Abstract
RNA splicing, the enzymatic process of removing segments of premature RNA to produce mature RNA, is a key mediator of proteome diversity and regulator of gene expression. Increased systematic sequencing of the genome and transcriptome of cancers has identified a variety of means by which RNA splicing is altered in cancer relative to normal cells. These findings, in combination with the discovery of recurrent change-of-function mutations in splicing factors in a variety of cancers, suggest that alterations in splicing are drivers of tumorigenesis. Greater characterization of altered splicing in cancer parallels increasing efforts to pharmacologically perturb splicing and early-phase clinical development of small molecules that disrupt splicing in patients with cancer. Here we review recent studies of global changes in splicing in cancer, splicing regulation of mitogenic pathways critical in cancer transformation, and efforts to therapeutically target splicing in cancer.
Keywords: RNA, SF3B1, splicing, SRSF2, U2AF1, ZRSR2
INTRODUCTION
RNA splicing, the process of removing nucleotide sequences from precursor RNA to form mature RNA, is a key regulator of gene expression and proteome diversity. Recent analyses of genetic alterations in cancer and how these alterations relate to the transcriptome and epigenome have uncovered a myriad of means by which splicing is altered in cancer cells. These include mutations in DNA that abolish or generate splicing regulatory sequences in cis, mutations in genes encoding RNA splicing regulators, changes in the expression of splicing factors by oncogenic processes, and alterations in chromatin state that modify splicing patterns (Figure 1). Splicing is an enzymatic process requiring numerous protein-RNA complexes and posttranslational modifications (PTMs) of splicing proteins, as well as protein-protein, protein-RNA, and RNA-RNA interactions, providing a variety of avenues for pharmacologic perturbation. Here we review patterns of cancer-associated alternative splicing (AS) events, how cancer cells generate novel splicing events to promote disease, and increasing modalities to therapeutically target splicing in cancer.
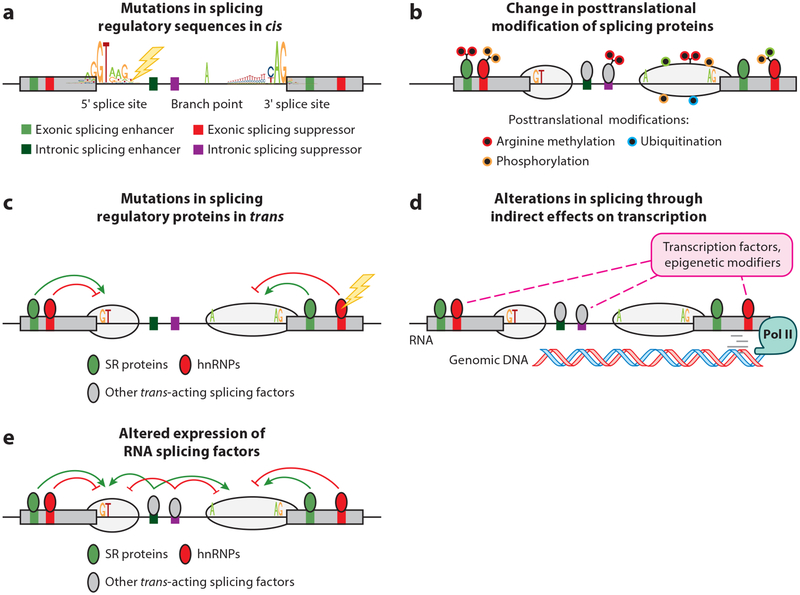
Figure 1.
Means by which RNA splicing is altered in cancer. As shown, genetic alterations affecting (a) critical sequences required for splicing of genes in cis have been described at splice sites, branch points, and splicing enhancers and silencers within exons and introns. In addition, altered (b) posttranslational modification of splicing proteins as well as (c) change-of-function and loss-of-function mutations in RNA splicing factors themselves occur in a variety of cancers. Given that splicing occurs cotranscriptionally, processes that (d) modify the transcription rate of RNA polymerase II (Pol II) may modify splicing. Finally, (e) altered expression of RNA splicing factors has been demonstrated to play a pathogenic role in a variety of cancers. Abbreviations: hnRNPs, heterogeneous nuclear ribonucleoproteins; SR proteins, serine- and arginine-rich proteins.
RNA SPLICING CATALYSIS AND REGULATION
Splicing is a catalytic process by which introns, noncoding segments of precursor messenger RNAs (pre-mRNAs), and long noncoding RNAs are removed to generate mature RNAs. Splicing is catalyzed by a large RNA-protein complex known as the spliceosome. Recent single-particle cryo-electron microscopy has allowed the characterization of spliceosomes that provide atomic model building. The biochemistry of splicing is covered in several recent reviews (Fica & Nagai 2017; Scheres & Nagai 2017; Shi 2017; Wahl & Luhrmann 2015a,b). Meanwhile, greater analyses of RNA sequencing (RNA-seq) data have elucidated sequences utilized by the spliceosome to differentiate introns from exons and identify branch point sequences. Consensus splice sites (ss) located at the boundaries between the upstream exon and intron (the 5′ ss) and the intron and downstream exon (the 3′ ss), as well as the branch point, are recognized by the spliceosome (Figure 1). The major spliceosome (which recognizes the majority of ss) consists of five small nuclear ribonucleoprotein (snRNP) complexes and more than 150 proteins, while the minor spliceosome (which recognizes a minority of introns after the snRNP complex specialized to recognize these sequences) contains five snRNPs, most of which are distinct from those of the major spliceosome. All ss are either constitutive or alternative, depending on whether they are always (constitutive) or only sometimes (alternative) recognized by the spliceosome.
Although splicing of constitutive and alternative ss is determined by the spliceosome, efficient recruitment of spliceosomal proteins to ss depends on the binding of additional trans-acting splicing factors [reviewed recently by Dvinge et al. (2016) and Scotti & Swanson (2016)]. Such regulatory splicing factors bind to motifs associated with the promotion (enhancers) or repression (silencers) of splicing (Figure 1). Enhancer and silencer motifs are recognized by two common families of splicing factors: serine- and arginine-rich splicing factors (SR proteins) (Long & Caceres 2009) and heterogeneous nuclear ribonucleoproteins (hnRNPs) (Krecic & Swanson 1999). In exons, SR proteins bind enhancer sequences to activate splicing, whereas hnRNPs bind silencer sites to inhibit splicing. However, the activities of splicing factors are context dependent (Fu & Ares 2014). In addition to SR proteins and hnRNPs, many other RNA-binding proteins regulate splicing. These include CELF (Dasgupta & Ladd 2012), MBNL (Konieczny et al. 2014), RBFOX, RBM (Sutherland et al. 2005), STAR, NOVA (Ule et al. 2006), epithelial splicing regulatory proteins, TIA1, TIAL1, and others (Fu & Ares 2014). Proteomic studies indicate that the spliceosome consists of more than 170 proteins (Jurica & Moore 2003), while computational studies of exon recognition suggest that hundreds of motifs contribute to the regulation of splicing (Barash et al. 2010). Although the total number of splicing factors is unknown, the above studies suggest that hundreds of proteins regulate splicing.
GLOBAL ALTERATIONS IN RNA SPLICING IN CANCER
Efforts to analyze the transcriptomes of tumor versus normal tissue, such as The Cancer Genome Atlas, have revealed that many cancers exhibit aberrant splicing (Climente-Gonzalez et al. 2017, Jayasinghe et al. 2018, Jung et al. 2015, Kahles et al. 2018, Supek et al. 2014), including changes in usage of annotated transcript isoforms and increased use of aberrant unannotated splicing events. One recent effort characterized the abundance of annotated transcript isoforms across 4,542 samples from 11 cancer types and identified that splicing changes in cancer impact the same protein-coding domains targeted by somatic mutations (Climente-Gonzalez et al. 2017). Moreover, the number of AS changes in a tumor was inversely correlated with the number of driver mutations, and AS switches displayed some mutual exclusion with driver mutations, suggesting that AS may serve as independent tumorigenic processes. Further efforts revealed that tumors exhibit a ~20% increase in novel splicing events and exon-exon junctions, many of which are specific to cancer type (Kahles et al. 2018).
Aberrant splicing in cancer has also been linked to DNA mutations that abolish ss or generate novel ss. The largest effort yet to characterize mutations altering ss in cis utilized whole-exome sequencing of more than 8,000 tumors across 33 cancer types and identified that many mutations that alter ss were previously misannotated as missense or silent mutations (Jayasinghe et al. 2018). Given that critical regulatory splice sequences are far from the consensus 5′ or 3′ ss, it is important to further integrate data from whole-genome sequencing with RNA-seq for a more comprehensive model of how cancer-associated mutations impact splicing in cis.
RECURRENT CHANGE-OF-FUNCTION MUTATIONS IN SPLICEOSOMAL GENES IN CANCER
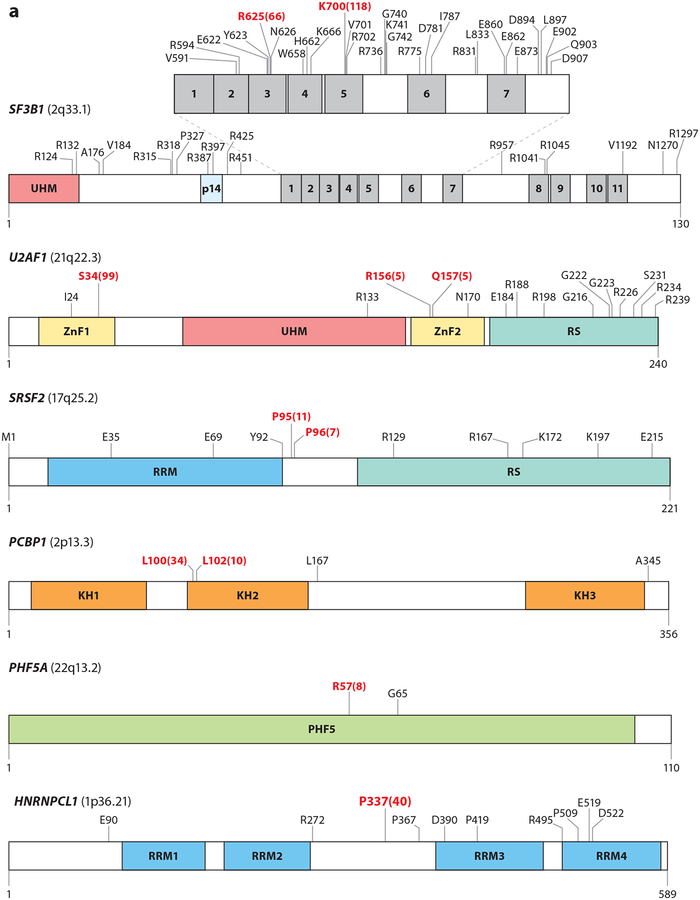
Recurrent heterozygous change-of-function mutations affecting specific residues (or hot spots) in splicing factors have been described in cancer (Figure 2) (Graubert et al. 2012, Papaemmanuil et al. 2011, L. Wang et al. 2011, Yoshida et al. 2011). SF3B1, a subunit of the U2 snRNP that recognizes the branch point, is the most commonly mutated splicing factor in cancer, occurring frequently in myelodysplastic syndromes (MDS), chronic lymphocytic leukemia (CLL), acute myeloid leukemia (AML), and uveal and mucosal melanoma (Alsafadi et al. 2016, Furney et al. 2013, Harbour et al. 2013, Martin et al. 2013), in addition to many carcinomas (Seiler et al. 2018a). Several SF3B1-mutant residues are enriched in specific disease subtypes. For example, mutations at R625 and E902 appear specific to uveal melanoma (UM) and bladder cancer, respectively (Seiler et al. 2018a). SF3B1 mutational hot spots occur within the HEAT domains [a repeated motif consisting of two alpha helices linked by a short loop found in Huntingtin, elongation factor 3 (EF3), PP2A, and yeast TOR1] and possibly affect protein-protein interactions. Global changes in splicing have been observed in cells harboring SF3B1 mutations and are also seen in mouse cells upon introduction of the Sf3b1K700E mutation (Mupo et al. 2017, Obeng et al. 2016). Cancer-associated SF3B1 mutations have repeatedly been found to alter 3′ ss via preference of intron-proximal cryptic 3′ ss over normal sites (Alsafadi et al. 2016, Darman et al. 2015, DeBoever et al. 2015). The aberrant 3′ ss utilized by SF3B1 are associated with shorter, weaker polypyrimidine tracts and are linked to aberrant branch point usage by the mutant SF3B complex. The mechanism governing change in SF3B1 3′ ss or the use of aberrant branch points is not fully understood. One possibility is that SF3B1 mutations result in altered interaction with other spliceosomal components required for branch point recognition. To this end, it has been demonstrated that the introduction of SF3B1 mutations in yeast (homolog Hsh155p) alters the physical interaction with Prp5p (DDX46 in humans), an ATP-dependent RNA helicase important in stabilizing the U2 snRNP/pre-mRNA interaction (Carrocci et al. 2017, Tang et al. 2016). Confirmation of physical interactions between mutant SF3B1 and DDX46 in mammalian homologs will be critical. Of note, the change in 3′ ss preference associated with mutant SF3B1 is distinct from splicing changes observed with genetic loss or pharmacologic inhibition of SF3B1 (Darman et al. 2015, Lee et al. 2016a, Seiler et al. 2018b).
Figure 2.
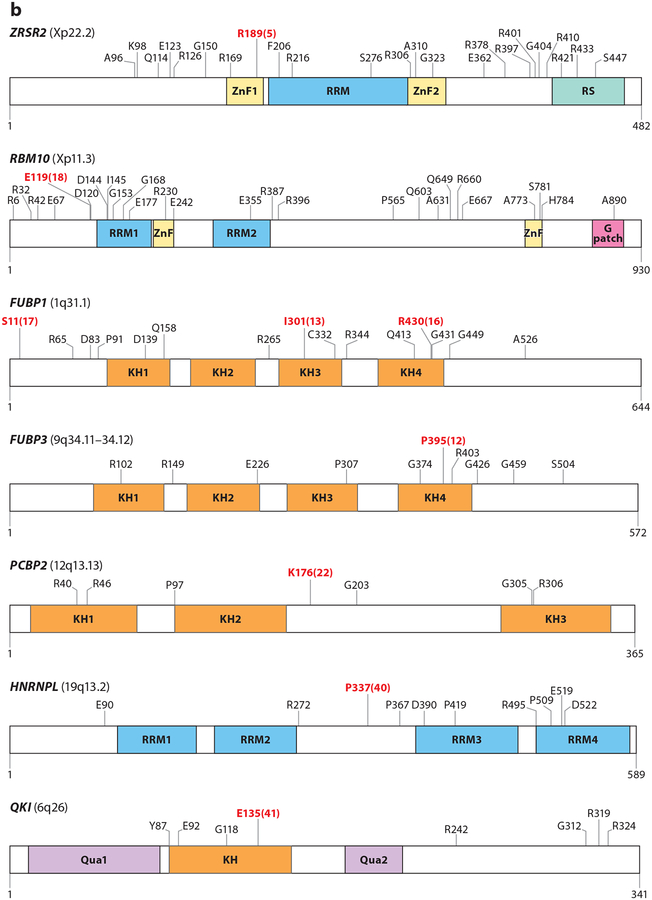
Mutations in RNA splicing factors in cancer. For each protein, mutations occurring in at least three samples are annotated, with the exceptions of FUBP1 and RBM10, where mutations occurring in at least four samples are annotated. Residues in red represent the most frequently reported hot spot mutations, and the total numbers of observed occurrences are displayed in parentheses. Residue and frequency data were mined from cBioPortal (Cerami et al. 2012) and the image was created with DOG 1.0 (Ren et al. 2009). Mutations in proteins in panel a are commonly thought to induce change-of-function mutations, whereas mutations in proteins in panel b typically result in loss-of-function mutations. Colored regions within each protein diagram represent known domains.
In MDS, SF3B1 mutations are highly enriched within a low-risk subtype known as MDS with ring sideroblasts (MDS-RS) (Papaemmanuil et al. 2011). Although the exact link between SF3B1 mutations and MDS-RS is unknown, clinical diagnostic criteria for MDS-RS incorporate mutation status (Arber et al. 2016). In contrast to the favorable outcome of SF3B1 mutations in MDS, SF3B1 mutations in CLL are associated with adverse outcome and chemoresistance (Quesada et al. 2012, L. Wang et al. 2011). Finally, in the context of UM, SF3B1 mutations are associated with disomy 3 and intermediate risk (Robertson et al. 2017).
SRSF2, which promotes exon splicing, is found to be mutated in chronic myelomonocytic leukemia (CMML), AML, high-risk MDS, myeloproliferative neoplasms, and a small percentage of patients with disomy 3 UM (Yoshida et al. 2011). SRSF2 normally binds to C- and G-rich exonic splicing enhancers (ESEs) to promote splicing (Daubner et al. 2012). SRSF2 mutations concentrate on residue P95 (Figure 2) and confer altered RNA-binding preference that favors recognition of C-rich CCNG over G-rich ESEs and leads to altered splicing of hundreds of mRNAs (Kim et al. 2015, Zhang et al. 2015). One key alteration in SRSF2-mutant cells is altered splicing of EZH2 mRNA, encoding a protein that regulates histone methylation that is also affected by loss-of-function mutations in myeloid neoplasms. The aberrant EZH2 mRNA produced by mutant SRSF2 is targeted for nonsense-mediated decay, and mutations in EZH2 and SRSF2 are significantly mutually exclusive in MDS (Papaemmanuil et al. 2013).
Hot spot mutations in RNA splicing factors occur with mutual exclusivity across the myeloid neoplasms. Initially, this pattern of mutual exclusivity was assumed to indicate redundant effects of these mutations; however, a unifying role for mutations across each mutated splicing factor has been elusive. Mutations across SF3B1 and SRSF2 confer distinct effects on RNA splicing. Likewise, mutations in U2AF1, required for recognition of the AG-dependent 3′ ss recognized by the major spliceosome, are exclusive to SF3B1 and SRSF2 mutations in myeloid neoplasms. U2AF1 mutations predominantly affect S34 and U157 in the zinc fingers (Figure 2). These mutations alter recognition of the 3′ ss, but mutations at each site are associated with differences in splicing events based on the nucleotide surrounding the 3′ AG dinucleotide (Ilagan et al. 2015).
The seemingly disparate effects of mutations in SF3B1, SRSF2, and U2AF1 on splicing led to a search for convergent effects of these mutations in processes unrelated to splicing. To this end, cells bearing mutations in U2AF1 (Nguyen et al. 2017) as well as SRSF2 (Chen et al. 2018) have been reported to have augmented the formation of R-loops, three-stranded nucleic acid structures composed of DNA-RNA hybrids. The increased generation of R-loops in SRSF2- or U2AF1-mutant cells is associated with increased DNA damage and activation of the ATR pathway. Although it is not clear how mutant U2AF1 augments R-loops, mutant SRSF2–induced increased transcriptional pausing appears to increase R-loop generation (Chen et al. 2018). These data provide a potential unifying effect of mutant U2AF1 and SRSF2 with important therapeutic implications. It will therefore be important to determine if mutant SF3B1 similarly impacts R-loop generation.
In addition to an effect on R-loops, one recent report suggested that the U2AF1 S34F mutation may alter interactions with the cleavage and polyadenylation (CP) machinery, resulting in increased use of a distal CP site and longer 3′ untranslated regions (UTRs) (Park et al. 2016). In particular, altered CP of the mRNA encoding the autophagy protein ATG7 was found to result in decreased ATG7, impaired autophagy, and accumulation of secondary mutations. Future efforts will need to confirm if other mutations in U2AF1 or other splicing factors similarly alter CP usage, 3′ UTR length, or autophagy.
Until recently, recurrent mutations in SF3B1, U2AF1, and SRSF2 were the only splicing factors known to harbor hot spot mutations. However, a recent reanalysis of whole-exome sequencing data from 119 patients with 33 solid tumor types has identified recurrent hot spot mutations in PHF5A (a key U2 snRNP component that interacts with SF3B1) and the hnRNP proteins hn-RNPCL1 and PCBP1 (Figure 2) (Seiler et al. 2018a). Effects of these mutations on splicing and tumorigenesis remain unknown.
RECURRENT LOSS-OF-FUNCTION MUTATIONS IN SPLICEOSOMAL GENES IN CANCER
In addition to the mutually exclusive mutations in SF3B1, SRSF2, and U2AF1 in myeloid malignancies, loss-of-function mutations in ZRSR2, essential for 3′ ss recognition in U12-type splicing, were also identified in early reports (Figure 2) (Yoshida et al. 2011). ZRSR2 mutations are enriched in MDS, a form of AML known as blastic plasmacytoid dendritic cell neoplasms, in a small percentage of T cell acute lymphoblastic leukemias, and in thyroid cancers. ZRSR2 is affected by nonsense or frameshift mutations, which presumably result in loss of ZRSR2. Coincident with the role of ZRSR2 in the minor spliceosome, mutation or suppression of ZRSR2 appears to result in retention of U12-type introns (Madan et al. 2015). Splicing events altered by ZRSR2 mutations appear to impact expression of MAPK pathway members and E2F transcription factors. Further work defining how ZRSR2 mutations relate to the mutually exclusive mutations in other splicing factors in MDS may provide novel clues to a shared disease mechanism.
Loss-of-function mutations also prominently affect the splicing factor RBM10, an RNA-binding protein that generally represses splicing (Figure 2). RBM10 mutations are present in lung and bladder adenocarcinomas as well as fatal nonanaplastic thyroid carcinomas (Ibrahimpasic et al. 2017, Imielinski et al. 2012, Seiler et al. 2018a). RBM10 mutations are associated with exon inclusion while RBM10 loss has been shown to promote the proliferation of mouse and human immortalized cells (Bechara et al. 2013, Hernandez et al. 2016). One critical splicing change regulated by RBM10 expression is AS of NUMB, an inhibitor of NOTCH signaling. RBM10 loss promotes expression of a form of NUMB with the inclusion of exon 9, which promotes NOTCH activity (Bechara et al. 2013, Hernandez et al. 2016). In lung and thyroid cancers, RBM10 is frequently mutated with commonly mutated kinases (KRAS, BRAF, EGFR, and PI3K), although the biological significance of these concurrent mutations remains unknown. Other splicing factors recurrently affected by loss-of-function mutations are shown in Figure 2. The spectrum and frequency of these mutations across cancer types are best described by Seiler et al. (2018a).
ABERRANT EXPRESSION OF SPLICING FACTORS AS DRIVERS OF CANCER
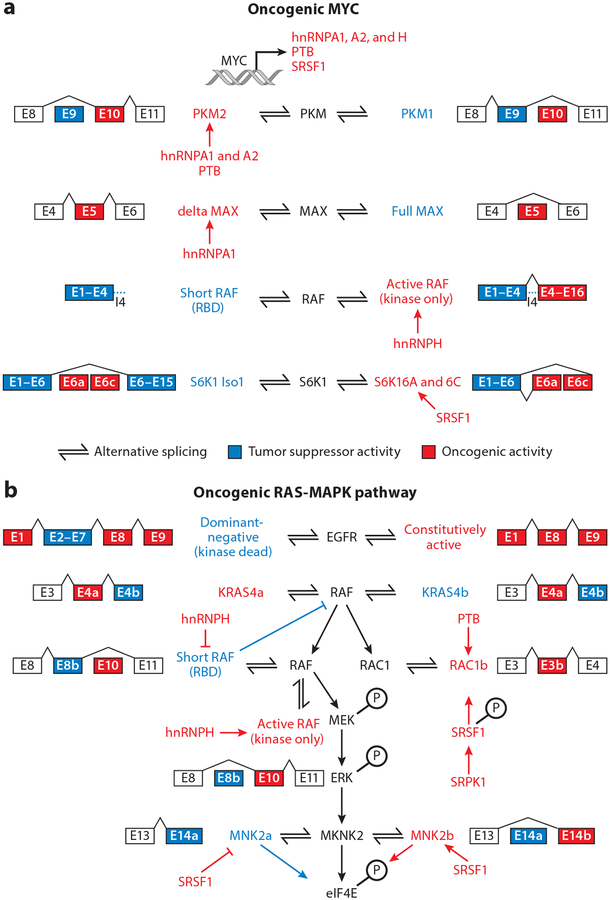
Although hot spot mutations have called attention to the concept of splicing factors as potential oncogenes, the expression of splicing factors in tumors changes frequently and may be driven by oncogenic signaling (Goncalves et al. 2017). For example, the transcription factor MYC, commonly amplified in cancers, upregulates the expression of multiple splicing factors and deregulates splicing. MYC’s involvement in splicing was first demonstrated in the regulation of pyruvate kinase (PKM). Two mutually exclusive isoforms of PKM exist: PKM2, which is almost universally upregulated in cancer and promotes aerobic glycolysis, and PKM1, which is expressed in most normal adult tissues and promotes oxidative phosphorylation. MYC enhances transcription of specific hnRNPs (hnRNPA1, hnRNPA2, and PTB), which in turn promote the expression of the cancer-associated embryonic PKM2 isoform and aerobic glycolysis in glioma (Clower et al. 2010, David et al. 2010). Interestingly, hnRNPA1 also regulates AS of MYC-associated factor X (MAX) to produce delta MAX, which further promotes MYC-dependent transformation and glycolytic gene expression (Figure 3) (Babic et al. 2013, Roy et al. 2017). MYC also controls expression of hnRNPH, which regulates splicing of ARAF kinase (Rauch et al. 2011), increasing the expression of the long isoform that promotes RAS-induced transformation.
Figure 3.
Alternative splicing regulation of the oncogenic MYC and RAS-MAPK pathways. (a) MYC increases the expression of splicing regulators PTB, SRSF1, and hnRNPA1, A2, and H, which in turn change the expression of isoforms of PKM, RAF, MAX, and S6K1. hnRNPA1, A2, and PTB promote the expression of PKM2, a variant of PKM that promotes aerobic glycolysis. hnRNPA1 has also been found to promote the expression of delta MAX, an isoform of MAX that further promotes MYC-dependent transformation and glycolytic gene expression. hnRNPH, under a MYC oncogenic background, promotes the expression of active oncogenic RAF while repressing the short RAF containing only the RBD, which inhibits RAS. The splicing factor SRSF1 can affect the splicing of S6K1, inducing oncogenic short isoforms of this kinase (h6A and h6C), which bind mTOR and enhance 4E-BP1 phosphorylation and cap-dependent translation.(b) Multiple splicing factor regulators change the expression of oncogenic isoforms of proteins involved in the RAS-MAPK pathway. Receptor tyrosine kinases, such as EGFR, are alternatively spliced to generate truncated isoforms, which act in a dominant-negative manner, or constitutively active isoforms (EGFRvIII), which are active regardless of ligand binding. RAS can be alternatively spliced to generate RAS4A, an isoform commonly found in cancers. RAS activates RAF, which can be alternatively spliced to generate a short isoform with RBDs that inhibit RAS or constitutively active isoforms containing only the kinase domain. hnRNPH inhibits the production of dominant-negative RAF isoforms. RAF phosphorylates MEK, which in turn phosphorylates ERK. ERK can phosphorylate MNK2, which is alternatively spliced and regulated by SRSF1. SRSF1 upregulates a pro-oncogenic MNK2B isoform and reduces the MNK2A isoform. Figure adapted from Siegfried et al. (2013). Abbreviations: E, exon; hnRNP, heterogeneous nuclear ribonucleoprotein; RBD, RAS-binding domain.
As evidence of the importance of aberrant splicing downstream of MYC (Anczuków et al. 2012, Das et al. 2012, David et al. 2010, Hsu et al. 2015, Koh et al. 2015), several studies have shown that MYC-transformed cells are exquisitely sensitive to perturbations of splicing. Through screening for synthetic lethal genes in MYC-driven human mammary epithelial cells, Hsu et al. (2015) identified several spliceosome components (SF3B1, U2AF1, SNRPF, EFTUD2, and BUD31) as preferentially required in MYC-transformed cells. BUD31 knockdown led to intron retention and cell death in MYC cells, but not in HER2- or EGFR-transformed cells. Consistent with this genetic dependence, pharmacological spliceosome inhibition impaired survival, tumorigenicity, and metastatic proclivity of MYC-dependent breast cancers.
SRSF1, SRSF3, and SRSF6 are also amplified among certain cancer types (Figure 3) and have been proposed as oncoproteins (Cohen-Eliav et al. 2013, Jensen et al. 2014, Jia et al. 2010). Increased expression of SRSF1 is sufficient to transform human and mouse mammary epithelial cells and regulates splicing of hundreds of transcripts (Anczuków et al. 2012; Karni et al. 2007, 2008). SRSF3 downregulation promoted p53-mediated cellular senescence in part by promoting the expression of p53β, an AS isoform of p53 that enforces p53-mediated senescence (Tang et al. 2013).
SPLICING REGULATION OF ONCOGENIC SIGNALING PATHWAYS IN CANCER
Alternative and aberrant splicing of numerous members of cancer-associated cell growth and death pathways (MAPK, PI3K-AKT, HIPPO, and apoptosis) have been described. These events either promote the expression of isoform proteins that enhance positive feedback signaling or confer resistance to inhibitors of this pathway (Figure 3). For example, the mRNA encoding KRAS undergoes AS, utilizing one of two mutually exclusive exons to generate the isoforms KRAS4A or KRAS4B (Tsai et al. 2015), which transform cells at different rates and exhibit variable targeting to the plasma membrane, the site of interaction with signaling effectors.
Downstream of KRAS, RAF splice variants have been described. For example, BRAF has two annotated variable exons, 8b and 10, generating four distinct isoforms (Papin et al. 1998). The variant that includes exon 10 enhances kinase activity and affinity for downstream kinases MEK1/2, while inclusion of exon 8b has the opposite effect. Aside from these annotated isoforms, several pathological aberrant forms of wild-type and mutant BRAF have been described. For example, thyroid carcinomas express BRAF splice variants that lack the N-terminal autoinhibitory domain, resulting in constitutive BRAF activity (Baitei et al. 2009). Similarly, the variant lacking exons 4 to 8 was identified in BRAFV600E-mutant melanoma cells, which exhibit acquired resistance to the ATP-competitive BRAF inhibitor vemurafenib (Poulikakos et al. 2011). This variant results in the production of a stable truncated BRAFV600E protein lacking the RAS-binding domain (RBD). In the absence of the RBD, BRAF dimerizes and confers resistance to vemurafenib. Truncated forms of BRAFV600E were identified in several primary melanoma patients with acquired resistance to vemurafenib.
Isoforms of ARAF, MEK1/2, and ERK1/2, as well as members of the PI3K-AKT-mTOR pathway, have been identified and shown to have distinct functions. For example, the tumor suppressor PTEN, which counteracts PI3K activity, is regulated by AS. The splice isoform PTEN-5b acts as a dominant negative to promote the activity of PI3K (Agrawal & Eng 2006). Downstream of AKT, mTOR can undergo AS to generate an active oncogenic form, mTORβ (Panasyuk et al. 2009). SRSF1 promotes the production of short S6K1 isoforms frequently upregulated in tumors, h6A and h6C (Ben-Hur et al. 2013, Karni et al. 2007), which enhance the transformation of cells via activation of the mTOR pathway in the absence of external stimuli (Ben-Hur et al. 2013). S6K1 short isoforms also cause 4E-BP1 inactivation and enhanced translation of oncogenes and antiapoptotic genes (Figure 3) (Ben-Hur et al. 2013).
In addition to AS of MAPK signaling intermediates, the upstream receptor tyrosine kinases activating MAPK signaling are also subject to dysregulation by changes in splicing. For example, the variant of EGFR known as variant EGFRvIII contains an in-frame deletion of exons 2–7 and can be generated by rearrangement or altered pre-mRNA processing (Nishikawa et al. 1994, Sugawa et al. 1990). EGFRvIII lacks part of the ligand-binding domain, is constitutively active, and confers growth advantage to cells (Nishikawa et al. 1994, Weidle et al. 2011). Another EGFR isoform produced by skipping exon 4, de4 EGFR, is also constitutively active and promotes metastases (Figure 3) (H. Wang et al. 2011). Selective expression of EGFR isoforms in several tumors makes them attractive cancer therapy targets (Weidle et al. 2011). Similarly, a variety of truncating mutations resulting in exclusion of exon 14 in MET inhibit degradation of MET, prolonging its oncogenic activity. MET exon 14 alterations have been detected in a variety of cancers and confer sensitivity to MET inhibitors (Frampton et al. 2015).
Activation of RAS pathways increases the expression of PTBP1, which, in turn, shifts AS of transcripts encoding the small GTPase RAC1, NUMB, and PKM, each of which are involved in tumorigenesis (Climente-Gonzalez et al. 2017, Hollander et al. 2016, Israelsen et al. 2013, Takahashi et al. 2015). In addition to transcriptional stimulation of PTBP1 downstream of RAS, ERK phosphorylates splicing factor SAM68 to induce its binding to SRSF1 pre-mRNA (Valacca et al. 2010) and promote retention of an intron required for production of full-length SRSF1, diverting the AS event that would cause SRSF1 degradation (Valacca et al. 2010). In turn, SRSF1 promotes AS of MNK2, producing the oncogenic isoform MNK2B (Figure 3) (Karni et al. 2007, Maimon et al. 2014, Scheper et al. 2003). Lastly, splice variants resulting from increased hnRNPA2 expression induce a positive-feedback loop that promotes MAPK signaling to maintain tumor cells (Shilo et al. 2014).
STRATEGIES TO TARGET SPLICING IN CANCER
Motivated by altered splicing in a variety of tumor types, compounds that impair splicing catalysis directly or through inhibition of PTMs of splicing factors have been developed (Figure 4). The rationale for these approaches is supported by the observation that cancer cells bearing heterozygous change-of-function mutations in SF3B1, SRSF2, and U2AF1 require the wild-type allele for survival and are expressed in a mutually exclusive manner, in part, due to a synthetic lethal interaction between these mutations (Lee et al. 2016a,b). Consistent with this, hematopoietic cells bearing mutations in these factors have been shown to be preferentially sensitive to compounds that bind to the SF3B complex and impede splicing (Lee et al. 2016a, Obeng et al. 2016, Shirai et al. 2017). Furthermore, given that SF3B1 is an essential protein, cancer cells with partial copy number loss of SF3B1 are preferentially sensitive to inhibition of residual SF3B1 (Paolella et al. 2017). Finally, there is also evidence that MYC-amplified tumors rely on increased splicing activity, rendering them sensitive to inhibition of splicing catalysis.
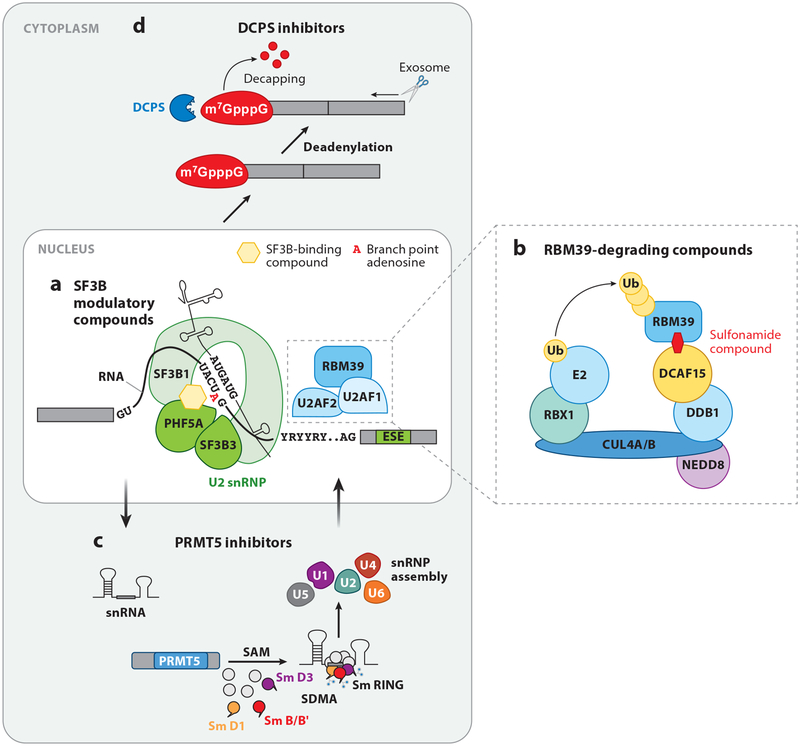
Figure 4.
Means to pharmacologically target splicing in cancer. (a) SF3B modulatory compounds bind to the SF3B complex and block its interaction with the branch point adenosine, a process essential for RNA splicing. (b) Recently, a series of molecules known as anticancer sulfonamides were described (Han et al. 2017, Uehara et al. 2017) that bridge the cellular CUL4-DDB1-DDA1 E3 ubiquitin ligase complex to RBM39 via the adaptor protein DCAF15, resulting in polyubiquitination and subsequent proteosomal degradation of RBM39. (c) Arginine methylation of a variety of splicing proteins by protein arginine methyltransferases (PRMTs) is required for normal spliceosome assembly and function. Symmetric dimethylation of arginines (SDMA) on Sm proteins is required for assembly of the small nuclear ribonucleoprotein (snRNP) complexes. (d) It has recently been shown that small-molecule inhibitors of the scavenger messenger (mRNA)-decapping enzyme DCPS [required for removal of the N7-methylated guanine cap (m7GpppG) on mRNAs] result in a change in dose-dependent effects on splicing (Yamauchi et al. 2018).
Among the first compounds known to alter splicing were natural compounds (spliceostatin A, pladienolide B, and herboxidiene) that bind the SF3B complex [reviewed recently by Agrawal et al. (2018) and Lee & Abdel-Wahab (2016)]. These agents informed the development of synthetic analogs E7107 and H3B-8800 (Seiler et al. 2018b). Structures of the SF3B complex bound to pladienolide B or E7107 have shown that these molecules bind in the branch point binding pocket of the SF3B complex and thereby block splicing (Figure 4a) (Cretu et al. 2018, Finci et al. 2018). Moreover, studies of cancer cells with acquired resistance to SF3B inhibitor compounds have identified mutations in SF3B1 as well as PHF5A that confer resistance to these compounds (Teng et al. 2017). The structure of SF3B1 reveals that these key residues are involved in binding compounds as well as branch points, explaining their role in drug resistance.
The SF3B modulator E7107 was previously studied in two phase I clinical trials in patients with advanced carcinomas (Eskens et al. 2013, Hong et al. 2014). However, development of ocular complications via an undefined mechanism halted further development of E7107. H3B-8800 is an orally bioavailable SF3B modulatory complex that similarly interferes with the interaction of the complex with branch points. A phase I dose-escalation study of H3B-8800 in AML, CMML, and MDS has recently opened and is recruiting patients (https://www.clinicaltrials.gov identifier NCT02841540).
Recently, a novel strategy to target splicing has emerged using anticancer sulfonamides, including the agents indisulam, tasisulam, and chloroquinoxaline sulfonamide. Phase I and II clinical trials of sulfonamides were previously conducted in a variety of cancer types (Assi et al. 2018, Dittrich et al. 2003, Talbot et al. 2007). Although target plasma concentrations were achieved and remarkable responses were observed in individual patients, consistent antitumor effects were not identified with single-agent sulfonamides. Recent studies, however, have elucidated the mechanism of action of these agents and provided insight into possible biomarkers for response to sulfonamides. Two publications showed that sulfonamides physically bridge the splicing factor RBM39 (also known as CAPERα) to the CUL4-DDB1-DDA1-DCAF15 E3 ubiquitin ligase complex, resulting in polyubiquitination and proteosomal degradation of RBM39 (Figure 4b) (Han et al. 2017, Uehara et al. 2017). Degradation of RBM39, an RNA-binding protein known to associate with the U2AF complex (Loerch et al. 2014, Stepanyuk et al. 2016), causes intron retention and exon skipping. Supporting evidence for on-target effects of sulfonamides for RBM39 includes the fact that mutations within RBM39 confer resistance to these compounds. At the same time, expression of DCAF15 (the substrate-specific receptor component of the Cullin-RING ligase complex responsible for RBM39 degradation) also correlates with sensitivity to sulfonamides. In the absence of DCAF15, RBM39 is not degraded by sulfonamides and cells are resistant to these compounds. In future studies, RBM39 mutational status and DCAF15 expression levels may therefore predict response or resistance to these agents.
In addition to the above approaches targeting proteins involved in RNA splicing, several inhibitors of enzymes that place critical PTMs on splicing proteins have been developed. These include inhibitors of protein arginine methyltransferases (PRMTs), SR protein kinases, CDC-like kinases, and dual-specificity tyrosine phosphorylation–regulated kinases (Figure 4c). At least one such molecule, an inhibitor of PRMT5, is already in phase I clinical trials in solid tumor patients (identifier NCT02783300). Rationale for the use of PRMT5 inhibitors in cancers sensitive to alterations in splicing comes from work by Koh et al. (2015), which identified several components of splicing machinery as key effectors of MYC in the Eμ-myc mouse model of lymphoma, exposing therapeutic vulnerabilities in MYC-driven cancers where existing therapeutic strategies are limited. Outside of spliceosomal proteins, a genome-wide CRISPR-based screen recently identified that inhibition of DCPS, an mRNA-decapping enzyme, also perturbs splicing and alters RNA degradation (Yamauchi et al. 2018). DCPS deletion or inhibition using RG3039, a DCPS inhibitor, decreased proliferation and induced differentiation of AML cells (Figure 4d). The basis for the specific antileukemic mechanism of DCPS inhibition is unknown. Nonetheless, prior use of RG3039 in clinical trials in spinal muscular atrophy patients will hopefully facilitate use of this compound in cancer patients soon.
SUMMARY AND FUTURE PERSPECTIVES
In the 40 years since the discovery of splicing and the 10 years since the initial use of RNA-seq, a great deal has been learned about how splicing is altered in cancer. While many individual pathogenic splicing events have been characterized, systematic studies of the functional impact of widespread splicing alterations in cancer have yet to be performed. Determining the functional impact of splicing changes with sufficient resolution at the proteome level would greatly help in this regard. Understanding the true effect of splicing changes on the cancer proteome has the potential to identify novel biomarkers and develop complementary means to therapeutically target altered RNA splicing. Moreover, refined use of RNA-seq and proteomic profiling will help address these outstanding questions and inform the development of a unified theme describing the effects of altered RNA splicing in cancer.
ACKNOWLEDGMENTS
L.E.H. has received research support from the Pancreatic Cancer Action Network-AACR (American Association for Cancer Research) Pathway to Leadership Grant, a National Pancreas Foundation grant, and the NIH (National Institutes of Health)/NCI (National Cancer Institute). O.A.-W. has received research support from NIH/NHLBI (National Heart, Lung, and Blood Institute) (R01 HL128239), the Department of Defense Bone Marrow Failure Research Program (W81XWH-16–1–0059), the Starr Foundation (I8–A8–075), the Henry and Marilyn Taub Foundation, The Edward P. Evans Foundation, the Leukemia and Lymphoma Society, the Pershing Square Sohn Cancer Research Alliance, and Cycle for Survival.
DISCLOSURE STATEMENT
O. A.-W. is a consultant for H3 Biomedicine Inc. and Merck and has received research support from H3 Biomedicine.
LITERATURE CITED
- Agrawal AA, Yu L, Smith PG, Buonamici S. 2018. Targeting splicing abnormalities in cancer. Curr. Opin. Genet. Dev 48:67–74 [DOI] [PubMed] [Google Scholar]
- Agrawal S, Eng C. 2006. Differential expression of novel naturally occurring splice variants of PTEN and their functional consequences in Cowden syndrome and sporadic breast cancer. um. Mol. Genet 15:777–87 [DOI] [PubMed] [Google Scholar]
- Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, et al. 2016. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun 7:10615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anczuków O, Rosenberg AZ, Akerman M, Das S, Zhan L, et al. 2012. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat. Struct. Mol. Biol 19:220–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, et al. 2016. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127:2391–405 [DOI] [PubMed] [Google Scholar]
- Assi R, Kantarjian HM, Kadia TM, Pemmaraju N, Jabbour E, et al. 2018. Final results of a phase 2, open-label study of indisulam, idarubicin, and cytarabine in patients with relapsed or refractory acute myeloid leukemia and high-risk myelodysplastic syndrome. Cancer 124:2758–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic I, Anderson ES, Tanaka K, Guo D, Masui K, et al. 2013. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 17:1000–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baitei EY, Zou M, Al-Mohanna F, Collison K, Alzahrani AS, et al. 2009. Aberrant BRAF splicing as an alternative mechanism for oncogenic B-Raf activation in thyroid carcinoma. J. Pathol 217:707–15 [DOI] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, et al. 2010. Deciphering the splicing code. Nature 465:53–59 [DOI] [PubMed] [Google Scholar]
- Bechara EG, Sebestyen E, Bernardis I, Eyras E, Valcarcel J. 2013. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol. Cell 52:720–33 [DOI] [PubMed] [Google Scholar]
- Ben-Hur V, Denichenko P, Siegfried Z, Maimon A, Krainer A, et al. 2013. S6K1 alternative splicing modulates its oncogenic activity and regulates mTORC1. Cell Rep. 3:103–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. 2017. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic Acids Res. 45:4837–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen JY, Huang YJ, Gu Y, Qiu J, et al. 2018. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol. Cell 69:412–25.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climente-Gonzalez H, Porta-Pardo E, Godzik A, Eyras E. 2017. The functional impact of alternative splicing in cancer. Cell Rep. 20:2215–26 [DOI] [PubMed] [Google Scholar]
- Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. 2010. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. PNAS 107:1894–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, et al. 2013. The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers. J. Pathol 229:630–39 [DOI] [PubMed] [Google Scholar]
- Cretu C, Agrawal AA, Cook A, Will CL, Fekkes P, et al. 2018. Structural basis of splicing modulation by antitumor macrolide compounds. Mol. Cell 70:265–73.e8 [DOI] [PubMed] [Google Scholar]
- Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, et al. 2015. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep. 13:1033–45 [DOI] [PubMed] [Google Scholar]
- Das S, Anczukow O, Akerman M, Krainer AR. 2012. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1:110–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta T, Ladd AN. 2012. The importance of CELF control: molecular and biological roles of the CUGBP, Elav-like family of RNA-binding proteins. WIREs RNA 3:104–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. 2012. A syn-anti conformational difference allowsSRSF2 to recognize guanines and cytosines equally well. EMBO J. 31:162–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463:364–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C, Ghia EM, Shepard PJ, Rassenti L, Barrett CL, et al. 2015. Transcriptome sequencing reveals potential mechanism of cryptic 3′ splice site selection in SF3B1-mutated cancers. PLOS Comput. Biol 11:e1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich C, Dumez H, Calvert H, Hanauske A, Faber M, et al. 2003. Phase I and pharmacokinetic study ofE7070, a chloroindolyl-sulfonamide anticancer agent, administered on a weekly schedule to patients with solid tumors. Clin. Cancer Res 9:5195–204 [PubMed] [Google Scholar]
- Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. 2016. RNA splicing factors as oncoproteins and tumour suppressors. Nat. Rev. Cancer 16:413–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskens FA, Ramos FJ, Burger H, O’Brien JP, Piera A, et al. 2013. Phase I pharmacokinetic and pharmaco-dynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clin. Cancer Res 19:6296–304 [DOI] [PubMed] [Google Scholar]
- Fica SM, Nagai K. 2017. Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat. Struct. Mol. Biol 24:791–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finci LI, Zhang X, Huang X, Zhou Q, Tsai J, et al. 2018. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev. 32:309–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, et al. 2015. Activation of MET via diverse exon14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 5:850–59 [DOI] [PubMed] [Google Scholar]
- Fu XD, Ares M Jr. 2014. Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet 15:689–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, et al. 2013. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 3:1122–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves V, Pereira JFS, Jordan P. 2017. Signaling pathways driving aberrant splicing in cancer cells. Genes 9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, et al. 2012. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat. Genet 44:53–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T, Goralski M, Gaskill N, Capota E, Kim J, et al. 2017. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 356:eaal3755. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. 2013. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat. Genet 45:133–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcarcel J. 2016. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 13:466–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander D, Donyo M, Atias N, Mekahel K, Melamed Z, et al. 2016. A network-based analysis of colon cancer splicing changes reveals a tumorigenesis-favoring regulatory pathway emanating from ELK1. Genome Res. 26:541–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong DS, Kurzrock R, Naing A, Wheler JJ, Falchook GS, et al. 2014. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Investig. New Drugs 32:436–44 [DOI] [PubMed] [Google Scholar]
- Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, et al. 2015. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525:384–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahimpasic T, Xu B, Landa I, Dogan S, Middha S, et al. 2017. Genomic alterations in fatal forms of non-anaplastic thyroid cancer: identification of MED12 and RBM10 as novel thyroid cancer genes associated with tumor virulence. Clin. Cancer Res 23:5970–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, et al. 2015. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 25:14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, et al. 2012. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150:1107–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, et al. 2013. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 155:397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasinghe RG, Cao S, Gao Q, Wendl MC, Vo NS, et al. 2018. Systematic analysis of splice-site-creating mutations in cancer. Cell Rep. 23:270–81.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Wilkinson JE, Krainer AR. 2014. Splicing factor SRSF6 promotes hyperplasia of sensitized skin Nat. Struct. Mol. Biol 21:189–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Li C, McCoy JP, Deng C-X, Zheng Z-M. 2010. SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int. J. Biol. Sci 6:806–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Lee D, Lee J, Park D, Kim YJ, et al. 2015. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat. Genet 47:1242–48 [DOI] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5–14 [DOI] [PubMed] [Google Scholar]
- Kahles A, Lehmann K-V, Toussaint NC, Huser M, Stark SG, et al. 2018. Comprehensive analysis of alternative splicing across tumors from 8,512 patients. Cell 34:211–224.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe S, Sinha R, Mu D, Krainer A. 2007. The gene encoding the splicing factorSF2/ASF is a proto-oncogene. Nat. Struct. Mol. Biol 14:185–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, Hippo Y, Lowe SW, Krainer AR. 2008. The splicing-factor oncoprotein SF2/ASF activates mTORC1. PNAS 105:15323–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, et al. 2015. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27:617–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, et al. 2015. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523:96–100 [DOI] [PubMed] [Google Scholar]
- Konieczny P, Stepniak-Konieczna E, Sobczak K. 2014. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 42:10873–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. 1999. hnRNP complexes: composition, structure, and function. Curr. Opin. Cell Biol 11:363–71 [DOI] [PubMed] [Google Scholar]
- Lee SC-W, Abdel-Wahab O. 2016. Therapeutic targeting of splicing in cancer. Nat. Med 22:976–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC-W, Dvinge H, Kim E, Cho H, Micol J-B, et al. 2016a. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat. Med 22:672–78. Erratum. 2016. Nat. Med. 22:692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC-W, Khrystyna D, Obeng EA, Kim E, Micol J-B, Yoshimi A, et al. 2016b. Synthetic lethal interactions of MDS-associated spliceosomal gene mutations identifies the basis for their mutual exclusivity. Blood 128:961 [Google Scholar]
- Loerch S, Maucuer A, Manceau V, Green MR, Kielkopf CL. 2014. Cancer-relevant splicing factor CAPERα engages the essential splicing factor SF3b155 in a specific ternary complex. J. Biol. Chem 289:17325–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J 417:15–27 [DOI] [PubMed] [Google Scholar]
- Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, et al. 2015. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun 6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon A, Mogilevsky M, Shilo A, Golan-Gerstl R, Obiedat A, et al. 2014. Mnk2 alternative splicing modulates the p38-MAPK pathway and impacts Ras-induced transformation. Cell Rep. 7:501–13 [DOI] [PubMed] [Google Scholar]
- Martin M, Masshofer L, Temming P, Rahmann S, Metz C, et al. 2013. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet 45:933–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mupo A, Seiler M, Sathiaseelan V, Pance A, Yang Y, et al. 2017. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia 31:720–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. 2017. Functions of replication protein A as a sensor of R loops and a regulator of RNaseH1. Mol. Cell 65:832–47.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, et al. 1994. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. PNAS 91:7727–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, et al. 2016. Physiologic expression of Sf3b1K700Ecauses impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell 30:404–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasyuk G, Nemazanyy I, Zhyvoloup A, Filonenko V, Davies D, et al. 2009. mTORβ splicing isoform promotes cell proliferation and tumorigenesis. J. Biol. Chem 284:30807–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolella BR, Gibson WJ, Urbanski LM, Alberta JA, Zack TI, et al. 2017. Copy-number and gene dependency analysis reveals partial copy loss of wild-type SF3B1 as a novel cancer vulnerability. eLife 6:e23268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, et al. 2011. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N. Engl. J. Med 365:1384–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, et al. 2013. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 122:3616–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papin C, Denouel-Galy A, Laugier D, Calothy G, Eychene A. 1998. Modulation of kinase activity and oncogenic properties by alternative splicing reveals a novel regulatory mechanism for B-Raf. J. Biol. Chem 273:24939–47 [DOI] [PubMed] [Google Scholar]
- Park SM, Ou J, Chamberlain L, Simone TM, Yang H, et al. 2016. U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3′ end formation. Mol. Cell 62:479–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, et al. 2011. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480:387–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Conde L, Villamor N, Ordóñez G, Jares P, et al. 2012. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet 44:47–52 [DOI] [PubMed] [Google Scholar]
- Rauch J, Moran-Jones K, Albrecht V, Schwarzl T, Hunter K, et al. 2011. c-Myc regulates RNA splicing of theA-Raf kinase and its activation of the ERK pathway. Cancer Res. 71:4664–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. 2009. DOG 1.0: illustrator of protein domain structures. Cell Res.19:271–73 [DOI] [PubMed] [Google Scholar]
- Robertson AG, Shih J, Yau C, Gibb EA, Oba J, et al. 2017. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32:204–20.e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Huang Y, Seckl MJ, Pardo OE. 2017. Emerging roles of hnRNPA1 in modulating malignant transformation. WIREs RNA 8:e1431. [DOI] [PubMed] [Google Scholar]
- Scheper GC, Parra JL, Wilson M, Van Kollenburg B, Vertegaal AC, et al. 2003. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol. Cell Biol 23:5692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Nagai K. 2017. CryoEM structures of spliceosomal complexes reveal the molecular mechanism of pre-mRNA splicing. Curr. Opin. Struct. Biol 46:130–39 [DOI] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. 2016. RNA mis-splicing in disease. Nat. Rev. Genet 17:19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, et al. 2018a. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 23:282–96.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M, Yoshimi A, Darman R, Chan B, Keaney G, et al. 2018b. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nat. Med 24:497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y 2017. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol Cell Biol 18:655–70 [DOI] [PubMed] [Google Scholar]
- Shilo A, Ben Hur V, Denichenko P, Stein I, Pikarsky E, et al. 2014. Splicing factor hnRNP A2 activates the Ras-MAPK-ERK pathway by controlling A-Raf splicing in hepatocellular carcinoma development. RNA 20:505–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai CL, White BS, Tripathi M, Tapia R, Ley JN, et al. 2017. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat. Commun 8:14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried Z, Bonomi S, Ghigna C, Karni R. 2013. Regulation of the Ras-MAPK and PI3K-mTOR signalling pathways by alternative splicing in cancer. Int. J. Cell Biol 2013:568931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanyuk GA, Serrano P, Peralta E, Farr CL, Axelrod HL, et al. 2016. UHM–ULM interactions in theRBM39–U2AF65 splicing-factor complex. Acta Crystallogr. D 72:497–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawa N, Ekstrand AJ, James CD, Collins VP. 1990. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. PNAS 87:8602–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Minana B, Valcarcel J, Gabaldon T, Lehner B. 2014. Synonymous mutations frequently act as driver mutations in human cancers. Cell 156:1324–35 [DOI] [PubMed] [Google Scholar]
- Sutherland LC, Rintala-Maki ND, White RD, Morin CD. 2005. RNA binding motif (RBM) proteins: A novel family of apoptosis modulators? J. Cell Biochem. 94:5–24 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Nishimura J, Kagawa Y, Kano Y, Takahashi Y, et al. 2015. Significance of polypyrimidine tract-binding protein 1 expression in colorectal cancer. Mol. Cancer Ther 14:1705–16 [DOI] [PubMed] [Google Scholar]
- Talbot DC, von Pawel J, Cattell E, Yule SM, Johnston C, et al. 2007. A randomized phase II pharmacokinetic and pharmacodynamic study of indisulam as second-line therapy in patients with advanced non-small cell lung cancer. Clin. Cancer Res 13:1816–22 [DOI] [PubMed] [Google Scholar]
- Tang Q, Rodriguez-Santiago S, Wang J, Pu J, Yuste A, et al. 2016. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev. 30:2710–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Horikawa I, Ajiro M, Robles AI, Fujita K, et al. 2013. Downregulation of splicing factor SRSF3 induces p53β, an alternatively spliced isoform of p53 that promotes cellular senescence. Oncogene 32:2792–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Tsai JH, Puyang X, Seiler M, Peng S, et al. 2017. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A-SF3b complex. Nat. Commun 8:15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FD, Lopes MS, Zhou M, Court H, Ponce O, et al. 2015. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. PNAS 112:779–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Minoshima Y, Sagane K, Sugi NH, Mitsuhashi KO, et al. 2017. Selective degradation of splicing factor CAPERα by anticancer sulfonamides. Nat. Chem. Biol 13:675–80 [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, et al. 2006. An RNA map predicting Nova-dependent splicing regulation. Nature 444:580–86 [DOI] [PubMed] [Google Scholar]
- Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, et al. 2010. Sam68 regulates EMT through alternative splicing–activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol 191:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Luhrmann R. 2015a. SnapShot: spliceosome dynamics I. Cell 161:1474.e1. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Luhrmann R. 2015b. SnapShot: spliceosome dynamics II. Cell 162:456.e1. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhou M, Shi B, Zhang Q, Jiang H, et al. 2011. Identification of an exon 4-deletion variant of epidermal growth factor receptor with increased metastasis-promoting capacity. Neoplasia 13:461–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lawrence M, Wan Y, Stojanov P, Sougnez C, et al. 2011. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med 365:2497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle UH, Maisel D, Klostermann S, Weiss EH, Schmitt M. 2011. Differential splicing generates new trans-membrane receptor and extracellular matrix-related targets for antibody-based therapy of cancer. Cancer Genom. Proteom 8:211–26 [PubMed] [Google Scholar]
- Yamauchi T, Masuda T, Canver MC, Seiler M, Semba Y, et al. 2018. Genome-wide CRISPR-Cas9 screen identifies leukemia-specific dependence on a pre-mRNA metabolic pathway regulated by DCPS. Cancer Cell 33:386–400.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, et al. 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478:64–69 [DOI] [PubMed] [Google Scholar]
- Zhang J, Lieu YK, Ali AM, Penson A, Reggio KS, et al. 2015. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. PNAS 112:E4726–34 [DOI] [PMC free article] [PubMed] [Google Scholar]