Abstract
The emergence of coronavirus disease 2019 (COVID-19) in December 2019 has resulted in over 20 million cases and 741,808 deaths globally, affecting more than 200 countries. COVID-19 was declared a pandemic on 11 March 2020 by the World Health Organization. The disease is caused by severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). There is limited information on COVID-19, and treatment has so far focused on supportive care and use of repurposed drugs. COVID-19 can be transmitted via person-to-person contact through droplet spread. Some of the recommended precautionary measures to reduce the rate of disease spread include social distancing, good hygiene practices, and avoidance of crowded areas. These measures are effective because the droplets are heavy and can only travel approximately 1 meter in the air, settling quickly on fixed surfaces. Promising strategies to combat SARS-CoV-2 include discovery of therapeutic targets/drugs and vaccines. In this review, we summarize the epidemiology, pathophysiology, and diagnosis of COVID-19. We also address the mechanisms of action of approved repurposed drugs for therapeutic management of the disease.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, 2019-nCoV, ACE-2, repurposed drugs
Introduction
COVID-19 has become one of the most dangerous pandemics in recent history. The pandemic has claimed more than 740,000 lives, with more than 20 million reported cases since the original outbreak. The disease is caused by SARS-CoV-2, a zoonotic pathogen that acquired mutations as it crossed the species barrier from bat to pangolin enabling it to infect humans.1 SARS-CoV-2 was confirmed as a novel coronavirus using molecular methods and initially named 2019 novel coronavirus (2019-nCoV).2 The disease caused by this virus was later renamed COVID-19 by the World Health Organization (WHO).3 SARS-CoV-2 is highly infectious and has spread to more than 200 countries in all continents. Hence, the virus was declared a global threat (pandemic) by the WHO. SARS-CoV-2 has a single-stranded positive sense RNA genome (+ssRNA) approximately 30 kb long. The SARS-CoV-2 genome is organized similarly to those of SARS-CoV-1 and Middle East respiratory syndrome (MERS)-CoV.4 Using phylogenetic analysis, the SARS-CoV-2 genome was demonstrated to share 96% sequence similarity with a bat CoV genome. SARS-CoV-2 belongs to the β-coronavirus genus that includes SARS-CoV-1 and MERS-CoV.5 The clinical symptoms of COVID-19 include fever, cough, and pneumonia, which makes the disease enormously dangerous with a high case fatality rate.6,7 In contrast to other β-coronaviruses, many SARS-CoV-2 deaths have resulted from multiple organ dysfunction syndrome (MSOF) rather than respiratory failure.8 This could be attributed to the widespread distribution of angiotensin converting enzyme-2 (ACE-2), the primary receptor for SARS coronaviruses, in multiple organs.9,10 ACE-2 is expressed as a cell-surface molecule in the respiratory tract (epithelium, arterial and venous endothelium), the small intestinal epithelium, and arterial smooth muscle cells.11
SARS-CoV-2 is morphologically spherical and is characterized by presence of a spike protein, lower pathogenicity and higher transmissibility between humans.12 Some of the primary measures taken to reduce the number of infections and prevent community transmission are to avoid crowds and gatherings and to practice good hygiene. Interestingly, countries with the highest reported prevalence and mortality such as the United States, Spain, Italy, the United Kingdom, Russia, Germany, Brazil, France, Turkey, Iran and China, are more concerned with flattening the curve through early case detection, isolation, and development of therapeutic drugs and vaccines. Because of the novel nature of this disease, there is limited information regarding risk factors for severe outcomes. Specific factors such as the serial interval, viral lifespan, incubation period, pathogenic mechanisms, clinical features and optimal disease management remain unclear. Therefore, this review aimed to summarize the epidemiology and pathophysiology of SARS-CoV-2 as well as the use of repurposed Food and Drug Administration (FDA) approved drugs for therapy. The entry of this virus into host cells and possible downstream complications deserve closer attention.
Coronaviruses
CoVs are members of the subfamily Othocoronavirinae of the family Coronaviridae. The subfamily consist of four genera: alpha, beta, gamma and delta CoVs.13 Both alpha- and beta-CoVs can infect mammals, including humans, while the gamma- and delta-CoVs only infect birds.14 About seven CoVs have been isolated from humans (Figure 1). These include two alpha-CoVs, human coronavirus 229E (HCoV-229E) and human coronavirus NL63 (HCoV-NL63), and five beta-CoVs: human coronavirus OC43 (HCoV-OC43), human coronavirus HKU1 (HCoV-HKU1), SARS-CoV-1, MERS-CoV, and SARS-CoV-2. SARS-CoV-1, MERS-CoV, and SARS-CoV-2 are pathogenic and cause severe infections in humans following contact with the respective intermediate hosts (bats) (Figure 2). However, HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1 do not appear to cause severe infections in humans.14
Figure 1.
Alpha- and beta-coronaviruses in humans.
Figure 2.
Mode of coronavirus transmission from carriers to humans.
CoVs are enveloped viruses with +ssRNA genomes. They have the largest genomes (approximately 26–33 kb) among RNA viruses. All CoVs possess non-segmented genomes with similar organization.15 Generally, about two-thirds of the genome consists of two large and overlapping open reading frames (ORF1a and ORF1b), which are translated into polyproteins pp1a and pp1ab and subsequently processed to yield 16 non-structural proteins (nsp1 to nsp16).16 The remaining one-third of the genome consists of ORFs encoding structural proteins including the spike (S) glycoprotein embedded in the envelope and the envelope (E), matrix (M), and nucleoproteins (N).17 There are short untranslated regions at both the 3ʹ and 5ʹ ends. The S protein plays a role in receptor-binding and entry of virus into host cells and is thus considered a major therapeutic target. The M and E proteins are important for viral assembly, while the N protein is necessary for synthesis of RNA.18
Overview of the SARS-CoV family
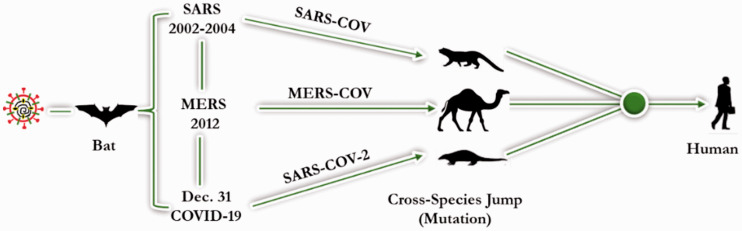
SARS-CoVs belongs to a global family of viruses causing respiratory disease and influenza-like symptoms such as fever, muscle pain, sore throat, headache, and cough.19 Figure 3 highlights the onset and progression of SARS-CoV-1, MERS-CoV, and SARS-CoV-2 infection. The first case of SARS-CoV-1, reported in China, resulted in an outbreak that caused hundreds of deaths and thousands of infected cases in the early 2000s.20 A pneumonia-like syndrome (MERS) was first discovered in Saudi Arabia and then spread to several countries, where it incurred a mortality rate of about 3% to 6%. Marra et al (21) observed that the MERS mortality rate increased with age and was as high as 43% to 55% in people older than 60 years. In December 2019, SARS-CoV-2, caused an outbreak in China and then spread worldwide. The resulting disease (COVID-19) is a serious threat mostly in people with compromised immune systems or underlying conditions such as lung disease, diabetes mellitus, and human immunodeficiency virus infection.22
Figure 3.
Development of the (a) SARS-CoV-1, (b) MERS-CoV, and (c) SARS-CoV-2 epidemics.
SARS-CoV-1
SARS-CoV-1 causes a viral respiratory disease and belongs to lineage B of the beta-CoVs.11 SARS-CoV-1 infection in humans was first reported in 2002 in China (Figure 3a). Within 1 year, it affected about 29 countries with about 8,422 cases globally.13 Bats are the primary hosts of this virus, with palm civets as intermediate hosts. Humans are infected via direct contact with intermediate hosts or through consumption of uncooked meat, milk or urine, as shown in Figure 2. Human-to-human infection also occurred through nosocomial transmission.11 Symptoms of human SARS-CoV-1 infections include headache, fever and respiratory complications such as cough, dyspnea, and pneumonia.23
The SARS-CoV-1 genome is 29,727 nucleotides in length. The 5ʹ end of the genome contains ORF1a and ORF1b. The polyproteins encoded by these ORF are auto-catalytically processed to yield a number of viral proteases as well as the RNA-dependent RNA polymerase (RdRp). The remainder of the genome encodes the viral structural proteins (S, E, M and N) as well as several accessory proteins.18
The receptor for SARS-CoVs is ACE-2, a surface molecule found on cells of the respiratory tract, the small intestinal epithelium, and smooth muscle. In the respiratory tract, ACE-2 is expressed on epithelial cells of the alveoli, bronchi, trachea, and bronchial serous glands, as well as on alveolar monocytes and macrophages. The ACE-2 enzyme plays an important role in protection against lung failure.11
MERS-CoV
MERS-CoV causes a viral respiratory disease and belongs to lineage C of the beta-CoVs.11 The first cases of human infection by MERS-CoV were reported in Saudi Arabia in 2012. Cases were subsequently reported in other countries including Qatar, Egypt, and the Republic of Korea following contact with infected camels (Figure 3b). Cases of MERS-CoV were identified in about 27 countries between 2012 and 2018.18 MERS-CoV RNA in camels showed more than 99% genomic sequence similarity to human MERS-CoV.23 Bats are the natural hosts of MERS-CoV (Figure 2), and a high prevalence of MERS-CoV infections in dromedary camels (intermediate hosts) was confirmed in the Middle East, Spain, and Africa. MERS-CoV infections were transmitted to humans following contact with camels infected with the virus.23
The MERS-CoV genome is about 30,119 nucleotides in length and has a 5ʹ terminal cap structure and a poly (A) tail at the 3ʹ end. The genome encodes 16 non-structural proteins (nsp1–nsp16) at the 5ʹ end, four structural proteins (S, E, M, and N) at the 3ʹ end,24 and five accessory proteins in ORF3, ORF4a, ORF4b, ORF5, and ORF8.18 Risk factors for severe MERS-CoV include age and the presence of underlying conditions such as diabetes, obesity, hypertension, chronic renal disease, and lung diseases.25
The receptor for MERS-CoV is dipeptidyl peptidase 4 (DPP4 or CD26). DPP4 is a multifunctional cell surface protein and is expressed on the epithelial cells of the respiratory tract, liver, kidney, small intestine, and prostate, as well as on activated white blood cells. DPP4 also plays a vital role in activation of T cells and providing co-stimulatory signals for immune responses of T cells.11,18 Consequently, MERS-CoV causes acute pneumonia and renal dysfunction with associated clinical symptoms such as cough, chest pain, sore throat, fever, diarrhea, vomiting, and abdominal pain. MERS-CoV can infect human dendritic cells and macrophages in vitro, thereby contributing to dysregulation of the immune system.18
SARS-CoV-2
SARS-CoV-2, a newly discovered coronavirus, has a +ssRNA genome and a spherical morphology when observed under the electron microscope.26,27 SARS-CoV-2 encodes a richly glycosylated spike protein responsible for binding to the ACE-2 receptor.28 The virion’s shape and the ability of spike proteins to form a crown-like structure gave coronaviruses their name.29,30 The SARS-CoV-2 genetic material is surrounded by a lipid-bilayer envelope. Other structural components include the nucleocapsids, membrane, envelope, and hemagglutinin. Unlike the envelope, the membrane exists in higher quantities within the virions.31 Among other functions, the envelope serves to release viral particles from the host cells.32 The nucleocapsid assist in RNA packaging during virion assembly.33,34 Hemagglutinin enhances the entry and pathogenesis of coronaviruses.33 Some of the characteristics of SARS-CoV-2 are summarized in Table 1. Many of these features are shared with SARS-CoV-1. More information on the epidemiology and general characteristics of SARS-CoV-1 can be found later in the review.
Table 1.
Biological characteristics of SARS-CoV-2 and COVID-19.
| References | ||
|---|---|---|
| Name of disease | COVID-19 | [35] |
| Caused by | SARS-CoV-2 | [35–37] |
| Virus genus | β-coronavirus | [14] |
| Virus composition | RNA, Spike protein, β-coronavirus, and hemagglutinin | [38] |
| Virus target | Respiratory system | |
| Functional receptor | ACE-2 | [28] |
| Mode of transmission | Contact, respiratory droplets, fecal oral route, and fomites | |
| Incubation period | 2–14 days | [39] |
| Average age affected | 46% of cases occur in adults | |
| Classic symptoms | Fever, cough, sore throat, and shortness of breath | [40] |
| Complications | Pneumonia, ARDS | |
| Mortality rate | >3%; increases with age and co-morbidities | [41] |
| Investigation/findings | CT chest, lymphocytopenia | |
| Diagnosis | Identification of viral RNA by PCR | [2] |
| Treatment | Supportive care including antiviral drugs | |
| Vaccines | Under clinical trials in selected countries | |
| Mode of reduction | Social distancing, avoidance of close contact, good personal hygiene, personal protective equipment, sheltering at home, isolation of identified cases |
Other human coronaviruses
Other important human CoVs include alpha‐CoVs (HCoV‐229E and HCoV‐NL63) and two beta‐CoVs (HCoV‐OC43 and HCoV‐HKU1). The receptor for HCoV‐229E is human aminopeptidase N (CD13), a cell‐surface metalloprotease present on cells of the kidney and lung, as well as on epithelial and intestinal cells. The receptor for HCoV‐NL63 is also ACE-2. The receptor for HCoV‐OC43 is 9‐O‐acetylated sialic acid, while no receptor has been identified for HCoV‐HKU1.42,43
Generally, the most common diagnostic tools for human CoVs are molecular detection techniques such as reverse transcription-polymerase chain reaction (RT‐PCR) using RNA extracted from respiratory tract samples as template. Other methods include serological tests and viral cultures.11 Although several agents against CoVs, including antibodies, antiviral peptides, and cell or viral protease inhibitors, have been shown to be effective both in vitro and/or in vivo, clinical trial outcomes have not been reported.11 Therefore, clinical treatments for CoVs are still lacking. Nevertheless, supportive and symptomatic therapy are used for treatment. Interferons, ribavirin, lopinavir/ritonavir, and cyclophilin inhibitors are the most commonly administered antiviral agents as elements of combination therapies.11,18 Type I interferons were reportedly used against MERS-CoV in various cell lines and in rhesus macaques. Interferon-alpha 2b and ribavirin have a synergistic effect on SARS-CoV-1 and MERS-CoV in cell cultures.44
Epidemiology of COVID-19
COVID-19, the disease caused by SARS-CoV-2, is similar to that caused by SARS-CoV-1. COVID-19 was first reported in Wuhan (China) in December 2019. The virus likely underwent mutation during zoonotic transmission through pangolin, and later infected humans. The case fatality rate (CFR) of COVID-19 was estimated at 5.93% (3 June 2020). At the time of writing, data from the Johns Hopkins University (https://coronavirus.jhu.edu/map.html) indicated a total of 20,394,078 confirmed cases of SARS-CoV-2 worldwide with 741,808 total deaths and over 13,283,665 recovered cases.
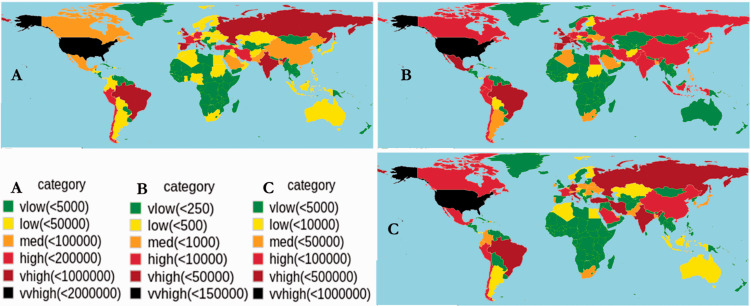
At present, 215 countries and territories are affected by the SARS-CoV-2 outbreak, including the United States, Italy, Germany, South Africa, and Nigeria. The distribution of SARS-CoV-2 confirmed cases, including mortality and recovery, is shown in Figure 4. The United States, China, and some European countries have high case numbers and mortality rates. Recovery rates are increasing worldwide with higher numbers reported in China.
Figure 4.
Prevalence of COVID-19 as of 3 June 2020 (GMT 14:52). The figure shows (a) total cases, (b) total deaths, and (c) the recovery rate per country since the time of the first case of COVID-19. Data were downloaded from worldometers (https://www.worldometers.info/coronavirus/) and John Hopkins University (https://github.com/CSSEGISandData/COVID-19). Graphs were produced using R-studio v3.4.4 and the 'rworldmap' package v1.3-6.
The COVID-19 CFR
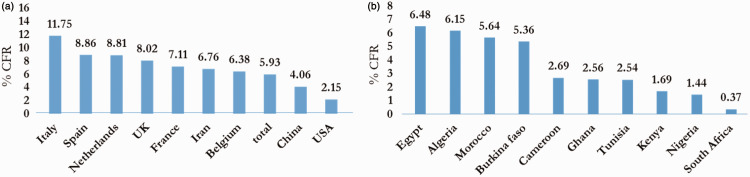
The CFR is defined as the percentage of deaths recorded among confirmed cases. As of 14 April 2020, the number of deaths among confirmed cases was estimated at 126,066 to 1,992,189, resulting in a CFR of 6.33%. This differs from the CFR of 3.70% calculated on 15 March 2020. However, several factors can prevent the accurate determination of the CFR. We compared the global CFR to the African CFR. The highest CFR was observed in Italy, followed by Spain and the Netherlands (Figure 5a). In Africa, countries such as Egypt, Algeria, Burkina Faso, and Morocco had CFRs above 5% (Figure 5b). A major challenge in the accurate calculation of the CFR is the denominator (the number of confirmed cases). Asymptomatic cases of COVID-19, patients with mild symptoms, or individuals who are misdiagnosed may be left out of the denominator, leading to underestimation or overestimation of the CFR.45 High CFRs reflect limited access to health care for the most vulnerable patients and limitations in health-care systems, including limited capacity of surveillance systems to trigger a timely response (WHO, 2020).
Figure 5.
Top 10 COVID-19 CFRs over a period of 3 months (onset to 1 April 2020). CFRs are presented (a) globally and (b) for Africa.
Estimation of the COVID-19 reproductive number
To date, over 20 million cases of COVID-19 have been reported globally. It is important to estimate the reproductive number for this virus to enable accurate predictions. Two primary factors, the reproductive ratio (Ro) and the serial interval (SI), are essential to estimate the rate of transmission of this disease.
The reproductive ratio (Ro) of COVID-19
Ro measures the degree of COVID-19 contagion or infectiousness. Ro represents the average number of people who will be infected by one infected individual. Zhang et al46 estimated that the median and 95% confidence interval (CI) of Ro were about 2.28 (2.06–2.52) for early outbreaks of COVID-19. This means that an infected person can transmit COVID-19 to an average of two to three other people. Various studies reported significant variations in Ro using different datasets.47,48
Serial interval (SI)
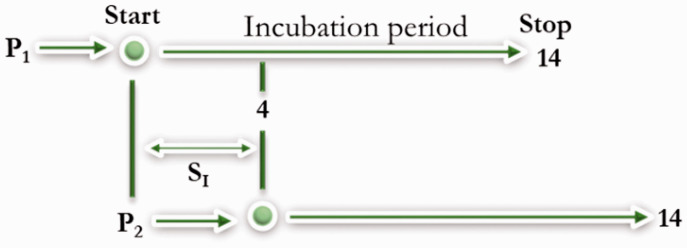
To understand the case turnover and transmissibility of COVID-19, the serial interval (SI) from the onset of illness in a primary case to the onset of illness in a secondary case is important.49 Recent studies estimated the average SI for COVID-19 as 3.77 (2.23–4.82) days.49–51 A shorter SI makes COVID-19 harder to contain and more likely to rapidly transmit within populations (Figure 6).
Figure 6.
The SI of COVID-19 as of 6 March 2020. The time from the onset of symptoms in patient one (P1) to the onset in patient two (P2) is equal to SI. The shorter the period the more dangerous and transmissible the infectious disease, and vice versa.
Taking the R0 and SI of COVID-19 into consideration, it can be inferred that approximately 4 days are required for an infected person to transmit COVID-19. It is highly likely that the individual can infect approximately two or three other persons, making the spread of COVID-19 extremely rapid and dangerous (Figure 7a). The extent of spread is totally dependent on the Ro value (Figure 7b). If R0 < 1, each existing infection causes less than one new infection. In this case, the disease will decline and eventually die out. If Ro = 1, each existing infection causes one new infection. The disease will remain stable in the population, but will not result in epidemic spread. If R0 > 1, each existing infection causes more than one new infection. The disease will spread between individuals and eventually lead to an outbreak. COVID-19 was regarded as an outbreak (30 January 2020) with a R0 > 1 (Figure 7b).
Figure 7.
The association between R0 and SI in COVID-19 estimators. The extent of spread is dependent on Ro.
Importantly, the R0 value of a disease only applies when everyone in a population is completely susceptible. This means that no one has been vaccinated or had the disease previously, and there is no way to control the spread of the disease using interventions such as drugs or vaccines.
Pathophysiology of COVID-19
There are currently two known modes of COVID-19 transmission: the fecal-oral route and respiratory droplets.6,52–56 Droplets have potential to come into contact and infect a healthy person within 3 to 6 feet (1 meter). Droplets that stick to surfaces can survive for more than 24 hours, remaining infectious. The virus can remain airborne for about 3 hours, long enough to permit transmission.
Upon infection with SARS-CoV-2, the virus infects type II pneumocytes of the alveoli.57 These pneumocytes are responsible for surfactant production. Surfactant decreases the surface tension within alveoli and reduces the collapsing pressure. The spike protein of the virus binds to ACE-2 on the pneumocytes (Figure 8) permitting virion entry into the host cell.58 The virus hijacks the host cell’s machinery (ribosomes) to enable translation of its +ssRNA genome into different protein molecules. The virus can also use its RdRp to produce additional copies of its +ssRNA genome (Figure 9). The translated polyproteins are further processed into different individual components within the host cell. These processes give rise to multiple virions, which are then released upon pneumocyte damage. In response to this process, type II pneumocytes releases specific inflammatory mediators that instruct macrophages to secrete interleukins 1 and 6 (IL-1 and IL-6) and tumor necrosis factor-alpha. These cytokines cause the endothelial cells lining blood vessels to dilate, leading to increased capillary permeability. In response, fluids accumulate in the alveoli leading to edema.59 As surface tension increases, the collapsing pressure of the alveoli increases. A decrease in gas exchange is also observed through this process, which in turn leads to hypoxia and difficulty breathing (dyspnea). This can progress to a critical condition such as acute respiratory distress syndrome (ARDS).
Figure 8.
Structure of SARS-COV-2 adapted from Encyclopaedia Britannica [60].
Figure 9.
Pathophysiology of COVID-19. The virus can be transmitted via person-to-person contact through (1) the fecal-oral route and or (2) respiratory droplets. (3) The virion enters through the mouth, nose or eyes and (4) infects the alveoli of the lungs. (5) The COVID-19 spike protein binds to ACE-2 on type 2 pneumocytes. (6) COVID-19 releases its genetic material (+ssRNA) for processing. (7) the host machinery, +ssRNA is translated into polyproteins (8), which are further cleaved by proteinases into the different components of the virus. (9) More RNA copies are synthesized by RNA-dependent RNA polymerase. (10) Proteins and RNA are assembled into a new virion in the Golgi apparatus, which is then (11) released. (12) These processes stimulate the release of cytokines and other proteolytic enzymes, which cause various complications such as consolidation and (13) pneumonia.
Inflammatory mediators further stimulate neutrophils, which release reactive oxygen species and proteases. This process damages the alveoli (both type 1 and 2), leading to consolidation and alveolar collapse. High levels of IL-1 and IL-6 travel through the blood to the central nervous system, instructing the hypothalamus to release prostaglandins and causing fever. Severe lung inflammation leads to systemic inflammatory respiratory syndrome. Progression can lead to increased capillary permeability. The overall blood volume decreases, and through a series of processes involving hypotension and decreased perfusion of multiple different organs, multiple system organ failure (MSOF) can occur. During MSOF, elevated levels of blood urea, nitrogen, and creatinine accrue in the kidney. The liver also releases specific inflammatory and acute phase response biomolecules (aspartate transaminase, alanine transaminase, bilirubin, C-reactive Protein [CRP], fibrinogen and IL-6) that can serve as biomarkers for patients with COVID-19.
Transmission of SARS-CoV-2 through the eyes
There is continued research by multiple groups into the mechanism of transmission of SARS-CoV-2 through the eyes. Investigations were necessary because health care providers, including a Perking University Physician, may have contracted the virus while not wearing eye protection when treating patients.61 Some researchers have asserted that avoidance of touching the eyes, nose or mouth with unwashed or unsterilized hands can reduce COVID-19 transmission. The mucous membranes that line various cavities in the body are generally most susceptible to viral transmission.62 Ocular symptoms such as viral conjunctivitis can result from SARS-CoV-2 upper respiratory tract infections.63 This was confirmed in 9 of 1,099 patients in China.64 Other research also found that 1 of 30 patients hospitalized with COVID-19 were diagnosed with conjunctivitis.65 In a study conducted by the American Optometric Association, COVID-19 was linked to ocular signs and symptoms including photophobia, irritation, conjunctival infection and ocular discharge.66 Thus, the superficial blood vessels of the conjunctiva are an alleged route of exposure and transmission of SARS-CoV-2.
Clinical symptoms of COVID-19
The clinical manifestations of SARS-CoV-2 are uncertain and change frequently. Some infections are asymptomatic. Symptoms can include respiratory distress syndrome, pneumonia of different levels of severity,67 and sometimes death. According to the WHO, the most common symptoms of COVID-19 are fever, fatigue, dry continuous cough, and shortness of breath.68,69 Some patients may have a runny nose, sore throat, nasal congestion, aches and pains, and diarrhea.70 Some patients report a loss of sense of smell and taste.71 In some cases, symptoms are mild and similar to those of the common cold; in such patients, recovery can occur without any treatment.72 The least commonly observed symptoms include nausea or vomiting, coughing up blood or bloody mucus, and viral conjunctivitis causing red eyes, watery discharge from the eyes, swollen eyelids and light sensitivity.3 Occasional upper respiratory and gastrointestinal symptoms, accompanied by changes vital signs such as increased respiration (heart rate) and low blood pressure may also be observed, especially in the elderly and among individuals suffering from heart disease, chronic respiratory conditions and diabetes.73 Additionally, patients critically ill with COVID-19 may present with increased venous thromboembolism including thrombocytopenia, elevated D-dimer, prolonged prothrombin time and disseminated intravascular coagulation (DIC). These coagulation abnormalities are associated with a systemic inflammatory response and an imbalance between pro-coagulant and anticoagulants homeostasis mechanisms. and increase the risk of mortality.74 Some of these clinical features are also observed in cases of DIC observed in septic patients. These features are very distinct in COVID-19 patients as their levels are higher than the standards for sepsis.75,76
Diagnosis, prognosis, treatment, and management of SARS-CoV-2
Diagnosis of SARS-CoV-2
SARS-CoV-2 causes various complications ranging from fever, dry cough, and pneumonia to decreased organ perfusion leading to MSOF. Early symptoms are similar to those of influenza, and the first step to differentiate COVID-19 from flu and pneumonia is a nasopharyngeal swab test (viral testing for influenza A/B).77 Quantitative polymerase chain reaction-based (qPCR) methods are the major diagnostic tests for SARS-CoV-2 using nasal swab, aspirate, sputum, or blood as samples. These method have some limitations as they are time consuming and have variable sensitivity (30%–80%).78–80 Another diagnostic method is the newly approved nucleic acid test, which is carried out based on the principle of fluorescence PCR.81 The main goal of SARS-CoV-2 diagnosis is to accurately detect the virus and to minimize further transmissions by timely isolation and treatment of infected patients. Other tests (not specific for SARS-CoV-2) used in conjunction with the methods above are based on clinical manifestations. These include blood tests such as complete blood count, comprehensive metabolic panels, basic metabolic panels, and assessment of liver/kidney markers and procalcitonin levels (for bacterial infections). Inflammatory markers may also be assessed including CRP, erythrocyte sedimentation rate, IL-6, lactate dehydrogenase, D-dimer, ferritin, troponin and creatine kinase-MB. Imaging investigations are typically computed tomography scans: in COVID-19 patients these often show glass opacities, areas of consolidation, and paving patterns in cases of severe and progressive disease. Ground glass opacities can also be observed on chest X-ray. Finally, ultrasound can show B-lines, pleural line thickening, and lung consolidation. Air bronchograms can also be used for assessment. These tests are non-specific but helpful to determine patients’ health status.
COVID-19 patients with severe ARDS could potentially present with pneumonia. Pneumonia may be severe, leading to ARDS and MSOF. It is therefore imperative to mechanically ventilate the lungs to avoid ARDS and MSOF.
Prognosis of SARS-COV-2
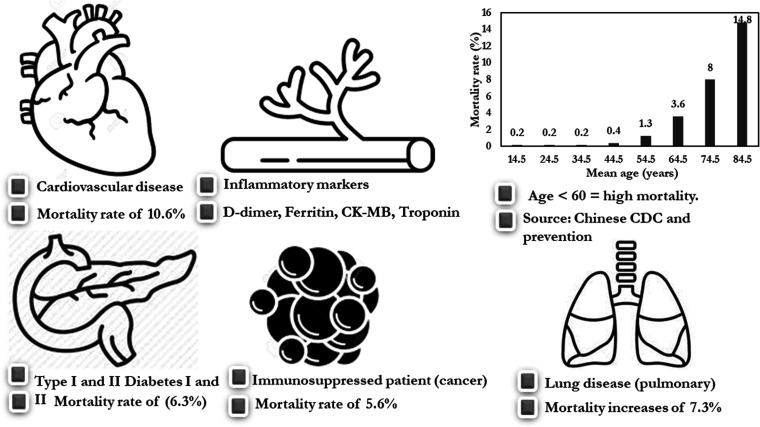
Although the risk factors for COVID-19 remain unclear, some risk factors put patients at a significantly higher risk of mortality, especially in individuals with underlying conditions.6,35,40,82 Some medical disorders are correlated with higher risk of mortality in COVID-19 patients. These include, cardiovascular diseases mortality risk 10.6%),41 lung disease (mortality risk 7.3%), type 1 and 2 diabetes (mortality risk 6.3%), immunosuppression (e.g., cancer patients; mortality risk 5.6%),83–86 and age (Figure 10).87 COVID-19 mortality rates increase with advancing age, and are especially high in those aged >60 years. Elevated inflammatory markers in response to tissue damage (elevated levels of D-dimer, ferritin, creatine kinase-MB, and troponin) have been associated with higher mortality rates.
Figure 10.
Potential prognosis of COVID-19 and associated mortality rates. Prognoses are shown for patients with (a) cardiovascular diseases, (b) inflammatory blood markers, (c) ageing, (d) pulmonary diseases, (e) immunosuppression (e,g., cancer patients), and (f) diabetes.
Potential treatment of COVID-19
The first line of treatment for patients presented with the symptoms of COVID-19 (fever, dry cough, and shortness of breath) is self-quarantine for at least 14 days. Cases are monitored for progression of symptoms such as increased fever (>40°C), significant difficulty breathing or shortness of breath, mouth breaks, constant coughing, and dehydration. If there is no significant improvement in symptoms, it is advisable to consult a clinician for confirmation of the diagnosis and to avoid further spread of the virus. The main treatment currently available is supportive care. Although there is limited information on COVID-19, it has been linked to ARDS. For patients with high fevers that could potentially lead to dehydration, intravenous fluids such as normal saline or lactated Ringer’s fluid can be administered sparingly to prevent lung overload. To reduce fever, antipyretic drugs such as paracetamol or acetaminophen can be administered. Drugs such as remdesivir, chloroquine, ritonavir, tocilizumab, corticosteroids have been repurposed for the treatment of COVID-19 (Figure 11), although their clinical effectiveness has not yet been confirmed.88–93 Unfortunately, the use of chloroquine and derivatives such as hydroxychloroquine, alone or in combination with other drugs, resulted in cardiac toxicity. Investigations of these drugs were recently suspended by the WHO.94
Figure 11.
Selected repurposed drugs for treatment of COVID-19.
Mechanism of action of selected repurposed drugs for treatment of COVID-19
The elucidation of potential targets could lead to COVID-19-specific treatments (Figure 11). Approaches to anti-SARS-CoV-2 drug development include (a) inhibition of SARS-CoV-2 entry, (b) disruption of SARS-CoV-2 +ssRNA synthesis after entry, (c) inflammatory response suppression, and (d) disruption of SARS-CoV-2 translation.
An FDA-approved immunosuppressive and anti-malarial parasite drug (chloroquine) can inhibit the entry of COVID-19 into the endosome after binding to ACE-2 receptors, thereby preventing the release of +ssRNA for translation.89,93,95,96 Studies also showed that hydroxychloroquine, an analog of chloroquine, was more potent and could be used in place of chloroquine.97 Remdesivir, a novel antiviral drug and nucleotide analog used to treat Ebola virus infection, is currently under clinical development. The mechanism of the drug is reported to be at the post viral entry stage.89 Its mode of action is to inhibit the RdRp, preventing synthesis of the viral +ssRNA (Figure 11 (2)). This drug is currently in phase 2 clinical trials. The protein inhibitor ritonavir has also been proposed for the treatment of COVID-19. This drug interferes with the protease enzymes (proteinases) by inhibiting the conversion of polyproteins into the mature components (spike proteins, nucleocapsids) needed by the virus for multiplication (Figure 11 (3)). Another immunosuppressive drug, tocilizumab, has also been repurposed for COVID-19 because of its ability to block IL-6 and inhibit inflammatory responses. Corticosteroids can also decrease inflammation by inhibiting phospholipases such as phospholipase A2, and thus suppress the excessive production of prostaglandins (Figure 11 (4)).
SARS-CoV-2 appears unfamiliar to the human innate immune system. Coupled with its emergence and its spread globally via human-to-human transmission, the development of vaccines for prevention is no longer a debate but a necessity. Several vaccine platforms are being developed and some have entered clinical trials. However, approval by regulatory agencies such as the FDA and manufacturing may take 12 to 18 months.98
Studies of the antiviral activity of host-directed drugs and compounds have identified two classes of molecules (protein biogenesis inhibitors and ligands of the Sigma1 and Sigma2 receptors) as effective inhibitors of viral infectivity. Molecules that target the Sigma1 and Sigma2 receptors perturb the virus through different mechanisms from translation inhibitors, potentially including modulation of cellular stress responses.99 The ligands haloperidol, PB28, PD-144418, and hydroxychloroquine are currently undergoing clinical trials in COVID-19 patients.100 These molecules exert their antiviral effects during viral replication by inhibiting nucleoprotein expression after viral entry has occurred. The lack of selectivity of chloroquine and hydroxychloroquine, including off-target effects on the human Ether-à-go-go-Related Gene (hERG) and other molecules, may be related to the adverse cardiac reactions that have limited their use.101 A recent study by Gordon et al identified 332 high-confidence SARS-CoV-2-human protein interactions connected to multiple biological processes, including protein trafficking, translation, transcription and ubiquitination regulation.102 The study identified 69 ligands, including FDA approved drugs, compounds in clinical trials, and preclinical compounds that might theoretically have therapeutic effects as host-directed interventions against COVID-19. To date, no known antiviral drugs nor vaccines against SARS-CoV-2 with proven clinical efficacy have been identified. Part of the reason for the absence of such agents is limited information regarding the molecular details of the infection. To develop therapeutic interventions against COVID-19, it is crucial to understand how the virus interactions with the host during infection.
ACE-2, a potential target for COVID-19 treatment
ACE-2 has been identified as the functional receptor for SARS-CoV-2. It is an outer membrane enzyme expressed in vascular endothelial cells, the renal tubular epithelium, and Leydig’s cells of the testes, lungs, heart, kidney, and gastrointestinal tract.59,103 It is a type-II transmembrane metallocarboxypeptidase with its enzymatically active domain exposed at the cell surface.104 ACE-2 is a key player in the renin-angiotensin system (RAS) and a target for treatment of hypertension.105 ACE-2 catalyzes the cleavage of angiotensin II, a vasoconstrictor, into angiotensin 1–7, a vasodilator, thereby lowering blood pressure by negatively regulating RAS.106 ACE-2 is a promising drug target for treating CoV infection as well as other cardiovascular diseases.5 ACE-2 confers a protective effect against lung injury induced by viruses by increasing the production of angiotensin 1–7. The virus presumably causes lung damage by reducing ACE-2 levels on cells through the process of degradation and internalization.107,108 Because studies have shown that ACE inhibitors and angiotensin II receptor blockers could be used to up-regulate the expression and activity of ACE-2 in hypertensive patients, similar strategies might reduce the severity of COVID-19.109,110 Recent studies have shown that the expression of human ACE-2 is associated with SARS‐CoV infection and that genetic variations of this receptor may contribute to susceptibility and/or resistance against infection.111 For instance, a single-cell RNA sequencing analysis of ACE-2 revealed that type II alveolar cells had higher expression of ACE-2 in eight individual lung tissues obtained from normal donors. ACE-2 expression was higher in the two male samples compared with the six female samples. Additionally, ACE-2 expression was higher in the only Asian male in the study compared with Caucasian and black Americans,59which might explain why the four German cases showed mild clinical symptoms with no severe illness.112 This implies that variation in ACE-2 expression in COVID-19 patients is likely to affect susceptibility, symptoms and intervention outcomes following SARS-CoV-2 infection.
Precautionary measures against COVID-19
The degree of spread of COVID-19 is currently about 5.3% and could potentially increase if precautionary measures are not considered. Global prevention of the spread of COVID-19 is therefore a crucial and urgent goal. To prevent further spread of this disease, detection and isolation of individuals with COVID-19 is of the utmost priority. Examples of measures to curb spread include self-quarantine, isolation of infected individuals, social distancing, good personal hygiene (frequent hand washing with soap and water/alcohol-based sanitizers and avoiding touching the eyes, nose and the mouth), and use of personal protective equipment. Certain classes of compounds, called surfactants, are contained in soap and have the ability to neutralize microbes such as SARS CoV-2.113This is because soap can assemble into bubble-like structures called micelles that trap viral matter and other biomaterials.114 Surfactants in soap lather have their hydrophilic parts pointing outwards and interacting with solvent and their hydrophobic heads pointing inwards. This opens the coronavirus outer membrane and encapsulates viral molecules within micelles, thus making insoluble viral molecules easily soluble in water and effectively removing them from hands, surfaces or other areas after about 20 s.115 Therefore surfactants in soap can disrupt and sequester viruses and other contaminants while sanitizer and disinfectants are designed to kill SARS-CoV-2.116
Conclusion
The outbreak of SARS-CoV-2 has become a global threat. However, information regarding this virus remains limited. The available information is inconsistent and there are constant data updates, which may contribute to variation between study results. For more consistent and accurate results, well-annotated data from clinical patients and subclinical subjects in normal populations could help to better understand the pandemic. The information provided in this review is based on data provided by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University during specific date ranges. Key insights into the prevalence, pathophysiology, diagnosis, and potential treatment of SARS-CoV-2 are herein summarized. Research efforts are being intensified to address the current challenges in the quest for adequate treatments, diagnostics, and vaccines. Clinical studies into the genetic variation of receptors such as ACE-2 in tissues and across populations will remain an active area of research until relevant targets and therapies are found. While the development of adequate treatments and vaccines for COVID-19 is underway, it is advisable that good hygiene practices including washing of hands and social distancing should be practiced, and government guidance/guidelines should be followed. This will help to reduce the spread of the disease. We hope that the insights gained from this review will enable researchers to help patients develop accommodating lifestyles and improve the efficiency of health care practitioners.
Availability of data and material
Data are freely available at the following sources. Centers for Disease Control and Prevention (https://www.cdc.gov/). Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (https://coronavirus.jhu.edu/map.html). Worldometers (https://www.worldometers.info/coronavirus/?).
Authors’ contributions
A.O.F., N.R.S.S., O.B.A., O.O.B., and A.K. carried out the literature review and drafted the manuscript. M.O.A., A.M.M. and M.M. critically evaluated the manuscript. All authors read and approved the final manuscript.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iDs
Adewale Oluwaseun Fadaka https://orcid.org/0000-0002-3952-2098
Nicole Remaliah Samantha Sibuyi https://orcid.org/0000-0001-7175-5388
Ashwil Klein https://orcid.org/0000-0002-5606-886X
References
- 1.Ye ZW, Yuan S, Yuen KS, et al. Zoonotic origins of human coronaviruses. Int J Biol Sci 2020; 16: 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 2020, 25: 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai CC, Shih TP, Ko WC, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents 2020; 55: 105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382: 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579: 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munster VJ, Koopmans M, Van Doremalen N, et al. A novel coronavirus emerging in China–key questions for impact assessment. N Engl J Med 2020, 382: 692–694. [DOI] [PubMed] [Google Scholar]
- 8.Yao W, Maitra A, Ying H. Recent insights into the biology of pancreatic cancer. EBioMedicine 2020; 53: 102655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas SK, Lee J, Beatty GL. Paracrine and cell autonomous signalling in pancreatic cancer progression and metastasis. EBioMedicine 2020; 53: 102662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christenson ES, Jaffee E, Azad NS. Current and emerging therapies for patients with advanced pancreatic ductal adenocarcinoma: a bright future. Lancet Oncol 2020; 21: e135–e145. Epub 2020/03/07. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology 2018; 23: 130–137. Epub 2017/10/21. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harcourt J, Tamin A, Lu X, et al. Isolation and characterization of SARS-CoV-2 from the first US COVID-19 patient. bioRxiv 2020. [Google Scholar]
- 13.Schwartz DA, Graham AL. Potential maternal and infant outcomes from coronavirus 2019-nCoV (SARS-CoV-2) infecting pregnant women: Lessons from SARS, MERS, and other human coronavirus infections. Viruses 2020; 12: 194. doi:10.3390/v12020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang N, Shang J, Jiang S, et al. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol 2020; 11: 298. English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020; 395: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa K, Lokugamage K, Makino S. Viral and cellular mRNA translation in coronavirus-infected cells. Adv Virus Res 2016; 96: 165–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forni D, Cagliani R, Clerici M, et al. Molecular evolution of human coronavirus genomes. Trends Microbiol 2017; 25: 35–48. Epub 2016/10/25. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses 2019; 11: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu Q, Yu M, Fan B, et al. A complete sequence and comparative analysis of a SARS-associated virus (isolate BJ01). Chin Sci Bull 2003; 48: 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldsmith CS, Tatti KM, Ksiazek TG, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis 2004; 10: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marra MA, Jones SJ, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science 2003; 300: 1399–1404. [DOI] [PubMed] [Google Scholar]
- 22.Pan X, Chen D, Xia Y, et al. Asymptomatic cases in a family cluster with SARS-CoV-2 infection. Lancet Infect Dis 2020; 20: 410–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu B, Ge X, Wang LF, et al. Bat origin of human coronaviruses. Virol J 2015; 12: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamin A, Queen K, Paden CR, et al. Isolation and growth characterization of novel full length and deletion mutant human MERS-CoV strains from clinical specimens collected during 2015. J Gen Virol 2019; 100: 1523–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fehr AR, Channappanavar R, Perlman S. Middle East Respiratory Syndrome: Emergence of a pathogenic human coronavirus. Annu Rev Med 2017; 68: 387–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bárcena M, Oostergetel GT, Bartelink W, et al. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc Natl Acad Sci USA 2009; 106: 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuman BW, Adair BD, Yoshioka C, et al. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol 2006; 80: 7918–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bosch BJ, Van Der Zee R, De Haan CA, et al. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 2003; 77: 8801–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izaguirre G. The proteolytic regulation of virus cell entry by furin and other proprotein convertases. Viruses 2019; 11: 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fehr AR, Perlman S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol Biol 2015; 1282: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nal B, Chan C, Kien F, et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J Gen Virol 2005; 86: 1423–1434. [DOI] [PubMed] [Google Scholar]
- 32.Siu Y, Teoh K, Lo J, et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol 2008; 82: 11318–11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klausegger A, Strobl B, Regl G, et al. Identification of a coronavirus hemagglutinin-esterase with a substrate specificity different from those of influenza C virus and bovine coronavirus. J Virol 1999; 73: 3737–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hurst KR, Koetzner CA, Masters PS. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein. J Virol 2009; 83: 7221–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020; 395: 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sallares R. plague. Oxford Research Encyclopedia of Classics 2016.
- 37.Clawson P. Review of A Modern Contagion: Imperialism and Public Health in Iran's Age of Cholera. Middle East Quarterly. 2020.
- 38.Rao H, Greve HR. Disasters and community resilience: Spanish flu and the formation of retail cooperatives in Norway. Acad Manage J 2018; 61: 5–25. [Google Scholar]
- 39.Jiang X, Rayner S, Luo MH. Does SARS‐CoV‐2 has a longer incubation period than SARS and MERS? J Med Virol 2020; 92: 476–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. MedRxiv 2020. [Google Scholar]
- 41.Yang J, Zheng Y, Gou X, et al. Prevalence of comorbidities and its effects in patients infected with SARS-CoV2: A systematic review and meta-analysis. Int. J Infect Dis 2020; 94: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hulswit RJG, Lang Y, Bakkers MJG, et al. Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A 2019; 116: 2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X, Dong W, Milewska A, et al. Human coronavirus HKU1 spike protein uses 9-O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol 2015; 89: 7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omrani AS, Saad MM, Baig K, et al. Ribavirin and interferon alfa-2a for severe Middle East respiratory syndrome coronavirus infection: A retrospective cohort study. Lancet Infect Dis 2014; 14: 1090–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajgor DD, Lee MH, Archuleta S, et al. The many estimates of the COVID-19 case fatality rate. Lancet Infect Dis 2020; 20: 776–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S, Diao M, Yu W, et al. Estimation of the reproductive number of Novel Coronavirus (COVID-19) and the probable outbreak size on the Diamond Princess cruise ship: A data-driven analysis. Int J Infect Dis 2020; 93: 201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kucharski AJ, Russell TW, Diamond C, et al. Early dynamics of transmission and control of COVID-19: a mathematical modelling study. Lancet Infect Dis 2020; 20: 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hellewell J, Abbott S, Gimma A, et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. Lancet Glob Health 2020; 8: e488–e4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishiura H, Linton NM, Akhmetzhanov AR. Serial interval of novel coronavirus (COVID-19) infections. Int J Infect Dis 2020; 93: 284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du Z, Xu X, Wu Y, et al. The serial interval of COVID-19 from publicly reported confirmed cases. medRxiv 2020: 2020.02.19.20025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y, Lu Q, Liu M, et al. Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China. medRxiv 2020. [Google Scholar]
- 52.Liu J, Liao X, Qian S, et al. Community transmission of Severe Acute Respiratory Syndrome Coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis 2020; 26: 1320–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020; 395: 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson R. Pandemic potential of 2019-nCoV. Lancet Infect Dis 2020; 20: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burke RM, Midgley CM, Dratch A, et al. Active monitoring of persons exposed to patients with confirmed COVID-19—United States, January–February 2020. MMWR Morb Mortal Wkly Rep 2020; 69: 245–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.World Health organization. Modes of transmission of virus causing COVID-19: implications for IPC precaution recommendations: Scientific brief, 29 March 2020, https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations (2020, accessed 1 August 2020).
- 57.Dan H, Maureen G, Richard B. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett 2002; 532: 107. [DOI] [PubMed] [Google Scholar]
- 58.Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020; 5: 562–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Zhao Z, Wang Y, et al. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv 2020. [Google Scholar]
- 60.Encyclopædia Britannica, Inc. Coronavirus, https://www.britannica.com/science/coronavirus-virus-group (2020, accessed 1 Aug 2020).
- 61.Xie J, Tong Z, Guan X, et al. Critical care crisis and some recommendations during the COVID-19 epidemic in China. Intensive Care Med 2020; 46: 837–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li JPO, Lam DSC, Chen Y, et al. Novel coronavirus disease 2019 (COVID-19): The importance of recognising early possible ocular manifestation and using protective eyewear. Br J Ophthamol 2020; 104: 297–298. [DOI] [PubMed] [Google Scholar]
- 63.Seah I, Agrawal R. Can the Coronavirus Disease 2019 (COVID-19) affect the eyes? A review of coronaviruses and ocular implications in humans and animals. Ocul Immunol Inflamm 2020; 28: 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020; 382: 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia J, Tong J, Liu M, et al. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS‐CoV‐2 infection. J Med Virol 2020; 92: 589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parrish RK, 2nd, Stewart MW, Powers SLD. Ophthalmologists are more than eye doctors–in memoriam Li Wenliang. Am J Ophthalmol 2020; 213: A1–A2.32169251 [Google Scholar]
- 67.Lai CC, Liu YH, Wang CY, et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J Microbiol Immunol Infect 2020; 53: 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.World Health Organization. Coronavirus disease 2019 (COVID-19) situation report 51, https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200311-sitrep-51-covid-19.pdf?sfvrsn=1ba62e57_10 (2020, accessed 1 August 2020).
- 69.Novel CPERE. The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China. Zhonghua Liu Xing Bing Xue Za Zhi 2020; 41: 145–151. [DOI] [PubMed] [Google Scholar]
- 70.Chen H, Guo J, Wang C, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: A retrospective review of medical records. Lancet 2020; 395: 809–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA 2020; 323: 1406–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020. [DOI] [PubMed] [Google Scholar]
- 73.Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health 2020; 25: 278–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiong M, Liang X, Wei YD. Changes in blood coagulation in patients with severe coronavirus disease 2019 (COVID‐19): A meta‐analysis. Br J Haematol 2020; 189: 1050–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Levi M, Thachil J, Iba T, et al. Coagulation abnormalities and thrombosis in patients with COVID-19. Lancet Haematol 2020; 7: e438–e440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Han H, Yang L, Liu R, et al. Prominent changes in blood coagulation of patients with SARS-CoV-2 infection. Clin Chem Lab Med 2020; 58: 1116–1120. [DOI] [PubMed] [Google Scholar]
- 77.Zeng Q, Khan K, Wu J, et al. The utility of preemptive mass influenza vaccination in controlling a SARS outbreak during flu season. Math Biosci Eng 2007; 4: 739. [DOI] [PubMed] [Google Scholar]
- 78.Wang N, Luo C, Liu H, et al. Characterization of a new member of alphacoronavirus with unique genomic features in rhinolophus bats. Viruses 2019; 11: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bhadra S, Jiang YS, Kumar MR, et al. Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV). PLoS One 2015; 10: e0123126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan JFW, Choi GKY, Tsang AKL, et al. Development and evaluation of novel real-time reverse transcription-PCR assays with locked nucleic acid probes targeting leader sequences of human-pathogenic coronaviruses. J Clin Microbiol 2015; 53: 2722–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu J, Zhao S, Teng T, et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses 2020; 12: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 2020; 323: 1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kamboj M, Sepkowitz KA. Nosocomial infections in patients with cancer. Lancet Oncol 2009; 10: 589–597. [DOI] [PubMed] [Google Scholar]
- 84.Li JY, Duan XF, Wang LP, et al. Selective depletion of regulatory T cell subsets by docetaxel treatment in patients with non-small cell lung cancer. J Immunol Res 2014; 2014: 286170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longbottom ER, Torrance HD, Owen HC, et al. Features of postoperative immune suppression are reversible with interferon gamma and independent of interleukin-6 pathways. Ann Surg 2016; 264: 370–377. [DOI] [PubMed] [Google Scholar]
- 86.Sica A, Massarotti M. Myeloid suppressor cells in cancer and autoimmunity. J Autoimmun 2017; 85: 117–125. [DOI] [PubMed] [Google Scholar]
- 87.COVID C, Team R. Severe outcomes among patients with coronavirus disease 2019 (COVID-19)—United States, February 12–March 16, 2020. MMWR Morb Mortal Wkly Rep 2020; 69: 343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends 2020; 14: 69–71. [DOI] [PubMed] [Google Scholar]
- 89.Wang M, Cao R, Zhang L, et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020; 30: 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med 2020; 382: 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ko WC, Rolain JM, Lee NY, et al. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int J Antimicrob Agents 2020; 55: 105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Colson P, Rolain JM, Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents 2020; 55: 105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kapoor KM, Kapoor A. Role of chloroquine and hydroxychloroquine in the treatment of COVID-19 infection – a systematic literature review. medRxiv 2020. [Google Scholar]
- 94.Mehra MR, Desai SS, Ruschitzka F, et al. Hydroxychloroquine or chloroquine with or without a macrolide for treatment of COVID-19: a multinational registry analysis. Lancet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Li C, Zhu X, Ji X, et al. Chloroquine, a FDA-approved drug, prevents Zika virus infection and its associated congenital microcephaly in mice. EBioMedicine 2017; 24: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu X, Chen G, Shang Y, et al. Efficacy of chloroquine versus lopinavir/ritonavir in mild/general COVID-19 infection: A prospective, open-label, multicenter, randomized controlled trial. Trials 2020; 21: 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yao X, Ye F, Zhang M, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020: ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu S. Timely development of vaccines against SARS-CoV-2. Emerg Microbes Infect 2020; 9: 542–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mitsuda T, Omi T, Tanimukai H, et al. Sigma-1Rs are upregulated via PERK/eIF2α/ATF4 pathway and execute protective function in ER stress. Biochem Biophys Res Commun 2011; 415: 519–525. [DOI] [PubMed] [Google Scholar]
- 100.Thompson BT. Outcomes related to COVID-19 treated with hydroxychloroquine among in-patients with symptomatic disease. https://clinicaltrialsgov/ct2/show/NCT04332991. 2020.
- 101.White NJ. Cardiotoxicity of antimalarial drugs. Lancet Infect Dis 2007; 7: 549–558. [DOI] [PubMed] [Google Scholar]
- 102.Gordon DE, Jang GM, Bouhaddou M, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020; 583: 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang H, Penninger JM, Li Y, et al. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: Molecular mechanisms and potential therapeutic target. Intensive Care Med 2020; 46: 586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Thompson-Nauman AE, Christie MG, McVenes RD. Substernal electrical stimulation system. Google Patents; 2020.
- 105.Wu Y. Compensation of ACE2 function for possible clinical management of 2019-nCoV-induced acute lung injury. Virol Sin 2020; 35: 256–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lei C, Fu W, Qian K, et al. Potent neutralization of 2019 novel coronavirus by recombinant ACE2-Ig. bioRxiv 2020. [Google Scholar]
- 107.Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci 2020; 63: 457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wan Y, Shang J, Graham R, et al. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J Virol 2020; 94: e00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181: 271–280.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gurwitz D. Angiotensin receptor blockers as tentative SARS‐CoV‐2 therapeutics. Drug Dev Res 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hussain M, Jabeen N, Raza F, et al. Structural variations in human ACE2 may influence its binding with SARS‐CoV‐2 spike protein. J Med Virol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cao Y, Li L, Feng Z, et al. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov 2020; 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khalaf MM, Tantawy AH, Soliman KA, et al. Cationic Gemini-surfactants based on waste cooking oil as new ‘green’ inhibitors for N80-steel corrosion in sulphuric acid: A combined empirical and theoretical approaches. J Mol Struct 2020; 1203: 127442. [Google Scholar]
- 114.Belhaj AF, Elraies KA, Mahmood SM, et al. The effect of surfactant concentration, salinity, temperature, and pH on surfactant adsorption for chemical enhanced oil recovery: A review. J Petrol Explor Prod Technol 2020; 10: 125–137. [Google Scholar]
- 115.Zheng Y, Lu X, Lai L, et al. The micelle thermodynamics and mixed properties of sulfobetaine-type zwitterionic Gemini surfactant with nonionic and anionic surfactants. J Mol Liq 2020; 299: 112108. [Google Scholar]
- 116.Wang J, Shi Y. Managing neonates with respiratory failure due to SARS-CoV-2–Authors' reply. Lancet Child Adolesc Health 2020. 4: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are freely available at the following sources. Centers for Disease Control and Prevention (https://www.cdc.gov/). Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (https://coronavirus.jhu.edu/map.html). Worldometers (https://www.worldometers.info/coronavirus/?).