Abstract
Pluripotent stem cells (PSCs) can immortally self-renew in culture with a high proliferation rate, and they possess unique metabolic characteristics that facilitate pluripotency regulation. Here, we review recent progress in understanding the mechanisms that link cellular metabolism and homeostasis to pluripotency regulation, with particular emphasis on pathways involving amino acid metabolism, lipid metabolism, the ubiquitin-proteasome system and autophagy. Metabolism of amino acids and lipids is tightly coupled to epigenetic modification, organelle remodeling and cell signaling pathways for pluripotency regulation. PSCs harness enhanced proteasome and autophagy activity to meet the material and energy requirements for cellular homeostasis. These regulatory events reflect a fine balance between the intrinsic cellular requirements and the extrinsic environment. A more complete understanding of this balance will pave new ways to manipulate PSC fate.
Keywords: autophagy, amino acid metabolism, lipid metabolism, pluripotent stem cell (PSC), ubiquitin-proteasome system (UPS)
Introduction
Pluripotent stem cells (PSCs), including embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), have the capacity to self-renew and differentiate into all cell types of our bodies (Liu et al., 2014; Martello and Smith, 2014). These properties depend on a series of pluripotency genes that are highly expressed and coordinately regulated in PSCs (Boyer et al., 2005; Orkin and Hochedlinger, 2011). At the same time, PSCs have developed unique cell cycle characteristics and a high proliferation rate to match the activity of pluripotency gene networks (Wang et al., 2008; Singh and Dalton, 2009). Emerging evidence shows that metabolic pathways are mediators of crosstalk between cellular degradation, cellular recycling, epigenetic regulation, signal transduction and stem cell fate determination (Folmes et al., 2012; Buck et al., 2016; Gascon et al., 2016; Zhang et al., 2016b; Zheng et al., 2016).
Cellular metabolism, including anabolism and catabolism, involves multiple complex biochemical processes, such as amino acid metabolism, nucleic acid metabolism, fatty acid metabolism, glycolysis, oxidative phosphorylation, the ubiquitin-proteasome degradation system, autophagy, and so on (Naujokat and Saric, 2007; Vessoni et al., 2012; Kilberg et al., 2016; Wang et al., 2017). Metabolism is not only involved in energy production and degradation and biosynthesis of cellular components, but also takes part in signal transduction for genetic and epigenetic regulation through its intermediate metabolites (Zhang et al., 2018). Cells utilize anabolism to produce biological macromolecules and organelles. Catabolism is responsible for degrading and recycling both normal and harmful substances and dysfunctional organelles by breaking them down into small molecules (Mathieu and Ruohola-Baker, 2017). Anabolism and catabolism are tightly coordinated and essential for maintaining cell proliferation and function (Zhang et al., 2014; Kaur and Debnath, 2015).
Existing studies have proposed that PSCs mainly rely on glycolysis for energy generation, while somatic cells prefer oxidative phosphorylation for ATP production; this issue has been vigorously discussed by several excellent reviews (Folmes et al., 2012; Zhang et al., 2012; Xu et al., 2013; Mathieu and Ruohola-Baker, 2017; Zhang et al., 2018). Here we mainly summarize recent research progress in understanding how pluripotency is regulated by metabolic pathways involving amino acids, fatty acids, the ubiquitin-proteasome system and autophagy.
Amino acid metabolism and pluripotency
The important role of amino acids in the later stages of preimplantation embryo development was firstly noted by Gwatkin while he was formulating mouse embryo culture medium. He initially found that adding amino acids into the culture medium induced the attachment and outgrowth of mouse blastocysts (Gwatkin, 1966). Following these observations, Spindle and Pedersen demonstrated that adding amino acids into culture medium not only improved attachment and outgrowth of blastocysts but also increased embryo hatching rate (Spindle and Pedersen, 1973). It was later found that preimplantation embryos have transporters for specific amino acids and can maintain an endogenous pool of amino acids (Schultz et al., 1981; Sellens et al., 1981; Van Winkle, 2001).
In cellular biosynthetic pathways, amino acids are essential nutrients that contribute directly to protein synthesis as well as providing compounds for chemical modifications. Although the amino acid metabolism and protein synthesis pathways are well defined in somatic cells, relatively little is known about the contribution of amino acid metabolism to pluripotency regulation in PSCs. Emerging studies provide evidence that amino acids regulate pluripotency by providing chemical groups for chromatin modifications (Kilberg et al., 2016; D’Aniello et al., 2019).
Threonine metabolism and pluripotency
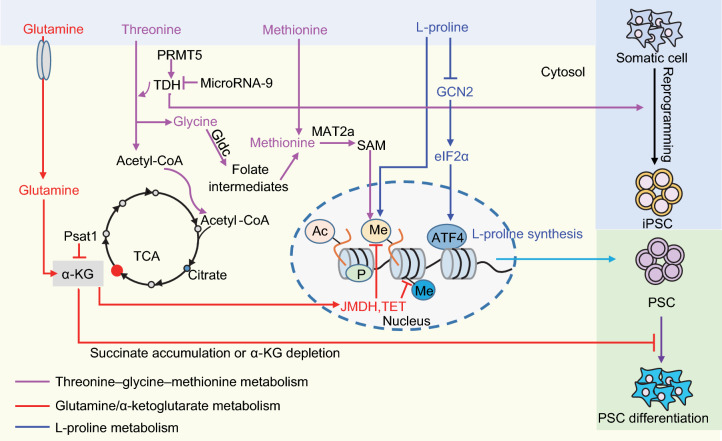
Threonine is an essential amino acid, which can be catabolized by threonine dehydrogenase (TDH) to glycine and acetyl-coenzyme A (acetyl-CoA). Glycine can be catabolized by glycine decarboxylase (Gldc) to generate folate intermediates to fuel one-carbon metabolism. The folate intermediates fuel nucleotide synthesis and remethylation of homocysteine to form methionine (Met) and S-adenosyl-methionine (SAM). SAM serves as a universal methyl donor for DNA and histone methylation reactions. Acetyl-CoA feeds the tricarboxylic acid (TCA) cycle as well as serving as the donor of acetyl groups (Fig. 1).
Figure 1.
Amino acid metabolism in pluripotency regulation. Threonine/methionine metabolism contribute to pluripotency regulation by providing SAM for DNA and histone methylation in PSCs. The threonine dehydrogenase TDH is highly expressed in mESCs, maintaining a high ratio of SAM/SAH that is correlated with high H3K4me3 levels. TDH expression is positively regulated by PRMT5 and negatively regulated by microRNA-9. Metabolism of glutamine and glucose regulates pluripotency through α-KG, which is a cofactor for Jumonji domain-containing histone demethylases (JMDH) and the ten-eleven translocation family of enzymes (TETs) that are involved in DNA demethylation. The cellular level of L-proline is fine-tuned by the amino acid starvation response (AAR) pathway Gcn2-Eif2α-Atf4. Excessive supplementation with L-proline leads to ESC differentiation. Appropriate intracellular synthesis of L-proline safeguards PSC pluripotency
Mouse ESCs (mESCs) exist in a high-flux metabolic state that depends on threonine catabolism. By depriving mESCs of each of the 20 amino acids individually, Wang et al. have shown that threonine catabolism is required for mESC identity maintenance (Wang et al., 2009). The underlying mechanism was revealed by Shyh-Chang et al.: threonine maintains mESC pluripotency by providing SAM for trimethylation of histone H3 lysine 4 (H3K4me3) (Shyh-Chang et al., 2013).
In addition, Alexander et al. have shown that mESCs rapidly discontinue DNA synthesis, arrest cell division and eventually die after threonine deprivation (Alexander et al., 2011). When threonine is withdrawn for 1 to 4 days, the expression of pluripotency marker genes decreases, accompanied by increased expression of trophoectodermal and mesodermal marker genes. Refeeding threonine one day after deprivation leads to increased expression of cyclin D1 and E in mESCs. They proposed that threonine regulates mESC proliferation by stimulating G1/S transition through lipid raft/caveolae-dependent PI3K/Akt, MAPKs, mTOR, p70S6K, and 4E-BP1 signaling pathways (Ryu and Han, 2011; Kilberg et al., 2016).
The enzyme L-threnonine-3-dehydrogenase (TDH) is highly expressed in mESCs and is the first and rate-limiting enzyme in the pathway that hydrolyzes threonine into glycine and acetyl-CoA in mitochondria. High activity and expression of TDH maintain a high SAM/SAH(S-adenosylhomocysteine) ratio in mESCs, which is tightly correlated with H3K4me3 levels (Wang et al., 2009; Ang et al., 2011; Shyh-Chang et al., 2013). During reprogramming, induction of TDH enhances, whereas knockdown of TDH inhibits, reprogramming efficiency (Han et al., 2013). TDH expression can be negatively regulated at the post-transcriptional level by microRNA-9, and positively regulated by protein arginine methyltransferase (PRMT5) (Han et al., 2013).
Methionine metabolism and pluripotency
Methionine metabolism is involved in epigenetic maintenance, redox homeostasis and organism development (Tang et al., 2017). Methionine can be directly converted to SAM in a reaction catalyzed by methionine adenosyltransferase 2a (MAT2a), which provides a threonine-independent route for generating SAM. SIRT1, the most conserved mammalian NAD+-dependent protein deacetylase, critically regulates the activity of MAT2a in coordination with Myc. Deletion of SIRT1 in mESCs increased the hypersensitivity of the cells to methionine restriction/depletion-induced differentiation and apoptosis (Tang et al., 2017).
Unlike the situation in mESCs, absence of threonine does not evidently affect the pluripotency of human ESCs (hESCs), as the TDH gene is a non-functional pseudogene in humans (Edgar, 2002). Instead, methionine directly produces SAM in hESCs. Depletion of methionine induces a rapid decrease of intracellular SAM in hPSCs, resulting in decreased global H3K4me3, activation of p53-p38 signaling, reduction of NANOG expression, and thereafter differentiation and apoptosis of hPSCs (Shiraki et al., 2014). Interestingly, the precise level of SAM, which is restricted by a nicotinamide N-methyltransferase (NNMT)-dependent SAM-consuming pathway, is critical for maintaining the naïve state of hESCs. NNMT consumes SAM to keep it at a low level to preserve the naïve state of hESCs, while a high level of SAM promotes the naïve-to-primed transition through H3K27me3 (Sperber et al., 2015).
L-proline (L-Pro) and pluripotency
The intracellular concentration of L-Pro in mESCs is around 4-fold lower than in mESCs in the L-Pro-induced primed state, and 10-fold lower than in mouse embryonic fibroblasts. The cellular level of L-Pro in mESCs is strictly regulated by the Gcn2-Eif2α-Atf4 amino acid starvation response (AAR) pathway (D’Aniello et al., 2015). The low level of intracellular L-Pro induces expression of Atf4 to enhance intracellular synthesis of L-Pro through activation of Aldh18a1/Pycr1; this maintains a naïve state of pluripotency that is less dependent on exogenous L-Pro supplementation. Excessive supplementation with L-Pro inactivates Gcn2-Eif2α-Atf4 AAR and thus inhibits intracellular L-Pro synthesis, resulting in mESC mesenchymal transition. This feedback regulatory loop precisely maintains the appropriate intrinsic L-Pro level, which restricts proliferation of tightly packed dome-like mESC colonies and safeguards mESC identity (Fig. 1).
Differentiation of pluripotent cells within the mammalian blastocyst starts with the formation of the primitive ectoderm, or epiblast, from the inner cell mass (ICM). This early differentiation step can be recaptured in vitro by culturing mESCs in MEDII conditioned medium, which leads to formation of primitive ectoderm-like cells. Supplementation with L-Pro promotes, whereas depletion of L-Pro uptake inhibits, the differentiation of mESCs toward primitive ectoderm (Washington et al., 2010; Kilberg et al., 2016). In support of this observation, another independent study identified that L-Pro induces mESCs toward an epiblast-like stem cell (EpiSCs) phenotype in a dose- and time- dependent manner. This EpiSC phenotype induced by L-Pro can be reversed by either withdrawal of L-Pro or addition of L-ascorbic acid (vitamin C, Vc) (Casalino et al., 2011).
Following on from these studies, Comes et al. showed that the phenotype remodeling in EpiSCs is reminiscent of a change toward a mesenchymal-like state. They proposed that L-Pro serves as a signaling molecule to promote mESC differentiation by stimulation of the epithelial-to-mesenchymal transition (Comes et al., 2013). Interestingly, this ESC-to-mesenchymal transition is accompanied by a genome-wide increase of DNA methylation and specific histone modifications (H3K9me2/H3K9me3 and H3K36me3) (Comes et al., 2013; D’Aniello et al., 2017). These changes in histone modification can be reversed by either removal of L-Pro or treatment with Vc. Conversely, somatic cell reprogramming to iPSCs involves a mesenchymal-to-epithelial transition, which can be promoted by Vc through the H3K36 demethylase Jhdm1a/1b (Wang et al., 2011). Thus, L-Pro antagonizes Vc in terms of DNA methylation and chromatin structure, which leads to opposing effects on pluripotency regulation.
Glutamine/α-Ketoglutarate and pluripotency
In mESCs, glutamine can be catabolized to α-ketoglutarate (α-KG) to support TCA cycle anaplerosis. α-KG is an important metabolic intermediate that acts as a cofactor for Jumonji domain-containing histone demethylases (JMDH) and the ten-eleven translocation family of enzymes (TETs) that are involved in DNA demethylation (Fig. 1). Glutamine contributes to pluripotency by generating α-KG to regulate cellular DNA/histone methylation states.
Naïve mESCs use both glucose and glutamine catabolism to generate a high level of cellular α-KG, which contributes to a low level of intracellular H3K27me3 and increases TET-dependent DNA demethylation (Carey et al., 2015). Direct manipulation of the intracellular α-KG level is sufficient to change the levels of cellular H3K27me3 and TET-dependent DNA demethylation, which are correlated with pluripotency gene expression. Cellular α-KG levels can be regulated by phosphoserine aminotransferase1 (Psat1), which is an Oct4/Sox2/Nanog target protein. Decreased expression of Psat1 in mESCs lowers the levels of DNA 5’-hydroxymethylcytosine and H3K9me3, resulting in accelerated differentiation of mESCs (Hwang et al., 2016).
Fatty acid synthesis and pluripotency
Lipids serve as the predominant components of plasma and organelle membranes, as secondary messengers for signal transduction, and as an important source of energy. In a search for new molecules that contribute to long-term hESC self-renewal, Garcia-Gonzalo and Izpisua Belmonte identified that the lipids in lipid-rich albumin from a chemically-defined medium stimulated hESC pluripotency (Garcia-Gonzalo and Izpisua Belmonte, 2008). This provided the first evidence to show that lipids contribute to pluripotency regulation. Using mass spectrometry-based metabolomics analysis, Yanes et al. identified that unsaturated lipid levels decreased upon mESC differentiation (Yanes et al., 2010). Inhibition of the eicosanoid signaling pathway maintained the levels of unsaturated fatty acids and improved pluripotency. In accordance with these observation, Wang et al. demonstrated that de novo synthesis of fatty acids, which is mediated by the rate-limiting enzyme ACC1 (acetyl-Coenzyme A carboxylase alpha), is required for pluripotency acquisition and maintenance (Wang et al., 2017). Activation of ACC1 leads to decreased acetyl-CoA production and increased cellular lipid generation, which promotes mitochondrial fission and cellular pluripotency (Fig. 2A). This pathway is conserved in both human and mouse ESCs (Wang et al., 2017). Conversely, Dunning et al. showed that promoting fatty acid beta-oxidation by carnitine supplementation during in vitro culture of early mouse embryos increased the blastocyst hatching rate and the ICM cell number (Dunning et al., 2010). The reason for this discrepancy is unclear. One possible interpretation is that the balance between fatty acid oxidation and fatty acid synthesis maintains an appropriate cellular fatty acid level in ESCs that facilitates pluripotency regulation.
Figure 2.
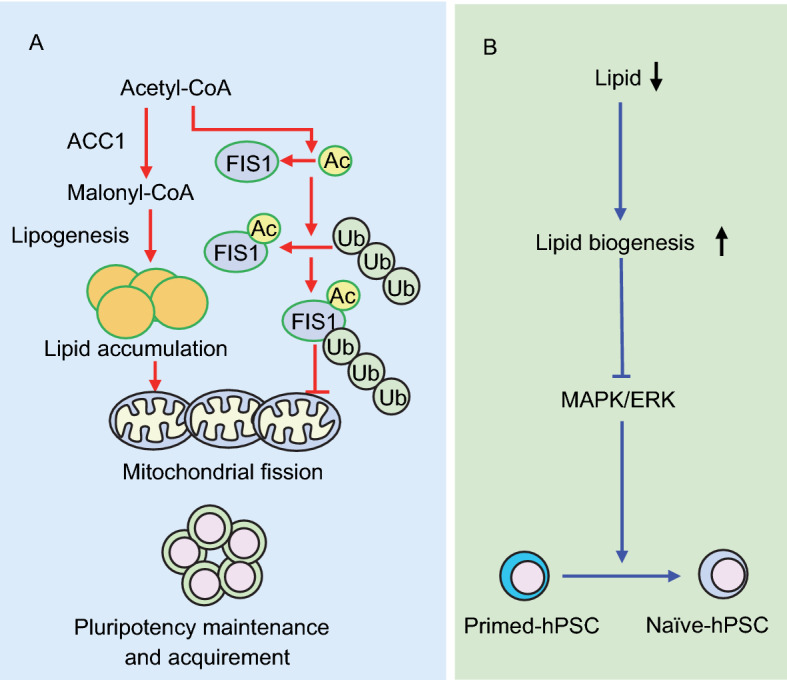
Fatty acid metabolism in pluripotency regulation. Appropriate cellular levels of lipids safeguard pluripotency. (A) De novo synthesis of fatty acids, initiated by the enzyme acetyl-Coenzyme A carboxylase alpha (ACC1), promotes pluripotency maintenance and acquisition by enhancing mitochondrial fission. This conserved pathway is antagonized by ubiquitin-proteasome mediated degradation of the acetylated mitochondrial fission mediator FIS1. (B) In human PSCs, exogenous lipid deficiency induces intracellular lipogenesis which ultimately inhibits endogenous ERK and promotes pluripotency
By comprehensive metabolic flux analysis of hESCs cultured either in mouse embryonic fibroblast (MEF)-conditioned medium or essential 8 minimal medium (E8, no lipids), Zhang et al. demonstrated that the lack of lipids in E8 medium promotes oxidative pentose phosphate pathway (PPP) flux for NADPH synthesis, de novo lipogenesis and reductive carboxylation, while simultaneously decreasing mitochondrial respiration. Supplementation of E8 medium with lipids resulted in decreased lipogenesis and increased oxidative mitochondrial metabolism in hESCs (Zhang et al., 2016a). Most recently, Cornacchia et al. demonstrated that hESCs cultured in E8 medium captured a naïve-to-primed intermediate state of pluripotency with de novo lipogenesis and endogenous ERK inhibition that mimics in vivo regulation during pre-implantation development (Fig. 2B) (Cornacchia et al., 2019).
Together, these data indicate that de novo lipogenesis and appropriate cellular levels of lipids play critical roles in pluripotency regulation in both mouse and human PSCs. In addition, these studies deliver supporting evidence that mitochondrial respiration contributes to pluripotency maintenance, which challenges the traditional notion that mitochondrial function is dispensable for pluripotent stem cell function.
The ubiquitin-proteasome system and pluripotency
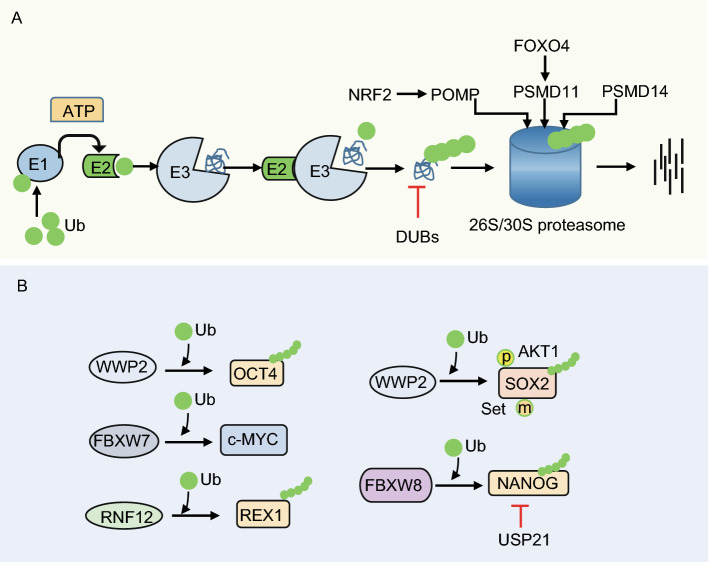
Ubiquitination is a cascade enzymatic reaction that involves covalent addition of ubiquitin (Ub) to target proteins (Fig. 3A). The E1 Ub-activating enzyme activates Ub in an ATP-dependent manner, and forms a thioester bond between a cysteine in the enzyme and the carboxyl terminus of Ub. Then, activated Ub is transferred to E2 (Ub carrier enzyme or Ub-conjugating enzyme). Delivery of the Ub by E2 to the target protein is dependent on the E3 Ub protein ligase. The E3 enzyme is responsible for substrate recognition and for promoting the elongation of Ub chains (Weissman, 2001). Deubiquitinating enzymes (DUBs) are required to specifically disassemble ubiquitin chains, thus balancing ubiquitination and deubiquitination (Chandrasekaran et al., 2017).
Figure 3.
Regulation of pluripotency by the ubiquitin-proteasome system (UPS). (A) PSCs exhibit high proteasome degradation activity, which is regulated by FOXO4-driven expression of the 19S proteasome subunit PSMD11 and corresponding enhanced assembly of 26S/30S proteasomes. (B) Levels of the pluripotency factors OCT4, c-MYC, REX1, SOX2 and NANOG are fine-tuned by UPS to maintain the precise quantity that facilitates pluripotency. p, phosphorylation; m, methylation
The 26S/30S proteasome consists of the 20S core structure (which contains the proteolytic active site) and 19S cap structures (which regulate the activity of the holo-complex 26S, single and 30S, double capped) (Finley, 2009). The proteasome has caspase-, trypsin-, and chymotrypsin-like activity that degrades ubiquitin-labeled target proteins into 2–24 amino acid peptides (Bedford et al., 2010; Dikic, 2017).
The ubiquitin-proteasome system (UPS) functions in a variety of cellular processes including proliferation, differentiation, apoptosis and senescence (Mayer, 2000). The cardinal role of ubiquitination is to generate poly-ubiquitinated proteins that are recognized and degraded by the 26S proteasome (Weissman, 2001).
Early studies have shown that ubiquitin ligases are involved in degradation of pluripotency factors (Fig. 3B). For example, Xu et al. demonstrated that WWP2, an E3 ubiquitin (Ub)-protein ligase, interacts with the pluripotency protein OCT4 and enhances its Ub modification in both mouse and human ESCs (Xu et al., 2004; Xu et al., 2009). Accordingly, disruption of WWP2-mediated OCT4 ubiquitination promotes reprogramming of mouse embryonic fibroblasts (Li et al., 2018). Interestingly, WWP2 specifically interacts with Set7-methylated SOX2 and induces SOX2 ubiquitination and degradation in mESCs, which can be antagonized by AKT1-mediated phosphorylation at T118. AKT1 activity dominates over Set7 to maintain SOX2 stability and pluripotency in mESCs (Fang et al., 2014). In addition, the X-encoded E3 ubiquitin ligase RNF12 has been identified to mediate degradation of REX1 in a dose-dependent manner in mESCs, thereby contributing to X-chromosome inactivation (Jonkers et al., 2009; Gontan et al., 2012). Furthermore, Ddb1, a component of the CUL4-DDB1 E3 ligase complex, was demonstrated to contribute to pluripotency regulation. Deletion of DDB1 results in mESC differentiation (Gao et al., 2015).
In addition to E3 ligases, the E2 ligase UBE2S (ubiquitin-conjugating enzyme E2S) directly interacts with SOX2 at the K123 residue to maintain a proper SOX2 protein level through K11-linked polyubiquitination, thus reinforcing the undifferentiated state of mESCs (Wang et al., 2016).
A high-throughput functional genetic screening study using a cDNA-based random RNA interference library in mESCs revealed that knockdown of ubiquitin increased the formation of ESC colonies in the absence of LIF (Jian et al., 2007). A more comprehensive study using UPS-targeted RNAi screens identified that the DUBs PSMD14 and USP9X; the E3 ligases RBX1, RFWD2, RNF12, UBR5, and DDB1; and the putative E3 ligases TRIM28 and PHF5a contribute to ESC self-renewal, whereas the E3 ligases FBXW7, RNF152, RNF31, RNF8, SOCS3, and TOPORS; the putative ligases RNF36 and TNFRSF25; and the ubiquitin-like protein UBL5 regulate early ESC differentiation (Buckley et al., 2012). The authors provided evidence to show that, mechanistically, the E3 ligase FBXW7 acts as a key regulator of ESC differentiation through regulation of c-MYC stability. Silencing of Fbxw7 expression inhibited ESC differentiation and enhanced cellular reprogramming through stabilization of c-MYC. In contrast, the DUB PSMD14 maintains mESC self-renewal by deubiquitination and subsequent degradation of target proteins. Knockdown of Psmd14 significantly inhibits mESC self-renewal and pluripotency (Buckley et al., 2012). In support of these findings, subsequent studies have shown that the DUB USP21 maintains mESC stemness via stabilization of NANOG (Jin et al., 2016; Liu et al., 2016b; Kwon et al., 2017). A motif rich in proline, glutamine, serine, and threonine from amino acids 47 to 72 in the N-terminus of NANOG was identified as the target for degradation (Ramakrishna et al., 2011). In addition, the prolyl isomerase PIN1 interacts with and stabilizes NANOG by inhibiting NANOG ubiquitination, thus maintaining pluripotency in both mouse and human ESCs (Moretto-Zita et al., 2010).
In parallel with these findings, Szutorisz et al. have demonstrated that proteasomes target transcription factors and RNA polymerase II bound at tissue-specific gene domains in mESCs to restrict permissive transcription and maintain these genes in a poised state, facilitating the following differentiation initiations (Szutorisz et al., 2006).
Interestingly, Vilchez et al. have shown that hESCs exhibit high proteasome activity, which is characterized by increased levels of the 19S proteasome subunit PSMD11 and corresponding enhanced assembly of 26S/30S proteasomes. The expression of PSMD11 is regulated by the insulin/insulin-like growth factor-I (IGF-I)-responsive transcription factor FOXO4 (Vilchez et al., 2012).
Together, these existing studies provide supporting evidence that enhanced proteasome activity in ESCs is essential for cellular proteostasis regulation and pluripotency maintenance. On one hand, components of the UPS interact with multiple pluripotency factors like OCT4, SOX2, NANOG, c-MYC, and REX1 to fine-tune their cellular levels to meet pluripotency requirements; on the other hand, the UPS regulates the degradation of transcription factors and RNA polymerase II bound to tissue-specific genes in ESCs to maintain the pluripotent state of ESCs.
Autophagic degradation and pluripotency
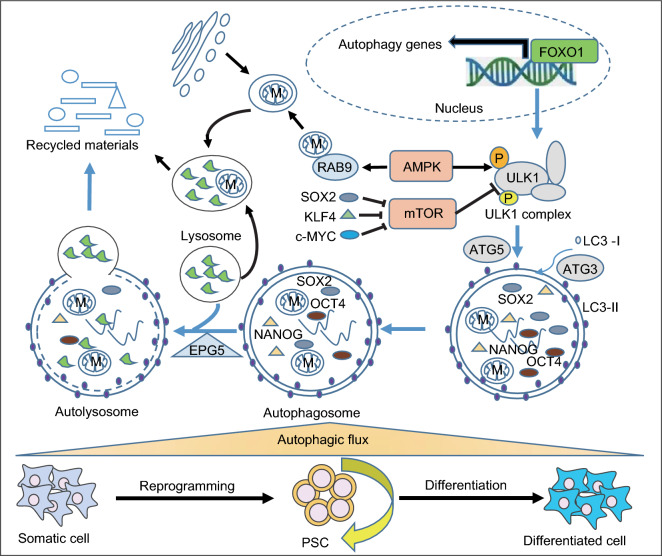
Autophagy is a lysosome-dependent catabolic process, in which cytosolic materials (including proteins, organelles, and lipids) are sequestrated into double-membrane vesicles (termed autophagosome) and delivered to lysosomes for degradation (Klionsky and Emr, 2000). Autophagy recycles damaged or superfluous cytoplasmic contents to maintain cellular homeostasis. Increasing numbers of studies suggest that autophagy plays important roles in regulating ESC cellular homeostasis to maintain self-renewal and pluripotency (Fig. 4).
Figure 4.
Regulation of pluripotency by autophagy. PSCs exhibit a high autophagic flux that is regulated by FOXO1, which coordinates the autophagy machinery gene program at the transcriptional level. High autophagic flux maintains appropriate levels of cellular pluripotency factors like OCT4, SOX2 and NANOG, and organelles like mitochondria (M). Inhibition of autophagy leads to accumulation of abnormal mitochondria and breakdown of pluripotency in spite of increased levels of pluripotency proteins. Activation of autophagy by AMPK is essential for both pluripotency maintenance and acquisition. Inactivation of mTOR by the pluripotency factors SOX2, KLK4 or c-MYC facilitates somatic cell reprogramming to pluripotency
Autophagy was first revealed to regulate cellular homeostasis in ESCs by Yoshimori’s group, who used Atg5-null mESCs to study autophagy mechanisms. They found that ATG5 is critical for autophagosome formation, and ATG5-dependent autophagy accounts for the majority of protein degradation by lysosomes in mESCs (Mizushima et al., 2001). Autophagic flux is maintained by a core molecular machinery which is composed of autophagy sensor systems, the induction complex, and autophagosome formation systems. mESCs possess a powerful autophagic infrastructure with enhanced expression of most of the core autophagy machinery genes compared to somatic fibroblasts (Liu et al., 2017). This substantial infrastructure, regulated by the master transcriptional factor FOXO1, endows mESCs with a high autophagic flux. Downregulation of autophagic flux compromised mESC self-renewal and pluripotency (Liu et al., 2017). Furthermore, deletion of the autophagy regulator Atg3 in mESCs resulted in increased accumulation of abnormal mitochondria and enhanced production of reactive oxygen species (ROS), together with decreased mitochondrial potential and ATP generation (Liu et al., 2016a).
In the search for molecules that protect pluripotency in mouse ESCs, Gu et al. identified EPG5, a eukaryotic-specific autophagy regulator which mediates autophagosome/lysosome fusion, as a guardian of ESC stemness. Deubiquitination of EPG5 by USP8 at Lys 252 consolidates the interaction between EPG5 and LC3 and thus sustains a high autophagic flux for stemness maintenance (Gu et al., 2019). In addition, the mammalian autophagy-initiating kinase ULK1 is highly expressed in mESCs, and constitutive activation of ULK1 by AMPK (AMP activated protein kinase) functions as an intrinsic signaling pathway in ESCs to regulate their identity under normal physiological conditions (Gong et al., 2018).
An appropriate cellular level of pluripotency proteins in ESCs is critical for pluripotency maintenance. The stability of the pluripotency proteins OCT4, SOX2 and NANOG in hESCs is regulated by autophagy in addition to regulation by the UPS as described above. Autophagy inhibition led to reduction of pluripotency despite accumulation of these pluripotency proteins (Cho et al., 2014).
PSCs are distinct from somatic cells in having a smaller volume and fewer mitochondria. During somatic cell reprogramming, treatment with mTOR (mammalian target of rapamycin) inhibitors like rapamycin or PP242, or the autophagy inducer spermidine, significantly improved the speed and efficiency of iPSC generation, which indicates that autophagy is involved in this cellular remodeling (Chen et al., 2011; Menendez et al., 2011). In support of this proposition, the reprogramming factor SOX2 was identified to directly bind to the promoter of mTOR to recruit the NuRD complex. This resulted in transcriptional repression of mTOR, thus inducing a transient activation of autophagy, which accounted for successful reprogramming (Wang et al., 2013). In addition, KLF4 and c-MYC were found to inhibit mTORC1 (mechanistic target of rapamycin complex 1) during pluripotency acquisition, leading to autophagy activation, mitochondrial remodeling and cell size reduction, which facilitated reprogramming (Wu et al., 2015). Consistent with this, silencing of canonical autophagy by Atg3 deletion inhibited mitochondrial remodeling during pluripotency induction, resulting in decreased reprogramming efficiency and more abnormal mitochondria in established mouse iPSCs (Liu et al., 2016a). Interestingly, in parallel with these findings, Ma et al. demonstrated that an AMPK-activated Atg5-independent autophagy pathway contributes to mitochondrial clearance, and facilitates the metabolic switch from mitochondrial oxidative phosphorylation to glycolysis during somatic cell reprogramming (Ma et al., 2015). Mitophagy, the specific clearance of mitochondria by autophagy, can be initiated by BNIP3L (BCL2/adenovirus E1B interacting protein 3-like). BNIP3L-mediated mitophagy contributes to mitochondrial clearance during reprogramming induced by OCT4/SO2/KLF4 but not OCT4/SO2/KLF4/c-MYC (Xiang et al., 2017).
Taken together, the existing studies deliver supporting evidence that a high level of autophagy activity is required in ESCs to maintain cellular homeostasis of proteins and mitochondria, thus safeguarding pluripotency. During somatic cell reprogramming, both canonical and non-canonical autophagy contribute to cellular remodeling (Fig. 4).
Future directions
PSCs can potentially provide unlimited resources to benefit regenerative medicine through in vitro-directed differentiation. Significant achievements have been made in understanding the mechanistic roles of transcription factors, epigenetic factors and signaling pathways in pluripotency regulation, which will facilitate the optimization of differentiation protocols. However, efficient generation of the desired specific cell type from PSCs remains a major challenge. This is partly because there are still gaps in our knowledge about pluripotency regulation. The recent findings described here—that cellular catabolism, anabolism and homeostasis pathways contribute to pluripotency maintenance—provide new insights into PSC fate determination and pave new ways for stem cell fate manipulation. These findings raise a number of questions: (1) What are the upstream signals for metabolic switching during cell fate transition? (2) How is the balance between anabolism and catabolism regulated in pluripotency acquisition and maintenance? (3) What is the role and mechanism of anabolism and catabolism in regulating reprogramming and differentiation? (4) How does the UPS cooperate with autophagy to regulate pluripotency? (5) How do intermediate metabolites play roles in cell fate transition? (6) Is it possible to optimize PSC differentiation protocols by manipulating anabolism, catabolism, UPS or autophagy? Given the rapid development of omics technologies like proteomics, transcriptomics, epigenomics and metabolomics, together with bioinformatics and artificial intelligence, our understanding of the metabolic regulation of pluripotency will rapidly move forward and thus significantly benefit regenerative medicine.
Acknowledgements
This research was funded by grants from the National Key R&D Program of China 2018YFA0108402, the Strategic Priority Research Program of the Chinese Academy of Sciences XDA16030302, the National Natural Science Foundation of China Program 31720103907, 31621004, 31570995, the Key Research Program of the Chinese Academy of Sciences ZDRW-ZS-2017-5 to T.Z, the Strategic Priority Research Program of the Chinese Academy of Science XDA16040501 to J.C and the China Postdoctoral Science Foundation (2018M630205) to K.L.
Abbreviations
ADP, adenosine diphosphate; ATP, adenosine triphosphate; ATPase, ATP synthase; AMPK, AMP-activated protein kinase; ATF4, stress-activated transcription factor 4; AAR, amino acid starvation response; DNMT, DNA methyltransferase; DUB, deubiquitinating enzyme; ETC, electron transport chain; Eif2α, eukaryotic initiation factor 2; G6P, glucose 6 phosphate; GCN2, general control nonrepressed 2; HAT, histone acetyltransferase; HMT, histone methyltransferase; JMDH, Jumonji-c domain histone demethylase; SAM, S-adenosyl methionine; SAH, S-adenosylhomocysteine; SIRT1, activating sirtuin 1; TCA, tricarboxylic acid; TET, Tet methylcytosine dioxygenase
Compliance with ethics guidelines
All the authors of this paper declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
Footnotes
Kun Liu, Jiani Cao and Xingxing Shi contributed equally.
References
- Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc Natl Acad Sci USA. 2011;108:15828–15833. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford L, Paine S, Sheppard PW, Mayer RJ, Roelofs J. Assembly, structure, and function of the 26S proteasome. Trends Cell Biol. 2010;20:391–401. doi: 10.1016/j.tcb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MD, O’Sullivan D, Geltink RIK, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJW, et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino L, Comes S, Lambazzi G, De Stefano B, Filosa S, De Falco S, De Cesare D, Minchiotti G, Patriarca EJ. Control of embryonic stem cell metastability by L-proline catabolism. J Mol Cell Biol. 2011;3:108–122. doi: 10.1093/jmcb/mjr001. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran AP, Suresh B, Kim HH, Kim KS, Ramakrishna S. Concise review: fate determination of stem cells by deubiquitinating enzymes. Stem Cells (Dayton, Ohio) 2017;35:9–16. doi: 10.1002/stem.2446. [DOI] [PubMed] [Google Scholar]
- Chen T, Shen L, Yu J, Wan H, Guo A, Chen J, Long Y, Zhao J, Pei G. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–911. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- Cho YH, Han KM, Kim D, Lee J, Lee SH, Choi KW, Kim J, Han YM. Autophagy regulates homeostasis of pluripotency-associated proteins in hESCs. Stem Cells (Dayton, Ohio) 2014;32:424–435. doi: 10.1002/stem.1589. [DOI] [PubMed] [Google Scholar]
- Comes S, Gagliardi M, Laprano N, Fico A, Cimmino A, Palamidessi A, De Cesare D, De Falco S, Angelini C, Scita G, et al. L-Proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem Cell Rep. 2013;1:307–321. doi: 10.1016/j.stemcr.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornacchia D, Zhang C, Zimmer B, Chung SY, Fan Y, Soliman MA, Tchieu J, Chambers SM, Shah H, Paull D, et al. Lipid deprivation induces a stable, naive-to-primed intermediate state of pluripotency in human PSCs. Cell Stem Cell. 2019;25:120–136. doi: 10.1016/j.stem.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aniello C, Cermola F, Palamidessi A, Wanderlingh LG, Gagliardi M, Migliaccio A, Varrone F, Casalino L, Matarazzo MR, De Cesare D, et al. Collagen prolyl hydroxylation-dependent metabolic perturbation governs epigenetic remodeling and mesenchymal transition in pluripotent and cancer cells. Cancer Res. 2019;79:3235–3250. doi: 10.1158/0008-5472.CAN-18-2070. [DOI] [PubMed] [Google Scholar]
- D’Aniello C, Fico A, Casalino L, Guardiola O, Di Napoli G, Cermola F, De Cesare D, Tate R, Cobellis G, Patriarca EJ, et al. A novel autoregulatory loop between the Gcn2-Atf4 pathway and (L)-Proline [corrected] metabolism controls stem cell identity. Cell Death Differ. 2015;22:1094–1105. doi: 10.1038/cdd.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aniello C, Habibi E, Cermola F, Paris D, Russo F, Fiorenzano A, Di Napoli G, Melck DJ, Cobellis G, Angelini C, et al. Vitamin C and l-proline antagonistic effects capture alternative states in the pluripotency continuum. Stem Cell Rep. 2017;8:1–10. doi: 10.1016/j.stemcr.2016.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I. Proteasomal and autophagic degradation systems. Annu Rev Biochem. 2017;86:193–224. doi: 10.1146/annurev-biochem-061516-044908. [DOI] [PubMed] [Google Scholar]
- Dunning KR, Cashman K, Russell DL, Thompson JG, Norman RJ, Robker RL. Beta-oxidation is essential for mouse oocyte developmental competence and early embryo development. Biol Reprod. 2010;83:909–918. doi: 10.1095/biolreprod.110.084145. [DOI] [PubMed] [Google Scholar]
- Edgar AJ. The human L-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet. 2002;3:18. doi: 10.1186/1471-2156-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Zhang L, Wei W, Jin X, Wang P, Tong Y, Li J, Du JX, Wong J. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol Cell. 2014;55:537–551. doi: 10.1016/j.molcel.2014.06.018. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Buckley SM, Cimmino L, Guillamot M, Strikoudis A, Cang Y, Goff SP, Aifantis I. The CUL4-DDB1 ubiquitin ligase complex controls adult and embryonic stem cell differentiation and homeostasis. eLife. 2015;4:e07539. doi: 10.7554/eLife.07539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Izpisua Belmonte JC. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS ONE. 2008;3:e1384. doi: 10.1371/journal.pone.0001384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon S, Murenu E, Masserdotti G, Ortega F, Russo GL, Petrik D, Deshpande A, Heinrich C, Karow M, Robertson SP, et al. Identification and successful negotiation of a metabolic checkpoint in direct neuronal reprogramming. Cell Stem Cell. 2016;18:396–409. doi: 10.1016/j.stem.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Gong J, Gu H, Zhao L, Wang L, Liu P, Wang F, Xu H, Zhao T. Phosphorylation of ULK1 by AMPK is essential for mouse embryonic stem cell self-renewal and pluripotency. Cell Death Dis. 2018;9:38. doi: 10.1038/s41419-017-0054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, Grootegoed JA, Gribnau J. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature. 2012;485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- Gu H, Shi X, Liu C, Wang C, Sui N, Zhao Y, Gong J, Wang F, Zhang H, Li W, et al. USP8 maintains embryonic stem cell stemness via deubiquitination of EPG5. Nat Commun. 2019;10:1465. doi: 10.1038/s41467-019-09430-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwatkin RBL. Amino acid requirements for attachment and outgrowth of the mouse blastocyst in vitro. J Cell Physiol. 1966;68:335–343. [Google Scholar]
- Han C, Gu H, Wang J, Lu W, Mei Y, Wu M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells (Dayton, Ohio) 2013;31:953–965. doi: 10.1002/stem.1335. [DOI] [PubMed] [Google Scholar]
- Hwang IY, Kwak S, Lee S, Kim H, Lee SE, Kim JH, Kim YA, Jeon YK, Chung DH, Jin X, et al. Psat1-dependent fluctuations in alpha-ketoglutarate affect the timing of ESC differentiation. Cell Metab. 2016;24:494–501. doi: 10.1016/j.cmet.2016.06.014. [DOI] [PubMed] [Google Scholar]
- Jian R, Cheng X, Jiang J, Deng S, Hu F, Zhang J. A cDNA-based random RNA interference library for functional genetic screens in embryonic stem cells. Stem Cells (Dayton, Ohio) 2007;25:1904–1912. doi: 10.1634/stemcells.2006-0448. [DOI] [PubMed] [Google Scholar]
- Jin J, Liu J, Chen C, Liu Z, Jiang C, Chu H, Pan W, Wang X, Zhang L, Li B, et al. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nat Commun. 2016;7:13594. doi: 10.1038/ncomms13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Barakat TS, Achame EM, Monkhorst K, Kenter A, Rentmeester E, Grosveld F, Grootegoed JA, Gribnau J. RNF12 is an X-encoded dose-dependent activator of X chromosome inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16:461–472. doi: 10.1038/nrm4024. [DOI] [PubMed] [Google Scholar]
- Kilberg MS, Terada N, Shan J. Influence of amino acid metabolism on embryonic stem cell function and differentiation. Adv Nutr (Bethesda, Md) 2016;7:780S–789S. doi: 10.3945/an.115.011031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SK, Lee DH, Kim SY, Park JH, Choi J, Baek KH. Ubiquitin-specific protease 21 regulating the K48-linked polyubiquitination of NANOG. Biochem Biophys Res Commun. 2017;482:1443–1448. doi: 10.1016/j.bbrc.2016.12.055. [DOI] [PubMed] [Google Scholar]
- Li S, Xiao F, Zhang J, Sun X, Wang H, Zeng Y, Hu J, Tang F, Gu J, Zhao Y, et al. Disruption of OCT4 ubiquitination increases OCT4 protein stability and ASH2L-B-mediated H3K4 methylation promoting pluripotency acquisition. Stem Cell Rep. 2018;11:973–987. doi: 10.1016/j.stemcr.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Song Y, Yu H, Zhao T. Understanding the roadmaps to induced pluripotency. Cell Death Dis. 2014;5:e1232. doi: 10.1038/cddis.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Zhao Q, Liu P, Cao J, Gong J, Wang C, Wang W, Li X, Sun H, Zhang C, et al. ATG3-dependent autophagy mediates mitochondrial homeostasis in pluripotency acquirement and maintenance. Autophagy. 2016;12:2000–2008. doi: 10.1080/15548627.2016.1212786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Liu K, Gu H, Wang W, Gong J, Zhu Y, Zhao Q, Cao J, Han C, Gao F, et al. High autophagic flux guards ESC identity through coordinating autophagy machinery gene program by FOXO1. Cell Death Differ. 2017;24:1672–1680. doi: 10.1038/cdd.2017.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yao Y, Ding H, Han C, Chen Y, Zhang Y, Wang C, Zhang X, Zhang Y, Zhai Y, et al. USP21 deubiquitylates Nanog to regulate protein stability and stem cell pluripotency. Signal Transduct Target Ther. 2016;1:16024. doi: 10.1038/sigtrans.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Li J, Xu Y, Yu C, Xu T, Wang H, Liu K, Cao N, Nie BM, Zhu SY, et al. Atg5-independent autophagy regulates mitochondrial clearance and is essential for iPSC reprogramming. Nat Cell Biol. 2015;17:1379–1387. doi: 10.1038/ncb3256. [DOI] [PubMed] [Google Scholar]
- Martello G, Smith A. The nature of embryonic stem cells. Annu Rev Cell Dev Biol. 2014;30:647–675. doi: 10.1146/annurev-cellbio-100913-013116. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Ruohola-Baker H. Metabolic remodeling during the loss and acquisition of pluripotency. Development. 2017;144:541–551. doi: 10.1242/dev.128389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer RJ. The meteoric rise of regulated intracellular proteolysis. Nat Rev Mol Cell Biol. 2000;1:145–148. doi: 10.1038/35040090. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Vellon L, Oliveras-Ferraros C, Cufi S, Vazquez-Martin A. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle (Georgetown, Tex) 2011;10:3658–3677. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretto-Zita M, Jin H, Shen Z, Zhao T, Briggs SP, Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci USA. 2010;107:13312–13317. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokat C, Saric T. Concise review: role and function of the ubiquitin-proteasome system in mammalian stem and progenitor cells. Stem Cells (Dayton, Ohio) 2007;25:2408–2418. doi: 10.1634/stemcells.2007-0255. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S, Suresh B, Lim KH, Cha BH, Lee SH, Kim KS, Baek KH. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011;20:1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- Ryu JM, Han HJ. L-threonine regulates G1/S phase transition of mouse embryonic stem cells via PI3K/Akt, MAPKs, and mTORC pathways. J Biol Chem. 2011;286:23667–23678. doi: 10.1074/jbc.M110.216283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz GA, Kaye PL, McKay DJ, Johnson MH. Endogenous amino acid pool sizes in mouse eggs and preimplantation embryos. J Reprod Fertil. 1981;61:387–393. doi: 10.1530/jrf.0.0610387. [DOI] [PubMed] [Google Scholar]
- Sellens MH, Stein S, Sherman MI. Protein and free amino acid content in preimplantation mouse embryos and in blastocysts under various culture conditions. J Reprod Fertil. 1981;61:307–315. doi: 10.1530/jrf.0.0610307. [DOI] [PubMed] [Google Scholar]
- Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19:780–794. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–226. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AM, Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5:141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, et al. The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat Cell Biol. 2015;17:1523–1535. doi: 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindle AI, Pedersen RA. Hatching, attachment, and outgrowth of mouse blastocysts in vitro: fixed nitrogen requirements. J Exp Zool. 1973;186:305–318. doi: 10.1002/jez.1401860308. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Tang S, Fang Y, Huang G, Xu X, Padilla-Banks E, Fan W, Xu Q, Sanderson SM, Foley JF, Dowdy S, et al. Methionine metabolism is essential for SIRT1-regulated mouse embryonic stem cell maintenance and embryonic development. EMBO J. 2017;36:3175–3193. doi: 10.15252/embj.201796708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkle LJ. Amino acid transport regulation and early embryo development. Biol Reprod. 2001;64:1–12. doi: 10.1095/biolreprod64.1.1. [DOI] [PubMed] [Google Scholar]
- Vessoni AT, Muotri AR, Okamoto OK. Autophagy in stem cell maintenance and differentiation. Stem Cells Dev. 2012;21:513–520. doi: 10.1089/scd.2011.0526. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Hou J, Qian X, Zhang H, Zhang Z, Li M, Wang R, Liao K, Wang Y, et al. Ube2s regulates Sox2 stability and mouse ES cell maintenance. Cell Death Differ. 2016;23:393–404. doi: 10.1038/cdd.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhang T, Wang L, Cai Y, Zhong X, He X, Hu L, Tian S, Wu M, Hui L, et al. Fatty acid synthesis is critical for stem cell pluripotency via promoting mitochondrial fission. EMBO J. 2017;36:1330–1347. doi: 10.15252/embj.201695417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Xia P, Ye B, Huang G, Liu J, Fan Z. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13:617–625. doi: 10.1016/j.stem.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9:575–587. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington JM, Rathjen J, Felquer F, Lonic A, Bettess MD, Hamra N, Semendric L, Tan BS, Lake JA, Keough RA, et al. L-Proline induces differentiation of ES cells: a novel role for an amino acid in the regulation of pluripotent cells in culture. Am J Physiol Cell Physiol. 2010;298:C982–992. doi: 10.1152/ajpcell.00498.2009. [DOI] [PubMed] [Google Scholar]
- Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–178. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- Wu Y, Li Y, Zhang H, Huang Y, Zhao P, Tang Y, Qiu X, Ying Y, Li W, Ni S, et al. Autophagy and mTORC1 regulate the stochastic phase of somatic cell reprogramming. Nat Cell Biol. 2015;17:715–725. doi: 10.1038/ncb3172. [DOI] [PubMed] [Google Scholar]
- Xiang G, Yang L, Long Q, Chen K, Tang H, Wu Y, Liu Z, Zhou Y, Qi J, Zheng L, et al. BNIP3L-dependent mitophagy accounts for mitochondrial clearance during 3 factors-induced somatic cell reprogramming. Autophagy. 2017;13:1543–1555. doi: 10.1080/15548627.2017.1338545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Wang W, Li C, Yu H, Yang A, Wang B, Jin Y. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 2009;19:561–573. doi: 10.1038/cr.2009.31. [DOI] [PubMed] [Google Scholar]
- Xu HM, Liao B, Zhang QJ, Wang BB, Li H, Zhong XM, Sheng HZ, Zhao YX, Zhao YM, Jin Y. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J Biol Chem. 2004;279:23495–23503. doi: 10.1074/jbc.M400516200. [DOI] [PubMed] [Google Scholar]
- Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, Trauger SA, Desponts C, Ding S, Siuzdak G. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–417. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014;20:526–540. doi: 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- Zhang H, Badur MG, Divakaruni AS, Parker SJ, Jager C, Hiller K, Murphy AN, Metallo CM. Distinct metabolic states can support self-renewal and lipogenesis in human pluripotent stem cells under different culture conditions. Cell Rep. 2016;16:1536–1547. doi: 10.1016/j.celrep.2016.06.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D’Amico D, Ropelle ER, Lutolf MP, Aebersold R, et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589–595. doi: 10.1016/j.stem.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhao J, Dahan P, Lu V, Zhang C, Li H, Teitell MA. Metabolism in pluripotent stem cells and early mammalian development. Cell Metab. 2018;27:332–338. doi: 10.1016/j.cmet.2018.01.008. [DOI] [PubMed] [Google Scholar]
- Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, Ma L, Hamm M, Gage FH, Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Life. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]