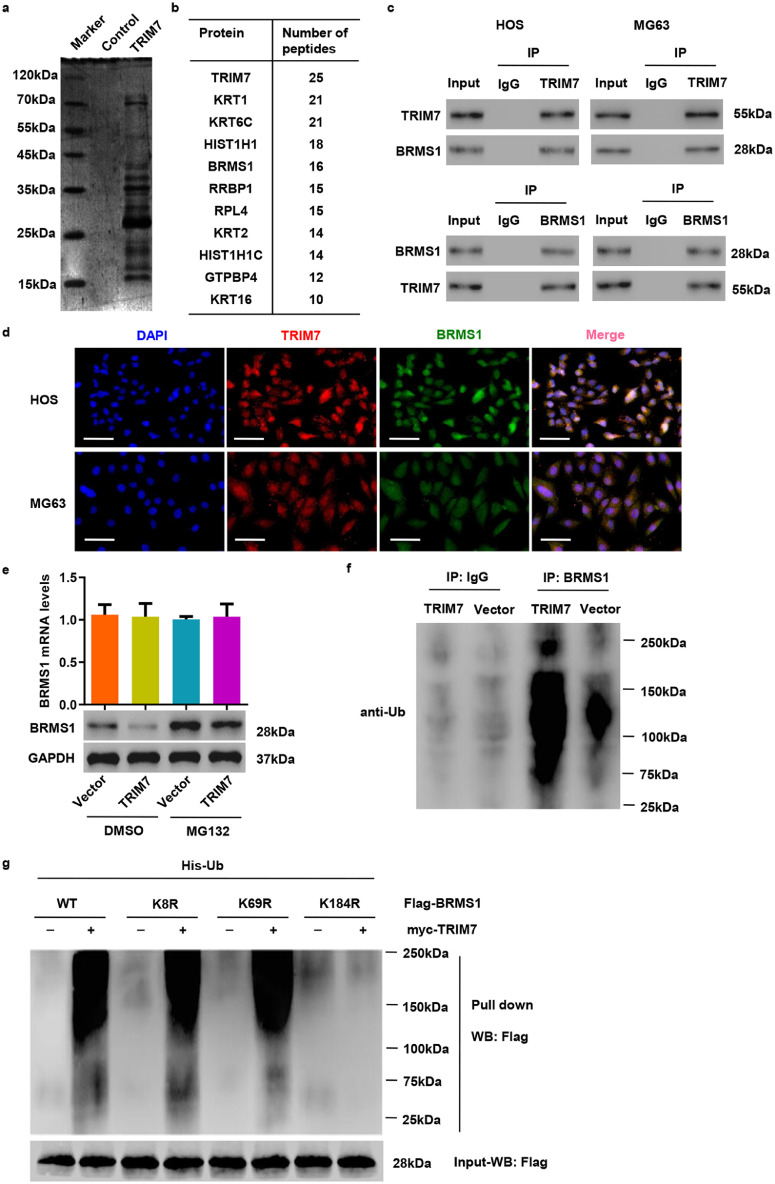
To investigate the molecular mechanism through which TRIM7 exerts its function, we performed a proteomics analysis to identify candidate proteins associated with TRIM7. The candidate proteins were identified by LC/MS and are shown in Fig. 4(a). The top ten proteins that contain more than 10 peptides are listed in Fig. 4(b). Among the identified interaction partners, BRMS1, which plays a tumour suppressor in osteosarcoma [15], was selected for further study. Co-IP and subsequent immunoblot analysis revealed that TRIM7 co-immunoprecipitated with BRMS1 in HOS and MG63 cells (Fig. 4(c)). Reciprocal Co-IP with BRMS1 antibodies also brought down TRIM7, confirming a physical interaction between the two proteins. In addition, immunofluorescence staining showed that both TRIM7 and BRMS1 could be localized to the nucleus and the cytoplasm in HOS and MG63 cells (Fig. 4(d)). These results further support the idea that TRIM7 and BRMS1 interact with each other physically.
Fig. 4.
TRIM7 interacts with BRMS1 and ubiquitinates it. (a) Purification of the TRIM7 complex was carried out according to the procedure described in Materials and Methods. Proteins were separated on SDS-PAGE and stained with Coomassie Blue. (b) List of TRIM7-associated proteins identified by MS analysis. (c) HOS and MG63 cell lysates were subjected to IP with anti-TRIM7, anti-BRMS1 or control IgG antibody. (d) The subcellular localization of TRIM7 (red) and BRMS1 (green) was measured by immunofluorescence. Nuclei were stained with DAPI (blue) for reference. Scale bar: 50 μm. (e) qRT-PCR and Western blot analysis showing the effect of TRIM7 overexpression on endogenous BRMS1 levels in SAOS2 cells in the presence of 10 μM proteasome inhibitor (MG132) or DMSO. (f) IP and western blot showing the effect of TRIM7 overexpression on the ubiquitination of BRMS1 in SAOS2 cells. (g) 293T cells were cotransfected with the Flag-BRMS1 (WT) or Flag-mutant BRMS1 constructs (K8R, K69R or K184R) along with myc-TRIM7 and His-Ub constructs, and the pull-down assay was carried out. All the experiments were repeated at least three times, and data are represented as mean ± SD.
Since TRIM7 is a ubiquitin ligase, we hypothesized that TRIM7 could regulate the protein level of BRMS1 through ubiquitin modification. Overexpression of TRIM7 overexpression in SAOS2 cells, which expressed lower levels of TRIM7 (Fig. S2(a)), significantly increased TRIM7 at both mRNA and protein levels (Fig. S2(d)). Moreover, TRIM7 overexpression-induced decrease in the BRMS1 protein expression could be reversed by the addition of proteasome inhibitor MG132 in SAOS2 cells (Fig. 4(e)). This indicates that TRIM7 regulates BRMS1 protein expression in a proteasome-dependent manner. Next, we examined whether TRIM7 regulates BRMS1 through ubiquitination. We found that TRIM7 overexpression significantly increased BRMS1 ubiquitination in SAOS2 cells (Fig. 4(f)). We also inserted various mutants of BRMS1 in a pCMV-Tag 2B vector, and co-transfected these constructs in 293T cells along with myc-TRIM7 and His-Ub constructs. As shown in Fig. 4(g), TRIM7 stimulated efficient ubiquitination of BRMS1 when K8- or K69-mutant BRMS1 was introduced; however, ubiquitination was markedly decreased when K184-mutant BRMS1 was introduced. Therefore, the K184 site of BRMS1 is both necessary and sufficient for the ubiquitination by TRIM7.
3.5. BRMS1 is a mediator for TRIM7-induced invasion and migration of osteosarcoma cells
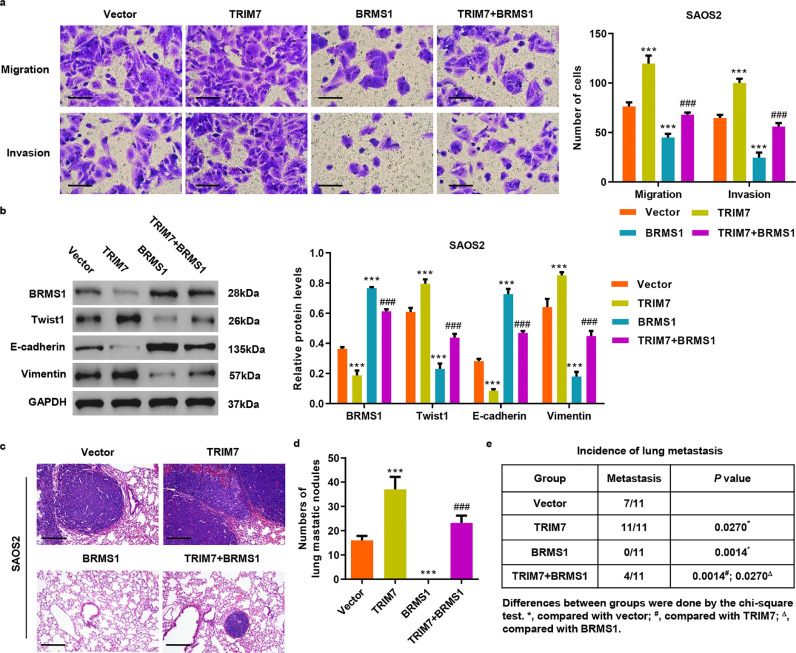
To pinpoint whether TRIM7 acts through BRMS1, we transduced SAOS2 cells with lentivirus expressing TRIM7 and BRMS1 (Fig. S3(a)). Transwell analysis demonstrated that TRIM7 overexpression significantly increases invasion and migration by 54.6% and 56.8% while BRMS1 overexpression significantly decreases invasion and migration by 61.9% and 41.1% compared with the vector group, respectively (Fig. 5(a)). BRMS1 overexpression also reversed the increased invasion and migration in SAOS2 cells with ectopic TRIM7 expression. Remarkably, TRIM7 overexpression enhanced Twist1 and Vimentin expression but reduced E-cadherin expression, which was also reversed by BRMS1 overexpression (Fig. 5(b)). To explore whether TRIM7 also exerted functions through BRMS1 in vivo, we also transduced SAOS2 cells with TRIM7, BRMS1, TRIM7 plus BRMS1 or control lentiviral expression vectors and injected these cells via the tail vein into nude mice. The number of lung metastasis nodules and the incidence of metastasis in mice were significantly increased by TRIM7 overexpression and additional BRMS1 overexpression was significantly blocked by TRIM7 overexpression (Fig. 5(c–e)). Taken together, these results show that BRMS1 mediates TRIM7-induced invasion and migration of cancer cells in osteosarcoma.
Fig. 5.
BRMS1 overexpression inhibits invasion and migration induced by TRIM7 in osteosarcoma cell lines. (a) Transwell and (b) western blot analysis showing the effect of pLVX-Puro-TRIM7 and/or pLVX-Puro-BRMS1 lentivirus transduction on the invasion, migration, and expression of BRMS1, Vimentin, Twist1 and E-cadherin in SAOS2 cells. Scale bar: 50 μm. (c) After 21 days of intravenous injection with SAOS2 cells that have been transduced with lentivirus expressing TRIM7 and/or BRMS1, a histological inspection was measured by H&E. Scale bar: 200 μm. (d) Quantification of lung microscopic nodules and (e) incidence of metastasis in mice of each group (n = 11). All the experiments were repeated at least three times, and data are represented as mean ± SD. (a, b) ***P 0.001 (two-way ANOVA followed by Bonferroni post-tests) compared with the vector group. (a, b) ###P < 0.001 (two-way ANOVA followed by Bonferroni post-tests) compared with the TRIM7 group. (d) ***P < 0.001 (one-way ANOVA followed by Bonferroni post-tests) compared with the vector group. (d) ###P < 0.001 (one-way ANOVA followed by Bonferroni post-tests) compared with the TRIM7 group.
3.6. TRIM7 regulates osteosarcoma cells’ response to chemotherapy through BRMS1
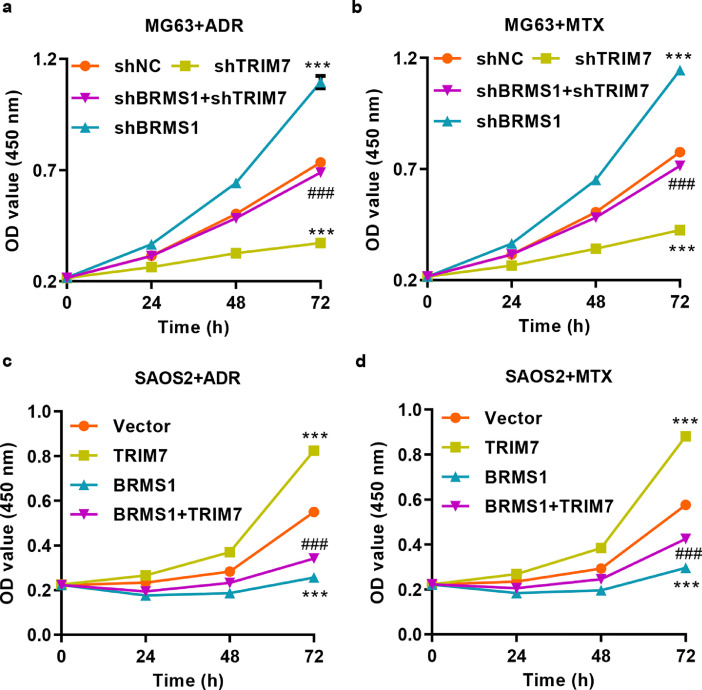
To examine whether TRIM7 plays a role in osteosarcoma response to chemotherapy, we treated MG63 cells that were transduced with pLKO.1-shTRIM7 and/or pLKO.1-shBRMS1 lentivirus with ADR or MTX. After BRMS1 knockdown, the expression of BRMS1 in MG63 cells was significantly reduced at both mRNA and protein levels (Fig. S3(b)). CCK-8 assay showed that MG63 cells with TRIM7 knockdown were more sensitive to ADR and MTX treatment (Fig. 6(a) and (b)). Conversely, MG63 cells with BRMS1 knockdown resulted in increased resistance to ADR and MTX treatment. Moreover, the BRMS1 knockdown significantly counteracted the chemosensitivity induced by TRIM7 knockdown (Fig. 6(a) and (b)). We also treated SAOS2 cells transduced with pLVX-Puro-TRIM7 and/or pLVX-Puro-BRMS1 lentivirus with ADR or MTX. SAOS2 cells with TRIM7 overexpression resulted in increased resistance to ADR and MTX treatment (Fig. 6(c) and (d)). Conversely, SAOS2 cells with BRMS1 overexpression were more sensitive to ADR and MTX treatment. Moreover, BRMS1 overexpression significantly reversed the chemosensitivity induced by TRIM7 overexpression (Fig. 6(c) and (d)).
Fig. 6.
TRIM7 regulates osteosarcoma cells’ response to chemotherapy by targeting BRMS1. CCK-8 showing the effect of pLKO.1-shTRIM7 and/or pLKO.1-shBRMS1 lentivirus transduction on the proliferation of MG63 cells treated with (a) 5 μg/ml ADR or (b) 2 ng/ml MTX. CCK-8 showing the effect of pLVX-Puro-TRIM7 and/or pLVX-Puro-BRMS1 lentivirus transduction on the proliferation of SAOS2 cells treated with (c) 5 μg/ml ADR or (d) 2 ng/ml MTX. All the experiments were repeated at least three times, and data are represented as mean ± SD. ***P < 0.001 (two-way ANOVA followed by Bonferroni post-tests) compared with the shNC or the vector group. ###P < 0.001 (two-way ANOVA followed by Bonferroni post-tests) compared with the shTRIM7 or the TRIM7 group.
3.7. TRIM7 and BRMS1 expressions show a negative correlation in clinical osteosarcoma samples
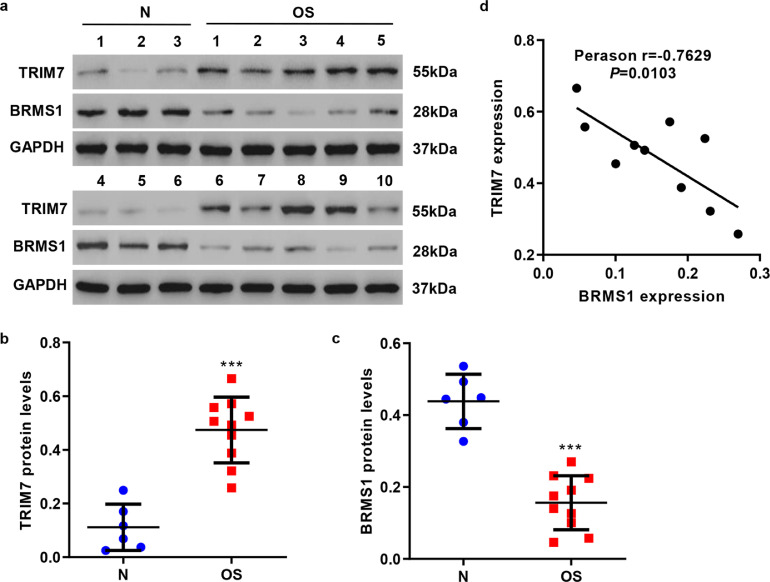
We assessed the expressions of TRIM7 and BRMS1 in clinical osteosarcoma samples by western blot. As shown in Fig. 7(a–c), TRIM7 showed high expression while BRMS1 showed low expression in osteosarcoma tissues (n = 10) compared with adjacent nontumorous tissues (n = 6). Thus, there is a negative correlation between TRIM7 and BRMS1 expressions in osteosarcoma tissues (Fig. 7(d)).
Fig. 7.
Correlation analyses in osteosarcoma samples. (a–c) Western blot analysis showing the expression of TRIM7 or BRMS1 expression in adjacent nontumorous (n = 6) and osteosarcoma samples (n = 10). ***P < 0.001 (Mann–Whitney U test) compared with adjacent nontumorous tissues (N). (d) Pearson correlation scatter plots in osteosarcoma tissues (n = 10). OS, osteosarcoma. All the experiments were repeated at least three times, and data are represented as mean ± SD..
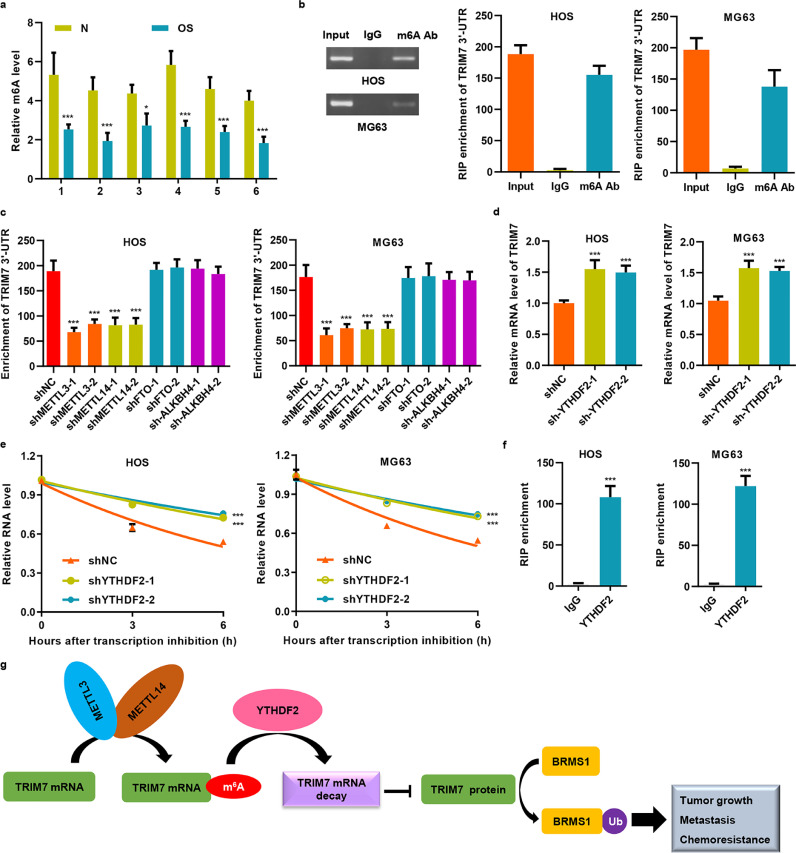
3.8. METTL3 and METTL14 promotes the m6A modification of TRIM7 via the m6A reader YTHDF2
Our bioinformatics analysis revealed that the motif of m6A modification is present in the 3′-UTR region of TRIM7. In addition, compared with cancer tissues, the m6A motif was significantly enriched in the 3′-UTR regions of TRIM7 in normal control tissues (Fig. 8(a)); we confirmed this in HOS and MG63 cells as well (Fig. 8(b)). Furthermore, qRT-PCR assays showed that knockdown of METTL3 and METTL14 significantly decreased the m6A level of TRIM7 in HOS and MG63 cells, but not in FTO and ALKBH4 cells (Fig. 8(c) and S4(a–d)), which indicates that METTL3 and METTL14 are responsible for the m6A modification of TRIM7. Next, we knocked down YTHDF2 (Fig. S4(e)), which is an m6A reader that decomposes the mRNA of the target gene, to see how YTHDF2 affects the stability of TRIM7 mRNA. Knocking down YTHDF2 significantly increased the mRNA level of TRIM7 in HOS and in MG63 cells (Fig. 8(d)), and half-life of TRIM7 mRNA was significantly extended in YTHDF2 down-regulated HOS and MG63 cells (Fig. 8(e)). Further RIP-qRT-PCR assay indicated that YTHDF2 might directly bind to the 3’-UTR of TRIM7 mRNA (Fig. 8(f)). Taken together, our data demonstrate that TRIM7 mRNA stability was regulated by the METTL3/14-YTHDF2-mRNA in a decay-dependent manner.
Fig. 8.
METTL3 and METTL14 promote the m6A modification of TRIM7 via the m6A reader YTHDF2. (a) The m6A levels in adjacent nontumorous (N) and osteosarcoma samples were measured. (b) Relative m6A levels of TRIM7 3′-UTR in HOS and MG63 cells were detected by RIP followed by qRT-PCR analysis. (c) TRIM7 3′-UTR enrichment was measured by RIP followed by qRT-PCR analysis in HOS and MG63 cells that were transfected with shRNAs targeting METTL3, METTL14, FTO, or ALKBH4. YTHDF2 silencing in HOS and MG63 cells increased TRIM7 mRNA levels (d) and stability (e). (f) RIP followed by qRT-PCR assay was performed to examine the interaction between YTHDF2 and 3′-UTR of TRIM7 mRNA. (g) A proposed molecular model of how increased TRIM7 expression, via inhibition of m6A modification, leads to the development and progression of osteosarcoma. All the experiments were repeated at least three times, and data are represented as mean ± SD. (a, e) *P < 0.05, ***P < 0.001 (two-way ANOVA followed by Bonferroni post-tests) compared with N or shNC. (c, d) ***P < 0.001 (one-way ANOVA followed by Bonferroni post-tests) compared with shNC. (f) ***P < 0.001 (unpaired Student's t-test) compared with IgG.
4. Discussion
Although metastasis and chemoresistance are the leading causes of death in osteosarcoma patients, the mechanisms responsible for metastasis and chemoresistance remain unclear. A large body of evidence shows that TRIM family member proteins can function as potential oncogenes or tumour suppressor genes and that these proteins could be used as potential biomarkers for chemoresistance, prediction of prognosis, and diagnosis in various cancers [7,8,28]. TRIM7 expression is markedly increased in patients with metastatic osteosarcoma than that in patients without metastasic osteosarcoma, and this increased expression is associated with survival time in patients with osteosarcoma patients. These studies suggest that TRIM7 is involved in osteosarcoma metastasis and that metastasis and chemoresistance in cancer are linked phenomena. Here, we investigated the effect(s) of TRIM7 on regulating cell metastasis and chemoresistance in osteosarcoma. We found that TRIM7 expression was significantly increased in patients with osteosarcoma and that TRIM7 expression was correlated with several clinicopathological indicators such as tumour size, lung metastasis, and Enneking's stage. Furthermore, we found that increased TRIMP7 expression was an independent prognostic factor. Functional studies illustrated that while TRIM7 overexpression induced invasion and migration in osteosarcoma cells, TRIMP7 silencing inhibited invasion and migration. Furthermore, data from both in vitro and in vivo studies show that high TRIM7-expression reduces the sensitivity to ADR and MTX treatment. Mechanistically, TRIM7 regulates metastasis and chemoresistance of osteosarcoma cells through the ubiquitination of BRMS1 at the K184 site. TRIM7 3′-UTR methylation was significantly decreased in osteosarcoma compared with normal samples, and analysis of osteosarcoma cell lines strongly suggested that TRIM7 3′-UTR methylation, mediated by METTL3/14-YTHDF2, was associated with decreased expression. Thus one of the oncogenic effects of aberrant expression of TRIM7 is to inhibit BRMS1, forming a vicious METTL3/14-YTHDF2-TRIM7-BRMS1 axis that promotes osteosarcoma metastasis and chemoresistance (Fig. 8(g)).
We found TRIM7 expression to be higher in osteosarcoma clinical samples compared with normal bone tissues. This suggests that TRIM7, as an oncogene, may be involved in osteosarcoma development. Similarly, TRIM7 is also up-regulated in mucinous colorectal carcinomas compared with non-mucinous colorectal carcinomas [10], and in lung adenocarcinoma tissues compared with normal lung tissues [11]. TRIM37 is upregulated in pediatric osteosarcoma tissues, and its overexpression induces cell proliferation and reduces chemosensitivity in osteosarcoma [7]. TRIM2 is increased in osteosarcoma tissues, and its downregulation induces increased cell apoptosis, decreased cell invasion and migration, and inhibition of metastasis in vivo nude mice model of osteosarcoma mice [8]. TRIM14 is upregulated in osteosarcoma tissues and cell lines, and its overexpression promotes tumour growth in vivo and increases osteosarcoma cell cycle procession, clone formation, as well as cell invasion, migration, and proliferation in vitro [9]. In line with the previous studies that show the involvement of other TRIM family proteins in tumorigenesis, we found that TRIM7 knockdown inhibited osteosarcoma cell invasion and migration in vitro and lung metastasis in vivo nude mice. Furthermore, TRIM7’s role in desensitizing osteosarcoma cells to ADR and MTX treatment may have important therapeutic implications. Because osteosarcoma chemotherapy is based primarily on the use of ADR and MTX, therapeutic interventions targeting TRIM7 can be combined with existing therapies to improve therapeutic efficacy. MG63 cells with higher TRIM7 levels reversed the chemosensitivity to ADR and MTX treatment compared with SAOS2 cells with lower TRIM7 levels both in vitro and in vivo. TRIM7, however, did not change the chemosensitivity of osteosarcoma cells to cisplatin (DDP), another effective chemotherapeutic drug (unpublished data).
We also observed high expression and localization of TRIM7 and BRMS1 to the nucleus and the cytoplasm in HOS and MG63 cells. A previous study also showed that TRIM7 is occasionally found in the nucleus and is distributed throughout the cytoplasm [29]. BRMS1 is an important component of the Sin3-HDAC chromatin remodeling complex and plays a key role in histone deacylation and chromatin condensation, which leads to reduced transcription of several genes [30]. BRMS1 has also been found in the nucleus and the cytoplasm, which is consistent with earlier reports that classified BRMS1 as a nucleo-cytoplasmic protein [30,31]. The Ring finger domain of the TRIM family proteins may mediate the ubiquitination and stabilization of different substrates through the E3 ubiquitin ligase activity [32,33]. TRIM7 drives lung tumorigenesis by mediating Lys63-linked ubiquitination of the AP-1 co-activator RACO-1, which leads to RACO-1 protein stabilization [11]. Not surprisingly, in this study, we saw that TRIM7 overexpression induced BRMS1 ubiquitination at the K184 site. A Recent report has characterized BRMS1 as a metastasis suppressor that has low expression in metastatic osteosarcoma tissues than that in non-metastatic osteosarcoma tissues, as well as in osteosarcoma tissues compared with non-tumour bone tissues. BRMS1 suppression, furthermore, has been shown to promote osteosarcoma cell invasion in vitro [15] and lung cancer cell metastasis in vivo [19]. Our results corroborate those findings. Pearson correlation scatter plots also demonstrated a negative correlation between TRIM7 and BRMS1 protein expression in osteosarcoma tissues. Moreover, TRIM7 expression was low in high BRMS1-expressing SAOS2 cells and the expression was high in low BRMS1-expressing U2-OS cells. Together, these results suggest a negative correlation between TRIM7 and BRMS1 in osteosarcoma cells.
Epithelial to mesenchymal transition (EMT) is an essential mechanism for development of malignant cells for invasion and metastasis. A previous study showed that BRMS1 knockdown is associated with decreased E-cadherin and increased Twist1 and Vimentin [19], and our data is consistent with those results. The study also showed that BRMS1 regulates the EMT phenotype via targeting Twist1 through NF-κB signaling, which is required for the activation of Twist1 promoter. BRMS1 contributes to the positive regulation of E-cadherin expression through the recruitment of HDAC1 binding to the E-cadherin promoter [34]. BRMS1 and LSD1 co-occupied the promoters of Vimentin as one functionally collaborated protein complex, and LSD1 knockdown led to a significant reduction in the binding of BRMS1 to the promoter of Vimentin, suggesting that LSD1 is required for the transcriptional repression of Vimentin induced by BRMS1 [35]. These accumulated data suggested that TRIM7 knockdown induced mesenchymal to epithelial transition (MET) via regulation of BRMS1 expression and that the MET resulted in loss of malignancy or waiver of malignancy, although it was suggested that cancer cells probably use this MET process during the later stages of metastasis [36]. In our study, TRIM7 overexpression-induced tumorigenesis of osteosarcoma was significantly inhibited by BRMS1 overexpression. This suggests that the molecular mechanism by which TRIM7 regulates osteosarcoma involves the targeting of BRMS1. Moreover, BRMS1 also associated with chemosensitivity in cancers [37]. BRMS1 mediates ubiquitination and destabilization of p300/CBP, which leads to p53 activation as transcription factors and ultimately triggers apoptosis, and contributes to DDP resistance in melanoma cells [38,39]. Similarly, BRMS1 significantly reversed the cell proliferation induced by TRIM7 in osteosarcoma cells after ADR and MTX treatment, suggesting there is ADR and MTX resistance in BRMS1-depleted cells as well as in TRIM7-expressing cells through the targeting of BRMS1. However, breast cancer MDA-MB-231 and MDA-MB-435 cells or lung cancer A549 cells with or without BRMS1 similarly reacted to ADR, vincristine, 5-Fluorouracil, and paclitaxel, which suggests that BRMS1 expression did not influence chemoresistance of these cells [30,37]. Therefore, the chemoresistance of BRMS1 and the underlying molecular mechanism involved need further investigation. Numerous studies have suggested that a reversal of the EMT process, namely MET, could potentially be a unique therapeutic method for inhibiting metastasis and sensitizing cancer cells to chemotherapeutics, and thus the mesenchymal and epithelial markers including Twist1, Vimentin and E-cadherin may regulate the chemosensitivity in cancer [40], [41], [42], [43], [44], [45]. These data indicate that BRMS1 may regulate TRIM7-induced chemosensitivity to ADR and MTX treatment in osteosarcoma via inducing MET process. In the future, we will screen and attempt to identify drug-resistant osteosarcoma cell lines, which may provide crucial mechanistic insights and a foundation for TRIM7-BRMS1-based therapeutics and biomarkers in osteosarcoma.
The modification of m6A in RNAs is a common epigenetic regulation involved in a variety of cellular processes, such as protein production, splicing, RNA stability; all of these processes, if aberrant, may cause diseases including malignancies [46]. The m6A methyltransferase METTL3 promoted osteosarcoma progression by regulating the m6A level of LEF1 [47]. Similarly, METTL3 and METTL14 promoted the m6A modification of TRIM7 3’-UTR via the m6A reader YTHDF2 in osteosarcoma cells. Moreover, METTL3 expression was increased in multidrug-resistant MG63/ADR cells compared with its progenitor MG63 cells and associated with worse metastasis-free survival in osteosarcoma patients [48]. However, further investigations on the link between aberrant m6A modification in TRIM7 and the development, progression, and chemoresistance of osteosarcoma are urgently needed.
In summary, we have identified an E3 ubiquitin ligase TRIM7 that regulates osteosarcoma cell invasion, migration and chemoresistance through ubiquitination of BRMS1. Upregulated TRIM7 was regulated by m6A modification via the METTL3/14-YTHDF2-mRNA decay-dependent manner. Given the roles of TRIM7 in regulating cell metastasis and chemoresistance, approaches to downregulate TRIM7 by reversing its upregulation may inhibit metastasis and resensitize tumours to chemotherapy as well as improve the outcome of chemotherapy to osteosarcoma patients.
Author contributions
C.Z., Z.Z., Z.S., and S.Z. designed studies, analyzed and wrote the manuscript. X.Z., G.Q., Y.Z., Y.S., W.Y., and J.W. conduct the in vitro and in vivo experiments and data analysis; C.Z., Z.Z., Z.S., and S.Z. collect the patient samples, follow-up information and performed the clinical data analysis. X.Z., H.L., and F.L. performed the bioinformatics analysis. All the authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Funding sources
This work was funded by National Natural Science Foundation of China (81001192, 81672658 and 81972521) and National Key Research Project of Science and Technology Ministry (2016YFC0106204). The funders had no role in the study design, data collection, data analysis, interpretation, and writing of the report.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.102955.
Contributor Information
Zan Shen, Email: sshenzzan@vip.sina.com.
Shuier Zheng, Email: 2297456501@qq.com.
Appendix. Supplementary materials
References
- 1.Yan GN, Lv YF, Guo QN. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016;370(2):268–274. doi: 10.1016/j.canlet.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Smeland S, Bielack SS, Whelan J, Bernstein M, Hogendoorn P, Krailo MD. Survival and prognosis with osteosarcoma: outcomes in more than 2000 patients in the EURAMOS-1 (European and American Osteosarcoma Study) cohort. Eur J Cancer. 2019;109:36–50. doi: 10.1016/j.ejca.2018.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang W, Zhao HF, Yao TF, Gong H. Advanced development of ErbB family-targeted therapies in osteosarcoma treatment. Investig New Drugs. 2018 doi: 10.1007/s10637-018-0684-8. [DOI] [PubMed] [Google Scholar]
- 4.Mayr L, Pirker C, Lotsch D, Van Schoonhoven S, Windhager R, Englinger B. CD44 drives aggressiveness and chemoresistance of a metastatic human osteosarcoma xenograft model. Oncotarget. 2017;8(69):114095–114108. doi: 10.18632/oncotarget.23125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tomar D, Singh R. TRIM family proteins: emerging class of RING E3 ligases as regulator of NF-kappaB pathway. Biol Cell. 2015;107(1):22–40. doi: 10.1111/boc.201400046. [DOI] [PubMed] [Google Scholar]
- 6.Forlani G, Tosi G, Turrini F, Poli G, Vicenzi E, Accolla RS. Tripartite motif-containing protein 22 interacts with class II transactivator and orchestrates its recruitment in nuclear bodies containing TRIM19/PML and Cyclin T1. Front Immunol. 2017;8:564. doi: 10.3389/fimmu.2017.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao Y, Xin M, Cheng H, Huang Z, Hu T, Zhang T. TRIM37 promotes tumor cell proliferation and drug resistance in pediatric osteosarcoma. Oncol Lett. 2017;14(6):6365–6372. doi: 10.3892/ol.2017.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin Y, Ye J, Zhao F, Hu S, Wang S. TRIM2 regulates the development and metastasis of tumorous cells of osteosarcoma. Int J Oncol. 2018;53(4):1643–1656. doi: 10.3892/ijo.2018.4494. [DOI] [PubMed] [Google Scholar]
- 9.Xu G, Guo Y, Xu D, Wang Y, Shen Y, Wang F. TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Sci Rep. 2017;7:42411. doi: 10.1038/srep42411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim HS, Kang SH, Park CH, Yang WI, Jeung HC, Chung HC. Genome-wide molecular characterization of mucinous colorectal adenocarcinoma using cDNA microarray analysis. Oncol Rep. 2011;25(3):717–727. doi: 10.3892/or.2010.1126. [DOI] [PubMed] [Google Scholar]
- 11.Chakraborty A, Diefenbacher ME, Mylona A, Kassel O, Behrens A. The E3 ubiquitin ligase Trim7 mediates c-Jun/AP-1 activation by Ras signalling. Nat Commun. 2015;6:6782. doi: 10.1038/ncomms7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu L, Qin C, Li T, Ma X, Qiu Y, Lin Y. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2019:1–13. doi: 10.1038/s41418-019-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Tang Z, Ma S, Yu Y, Chen X, Zang G. Tripartite motif-containing protein 7 regulates hepatocellular carcinoma cell proliferation via the DUSP6/p38 pathway. Biochem Biophys Res Commun. 2019;511(4):889–895. doi: 10.1016/j.bbrc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Aoe K, Amatya VJ, Fujimoto N, Ohnuma K, Hosono O, Hiraki A. CD26 overexpression is associated with prolonged survival and enhanced chemosensitivity in malignant pleural mesothelioma. Clin Cancer Res. 2012;18(5):1447–1456. doi: 10.1158/1078-0432.CCR-11-1990. [DOI] [PubMed] [Google Scholar]
- 15.Li G, Li L, Sun Q, Wu J, Ge W, Lu G. MicroRNA-3200-5p promotes osteosarcoma cell invasion via suppression of BRMS1. Mol Cells. 2018;41(6):523–531. doi: 10.14348/molcells.2018.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang HM, Qiao QD, Xie HF, Wei JX. Breast cancer metastasis suppressor 1 (BRMS1) suppresses prostate cancer progression by inducing apoptosis and regulating invasion. Eur Rev Med Pharmacol Sci. 2017;21(1):68–75. [PubMed] [Google Scholar]
- 17.Cao Y, Tan S, Tu Y, Zhang G, Liu Y, Li D. MicroRNA-125a-5p inhibits invasion and metastasis of gastric cancer cells by targeting BRMS1 expression. Oncol Lett. 2018;15(4):5119–5130. doi: 10.3892/ol.2018.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucciarelli PR, Tan KS, Chudgar NP, Brandt W, Montecalvo J, Eguchi T. BRMS1 expression in surgically resected lung adenocarcinoma predicts future metastases and is associated with a poor prognosis. J Thorac Oncol. 2018;13(1):73–84. doi: 10.1016/j.jtho.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Mayo MW, Xiao A, Hall EH, Amin EB, Kadota K. Loss of BRMS1 promotes a mesenchymal phenotype through NF-kappaB-dependent regulation of Twist1. Mol Cell Biol. 2015;35(1):303–317. doi: 10.1128/MCB.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo X, Li S, Shi T, Suo A, Ruan Z, Guo H. Cullin3 promotes breast cancer cells metastasis and epithelial-mesenchymal transition by targeting BRMS1 for degradation. Oncotarget. 2015;6(39):41959–41975. doi: 10.18632/oncotarget.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulzico D, Pires BRB, PAS DEF, Neto LV, Abdelhay E. Twist1 correlates with epithelial-mesenchymal transition markers fibronectin and vimentin in adrenocortical tumors. Anticancer Res. 2019;39(1):173–175. doi: 10.21873/anticanres.13094. [DOI] [PubMed] [Google Scholar]
- 22.Zheng S, Qiao G, Min D, Zhang Z, Lin F, Yang Q. Heterogeneous expression and biological function of ubiquitin carboxy-terminal hydrolase-L1 in osteosarcoma. Cancer Lett. 2015;359(1):36–46. doi: 10.1016/j.canlet.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Shen Y, Xu L, Ning Z, Liu L, Lin J, Chen H. ARHGAP4 regulates the cell migration and invasion of pancreatic cancer by the HDAC2/beta-catenin signaling pathway. Carcinogenesis. 2019 doi: 10.1093/carcin/bgz067. [DOI] [PubMed] [Google Scholar]
- 24.Tang S, Bai C, Yang P, Chen X. 14-3-3epsilon boosts bleomycin-induced DNA damage response by inhibiting the drug-resistant activity of MVP. J Proteome Res. 2013;12(6):2511–2524. doi: 10.1021/pr301085c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J, Liu Y, Xu X, Zhang W, Yu T, Jia J. Deubiquitinase USP47/UBP64E regulates beta-catenin ubiquitination and degradation and plays a positive role in Wnt signaling. Mol Cell Biol. 2015;35(19):3301–3311. doi: 10.1128/MCB.00373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Chen Y, Xiang F, Li M, Li H, Chi J. Suppression of TGF-beta1 enhances chemosensitivity of cisplatin-resistant lung cancer cells through the inhibition of drug-resistant proteins. Artif Cells Nanomed Biotechnol. 2018;46(7):1505–1512. doi: 10.1080/21691401.2017.1374285. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Guo Y, Yang H, Shi G, Xu G, Shi J. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget. 2015;6(27):23708–23719. doi: 10.18632/oncotarget.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montori-Grau M, Pedreira-Casahuga R, Boyer-Diaz Z, Lassot I, Garcia-Martinez C, Orozco A. GNIP1 E3 ubiquitin ligase is a novel player in regulating glycogen metabolism in skeletal muscle. Metabolism. 2018;83:177–187. doi: 10.1016/j.metabol.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 30.Li Q, Xiao L, Harihar S, Welch DR, Vargis E, Zhou A. In vitro biophysical, microspectroscopic and cytotoxic evaluation of metastatic and non-metastatic cancer cells in responses to anti-cancer drug. Anal Methods. 2015;7(24):10162–10169. doi: 10.1039/C5AY01810B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodenstine TM, Vaidya KS, Ismail A, Beck BH, Cook LM, Diers AR. Homotypic gap junctional communication associated with metastasis suppression increases with PKA activity and is unaffected by PI3K inhibition. Cancer Res. 2010;70(23):10002–10011. doi: 10.1158/0008-5472.CAN-10-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Deng L, Zhao X, Li B, Ren D, Yu L, et al. Tripartite motif-containing 37 (TRIM37) promotes the aggressiveness of non-small-cell lung cancer cells by activating the NF-kappaB pathway. 2018;246(3):366-78. [DOI] [PubMed]
- 33.Zhang Y, Tao R, Wu SS, Xu CC, Wang JL, Chen J. TRIM52 up-regulation in hepatocellular carcinoma cells promotes proliferation, migration and invasion through the ubiquitination of PPM1A. J Exp Clin Cancer Res. 2018;37(1):116. doi: 10.1186/s13046-018-0780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou Z, Zeng F, Xu W, Wang C, Ke Z, Wang QJ. PKD2 and PKD3 promote prostate cancer cell invasion by modulating NF-kappaB- and HDAC1-mediated expression and activation of uPA. J Cell Sci. 2012;125(Pt 20):4800–4811. doi: 10.1242/jcs.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu R, Shi H, Wang S, Leng S, Liu R, Zheng Y. BRMS1 coordinates with LSD1 and suppresses breast cancer cell metastasis. Am J Cancer Res. 2018;8(10):2030–2045. [PMC free article] [PubMed] [Google Scholar]
- 36.Niinaka Y, Harada K, Fujimuro M, Oda M, Haga A, Hosoki M. Silencing of autocrine motility factor induces mesenchymal-to-epithelial transition and suppression of osteosarcoma pulmonary metastasis. Cancer Res. 2010;70(22):9483–9493. doi: 10.1158/0008-5472.CAN-09-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vaidya KS, Sanchez JJ, Kim EL, Welch DR. Expression of the Breast Cancer Metastasis Suppressor 1 (BRMS1) maintains in vitro chemosensitivity of breast cancer cells. Cancer Lett. 2009;281(1):100–107. doi: 10.1016/j.canlet.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer D, Schon M, Bayerlova M, Bleckmann A, Schon MP, Zornig M. A pro-apoptotic function of iASPP by stabilizing p300 and CBP through inhibition of BRMS1 E3 ubiquitin ligase activity. Cell Death Dis. 2015;6:e1634. doi: 10.1038/cddis.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rada M, Vasileva E, Lezina L, Marouco D, Antonov AV, Macip S. Human EHMT2/G9a activates p53 through methylation-independent mechanism. Oncogene. 2017;36(7):922–932. doi: 10.1038/onc.2016.258. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Ling MT, Guan XY, Tsao SW, Cheung HW, Lee DT. Identification of a novel function of TWIST, a bHLH protein, in the development of acquired taxol resistance in human cancer cells. Oncogene. 2004;23(2):474–482. doi: 10.1038/sj.onc.1207128. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, Huang Z, Wu S, Zang X, Liu M, Shi J. miR-33a is up-regulated in chemoresistant osteosarcoma and promotes osteosarcoma cell resistance to cisplatin by down-regulating TWIST. J Exp Clin Cancer Res. 2014;33:12. doi: 10.1186/1756-9966-33-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Yu H, Shen M, Wei W, Xia L, Zhao P. N1-guanyl-1,7-diaminoheptane sensitizes bladder cancer cells to doxorubicin by preventing epithelial–mesenchymal transition through inhibition of eukaryotic translation initiation factor 5A2 activation. Cancer Sci. 2014;105(2):219–227. doi: 10.1111/cas.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang B. miRNA-200c increases the sensitivity of breast cancer cells to doxorubicin through the suppression of E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 2013;7(5):1579–1584. doi: 10.3892/mmr.2013.1403. [DOI] [PubMed] [Google Scholar]
- 44.Tezcan O, Gunduz U. Vimentin silencing effect on invasive and migration characteristics of doxorubicin resistant MCF-7 cells. Biomed Pharmacother = Biomed Pharmacother. 2014;68(3):357–364. doi: 10.1016/j.biopha.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Lehtinen L, Ketola K, Makela R, Mpindi JP, Viitala M, Kallioniemi O. High-throughput RNAi screening for novel modulators of vimentin expression identifies MTHFD2 as a regulator of breast cancer cell migration and invasion. Oncotarget. 2013;4(1):48–63. doi: 10.18632/oncotarget.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Bai R, Li M, Ye H, Wu C. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. 2019;10(1):1858. [DOI] [PMC free article] [PubMed]
- 47.Miao W, Chen J, Jia L, Ma J, Song D. The m6A methyltransferase METTL3 promotes osteosarcoma progression by regulating the m6A level of LEF1. Biochem Biophys Res Commun. 2019;516(3):719–725. doi: 10.1016/j.bbrc.2019.06.128. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Zeng L, Liang C, Zan R, Ji W, Zhang Z. Integrated analysis of transcriptome-wide m(6)A methylome of osteosarcoma stem cells enriched by chemotherapy. Epigenomics. 2019 doi: 10.2217/epi-2019-0262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Proteomics data have been deposited in the PeptideAtlas (http://www.peptideatlas.org/) under accession number PASS01605. All relevant data supporting the key findings are available from the corresponding author upon reasonable request.