Abstract
Cellular homeostais, that is normally maintained through autophagy, is disrupted in alcoholic liver disease (ALD). Because autophagy and exosome biogenesis share common elements, we hypothesized that increased exosome production in ALD may be linked to disruption of autophagic function. We found impaired autophagy both in ALD and alcoholic hepatitis (AH) mouse models and human livers with ALD as indicated by increased hepatic p62 and LC3-II levels. Alcohol reduced autophagy flux in vivo in chloroquine-treated mice as well as in vitro in hepatocytes and macrophages treated with bafilomycin A. Our results revealed that alcohol targets multiple steps in the autophagy pathway. Alcohol-related decrease in mechanistic target of rapamycin (mTOR) and Ras homolog enriched in brain (Rheb), that initiate autophagy, correlated with increased Beclinl and autophagy-related protein 7 (Atg7), proteins involved in phagophore-autophagosome formation, in ALD. We found that alcohol disrupted autophagy function at the lysosomal level through decreased lysosomal-associated membrane protein 1 (LAMP1) and lysosomal-associated membrane protein 2 (LAMP2) in livers with ALD. We identified that micro-RNA 155 (miR-155), that is increased by alcohol, targets mTOR, Rheb, LAMP1, and LAMP2 in the authophagy pathway. Consistent with this, miR-155-deficient mice were protected from alcohol-induced disruption of autophagy and showed attenuated exosome production. Mechanistically, down-regulation of LAMP1 or LAMP2 increased exosome release in hepatocytes and macrophages in the presence and absence of alcohol. These results suggested that the alcohol-induced increase in exosome production was linked to disruption of autophagy and impaired autophagosome and lysosome function. Conclusion: Alcohol affects multiple genes in the autophagy pathway and impairs autophagic flux at the lysosome level in ALD. Inhibition of LAMP1 and LAMP2 promotes exosome release in ALD. We identified miR-155 as a mediator of alcohol-related regulation of autophagy and exosome production in hepatocytes and macrophages. (Hepatology 2019;70:2123–2141).
Alcoholic liver disease (ALD) affects millions of individuals worldwide. ALD is driven by significant cellular stress mediated by the direct effects of alcohol, alcohol metabolites, gut-derived endotoxins, and alcohol-induced danger molecules.(1,2) Recent evidence suggests that alcohol has differential effects on autophagy that is a critical pathway for maintaining homeostasis by eliminating cellular waste. Acute alcohol treatment activates autophagy,(2) whereas impaired autophagic flux has been reported in chronic mouse models of ALD.(3–7) However, the molecular mechanisms underlying the effect of chronic alcohol on autophagic flux and lysosome integrity are still unclear. Therefore, in this study, we aimed to elucidate the effects of chronic alcohol at different stages of the autophagic pathway.
The autophagy lysosomal pathway maintains cell viability by targeted degradation of damaged cellular organelles in autolysosomes.(3,8) Efficient autophagy involves a coordinated cascade of molecular events involving mechanistic target of rapamycin (mTOR) kinase activation, Ras homolog enriched in brain (Rheb), and lipid conjugation of soluble microtubule-associated protein light chain 3 (LC3)-I to LC3-II, which attaches to the autophagosome membrane.(9) In this process, p62 is a substrate for LC3-ll before multivesicular bodies (MVBs) are formed.(10) The autophagosome matures by fusing with lysosomes to create autophagolysosomes where its selected cargo is degraded.(11) Lysosome-associated membrane protein 2 (LAMP2) is essential for autophagosome lysosome fusion during autophagy.(11)
Recent studies indicated that, in addition to autophagy, the exosomal pathway can also eliminate pathogenic or damaged proteins and nucleic acids.(10,12) Exosomes are derived from MVBs and play functional roles in cellular communication through transfer of proteins/nucleic acids.(13–16) Recent studies suggest that autophagy and MVB pathways may intersect with exosome biogenesis.(14)
We previously showed that micro-RNA 155 (miR-155) is induced by alcohol and that miR-155 knockout (KO) mice were protected from ALD.(17,18) miR-155 was shown to target Rheb and mTOR to induce autophagy under hypoxic conditions.(19) In this study, we hypothesized that miR-155 regulates autophagy in ALD. We found impaired autophagy at the lysosomal functional level in ALD. We also show that miR-155 KO mice are protected from alcohol-induced impairment in autophagy and that miR-155 regulates exosome release. Our findings highlight the role of miR-155 in the crosstalk between autophagy and exosome production and define a mechanistic role for alcohol-related dysregulation of autophagy in ALD.
Materials and Methods
ANIMAL STUDIES
C57BL/6 wild-type (WT) and miR-155 KO mice were purchased from The Jackson Laboratory (Bar Harbor, ME).(17,18) Mice were housed at the University of Massachusetts Medical School (UMMS) animal facility in compliance with all Institutional Animal Use and Care Committee directives. For ALD, 8- to 10-week-old WT and miR-155 KO female mice (n = 8–10) received either a Lieber-DeCarli diet containing 5% (v/v) alcohol or an isocaloric dextrin maltose diet for 5 weeks.(17) Alcoholic hepatitis (AH) in mice was induced using the National Institute on Alcohol Abuse and Alcoholism (NIAAA) model.(20) After experimental treatments, blood and liver samples were collected, processed immediately, and stored at −80°C for further analyses.(12,17,18)
CELL LINES AND ISOLATION OF PRIMARY HEPATOCYTES AND KUPFFER CELLS FROM MOUSE LIVERS
For in vitro studies, mouse Hepa1–6 hepatocytes and RAW264.7 macrophage cell lines were used. Primary hepatocytes and Kupffer cells (KCs) were harvested from mice following liver perfusions as we described.(12,17,18)
PATIENT SAMPLES
Human liver sample was obtained from the National Institutes of Health Liver Tissue Cell Distribution System (Minneapolis, MN; Pittsburgh, PA; Richmond, VA). Liver samples were from 3–5 control subjects and 6–8 patients with cirrhosis, superimposed with AH. Number of exosomes were analyzed from the healthy and patient serum with informed consent from 6 healthy donors and 8 patients with acute AH with approval from the UMMS Institutional Review Board for Protection of Human Subjects in Research. The criteria to define patients with AH were based on “Recommendation from the NIAAA Alcoholic Hepatitis Consortia.”(21)
WESTERN BLOTTING ANALYSIS AND RT-qPCR
Western blotting was performed on total proteins extracted from mouse livers, human livers, exosomes, and cell lines. Equal amounts of total proteins were resolved on a 10% acrylamide gel and transferred to nitrocellulose membranes. Membranes were then probed with specific antibodies as indicated. For gene expression analysis, we used RT-qPCR methods as described.(17) Total RNA was extracted using the miRNeasy isolation kit (Qiagen, USA).
EXOSOME ISOLATION AND QUANTIFICATION
Exosomes from serum and cell-culture supernatants were isolated as described.(16) Briefly, serum samples were centrifuged at 1,500g for 10 minutes to remove cellular debris and 10,000g for 20 minutes. Samples were filtered through 0.8- and 0.22-μm filters (Millipore, USA). To precipitate exosomes, we used Exoquick-TC (cell culture) or Exoquick (serum samples) reagent according to the manufacturer’s guidelines (SBI, USA). After exosome precipitation, exosome pellets were resuspended in phosphate-buffered saline then quantified using NanoSight Tracking Analysis (NTA).
TRANSFECTION
For miR-155 overexpression and knockdown of lysosomal-associated membrane protein 1 (LAMP1) and lysosomal-associated membrane protein 2 (LAMP2) RAW 264.7 macrophages and Hepa 1–6 hepatocytes were transfected with a negative control, a miR-155 mimic, or small interfering RNA (siRNA) targeting for LAMP1 and LAMP2 using the methods described.(17)
BAFILOMYCIN A TREATMENT
RAW macrophages and Hepa1–6 hepatocytes were treated with 50 mM of alcohol for 24 hours. In the last 12 hours, cells were treated in the presence or absence of 100 nM of bafilomycin A. Exosome-depleted fetal bovine serum was used. Cells were harvested for western blotting analysis, and the cell-free supernatant was analyzed by NTA for exosomes.
STATISTICAL ANALYSIS
Statistical significance was determined using analysis of variance. Each experiment was repeated at least three times to determine the biological significance. Data are shown as mean ± SEM and were considered statistically significant at P < 0.05. GraphPad Prism software (version 7; GraphPad Software Inc.) was used for analysis.
Additional details on all the methods are provided in the Supporting Methods.
Results
INCREASED ACCUMULATION OF LC3-II AND p62 SUGGEST IMPAIRED AUTOPHAGY IN ALD AND AH
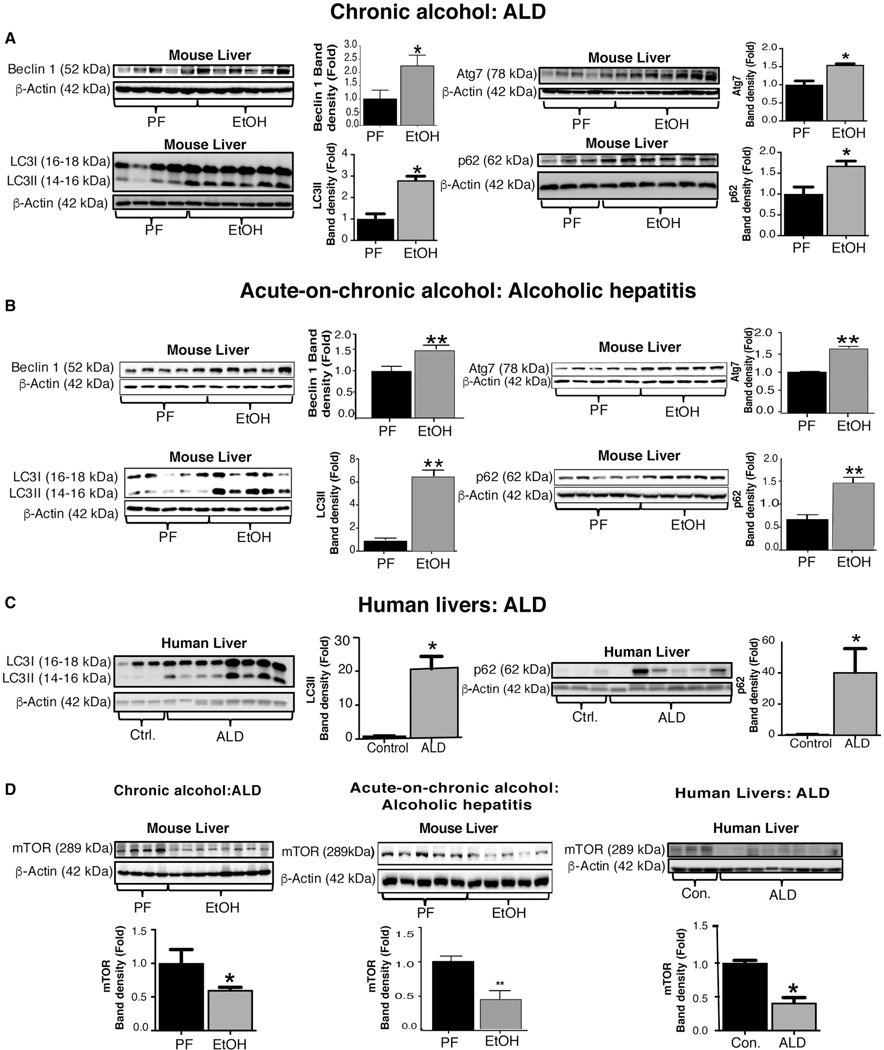
The role of autophagy in ALD is yet to be fully understood.(3,4) Here, we evaluated the different stages of the autophagy pathway in an ALD model (after chronic alcohol treatment) and in an AH model (after acute-on-chronic feeding) in mice. We found that Beclin-1 protein, an upstream activator of autophagy, was increased in ALD and AH in mice compared to controls (Fig. 1A,B). LC3 lipidation is crucial for autophagosome formation and maturation; hence, we assessed the expression of two LC3 lipidation proteins, autophagy-related protein 3 (Atg3) and autophagy-related protein 7 (Atg7). We found no significant changes in Atg3 protein levels (Supporting Fig. S1A); however, Atg7 was significantly increased in alcohol-fed mice in both models (Fig. 1A,B). This correlated with significantly increased LC3-II protein expression in both models (Fig. 1A,B). According to the autophagy guidelines, LC3 measures cannot distinguish between activation or impairement of the autophagy flux.(22) Therefore, we checked p62 levels and found significant accumulation of p62 in both ALD and AH mouse models, indicating a potential impairement of autophagy (Fig. 1A,B).
FIG. 1.
ALD in mice and humans is associated with impaired autophagic flux. (A,B) Total liver proteins were extracted from pair-fed (PF), chronic alcohol diet (EtOH), and acute-on-chronic alcohol (EtOH) fed mice (n = 8–10/group) and analyzed by western blotting using β-actin as a loading control. Protein levels of Beclinl, Atg7, LC3-I, LC3-II, and p62 in chronic alcohol (A) and in acute-on-chronic alcohol-fed (B) mice. Western blotting analysis for LC3-I, LC3-II, and p62 from livers of control subjects and ALD patients (C). mTOR protein levels in mouse livers in chronic alcohol- and in acute-on-chronic alcohol-fed mice and in ALD patients (D). The densitometry analysis is shown as bar diagrams. Human liver sample data are representative of 3 control donors and 6–8 patients with cirrhosis and superimposed AH. *P < 0.05; **P < 0.01. Abbreviations: Con., control; ctrl., control.
To validate our findings from the mouse models, we next tested liver samples from ALD patients and found a significant increase in both LC3-II and p62 (Fig. 1C) protein levels compared to controls, suggesting disruption of autophagy.
THE mTOR PATHWAY IS INHIBITED IN MICE AND IN ALD PATIENTS AFTER CHRONIC ALCOHOL CONSUMPTION
Autophagy is a complex cascade of events with multiple levels of regulation 3–5 Various stresses, such as nutrient deprivation, trigger autophagy flux initiated by inhibition of the mTOR pathway.(23) Given the observation of increased Beclin1 and Atg7, we tested upstream components of the autophagic cascade and found significantly lower mTOR (Fig. 1D) and Rheb (Supporting Fig. S1B) protein expression in alcohol-fed mice compared to controls in the ALD and AH mouse models. Consistent with the results in mice, protein levels of mTOR (Fig. 1D) and Rheb (Supporting Fig. S1B) were also decreased in patients with ALD. To further assess whether mTOR activity was inhibited in ALD, we evaluated phosphorylation levels of mTOR substrate proteins, S6p70 and 4E-binding protein 1 (4EBP1). We found a significant decrease in phosphorylation of S6p70 (serine 235/236; Supporting Fig. S1C) and 4EBP1 (serine 65; Supporting Fig. S1D) proteins in alcohol-fed mice and in patients with ALD compared to controls. These results indicate inhibition of the mTOR pathway, an early event in autophagy, in ALD in both mice and humans.
miR-155 DEFICIENCY PREVENTS THE ALCOHOL-INDUCED mTOR INHIBITION AND p62 AND LC3-II ACCUMULATION IN ALD
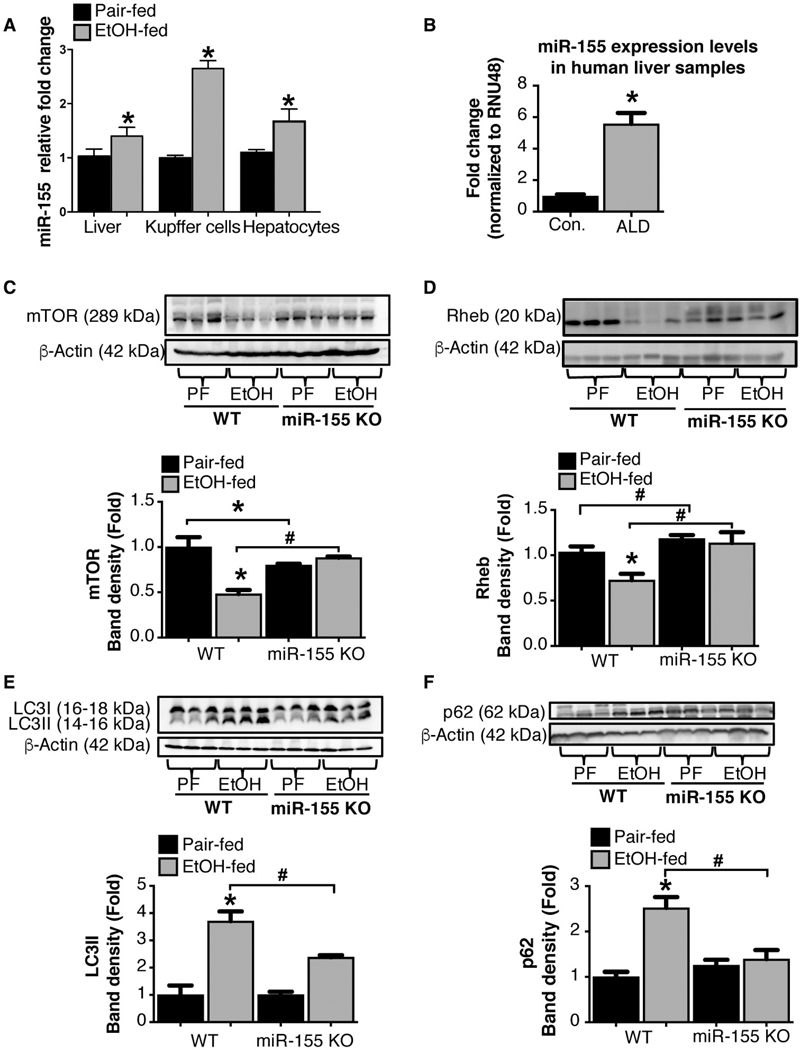
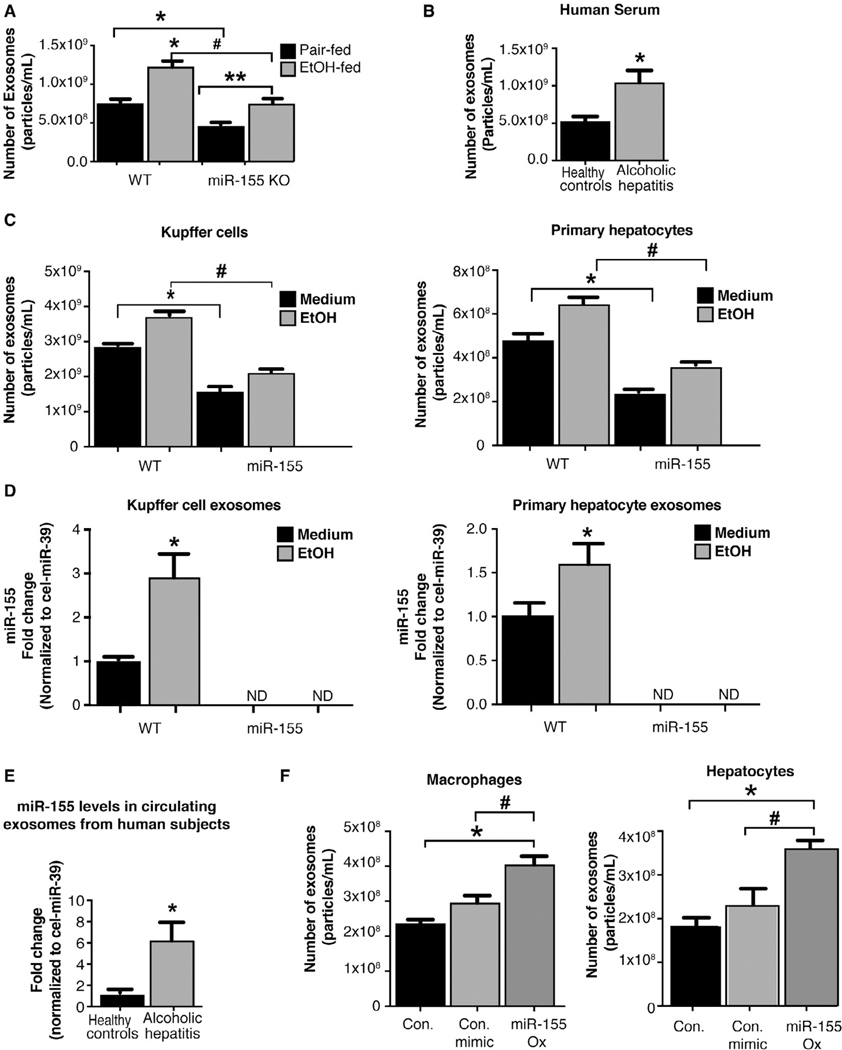
Our previous studies demonstrated a pathogenic role for miR-155 in ALD.(17) Consistent with our earlier studies, we observed an increase in miR-155 expression in total liver, KCs, and hepatocytes isolated from alcohol-fed mice compared to control mice (Fig. 2A). Next, we evaluated miR-155 in human ALD and found a significant increase in hepatic miR-155 expression with ALD compared to control subjects (Fig. 2B).
FIG. 2.
miR-155 deficiency prevents alcohol-induced impairment in autophagic flux in mice. Total RNA from livers, KCs, and hepatocytes from mice as well as liver samples from ALD patients (n = 8–10) analyzed for miR-155 expression by RT-qPCR (A,B). qCT values were normalized to snoRNA-202 for mouse samples and to RNU48 for human samples. Total liver protein was extracted from pair-fed (PF) or chronic alcohol-fed (EtOH) WT and miR-155 KO mice (n = 8–10) and analyzed by western blotting using β-actin as a loading control. Immunoblottings were probed with antibodies for mTOR (C), Rheb (D), LC3-I and LC3-II (E), and p62 (F). The densitometry analysis is shown as bar diagrams. *,#P < 0.05. Abbreviation: Con., control.
A recent study indicating that miR-155 regulates mTOR and Rheb, key molecules in autophagy,(19) prompted us to delineate the role of miR-155 in autophagy in ALD. We found that miR-155 deficiency prevented alcohol-induced reduction in mTOR (Fig. 2C) and Rheb (Fig. 2D) protein levels compared to WT mice. Furthermore, alcohol-induced increase in hepatic LC3-II (Fig. 2E) and p62 (Fig. 2F) protein expression were attenuated in miR-155 KO compared to WT mice. Because p62 levels are regulated both at the transcriptional and posttranslational levels, we evaluated mRNA levels and found an increase in p62 mRNA levels only in WT and not in miR-155 KO mice after alcohol feeding (Supporting Fig. S2A). The increased accumulation of LC3- and p62-positive autophagosomes in livers of WT mice after chronic alcohol diet was confirmed by fluorescent immunohistochemistry (IHC; Supporting Fig. S2B). There was a reduction in p62-positive autophagosomes in livers of miR-155 KO compared to WT mice, indicative of normal autophagy in miR-155 KO mice after alcohol feeding (Supporting Fig. S2B). We also found that miR-155 KO mice were protected from alcohol-induced liver damage (ala-nine aminotransferase; ALT) and steatohepatitis (SH) indicated by reduced fat accumulation and attenuated tumor necrosis factor alpha, interleukin-1p, and monocyte chemoattractant protein 1 gene induction by alcohol treatment (Supporting Fig. S3A–E).
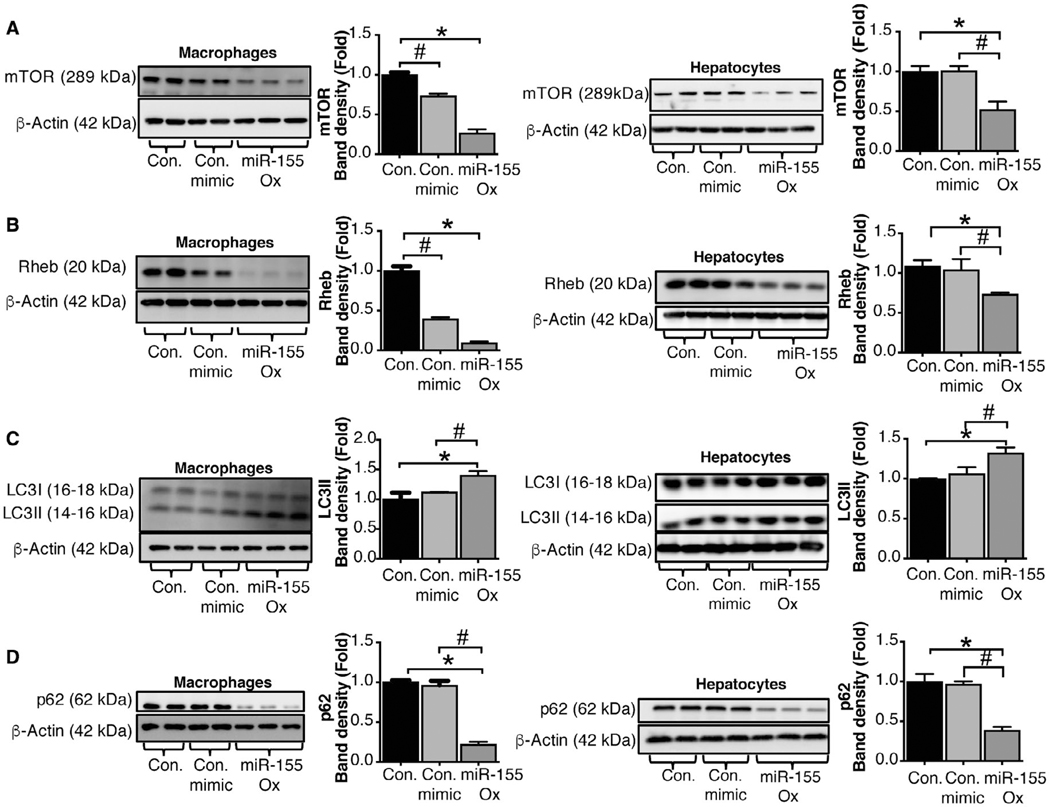
OVEREXPRESSION OF miR-155 INHIBITS THE mTOR PATHWAY BOTH IN MACROPHAGES AND HEPATOCYTES
To delineate the role of miR-155 in mTOR and early autophagy pathways at the cellular level, we performed simulation experiments in macrophages (RAW264.7) and hepatocytes (Hepa1–6). Transfection of an miR-155 mimic into macrophages and hepatocytes significantly increased miR-155 levels compared to a control miRNA mimic (Supporting Fig. S4A,B) leading to significantly decreased protein levels of mTOR (Fig. 3A) and Rheb (Fig. 3B). We also found a significant increase in LC3-II (Fig. 3C) and a decrease in p62 (Fig. 3D) protein levels in macrophages and hepatocytes treated with a miR-155 mimic and not with a control miRNA mimic. These results demonstrate a role for miR-155 in regulation mTOR and Rheb in both macrophages and hepatocytes.
FIG. 3.
miR-155 regulates autophagy in macrophages and hepatocytes by targeting the mTOR pathway. Macrophages (RAW 264.7) or hepatocytes (Hepa1–6) were untreated or transfected with either a miR-155 mimic or a control mimic. Protein levels from untreated, control mimic, and miR-155 mimic transfected macrophages and hepatocytes were analyzed by western blotting for mTOR (A), Rheb (B), LC3-I and LC3-II (C), and p62 (D) using β-actin as a loading control (n = 7). The densitometry analysis is shown as bar diagrams. *,#P < 0.05. Abbreviations: Con., control; ox, overexpression.
CHRONIC ALCOHOL IMPAIRS AUTOPHAGY AT THE LEVEL OF LYSOSOME-ASSOCIATED MEMBRANE PROTEINS THROUGH miR-155
Our observation that alcohol reduces both mTOR and Rheb that normally increases autophagy seemed to be contradictory to the finding of increased LC3-II and p62 that indicates disruption of autophagy.
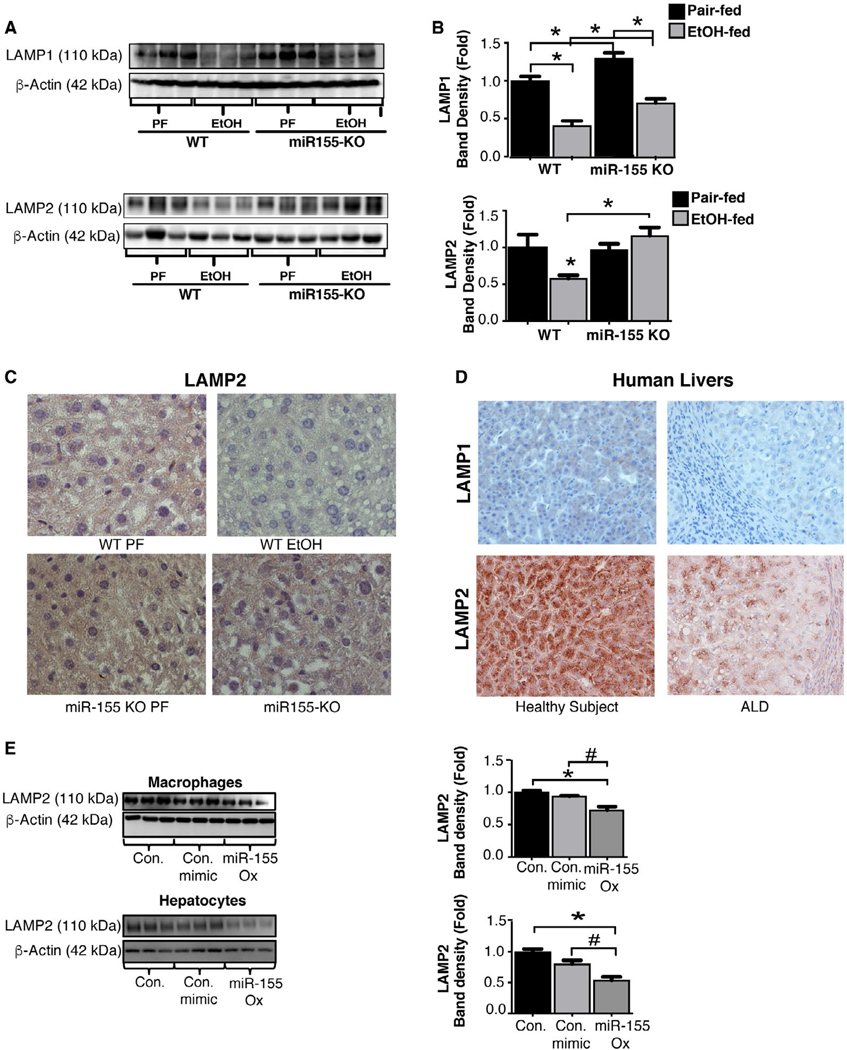
However, alcohol and/or miR-155 may independently modulate different steps in autophagy and still result in an overall disruption of the autophagy process. Therefore, we evaluated the effect of alcohol on the step of fusion of lysosomes with autophagosomes, which is mediated by lysosome-associated membrane proteins (LAMP1 And LAMP2).(24) Our bioinformatic analyses revealed that miR-155 has binding sites at the 3’ unstranslated region (3’UTR) of LAMP1 and LAMP2 genes (Supporting Fig. S5A,B). Thus, we hypothesized that alcohol impairs autophagy by affecting lysosome function through miR-155-targeted suppression of LAMP1 and LAMP2. Consistent with this hypothesis, we found significantly decreased protein levels of LAMP1 and LAMP2 (Fig. 4A,B) in livers of alcohol-fed WT mice. Compared to WT mice, alcohol-fed miR-155 KO mice showed no reduction in hepatic LAMP2 protein expression by western blotting (Fig. 4A) and IHC (Fig. 4C) after alcohol feeding. The alcohol-induced reduction in LAMP1 protein levels observed in WT mice was partially prevented in miR-155 KO mice (Fig. 4B). Finally, livers of patients with ALD also exhibited decreased expression of LAMP1 and LAMP2 compared to control subjects as determined by IHC (Fig. 4D). The finding that miR-155 KO mice had preserved normal LAMP2 protein expression in ALD suggests normal autolysosomal degradation in these mice during autophagy. To determine whether miR-155 directly affects LAMP2 in macrophages and hepatocytes, we performed in vitro mechanistic studies. We found that miR-155 overexpression in macrophages (RAW264.7) or hepatocytes (Hepa1–6) significantly decreased LAMP2 protein expression (Fig. 4E), suggesting that miR-155 targets LAMP2 to modulate lysosome function.
FIG. 4.
Chronic alcohol-induced increase in miR-155 regulates autophagy at the autophagosome formation stage by targeting lysosome-associated membrane proteins. Total liver protein was extracted from pair-fed (PF) or chronic alcohol-fed (EtOH) WT and miR-155 KO mice (n = 8–10) and analyzed for LAMP2 and LAMP1 (A,B) by western blotting using β-actin as a loading control. Representative IHC images of LAMP2 from WT and miR-155 KO mice in livers (C). LAMP1 and LAMP2 IHC images from livers of healthy subjects and from patients with ALD (n = 64) (D). Macrophages (RAW 264.7) or hepatocytes (Hepa1–6) were untreated or transfected with either a miR-155 mimic or a control mimic and the LAMP2 protein levels determined by western blotting (E). *,#P < 0.05. Abbreviations: Con., control; ox, overexpression.
We also evaluated how alcohol affects other modulators of lysosomal function. Transcription factor EB (TFEB), a master regulator of lysosomal biogenesis,(7) was significantly decreased in alcohol-fed mice and in patients with ALD compared to controls (Supporting Fig. S6A,B). Furthermore, mRNA and protein levels of TFEB in miR-155 KO mice were significantly higher as compared to WT mice after chronic alcohol feeding (Supporting Fig. S6C,D). To further assess TFEB regulation through miR-155, we performed in silico analysis and found no miR-155 direct binding site at the 3’UTR region of TFEB (Target scan, and miRBase), suggesting that miR-155 regulates TFEB indirectly. A nonsignificant decrease in expression of Ras-related protein Rab-7a (Rab7), another lysosomal protein involved in the biogenesis of lysosome compartments,(5) was observed in livers of alcohol-fed mice (Supporting Fig. S6E). Together, these results showed that alcohol inhibits expression of key lysosomal proteins, (LAMP1, LAMP2, and TFEB) and miR-155 is involved in LAMP1 and LAMP2 regulation.
miR-155 KO MICE EXHIBIT ATTENUATED EXOSOME RELEASE IN ALD
Our findings indicate that alcohol-induced impairment of autophagy occurs at the level of lysosomal proteins and is mediated by miR-155. Emerging studies suggest a potential crosstalk between the autophagic and exosomal secretory pathways.(10) We found increased circulating exosomes after alcohol feeding in WT mice compared to pair-fed controls; however, this was significantly attenuated in miR-155 KO mice even after chronic alcohol feeding (Fig. 5A). The alcohol-related increase in number of circulating exosomes was also observed in sera of human AH patients (Fig. 5B).
FIG. 5.
Alcohol-induced increased miR-155 regulates the cellular release of exosomes. NTA on the number of exosomes from sera of WT and miR-155 KO mice (n = 19; A) and the number of serum exosomes from AH patients (n = 8; B). NTA quantification of the number of exosomes in supernatants from KCs and hepatocytes (n = 15) after alcohol (50 mM, 24 hours) treatment (C). qPCR on miR-155 expression levels in exosomes isolated from KCs and primary hepatocytes (D) and in exosomes from sera of healthy subjects and patients with AH (n = 5–7; E); miR-155 levels were normalized to a spiked-in synthetic Cel-miR-39 used as an internal control. Macrophages (RAW 264.7) or hepatocytes (Hepa1–6) were untreated or transfected with either a miR-155 mimic or a control mimic and the number of exosomes in the supernatants quantified by NTA (n = 9) (F). *,#P < 0.05; **P < 0.01. Abbreviations: Con., control; ND, not detected; ox, overexpression.
Based on these findings, we hypothesized that alcohol-induced miR-155 regulates both autophagy and exosome release. To evaluate the mechanistic relationship between miR-155 and exosome production, we utilized in vitro primary hepatocyte and KC culture systems. Alcohol treatment resulted in a significant increase in exosome release in both KCs and hepatocytes isolated from WT mice, but not from miR-155 KO mice, compared to untreated cells (Fig. 5C). The baseline exosome release in KCs and hepatocytes isolated from miR-155 KO mice was also lower compared to WT mice (Fig. 5C).
Exosome production is a dynamic process, and cells can actively sort molecules, including miRNAs, into exosomes.(16,25) Previously, we showed that miR-155 was associated with exosome-rich fractions in the circulation in ALD(12); however, the cellular source of exosomes was unknown. We found increased miR- 155 levels in exosomes isolated from KCs and hepatocytes from alcohol-fed WT mice (Fig. 5D) and did not detect miR-155 expression in exosomes isolated from miR-155 KO—KCs or hepatocytes (Fig. 5D). Circulating exosomes isolated from AH patients were also enriched in miR-155 compared to exosomes from control subjects (Fig. 5E).
We further evaluated the mechanistic role of miR-155 on exosome release in macrophages and hepatocytes in vitro. Overexpression of miR-155 in macrophages and hepatocytes using a miR-155 mimic significantly increased exosome release compared to cells treated with a control miRNA mimic (Fig. 5F). These results suggest that miR-155 plays a role in exosome production in general as well as in alcohol-induced increase in exosome production.
Size and purity of exosomes were determined by NanoSight and western blotting, respectively. Exosomes from hepatocytes (Supporting Fig. S7A) and KCs (Supporting Fig. S7C) had an average size of ~84 nm and were enriched in the exosome marker, CD63 (Supporting Fig. S7B,D). Calnexin, an endoplasmic reticulum protein, was present in cells and absent in exosomes (Supporting Fig. S7B,D).
INHIBITION OF AUTOPHAGY AT THE AUTOPHAGOSOME-LYSOSOME FUSION STEP INCREASES LIVER INJURY AND EXOSOME PRODUCTION IN THE ACUTE-ON-CHRONIC ALCOHOL FEEDING MODEL
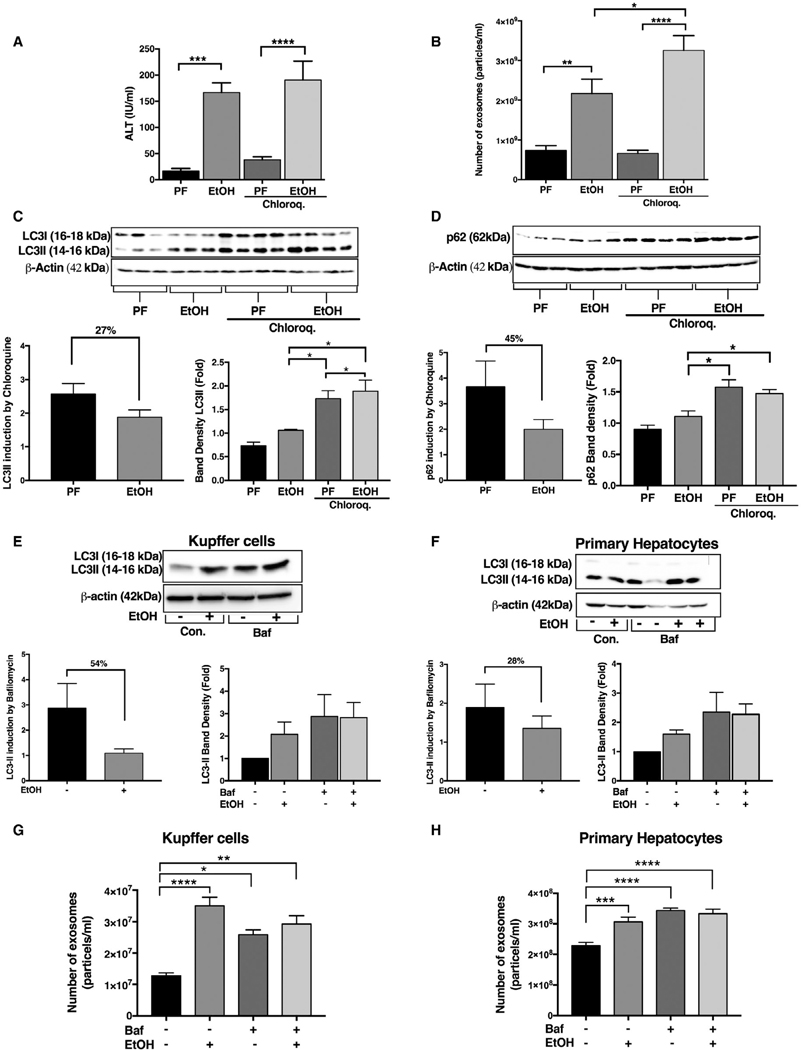
To mechanistically assess whether impaired autophagy in ALD depends on lysosomal function and involves crosstalk with exosome release, we performed in vivo studies using chloroquine, a lysosome function inhibitor.(26) Given that we found similar results in mouse models of ALD and AH (increases in Beclin1, Atg7, LC3-II, and p62 and decreases in mTOR protein levels), we focused on the acute-on-chronic alcohol feeding (AH) model. Alcohol feeding in the presence or absence of chloroquine increased serum ALT levels in mice (Fig. 6A). We found a significant increase in circulating exosomes in acute-on-chronic alcohol-fed compared to control-diet-fed mice, and this increase was further augmented by chloroquine administration (Fig. 6B). Blockade of lysosome function with chloroquine(26) significantly increased LC3-II and p62 levels in pair-fed mice, and these increases were further augmented in chloroquine-treated acute-on-chronic alcohol-fed mice (Fig. 6C,D). The LC3-II ratio in chloroquine treated to that of untreated pair-and alcohol-fed mice showed a decreasing trend, indicating impaired autophagy flux (27% reduction in alcohol fed mice; Fig. 6C,D).
FIG. 6.
Functional block of autophagy at the autophagosome-lysosome fusion step results in increased liver injury and exosome production in acute-on-chronic alcohol feeding. Serum ALT levels were used to determine the level of liver damage in acute-on-chronic alcohol feeding (A). The number of exosomes from sera of pair-fed (PF)- and acute-on-chronic alcohol-fed (EtOH)-fed mice with or without chloroquine injection was quantified by NTA (n = 8; B). Total liver protein was extracted from PF or EtOH mice (n = 6–8) with or without 65 mg/kg of chloroquine injected intraperitonealy and analyzed by western blotting using β-actin as a loading control. Immunoblottings were probed with antibodies for LC3-I and LC3-II (C) and p62 (D). The densitometry analysis is shown as bar diagrams. The LC3-II and P62 ratio in chloroquine treated to that of untreated alcohol-fed and pair-fed animals was calculated. (C,D) KCs and primary hepatocytes were treated with and without 50 mM of alcohol for 24 hours, and bafilomycin was added (in last 12 hours) as indicated. After 24 hours, the protein levels from KCs and primary hepatocytes were analyzed by western blotting for LC3-I andLC3-II (E,F) using β-actin as a loading control. The densitometry analysis is shown as bar diagrams. The LC3-II ratio was calculated (E,F). The number of exosomes from KCs and primary hepatocytes supernatants after 24 hours was quantified by NTA (G,H). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Abbreviations: Baf, bafilomycin; Chloroq., chloroquine; Con., control.
Next, we isolated primary hepatocytes and KCs from WT mice and treated them with alcohol in the presence and absence of bafilomycin. Previous reports and our pilot experiments inidcated that bafilomycin is more consistent than chloroquine for lysosomal inhibition in vitro.(27) We found significant increases in protein levels of LC3-II in both cell types after bafilomycin or alcohol treatment (Fig. 6E,F). The LC3-II ratios between samples with and without bafilomycin treatment were decreased in alcohol-treated hepatocytes (28% decrease) and KCs (54% decrease) compared to no alcohol treatment (Fig. 6E,F). We also found a significant increase in exosome numbers from ethanol- or bafilomycin-treated cells, suggesting that disruption of lysosomal function increases exosome production (Fig. 6G,H). The combination of bafilomycin and alcohol resulted in no further increase in exosome release compared to bafilomycin or alcohol alone in exosome release (Fig. 6G,H).
LYSOSOMAL DYSFUNCTION INCREASES EXOSOME RELEASE IN MACROPHAGES AND HEPATOCYTES
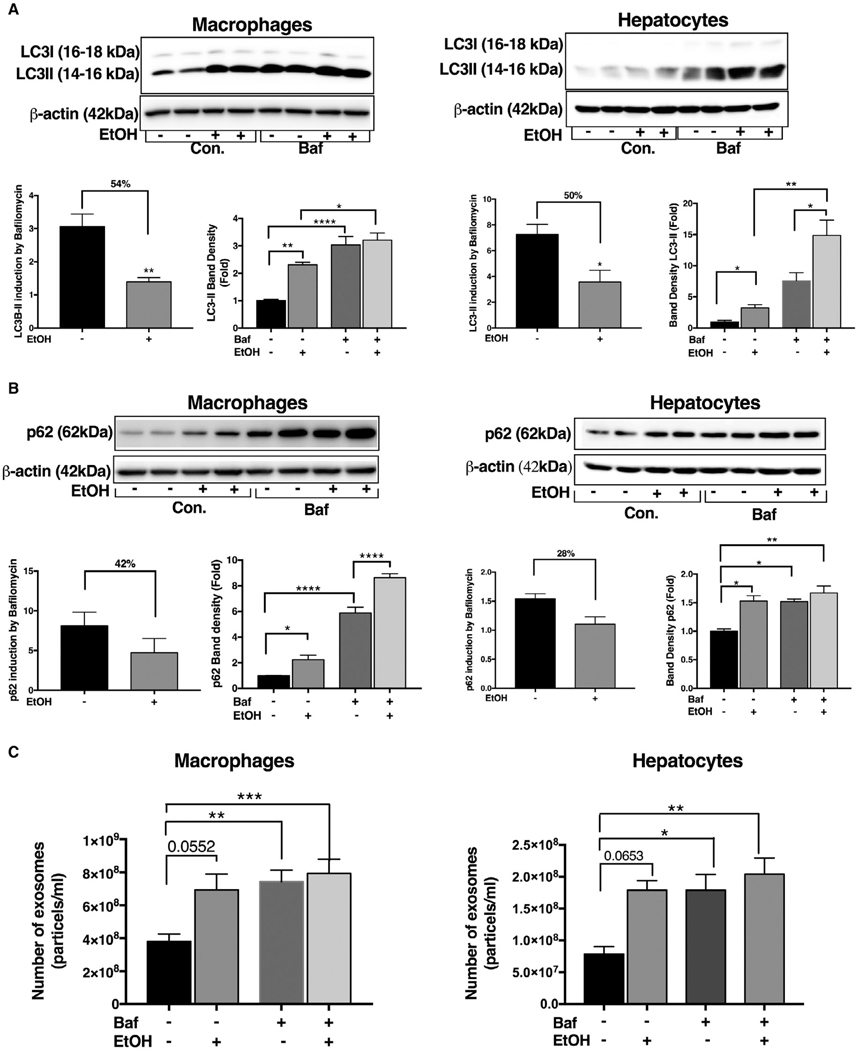
To further elucidate the crosstalk between disruption of autophagy and exosome secretory pathways, we performed a series of simulation experiments where we treated macrophages (RAW 264.7) or hepatocytes (Hepa1–6) with bafilomycin A, an inhibitor of vacular type ATPase that disrupts lysosome fusion with auto-phagosome,(28,29) in the presence or absence of alcohol. Alcohol or bafilomycin A alone significantly increased LC3-II protein expression both in macrophages and hepatocytes (Fig. 7A). The LC3-II ratio between cells without and with bafilomycin was decreased in alcohol-treated cells compared to controls, indicating a reduction in autophagy flux by alcohol in macrophages (54% reduction) and in hepatocytes (50% reduction; Fig. 7A). We also found significant cellular accumulation of p62 after alcohol or bafilomycin treatment, respectively, in both cell types (Fig. 7B). Furthermore, we observed a decreasing trend in the ratio of p62 in alcohol-treated cells in the presence of bafilomycin compared to no bafilomycin control (Fig. 7B).
FIG. 7.
Inhibition of autophagy at the lysosomal level results in increased exosome production in macrophages and hepatocytes. Macrophages (RAW 264.7) and hepatocytes (Hepal-6) were treated with or without 50 mM of alcohol (EtOH) for 24 hours, and bafilomycin A (100 nM) was added in the last 12 hours of the experiment. After 24 hours, protein levels from macrophages and hepatocytes were analyzed by western blotting for LC3-I and L3-II (A) and p62 (B) using β-actin as a loading control. The densitometry analysis is shown as bar diagrams. The LC3-II and p62 ratio in bafilomycin-treated macrophages and hepatocytes to that of untreated macrophages and hepatocytes was calculated to show reduction in autophagic flux (B). The number of exosomes from macrophages and hepatocytes supernatants after 24 hours was quantified by NTA (C; n = 12). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Abbreviations: Baf, bafilomycin; Con., control.
To test whether functional inhibition of autopha-gosome-lysosome fusion triggers exosome release, we analyzed cellular supernatants. We found that bafilomycin A or alcohol alone increased exosome release in macrophages and hepatocytes (Fig. 7C). There was no increase in exosome numbers in cells treated with both bafilomycin A and alcohol compared to single-agent treatment. The bafilomycin A treatment did not cause any cellular toxicity, as demonstrated by lactate dehydrogenase assay (Supporting Fig. S8A).
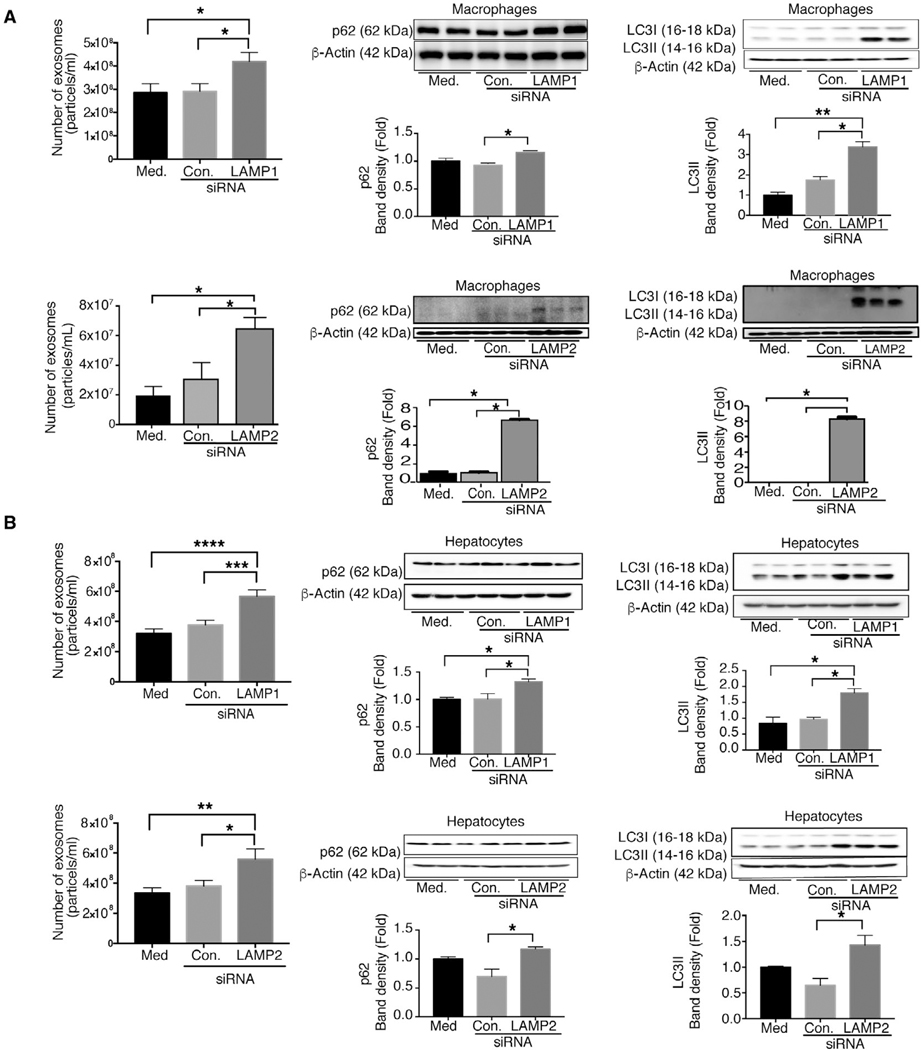
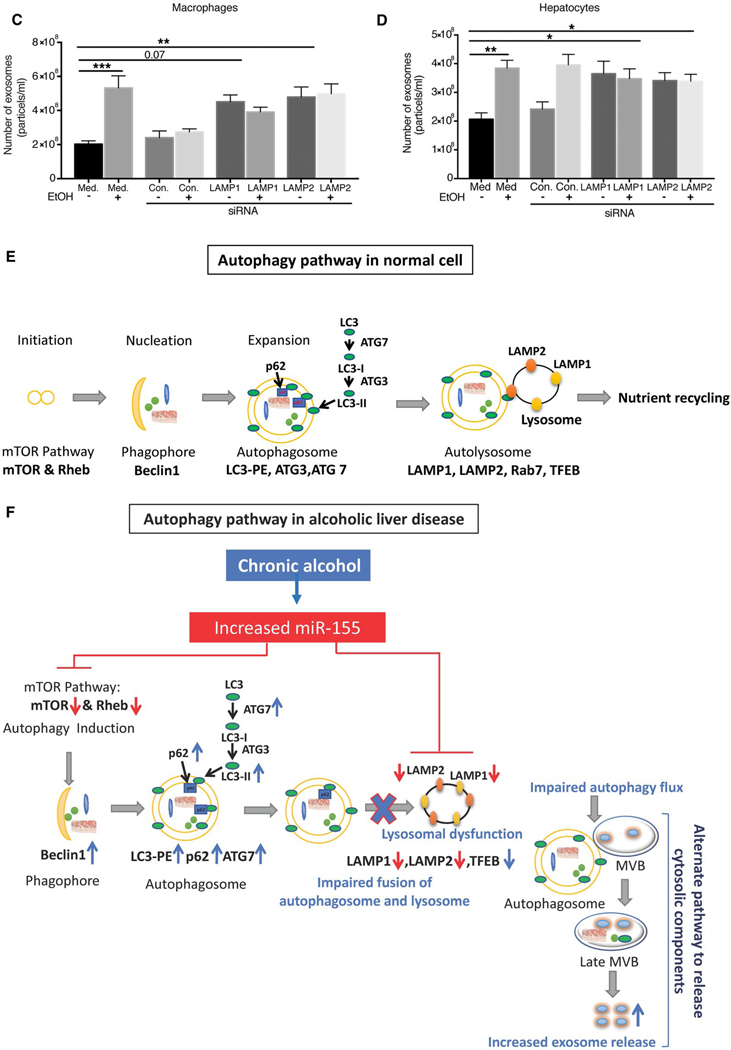
SILENCING OF LAMP1 AND LAMP2 INCREASES EXOSOME RELEASE AND p62 ACCUMULATION
Because we found a significant decrease in lysosomal protein levels after chronic alcohol feeding, next we evaluated the mechanistic role of LAMP1 and LAMP2 on exosome production. Knockdown of LAMP1 and LAMP2 protein expression in macrophages (RAW264.7 cells) and in hepatocytes (HEPAl-6) was achieved using specific siRNA (Supporting Fig. S9A,B). We found that LAMP1 and LAMP2 knockdown, respectively, resulted in increased exosome release in macrophages (Fig. 8A) and hepatocytes (Fig. 8B). We also found increased expression of both p62 and LC3-II protein levels in LAMPI and LAMP2 siRNA transfected cells compared to control siRNA-treated cells (Fig. 8A,B), indicating that down-regulation of LAMPI or LAMP2 dirupts autophagy in macrophages and hepatocytes. Finally, we tested the effects of alcohol on exosome release in combination with LAMP inhibitition and found no further increase in exosome secretion in LAMPI or LAMP2 knockdown macrophages (Fig. 8C) or hepatocytes (Fig. 8D) after alcohol treatment. These results suggest that both alcohol and LAMP knockdown promotes exosome release, but have no additive effects.
FIG. 8.
Knockdown of LAMP1 and LAMP2 increases exosome production in macrophages and hepatocytes. Macrophages (RAW 264.7) and hepatocytes (HEPA 1.6) were untreated or transfected with control or LAMP1 or LAMP2 siRNA, and the number of exosomes from supernatants of LAMP1/LAMP2 siRNA-treated macrophages and hepatocytes was quantified by NTA (A,B; n = 6). p62 (A,B) and LC3-I and LC3-II (A,B) were analyzed by western blotting using β-actin as a loading control (n = 6). The densitometry analysis is shown as bar diagrams. The number of exosomes from supernatants of LAMP1 and LAMP2 siRNA-treated macrophages and hepatocytes in the presence and absence of alcohol was quantified by NTA (n = 9; C,D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Schematic representation of miR-155-autophagy-exosome axis in ALD (E,F). Chronic alcohol induces miR-155, which then decreases mTOR and Rheb, thus inhibiting the mTOR pathway (E,F). Chronic alcohol increases Beclin1, Atg7, and LC3II expression. However, alcohol impairs autophagic flux by affecting LAMPs (LAMP1 and LAMP2) through miR-155. miR-155 regulates lysosome function by targeting LAMP1 and LAMP2. Because of impaired lysosome function, autophagosomes merge with MVBs to release their components outside the cell, and, as a consequence, there is increased exosomes secretion. Abbreviation: Med, medium.
Discussion
A major factor in ALD pathogenesis is cellular stress and recent evidence suggests that this could be attributed to the effects of alcohol on autophagy/3–6 Autophagy is a regulated, dynamic, multistep process aiming to maintain cellular homeostasis. Exosome production is another mechasnism by which cells excrete harmful cargo to maintain homeostasis.(12–15,30) Using mouse models and human liver samples, we investigated how alcohol and alcohol- induced miR-155 affect autophagy and exosome production in AH and ALD. We found that expression of several key molecules of the autophagy pathway was changed in livers of mice and humans with ALD or AH compared to heathy livers. Protein levels of mTOR and Rheb, involved in initiation of autophagy, were decresased and levels of Beclinl, Atg7, LC3-II, and p62, involved in phagophore and autophagosome formation, were significantly increased by alcohol. We identified that alcohol, both in vitro and in vivo, inhibits lysosomal-associated proteins 1 and 2, and this is associated with accumulation of LC3-II and p62 as well as increased exosome production. Decreased ratios of LC3-II in alcohol-fed versus pair-fed mice and alcohol-treated cells in vitro suggested a decrease in autophagy flux. Our study shows that miR-155 is a mediator of alcohol-induced reduction of mTOR and Rheb as well as LAMP1 and LAMP2 decreases whereas increases in Beclin 1, Atg7, LC3-II, and p62 were not directly related to miR-155 in ALD (Fig. 8E). Together, our data reveal that inhibition of LAMP1 and LAMP2 by alcohol critically affects both autophagy and exosome production in the liver in hepatocytes and macrophages.
Using the ALD mouse model and patient samples, we revealed a connection between alcohol and autophagy involving miR-155. A recent study demonstrated that miR-155 regulates autophagy by targeting mTOR and Rheb in hypoxic conditions.(19) We previously reported that miR-155 KO mice are protected from ALD and that chronic alcohol regulates miR- 155 at the transcriptional and posttranscriptional levels.(17) Altogether, these studies prompted us to ask whether miR-155 regulates autophagy in ALD. We demonstrate that miR-155 regulates autophagy by targeting both the mTOR pathway and lysosomal proteins (LAMP1 and LAMP2) in vivo and at the cellular level. Our findings revealed that miR-155 is a significant regulator of autophagy.
Our studies revealed inhibition of mTOR and its activity in both mouse models of ALD and in humans. Inhibition of mTOR activity was shown to increase autophagy.(31) We found disruption of autophagy at the lysosomal level. Apart from its role in autophagy, mTOR is a central regulator of cell growth, proliferation, and survival in response to nutritional status, growth factor, and stress signals.(31) A recent study showed a role of mTOR in the regulation of exosome release. It was demonstrated that inhibition of mTOR complex 1 (mTORC1; mTOR) by rapamycin or nutrient and growth factor deprivation stimulates exosome release, which occurs concomitantly with autophagy.(32) We also demonstrate that both mTOR and phosphorylation levels of its substrate proteins (S6p70 and 4EBP1) were decreased in alcohol-fed mice. mTOR was shown to be involved in activation of autophagy in hepatocytes after in vitro alcohol treatment.(3) Recently, acute-on-chronic alcohol administration in mice was shown to activate mTORC1(7); however, we found that this pathway was inhibited in ALD. This could be attributed to differences in duration of alcohol feeding, route of alcohol administration, and perhaps the cellular activation stage at the endpoint.(3–5,33)
We found an LC3-II increase and accumulation of p62 in ALD and AH mouse models and in ALD patient livers. Consistent with our results, Cho et al. also reported increases in p62 levels and impaired autophagic flux after chronic alcohol administration in ALD.(5) In contrast, we found that miR-155 KO mice exhibited normal autophagic degradation and attenuation of alcohol-induced liver injury and SH. In addition, miR-155 KO mice were protected from alcohol-induced LC3-II and p62 accumulation in vivo compared to their WT, alcohol-fed counterparts. These results indicate that miR-155 plays a role in chronic alcohol-induced impairment of the autophagy process.
A recent study by Chao et al., using the acute-on-chronic alcohol mouse model of AH, demonstrated that impaired autophagic flux was caused by a decrease in lysosome numbers leading to insufficient autophagic degradation.(7) They determined that TFEB, a master regulator of lysosomal biogenesis, was involved in the alcohol-induced decrease in lysosome biogenesis. TFEB promotes the formation of autophagosomes and their fusion with lysosomes.(34) We found decreased TFEB in our mouse model and in patients with ALD. Interestingly, we also found increased TFEB levels in miR-155 KO mice after chronic alcohol, implying optimal autophago-some-lysosome fusion and hence normal autophagic degradation in miR-155 KO mice. Our results, however, suggest that miR-155 does not regulate TFEB directly, and we cannot rule out indirect regulation.
We identified that the autophagic degradation capacity of lysosomes is impaired because of an alcohol-related reduction in lysosomal proteins (LAMP1 and LAMP2) in chronic alcohol-fed mice and in ALD patients. Consistent with our findings, Cho et al. reported decreased LAMP2 levels and increased accumulation of autophagosomes after chronic alcohol diet.(5) LAMP1 and LAMP2 have crucial roles in the degradation of autophagic contents.(9,24) Fusion of lysosomes with autophagosomes produces autolysosomes and LAMPs are responsible for lysosomal proteolytic activity.(11) Impotantly, we discovered that miR-155 regulates LAMP1 and LAMP2 in ALD. We confirmed the causal role of miR-155 in ALD by LAMP2 regulation in our in vitro studies and showed that silencing of LAMP1 or LAMP2 increased both LC3-II and p62 in macrophages and in hepatocytes. Decreased autophagy was also found by Ilyas et al. in macrophages that promoted liver injury and inflammation in alcohol-fed mice.(35)
Silencing of LAMP1 and LAMP2 in our experiments was associated with a significant increase in exosome release both in macropahges and hepatocytes. Various studies have shown a causal relationship between lysosomal dysfunction and exosome secretion.(36,37) Another study reported that increased secretion of atypical exosomes is a homeostatic response to counter lysosomal dysfunction to eliminate lysosomal waste.(38) Our results confirm that lysosomal dysfunction results in significantly higher exosome release in ALD, suggesting a crosstalk between lysosomal dysfunction and the exosomal pathway. We found impaired autophagy and increased exosome production in the mouse model of ALD and also in patients with ALD. In contrast, miR-155 KO mice had significantly lower levels of exosomes compared to their WT, alcohol-fed counterparts. These results suggest that miR-155 is involved in impairing autophagy at the level of lysosome function. Collectively, our results revealed a previously undiscovered role for miR-155 in regulating lysosome function through effects on LAMPs in ALD.
We also show that miR-155 was increased in exosomes of ALD patients and in sera of chronic alcohol-fed mice. Our previous study showed that miRNAs in exosomes can reprogram recipient cells and modulate their function.(16) Thus, it is likely that exosomal miR-155 has a biological function in ALD. We found that miR-155 levels were increased in exosomes of KCs and hepatocytes in alcohol-treated cells, suggesting that exosomal sorting and release of miR-155 may represent a “rescue” mechanism for cells to reduce cellular miR-155 levels. Our results provide an aspect of miR-155 in the autophagy-exosome axis and highlight the importance of miR-155 in both hepatocytes and macrophages.
In addition, we discovered a potential alcohol-independent role for miR-155 in exosome biogenesis. Alcohol-naive macrophages and hepatocytes from miR-155 KO mice had decreased exosome production, and these findings were further supported by experiments where miR-155 overexpression in macrophages and hepatocytes decreased mTOR, Rheb, and LAMP2 followed by an increase in exosome production. Altogether, our findings suggest a crosstalk between autophagy and exosomal secretory pathways in ALD and reveal a central mechanistic role of miR-155 in this process (Fig. 8E).
To further examine the effects of lysosomal disruption on the autophagy pathway, we performed experiments with chloroquine in vivo. Chloroquine changes the lysosomal pH, thereby inhibiting autophagic degradation in lysosomes.(26,29) Dysruption of lysosome function with chloroquine treatment caused increased levels of p62 and LC3-II and a higher increase in exo-some secretion in acute-on-chronic alcohol-fed mice. In vitro experiments using bafilomycin A (autophagy inhibitor, by preventing the fusion of autophagosome with lysosome) treatment resulted in impaired autophagic flux, as demonstrated by significant cellular accumulation of p62. We demonstrated that blocking the fusion of autophagosomes and lysosomes inhibits autophagic flux and promotes exosome secretion, as an alternative mechanism for the clearance of toxic proteins; our results are consistent with a recent study.(39) Collectively, our results suggest that chronic alcohol administration affects lysosomes, thus disrupting autophagosome and lysosome fusion and exosome secretion. Autophagy-exosome crosstalk is a complex and context-dependent process. Emerging evidence suggests direct links between autophagy and exosome biogenesis through shared molecular machinery and crosstalk between these two processes.(40) Exosome biogenesis and autophagy are linked by the endolyso-somal pathway.(40) A recent study demonstrated that knockout of autophagy-related protein 5 (Atg5) or autophagy related 16-like 1 significantly reduces exosome release.(41) Atg5 specifically decreases acidification of late endosomes where exosomes are produced,(41) suggesting that autophagy related proteins may directly regulate the fate of MVBs and exosome biogenesis.(41) Our findings discover mechanisms controlling exosome release and identify processes where autophagy-related genes crosstalk with the exosome pathway in ALD.
In conclusion, our findings suggest that lysosomal function is defective in chronic ALD, resulting in disruption of autophagy and increased exosome secretion. Additionally, we identified miR-155 as a key factor regulating autophagy and exosome production in liver macrophages and hepatocytes. Here, we show that there is, in part, association between autophagy, miR-155, and exosome secretion in ALD.
Acknowledgment: Human liver samples were provided by the NIH-Funded Liver Tissue Procurement and Cell Distribution System (N01-DK-7–0004/HHSN26700700004C). The authors thank Sahin Coban and Andras Budai for their technical assistance as well as Candice Dufour and Melanie Trombly for their assistance in preparing the manuscript.
Supplementary Material
Acknowledgments
Supported by NIHgrant AA017729, U01 translational (AA021907), and U01 clinical (AA021893) grants to G.S. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations:
- 4EBP1
4E-binding protein 1
- AH
alcoholic hepatitis
- ALD
alcoholic liver disease
- ALT
alanine aminotransferase
- Atg3
autophagy-related protein 3
- Atg7
autophagy-related protein 7
- IHC
immunohistochemistry
- KCs
Kupffer cells
- KO
knockout
- LAMP1
lysosomal-associated membrane protein 1
- LAMP2
lysosomal-associated membrane protein 2
- LC3 (MAP1LC3B)
microtubule-associated proteins 1A/1B light chain 3B
- miR-155
micro-RNA 155
- mTOR
mechanistic target of rapamycin
- MVBs
multivesicular bodies
- NADH
reduced nicotinamide adenine dinucleotide
- NTA
NanoSight Tracking Analysis
- Rheb
Ras homolog enriched in brain
- siRNA
small interfering RNA
- TFEB
transcription factor EB
- WT
wild type
Footnotes
Potential conflict of interest: Dr. Szabo consults for and received grants from Allergan. She consultsfor Terra Firma, Glympse Bio, Quest, Arrow, GLG, Salix, and Tobira. She received grants from Gilead, Genfit, Intercept, Verlyx, Novartis, SignaBlok, and Shire. She holds intellectual property rights with Up to Date.
Supporting Information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.30766/suppinfo
REFERENCES
- 1).Szabo G, Bala S. Alcoholic liver disease and the gut-liver axis. World J Gastroenterol 2010;16:1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Nagy LE. The role of innate immunity in alcoholic liver disease. Alcohol Res 2015;37:237–250. [PMC free article] [PubMed] [Google Scholar]
- 3).Ding WX, Li M, Chen X, Ni HM, Lin CW, Gao W, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 2010;139:1740–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4).Lin CW, Zhang H, Li M, Xiong X, Chen X, Chen X, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol 2013;58:993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5).Cho HI, Choi JW, Lee SM. Impairment of autophagosome- lysosome fusion contributes to chronic ethanol-induced liver injury. Alcohol 2014;48:717–725. [DOI] [PubMed] [Google Scholar]
- 6).Ni HM, Du K, You M, Ding WX. Critical role of FoxO3a in alcohol-induced autophagy and hepatotoxicity. Am J Pathol 2013;183:1815–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7).Chao X, Wang S, Zhao K, Li Y, Williams JA, Li T, et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology 2018;155:865–879.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Ding WX, Manley S, Ni HM. The emerging role of autophagy in alcoholic liver disease. Exp Biol Med (Maywood) 2011;236:546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Hansen TE, Johansen T. Following autophagy step by step. BMC Biol 2011;9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10).Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ 2009;16:70–78. [DOI] [PubMed] [Google Scholar]
- 11).Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell 2002;13:3355–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012;56:1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol 2002;2:569–579. [DOI] [PubMed] [Google Scholar]
- 14).Baixauli F, Lopez-Otin C, Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol 2014;5:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15).Szabo G, Momen-Heravi F. Extracellular vesicles in liver disease and potential as biomarkers and therapeutic targets. Nat Rev Gastroenterol Hepatol 2017;14:455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep 2015;5:9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17).Bala S, Csak T, Saha B, Zatsiorsky J, Kodys K, Catalano D, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol 2016;64:1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor alpha (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem 2011;286:1436–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N, et al. Hypoxia- induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy 2014;10:70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20).Bukong TN, Iracheta-Vellve A, Saha B, Ambade A, Satishchandran A, Gyongyosi B, et al. Inhibition of spleen tyrosine kinase activation ameliorates inflammation, cell death, and steatosis in alcoholic liver disease. Hepatology 2016;64:1057–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Crabb DW, Bataller R, Chalasani NP, Kamath PS, Lucey M, Mathurin P, et al. Standard definitions and common data elements for clinical trials in patients with alcoholic hepatitis: recommendation from the NIAAA Alcoholic Hepatitis Consortia. Gastroenterology 2016;150:785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22).Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012;8:445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23).Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett 2010;584:1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24).Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 2009;10:623–635. [DOI] [PubMed] [Google Scholar]
- 25).Bala S, Csak T, Momen-Heravi F, Lippai D, Kodys K, Catalano D, et al. Biodistribution and function of extracellular miRNA-155 in mice. Sci Rep 2015;5:10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol 2009;625:220–233. [DOI] [PubMed] [Google Scholar]
- 27).Yoshii SR, Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Yuan N, Song L, Zhang S, Lin W, Cao Y, Xu F, et al. Bafilomycin A1 targets both autophagy and apoptosis pathways in pediatric B-cell acute lymphoblastic leukemia. Haematologica 2015;100:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Moulis M, Vindis C. Methods for measuring autophagy in mice. Cells 2017;6:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Zhou X, Jiao Z, Ji J, Li S, Huang X, Lu X, et al. Characterization of mouse serum exosomal small RNA content: the origins and their roles in modulating inflammatory response. Oncotarget 2017;8:42712–42727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).paquette M, El-Houjeiri L, Pause A. mTOR pathways in cancer and autophagy. Cancers (Basel) 2018;10:E16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32).Zou W, Lai M, Zhang Y, Zheng L, Xing Z, Li T, et al. Exosome release is regulated by mTORC1. Adv Sci (Weinh) 2019;6:1801313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Khambu B, Wang L, Zhang H, Yin XM. The activation and function of autophagy in alcoholic liver disease. Curr Mol Pharmacol 2017;10:165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Sardiello M Transcription factor EB: from master coordinator of lysosomal pathways to candidate therapeutic target in degenerative storage diseases. Ann N Y Acad Sci 2016;1371:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35).Ilyas G, Cingolani F, Zhao E, Tanaka K, Czaja MJ. Decreased macrophage autophagy promotes liver injury and inflammation from alcohol. Alcohol Clin Exp Res 2019. April 9 10.1111/acer.14041. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Alvarez-Erviti L, Seow Y, Schapira AH, Gardiner C, Sargent IL, Wood MJ, Cooper JM. Lysosomal dysfunction increases exosome-mediated alpha-synuclein release and transmission. Neurobiol Dis 2011;42:360–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernandez-Delgado I, Torralba D, Moreno-Gonzalo O, et al. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun 2016;7:13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38).Miranda AM, Lasiecka ZM, Xu Y, Neufeld J, Shahriar S, Simoes S, et al. Neuronal lysosomal dysfunction releases exosomes harboring APP C-terminal fragments and unique lipid signatures. Nat Commun 2018;9:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39).Yang Y, Qin M, Bao P, Xu W, Xu J. Secretory carrier membrane protein 5 is an autophagy inhibitor that promotes the secretion of alpha-synuclein via exosome. PLoS One 2017; 12:e0180892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40).Xu J, Camfield R, Gorski SM. The interplay between exosomes and autophagy—partners in crime. J Cell Sci 2018;131: jcs215210. [DOI] [PubMed] [Google Scholar]
- 41).Guo H, Chitiprolu M, Roncevic L, Javalet C, Hemming FJ, Trung MT, et al. Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev Cell 2017;43: 716–730.e7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.