Abstract
Lung adenocarcinoma (LUAD), a major subtype of lung cancer, is the leading cause of cancer-related mortality worldwide. Previous studies have determined the role of the protein arginine methyltransferases (PRMTs) in the physiology and pathology of LUAD. However, to the best of our knowledge, no empirical studies have been performed determining the association between protein arginine methyltransferase 6 (PRMT6) and LUAD. The present study aimed to determine the expression levels of PRMT6 in LUAD and its association with the clinicopathological characteristics. The effect of PRMT6 knockdown on cell growth was analyzed and chromatin immunoprecipitation (ChIP) assay was used to investigate the regulatory mechanisms of PRMT6 on downstream gene expression. In addition, a xenograft model was used to determine whether the PRMT6-regulated expression levels of p18 in vitro could be validated in vivo. PRMT6 overexpression in LUAD is associated with high clinical stage, lymph node metastasis and poor clinical outcomes. Furthermore, the silencing of PRMT6 significantly reduced the enrichment of Histone H3 asymmetric demethylation at arginine 2 in the promoter region of the p18 gene, thereby activating the expression of the gene. This, in turn, induced G1/S phase cell cycle arrest, resulting in the inhibition of cell proliferation. The xenograft model also suggested that PRMT6 suppressed LUAD development by activating p18 expression in vivo. In conclusion, the findings of the present study suggested that PRMT6 may serve as an oncogene in the progression of LUAD through epigenetically suppressing p18 expression. Thus, PRMT6 may represent a novel potential therapeutic target for LUAD.
Keywords: lung adenocarcinoma, protein arginine methyltransferase 6, cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4), cell proliferation
Introduction
Non-small cell lung carcinoma (NSCLC) accounts for ~85% of all lung cancer cases (1); lung adenocarcinoma (LUAD) is the main pathological type of NSCLC (2). The prognosis of lung cancer is related to the rate of recurrence, metastasis and chemotherapy resistance (3). The 5-year survival rate is 51.4% for patients with adenocarcinoma (4). The poor prognosis of lung cancer highlights the requirement for the development of novel biomarkers for the early diagnosis of the disease (5,6). Thus, there is an urgent need to improve the diagnosis and management of NSCLC.
Protein arginine methyltransferases (PRMTs) can be divided into three types: Asymmetrically, symmetrically and monomethylate protein arginines (type I/II/III, respectively) (7–9). Histone protein arginine methylation, catalyzed by PRMTs serves a crucial role in gene regulation (10). In addition, several non-histone substrates have also been discovered to be involved in gene transcription and protein translation (11). PRMTs were revealed to be widely expressed and activated in gastric and prostate cancer, as well as myeloid leukemia, where they were involved in cell growth, differentiation and apoptosis (12–15). In fact, the disruption of the modification catalyzed by PRMTs suppressed tumor development, indicating that PRMTs may be used as a potential therapeutic target for cancer (16). However, to the best of our knowledge, only a few studies have reported the dysregulation of PRMTs in lung cancer. For example, PRMT1 and PRMT4 were identified to be involved in the regulation of proliferation in lung cancer (17); and PRMT1 and PRMT5 were discovered to regulate apoptosis induced by doxorubicin or pemetrexed by affecting cellular FADD-like IL-1β-converting enzyme-inhibitory protein in NSCLC cells (18). In addition, enolase 1 methylation by PRMT5 was discovered to be critical for lung cancer cell invasion (19). Interestingly, to the best of our knowledge, no other PRMT members and their dysregulation were reported to be associated with lung cancer.
PRMT6, a type I arginine methyltransferase, has high affinity for the arginine-2 of Histone H3, specifically catalyzing Histone H3 asymmetric demethylation at arginine 2 (H3R2me2a) (10). PRMT6 was first identified to modify the glycine-and arginine-rich motifs (13), and subsequently reported to target histones and non-histones (20). However, the role of PRMT6 in human cancer remains controversial. The downregulation of PRMT6 expression levels has been reported in melanoma (21), while the upregulation of PRMT6 expression levels was reported in the bladder (13), liver (22) and prostate (14). Interestingly, in one study, PRMT6 upregulation contributed to global DNA hypomethylation in colorectal and lung adenocarcinoma (23).
At present, studies on PRMT6 are mainly focused on its function in the nucleus, while the biological function and important target proteins of PRMT6 in human cancer remain unclear. The present study aimed to determine the association between PRMT6 expression levels and clinicopathological characteristics of LUAD via analyzing the putative oncogenic role and the potential underlying mechanism of PRMT6 in LUAD. The present study demonstrated that PRMT6 expression levels were markedly upregulated LUAD.
Materials and methods
Tissue microarray
The LUAD tissue microarray (cat. no. HlugA180Su05), including 85 pairs of LUAD tissues and matched normal adjacent tissues (NAT) with clinicopathological data, was provided by Shanghai Outdo Biotech Co. Ltd. (Outdo Biotech). All patients were classified according to the tumor-node-metastasis (TNM) classification by the American Joint Commission of Cancer (24). Lymph node metastasis and the depth of invasion were classified using the 7th edition of the International Union Against Cancer TNM staging system (25). The survival time was set as the time from the day of pathological diagnosis to the day of last contact or the date of death.
Patient studies
Fresh LUAD tissues and matched NAT from 7 LUAD patients [3 male and 4 female; age, 47.4±5.2 years (mean ± SD)] were obtained from The Second Affiliated Hospital of Nanjing University of Chinese Medicine (Nanjing, China) from March 2018 to February 2019. Patients included in the study had neither received chemotherapy nor undergone surgery. Combined with lung disease already known, other tumor and autoimmune diseases as exclusion criteria. The study was approved by The Ethics Committee of The Second Affiliated Hospital of Nanjing University of Chinese Medicine (Nanjing, China). Written informed consent was obtained from the patients for the use of fresh lung tissues.
Hematoxylin & Eosin (H&E) staining and immunohistochemistry
Lung tissues were fixed in 4% paraformaldehyde for 24 h at room temperature before paraffin embedding. Sections (3.5 µm) cut from paraffin-embedded specimens were deparaffinised in xylene and rehydrated through graded alcohol series. The sections were first processed for hematoxylin and eosin (H&E) staining according to standard method (26). Then, antigen retrieval was conducted using the 1X Diva Decloaker antigen retrieval solution (Biocare Medical, LLC). at 95°C for 15 min, and then blocking non-specific sites with 10% goat serum (cat. no. C0265; Beyotime Institute of Biotechnology) in PBS for 1 h at room temperature, Subsequently, the sections were incubated with an anti-PRMT6 primary antibody (1:250; cat. no. 14641; Cell Signaling Technology, Inc.), anti-p18 (1:250; cat. no. A8751; Abclonal Biotech Co., Ltd.) and anti-Ki67 (1:500; cat. no. A2094; Abclonal Biotech Co., Ltd.) at 4°C for overnight. Following the primary antibody incubation, the slides were then incubated with a horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (1:1,000; cat. no. AS038; Abclonal Biotech Co., Ltd.) at 25°C for 1 h. The slides were subsequently stained with a DAB substrate kit (Dako; Agilent Technologies, Inc.) and counterstained with hematoxylin at 25°C for 20 sec. The immunostaining was detected using an Aperio Digital Pathology Slide scanner (Leica Biosystems, Wetzlar, Germany).
The nuclear staining of PRMT6/p18/Ki67 was analyzed using the H-score system. Nuclear staining results were analyzed using Hscore using Zeiss microscope at a ×100 magnification. Positive cells were analyzed according to the staining intensity on a scale of 0–3 (0 = negative, 1 = weak, 2 = moderate, 3 = strong). H-scores were calculated as the sum of the intensity score (i) multiplied by the percentage of cells at each intensity (Pi). H-score =Σ [Pi(i)] ×100. Score values range between 0 and 300.
Cell culture
A549 cells were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and cultured in high glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin streptomycin combination (Gibco; Thermo Fisher Scientific, Inc.). Cells were maintained at 37°C in an atmosphere containing 5% CO2.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the adherent cells and tissues using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and cDNA was synthesized using HiScript III 1st Strand cDNA Synthesis Kit for qPCR (cat. no. R312; Vazyme Biotech Co., Ltd.). The thermocycling conditions of the RT were as follows: Remove genomic DNA at 42°C for 2 min; first strand cDNA synthesised at 25°C for 5 min, 37°C for 45 sec and 85°C for 5 sec. qPCR was subsequently performed using the ChamQ Universal SYBR qPCR Master Mix (cat. no. Q711 Vazyme Biotech Co., Ltd.) and the primers provided in Table I on an ABI 7500 Real-Time PCR machine (Applied Biosystems Inc.). The thermocycling conditions of the qPCR were as follows: Denaturation at 95°C for 5 min; 40 cycles at 95°C for 10 sec and 60°C for 30 sec; and a final dissociation stage (95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec) was added at the end of the amplification procedure. The data were analyzed using the ABI 7500 SDS software (Version 2.0.6, Applied Biosystems Inc.). The relative mRNA expression levels were calculated using the 2−ΔΔCq method (27) and normalized to the GAPDH reference gene.
Table I.
Sequences of the primers used for reverse transcription-quantitative PCR.
| Gene | Primer sequence (5′→3′) |
|---|---|
| Protein arginine methyltransferase 6 | F: ACGAGTGCTACTCGGACGTT |
| R: AGTTCCGAAGGATACCCAGG | |
| p21 | F: TACCCTTGTGCCTCGCTCAG |
| R: CGGCGTTTGGAGTGGTAGA | |
| p27 | F: GGAGCAATGCGCAGGAATAA |
| R: TGGGGAACCGTCTGAAACAT | |
| p18 | F: ACTGGTTTCGCTGTCATTCA |
| R: GCAGGTTCCCTTCATTATCC | |
| CDK inhibitor 3 | F: TCCGGGGCAATACAGACCAT |
| R: CAGCTAATTTGTCCCGAAACTC | |
| CDK4 | F: ATGGCTACCTCTCGATATGAGC |
| R: CATTGGGGACTCTCACACTCT | |
| CDK6 | F: GCTGACCAGCAGTACGAATG |
| R: GCACACATCAAACAACCTGACC | |
| Cyclin D1 | F: GCTGCGAAGTGGAAACCATC |
| R: CCTCCTTCTGCACACATTTGAA | |
| Cyclin E1 | F: ACTCAACGTGCAAGCCTCG |
| R: GCTCAAGAAAGTGCTGATCCC | |
| S-phase kinase-associated protein 2 | F: ATGCCCCAATCTTGTCCATCT |
| R: CACCGACTGAGTGATAGGTGT | |
| GAPDH | F: GAGCCACATCGCTCAGACAC |
| R: CATGTAGTTGAGGTCAATGAAGG |
F, forward; R, reverse.
Western blotting
Total protein was extracted from tissues and LUAD cells using RIPA lysis buffer (Beyotime Institute of Biotechnology), supplemented with protease and phosphatase inhibitor cocktails (CST Biological Reagents Co., Ltd.). Nuclear protein extracts were obtained using the Nuclear Extract kit (cat. no. P0027; Beyotime Institute of Biotechnology). Core histones were extracted from the nuclear extracts of the LUAD cells using an acid-extraction method as previously described (28). Total protein was quantified using a bicinchoninic acid assay kit (cat. no. 23227; Pierce; Thermo Fisher Scientific, Inc.) and 25 µg total protein extracts/lane and 10 µg nuclear extracts/lane were separated via 12% SDS-PAGE. Subsequently, the separated proteins were transferred onto PVDF membranes (Roche Diagnostics) and probed with specific primary antibodies in 5% skimmed milk in PBST (PBS with 0.1% Tween-20) overnight at 4°C. The membranes were then incubated with rabbit-or mouse-specific HRP-conjugated secondary antibodies for 2 h at room temperature. The following primary antibodies were used: Anti-GAPDH (1:10,000; cat. no. M171-3; MBL Co., Ltd.), anti-PRMT6 (1:1,000; cat. no. A7814; Abclonal Biotech Co., Ltd.), anti-p18 (1:1,000; cat. no. A8751; Abclonal Biotech Co., Ltd.), anti-Lamin B1 (1:1,000; cat. no. A11495 Abclonal Biotech Co., Ltd.), anti-Histone H3 (1:1,000; cat. no. A2348; Abclonal Biotech Co., Ltd.) and anti-Histone H3R2me2a (1:1,000; cat. no. A3155; Abclonal Biotech Co., Ltd.). Goat HRP-conjugated anti-rabbit immunoglobulin G (IgG; 1:1,000; cat. no. AS014; Abclonal Biotech Co., Ltd.) and HRP-conjugated goat anti-mouse IgG (1:1,000; cat. no. AS003; Abclonal Biotech Co., Ltd.) secondary antibodies were used. Lamin B1 was used as a loading control for nuclear proteins and GAPDH was used as a loading control for total proteins. Antibody binding was detected using an ECL detection system (cat. no. 32106; Thermo Fisher Scientific, Inc.).
Chromatin immunoprecipitation (ChIP)
The ChIP assay was performed as previously described (29). Briefly, cells were crosslinked by 1% formaldehyde (Sigma-Aldrich) in PBS for 10 min at 25°C. Formaldehyde was quenched by the addition of glycine (Beijing Solarbio Science & Technology Co., Ltd.) to a final concentration of 125 µM. Then, 1×106 cells were collected by centrifugation at 300 × g for 3 min at 25°C and washed with pre-cooled PBS twice. The immunoprecipitation of crosslinked 100 µg DNA (using a spectrophotometer at 260 nm)/protein was performed using 2 µg anti-H3R2me2a (1 µg/µl, H3R2me2a; cat. no. A3155; Abclonal Biotech Co., Ltd.), anti-histone H3 lysine 4 trimethylation (1 µg/µl, H3K4me3; cat. no. A2357; Abclonal Biotech Co., Ltd.) or anti-mouse/rabbit IgG (1 µg/µl, cat. no. A7028 and A7016; Beyotime Institute of Biotechnology) antibodies for a 2-h incubation at 4°C. The immunoprecipitated DNA was purified using a ChIP DNA purification kit (cat. no. D0033; Beyotime Institute of Biotechnology) and amplified by qPCR as described above. The chip primers for the detection of H3R2me2a/H4K4me3 enrichment on p18 promotor as follows: Forward, 5′-GTCTTAAATAACAAACCCCTGTC-3′ and reverse, 5′-CTCCTCCCGTCAAGTCTCTCGCG-3′.
Vectors, transfections and infections
pLKO lentiviral vectors for gene knockdown and the pLKO-scrambled (Scr) short hairpin (sh)RNA vector (control) were obtained from Sigma-Aldrich; Merck KGaA. The shRNA sequences for the pLKO lentiviral vector constructions are listed in Table II. A total of 6×106 293T cells were seeded into 100-mm cell culture dishes and incubated at 37°C and 5% CO2 for 16 h. When the cultured cells reached 85% confluence, cells were co-transfected with 3 µg of lentiviral expression constructs pLKO.1-shRNA, pMD2.G and psPAX2 (Sigma- Aldrich; Merck KGaA) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Viral supernatants were collected by centrifugation at 800 × g for 5 min at 25°C and filtered through a 0.22-µm membrane filter 48 h post-transfection and stored at −80°C. A549 cells (5×106 cells per 100-mm culture dish) were seeded and incubated overnight at 37°C prior to infection. Medium was then replaced with 1:1 diluted viral supernatant supplemented with 8 µg/ml polybrene and incubated for 24 h at 37°C, followed by replacement with normal growth medium. Stable cell lines with shRNA were selected by puromycin (Clontech Laboratories, Inc.) at a final concentration of 2 µg/ml in A549 cells.
Table II.
shRNA sequence used for pLKO lentiviral vectors construction.
| shRNA | Sequence (5′→3′) |
|---|---|
| PRMT6 sh1 | CCGGCACCGGCATTCTGAGCATCTTCTCGAGAAGATGCTCAGAATGCCGGTGTTTTTG |
| PRMT6 sh2 | CCGGCACGGACGTTTCAGGAGAGATCTCGAGATCTCTCCTGAAACGTCCGTGTTTTTG |
| p18 sh1 | CCGGTGGATTTGGAAGGACTGCGCTCTCGAGAGCGCAGTCCTTCCAAATCCATTTTTG |
| p18 sh2 | CCGGACTGGTTTCGCTGTCATTCATCTCGAGATGAATGACAGCGAAACCAGTTTTTTG |
| Scr | CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG |
sh/shRNA, short hairpin; PRMT6, protein arginine methyltransferase 6; Scr, scramble.
Cell proliferation assay
For the cell proliferation assay, 2×103 A549 cells/well were seeded into 96-well plates in triplicate. Cell proliferation was analyzed using a Cell Counting Kit-8 (CCK-8; cat. no. A311; Vazyme Biotech Co., Ltd.) assay, according to manufacturer's protocol. Briefly, cells were plated and incubated for 24 h in 96-well plates prior to test, 10 µl CCK8 solution was added to each well and incubated at 37°C for 2 h. The optical density (OD) was read at an absorbance of 450 nm using a multifunction microplate reader (Safire, TECAN) for 4 continuous days.
For the colony formation assay, 500 viable A549 cells per well were seeded into 6-well plates in triplicate. Following incubation at 37°C for 10 days, the colonies were fixed with methanol at room temperature for 30 min, stained with 0.05% crystal violet (Beyotime Institute of Biotechnology) for 60 min at 25°C, washed with running water to remove the excessive dye and imaged with an Epson Perfection V550 Photo scanner (Seiko Epson Corporation). Number of colonies (>50 cells) was calculated using ImageJ software version 1.45 (National Institutes of Health, Bethesda, MD, USA).
Cell cycle assay
A total of 1×104 cells were collected by centrifugation at 300 × g for 3 min at 25°C and permeabilized with ice-cold 70% ethanol overnight at 4°C. Then, the cells were collected by centrifugation at 300 × g for 3 min at 4°C and stained with 50 ug/ml propidium iodide in ice-cold PBS supplemented with 0.25 mg/ml RNase A at 4°C for 30 min (cat. no. KGA214-10; Nanjing KeyGen Biotech Co., Ltd.). The cells were analyzed using a FACSCalibur (BD Biosciences) flow cytometer and FlowJo X V10.0.7 software (FlowJo LLC) was used to analyze the data.
In vivo tumor models
The animal studies were performed according to the guidelines of the National Institutes of Health for the Care and Use of Laboratory Animals (30). The protocols were approved by The Institute of Animal Care and Use Committee of Nanjing University of Chinese Medicine (Nanjing, China).
In order to establish a subcutaneous tumor model, 18 female BALB/c nu/nu mice (6-week-old; weight, 18–20 g) were obtained from the Model Animal Research Center of Nanjing University of Chinese Medicine, and maintained under specific pathogen-free conditions at The Animal Experiment Center of The Second Affiliated Hospital of Nanjing University of Chinese Medicine (Nanjing, China). Mice were housed 6 per cage at 25±2°C with 50±10% humidity and on a 12-hour light–dark cycle with free access to pellet food and water. They were given a minimum acclimation period of 1 week before subcutaneous tumor implantation. A549 cells were first transduced with either Scr, PRMT6 sh1 or combined PRMT6 sh1 and p18 sh1 lentiviruses to establish stable cell lines for in vivo studies. Subsequently, 2×106 cells in 200 µl DMEM supplemented with 50% Matrigel (2 mg/ml; BD Biosciences) were inoculated subcutaneously into the right flank of 8-week-old BALB/c nude mice (6 mice/group). Tumor growth rate was monitored by measuring tumor diameters every 4 days. Both length and width (W) of the tumor were measured using a slide caliper, and the tumor volume was calculated as (Lengthxwidthxwidth)/2.
The sizes of the tumors were measured every 3 days from injection and tumor volumes were calculated using the formula: Volume (cm3)=0.52×(lengthxwidth2). All animals were euthanized under general anesthesia with carbon dioxide when max tumor volumes reached humane endpoints (~1,000 mm3). The flow rate displaced 10–30% of the chamber volume/minute. The animals that lost consciousness and muscle activity were identified as deceased.
Statistical analysis
Data are presented as the mean ± SD of three independent experiments. Statistical significances were determined using a two-tailed Student's paired t-test, or a one-way ANOVA, followed by a Bonferroni's multiple comparisons post hoc test. Receiver operating characteristic (ROC) curve analysis was performed to determine the diagnostic value of PRMT6 expression levels in LUAD. Kaplan-Meier estimates for the primary end point were calculated and compared using a log-rank (Mantel-Cox) test of equality. The correlation between PRMT6 and p18 expression levels was analyzed using Pearson's correlation analysis. All the statistical analyses were conducted using GraphPad Prism 7 software (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
PRMT6 is overexpressed in LUAD
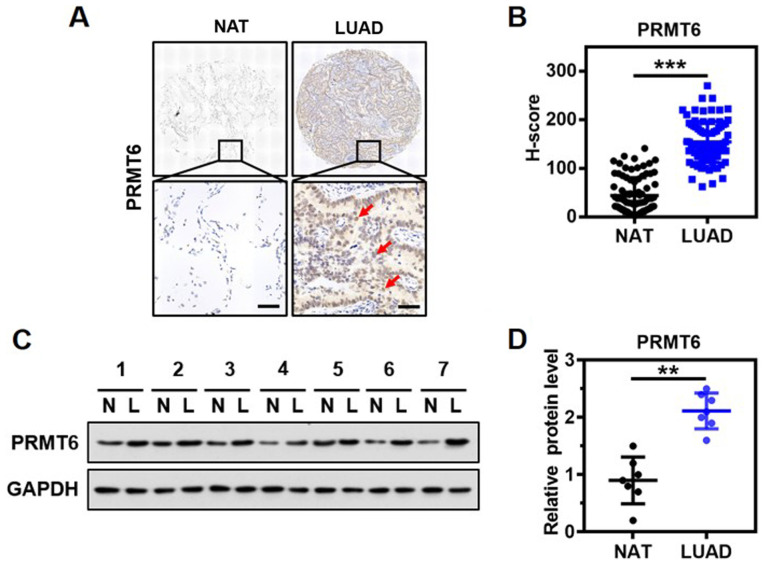
To determine the clinical significance of PRMT6 expression in LUAD, the expression levels of PRMT6 in the lung tissue from a cohort of 85 patients with LUAD were investigated using immunohistochemistry and a specific anti-PRMT6 antibody. The protein was discovered to be predominantly localized in the nucleus of the glandular epithelium of malignant tissues (Fig. 1A). Notably, significantly upregulated expression levels of PRMT6 were observed in the tumor tissues of patients with LUAD compared with the matched NAT (Fig. 1B). Western blotting results confirmed that the expression levels of PRMT6 in tumor tissues from 7 patients with LUAD were significantly upregulated compared with in NAT (Fig. 1C and D). Thus, these findings suggested that PRMT6 expression levels may be significantly upregulated in LUAD.
Figure 1.
PRMT6 expression levels are overexpressed in LUAD. (A) Immunohistochemistry analysis of PRMT6 expression levels in LUAD tissues and NAT from 85 patients with LUAD. PRMT6 are mainly distributed in the nucleus of glandular epithelium, as shown by the red arrow. Scale bar, 50-µm. (B) Quantitative analyses of immunohistochemistry scores of PRMT6 protein expression levels from part (A). ***P<0.001. (C) Representative western blots of PRMT6 expression levels in LUAD tissues and matched NAT (n=7). GAPDH served as the normalization control. (D) PRMT6 protein expression levels from part (C) were semi-quantified. **P <0.01. PRMT6, protein arginine methyltransferase 6; LUAD/L, lung adenocarcinoma; NAT/N, matched normal adjacent tissue.
Association between PRMT6 expression levels and clinicopathological features
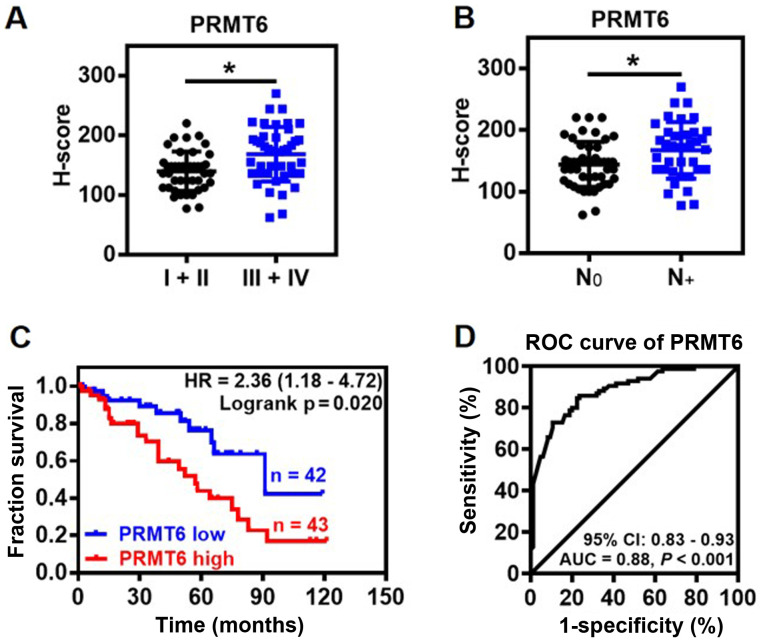
The association between PRMT6 expression levels and clinicopathological features of LUAD was further investigated. The expression levels of PRMT6 protein were significantly upregulated in patients with advanced clinical stages (III and IV) and lymph node metastasis compared with the patients with non-advanced clinical stages (I and II) and no lymph nodes metastasis, respectively (Fig. 2A and B). Notably, the expression levels of PRMT6 were not related to the age, sex, tumor size, differentiation or local invasion of patients with LUAD (data not shown). Kaplan-Meier survival analysis indicated that high expression levels of PRMT6 protein were linked to a significantly poorer prognosis in patients with LUAD compared with patients with low expression levels of PRMT6 (Fig. 2C). Furthermore, the predictive value of PRMT6 was evaluated using ROC curve analysis. The results indicated that the area under the curve (AUC) was 0.88 [95% confidence interval (CI), 0.83-0.93] between LUAD tissues and NAT (Fig. 2D). In the ROC curve analysis, H-score 97 was set as the cut-off for the expression levels, based on which, the tumor tissues were discriminated from NAT with high sensitivity (84.71%) and specificity (76.47%; Fig. 2D). These findings suggested that PRMT6 may be used as a novel diagnostic biomarker for LUAD.
Figure 2.
Association between PRMT6 expression levels and clinicopathological characteristics of LUAD. (A) Association between the expression levels of PRMT6 and the clinical stage of patients with LUAD (n=85). *P<0.05. (B) Expression levels of PRMT6 in tumor tissue were associated with lymph node metastasis (n=85). *P< 0.05. (C) Kaplan-Meier survival curve determining the overall survival according to low (n=42) and high (n=43) expression levels of PRMT6 using immunohistochemistry scores from 85 patients with LUAD. (D) Performance of PRMT6 protein expression levels as a molecular biomarker for LUAD was determined by ROC analysis. PRMT6, protein arginine methyltransferase 6; LUAD, lung adenocarcinoma; ROC, receiver operating characteristic; N, lymph node metastasis; AUC, area under the curve; CI, confidence interval.
Knockdown of PRMT6 suppresses LUAD cell growth in vitro through G1/S arrest
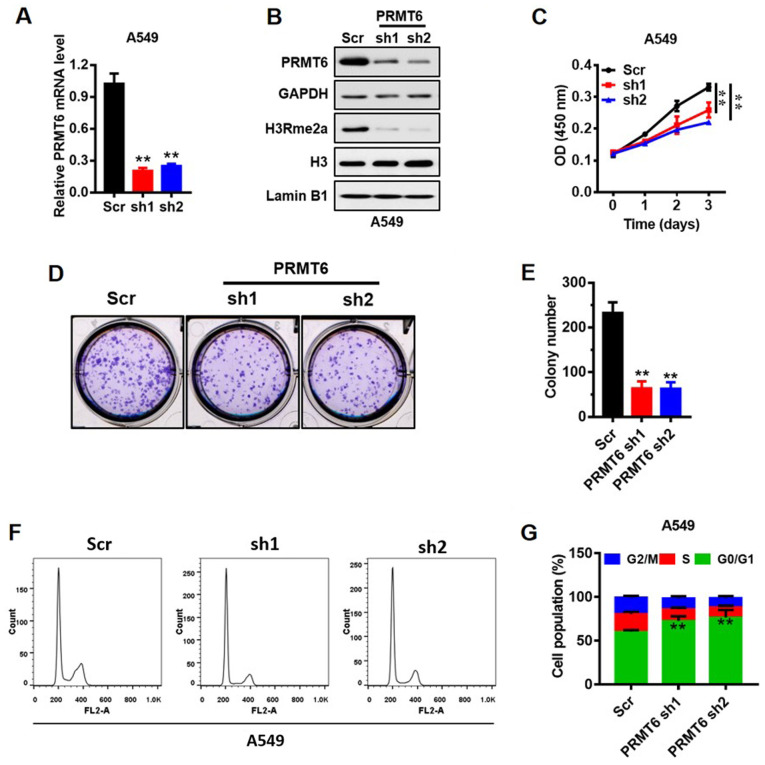
A549 cells are the most frequently used cells to study LUAD (31), thus, the present study used the A549 cell line to represent LUAD. The expression of PRMT6 in mRNA (Fig. 3A) and protein (Fig. 3B) levels was reduced in A549 cell lines using shRNAs (sh1 and sh2) mediated by lentivirus. PRMT6 was previously demonstrated to mediate the H3R2me2a modification (9). Herein, the expression levels of global H3R2me2a were markedly downregulated in PRMT6 sh1/2-transfected cells compared with Scr-transfected cells (Fig. 3B). Subsequently, the effects of PRMT6 on the proliferation of A549 cells in vitro were analyzed using a CCK-8 assay. Notably, the stable knockdown of PRMT6 significantly suppressed the proliferation of A549 cells compared with the Scr cells (Fig. 3C). In addition, the number of cell colonies formed were significantly decreased in the knockdown cells compared with in the Scr-transfected cells (Fig. 3D and E).
Figure 3.
Knockdown of PRMT6 expression levels suppresses LUAD cell growth in vitro through G1/S arrest. (A) mRNA expression levels of PRMT6 in A549 cells were analyzed using reverse-transcription-quantitative PCR following the knockdown of PRMT6 expression. **P <0.01 vs Scr group. (B) Protein expression levels of PRMT6, H3R2me2a and Histone H3 in PRMT6 knockdown cells were analyzed using western blotting. (C) Cell Counting Kit-8 assay was used to determine the effects of PRMT6 knockdown on the proliferative rate of A549 cells in vitro. **P<0.01. (D) Colony formation assay was used to evaluate the effect of PRMT6 knockdown on the proliferation of A549 cells. (E) Number of colonies formed from part (D) were counted using ImageJ software. (F) Cell cycling patterns of Scr and PRMT6 knockdown cells were determined by flow cytometry. (G) Quantitative analysis of the cell cycle distribution of LUAD cells transfected with PRMT6 sh1 or sh2, or Scr from part (F). **P <0.01 vs. Scr group. PRMT6, protein arginine methyltransferase 6; LUAD, lung adenocarcinoma; sh; short hairpin RNA; Scr, scramble; H3R2me2a, Histone H3 asymmetric demethylation at arginine 2; OD, optical density.
To further investigate the molecular mechanism underlying the action of PRMT6 in the proliferation of LUAD cells, the cell cycling patterns of Scr- and PRMT6 sh1/2-transfected cells were determined using flow cytometry (Fig. 3F and G). The number of PRMT6 knockdown cells in the G0/1 phase was significantly increased compared with the Scr-transfected cells. By contrast, the number of PRMT6 knockdown cells in the S and G2/M phases was significantly decreased compared with the Scr-transfected cells (Fig. 3F and G). These results suggested that the downregulation of PRMT6 may induce G1/S phase arrest in A549 cells, which may subsequently inhibit PRMT6-mediated proliferation.
p18 is a direct target of PRMT6 and interferes with G1/S phase arrest in LUAD
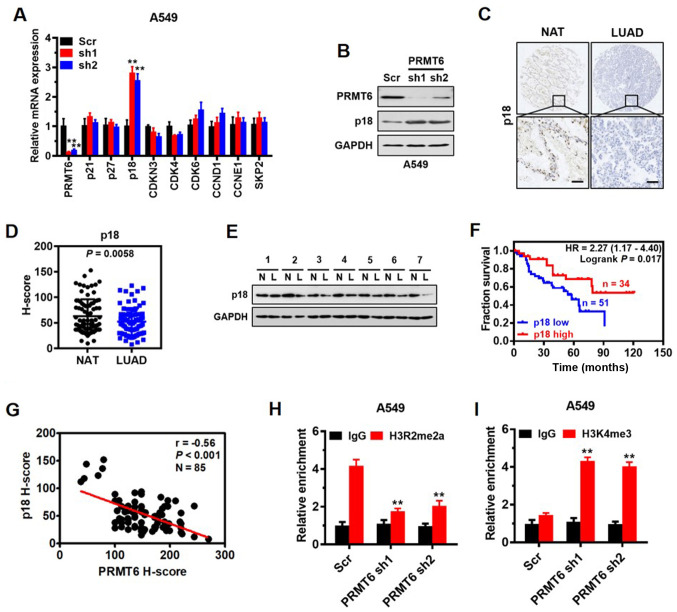
To investigate the mechanism underlying cell cycle arrest induced by the knockdown of PRMT6 in LUAD cells, the mRNA expression levels of important regulatory genes involved in the G1/S transition or switch, including p21, p27, p18, CDK inhibitor 3 (CDKN3), CDK4, CDK6, cyclin D1 (CCND1), cyclin E1 (CCNE1) and S-phase kinase-associated protein 2 (SKP2), were analyzed. RT-qPCR results demonstrated that the expression levels of p18 were significantly upregulated in PRMT6 knockdown cells compared with in Scr-transfected cells, whereas no significant differences were observed in the remaining genes between the groups (Fig. 4A). The effect of PRMT6 on p18 protein expression levels was also confirmed by western blotting, where a similar trend was observed (Fig. 4B). Altogether, these findings indicated that PRMT6 may negatively regulate p18 gene expression levels.
Figure 4.
p18 is a direct target of PRMT6 and interferes with G1/S phase arrest in LUAD. (A) Reverse transcription-quantitative PCR analysis was used to determine the expression levels of important regulatory genes involved in the G1/S phase in A549 cells infected with Scr, or PRMT6 sh1or sh2. **P <0.01 vs. Scr group. (B) Western blotting was used to analyze p18 and PRMT6 expression levels in A549 cells following PRMT6 knockdown. (C) Representative immunohistochemistry images of p18 expression levels in LUAD tissues and matched NAT from 85 patients with LUAD. Scale bar, 50 µm. (D) Quantitative analysis of immunohistochemistry scores of p18 protein expression levels in LUAD tissues and matched NAT from part (C). (E) Western blotting was used to determine the expression levels of p18 in LUAD tissues and matched NAT (n=7). GAPDH served as the normalization control. (F) Kaplan-Meier survival curves were used to determine the overall survival according to low (n=51) and high (n=34) expression levels of PRMT6 using immunohistochemistry scores from 85 patients with LUAD. (G) Correlation between PRMT6 and p18 expression levels was evaluated using Pearson's correlation analysis. Chromatin immunoprecipitation assays were used to determine the effects of PRMT6 knockdown on (H) H3R2me2a and (I) H3K4me3 enrichment in the p18 promoter of A549 cells. Normalized inputs of A549 chromatin DNA were pulled down by antibodies against H3R2me2a, H3K4me3 or negative IgG. **P <0.01 vs. IgG group. PRMT6, protein arginine methyltransferase 6; LUAD/L, lung adenocarcinoma; NAT/N, matched normal adjacent tissues; sh; short hairpin RNA; Scr, scramble; IgG, immunoglobulin G; CCND1, cyclin D1; CCNE1, cyclin E1; SKP2, S-phase kinase-associated protein 2; CDKN3, CDK inhibitor 3; H3R2me2a, Histone H3 asymmetric demethylation at arginine 2; H3K4me3, histone H3 lysine 4 trimethylation.
p18 INK4c (commonly referred to as p18) is a member of the INK4 family of CDK inhibitors, which interacts with CDK4/6 and suppresses its activation, functions as a cell growth regulator of G1/S cell cycle progression and serves as a tumor suppressor (32,33). However, to the best of our knowledge, the expression pattern of p18 in LUAD and its association with patient prognosis remains to be determined. Thus, the present study investigated the expression levels of p18 in LUAD tissue arrays using immunohistochemistry. The results demonstrated that p18 was mainly localized in the nucleus of the glandular epithelium of LUAD tissues (Fig. 4C) and the protein expression levels of p18 were significantly downregulated in the LUAD tissues compared with the NAT (Fig. 4D). Western blotting also confirmed that the expression levels of p18 in tumor tissues from seven patients with LUAD were downregulated compared with the NAT (Fig. 4E). Kaplan-Meier survival analysis demonstrated that lower expression levels of p18 protein were associated with a significantly poorer prognosis in patients with LUAD (Fig. 4F). In addition, a significantly negative correlation was established between PRMT6 and p18 expression levels in vivo by Pearson's correlation analysis (r=−0.56; Fig. 4G).
PRMT6 functions as a transcriptional repressor by generating H3R2me2a (34). Thus, to determine whether PRMT6 directly regulated p18, the enrichment of H3R2me2a on the p18 promoter was analyzed using a ChIP assay. A prominent enrichment of H3R2me2a was noted in the gene promoter of p18 in Scr-transfected cells, which was significantly decreased when PRMT6 was knocked down in A549 cells (Fig. 4H). The results of ChIP assay are consistent with the inhibitory effect of PRMT6 on p18 gene expression. Also, a significant increase was observed in the enrichment of H3K4me3 in the promoter of p18 when PRMT6 was knocked down in A549 cells (Fig. 4I), indicating a potential crosstalk between H3R2me2a and H3K4me3 to enhance p18 gene expression. These findings indicated that p18 may be a downstream target of PRMT6 and interfere with G1/S phase arrest in LUAD cells.
PRMT6 knockdown suppresses LUAD cell growth by activating p18 expression in vitro
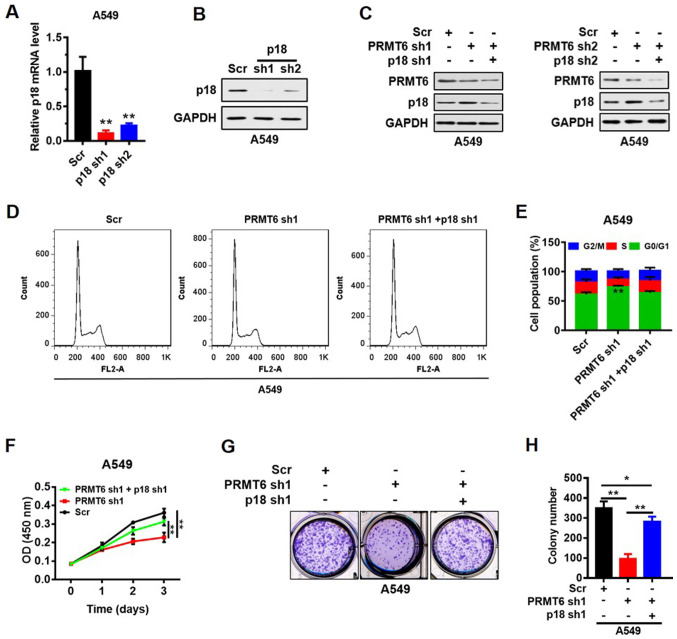
p18 was knocked down by shRNAs (sh1 and sh2) in the A549 cell line and the transfection efficiency was analyzed using RT-qPCR and western blotting. The results indicated that the expression levels of p18 mRNA (Fig. 5A) and protein (Fig. 5B) were downregulated in p18 shRNA-transfected cells compared with the Scr shRNA-transfected cells. To verify the suppression of LUAD cell proliferation mediated by p18 re-activation, stable cells with p18 and PRMT6 double knockdown were constructed. Western blot analyses indicated that p18 protein expression levels were not recovered by the knockdown of p18 and PRMT6 simultaneously (Fig. 5C). Furthermore, the G0/1 phase arrest induced by PRMT6 knockdown was abrogated by further knockdown of p18 (Fig. 5D and E). In addition, the PRMT6 knockdown-induced suppression of cell proliferation (Fig. 5F) and colony formation (Fig. 5G and H) was significantly reversed by p18 knockdown. These results indicated that PRMT6 knockdown may suppress LUAD cell growth by activating p18 expression in vitro.
Figure 5.
Knockdown of PRMT6 suppresses LUAD cell growth by activating p18 expression levels in vitro. (A) Transfection efficiency of PRMT6 knockdown was analyzed at the mRNA expression level using reverse transcription-quantitative PCR. **P<0.01 vs. Scr group. (B) Transfection efficiency of PRMT6 knockdown was detected at the protein level using western blotting. (C) Western blotting analysis of p18 protein expression levels following PRMT6 knockdown or the double knockdown of PRMT6 and p18 in A549 cells. (D) Flow cytometric analysis was used to determine the cell cycle distribution of PRMT6 knockdown or the double knockdown of PRMT6 and p18 in LUAD cells. (E) Quantification of the cell cycle distribution of PRMT6 knockdown or double knockdown of PRMT6 and p18 in LUAD cells from part (D). (F) Cell Counting Kit-8 assay was used to determine the effects on proliferation of PRMT6 knockdown or double knockdown of PRMT6 and p18 in A549 cells. (G) Colony formation assay was used to evaluate the effect on proliferation of PRMT6 knockdown or double knockdown of PRMT6 and p18 in A549 cells. (H) Number of colonies formed in part (G) was counted using Image J software. *P<0.05, **P< 0.01. PRMT6, protein arginine methyltransferase 6; LUAD, lung adenocarcinoma; sh; short hairpin RNA; Scr, scramble; OD, optical density.
PRMT6 knockdown suppresses LUAD development by activating p18 expression in vivo
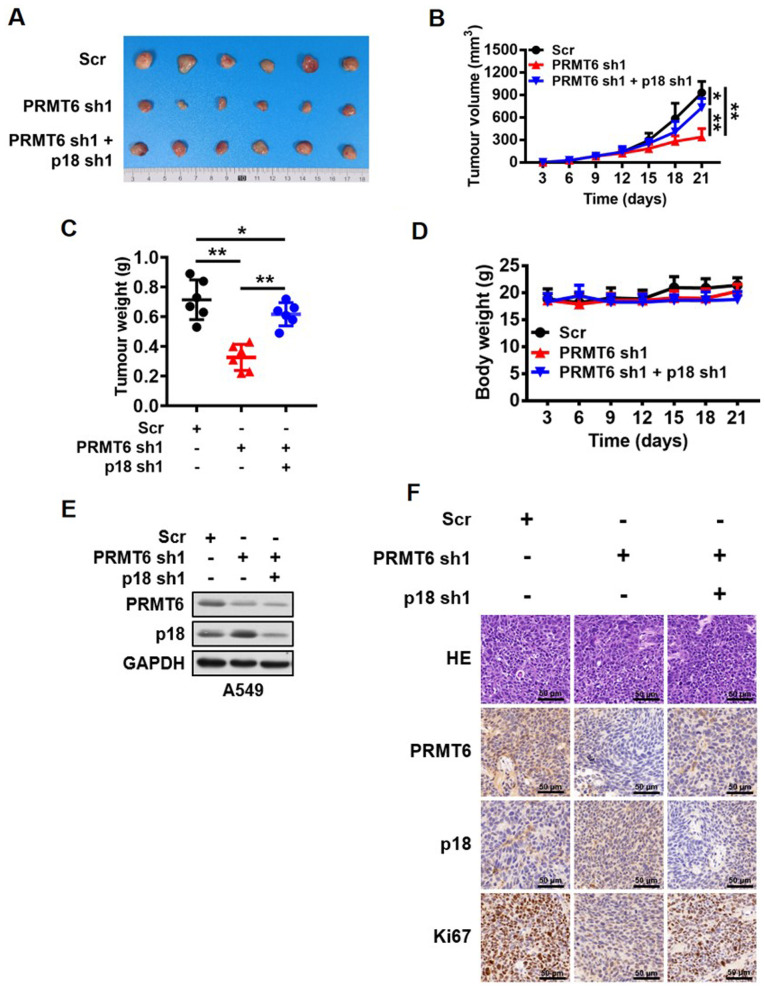
So far, the present study demonstrated that PRMT6 served an important role in the regulation of LUAD cell growth in vitro by regulating the expression levels of p18. However, whether this regulatory mechanism was reciprocated in vivo remained to be clarified. Stable cell lines transfected with Scr, PRMT6 sh1 or PRMT6 sh1 and p18 sh1 were used to create subcutaneous xenografts in BALB/c nude mice. The present study observed that the knockdown of PRMT6 expression levels significantly inhibited tumor growth compared with the Scr group, whereas the combined downregulation with p18 significantly reversed the inhibition of tumor growth mediated by PRMT6 knockdown in vivo (Fig. 6A-C). However, the body weight of the nude mice was not altered between the groups (Fig. 6D). In addition, the present study further analyzed the changes in the PRMT6 and p18 protein expression levels in the xenograft tumor tissues. Western blotting data revealed that the protein expression levels of p18 were markedly upregulated in the PRMT6 knockdown group compared with the Scr group; however, this regulation was abrogated by further knockdown of p18 (Fig. 6E). Similar results were obtained through using immunohistochemistry analysis to determine the levels of Ki67, PRMT6 and p18 in xenograft tumor tissues (Fig. 6F). These results suggested that the knockdown of PRMT6 may suppress LUAD development by activating p18 expression in vivo.
Figure 6.
Knockdown of PRMT6 suppresses lung adenocarcinoma development by activating p18 expression in vivo. (A) Images of tumors excised from BALB/c nude mice in the Scr, PRMT6 knockdown, and PRMT6 and p18 knockdown groups. (B) Tumor volume of xenograft tumors from mice in the Scr, PRMT6 knockdown, and PRMT6 and p18 knockdown groups were measured every 3 days. *P<0.05, **P<0.01. (C) Weight of xenograft tumors excised from mice in the Scr, PRMT6 knockdown, and PRMT6 and p18 knockdown groups were measured at the end of the experiment. *P<0.05, **P<0.01. (D) Body weight of A549 tumor-bearing mice were measured every 3 days. (E) Western blotting analysis of PRMT6 and p18 protein expression levels in xenograft tumors excised from mice in the Scr, PRMT6 knockdown and, PRMT6 and p18 knockdown groups. (F) Representative pathological images of PRMT6, p18 and Ki67 expression levels in xenograft tumors excised from mice in the Scr, PRMT6 knockdown and, PRMT6 and p18 knockdown groups. Scale bar, 50 µm. PRMT6, protein arginine methyltransferase 6; sh; short hairpin RNA; Scr, scramble; HE, hematoxylin and eosin; NS, non-significant.
Discussion
The current study aimed to investigate the biological effects of PRMT6 and its potential mechanism of action in LUAD. The results of the present study demonstrated that the proliferation of LUAD cells was significantly suppressed by silencing PRMT6 expression both in vitro and in vivo. In addition, PRMT6 knockdown decreased the enrichment of H3R2me2a in the promoter region of the p18 gene, thereby activating the expression of the gene. G1/S phase arrest was also induced, resulting in the inhibition of cell proliferation. These results strongly indicated that PRMT6 may serve a prooncogenic role in the progression of LUAD through the epigenetic suppression of p18 expression. Thus, these findings study may provide a novel potential target for the treatment of LUAD.
PRMTs, which specialize in methylating both histone and non-histone proteins, have been discovered to be involved in numerous biological processes, such as cell growth, metabolism and signal transduction, among others (35,36). However, to the best of our knowledge, the role of PRMT6 in human LUAD remains unknown. Considering that the detection of protein expression levels in tissue microarrays containing clinical samples are more accurate and reliable than the gene expression levels found in RNA-seq databases, and the observation time of the patients in tissue microarrays was longer compared with the patients with LUAD listed in the RNA-seq database, RNA-seq database analysis was not performed in the present study. In fact, to the best of our knowledge, the investigations of the current study were the first to analyze the expression levels of the proteins in fresh clinical tissues samples. In the present research, the expression levels of PRMT6 were discovered to be negatively associated with the clinical staging, lymph node metastasis and clinical prognosis of patients with LUAD, indicating that PRMT6 may serve an oncogenic role in LUAD development. Furthermore, silencing PRMT6 suppressed the cell proliferation of LUAD cells in vivo and in vitro, which was ascribed to G1/S cell cycle arrest. These data suggested that PRMT6 may serve a pivotal role in the G1/S phase transition of LUAD cells. Interestingly, differences were noted in the shRNA transfection efficiency in A549 cells between Figs. 3B and 4B; in Fig. 3B, sh2 appeared more effective, whereas in Fig. 4B, sh1 exhibited nearly 100% efficiency. The differences in the shRNA transfection efficiency may be related to the semi-quantification of protein samples and/or the transfer efficiency of western blotting. Nevertheless, PRMT6 was effectively knocked down in the present study and the differences in the shRNA transfection efficiencies did not affect the experimental conclusion.
It is well known that cancers are considered to be a disease of cell cycle disorder, which is accompanied by the abnormal regulation of cell cycle regulatory proteins (37). The arginine methylation of a protein is a post-translational modification, which has been contributed to the disorder of the cell cycle in melanoma (38). PRMTs have been reported to methylate several regulatory proteins of the cell cycle, such as p21, p53, cyclin D1 and phosphorylated Rb (39). Previous studies have also identified that p21 and p27 were direct target genes of PRMT6, which received H3R2me2a modifications and promoted cell cycle progression through CDK1/2 in U2OS and breast cancer cells (34,40,41). In the present study, it was hypothesized that the downregulation of PRMT6 expression levels may inhibit the proliferation of LUAD cells through p18 activation. Thus, to investigate whether p18 was a direct target gene of PRMT6. ChIP analysis was performed in LUAD cells, which confirmed that H3R2me2a was significantly enriched in the promoter of the p18 gene. These findings indicated that PRMT6 may serve a prooncogenic function in the development of LUAD via the epigenetic suppression of p18 expression.
PRMT6 has been reported to be responsible for H3R2me2a; it has been associated with the inactive promoters of mammalian (42). For example, the H3R2me2a modification has been identified to prevent MLL/SET lysine methyltransferase complexes from binding to H3 (43) and PRMT6 action was discovered to impede the deposition of H3K4me3 (44). In the present study, H3R2me2a was enriched at the p18 gene promoter in the control cells. Correspondingly, the study further confirmed that the enrichment of H3K4me3 on the p18 promoter was significantly increased following the knockdown of PRMT6, indicating crosstalk between H3R2me2a and H3K4me3 and the enhancement of p18 gene repression.
In conclusion, the findings of the current study suggested that PRMT6 may serve as a novel potential diagnostic biomarker for LUAD through acting as an oncogene in the disease, epigenetically suppressing p18 expression. Taken together, these findings may offer novel opportunities for the treatment, diagnosis and management of this major subtype of NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Project of The Second Affiliated Hospital of Nanjing University of Chinese Medicine (grant no. SEZJJP2018039), The General Project of Jiangsu Natural Science Foundation (grant no. BK20161605) and The Science and Technology Bureau of Nanjing City (grant no. 201803060).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JT performed the experiments; QM and RS analyzed the data; and YX designed the experiments, interpreted the data and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by The Ethics Committee of the Second Affiliated Hospital of Nanjing University of Chinese Medicine (approval no. 2019-010-054). Written informed consent was obtained from the patients for the use of lung tissue. The animal experiments were approved by The Institute of Animal Care and Use Committee of Nanjing University of Chinese Medicine.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:3161–594. doi: 10.1016/S0025-6196(11)60735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40:558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 4.de Perrot M, Fadel E, Mussot S, de Palma A, Chapelier A, Dartevelle P. Resection of locally advanced (T4) non-small cell lung cancer with cardiopulmonary bypass. Ann Thorac Surg. 2005;79:1691–1697. doi: 10.1016/j.athoracsur.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 5.Wu H, Zhou J, Mei S, Wu D, Mu Z, Chen B, Xie Y, Ye Y, Liu J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J Cell Mol Med. 2017;21:1228–1236. doi: 10.1111/jcmm.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joshi P, Jeon YJ, Laganà A, Middleton J, Secchiero P, Garofalo M, Croce CM. MicroRNA-148a reduces tumorigenesis and increases TRAIL-induced apoptosis in NSCLC. Proc Natl Acad Sci USA. 2015;112:8650–8655. doi: 10.1073/pnas.1500886112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedoriw A, Rajapurkar SR, O'Brien S, Gerhart SV, Mitchell LH, Adams ND, Rioux N, Lingaraj T, Ribich SA, Pappalardi MB, et al. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell. 2019;36:100–114.e25. doi: 10.1016/j.ccell.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pahlich S, Zakaryan RP, Gehring H. Protein arginine methylation: Cellular functions and methods of analysis. Biochim Biophys Acta. 2006;1764:1890–1903. doi: 10.1016/j.bbapap.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Blanc RS, Richard S. Arginine methylation: The coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 11.Bowitch A, Michaels KL, Yu MC, Ferkey DM. The protein arginine methyltransferase PRMT-5 regulates SER-2 tyramine receptor-mediated behaviors in caenorhabditis elegans. G3 (Bethesda) 2018;8:2389–2398. doi: 10.1534/g3.118.200360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altan B, Yokobori T, Ide M, Mochiki E, Toyomasu Y, Kogure N, Kimura A, Hara K, Bai T, Bao P, et al. Nuclear PRMT1 expression is associated with poor prognosis and chemosensitivity in gastric cancer patients. Gastric Cancer. 2016;19:789–797. doi: 10.1007/s10120-015-0551-7. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimatsu M, Toyokawa G, Hayami S, Unoki M, Tsunoda T, Field HI, Kelly JD, Neal DE, Maehara Y, Ponder BA, et al. Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int J Cancer. 2011;128:562–573. doi: 10.1002/ijc.25366. [DOI] [PubMed] [Google Scholar]
- 14.Almeida-Rios D, Graça I, Vieira FQ, Ramalho-Carvalho J, Pereira-Silva E, Martins AT, Oliveira J, Gonçalves CS, Costa BM, Henrique R, Jerónimo C. Histone methyltransferase PRMT6 plays an oncogenic role of in prostate cancer. Oncotarget. 2016;7:53018–53028. doi: 10.18632/oncotarget.10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenblatt SM, Man N, Hamard PJ, Asai T, Karl D, Martinez C, Bilbao D, Stathias V, Jermakowicz AM, Duffort S, et al. CARM1 is essential for myeloid leukemogenesis but dispensable for normal hematopoiesis. Cancer Cell. 2019;35:156. doi: 10.1016/j.ccell.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Cheung N, Chan LC, Thompson A, Cleary ML, So CW. Protein arginine-methyltransferase-dependent oncogenesis. Nat Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 17.Elakoum R, Gauchotte G, Oussalah A, Wissler MP, Clément-Duchêne C, Vignaud JM, Guéant JL, Namour F. CARM1 and PRMT1 are dysregulated in lung cancer without hierarchical features. Biochimie. 2014;97:210–218. doi: 10.1016/j.biochi.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Li M, An W, Xu L, Lin Y, Su L, Liu X. The arginine methyltransferase PRMT5 and PRMT1 distinctly regulate the degradation of anti-apoptotic protein CFLARL in human lung cancer cells. J Exp Clin Cancer Res. 2019;38:64. doi: 10.1186/s13046-019-1064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakrzewicz D, Didiasova M, Krüger M, Giaimo BD, Borggrefe T, Mieth M, Hocke AC, Zakrzewicz A, Schaefer L, Preissner KT, Wygrecka M. Protein arginine methyltransferase 5 mediates enolase-1 cell surface trafficking in human lung adenocarcinoma cells. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1816–1827. doi: 10.1016/j.bbadis.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Frankel A, Yadav N, Lee J, Branscombe TL, Clarke S, Bedford MT. The novel human protein arginine N-methyltransferase PRMT6 is a nuclear enzyme displaying unique substrate specificity. J Biol Chem. 2002;277:3537–3543. doi: 10.1074/jbc.M108786200. [DOI] [PubMed] [Google Scholar]
- 21.Limm K, Ott C, Wallner S, Mueller DW, Oefner P, Hellerbrand C, Bosserhoff AK. Deregulation of protein methylation in melanoma. Eur J Cancer. 2013;49:1305–1313. doi: 10.1016/j.ejca.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 22.Chan LH, Zhou L, Ng KY, Wong TL, Lee TK, Sharma R, Loong JH, Ching YP, Yuan YF, Xie D, et al. PRMT6 regulates RAS/RAF binding and MEK/ERK-mediated cancer stemness activities in hepatocellular carcinoma through CRAF methylation. Cell Rep. 2018;25:690–701 e698. doi: 10.1016/j.celrep.2018.09.053. [DOI] [PubMed] [Google Scholar]
- 23.Veland N, Hardikar S, Zhong Y, Gayatri S, Dan J, Strahl BD, Rothbart SB, Bedford MT, Chen T. The arginine methyltransferase PRMT6 regulates DNA methylation and contributes to global DNA hypomethylation in cancer. Cell Rep. 2017;21:3390–3397. doi: 10.1016/j.celrep.2017.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sellers AH. The clinical classification of malignant tumours: The TNM system. Can Med Assoc J. 1971;105:836. passim. [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlschläger J, Wittekind C, Theegarten D. New TNM classification of malignant lung tumours. Pathologe. 2010;31:355–360. doi: 10.1007/s00292-010-1303-4. (In German) [DOI] [PubMed] [Google Scholar]
- 26.Feldman AT, Wolfe D. Tissue processing and hematoxylin and eosin staining. Methods Mol Biol. 2014;1180:31–43. doi: 10.1007/978-1-4939-1050-2_3. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark JD, Gebhart GF, Gonder JC, Keeling ME, Kohn DF. Special report: The 1996 Guide for the Care and Use of Laboratory Animals. ILAR J. 1997;38:41–48. doi: 10.1093/ilar.38.1.41. [DOI] [PubMed] [Google Scholar]
- 31.Gong S, Qu X, Yang S, Zhou S, Li P, Zhang Q. RFC3 induces epithelialmesenchymal transition in lung adenocarcinoma cells through the Wnt/β-catenin pathway and possesses prognostic value in lung adenocarcinoma. Int J Mol Med. 2019;44:2276–2288. doi: 10.3892/ijmm.2019.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 33.Guan KL, Jenkins CW, Li Y, Nichols MA, Wu X, O'Keefe CL, Matera AG, Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 34.Stein C, Riedl S, Ruthnick D, Nötzold RR, Bauer UM. The arginine methyltransferase PRMT6 regulates cell proliferation and senescence through transcriptional repression of tumor suppressor genes. Nucleic Acids Res. 2012;40:9522–9533. doi: 10.1093/nar/gks767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auclair Y, Richard S. The role of arginine methylation in the DNA damage response. DNA Repair (Amst) 2013;12:459–465. doi: 10.1016/j.dnarep.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Biggar KK, Li SS. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16:5–17. doi: 10.1038/nrm3915. [DOI] [PubMed] [Google Scholar]
- 37.Malumbres M, Carnero A. Cell cycle deregulation: A common motif in cancer. Prog Cell Cycle Res. 2003;5:5–18. [PubMed] [Google Scholar]
- 38.AbuHammad S, Cullinane C, Martin C, Bacolas Z, Ward T, Chen H, Slater A, Ardley K, Kirby L, Chan KT, et al. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc Natl Acad Sci USA. 2019;116:17990–18000. doi: 10.1073/pnas.1901323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raposo AE, Piller SC. Protein arginine methylation: An emerging regulator of the cell cycle. Cell Div. 2018;13:3. doi: 10.1186/s13008-018-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinschmidt MA, de Graaf P, van Teeffelen HA, Timmers HT. Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin-dependent kinase inhibitors. PLoS One. 2012;7:e41446. doi: 10.1371/journal.pone.0041446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phalke S, Mzoughi S, Bezzi M, Jennifer N, Mok WC, Low DH, Thike AA, Kuznetsov VA, Tan PH, Voorhoeve PM, Guccione E. p53-independent regulation of p21Waf1/Cip1 expression and senescence by PRMT6. Nucleic Acids Res. 2012;40:9534–9542. doi: 10.1093/nar/gks858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guccione E, Bassi C, Casadio F, Martinato F, Cesaroni M, Schuchlautz H, Lüscher B, Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 43.Hyllus D, Stein C, Schnabel K, Schiltz E, Imhof A, Dou Y, Hsieh J, Bauer UM. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.