Abstract
MicroRNA (miR)-130a has been reported to promote cancer growth; however, its role during acute myeloid leukemia (AML) is not completely understood. In the present study, the effects of miR-130a on the sensitivity of AML cells to Adriamycin (Adr) were investigated. 5-Aza-2′-deoxycytidine (5-Aza-dC) was used to stimulate Adr resistance in AML cells, and cell viability and miR-130a expression were determined using the Cell Counting Kit-8 (CCK-8) assay and reverse transcription-quantitative PCR, respectively. miR-130a overexpression and knockdown in Adr-resistant AML cells was performed to investigate the proliferative and invasive abilities of the cells using CCK-8 and Transwell assays, respectively. Furthermore, the effects of miR-130a on the expression of epithelial-mesenchymal transition (EMT)-related proteins in Adr-resistant AML cells were detected using western blot analysis. Pre-treatment with 5-Aza-dC enhanced the cell viability and miR-130a expression of Adr-treated AML cells. Adr and miR-130a expression showed a dose-dependent relationship, with miR-130a expression decreasing with increasing Adr concentrations. Moreover, miR-130a overexpression alleviated the inhibitory effects of Adr on cell viability and invasion, while miR-130a knockdown enhanced the sensitivity of AML cells to Adr. Furthermore, Adr exerted an inhibitory effect on EMT in AML cells, which was rescued by miR-130a overexpression and enhanced by miR-130a knockdown. miR-130a knockdown also increased the sensitivity of AML cells to Adr by decreasing cell viability, invasion and EMT. Therefore, miR-130a knockdown is a potential therapeutic strategy for Adr-resistant AML.
Keywords: acute myeloid leukemia, Adriamycin, drug resistance, gene expression, cell invasion
Introduction
Acute myeloid leukemia (AML) is a frequently diagnosed malignant clonal disease of hematopoietic stem cells that shows high heterogeneity (1). The disease primarily manifests via differentiation and maturation disorders of myeloid progenitor cells, abnormal proliferation, inhibited malignant clonal leukemia cell apoptosis and abnormal hematopoiesis (2). AML is the most common type of acute leukemia in adults, accounting for approximately 80% of cases, and the incidence of AML is increasing annually, thus seriously affecting human health (3,4). According to statistics obtained between 2006 and 2010, the 5-year overall survival rate of patients with AML was approximately 21.4% and the 5-year overall survival rate for elderly patients was <10%, due to poor tolerability to chemotherapy (5,6). Therefore, it is important to identify novel treatment strategies to improve complete remission rates and prolong disease-free survival time for patients with AML. In the past few years, AML treatment strategies have progressed slowly, which may be associated with the increased resistance of AML cells to chemotherapeutic drugs (7). However, reducing the resistance of AML cells to chemotherapy is difficult and has become a key focus of the treatment of the disease.
MicroRNAs (miRNAs) are a class of highly conserved non-coding endogenous small molecules, which can bind to the 3′-untranslated regions (UTRs) of mRNAs to interfere with translation. As a result, miRNAs participate in a series of biological processes including cell proliferation, differentiation and apoptosis (8,9). Previous studies have reported that miRNAs are differentially expressed in tumor cells, and miRNA expression is associated with the regulation of cancer cell biological processes and the modulation of oncogene/anti-oncogene expression during tumor formation and development (10,11). Similarly, miRNAs also play crucial roles during the pathogenesis, diagnosis and treatment of leukemia (12). A previous study reported that multiple factors are involved in the regulation of multidrug resistance in leukemia (13). Szymczyk et al (14) revealed that low miR-34a expression was closely related to the sensitivity of patients with chronic lymphocytic leukemia to purine nucleoside analogs, including cladribine and fludarabine. Another previous study reported that miR-130a expression was upregulated in patients with t(8;21) AML and AML cell lines, but the expression of miR-130a decreased significantly once patients with t(8;21) AML achieved complete remission (15). Therefore, the aforementioned studies suggested that low miR-34a expression contributes to the decrease of clonal leukemic cells.
miR-130a has been reported to promote cancer growth (16). Yin et al (17) reported that the upregulation of miR-130a was highly correlated with advanced clinical stage and lymph node metastasis in cervical cancer. Therefore, inhibition of miR-130a expression may represent a treatment strategy with potentially antileukemic effects. At present, few studies have investigated the role of miR-130a in AML cell drug resistance. In the present study, the effects of miR-130a on the sensitivity of AML cells to Adriamycin (Adr) were investigated. AML cell viability and invasion were assessed following miR-130a overexpression and knockdown. The aim of the present study was to identify a potential therapeutic target for drug-resistant AML, to promote the sensitivity of AML cells to Adr and improve the prognosis of patients with AML.
Materials and methods
Cell culture
The AML cell line HL-60 was purchased from the German Collection of Microorganisms and Cell Cultures GmbH (DSMZ no. ACC-3). HL-60 cells (1×106 cells/ml) were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% heat-inactivated fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 µg/ml penicillin (Invitrogen; Thermo Fisher Scientific, Inc.) in a humidified incubator at 37°C with 5% CO2. Cells in the logarithmic phase of growth were used for subsequent experiments.
Cell treatment
HL-60 cells were stimulated using 5-aza-2′-deoxycytidine (5-Aza-dC; 2.5 µmol/l; Sigma-Aldrich; Merck KGaA). Control cells were incubated with DMSO (Sigma-Aldrich; Merck KGaA). After incubation for 3 days at 37°C, cells were treated with Adriamycin (Adr; Sigma-Aldrich; Merck KGaA) at different concentrations (0.00, 0.01, 0.10, 1.00, 5.00 or 10.00 µmol/l) for 24 h at 37°C.
Cell transfection
miR-130a mimic (50 nM; 5′-CAGUGCAAUGUUAAAAGGGCAU-3′; cat. no. 4464066; Thermo Fisher Scientific, Inc.) and miR-130a inhibitor (50 nM; 5′-AUGCCCUUUUAACAUUGCACUG-3′; cat. no. 4464084; Thermo Fisher Scientific, Inc.) were transfected into HL-60 cells (2×104 cells/well) alone or in combination with Adr (0.1 µmol/l) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, inc.), according to the manufacturer's protocol. Negative control cells were transfected with 50 nM negative inhibitor (5′-UUUGUACUACACAAAAGUACUG-3′; cat. no. AM17010; Thermo Fisher Scientific, Inc.) and 50 nM negative mimic (5′-CAGUACUUUUGUGUAGUACAAA-3′; cat. no. 4464058; Thermo Fisher Scientific, Inc.), while untransfected cells served as controls. At 24 h post-transfection, the cells were collected for subsequent experiments.
Cell Counting Kit-8 (CCK-8) assay
HL-60 cell viability following treatment and transfection was determined using the CCK-8 assay (Beyotime Institute of Biotechnology), according to the manufacturer's protocol. Briefly, cells in the logarithmic phase of growth were seeded (1×104 cells/well) into 96-well plates. After transfection or treatment for 24 h, 10 µl CCK-8 reagent was added to each well and incubated for 2 h at 37°C with 5% CO2. The optical density of each well was assessed at a wavelength of 450 nm using an ELX800 microplate reader (BioTek Instruments, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was performed to determine the expression level of miR-130a in HL-60 cells after treatment and transfection. Total RNA was extracted from HL-60 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Total RNA quality and integrity were evaluated using a NanoDrop-2000c spectrophotometer (Thermo Fisher Scientific, Inc.) and 1% modified agarose gel electrophoresis. Subsequently, total RNA (1 µg) was reverse transcribed into cDNA using the SuperScript III First-Strand Synthesis system (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. qPCR was performed using the SYBR Premix Ex Taq™ II (Takara Biotechnology) in the ABI Prism 7500 Fast Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The following primer pair was synthesized by Shanghai GenePharma Co., Ltd. and used for qPCR: miR-130a forward, 5′-GATGCTCTCAGTGCAATGTTA-3′ and reverse, 5′-CTCTGTCTCTCGTCTTGTTGGTAT-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 5 min, 40 cycles at 95°C for 10 sec and 60°C for 45 sec, and a final extension step at 72°C for 10 min. miRNA levels were quantified using the 2−ΔΔCq method (18) and normalized to the internal reference gene U6.
Transwell invasion assay
HL-60 cell invasion after transfection was analyzed using Transwell invasion chambers (Costar; Corning, Inc.). Transfected cells (1×104 cells/well) were suspended in serum-free RPMI-1640 media, and plated into the upper chambers. Transwell membranes were pre-coated with Matrigel® (BD Biosciences) at 37°C for 4 h. RPMI-1640 media containing 10% FBS was plated in the lower chambers. After incubation for 24 h at 37°C, non-invading cells were removed from the Transwell membrane using cotton swabs, and invading cells were fixed in 4% paraformaldehyde for 15 min at room temperature. Subsequently, invading cells were stained using 0.1% crystal violet solution for 20 min at room temperature and observed using an Eclipse TS-100 inverted microscope (magnification, ×200; Nikon Corporation).
Western blotting (WB)
WB was performed to determine the expression levels of EMT-related proteins in HL-60 cells after transfection. Total protein was extracted from transfected cells using RIPA lysis buffer (Beijing Solarbio Biotech Co., Ltd.) according to the manufacturer's protocol. Total protein was quantified using the bicinchoninic protein assay kit (Pierce; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Protein (50 µg per lane) was separated via 12% SDS-PAGE and transferred onto PVDF membranes (EMD Millipore). After blocking with 5% non-fat milk for 1 h at room temperature, the membranes were incubated overnight at 4°C with primary antibodies targeted against: E-Cadherin (1:20,000; cat. no. ab40772; Abcam), N-Cadherin (1:1,000; cat. no. ab18203; Abcam), Vimentin (1:2,000; cat. no. ab92547; Abcam) and GAPDH (1:1,000; cat. no. ab8245; Abcam). Subsequently, the membranes were incubated at 37°C for 2 h with horseradish peroxidase-conjugated rabbit anti-mouse IgG H&L (1:7,000; cat. no. ab3728; Abcam) and horseradish peroxidase-conjugated goat anti-rabbit IgG H&L (1:7,000; cat. no. ab6721; Abcam) secondary antibodies. Protein bands were visualized using an ECL detection reagent (EMD Millipore). GAPDH was used as the loading control. The data were analyzed via densitometry using ImageJ software (version 1.46; National Institutes of Health) and normalized to the expression level of the internal control.
Statistical analysis
SPSS software (version 20; IBM Corp.) was used to perform statistical analyses. Data are presented as the mean ± standard deviation. Comparisons among groups were analyzed using one-way or two-way ANOVA followed by Tukey's post-hoc test. All experiments were performed in triplicate. P<0.05 was considered to indicate a statistically significant difference.
Results
5-Aza-dC increases miR-130a expression levels in Adr-treated AML cells
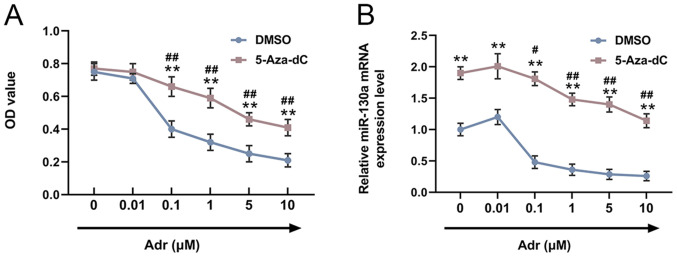
In order to explore the effects of 5-Aza-dC on the viability of Adr-treated AML cells, HL-60 cells were pre-stimulated with 5-Aza-dC or DMSO, and subsequently treated with different concentrations of Adr (0.00, 0.01, 0.10, 1.00, 5.00 or 10.00 µmol/l). Compared with the control cells, pre-treatment with 5-Aza-dC increased the viability of Adr-treated HL-60 cells, and cell viability decreased in a dose-dependent manner with increasing Adr concentrations (P<0.001; Fig. 1A). miR-130a expression in HL-60 cells was measured by RT-qPCR. The results suggested that miR-130a expression in Adr-treated cells was increased by 5-Aza-dC, and similarly, miR-130a expression levels decreased in a dose-dependent manner with increasing Adr concentrations (P<0.05; Fig. 1B).
Figure 1.
5-Aza-dC increases miR-130a expression levels in Adr-treated acute myeloid leukemia cells. (A) The Cell Counting Kit-8 assay was performed to detect the viability of HL-60 cells. (B) Reverse transcription-quantitative PCR was performed to measure the expression levels of miR-130a in HL-60 cells. **P<0.001 vs. DMSO group treated with same Adr concentration. ##P<0.001 and #P<0.05 vs. 5-Aza-dC group treated with 0 µmol/l Adr. 5-Aza-dC, 5-aza-2′-deoxycytidine; miR, microRNA; Adr, Adriamycin; OD, optical density.
miR-130a overexpression reduces the sensitivity of AML cells to Adr
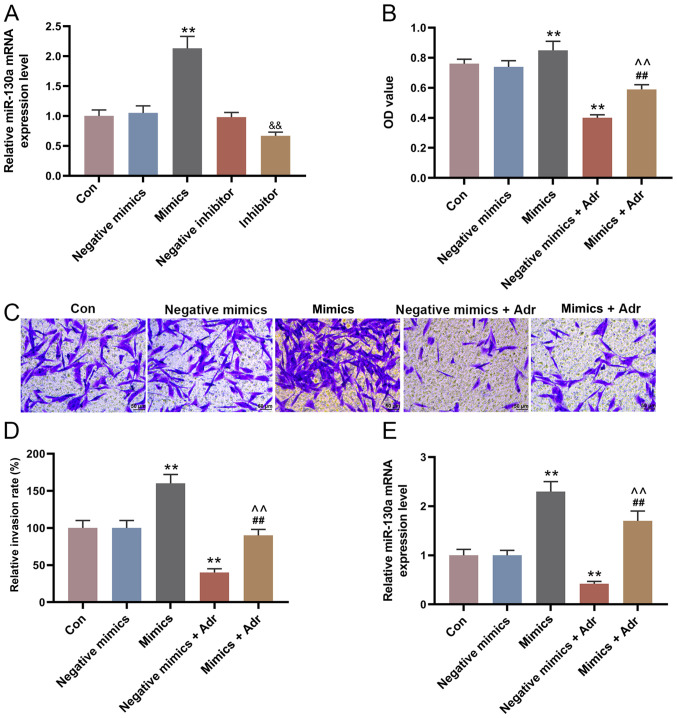
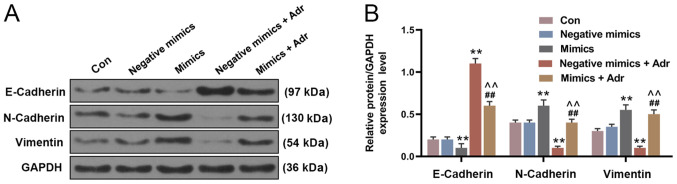
The expression of miR-130a in the mimic group was higher compared with the negative mimic group, and miR-130a expression was decreased in the miR-130a inhibitor group compared with the negative inhibitor group (P<0.001; Fig. 2A). To investigate the effect of miR-130a overexpression on the sensitivity of AML cells to Adr, miR-130a mimic and negative mimic were transfected into HL-60 cells alone or in combination with Adr (0.1 µmol/l). The results suggested that miR-130a overexpression increased HL-60 cell viability, compared with the control group, and alleviated the inhibitory effect of Adr on cell viability (P<0.001; Fig. 2B). Moreover, the Transwell assay results indicated that Adr significantly decreased the number of invasive HL-60 cells in the negative mimic group, which was partially reversed by miR-130a overexpression (P<0.001; Fig. 2C and D). The RT-qPCR results suggested that the expression of miR-130a in HL-60 cells transfected with negative mimic was significantly inhibited by Adr (P<0.001; Fig. 2E). Moreover, miR-130a overexpression significantly decreased the expression of E-Cadherin, and increased the expression of N-Cadherin and Vimentin in HL-60 cells compared with the negative mimic control group. However, Adr treatment displayed the opposite effect to miR-130a overexpression on EMT-related protein expression. Adr treatment following miR-130a overexpression reversed the effects of Adr on EMT-associated protein expression levels (P<0.001; Fig. 3).
Figure 2.
miR-130a overexpression decreases the sensitivity of acute myeloid leukemia cells to Adr. (A) miR-130a expression in HL-60 cells after transfection was detected by RT-qPCR. (B) The Cell Counting Kit-8 assay was used to detect the viability of transfected HL-60 cells. The invasive ability of HL-60 cells after transfection was (C) determined using Transwell assays (magnification, ×200) and (D) quantified. (E) RT-qPCR was performed to measure the expression of miR-130a in HL-60 cells after transfection and treatment. **P<0.001 vs. the negative mimic group. &&P<0.001 vs. the negative inhibitor group. ##P<0.001 vs. the mimic group. ^^P<0.001 vs. the negative mimics + Adr group. miR, microRNA; Adr, Adriamycin; RT-qPCR, reverse transcription-quantitative PCR; Con, control; OD, optical density.
Figure 3.
MicroRNA-130a overexpression reverses the effects of Adr on the expression of epithelial-mesenchymal transition-related proteins. Protein expression levels in HL-60 cells (A) determined by western blot analysis and (B) quantified. **P<0.001 vs. negative mimic group. ##P<0.001 vs. mimic group. ^^P<0.001 vs. negative mimic + Adr group. Adr, Adriamycin; Con, control.
miR-130a knockdown increases the sensitivity of AML cells to Adr
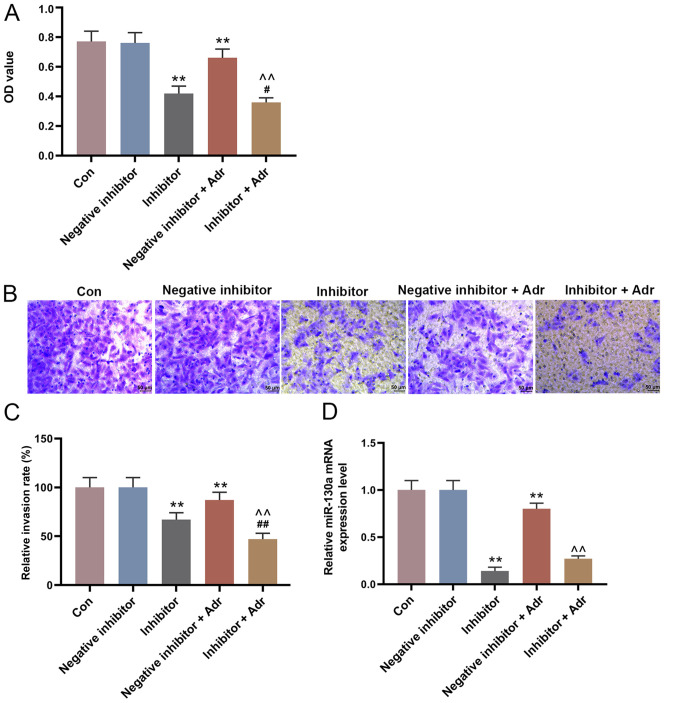
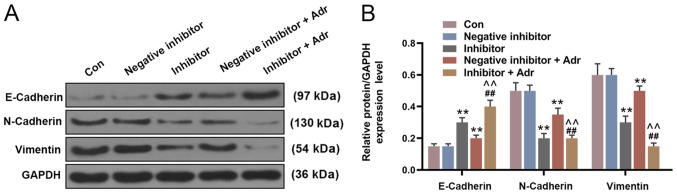
To investigate the effect of miR-130a knockdown on the sensitivity of AML cells to Adr, miR-130a inhibitor and negative inhibitor were transfected into HL-60 cells alone or in combination with Adr (0.1 µmol/l). miR-130a knockdown not only directly reduced the viability of HL-60 cells compared with the negative inhibitor group, but also enhanced the inhibitory effect of Adr on cell viability (P<0.05; Fig. 4A). Additionally, miR-130a knockdown further promoted Adr-induced inhibition of cell invasion, intensifying the anti-cell invasion function of Adr (P<0.001; Fig. 4B and C). Furthermore, miR-130a inhibitor promoted the inhibitory effect of Adr on miR-130a expression (P<0.001; Fig. 4D). The WB results showed similar Adr-induced effects on EMT-related protein expression. miR-130a knockdown increased E-Cadherin expression levels and decreased the expression levels of N-Cadherin and Vimentin in HL-60 cells compared with the negative inhibitor group. In addition, Adr in combination with miR-130a inhibitor displayed increased regulatory effects on EMT-related protein expression compared with either treatment alone (P<0.001; Fig. 5).
Figure 4.
miR-130a knockdown increases the sensitivity of acute myeloid leukemia cells to Adr. (A) The Cell Counting Kit-8 assay was performed to detect the viability of transfected HL-60 cells. The invasive ability of HL-60 cells was (B) determined using Transwell assays (magnification, ×200) and (C) quantified. (D) Reverse transcription-quantitative PCR was performed to measure the expression of miR-130a in HL-60 cells. **P<0.001 vs. the negative inhibitor group. ##P<0.001 and #P<0.05 vs. inhibitor group. ^^P<0.001 vs. negative inhibitor + Adr group. miR, microRNA; Adr, Adriamycin; Con, control; OD, optical density.
Figure 5.
MicroRNA-130a knockdown promotes the Adr-induced effects on the expression of epithelial-mesenchymal transition-related proteins. Protein expression levels were (A) determined by western blotting and (B) quantified. **P<0.001 vs. negative inhibitor group. ##P<0.001 vs. inhibitor group. ^^P<0.001 vs. negative inhibitor + Adr group. Adr, Adriamycin; Con, control.
Discussion
AML is a malignant clonal disease of hematopoietic stem cells, and is often accompanied by a variety of genetic alterations, such as patients with t(8;21) (q22;q22) [RUNX1/RUNX1T1], inv(16)(p13q22) [CBFB/MYH11] and t(15;17)(q24;q21) [PML/RARA] have a favorable prognosis with good response to treatment and complete remissions (19); IDH1 and IDH2 mutations are recurring genetic changes in AML, they constitute a poor prognostic factor in CN-AML with mutated NPM1 without FLT3-ITD (20). Furthermore, the poor prognosis of patients with AML is usually related to the presence of multidrug-resistant leukemia cells (21,22). Adr, a topoisomerase inhibitor, shows an anti-tumor role during AML by restraining the synthesis of double-stranded DNA to induce leukemic cell apoptosis (23,24). In the present study, Adr treatment downregulated the expression level of miR-130a in AML cells. miR-130a overexpression increased the viability and invasion of Adr-treated AML cells, while miR-130a knockdown enhanced the inhibitory effects of Adr on AML cells. In addition, Adr displayed a dose-dependent relationship with miR-130a expression, such that increasing concentrations of Adr decreased the expression levels of miR-130a. The results suggested that miR-130a upregulation may be an important cause of Adr resistance in AML, whereas miR-130a downregulation may serve an effective therapeutic strategy to increase the sensitivity of AML cells to Adr.28
In previous studies, miR-130a has been reported to promote or inhibit tumor growth in different types of cancer. For example, Kong et al (25) reported that miR-130a-3p expression levels were reduced in human breast cancer tissues, and miR-130a-3p overexpression decreased the proliferation and metastasis of breast cancer cells. Previous findings suggested that miR-130a is closely related to drug resistance and that it serves as an intermediary in drug resistance-related signaling pathways (26). For example, Liu et al (27) reported that miR-130a-3p activation decreased the migration and invasion of gemcitabine-resistant hepatoma cells. In the present study, 5-Aza-dC was used to induce Adr resistance in AML cells, and miR-130a expression was upregulated as a result, suggesting that miR-130a may be involved in tumor cell resistance to Adr. The results also suggested that miR-130a overexpression increased the viability and invasion of AML cells, indicating that miR-130a overexpression reversed the anti-AML effects of Adr. Consistent with the present study, Ding et al (15) reported that miR-130a was abnormally upregulated in adult patients with AML, and miR-130a knockdown increased the sensitivity of AML cells to etoposide. Moreover, another study reported that miR-130a overexpression stimulated the propagation of ovarian cancer cells to accelerate disease progression (16).
The effects of miR-130a knockdown on Adr-resistant AML cells were also investigated. Contrasting to the effect of miR-130a overexpression, miR-130a knockdown significantly decreased the viability of AML cells, and promoted the anti-AML effects of Adr. Based on the results, it was hypothesized that miR-130a knockdown increased the sensitivity of AML cells to Adr. A previous study demonstrated that the expression level of miR-130a was significantly reduced in patients with CML with poor prognosis, and that a low expression was associated with a short overall survival time (28). Moreover, Feng et al (29) reported that miR-130a overexpression reduced cisplatin-sensitivity of cervical cancer cells, and a low expression of miR-130a restored the chemoresistance of the cancer cells to cisplatin. The present study and the aforementioned previous studies suggested that miR-130a upregulation was related to the pathogenesis and progression of AML, and also contributed to the occurrence of Adr resistance.
To further verify the effects of miR-130a on AML chemoresistance, the present study detected the expression of EMT-related proteins, including E-Cadherin, N-Cadherin and Vimentin. EMT is a phenotypic transition, during which cells transform from an epithelial state to a mesenchymal state, in terms of cell functional characteristics and morphology (30,31). The transition of epithelial cells into mesenchymal cells results in increased invasive and migratory abilities, which can lead to metastasis during cancer progression (32,33). Cancer cells have shown increased levels of N-cadherin and Vimentin, and decreased levels of E-cadherin during the transition from epithelial cell to mesenchymal cell (34,35). In the present study, Adr promoted E-cadherin expression, and downregulated N-cadherin and Vimentin expression in AML cells. However, miR-130a overexpression inhibited Adr-induced E-cadherin expression, and upregulated N-cadherin and Vimentin expression. By contrast, miR-130a knockdown indicated the opposite effect and promoted Adr-induced effects. The results suggested that miR-130a overexpression increased AML cell Adr resistance by promoting EMT, whereas miR-130a downregulation increased the sensitivity of AML cells to Adr by regulating the expression of EMT-related proteins. However, the present study had a number of limitations. A key limitation of the present study was that investigations were only conducted using one cell line; therefore, the results of the present study need to be verified in multiple cell lines and in in vitro models.
In conclusion, the expression level of miR-130a was increased in Adr-resistant AML cells, and miR-130a knockdown promoted the inhibitory effects of Adr on AML cell viability, invasion and EMT. Therefore, miR-130a knockdown increased the sensitivity of AML cells to Adr and may serve as a therapeutic target for Adr-resistant AML.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- Adr
Adriamycin
- AML
acute myeloid leukemia
- CCK-8
Cell Counting Kit-8
- RT-qPCR
reverse transcription-quantitative PCR
- EMT
epithelial-mesenchymal transition
- WB
western blotting
- miR
microRNA
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HL conceived and designed the study. ML, JZ and YL acquired, analyzed and interpreted the data. HL drafted and critically revised the manuscript for important intellectual content. All authors provided final approval of the version to be published. ML agreed to be held accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Li S, Mason CE, Melnick A. Genetic and epigenetic heterogeneity in acute myeloid leukemia. Curr Opin Genet Dev. 2016;36:2810–106. doi: 10.1016/j.gde.2016.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juliusson G, Hough R. Leukemia. Prog Tumor Res. 2016;43:87–100. doi: 10.1159/000447076. [DOI] [PubMed] [Google Scholar]
- 3.De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016;6:e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto JF, Goodman MT. Patterns of leukemia incidence in the United States by subtype and demographic characteristics, 1997–2002. Cancer Causes Control. 2008;19:379–390. doi: 10.1007/s10552-007-9097-2. [DOI] [PubMed] [Google Scholar]
- 5.Pulte D, Gondos A, Brenner H. Expected long-term survival of patients diagnosed with acute myeloblastic leukemia during 2006–2010. Ann Oncol. 2010;21:335–341. doi: 10.1093/annonc/mdp309. [DOI] [PubMed] [Google Scholar]
- 6.Meyers J, Yu Y, Kaye JA, Davis KL. Medicare fee-for-service enrollees with primary acute myeloid leukemia: An analysis of treatment patterns, survival, and healthcare resource utilization and costs. Appl Health Econ Health Policy. 2013;11:275–286. doi: 10.1007/s40258-013-0032-2. [DOI] [PubMed] [Google Scholar]
- 7.Bose P, Vachhani P, Cortes JE. Treatment of relapsed/refractory acute myeloid leukemia. Curr Treat Options Oncol. 2017;18:17. doi: 10.1007/s11864-017-0456-2. [DOI] [PubMed] [Google Scholar]
- 8.McCall MN, Kim MS, Adil M, Patil AH, Lu Y, Mitchell CJ, Leal-Rojas P, Xu J, Kumar M, Dawson VL, et al. Toward the human cellular microRNAome. Genome Res. 2017;27:1769–1781. doi: 10.1101/gr.222067.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35:3–11. doi: 10.1055/s-0034-1397344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 11.Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-a brief overview. Adv Biol Regul. 2015;57:1–9. doi: 10.1016/j.jbior.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Chen H, Bai J, He A. MicroRNA: An important regulator in acute myeloid leukemia. Cell Biol Int. 2017;41:936–945. doi: 10.1002/cbin.10770. [DOI] [PubMed] [Google Scholar]
- 13.Li Q, Song W, Wang J. TUG1 confers Adriamycin resistance in acute myeloid leukemia by epigenetically suppressing miR-34a expression via EZH2. Biomed Pharmacother. 2019;109:1793–1801. doi: 10.1016/j.biopha.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Szymczyk A, Chocholska S, Macheta A, Szczepanek D, Hus M, Podhorecka M. Assessment of microRNA expression in leukemic cells as predictors of sensitivity to purine nucleoside analogs, fludarabine and cladribine, in chronic lymphocytic leukemia patients. Cancer Manag Res. 2019;11:5021–5031. doi: 10.2147/CMAR.S191311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding C, Chen SN, Macleod RAF, Drexler HG, Nagel S, Wu DP, Sun AN, Dai HP. miR-130a is aberrantly overexpressed in adult acute myeloid leukemia with t(8;21) and its suppression induces AML cell death. Ups J Med Sci. 2018;123:19–27. doi: 10.1080/03009734.2018.1440037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei H, Cui R, Bahr J, Zanesi N, Luo Z, Meng W, Liang G, Croce CM. miR-130a deregulates PTEN and stimulates tumor growth. Cancer Res. 2017;77:6168–6178. doi: 10.1158/0008-5472.CAN-17-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin S, Zhang Q, Wang Y, Li S, Hu R. MicroRNA-130a regulated by HPV18 E6 promotes proliferation and invasion of cervical cancer cells by targeting TIMP2. Exp Ther Med. 2019;17:2837–2846. doi: 10.3892/etm.2019.7226. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Lagunas-Rangel FA, Chavez-Valencia V, Gomez-Guijosa MA, Cortes-Penagos C. Acute myeloid leukemia-genetic alterations and their clinical prognosis. Int J Hematol Oncol Stem Cell Res. 2017;11:328–339. [PMC free article] [PubMed] [Google Scholar]
- 20.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Krönke J, Bullinger L, Späth D, Kayser S, Zucknick M, Götze K, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 21.Kater L, Claffey J, Hogan M, Jesse P, Kater B, Strauss S, Tacke M, Prokop A. The role of the intrinsic FAS pathway in Titanocene Y apoptosis: The mechanism of overcoming multiple drug resistance in malignant leukemia cells. Toxicol In Vitro. 2012;26:119–124. doi: 10.1016/j.tiv.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Madanat YF, Kalaycio ME, Nazha A. Advances in acute myeloid leukemia genomics, where do we stand in 2018? Acta Med Acad. 2019;48:35–44. doi: 10.5644/ama2006-124.240. [DOI] [PubMed] [Google Scholar]
- 23.Rivankar S. An overview of doxorubicin formulations in cancer therapy. J Cancer Res Ther. 2014;10:853–858. doi: 10.4103/0973-1482.139267. [DOI] [PubMed] [Google Scholar]
- 24.Han YQ, Hong Y, Su XL, Wang JR. Quercetin enhances the anti-leukemic effect of adriamycin. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2014;22:1555–1560. doi: 10.7534/j.issn.1009-2137.2014.06.011. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 25.Kong X, Zhang J, Li J, Shao J, Fang L. miR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem Biophys Res Commun. 2018;501:486–493. doi: 10.1016/j.bbrc.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HD, Jiang LH, Sun DW, Li J, Ji ZL. The role of miR-130a in cancer. Breast cancer. 2017;24:521–527. doi: 10.1007/s12282-017-0776-x. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Li Y, Wang R, Qin S, Liu J, Su F, Yang Y, Zhao F, Wang Z, Wu Q. miR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res. 2016;35:19. doi: 10.1186/s13046-016-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, Zhao H, Lin Z, Zhang G. Functional studies of miR-130a on the inhibitory pathways of apoptosis in patients with chronic myeloid leukemia. Cancer Gene Ther. 2015;22:573–580. doi: 10.1038/cgt.2015.50. [DOI] [PubMed] [Google Scholar]
- 29.Feng C, Ma F, Hu C, Ma JA, Wang J, Zhang Y, Wu F, Hou T, Jiang S, Wang Y, Feng Y. SOX9/miR-130a/CTR1 axis modulates DDP-resistance of cervical cancer cell. Cell Cycle. 2018;17:448–458. doi: 10.1080/15384101.2017.1395533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen T, You Y, Jiang H, Wang ZZ. Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. J Cell Physiol. 2017;232:3261–3272. doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dominguez C, David JM, Palena C. Epithelial-mesenchymal transition and inflammation at the site of the primary tumor. Semin Cancer Biol. 2017;47:177–184. doi: 10.1016/j.semcancer.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao TT, Yang MH. Revisiting epithelial-mesenchymal transition in cancer metastasis: The connection between epithelial plasticity and stemness. Mol Oncol. 2017;11:792–804. doi: 10.1002/1878-0261.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HH, Jung J, Moon A, Kang H, Cho H. Antitumor and anti-invasive effect of apigenin on human breast carcinoma through suppression of IL-6 expression. Int J Mol Sci. 2019;20:3143. doi: 10.3390/ijms20133143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.