Abstract
A fine-tuned activation and deactivation of proteases and their inhibitors are involved in the execution of the inflammatory response. The zymogen/proenzyme plasminogen is converted to the serine protease plasmin, a key fibrinolytic factor by plasminogen activators including tissue-type plasminogen activator (tPA). Plasmin is part of an intricate protease network controlling proteins of initial hemostasis/coagulation, fibrinolytic and complement system. Activation of these protease cascades is required to mount a proper inflammatory response. Although best known for its ability to dissolve clots and cleave fibrin, recent studies point to the importance of fibrin-independent functions of plasmin during acute inflammation and inflammation resolution. In this review, we provide an up-to-date overview of the current knowledge of the enzymatic and cytokine-like effects of tPA and describe the role of tPA and plasminogen receptors in the regulation of the inflammatory response with emphasis on the cytokine storm syndrome such as observed during coronavirus disease 2019 or macrophage activation syndrome. We discuss tPA as a modulator of Toll like receptor signaling, plasmin as an activator of NFkB signaling, and summarize recent studies on the role of plasminogen receptors as controllers of the macrophage conversion into the M2 type and as mediators of efferocytosis during inflammation resolution.
Keywords: Plasminogen, Plasmin, Plasminogen receptor, Matrix metalloproteinase, LRP1, Toll like recepotor, Cytokine storm syndrome, Cytokine, tPA, COVID-19, DIC, Coagulation, Complement, Macrophage activation syndrome, NFkB, PAR
Graphical abstract
1. Introduction
Tissue damage with vascular leakage occurs after inflammation [1] and requires a fine-tuned activation and deactivation of proteases and their inhibitors released from recruited leukocytes and tissue-resident endothelial cells or fibroblasts to initiate hemostasis executed by the activation of the coagulation system, followed by the activation of the fibrinolytic system driven by plasmin, and the concomitant complement activation. Coagulation factors enhance the formation of blood clots (thrombi) that stop hemorrhage (hemostasis) due to damaged vessels. As blood flow needs to resume for proper tissue repair, thrombi are dissolved, a task taken on by the serine protease plasmin, the key serine protease of the fibrinolytic pathway.
Endothelial damage leading to excessive thrombin generation or proinflammatory cytokines, including tumor necrosis factor-alpha (TNFa) or interleukins (ILs), like IL-6 [2] enhance the release of tissue-type plasminogen activator (tPA) from storage granules in endothelial cells. tPA and other plasminogen activators enhance the conversion from the proenzyme/zymogen plasminogen into the active enzyme plasmin.
Aside from clot dissolution, recent studies demonstrate plasmin or tPA can modify the inflammatory response on various levels: They can activate proteases like e.g. matrix metalloproteinases (MMPs) thereby modifying the extracellular matrix composition [3], promote macrophage and dendritic cell (DC) migration, function as a cytokine, and control nuclear factor kappaB (NFkB) activation due to their ability to engage with cellular receptors conveying either pro- or anti-inflammatory cellular responses including the response to Toll-like receptor (TLRs) activation.
While tPA binding to some of these receptors can enhance Plg activation (proteolytic activity), growing evidence suggests that tPA can directly modulate cytokine signaling pathways through a non-enzymatic mechanism. Finally, plasmin(ogen) and its cellular receptors contribute to the resolution of inflammation.
Depending on the experimental model, the stage of inflammatory disease, and the receptors involved, plasmin(ogen) and tPA orchestrate processes that protect against or are in favor of exacerbation of inflammatory processes, underscoring the complexity of the network induced by fibrinolytic factors at the molecular level.
2. The intricate network of initial coagulation, fibrinolysis and complement activates the immune system
Vascular damage activates the coagulation system that generates fibrin and supports the formation of a thrombus. The thrombus establishes a physical barrier for pathogens or non-self-pathogens (part of the innate immune system) and stopping bleeding (hemostasis). Inflammation shifts the hemostatic balance towards a prothrombotic and antifibrinolytic state that causes – if not properly shut down – causes the devastating consumptive coagulopathy and disseminated coagulation [4].
Tissue damage can activate the complement cascade during the inflammatory response, another cascade of enzymes of the innate immune system that enhances (complements) the ability of antibodies and phagocytic cells to clear microbes and damaged cells, induce inflammation to attract more macrophages, and activates the cell-killing membrane attack complex (MAC) to destroy the pathogen's cell membrane (Fig. 1 ).
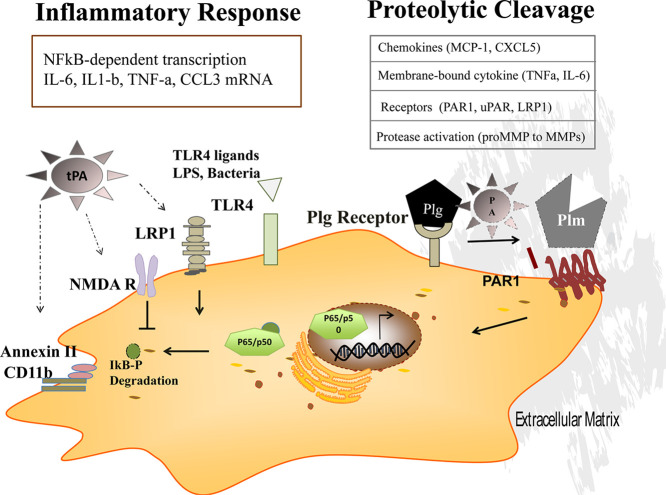
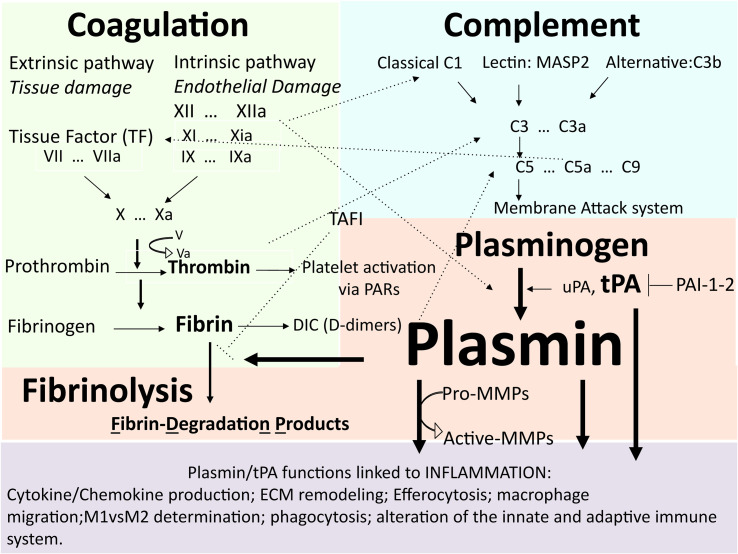
Fig. 1.
The inflammatory response: when the fibrinolytic system, coagulation, and the complement system talk to each other.
Factors of the coagulation, complement, and fibrinolytic system interact. The imbalance of these proteins can be the cause or the reason for thrombosis and inflammation. During inflammation, tissue damage inflicted by microbes or non-microbial stressors activates the coagulation system. Thrombin generates fibrin and enhances platelet activation and their growth factor release through protease activated receptors (PARs). Tissue-type plasminogen activator (tPA) and urokinase plasminogen activator (uPA) activate the inactive proenzyme/zymogen plasminogen (Plg) into active plasmin (Plm). Plasminogen activators are inhibited by plasminogen activator inhibitor-1, −2 (PAI-1, PAI-2), and protein C inhibitor (PCI). Plm functions include its ability to activate complement C3a and C5a, degrade fibrin into fibrinogen degradation products leading to fibrin fragments like the D-dimers, convert the proenzymes of matrix metalloproteinases (MMPs) to their active forms, regulate extracellular matrix (ECM) turnover (remodeling), and contribute to inflammation resolution (efferocytosis, M1 to M2 switch).
Coagulation Cascade. While the intrinsic, contact coagulation pathway is mainly activated through exposed endothelial collagen and XII, the extrinsic pathway is activated through the release of the transmembrane receptor tissue factor (TF) from endothelial cells and its plasma cofactor Factor VII/VIIa (Fig. 1). Extrinsic and intrinsic pathways activate Factor X, which renders the zymogen prothrombin into thrombin. Factor XII (FXII, also known as Hageman factor) is converted to its active enzyme (FXIIa) by plasma kallikrein (PKa) or by its unique ability to auto-activate following binding to artificial or biologic surfaces [5]. In disease states of infection, the surface of bacterial pathogens or polyphosphates released by them promotes the autoactivation of FXII [5]. FXIIa activation of PK forms plasma kallikrein that reciprocally activates FXII and liberates bradykinin (BK) from high molecular weight kininogen (HK). Bradykinin enhances vasodilation and increases capillary permeability, leading to edema and changes in arterial blood pressure. FXII deficiency is linked to decreased infiltration of inflammatory cells into skin windows [6]. FXII facilitates the recruitment of neutrophils, binds to the surface of pathogens (bacteria, fungi, viruses, and neutrophil extracellular traps (NETs) where it autoactivates [7]. FXII promotes neutrophil degranulation [8].
Factor XIIa also activates FXI and C1 esterases (C1r, C1s), the first components of the macromolecular complex of C1, and the classic complement cascade (Fig. 1) [9].
The complement cascade is part of the innate immune system. Complement factors (C1–9) are small proteins that enhance (complement) the ability of antibodies and macrophages to clear microbes following proteolytic activation (C1a-C9a) (Fig. 1): Complement enhances (complements) the antibody's ability and macrophages to clear microbes. Complement factors enhance macrophage and neutrophil chemoattraction and establish a membrane attack complex (MAC, C5b-C9) necessary for the rupture of the cell wall. Thrombin and plasmin enhance the formation of certain complement proteins (namely C3 and C5). Activated complement fragments C3a/C5a can recruit leukocytes and induced the MAC complex on macrophages [10]. One inhibitor of the classical C1 pathway of the complement is C1 inhibitor. Mutations in the C1 inhibitor gene can cause hereditary angioedema due to a reduced regulation of bradykinin by C1 inhibitor. HIV infection can alter the complement system causing further tissue damage [11].
Thrombin converts fibrinogen into fibrin. Factor XIII, another thrombin-generated enzyme, then crosslinks the fibrin protofibrils leading to an insoluble gel that serves as a scaffold for a blood clot (thrombus). Endothelial cells that cover the inner vessel wall modulate the balance between coagulation and fibrinolysis by counteracting coagulation through binding of anti-thrombin III, releasing tissue factor pathway inhibitor, expressing thrombomodulin, activating protein C, and producing and secreting tPA [12,13]. Protein C - produced in the liver in a Vitamin K-dependent manner - after binding to the endothelial cell protein C receptor in the presence of thrombomodulin and thrombin leads to the generation of activated protein C (APC) [14]. APC irreversibly inactivates Factor Va (Leiden) and VIIIa and thereby counteracts thrombin activity.
Thrombin activates the G-protein coupled receptors called protease activated receptors (PARs). PARs (PAR1 and PAR4) are found on platelets. The activation of PARs by thrombin and adenosine diphosphate, and collagen activates platelets, with changes in shape and mobility, and the releases chemokines and cytokines stored in alpha platelet granules (including CCL3 (MIP-1alpha), CCL5 (RANTES), CCL7 (monocyte chemoattractant protein3 (MCP-3), CCL17, CXCL1 (growth-regulated oncogene-alpha), CXCL5 (ENA-78), and CXCL8 (IL-8)). These factors can attract leukocytes and further activate other platelets. PAR activation on endothelial cells, smooth muscle cells, fibroblasts, epithelial cells, and mononuclear cells, -stimulates the production of cytokines and chemokines like interleukin1,6,8, granulocyte-macrophage colony-stimulating factor (GM-CSF), and MCP1 [[15], [16], [17]].
3. The physiological function of plasminogen and its associated factors
The fibrinolytic cascade, with plasmin as its central player, is required for fibrin degradation and blood clot/thrombus dissolution to regain vascular patency after initial hemostasis is achieved [18,19].
Vascular endothelial cells keep tPA and Plg on their surfaces. tPA (encoded by PLAT), urokinase plasminogen activator (uPA, encoded by PLAU), or other enzymes, e.g., plasma kallikrein (PKa) and coagulation factor XIIa convert the inactive zymogen Plg into the active enzyme plasmin [20]. Plg is produced by the liver and circulates in the blood. Plg is a seven-domain protein comprising an N-terminus PAN-apple domain, five tandem kringle domains, and a C-terminus catalytic domain [21]. The PAN/apple domain is proteolytically removed, and the kringle domains are responsible for fibrin binding. The PAN/apple domain-free plasmin can be further degraded by phosphoglucerate kinase to remove the catalytic domain. The remaining five kringle domains are referred to as angiostatin, an endogenous angiogenesis inhibitor [22].
Under steady-state conditions, plasmin activity is low as there is no need for fibrin removal. When fibrin had been generated, Plg and tPA bind to fibrin whit the cleavage of Plg (at the Arg561-Val562 peptide bond) into the two-chain enzyme plasmin. Plasmin is composed of an N-terminal heavy chain (12–65 kDa) and a C-terminal light chain (25 kDa) that contains the proteolytic active site [13]. Thrombin-activatable fibrinolysis inhibitor (TAFI) prevents Plg binding to fibrin. Soluble, but not fibrin-bound plasmin can be inactivated by its main inhibitor alpha2-antiplasmin [23], and by alpha2-anti-macroblogulin.
Early studies demonstrated that Lysine-Plg attaches with higher affinity to macrophages found in chronic inflammatory lesions than the native Glu-Plg [24]. Plg on cell surfaces enables a concentrated mode of fibrinolytic activity on the cell surface. Plg binds via Plg receptors to the plasma membrane. Plg receptors are typically proteins exposing a carboxy-terminal lysine. Carboxypeptidase B removes Carboxy-terminal lysines from proteins and therefore is used to identify novel Plg binding receptors [25]. The lysine binding sites within the kringle domains of Plg bind to the carboxy-terminal lysine of Plg receptors (reviewed [26,27]). Plg receptors include the heterotetrameric complex Annexin A2-S100A10, alpha-enolase-1, histone H2B, and the plasminogen receptor With A C-Terminal Lysine (Plg-RKT) [28], TBP-interacting protein 49a [29] or the high mobility group box-1 protein (HMGB-1) [30]. In chapter 8, we will discuss recent studies on the role of Plg-RKT for macrophage migration, efferocytosis, and macrophage subtype determination [31] (see chapter 8).
tPA is synthesized as a single-chain molecule with low plasmin binding affinity [32] ensuring that in the absence of fibrin the fibrinolytic activity is low. Plasmin cleaves tPA into its two-chain form, a form that has a 10-fold higher affinity for converting Plg into plasmin than the single-chain molecule [32]. The two chain tPA has a heavy A-chain and the light B-chain. The heavy A-chain is composed of the fibronectin finger domain, an epidermal growth factor analog domain, and Kringle 1 and 2 domains [13]. The finger domain of tPA is responsible for the binding of fibrin, Annexin II, and low-density lipoprotein receptor-related protein-1 (LRP1) [33,34]. The kringle 1 domain of tPA is involved in the uptake of tPA in the liver [35], while the Kringle 2 domain is important for interactions with N-methyl-D-aspartic acid receptor (NMDA-R) and platelet-derived growth factors (PDGFs) [36]. tPA also associates with the annexinA2S100A10 complex, Plg-RKT, and mannose receptors [[37], [38], [39], [40], [41]]. Natural inhibitors of tPA's fibrinolytic activity are plasminogen activator inhibitor-1 (PAI-1), a circulating serpin (serine protease inhibitor), and PAI-2 [32,42].
The specific binding of Plg and tPA to lytic edges of partly degraded fibrin via newly generated C-terminal lysine residues amplifies fibrin digestion generating diffusible and soluble fibrin fragments. Such fibrin degradation products (FDPs) include fragments D and E and D-dimers. D-dimers are two D fragments joined by a cross-link [43] often used as a measure for the presence of systemic inflammation in clinical settings.
Gene deficient mice for Plg show fibrin deposition in various tissues leading to high mortality, wasting, spontaneous gastrointestinal ulceration, rectal prolapse, severe thrombosis, and delayed wound healing [44]. Dependent on the disease model tested, mouse mutants with Plg deficiency can suppress or exacerbate inflammatory diseases. Plg deficiency exacerbates collagen-induced arthritis [45], and peripheral nerve injury [46]. The phenotypic abnormalities of Plg deficiency can be rescued in mice genetically deficient in both Plg and fibrinogen [47] or following pharmacological fibrin depletion in Plg knockout mice [45,46]. On the other hand, Plg deficiency (pharmacological or genetic) improves the outcome of inflammatory diseases in murine models of inflammatory bowel disease [48] or diseases where immune cells release vast amounts of pro-inflammatory factors resulting in a cytokine storm syndrome, like during Graft versus Host Disease (GvHD) and macrophage activation syndrome (MAS) [49,50]. The specific role of plasmin during the cytokine storm syndrome will be further discussed in chapter 7. These studies suggested that a major function of Plg is fibrinolysis.
Aside from the established role of fibrinolytic factors for the recruitment of macrophages, fibrinolytic factors also can change the cellular, immunomodulatory environment through the expansion of mesenchymal stem cell (MSC) expansion. MSCs orchestrate local and systemic innate and adaptive immune responses through the release of immunosuppressive molecules, growth factors, exosomes, chemokines, and complement components. We showed that MSCs express fibrinolytic factors (reviewed in [51]), and that tPA administration promotes MSC recruitment and expansion [51,52]. Mechanistically, the tPA-mediated release of kit ligand from endothelial cells resulted in the expansion of MSCs due to the release of basic fibroblast growth factor and PDGF-BB from c-kit+ MSCs [53].
4. Inflammation
Tissue damage initiates an inflammatory response. Adhesion molecule upregulation on leukocytes and endothelial cells enhances the influx of polymorph mononuclear cells (nonspecific inflammation) or eosinophils in case of allergic inflammatory reactions from the circulation. Recruited inflammatory cells, including macrophages, (DCs), lymphocytes, endothelial cells, fibroblasts, and mast cells locally release coagulation factors, fibrinolytic factors, complement, chemo−/cytokines, free radicals, vasoactive amines, and eicosanoids. Released chemo−/cytokines ensure the recruitment of more inflammatory cells to fight inflammatory intruders.
Damage-associated molecular patterns (DAMPs) are released from invading microorganisms and/or tissue damage-inducing agents resulting in the overexpression of pattern-recognition receptors (PRRs) on macrophages, monocytes, DCs, neutrophils, and epithelial cells. Microbe-specific molecules that can be sensed by PRRs include bacterial carbohydrates (such as lipopolysaccharide (LPS), mannose), bacterial or viral DNA or RNA, bacterial peptides, lipoproteins, and chitin. TLRs are typical membrane-bound PRRs that trigger the synthesis and secretion of cytokines following ligand binding thereby activating host defense programs that are necessary for both innate and adaptive immune responses. Following the acute inflammation phase, macrophages or dendritic cells help to clear the tissue debris starting the resolution phase [54].
5. The endocytosis receptor LRP1 controls the inflammatory response
The endocytosis receptor low-density lipoprotein LRP-1 (CD91) clears proteins like plasmin, free tPA or tPA in complex with PAI-1, a2-macroglobulin, MMP9, tissue inhibitors of metalloproteinases (TIMPs), lipoproteins, ECM proteins, complement factor-like C1r and C1s, and growth factors [[55], [56], [57], [58]] [59]. LRP1-binding antagonist receptor-associated protein (RAP) prevents LRP1-mediated endocytosis [60] by completely blocking interactions with all known LRP1 ligands. By controlling the uptake of proteases, LRP1-mediated endocytosis controls the amount of extracellular proteases, the “proteolytic niche”.
The mature type I transmembrane protein LRP1 consists of an a- and a b-chain. The structure of LRP1 includes cysteine-rich complement-type repeats, EGF repeats, b-propeller domains, and transmembrane and cytoplasmic domains. Proteolytic cleavage of the ectodomain of LRP1 by a disintegrin and metalloproteinase 10 (ADAM10), ADAM17, membrane type1-MMP, and tPA releases free LRP1 that can serve as a decoy receptor trapping ligands (like plasmin, tPA, and MMPs) in the extracellular space [61].
LRP1 controls cellular cytokine signaling. Earlier studies demonstrated that LRP1 suppresses TNF receptor (TNFR1) signaling [62]. LRP1 deficient cells showed increased proinflammatory factor expression like MCP1/CCL2), IL-6, and inducible nitric oxide synthase (iNOS). The observed anti-inflammatory LRP1 phenotype of membrane-anchored LRP1 was attributed to its ability to decrease TNF receptor1 (TNFR1) expression, resulting in impaired NFkB activity. NFkB nuclear translocation induces gene transcription of pro-inflammatory factors, including TNFa, IL-1, IL-6, IL-10, or type 1interferons (IFNs) (reviewed in [1]).
Furthermore, the extracellular amount of soluble TNFa, the ligand of TNFR1, can be regulated by LRP1. Membrane TNFa is shed by ADAM17 into its soluble form. LRP1 by controlling the activity of ADAM17 through endocytotic uptake of its natural inhibitor TIMP3, therefore, controls TNF release. Once LRP1 is shed by other proteases during LPS-mediated TLR4 activation endocytosis of TIMP3 ceases and TIMP3 accumulates on the cell surface where it inhibits ADAM17 resulting in the accumulation of membrane TNFa. As a consequence, TIMP accumulates on the cell surface, blocking ADAM17 activity and TNFa shedding [63].
The onset of the ischemic insult in a murine model of middle cerebral artery occlusion induces NFkB activation in astrocytes. Early studies showed that NFkB activation required tPA and LRP1 and was independent of Plg as shown using Plg deficient mice [64]. Brifault et al. demonstrated that RAP and LPS induced shedding of the 85-kDa b-chain of LRP1 on microglial cells (resident macrophages of the central nervous system) in an MMP dependent manner generating soluble LRP1 consisting of the intact 515-kD a-chain attached with a truncated small ectodomain of the b-chain. The sLRP1 induces the release of pro-inflammatory cytokines like TNFa, IL-6, IL1-b, and iNOS in microglial cells, and stereotactic injection into the brain resulted in neuroinflammation [65]. These studies show that soluble or membrane-type LRP1 can influence the inflammatory response.
6. tPA receptors modulates TLR-mediated cytokine signaling
Earlier studies established a link between clotting/fibrinolysis related-proteases and TLR signaling. One study reported that antithrombin III inhibited the response to LPS in the monocytic cell line THP-1 [66]. C1 inhibition was shown to prevent LPS shock [67], and fibrinogen was shown to activate TLR4 [68]. Ward et al. reported that plasmin exerted proinflammatory functions in monocytes, and potentiated TLR2 and TLR4 signaling [69]. These studies indicated that coagulation factors modulate the inflammatory TLR response.
TLR activation enhances the nuclear translocation of transcription factors, like activator protein-1, NFkB, or interferon regulatory factor 3 (IFR3). Of interest, the LRP1 β chain is processed by the presenilin-dependent γ-secretase to generate a small fragment-LRP1 C-terminal intracellular domain, which promotes the nuclear export of inflammatory transcription factor IFR3 blocking the transcription of inflammatory genes [70].
To further understand how fibrinolytic factors can alter TLR signaling, it is important to note that LRP1 is linked to uPA receptor urokinase-type plasminogen activator receptor (uPAR) or N-Methyl-d-Aspartate receptor (NMDA-R) via the adapter protein postsynaptic density protein 95. Due to this linkage, LRP1 can internalize these receptors after ligand binding [71], and control the presence and signaling of these cellular receptors [37].
Earlier studies demonstrated that a2-macroglobulin decreases calcium responses to the glutamate receptor NMDA-R by the expression of NMDA-R1 in rat hippocampal neurons [72]. Later, it was shown that enzymatically inactive tPA (El-tPA) and a2-macroglobulin treatment caused the formation of a complex between LRP1, PSD-95, and NMDA-Rs that activates tyrosine receptor kinase (Trk) receptors and stimulates ERK1/2 activity in neurons [73]. ERK1/2 activation can enhance MMP9 expression [52,56,58]. These studies indicated El-tPA modifies cellular signaling via LRP1.
Recent studies elegantly showed that tPA-mediated LRP1 signaling modulates the inflammatory response after TLR stimulation (Fig. 2 ) due to the unique function of LRP1 to link to other receptors like NMDA-R. Mantuano et al. demonstrated that the pro-inflammatory cytokine profile of LRP1 deficient macrophages is reversed in TLR4 activated macrophages [71]. In the presence of LPS-stimulated TLR4, EI-tPA that cannot promote plasmin generation and a2-macroglobulin suppresses NFkB signaling and TNFa, IL-6, CCL2/3 production by binding to the NFkB suppressing NMDA-R [61,74].
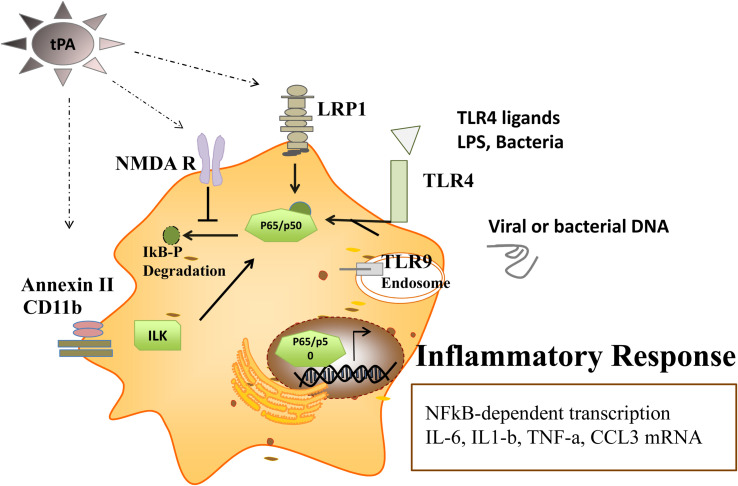
Fig. 2.
Receptor-mediated tPA signaling modulates the TLR-induced inflammatory response. TLR stimulation by bacterial LPS (TLR4) or viral or bacterial DNA/RNA (TLR9) induces transcription factor NFkB signaling in macrophages. NFkB translocation into the nucleus promotes the transcription of inflammatory response genes like interleukin-6, IL-1b, TNFa, chemokines like IP-10, MCP1 (CCL2), and MIP1 (CCL3). tPA can modify the TLR-mediated inflammatory response in a manner dependent on its interaction with tPA-associated receptors like low-density lipoprotein receptor-related protein-1 (LRP1) or the N-methyl-D-aspartic acid receptor (NMDA R), or AnnexinII/11b complex.
Similarly, EI-tPA and activated α2-macroglobulin binding to NMDA-R blocks ligand-induced TLR2 and TLR9 and NFkB signaling, but not that of PPRs like NOD1 or NOD2 [75].
Endothelial cells, fibroblasts, osteoblasts, smooth muscle, and macrophages produce the macrophage-colony stimulating factor (M-CSF; also known as CSF1). M-CSF signals through the M-CSF receptor (reviewed by Hamilton [76]), and is required for macrophage survival, proliferation, monocytic cell differentiation, and myeloid cell mobilization into circulation. M-CSF enhances NMDA-R expression in macrophages [75]. This study demonstrated EI-tPA engagement with NMDA-R on macrophages keeps cells in a resting, non-inflammatory stage, and that M-CSF could alter the response to fibrinolytic factors.
While TLR-mediated effects of tPA on NFkB signaling did not require plasmin, active plasmin can mount a pro-inflammatory response in macrophages or astrocytes through protease-activated receptor-1 (PAR1) (Fig. 3 ). PAR1 is a key thrombin receptor on platelets and mediates thrombin-induced platelet aggregation. Plg receptors like uPAR or a-enolase present Plg to extracellular tPA, or uPA or intracellular uPA causing the conversion of Plg into active plasmin. In turn, plasmin similar to thrombin, MMP1 [77], MMP13 [78], Cathepsin G, and neutrophil elastase [79] can cleave the N-terminal ectodomain part of PAR-1 and activate intracellular signaling pathways [80] like the NFkB pathway [81]. Plasmin-mediated PAR-1 activation induces IL-8 expression in human dental pulp fibroblast-like cells [82] and activates NFkB, ERK1/2, and p38 mitogen-activated protein kinase in bone marrow-derived macrophages [61].
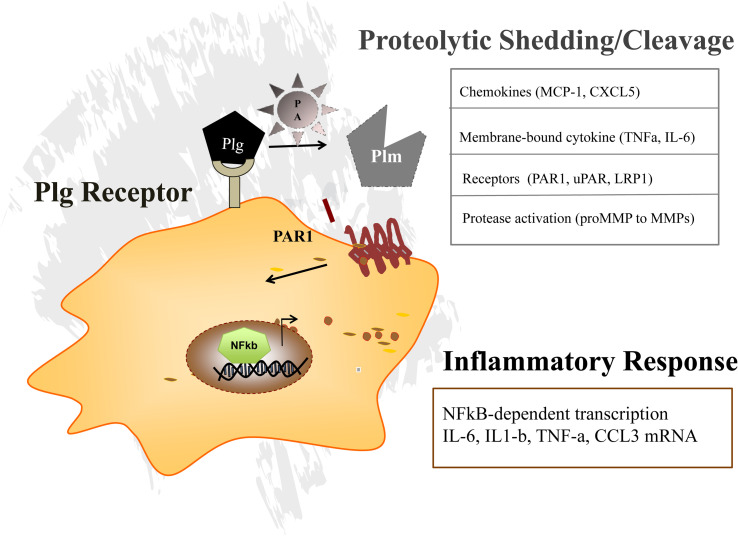
Fig. 3.
Plasminogen receptors localize plasmin generation to the pericellular space, thereby promoting signaling receptor shedding (e.g. PAR1). Plg receptors present Plg and enhance plasminogen activator (PA) mediated pericellular plasmin (Plm) generation. The proteolytic activity of Plm is required to activate PAR1 signaling or for shedding or cleavage of other proteases (MMPs) as well as in signaling by other molecules like cytokines or cellular receptors.
Our group demonstrated that plasmin treatment of TLR4 or TLR9 ligand-stimulated macrophages showed enhanced NFkB activation [49,50] supporting the notion of plasmin as a proinflammatory mediator. We showed that plasmin inhibition prevents the progression of inflammatory bowel disease in murine models in part through MMP9 activation [48].
Given the proinflammatory role of plasmin, studies evaluated the effect of plasmin inhibition to prevent immunosuppression after traumatic brain injury, ischemic stroke, and cardiac surgery. In a model of brain injury, treatment with tranexamic acid (TXA), the lysine analog that blocks the binding of Plg and the ultimate generation of plasmin on fibrin, increased migration and proliferation of conventional dendritic cells (cDCs), antigen-presenting cells and T cells in the draining cervical lymph nodes [83].
Acute ischemic stroke similarly can trigger immunosuppression. Using a mouse model of middle cerebral artery occlusion tPA administration worsened the already due to inflammation established immunosuppressive state by reducing lymphocyte and monocyte counts in circulation, enhancing plasma IL-10 and TNFa levels and decreasing DC subtypes [84].
Extensive surgery is fraught with a high risk of infection, depicting a state of immunosuppression. In patients undergoing cardiac surgery, the administration of TXA reduced surgery-induced infection rates and surgery-associated immunosuppression [85]. TXA treatment in patients after surgery and in healthy volunteers impaired the number of CD4+ and CD8 memory cells, and CCR7+ regulatory T cells, but increased CCR7 expression on NK cells, attenuated TNFa, IL6 and IL-1b upregulation after surgery. Finally, TXA increased the monocyte/dendritic activation marker CD83 but reduced the maturation marker HLA-DR. In summary, plasmin inhibition can mitigate immunosuppression after certain ischemic events including surgery.
7. Role of plasmin during the cytokine storm or cytokine release syndrome
Infections cause tissue damage not only because of the virulence of the germ or toxin but also because of the immunological host response to the infectious stimulant. Activated macrophages, T cells and endothelial cells can mount such a host response and cytokines are key elements of the derailed immunological host response. The clinical syndrome is therefore called the cytokine storm or the cytokine release syndrome. Cytokines itself can cause typical clinical symptoms: Fever was attributed to TNFa and IL-6 rises. IL-2 can induce capillary leakage syndrome with hypotension and kidney failure. Forward amplification by these cytokines occurs. Released proinflammatory cytokines further activate newly recruited or resident macrophages, DCs, or endothelial cells to secrete more pro-inflammatory cytokines like IL-1 [86,87].
A common feature of severe cytokine storm is endothelial cell death. Vascular damage requires the coagulation system for tissue repair. But over-activation of the coagulation system with the generation of massive thrombi can establish DIC leading to the depletion of platelets (thrombopenia) and coagulation factors. Hemostatic imbalance also creates lung and kidney damage due to complement-mediated injury.
In 1993, Ferrara's group first introduced the term cytokine storm to describe the derailed immunological host response during graft-versus-host disease (GvHD), a condition occurring after an allogeneic transplant [88]. In GvHD, the donated bone marrow or peripheral blood stem cells view the recipient's body as foreign, and the donated cells attack the recipient's body. Our group observed increased circulating plasmin levels in mice and men with acute GvHD [49]. Pharmacological plasmin inhibition protected against acute GVHD-associated lethality in mice by reducing the infiltration of inflammatory cells in part due to enhanced MCP1 signaling and augmented the release of membrane-associated pro-inflammatory cytokines including TNFa and Fas-ligand partially due to the activation of MMPs.
Other diseases can cause CRS ranging from rheumatological disorders [89], immune-related chimeric antigen receptors (CARs)-T cell therapy in a subset of patients with B cell malignancies, respiratory viral infections caused by influenza or coronavirus disease-2019 (COVID-19) [90,91]. Patients infected by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can develop a cytokine storm syndrome. Aside from a massive cytokine elevation of typical proinflammatory cytokines like TNFa, IL-1, IL-6, and GM-CSF, these patients show typical derailment of the coagulation system. The cytokine storm data found in COVID-19 patients seem to indicate increased levels of thrombi, and damage to the vascular endothelium, the involvement of cytokines and immuno-thrombosis [92,93]. Pathology analysis of COVID-19 patients' lungs shows widespread thrombosis with microangiopathy. It is yet to be determined whether plasmin in COVID-19 is a friend or foe as recently summarized in a review by Medcalf et al. [94].
Cytokine storm syndrome resembles hemophagocytic lymphohistiocytosis or macrophage activation syndrome (MAS). Typical clinical symptoms of MAS patients are: fever, organomegaly, multi-organ failure, hyperferritinemia, pancytopenia, increases in triglycerides, fibrinogen, ferritin, serum aspartate aminotransferase, and hemophagocytosis (the uptake of erythrocytes into macrophages) in bone marrow aspirates [91]. Respiratory symptoms range from mild cases with cough and tachypnea to acute respiratory distress syndrome with dyspnea, hypoxemia, and bilateral opacities on chest X-ray. Organ failure includes the kidney or heart, and accompanying hemodynamic instability, capillary leak, and consumptive coagulopathy are also often reported [95]. MAS can occur following severe viral inflammation. Viral RNA/DNA can activate TLR9. We established a murine MAS model by repeatedly administering the TLR9 activating dinucleotide CpG combined with D-galactosamine, a substance triggering hepatocyte apoptosis and TNFa activation, to test the potential involvement of plasmin in the pathogenesis of MAS [50]. Plasmin levels were increased during MAS progression in mice. We found that plasmin inhibition prevented tissue destruction and lethality in MAS mice while suppressing the cytokine peak [50]. Plasminogen activator administration aggravated the disease. In vitro, the addition of plasmin and TLR9 ligands activated NFkB signaling in macrophages with enhanced production of transmembrane TNFa, CCL2, IL-1, IL-6, and Fas ligand. This in turn augmented uPA [96], and PAI-1 expression [97]. Most importantly, our study indicated that plasmin inhibition blocked the vicious cycle of cytokine storm syndrome and hyperfibrinolysis in tandem with coagulation in the MAS model [50].
8. Role of Plg/plasmin during inflammation resolution
During inflammation resolution, apoptotic cells need to be cleaned up in a process known as efferocytosis. Apoptotic cells activate serum-derived Plg. The generated plasmin promotes apoptotic cell clearance [98,99]. Das et al. showed that Plg deficient cells showed impaired phagocytic activity [99].
Pleurisy is the inflammation of the lung covering membranes found as a complication of viral infections (Coxsackie B virus, adenovirus, influenza, and COVID-19). Sugimoto et al. studied the effects of plasmin in a pleurisy model [100] and found that plasmin enhances neutrophil apoptosis and increases the efferocytosis capacity of macrophages. Mechanistically, plasmin-mediated upregulation of annexin A1 (AnxA1) in macrophages was required to bridge phagocytes with the phosphatidylserine on the dying cell. Another critical step during inflammation resolution is mediated by plasmin, the M1 to M2 switch of macrophages. M1 macrophages activated by IFNg or LPS were characterized by a proinflammatory profile dominated by IL-1b/6/12/23 and TNFa cytokines. Plg injections into the murine pleura space led to the infiltration of M2 macrophages expressing IL-10 and TGFb, and chemokine ligands (CCL1/2/17/22) [100].
Recent studies showed that the transmembrane Plg receptor Plg-RKT [101] and Plg induce macrophage polarization from the M1 to the M2 type and efferocytosis in the pleurisy model as shown using Plg-RKT and Plg gene-deficient mice [102].
While fibrin depositions are part of the normal acute inflammatory reaction, provisional laid down fibrin needs to be removed by MMPs and plasmin to restore tissue homeostasis. Fibrin laid down within exudates tends to form adhesions between layers of membranes that cover inflamed organs. The balance between fibrin deposition and degradation is a critical factor in adhesion formation after surgery leading to adhesive small bowel obstruction. Scars can form that lead to adhesive small bowel obstruction, a common problem known by surgeons after intestinal surgery. Increases in activated resident macrophages were found at the surgical site [103]. These activated macrophages secrete epidermal growth factor (EGF), which induced the upregulation of its receptor EGF receptor (HER1) on peritoneal mesothelial cells. HER1-activated mesothelial cells secreted more PAI-1, which together with tPA recruited more macrophages to the surgical site establishing a vicious cycle of impaired fibrinolysis and macrophage accumulation at the adhesive site that can cumulate in adhesive small bowel obstruction. These studies demonstrate that plasmin contributes to the resolution phase of inflammation by its proteolytic ability (fibrin degradation), its ability to enhance the M1 into M2 macrophage switch, and accelerate the cleaning up of apoptotic neutrophils (efferocytosis).
9. Future perspectives
Microbe-driven inflammation as shown with COVID-19 and aseptic inflammation (e.g. atopic disease) is still a real challenge for physicians, medical scientists, and pharmacologists. Novel targets for the treatment of these inflammatory processes are necessary. We have outlined the various facets of plasmin during acute inflammation showing how it alters the activation status of coagulation and complement factors. Furthermore, plasmin(ogen) or tPA and their receptors influence inflammation resolution by controlling leukocyte recruitment, regulating the production of early and late phase cytokines and chemokines. Plg receptors like uPAR and Plg-RKT control macrophage migration and enhance efferocytosis. LRP1 binding of tPA can modulate the extracellular protease landscape, the proteolytic niche, but as discussed in this review also can titrate the TLR response. It will be interesting to see whether aside from LRP1, other Plg receptors also can modulate the TLR response.
Colony stimulating factors are major drivers of the inflammatory response. M-CSF upregulates certain tPA receptors thereby contributing to the inflammatory response. Given the known involvement of other CSFs in the regulation of fibrinolysis, it will be interesting to determine if and which other CSFs change PA or Plg receptor expression thereby influencing the inflammatory response.
Few specific treatment options exist for the derailed immune response underlying MAS syndrome-associated diseases, and cytokine storm syndrome. Future studies will show whether agents targeting plasmin are effective in disrupting the vicious and lethal cycle of hyperfibrinolysis, coagulation, and inflammation.
Acknowledgments
We thank Robert Whittier for proofreading the manuscript. This study was supported (partly) by MEXT KAKENHI Grant no. 16K09821 [B.H.] 23790878 (K.H.), 17K09941 (K.H.), 18K08657 (T.O.), 19K08857 (S.T.), a Grant from International Joint Usage/Research Center (2019-2058), the Institute of Medical Science, the University of Tokyo, The Japanese Society of Hematology and a grant from the Nakatani Foundation (K.H.).
References
- 1.Chen L., Deng H., Cui H., Fang J., Zuo Z., Deng J., Li Y., Wang X., Zhao L. Oncotarget. 2017;9:7204–7218. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hosoya S., Ohbayashi E., Matsushima K., Takeuchi H., Yamazaki M., Shibata Y., Abiko Y. J. Endod. 1998;24:331–334. doi: 10.1016/S0099-2399(98)80129-5. [DOI] [PubMed] [Google Scholar]
- 3.Heissig B., Eiamboonsert S., Salama Y., Shimazu H., Dhahri D., Munakata S., Tashiro Y., Hattori K. Adv. Drug Deliv. Rev. 2016;99:172–179. doi: 10.1016/j.addr.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Levi M. Int. J. Lab. Hematol. 2018;40(Suppl. 1):15–20. doi: 10.1111/ijlh.12830. [DOI] [PubMed] [Google Scholar]
- 5.Renné T., Stavrou E.X. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.02011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rebuck J.W. Am. J. Clin. Pathol. 1983;79:405–413. doi: 10.1093/ajcp/79.4.405. [DOI] [PubMed] [Google Scholar]
- 7.Maas C., Oschatz C., Renne T. Semin. Thromb. Hemost. 2011;37:375–381. doi: 10.1055/s-0031-1276586. [DOI] [PubMed] [Google Scholar]
- 8.Wachtfogel Y.T., Pixley R.A., Kucich U., Abrams W., Weinbaum G., Schapira M., Colman R.W. Blood. 1986;67:1731–1737. [PubMed] [Google Scholar]
- 9.Stavrou E., Schmaier A.H. Thromb. Res. 2010;125:210–215. doi: 10.1016/j.thromres.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foley J.H., Walton B.L., Aleman M.M., O’Byrne A.M., Lei V., Harrasser M., Foley K.A., Wolberg A.S., Conway E.M. EBioMedicine. 2016;5:175–182. doi: 10.1016/j.ebiom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datta P.K., Rappaport J. Biomed. Pharmacother. 2006;60:561–568. doi: 10.1016/j.biopha.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 12.Yau J.W., Teoh H., Verma S. BMC Cardiovasc. Disord. 2015;15:130. doi: 10.1186/s12872-015-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesarman-Maus G., Hajjar K.A. Br. J. Haematol. 2005;129:307–321. doi: 10.1111/j.1365-2141.2005.05444.x. [DOI] [PubMed] [Google Scholar]
- 14.Esmon C.T. Thromb. Res. 2004;114:321–327. doi: 10.1016/j.thromres.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 15.Riewald M., Petrovan R.J., Donner A., Mueller B.M., Ruf W. Science. 2002;296:1880–1882. doi: 10.1126/science.1071699. [DOI] [PubMed] [Google Scholar]
- 16.d’Audigier C., Cochain C., Rossi E., Guérin C.L., Bièche I., Blandinières A., Marsac B., Silvestre J.S., Gaussem P., Smadja D.M. Angiogenesis. 2015;18:347–359. doi: 10.1007/s10456-015-9471-8. [DOI] [PubMed] [Google Scholar]
- 17.Naldini A., Pucci A., Carney D.H., Fanetti G., Carraro F. Cytokine. 2002;20:191–199. doi: 10.1006/cyto.2002.2001. [DOI] [PubMed] [Google Scholar]
- 18.Bugge T.H., Kombrinck K.W., Flick M.J., Daugherty C.C., Danton M.J., Degen J.L. Cell. 1996;87:709–719. doi: 10.1016/s0092-8674(00)81390-2. [DOI] [PubMed] [Google Scholar]
- 19.Nesheim M. Chest. 2003;124:33S–39S. doi: 10.1378/chest.124.3_suppl.33s. [DOI] [PubMed] [Google Scholar]
- 20.Ramachandran R., Altier C., Oikonomopoulou K., Hollenberg M. Pharmacol. Rev. 2016;68:1110–1142. doi: 10.1124/pr.115.010991. [DOI] [PubMed] [Google Scholar]
- 21.Law R.H., Caradoc-Davies T., Cowieson N., Horvath A.J., Quek A.J., Encarnacao J.A., Steer D., Cowan A., Zhang Q., Lu B.G., Pike R.N., Smith A.I., Coughlin P.B., Whisstock J.C. Cell Rep. 2012;1:185–190. doi: 10.1016/j.celrep.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Cao Y., Xue L. Semin. Thromb. Hemost. 2004;30:83–93. doi: 10.1055/s-2004-822973. [DOI] [PubMed] [Google Scholar]
- 23.Schneider M., Nesheim M. J. Biol. Chem. 2004;279:13333–13339. doi: 10.1074/jbc.M313164200. [DOI] [PubMed] [Google Scholar]
- 24.Silverstein R.L., Friedlander R.J., Jr., Nicholas R.L., Nachman R.L. J. Clin. Invest. 1988;82:1948–1955. doi: 10.1172/JCI113814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miles L.A., Dahlberg C.M., Plescia J., Felez J., Kato K., Plow E.F. Biochemistry. 1991;30:1682–1691. doi: 10.1021/bi00220a034. [DOI] [PubMed] [Google Scholar]
- 26.Plow E.F., Doeuvre L., Das R. J. Biomed. Biotechnol. 2012;2012:141806. doi: 10.1155/2012/141806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miles L.A., Parmer R.J. Semin. Thromb. Hemost. 2013;39:329–337. doi: 10.1055/s-0033-1334483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flick M.J., Bugge T.H. J. Thromb. Haemost. 2017;15:150–154. doi: 10.1111/jth.13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawley S.B., Tamura T., Miles L.A. J. Biol. Chem. 2001;276:179–186. doi: 10.1074/jbc.M004919200. [DOI] [PubMed] [Google Scholar]
- 30.Parkkinen J., Rauvala H. J. Biol. Chem. 1991;266:16730–16735. [PubMed] [Google Scholar]
- 31.Wygrecka M., Marsh L.M., Morty R.E., Henneke I., Guenther A., Lohmeyer J., Markart P., Preissner K.T. Blood. 2009;113:5588–5598. doi: 10.1182/blood-2008-08-170837. [DOI] [PubMed] [Google Scholar]
- 32.Urano T., Castellino F.J., Suzuki Y. J. Thromb. Haemost. 2018;16:1487–1497. doi: 10.1111/jth.14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagitani H., Tagawa M., Hatanaka K., Ikari T., Saito A., Bando H., Okada K., Matsuo O. FEBS Lett. 1985;189:145–149. doi: 10.1016/0014-5793(85)80860-7. [DOI] [PubMed] [Google Scholar]
- 34.Bu G., Williams S., Strickland D.K., Schwartz A.L. Proc. Natl. Acad. Sci. U. S. A. 1992;89:7427–7431. doi: 10.1073/pnas.89.16.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuiper J., Otter M., Rijken D.C., van Berkel T.J. J. Biol. Chem. 1988;263:18220–18224. [PubMed] [Google Scholar]
- 36.Lesept F., Chevilley A., Jezequel J., Ladepeche L., Macrez R., Aimable M., Lenoir S., Bertrand T., Rubrecht L., Galea P., Lebouvier L., Petersen K.U., Hommet Y., Maubert E., Ali C., Groc L., Vivien D. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonias S.L., Gaultier A., Jo M. Curr. Pharm. Des. 2011;17:1962–1969. doi: 10.2174/138161211796718224. [DOI] [PubMed] [Google Scholar]
- 38.Lin L., Wu C., Hu K. J. Am. Soc. Nephrol. 2012;23:1329–1338. doi: 10.1681/ASN.2011111123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai H., Baik N., Kiosses W.B., Krajewski S., Miles L.A., Parmer R.J. J. Biol. Chem. 2011;286:33125–33133. doi: 10.1074/jbc.M111.218693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeanneret V., Yepes M. Neural Regen. Res. 2017;12:362–365. doi: 10.4103/1673-5374.202924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mican J., Toul M., Bednar D., Damborsky J. Comput. Struct. Biotechnol. J. 2019;17:917–938. doi: 10.1016/j.csbj.2019.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katz J., Tadi P. StatPearls Publishing LLC; Treasure Island FL: 2020. StatPearls. © 2020. [Google Scholar]
- 43.Sidelmann J.J., Gram J., Jespersen J., Kluft C. Semin. Thromb. Hemost. 2000;26:605–618. doi: 10.1055/s-2000-13216. [DOI] [PubMed] [Google Scholar]
- 44.Bugge T.H., Flick M.J., Daugherty C.C., Degen J.L. Genes Dev. 1995;9:794–807. doi: 10.1101/gad.9.7.794. [DOI] [PubMed] [Google Scholar]
- 45.Busso N., Peclat V., Van Ness K., Kolodziesczyk E., Degen J., Bugge T., So A. J. Clin. Invest. 1998;102:41–50. doi: 10.1172/JCI2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Akassoglou K., Kombrinck K.W., Degen J.L., Strickland S. J. Cell Biol. 2000;149:1157–1166. doi: 10.1083/jcb.149.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bugge T.H., Kombrinck K.W., Flick M.J., Daugherty C.C., Danton M.J.S., Degen J.L. Cell. 1996;87:709–719. doi: 10.1016/s0092-8674(00)81390-2. [DOI] [PubMed] [Google Scholar]
- 48.Munakata S., Tashiro Y., Nishida C., Sato A., Komiyama H., Shimazu H., Dhahri D., Salama Y., Eiamboonsert S., Takeda K., Yagita H., Tsuda Y., Okada Y., Nakauchi H., Sakamoto K., Heissig B., Hattori K. Gastroenterology. 2015;148:565–578. doi: 10.1053/j.gastro.2014.12.001. (e564) [DOI] [PubMed] [Google Scholar]
- 49.Sato A., Nishida C., Sato-Kusubata K., Ishihara M., Tashiro Y., Gritli I., Shimazu H., Munakata S., Yagita H., Okumura K., Tsuda Y., Okada Y., Tojo A., Nakauchi H., Takahashi S., Heissig B., Hattori K. Leukemia. 2015;29:145–156. doi: 10.1038/leu.2014.151. [DOI] [PubMed] [Google Scholar]
- 50.Shimazu H., Munakata S., Tashiro Y., Salama Y., Dhahri D., Eiamboonsert S., Ota Y., Onoda H., Tsuda Y., Okada Y., Nakauchi H., Heissig B., Hattori K. Blood. 2017;130:59–72. doi: 10.1182/blood-2016-09-738096. [DOI] [PubMed] [Google Scholar]
- 51.Heissig B., Dhahri D., Eiamboonsert S., Salama Y., Shimazu H., Munakata S., Hattori K. Cell. Mol. Life Sci. 2015;72:4759–4770. doi: 10.1007/s00018-015-2035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salama Y., Lin S.Y., Dhahri D., Hattori K., Heissig B. FASEB J. 2019;33:3465–3480. doi: 10.1096/fj.201801339RRR. [DOI] [PubMed] [Google Scholar]
- 53.Dhahri D., Sato-Kusubata K., Ohki-Koizumi M., Nishida C., Tashiro Y., Munakata S., Shimazu H., Salama Y., Eiamboonsert S., Nakauchi H., Hattori K., Heissig B. Blood. 2016;128:1063–1075. doi: 10.1182/blood-2015-10-673103. [DOI] [PubMed] [Google Scholar]
- 54.Fullerton J.N., Gilroy D.W. Nat. Rev. Drug Discov. 2016;15:551–567. doi: 10.1038/nrd.2016.39. [DOI] [PubMed] [Google Scholar]
- 55.Hayashi H., Campenot R.B., Vance D.E., Vance J.E. J. Neurosci. 2007;27:1933–1941. doi: 10.1523/JNEUROSCI.5471-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu K., Yang J., Tanaka S., Gonias S.L., Mars W.M., Liu Y. J. Biol. Chem. 2006;281:2120–2127. doi: 10.1074/jbc.M504988200. [DOI] [PubMed] [Google Scholar]
- 57.Mao H., Xie L., Pi X. Front. Cardiovasc. Med. 2017;4 doi: 10.3389/fcvm.2017.00034. (34–34) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Y., Mantuano E., Inoue G., Campana W.M., Gonias S.L. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000188. (ra18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orth K., Willnow T., Herz J., Gething M.J., Sambrook J. J. Biol. Chem. 1994;269:21117–21122. [PubMed] [Google Scholar]
- 60.Prasad J.M., Migliorini M., Galisteo R., Strickland D.K. J. Biol. Chem. 2015;290:17262–17268. doi: 10.1074/jbc.M115.660084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zalfa C., Azmoon P., Mantuano E., Gonias S.L. J. Leukoc. Biol. 2019;105:729–740. doi: 10.1002/JLB.3A0818-329RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaultier A., Arandjelovic S., Niessen S., Overton C.D., Linton M.F., Fazio S., Campana W.M., Cravatt B.F., 3rd, Gonias S.L. Blood. 2008;111:5316–5325. doi: 10.1182/blood-2007-12-127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schubert K., Collins L.E., Green P., Nagase H., Troeberg L. J. Immunol. 2019;202:1501. doi: 10.4049/jimmunol.1800834. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X., Polavarapu R., She H., Mao Z., Yepes M. Am. J. Pathol. 2007;171:1281–1290. doi: 10.2353/ajpath.2007.070472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brifault C., Gilder A.S., Laudati E., Banki M., Gonias S.L. J. Biol. Chem. 2017;292:18699–18712. doi: 10.1074/jbc.M117.798413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mansell A., Reinicke A., Worrall D.M., O’Neill L.A. FEBS Lett. 2001;508:313–317. doi: 10.1016/s0014-5793(01)03077-0. [DOI] [PubMed] [Google Scholar]
- 67.Liu D., Cai S., Gu X., Scafidi J., Wu X., Davis A.E., 3rd J. Immunol. (Baltimore, Md. : 1950) 2003;171:2594–2601. doi: 10.4049/jimmunol.171.5.2594. [DOI] [PubMed] [Google Scholar]
- 68.Smiley S.T., King J.A., Hancock W.W. J. Immunol. (Baltimore, Md. : 1950) 2001;167:2887–2894. doi: 10.4049/jimmunol.167.5.2887. [DOI] [PubMed] [Google Scholar]
- 69.Ward J.R., Dower S.K., Whyte M.K., Buttle D.J., Sabroe I. Biochem. Biophys. Res. Commun. 2006;341:299–303. doi: 10.1016/j.bbrc.2005.12.188. [DOI] [PubMed] [Google Scholar]
- 70.Zurhove K., Nakajima C., Herz J., Bock H.H., May P. Sci. Signal. 2008;1 doi: 10.1126/scisignal.1164263. (ra15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mantuano E., Brifault C., Lam M.S., Azmoon P., Gilder A.S., Gonias S.L. Proc. Natl. Acad. Sci. U. S. A. 2016;113:1369–1374. doi: 10.1073/pnas.1515480113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu Z., Strickland D.K., Hyman B.T., Rebeck G.W. J. Biol. Chem. 2002;277:14458–14466. doi: 10.1074/jbc.M112066200. [DOI] [PubMed] [Google Scholar]
- 73.Mantuano E., Lam M.S., Gonias S.L. J. Biol. Chem. 2013;288:34009–34018. doi: 10.1074/jbc.M113.509133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mantuano E., Azmoon P., Brifault C., Banki M.A., Gilder A.S., Campana W.M., Gonias S.L. Blood. 2017;130:1364–1374. doi: 10.1182/blood-2017-04-780205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Das L., Azmoon P., Banki M.A., Mantuano E., Gonias S.L. PLoS One. 2019;14 doi: 10.1371/journal.pone.0224738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamilton J.A. Nat. Rev. Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 77.Boire A., Covic L., Agarwal A., Jacques S., Sherifi S., Kuliopulos A. Cell. 2005;120:303–313. doi: 10.1016/j.cell.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 78.Austin K.M., Covic L., Kuliopulos A. Blood. 2013;121:431–439. doi: 10.1182/blood-2012-09-355958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heuberger D.M., Schuepbach R.A. Thromb. J. 2019;17:4. doi: 10.1186/s12959-019-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuliopulos A., Covic L., Seeley S.K., Sheridan P.J., Helin J., Costello C.E. Biochemistry. 1999;38:4572–4585. doi: 10.1021/bi9824792. [DOI] [PubMed] [Google Scholar]
- 81.Pontecorvi P., Banki M.A., Zampieri C., Zalfa C., Azmoon P., Kounnas M.Z., Marchese C., Gonias S.L., Mantuano E. J. Neuroinflammation. 2019;16:257. doi: 10.1186/s12974-019-1657-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kamio N., Hashizume H., Nakao S., Matsushima K., Sugiya H. Biochem. Pharmacol. 2008;75:1974–1980. doi: 10.1016/j.bcp.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 83.Draxler D.F., Daglas M., Fernando A., Hanafi G., McCutcheon F., Ho H., Galle A., Gregory J., Larsson P., Keragala C., Wright D.K., Tavancheh E., Au A.E., Niego B., Wilson K., Plebanski M., Sashindranath M., Medcalf R.L. J. Thromb. Haemost. 2019;17:2174–2187. doi: 10.1111/jth.14603. [DOI] [PubMed] [Google Scholar]
- 84.Draxler D.F., Lee F., Ho H., Keragala C.B., Medcalf R.L., Be Niego. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Draxler D.F., Yep K., Hanafi G., Winton A., Daglas M., Ho H., Sashindranath M., Wutzlhofer L.M., Forbes A., Goncalves I., Tran H.A., Wallace S., Plebanski M., Myles P.S., Medcalf R.L. Blood Adv. 2019;3:1598–1609. doi: 10.1182/bloodadvances.2019000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Norelli M. Nat. Med. 2018;24 doi: 10.1038/s41591-018-0036-4. [DOI] [PubMed] [Google Scholar]
- 87.Giavridis T. Nat. Med. 2018;24 doi: 10.1038/s41591-018-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferrara J.L., Abhyankar S., Gilliland D.G. Transplant. Proc. 1993;25:1216–1217. [PubMed] [Google Scholar]
- 89.Behrens E.M., Koretzky G.A. Arthritis Rheumatol. (Hoboken, N.J.) 2017;69:1135–1143. doi: 10.1002/art.40071. [DOI] [PubMed] [Google Scholar]
- 90.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;1(368):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 91.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Vanstapel A., Werlein C., Stark H., Tzankov A., Li W.W., Li V.W., Mentzer S.J., Jonigk D. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fox S.E., Akmatbekov A., Harbert J.L., Li G., Quincy Brown J., Vander Heide R.S. Lancet Respir. Med. 2020;8:681–686. doi: 10.1016/S2213-2600(20)30243-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Medcalf R.L., Keragala C.B., Myles P.S. J. Thromb. Haemost. 2020 [Google Scholar]
- 95.Hay K.A., Hanafi L.A., Li D., Gust J., Liles W.C., Wurfel M.M., López J.A., Chen J., Chung D., Harju-Baker S., Cherian S., Chen X., Riddell S.R., Maloney D.G., Turtle C.J. Blood. 2017;130:2295–2306. doi: 10.1182/blood-2017-06-793141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Novak U., Cocks B.G., Hamilton J.A. Nucleic Acids Res. 1991;19:3389–3393. doi: 10.1093/nar/19.12.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Swiatkowska M., Szemraj J., Cierniewski C.S. FEBS J. 2005;272:5821–5831. doi: 10.1111/j.1742-4658.2005.04979.x. [DOI] [PubMed] [Google Scholar]
- 98.Rosenwald M., Koppe U., Keppeler H., Sauer G., Hennel R., Ernst A., Blume K.E., Peter C., Herrmann M., Belka C., Schulze-Osthoff K., Wesselborg S., Lauber K. J. Immunol. (Baltimore, Md. : 1950) 2012;189:5722–5728. doi: 10.4049/jimmunol.1200922. [DOI] [PubMed] [Google Scholar]
- 99.Das R., Ganapathy S., Settle M., Plow E.F. Blood. 2014;124:679–688. doi: 10.1182/blood-2014-01-549659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sugimoto M.A., Ribeiro A.L.C., Costa B.R.C., Vago J.P., Lima K.M., Carneiro F.S., Ortiz M.M.O., Lima G.L.N., Carmo A.A.F., Rocha R.M., Perez D.A., Reis A.C., Pinho V., Miles L.A., Garcia C.C., Teixeira M.M., Sousa L.P. Blood. 2017;129:2896–2907. doi: 10.1182/blood-2016-09-742825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Andronicos N.M., Chen E.I., Baik N., Bai H., Parmer C.M., Kiosses W.B., Kamps M.P., Yates J.R., 3rd, Parmer R.J., Miles L.A. Blood. 2010;115:1319–1330. doi: 10.1182/blood-2008-11-188938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vago J.P., Sugimoto M.A., Lima K.M., Negreiros-Lima G.L., Baik N., Teixeira M.M., Perretti M., Parmer R.J., Miles L.A., Sousa L.P. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Honjo K., Munakata S., Tashiro Y., Salama Y., Shimazu H., Eiamboonsert S., Dhahri D., Ichimura A., Dan T., Miyata T., Takeda K., Sakamoto K., Hattori K., Heissig B. FASEB J. 2017;31:2625–2637. doi: 10.1096/fj.201600871RR. [DOI] [PubMed] [Google Scholar]