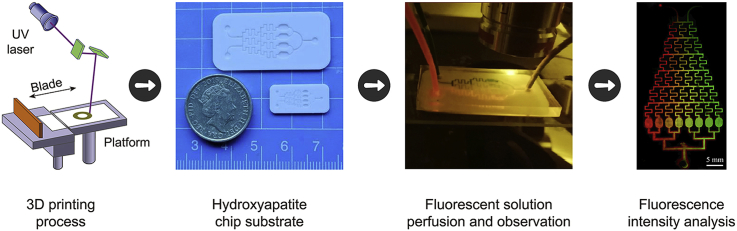
Abstract
Bone is an important part of the human body structure and plays a vital role in human health. A microfluidic chip that can simulate the structure and function of bone will provide a platform for bone-related biomedical research. Hydroxyapatite (HA), a bioactive ceramic material, has a similar structure and composition to bone mineralization products. In this study, we used HA as a microfluidic chip component to provide a highly bionic bone environment. HA substrates with different microchannel structures were printed by using ceramic stereolithography (SLA) technology, and the minimum trench width was 50 μm. The HA substrate with microchannels was sealed by a thin polydimethylsiloxane (PDMS) layer to make a HA-PDMS microfluidic chip. Cell culture experiments demonstrated that compared with PDMS, HA was more conducive to the proliferation and osteogenic differentiation of the human foetal osteoblast cell line (hFOB). In addition, the concentration gradient of the model drug doxorubicin hydrochloride (DOX) was successfully generated on a Christmas tree structure HA-PDMS chip, and the half maximal inhibitory concentration (IC50) of DOX was determined. The findings of this study indicate that the HA-PDMS microfluidic chip has great potential in the field of high-throughput bone-related drug screening and bone-related research.
Keywords: Hydroxyapatite (HA), Ceramic microfluidic chip, Drug screening, Biomimetic, Bone-on-a-chip
Graphical abstract
Highlights
-
•
3D printing of the hydroxyapatite (HA) substrate with microchannel networks.
-
•
Fabrication of HA-PDMS microfluidic chips. (3) Provided a new microfluidic platform for studying bone and bone-related diseases.
1. Introduction
Bones are the essential part of hard tissues and play a decisive role in the health of the human body. They provide a framework for the body, anchor muscle and also contribute to protecting organs and storing calcium. Clinically, many diseases are related to bone metabolism, such as postmenopausal osteoporosis, autoimmune diseases, allergic diseases, and rheumatoid arthritis [[1], [2], [3]]. In addition, breast cancer, prostate cancer, lung cancer, and melanoma often clinically prefer to home in bones, and the frequency of bone metastases is as high as approximately 70% in these cancers [4]. Therefore, it is of great significance to construct a bone bionic system for the studies of bone and bone-related diseases in vitro.
In recent years, organ-on-a-chip technology has developed rapidly. It refers to a microengineered biomimetic system built on a microfluidic chip, which can replicate key functions of living organs [[5], [6], [7]]. Through the development and utilization of these microfluidic systems, in vitro disease models can be established for drug screening and can help to understand the biological mechanism, which shows that these microfluidic systems have great potential in biomedical, pharmaceutical, and toxicological applications [8]. The most commonly used material for the fabrication of microfluidic chips is polydimethylsiloxane (PDMS), since it is easy to fabricate, inexpensive, highly biocompatible, and transparent. However, the rigidity and hardness of PDMS are far from those of the real bones [[9], [10], [11]].
In recent years, scientists have been working hard to find devices and materials suitable for the fabrication of bone-on-a-chip and provide a bionic bone microenvironment. Hao et al. developed a microfluidic device that mimicked the conditioned microenvironment of bone cells to study breast cancer metastasis to bone and cancer extravasation [12]. Ahn et al. combined fibrin as a model extracellular matrix (ECM) with hydroxyapatite (HA) nanocrystals and filled microfluidic chip channels with this material to simulate the real bone tissue matrix and induce blood vessel germination [13]. As the main inorganic component of human bones, HA can participate in metabolism in the body and is a very commonly used bioactive ceramic material [14]. Compared with PDMS, the stiffness and hardness of HA are much more similar to those of real bone. Moreover, the structure and composition of HA are similar to the products of bone mineralization. Hence, we considered directly using HA to fabricate the bone-on-a-chip. However, fabricating micro-structures on HA remains a challenge. The traditional microfluidic processing method, such as photolithography, cannot be applied to fabricate HA ceramics. Ordinary ceramic processing methods, such as cutting and laser processing, cannot meet the accuracy requirements of microfluidic chips. In 2017, Lantada et al. developed an alumina ceramic microfluidic chip by using photolithography-based ceramic manufacturing (LCM) technology [15]. They used this device to imitate the typical configuration of transwell assays. Different from their focus on structural design, we aimed to prepare a microfluidic chip with the materila HA that is more suitable for bone related research.
In this paper, we proposed a technological process to fabricate HA-PDMS microfluidic chips. HA substrates with microchannels were printed using the stereolithography (SLA) method. We compared the proliferation of osteoblasts on PDMS and HA substrates and demonstrate that HA substrates have excellent biocompatibility and can promote osteoblast differentiation. In addition, we formed a concentration gradient of the model drug DOX on the HA-PDMS chip with the Christmas tree-shaped networks and monitored the growth of cells at different drug concentrations to determine the optimal DOX concentration for osteosarcoma treatment. These results suggest that the HA-PDMS chip could serve as a promising choice to build a bone-on-a-chip for bone-related drug screening and bone biology research.
2. Materials and methods
2.1. Chemicals and materials
HA paste was purchased from 3DCeram Sinto, France. Sylgard 184 silicone elastomer and curing agent were purchased from Dow Corning Toray, Japan. Doxorubicin hydrochloride (DOX) was purchased from Sigma-Aldrich and was of analytical grade and used without further purification. Fluorescein isothiocyanate isomer I (FITC) and rhodamine were purchased from Sigma-Aldrich, USA. Cell Tracker Green CMFDA (5-chloromethylfluorescein diacetate) and other cellular fluorescent stains were purchased from Thermo Fisher, USA. Dulbecco's modified Eagle's medium (DMEM), phenol red-free DMEM/F12 (DMEM-F12), foetal bovine serum (FBS), phosphate-buffered saline (PBS) and trypsin-EDTA were purchased from Gibco, Invitrogen. The UMR-106 cell line and the hFOB cell line were purchased from the American Type Culture Collection (ATCC).
2.2. Preparation of the ceramic substrate
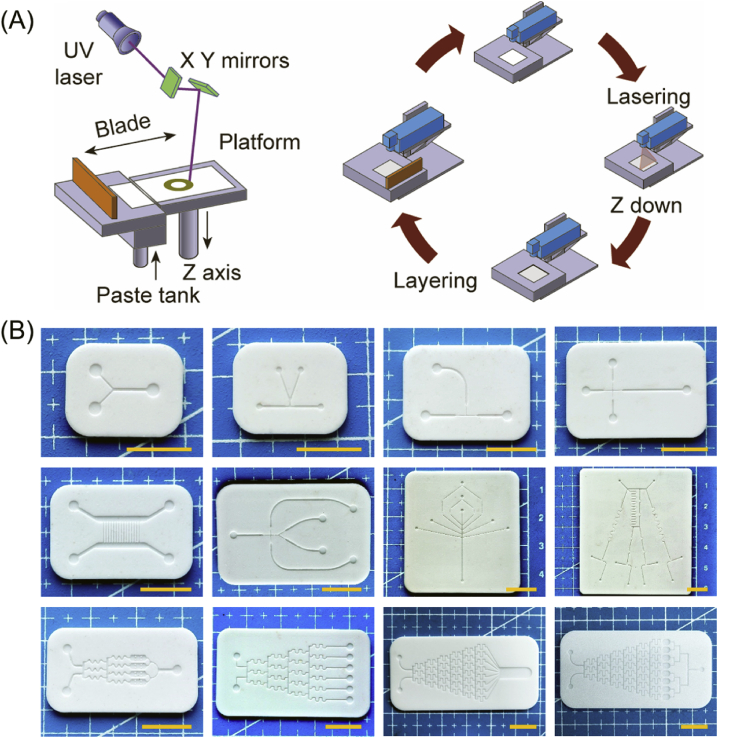
HA paste (55 wt%) was stirred uniformly and added to the tank of a ceramic printer (3DCeram Sinto, France). According to a designed model slicing procedure, we used a scraper to spread a 0.1 mm layer of paste onto the sample platform and then scanned and cured that layer of paste with a 48 mW ultraviolet laser beam at a scanning speed of 4500 mm/s. After one layer of the pattern was cured and formed, the sample platform was moved down the height of one layer, and a subsequent layer of the reactive suspension was deposited and polymerized according to the next cross-sectional pattern. The process was repeated until the printed solid ceramic sample (HA green bodies) was completed (Fig. 1A).
Fig. 1.
Printed schematic and photos of ceramic substrates. (A) Schematic diagram of the ceramic stereolithography printing process. (B) Photographs of the printed ceramic substrates with various microchannel structures, scale bar = 1 cm.
After printing, the HA green bodies were carefully and thoroughly cleaned with neopentylglykol diacrylat (a cleaning solvent) until the non-polymerized slurry was completely removed from the cavities. The HA green bodies were subsequently subjected to heat treatment including debinding and sintering to obtain a ceramic substrate that did not contain any organic materials. As shown in Fig. S1, the solvent and the crosslinked resin were removed slowly according to the pre-set debinding curve. The debinding curve was determined by thermogravimetric (TG) analysis and differential scanning calorimetry (DSC). Then, the HA green bodies were heated to 1280 °C at a rate of 3 °C/min for 1 h to sinter the ceramic particles together.
2.3. Characterization of the ceramic substrate
A field emission scanning electron microscope (MERLIN, Carl Zeiss AG, Germany) was used to observe the microstructure of the ceramic matrix, and at the same time, the elemental composition of the chip surface was obtained by energy dispersive spectroscopy. Then, we ground the ceramic substrate into a powder for analysis. The X-ray photoelectron spectroscopy (XPS) instrument (Axis uHru DCD, Krates, UK) used was equipped with a monochromatic X-ray source (Al Kα anode) operating at 15 kV. The diameter of the analysed spot was approximately 900 mm. The binding energy was calibrated with C1s = 284.85 eV. X-ray diffraction (XRD, Empyrean, Malvern Panalytical, Netherlands) was performed at room temperature using Cu Kα radiation for 2θ values of 10–70° with a 0.026° step size. The FT-IR spectrum (Vector 33-MIR, Brukev Optik, Germany) was recorded between 400 cm−1 and 4000 cm−1 with a resolution of 2 cm−1.
2.4. In vitro biological evaluation of ceramic substrates and PDMS
The human foetal osteoblast cell line (hFOB), which is commonly used in various osteogenic differentiation experiments, was cultured in DMEM-F12 containing 10% FBS [16]. Cells were maintained in an incubator at 34 °C and 5% CO2 until reaching confluence, and then the cells were trypsinized for use in subsequent experiments.
Prior to seeding the cells, the ceramic substrate and PDMS were autoclaved for 20 min at 121 °C. Additionally, a fibronectin solution with a concentration of 50 μg/mL was used to coat PDMS to promote cell adhesion [17]. The PDMS pieces were then incubated for 1 h in a 37 °C incubator, after which we removed the fibronectin solution and dried the PDMS in a clean hood at room temperature (RT). The HA substrate did not require additional surface modification treatment.
We used a cell counting kit (CCK-8, Dojindo Laboratories, Japan) to determine the proliferation of hFOB cells cultured on the HA substrate, PDMS and multi-well plates. A total of 0.5 mL of cell suspension containing 5000 cells was seeded onto the surfaces of the three materials. The medium was changed every 2 days. After 1, 4 and 7 days of incubation, 200 μL of CCK-8 working solution was added followed by incubation for 1 h at 37 °C. Subsequently, the supernatant was extracted and subjected to absorbance detection at 450 nm using a multifunctional microplate reader (Thermo Scientific 3001, USA). In addition, cell viability was analysed using a live/dead assay (Invitrogen, green (live)/red (dead)).
Quantitative real-time PCR (qRT-PCR) was used to detect the PCR marker genes RUNX2, COL-1, ALP, OPN and OCN for osteogenic differentiation in hFOB cells. hFOB cells were seeded on the ceramic substrate and PDMS at a density of 1 × 104/cm2 and then cultured at 37 °C for 7 days. Total RNA was isolated using a HiPure Total RNA kit (Magnetic, China). The RNA was then reverse transcribed into cDNA using a PrimeScript RT kit (TaKaRa Biotechnology, Japan) with a gDNA eraser. Finally, qRT-PCR analysis was performed on a qRT-PCR instrument (QuantStudio 6 Flex, Life Technologies) using a SYBR Green system (GeneCopoeia). The relative quantification of the target genes was normalized to beta actin and calculated using the 2−ΔΔCt method.
Immunofluorescence staining (IF) was used to detect the marker protein of osteogenic differentiation in hFOB cells. hFOB cells were seeded on the ceramic substrate and PDMS at a density of 2500 cells/cm2. After 7 days of incubation, the cells were fixed in a 4% neutral formaldehyde solution for 30 min, permeabilized with 0.1% Triton X-100 for 20 min, and then incubated for 1 h in 3% BSA to block nonspecific proteins. The cells were incubated with primary antibodies against ALP ((C-8): sc-373737, Santa Cruz), COL1A ((COL-1): sc-59772, Santa Cruz), RUNX2 ((27-K): sc-101145, Santa Cruz), OCN ((ABOC-5021): sc-73464, Santa Cruz) or OPN ((LFMb-14): sc-73631, Santa Cruz) at 4 °C overnight and then incubated with a donkey anti-mouse IgG H&L secondary antibody (Alexa Fluor® 647, ab150107) for 1 h at room temperature. F-actin was allowed to stain the cytoskeleton (green) for 1 h, and DAPI was allowed to stain the nucleus (blue) for 5 min at room temperature. The cells were washed gently with PBS and then observed under a laser confocal microscope (TCS SP8, Leica, Germany).
2.5. Fabrication of the Christmas tree-type HA-PDMS microfluidic chip
The HA-PDMS microfluidic chip contained two layers, a ceramic substrate with microchannels and a PDMS cover. The fabrication process of the ceramic substrate with the Christmas tree structure was the same as that described in section 2.2. The number of branches on the Christmas tree was designed as 8, and the size of each branch was 200 μm deep and 150 μm wide. The fabrication of the PDMS layer followed a basic process [17]. Briefly, the base elastomer and the curing agent were uniformly mixed at a mass ratio of 10:1 to obtain a mixed solution, and the bubbles in the mixed solution were removed by vacuuming. The mixed solution was poured into a dish that was flat on the bottom and heated in an oven at 65 °C for 2 h to obtain a smooth PDMS sheet. The PDMS sheet was peeled off from the Petri dish, cut according to the shape of the ceramic substrate, and perforated with a punch at the inlet and outlet positions.
The two layers were glued together to form sealed microfluidic channels. A Japanese OSP-10/250 inkjet bar was used to apply a layer of uncured PDMS prepolymer onto the PDMS sheet, and then the sheet was attached to the polished ceramic substrate. The assembly was placed in an oven at 75 °C for 2 h, and the two parts ultimately stuck together tightly.
2.6. Gradient generation and verification
The FITC solution was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 1 mg/mL and diluted to 0.1 mg/mL by using 0.01 M PBS, and then a few drops of blue ink were added. Similarly, rhodamine solution was dissolved in methanol at a concentration of 2 mg/mL and diluted to 0.2 mg/mL with 0.01 M PBS. The two fluorescent solutions were injected into the two inlets of the Christmas tree-type ceramic chip at a perfusion rate of 0.2 mL/h. After the flow was stable, the chip was placed under a laser confocal microscope (TCS SP8, Leica, Germany) for observation and inspection.
Semiquantitative results were obtained from the fluorescence photographs. The concentration distribution of the drug in the Christmas tree chip was measured by using the ultraviolet spectrophotometer method. DOX solution was dissolved in DMSO at a concentration of 1 mg/mL and diluted to 70 μg/mL using 0.01 M PBS. Subsequently, the prepared DOX solution was injected into one inlet of the ceramic chip, and the other inlet was simultaneously injected with drug-free PBS at a perfusion rate of 0.2 mL/h. After waiting for 5 min for the fluid to stabilize, the solution was collected from the eight chambers in the dark; the solution was collected for 1.5 h, a total of 0.2 mL was taken, and PBS was added to dilute the solution to 2 mL. The absorbance values of the DOX solution were determined at different standard concentrations by an ultraviolet spectrophotometer (UV-2600, SHIMADZU, Japan). Linear regression was performed on the absorbance values (A) with the DOX concentration (C) to obtain a standard curve. According to the standard curve, the measured absorbance of each chamber solution was converted into a concentration to determine the concentration distribution of the drug in the HA-PDMS chip.
2.7. Determination of the IC50 value of DOX
Mouse osteosarcoma cells (UMR-106), which are commonly used in osteosarcoma disease research, were cultured in DMEM supplemented with 10% FBS in a cell incubator at 37 °C with 5% CO2 [18]. To facilitate subsequent real-time observation, live cell staining was performed after the cells reached 80–90% confluence using Cell Tracker Green CMFDA dye at a concentration of 10 μM. After half an hour, the dye was washed off with PBS and re-added to the cell culture medium of choice.
Before cell seeding, we first autoclaved the HA-PDMS microfluidic chip. The stained cells were digested with trypsin and diluted to a density of 1 × 106 cells/mL. Then, we injected the cell suspension into the microchannels. After 1 day of culturing, 0.7 μg/mL DOX solution was added to one of the inlets, and the other junction was cell culture medium. The perfusion speed of both channels was 0.2 mL/h. One and three days after seeding the cells, the morphology and number of cells were observed by laser confocal microscopy and analysed by ImageJ software.
Additionally, the IC50 value of DOX was determined in a 48-well plate with the same batch of cells. UMR-106 cells were diluted to a concentration of 5000 cells/mL, and 0.5 mL of the cell suspension was seeded in a 48-well plate at 37 °C and 5% CO2 for 24 h. The DOX solution was diluted with cell culture medium into several drug solutions with concentrations ranging from 0 to 2 μg/mL and then added to the plate containing the cells from the day before. After 2 days of incubation, 200 μL of CCK-8 working solution was added followed by incubation for 1 h at 37 °C. The supernatants were also subsequently extracted and examined for their absorbance at 450 nm. Finally, the data were analysed, the IC50 curve was constructed, and the IC50 value of DOX was determined.
3. Results and discussion
3.1. Preparation of HA microfluidic chip substrates
Since it is difficult to construct high-precision microchannels on a hard and brittle ceramic surface, we used the ceramic SLA technique to directly print microchannel structures. SLA works on selective curing of photosensitive resins containing uniformly dispersed ceramic particles layer-by-layer. Although SLA is a relatively mature technology, for ultra-fine microchannel structures, the printing parameters need to be optimized. Inappropriate printing parameters, such as excessive laser power, will result in unclear grooves (Fig. S2) [19]. During printing, the photopolymer acts as a binder between the ceramic particles, making it possible to shape the part. After forming, in order to obtain a relatively high-density sample (99.45–99.95%), the part must be further processed to remove the photosensitive resin. These post-processing steps include debinding (thermal decomposition of the binder) and sintering into dense ceramic parts. However, incorrect heat treatment methods, such as an excessive degreasing rate, can cause the sample to crack. In order to determine the removal rate of organic molecules during the debinding process, thermogravimetric (TG) analysis and differential scanning calorimetry (DSC) were performed on the HA green bodies using a simultaneous thermal analyser. In Fig. S3, the thermal analysis results showed that sample degreasing could be divided into four stages, with large endothermic peaks and exothermic peaks appearing at 240 °C, 460 °C and 800 °C, and these temperatures were set as the temperature change rate transition points. When the temperature was below 800 °C, it was slowly heated under the protection of an inert atmosphere, and the organic matter volatiles were removed. After maintaining the temperature at 800 °C for 2 h, it entered the sintering stage of the HA green body at a later stage, increasing the heating rate and replacing it with an air atmosphere so that the carbonized residue was completely burned and removed. In addition, high-temperature sintering will also cause shrinkage of the sample. After several measurements, the XYZ axis scaling factors were 1.2150, 1.2344, and 1.1758, respectively, and the average linear shrinkage of the sample was 82.5%. Therefore, before printing the sample, the model needs to be enlarged according to this ratio to obtain the pre-set size.
Ultimately, through the exploration of preliminary experiments, the printing parameters of the HA slurry and the debinding and sintering curve of the HA green body were determined, which ensure that the microfluidic chip substrate with a complex morphological structure can be printed. As shown in Fig. 1B, we printed several representative microfluidic chip substrates, mainly Y-type, T-junction and Christmas tree-shaped structures. The sizes of the chips were less than 60 mm, the thicknesses of the substrates were 1253.6 ± 14.9 μm, and the depths of the grooves were 208.4 ± 3.6 μm. According to actual needs, the width of the groove was controlled between 50 and 200 μm, and the positive and negative deviations did not exceed 20 μm. For the eight-branch Christmas tree-type HA substrate, the spacing between each branch was only 0.8 mm, and the average width of the groove was 149.9 ± 12.5 μm.
3.2. Physical and chemical characterization of the ceramic substrate
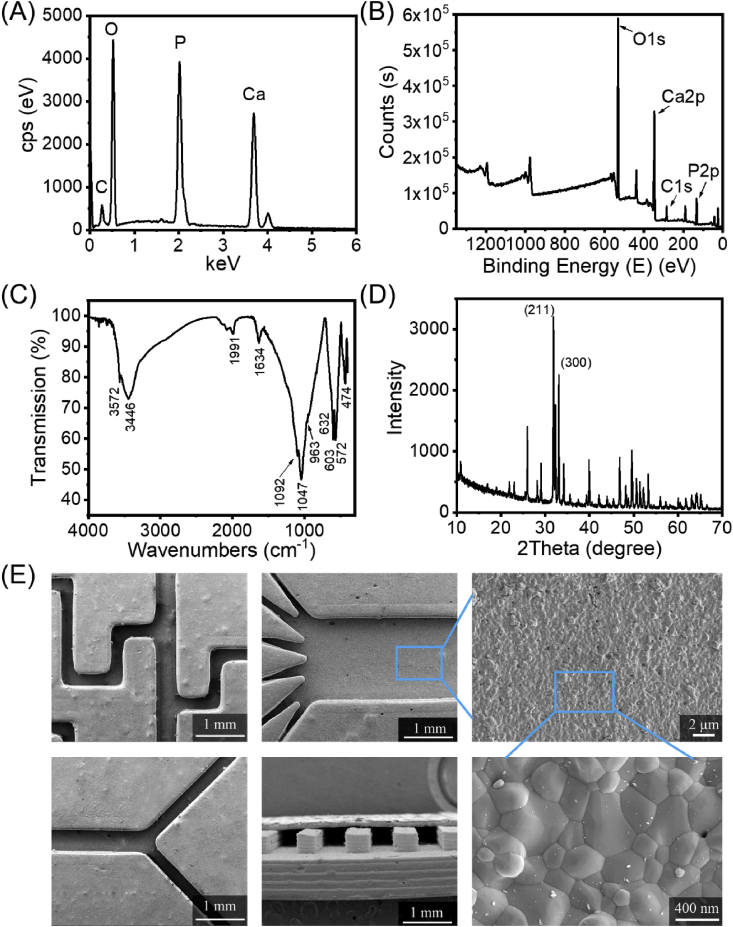
The ceramic substrate used for cell culture must be biocompatible, so it is necessary to completely remove the toxic organic resin component, leaving only high-purity HA components that are conducive to cell culture and can mimick the calcium and phosphorus environment of human bones. Here, we used a series of characterization methods to analyse the composition of the ceramic substrate to evaluate the sintered product and verify that the resin was completely removed. Energy dispersive X-ray spectroscopy (EDS) and X-ray photoelectron spectroscopy (XPS) results (Fig. 2A and B) showed that the substrate mainly contained Ca, P, and O, and the Ca/P ratio was 1.61. Hence, the presence of HA ceramics was confirmed. The ceramic substrate was ground into a powder, which was then analysed by means of Fourier transform infrared spectroscopy (FT-IR) and X-ray diffraction (XRD). The infrared spectrum (Fig. 2C) showed that the product was largely free from other impurity ions, demonstrating vibration peaks from PO43− located at 1092 (γ3), 1047 (γ3), 963 (γ1), 603 (γ4), 572 (γ4), and 474 (γ2) cm−1. As shown in Fig. 2D, the XRD spectrum of the ceramic powder sample was consistent with the standard spectrum of HA. Characteristic diffraction peaks appeared representing the crystal faces of HA, such as (002), (211), (112), (300), (202), and (310). The diffraction peaks of the (211) and (300) crystal planes were the main diffraction peaks of HA, which indicated that the sample composition was precisely HA. Moreover, there were no diffraction peaks from the other phases in the XRD spectrum, indicating that our ceramic substrate was composed of high-purity HA. In addition, sintering can also increase the strength (Fig. S4) and density of a ceramic substrate, which is conducive to the subsequent sealing process for the fabrication of ceramic chips with closed microchannels. As shown in Fig. 2E, the groove is dense and nonporous, has a good appearance with no clogging, and the widths of the grooves were consistent with the pre-set value.
Fig. 2.
Characterization of the physicochemical properties of the ceramic substrate. (A) EDS spectrum, (B) XPS spectrum, (C) FT-IR spectrum, (D) XRD spectrum and (E) scanning electron micrographs of the ceramic substrate.
3.3. Comparison of cell cultures on HA and PDMS
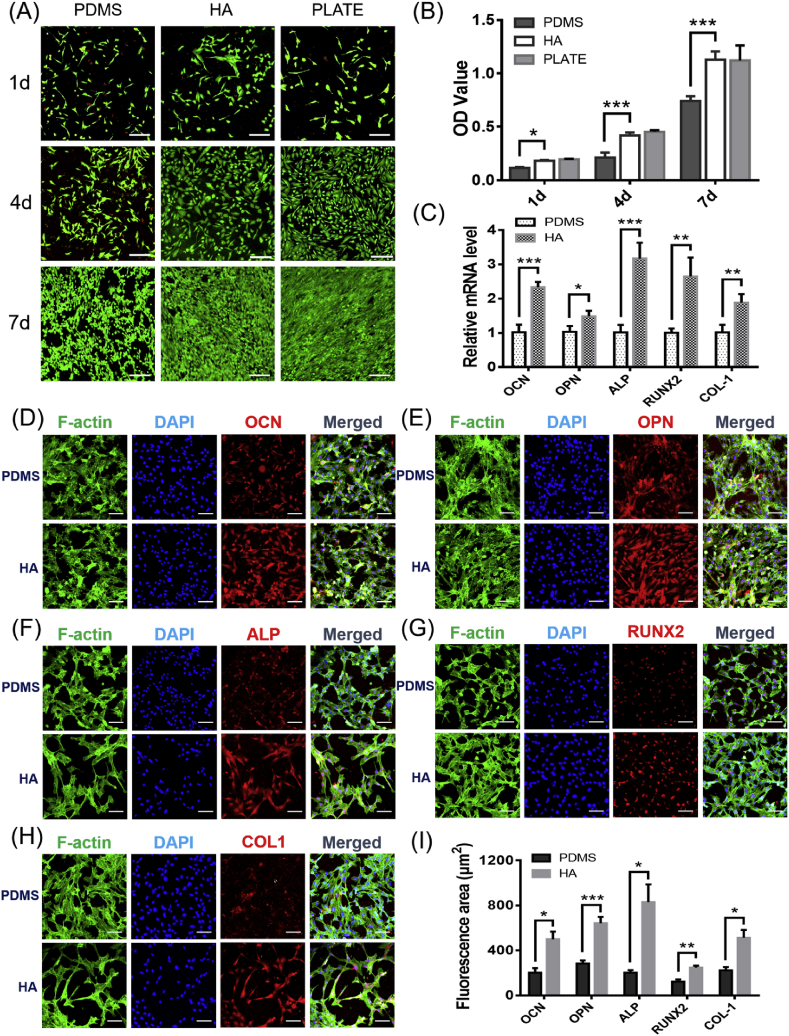
HA is compatible with the inorganic components of natural bone tissue. In terms of biological properties, HA has good biocompatibility and osteoconductivity and can provide a physiological scaffold for the formation of new bone [[20], [21], [22]]. Compared with inert PDMS, the surface of HA is polar. HA can hydrogen bond with human cells, polysaccharides and proteins and has a strong affinity for human tissues. Cell toxicity was evaluated by live/dead staining of hFOB cells, and the results are shown in Fig. 3A. The number and morphology of the cells cultured on the surface of HA were clearly better than those on the PDMS surface and not inferior to the cell multi-well plates, which indicated that HA was highly biocompatible and suitable for hFOB proliferation. The results of the CCK-8 assay of hFOB cells cultured on the surfaces of the three materials for 1, 4, and 7 days also supported this conclusion (Fig. 3B). The optical density (OD) values of the HA group was significantly higher than that of the PDMS group at 1, 4, and 7 days, and were close to the results of the multi-well plate group.
Fig. 3.
Biological evaluation of the microfluidic chip substrate. (A) Live/dead staining of hFOB cells cultured for 1, 4 and 7 days. Live cells are stained green, and dead cells are stained red. Scale bar = 200 μm. (B) CCK-8 assay of hFOB cells cultured on different materials for 1, 4 and 7 days. (C) qRT-PCR analysis of mRNA levels of osteogenic genes. Relative gene expression levels of RUNX2, ALP, COL-1, OCN and OPN in hFOB cells after 7 days of culture. Immunofluorescence staining of hFOB cells cultured on different materials for 7 days: (D) OCN, (E) OPN, (F) ALP, (G) RUNX2, and (H) COL-1. Scale bar = 100 μm. (I) Area statistics of specific red fluorescence per cell, obtained with the help of ImageJ software (total red fluorescence area divided by the number of cells). Data are presented as the mean ± standard error, *p < 0.05, **p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In order to evaluate the promotion effect of the HA substrate on the osteogenic differentiation of hFOB cells, qRT-PCR technology was selected to detect the expression levels of osteogenic differentiation-related marker genes of hFOB cells cultured on the material for 7 days. Alkaline phosphatase (ALP) is an early marker gene for cellular osteogenesis. High expression of ALP can promote the formation of mineralized active bone [23]. Insufficient expression of collagen-1 (COL-1) and structural changes can lead to congenital osteoporosis, osteogenesis imperfecta and other diseases [24]. Runx-related transcription factor-2 (RUNX2), an osteoblast transcription activator, plays an important role in regulating osteoblast differentiation and bone formation [25,26]. Osteocalcin (OCN) and osteopontin (OPN) are important bone-specific marker genes in the late stage of cellular osteogenic differentiation and are considered to be the main non-collagen components related to bone matrix deposition and mineralization, promoting the start of extracellular matrix deposition [27,28]. As shown in Fig. 3C, after 7 days of cultivation, the relative mRNA levels of OCN, OPN, ALP, RUNX2, and COL-1 in the HA group were 2.33 ± 0.15, 1.49 ± 0.16, 3.17 ± 0.46, 2.65 ± 0.55 and 1.88 ± 0.25 times than those in the PDMS group, respectively. The expression levels of the five marker genes related to osteogenic differentiation in the HA group were all higher than those in the PDMS group, indicating that the surface of the hydroxyapatite material was more conducive to the osteogenic differentiation of hFOB cells.
We also used immunofluorescence staining to specifically label the osteogenic differentiation-related proteins secreted by the cells and directly observed the morphology of the cells on the surface of the two materials and the secretion levels of the osteogenic-related proteins by means of laser confocal microscopy. As shown in Fig. 3D–H, the red fluorescent signal of the osteogenic differentiation marker protein was more visible and slightly stronger than that in the PDMS group at 7 days, which also confirmed that HA upregulated the expression of osteogenic differentiation genes. Using ImageJ software to calculate the average staining area of each cell, the red fluorescence area of the cells in the HA group was larger than that in the PDMS group (Fig. 3I), which was basically consistent with the results of qRT-PCR, indicating that HA can better promote the expression of osteogenic proteins.
Therefore, when bone-related cells are cultured in a microfluidic chip, the substrate HA can provide a more biomimic bone microenvironment than PDMS. In other words, cells cultured on HA, which contains the same components as bone, are closer to the state of the human bone microenvironment than cells cultured on PDMS.
3.4. Fabrication of the HA-PDMS microfluidic chip
The composition of HA is consistent with bone; however, if we culture cells on a pure ceramic chip, it may have several shortcomings. First, the ceramic chip is too dense, which is not conducive to cell respiration. Second, ceramic is opaque, which affects optical observation. Third, ceramic is difficult to process; that is, uncured resin is difficult to remove from the channels, which limits the channel size. Therefore, we used a composite structure of HA and PDMS. The lower layer is a HA substrate with an open groove structure that can be used for cell culture and is compatible with a bionic bone microenvironment. The upper layer is a flat PDMS piece which is transparent and gas permeable. It is conducive to the transportation of oxygen and carbon dioxide during the long-term cultivation of cells.
To obtain a ceramic-based chip with a sealed microchannel, we attached a PDMS layer onto the ceramic substrate. To bind the two layers together, we first tried the plasma oxidation treatment, which is a general bonding method for PDMS [9]. Unfortunately, this treatment did not work for the bonding between PDMS and the ceramic substrate. Therefore, we had to find an alternative method. In this study, we applied a layer of uncured PDMS prepolymer to the PDMS sheet and then attached the PDMS sheet to the polished ceramic substrate. The thickness of the PDMS adhesive layer is about 10 μm. After putting the assembly in an oven at 75 °C for 2 h, the two parts were heat-cured and ultimately glued together (Fig. 4A). As shown in Fig. S5, the PDMS sheet adhered well to the ceramic substrate without clogging of the groove. Experiments showed that higher oven temperatures could achieve stronger bonding and polishing the surface of the ceramic substrate could provide a larger contact area, resulting in a tighter bond.
Fig. 4.
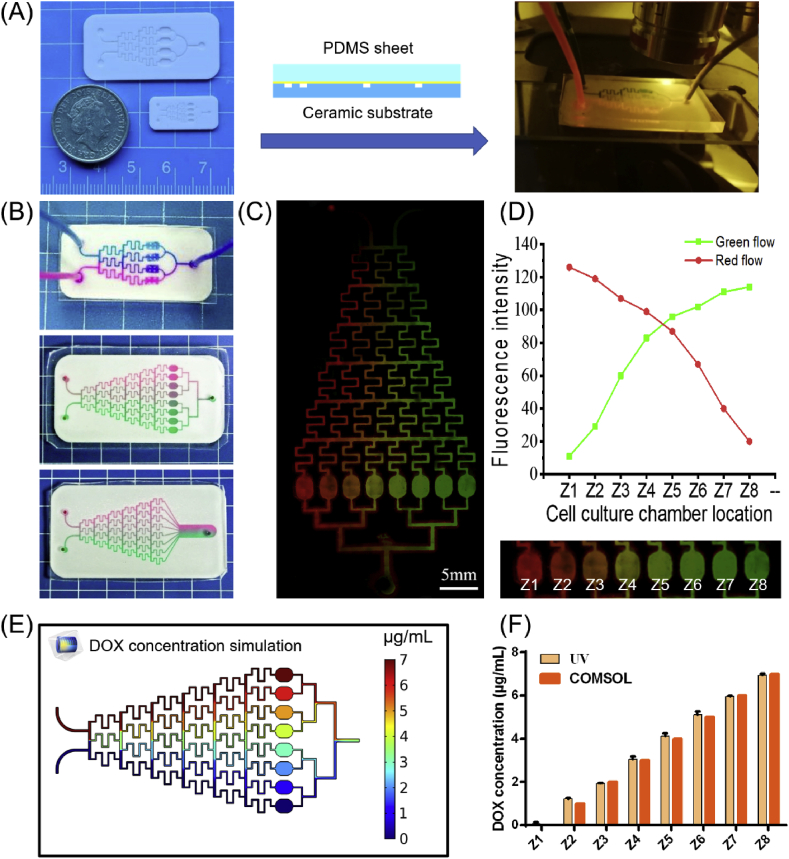
Generation and verification of concentration gradients. (A) Sealing and observation of the ceramic chips. The ceramic substrate was heat-cured to the PDMS sheet, and the two inlets were injected with different concentrations of fluorescent ink or drug and observed under a fluorescence microscope. (B) Photographs of the Christmas tree-type HA-PDMS chips. (C) Laser confocal photographs after the fluorescent inks were injected. (D) The fluorescence intensity of fluorescent inks in eight cell culture chambers. (E) Distribution of DOX drug concentration in HA-PDMS chips fitted using COMSOL simulation. (F) Comparison of the measured concentrations of DOX in eight cell culture chambers with COMSOL simulation results.
3.5. Drug screening for osteosarcoma on the HA-PDMS chip with a Christmas tree structure
To further demonstrate that the HA-PDMS chip can be used in bone-related studies, we fabricated a Christmas tree structure HA-PDMS chip and performed a drug concentration screening experiment with DOX and osteosarcoma cells.
Osteosarcoma is the most common bone-derived solid tumour, accounting for approximately 20% of primary osteosarcoma. The incidence of osteosarcoma is approximately 3/10 million people, which means that approximately 20,000 new cases are diagnosed every year worldwide [29,30]. DOX is an antitumour drug that inhibits DNA synthesis in cancer cells and is mainly used in the treatment of osteosarcoma [[31], [32], [33]]. Christmas tree structure microchannels repeatedly split streams and mix the streams by diffusion [[34], [35], [36], [37], [38]]. Using the Christmas tree-type HA-PDMS chip, we developed a high-throughput gradient drug screening of the HA-PDMS chip to quickly determine the half maximal inhibitory concentration (IC50) of DOX.
We first observed the generation of a concentration gradient on the HA-PDMS Christmas tree structure microfluidic chip. Downstream of the chip, several cell culture chambers are vertically distributed and different concentrations of liquid can flow in the culture chamber. Ideally, these concentrations are gradient distributed. To investigate the generation of the gradient, we constructed a two-span gradient by injecting two different fluorescent dye solutions into the ceramic chip, namely, 0.2 mg/mL rhodamine solution (red) and 0.1 mg/mL FITC solution (a few drops of blue ink were added to make the solution green). A good gradient distribution was formed in the HA-PDMS chip (Fig. 4B, Video V1, and Video V2). From left to right, the cell culture chambers on the HA-PDMS chips were called Z1, Z2, Z3, Z4, Z5, Z6, Z7, and Z8. Then, the chips were observed and photographed by means of a laser confocal microscope (Fig. 4C). In the case of no exposure, the fluorescence intensity was defined according to the RGB value of the photo. As shown in Fig. 4D, the red fluorescence of rhodamine gradually weakened from left to right, and the FITC solution showed a reverse trend. In fact, formation of the isocratic concentration gradient was very demanding on the fluid flow rate. Excessive flow rates would cause the solution to have insufficient time to diffuse and mix evenly, and the concentration on a single channel was not uniform. When the flow rate was too low, there would be insufficient thrust to push the fluid forward, and the fluid on both sides of the edge would no longer maintain the original concentration due to diffusion (Fig. S6). Fluid fitting analysis was performed by COMSOL software to determine the appropriate concentration range. The results showed that the perfusion flow rates of 0.0025 mL/h-0.5 mL/h could obtain an ideal isocratic gradient (Fig. 4E). To balance the experimental efficiency and prevent excessive shear rate damage to the cells, we used a perfusion rate of 0.2 mL/h for subsequent experiments.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.bioactmat.2020.07.016
The following are the supplementary data related to this article:
Then, we used this chip to generate a concentration gradient of DOX solution. Spectroscopic scanning of DOX solution by a UV spectrophotometer showed that the maximum absorption wavelength of DOX was 480 nm, and the absorbance value linearly regressed with DOX concentration in the range of 1–7 μg/mL (Fig. S7). We measured the absorbance of DOX solutions in different cell culture chambers and converted the absorbance into concentration through a standard curve. The results were similar to the concentrations obtained by simulation in COMSOL and showed an ideal gradient (Fig. 4F).
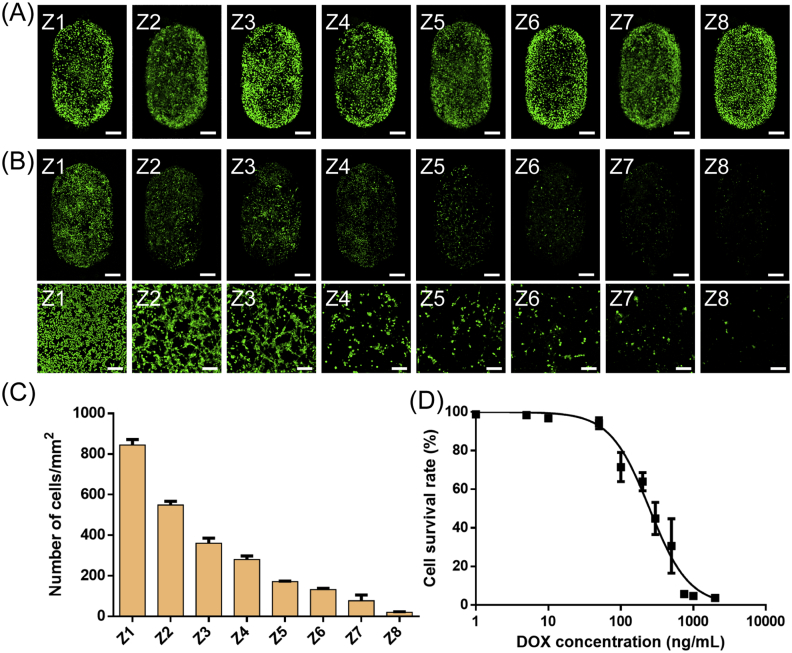
For the DOX IC50 measurement, we cultured UMR-106 cells and generated a concentration gradient of DOX on the Christmas tree structure HA-PDMS chip. As shown in Fig. 5A, after culture for one day in the chip, the UMR-106 cells spread well, and the number of cells in each chamber did not differ much. Then, one inlet of the HA-PDMS chip was injected with 0.7 μg/mL DOX solution, and the other inlet was only injected with cell culture medium. After perfusion culturing for 48 h, the chip was again placed under a laser confocal microscope. The number of cells decreased from left to right (Fig. 5B). The culture chamber Z8 had almost no cells, and the cells in the culture chamber Z1 were not affected by the drug. The number of cells in each culture chamber was counted using ImageJ software. As shown in Fig. 5C, the number of cells in culture chamber Z3 was approximately half that of chamber Z1. It could be inferred that the IC50 of DOX was the drug concentration present in chamber Z3, i.e., approximately 0.2 μg/mL. Subsequently, the IC50 curve of DOX was also determined by a conventional method in a 96-well plate. As shown in Fig. 5D, after 48 h of drug action, when the apoptotic rate of UMR-106 was 50%, the corresponding DOX concentration was 248.5 ng/mL. The results obtained by the two methods are relatively similar, which demonstrated that the HA-PDMS microfluidic chip can be used as an alternative platform for bone-related research.
Fig. 5.
Cell viability after drug addition. (A) Laser confocal photographs of cells cultured on a chip for 24 h before drug addition. Scale bar = 500 μm. (B) Laser confocal photograph of cells 48 h after drug addition. The scale bar in the first row is 500 μm, the second row is 200 μm. (C) Statistics on the number of cells in different culture chambers after three days of cultivation. (D) The IC50 curve obtained by the conventional method was measured; that is, the cells were cultured on a well plate, and the OD value was determined by adding the CCK-8 reagent.
4. Conclusion
In general, we proposed utilizing HA for the fabrication of engineered microfluidic chips to provide a highly bionic bone microenvironment and enable high-throughput drug screening. With the ceramic SLA technology, we printed a variety of ceramic substrates with clear channels and controllable sizes. Through gluing the ceramic substrate and the PDMS cover, we developed the HA-PDMS microfluidic chip with Christmas tree-like microchannel geometry. Within this chip, we successfully generated concentration gradients and determined the IC50 value of the antitumour drug DOX, which demonstrated that the HA-PDMS chip can be a potential platform for bone cell culture and bone diseases-related drug screening.
CRediT authorship contribution statement
Qiangqiang Tang: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data c. Xiaoyu Li: Validation, Formal analysis, Investigation, Data c. Chen Lai: Validation, Formal analysis. Lei Li: Supervision, Resources, Writing - review & editing, Funding acquisition. Hongkai Wu: Writing - review & editing. Yingjun Wang: Supervision, Resources, Funding acquisition. Xuetao Shi: Supervision, Resources, Writing - review & editing, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
The authors gratefully acknowledge funding from the Shenzhen Science and Technology Program (JCYJ20170815153105076, GJHZ20180411143347603), National Natural Science Foundation of China (31770107 and 21874116), Science and Technology Program of Guangdong Province (2019B010941002, 2017B090911008), Guangdong Natural Science Funds for Distinguished Young Scholars (2016A030306018), Science and Technology Program of Guangzhou (201804020060, 202007020002) and Outstanding Scholar Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR110102001).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2020.07.016.
Contributor Information
Lei Li, Email: lileilei@mail.ipc.ac.cn.
Xuetao Shi, Email: shxt@scut.edu.cn.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Walsh M.C., Kim N., Kadono Y., Rho J., Lee S.Y., Lorenzo J., Choi Y. Osteoimmunology: interplay between the immune system and bone metabolism. Annu. Rev. Immunol. 2006;24:33–63. doi: 10.1146/annurev.immunol.24.021605.090646. [DOI] [PubMed] [Google Scholar]
- 2.Takayanagi H. Osteoimmunology and the effects of the immune system on bone. Nat. Rev. Rheumatol. 2009;5:667–676. doi: 10.1038/nrrheum.2009.217. [DOI] [PubMed] [Google Scholar]
- 3.Tsukasaki M., Takayanagi H. Osteoimmunology: evolving concepts in bone-immune interactions in health and disease. Nat. Rev. Immunol. 2019;19:626–642. doi: 10.1038/s41577-019-0178-8. [DOI] [PubMed] [Google Scholar]
- 4.Patel L.R., Camacho D.F., Shiozawa Y., Pienta K.J., Taichman R.S. Mechanisms of cancer cell metastasis to the bone: a multistep process. Future Oncol. 2011;7:1285–1297. doi: 10.2217/fon.11.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shrestha J., Razavi Bazaz S., Aboulkheyr Es H., Yaghobian Azari D., Thierry B., Ebrahimi Warkiani M., Ghadiri M. Lung-on-a-chip: the future of respiratory disease models and pharmacological studies. Crit. Rev. Biotechnol. 2020:1–18. doi: 10.1080/07388551.2019.1710458. [DOI] [PubMed] [Google Scholar]
- 6.McAleer C.W., Pointon A., Long C.J., Brighton R.L., Wilkin B.D., Bridges L.R., Sriram N.N., Fabre K., McDougall R., Muse V.P., Mettetal J.T., Srivastava A., Williams D., Schnepper M.T., Roles J.L., Shuler M.L., Hickman J.J., Ewart L. On the potential of in vitro organ-chip models to define temporal pharmacokinetic-pharmacodynamic relationships. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-45656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Mello C.P.P., Rumsey J., Slaughter V., Hickman J.J. A human-on-a-chip approach to tackling rare diseases. Drug Discov. Today. 2019;24:2139–2151. doi: 10.1016/j.drudis.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 9.Pinto V., Sousa P., Catarino S.O., Correia-Neves M., Minas G. Microfluidic immunosensor for rapid and highly-sensitive salivary cortisol quantification. Biosens. Bioelectron. 2017;90:308–313. doi: 10.1016/j.bios.2016.11.067. [DOI] [PubMed] [Google Scholar]
- 10.Kim M., Moon B.U., Hidrovo C.H. Enhancement of the thermo-mechanical properties of PDMS molds for the hot embossing of PMMA microfluidic devices. J. Micromech. Microeng. 2013;23 doi: 10.1088/0960-1317/23/9/095024. [DOI] [Google Scholar]
- 11.Dall'Ara E., Schmidt R., Zysset P. Microindentation can discriminate between damaged and intact human bone tissue. Bone. 2012;50:925–929. doi: 10.1016/j.bone.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Hao S., Ha L., Cheng G., Wan Y., Xia Y., Sosnoski D.M., Mastro A.M., Zheng S.-Y. A Spontaneous 3D bone‐on‐a‐chip for bone metastasis study of breast cancer cells. Small. 2018;14 doi: 10.1002/smll.201702787. [DOI] [PubMed] [Google Scholar]
- 13.Ahn J., Lim J., Jusoh N., Lee J., Park T.-E., Kim Y., Kim J., Jeon N.L. 3D microfluidic bone tumor microenvironment comprised of hydroxyapatite/fibrin composite. Frontiers in Bioengineering and Biotechnology. 2019;7 doi: 10.3389/fbioe.2019.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fathi M.H., Hanifi A., Mortazavi V. Preparation and bioactivity evaluation of bone-like hydroxyapatite nanopowder. J. Mater. Process. Technol. 2008;202:536–542. doi: 10.1016/j.jmatprotec.2007.10.004. [DOI] [Google Scholar]
- 15.Diaz Lantada A., de Blas Romero A., Schwentenwein M., Jellinek C., Homa J., Predestinacion Garcia-Ruiz J. Monolithic 3D labs- and organs-on-chips obtained by lithography-based ceramic manufacture. Int. J. Adv. Manuf. Technol. 2017;93:3371–3381. doi: 10.1007/s00170-017-0443-6. [DOI] [Google Scholar]
- 16.Donahue H.J., Li Z.Y., Zhou Z.Y., Yellowley C.E. Differentiation of human fetal osteoblastic cells and gap junctional intercellular communication. Am. J. Physiol. Cell Physiol. 2000;278(2):C315–C322. doi: 10.1152/ajpcell.2000.278.2.C315. [DOI] [PubMed] [Google Scholar]
- 17.Li L., Lv X., Ostrovidov S., Shi X., Zhang N., Liu J. Biomimetic microfluidic device for in vitro antihypertensive drug evaluation. Mol. Pharm. 2014;11:2009–2015. doi: 10.1021/mp5000532. [DOI] [PubMed] [Google Scholar]
- 18.Hickman J., McElduff A. Insulin promotes growth of the cultured rat osteosarcoma cell line UMR-106-01: an osteoblast-like cell. Endocrinology. 1989;124(2):701–706. doi: 10.1210/endo-124-2-701. [DOI] [PubMed] [Google Scholar]
- 19.Chartier T., Dupas C., Lasgorceix M., Brie J., Champion E., Delhote N., Chaput C. Additive manufacturing to produce complex 3D ceramic parts. Journal of Ceramic Science and Technology. 2015;6:95–104. doi: 10.4416/jcst2014-00040. [DOI] [Google Scholar]
- 20.Putlyaev V.I., Safronova T.V. A new generation of calcium phosphate biomaterials: the role of phase and chemical compositions. Glass Ceram. 2006;63:99–102. doi: 10.1007/s10717-006-0049-1. [DOI] [Google Scholar]
- 21.van Blitterswijk C.A., Grote J.J., Kuypers W., Blok-van Hoek C.J., Daems W.T. Bioreactions at the tissue/hydroxyapatite interface. Biomaterials. 1985;6:243–251. doi: 10.1016/0142-9612(85)90020-1. [DOI] [PubMed] [Google Scholar]
- 22.Szczes A., Holysz L., Chibowski E. Synthesis of hydroxyapatite for biomedical applications. Adv. Colloid Interface Sci. 2017;249:321–330. doi: 10.1016/j.cis.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Pan T., Song W., Cao X., Wang Y. 3D bioplotting of gelatin/alginate scaffolds for tissue engineering: influence of crosslinking degree and pore architecture on physicochemical properties. J. Mater. Sci. Technol. 2016;32:889–900. doi: 10.1016/j.jmst.2016.01.007. [DOI] [Google Scholar]
- 24.Steinmetz N.J., Bryant S.J. The effects of intermittent dynamic loading on chondrogenic and osteogenic differentiation of human marrow stromal cells encapsulated in RGD-modified poly(ethylene glycol) hydrogels. Acta Biomater. 2011;7:3829–3840. doi: 10.1016/j.actbio.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X., Li H., Lin C., Ning C., Lin K. Synergetic topography and chemistry cues guiding osteogenic differentiation in bone marrow stromal cells through ERK1/2 and p38 MAPK signaling pathway. Biomaterials Science. 2018;6:418–430. doi: 10.1039/c7bm01044c. [DOI] [PubMed] [Google Scholar]
- 26.Jiang K., Huang D., Zhang D., Wang X., Cao H., Zhang Q., Yan C. Investigation of inulins from the roots of Morinda officinalis for potential therapeutic application as anti-osteoporosis agent. Int. J. Biol. Macromol. 2018;120:170–179. doi: 10.1016/j.ijbiomac.2018.08.082. [DOI] [PubMed] [Google Scholar]
- 27.Wang T., Qian S., Zha G.-C., Zhao X.J., Ding L., Sun J.Y., Li B., Liu X.Y. Synergistic effects of titania nanotubes and silicon to enhance the osteogenic activity. Colloids Surf. B Biointerfaces. 2018;171:419–426. doi: 10.1016/j.colsurfb.2018.07.052. [DOI] [PubMed] [Google Scholar]
- 28.Takafuji Y., Tatsumi K., Ishida M., Kawao N., Okada K., Matsuo O., Kaji H. Plasminogen activator inhibitor-1 deficiency suppresses osteoblastic differentiation of mesenchymal stem cells in mice. J. Cell. Physiol. 2019;234(6):9687–9697. doi: 10.1002/jcp.27655. [DOI] [PubMed] [Google Scholar]
- 29.Kansara M., Teng M.W., Smyth M.J., Thomas D.M. Translational biology of osteosarcoma. Nat. Rev. Canc. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 30.Kager L., Tamamyan G., Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017;13:357–368. doi: 10.2217/fon-2016-0261. [DOI] [PubMed] [Google Scholar]
- 31.Kamba S.A., Ismail M., Hussein-Al-Ali S.H., Ibrahim T.A.T., Zakaria Z.A.B. In vitro delivery and controlled release of doxorubicin for targeting osteosarcoma bone cancer. Molecules. 2013;18:10580–10598. doi: 10.3390/molecules180910580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown H.K., Schiavone K., Gouin F., Heymann M.-F., Heymann D. Biology of bone sarcomas and new therapeutic developments. Calcif. Tissue Int. 2018;102:174–195. doi: 10.1007/s00223-017-0372-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buondonno, Gazzano E., Tavanti E., Chegaev K., Kopecka J., Fanelli M., Rolando B., Fruttero R., Gasco A., Hattinger C., Serra M., Riganti C. Endoplasmic reticulum-targeting doxorubicin: a new tool effective against doxorubicin-resistant osteosarcoma. Cell. Mol. Life Sci. 2019;76:609–625. doi: 10.1007/s00018-018-2967-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeon N.L., Dertinger S.K.W., Chiu D.T., Choi I.S., Stroock A.D., Whitesides G.M. Generation of solution and surface gradients using microfluidic systems. Langmuir. 2000;16:8311–8316. doi: 10.1021/la000600b. [DOI] [Google Scholar]
- 35.Dertinger S.K.W., Chiu D.T., Jeon N.L., Whitesides G.M. Generation of gradients having complex shapes using microfluidic networks. Anal. Chem. 2001;73:1240–1246. doi: 10.1021/ac001132d. [DOI] [Google Scholar]
- 36.Shi X., Ostrovidov S., Shu Y., Liang X., Nakajima K., Wu H., Khademhosseini A. Microfluidic generation of polydopamine gradients on hydrophobic surfaces. Langmuir. 2014;30:832–838. doi: 10.1021/la4041216. [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Liu Z., Pang Y. Concentration gradient generation methods based on microfluidic systems. RSC Adv. 2017;7:29966–29984. doi: 10.1039/c7ra04494a. [DOI] [Google Scholar]
- 38.Hong B., Xue P., Wu Y., Bao J., Chuah Y.J., Kang Y. A concentration gradient generator on a paper-based microfluidic chip coupled with cell culture microarray for high-throughput drug screening. Biomed. Microdevices. 2016;18:21. doi: 10.1007/s10544-016-0054-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.