Abstract
Long non-coding RNA high expression in hepatocellular carcinoma (lncRNA HEIH) acts as an oncogene in multiple tumors, including hepatocellular carcinoma, colorectal cancer, melanoma and non-small cell lung cancer. However, the role of HEIH in breast cancer remains unknown. The present study focused on the clinical significance and biological function of HEIH in breast cancer. Specifically, the expression levels of HEIH in breast cancer tissues and breast cancer cell lines were investigated. The results indicated high expression levels of HEIH in human breast cancer tissues, and its expression was positively associated with malignancy status and poor disease prognosis. High expression levels of HEIH were detected in the breast cancer cell lines, including MCF-7, SK-BR-3, MDA-MB-231 and MDA-MB-468. These data were consistent with those derived from the in vivo study. Therefore, small interfering RNA was used to knockdown HEIH expression in order to explore whether HEIH exhibits an oncogenic function in breast cancer. Following HEIH knockdown, the proliferative and metastatic activity of MDA-MB-231 cells was decreased, whereas the induction of cell apoptosis was increased. These results suggested the oncogenic role of HEIH in breast cancer and the potential application of HEIH as an index of malignancy and poor prognosis in breast cancer.
Keywords: long non-coding RNA, high expression in hepatocellular carcinoma, breast cancer, oncogene
Introduction
Breast cancer accounts for 25% of cancer cases in women and is one of the four most common cancers in women worldwide (1). The majority of individuals affected include patients from developing countries, which represent 53% of the total breast cancer cases (1). Therefore, breast cancer has emerged as the second leading cause of cancer-related deaths among women in developing countries, following lung cancer (2). It has been reported that >80% of the genome is actively transcribed to non-coding RNAs (ncRNAs). This includes RNA molecules that serve no protein-coding function. Based on their size, ncRNAs are classified into long ncRNAs (lncRNAs) and small ncRNAs (3). Aberrant expression of lncRNAs can be observed in almost all tumors and can affect tumor development via an oncogenic or tumor suppressive mechanism of action. Moreover, the expression levels of lncRNAs closely correlate with malignancy status and disease prognosis (4).
High expression in hepatocellular carcinoma (HEIH) is a lncRNA that was initially identified in hepatitis B virus-induced hepatocellular carcinoma (5). High HEIH expression is associated with an increased risk of recurrence and significantly reduced overall postoperative survival in patients with hepatocellular carcinoma. Moreover, HEIH acts as a tumor promoter by accelerating the cell cycle progression of hepatocellular carcinoma cells (5,6). Recent studies have suggested that HEIH is highly expressed in other cancer types, including colorectal cancer (7), melanoma (8) and non-small cell lung cancer (9). However, the expression, pathophysiological roles and clinical significance of HEIH in breast cancer remain unknown.
In the present study, the expression pattern of HEIH was explored in breast cancer and the data revealed the upregulation of HEIH expression in breast cancer tissues compared with that noted in the paired adjacent normal tissues. The association between HEIH expression and clinical variables in patients with breast cancer was further investigated. To determine the functional role of HEIH in the development of breast cancer, the effects of HEIH knockdown on the proliferation, apoptosis and metastasis of breast cancer cells were investigated. The present study provides a more in-depth understanding of HEIH function, which may aid the diagnosis and targeted treatment of breast cancer.
Materials and methods
Patient samples
Cancer tissues and paired adjacent normal tissues (~5 cm from tumor) were obtained from 160 female patients (age, 40–68 years) with breast cancer who underwent surgical operation at the First Hospital of Lanzhou University (Gansu, China) between January 2013 and December 2016. The clinical information of the patients, including age, family history, tumor grade, tumor-node-metastasis (TNM) stage, lymph node status, estrogen receptor (ER) status, progesterone receptor (PR) status and human epidermal growth factor receptor-2 (HER2) status, were collected (Table I). Following surgery, the tissue samples obtained were stored immediately in liquid nitrogen, and then transferred into a freezer at an ultra-low temperature (−80°C) until further processing for reverse transcription-quantitative PCR (RT-qPCR).
Table I.
Association between lncRNA HEIH and clinical characteristics.
| HEIH expression | ||||
|---|---|---|---|---|
| Factors | Number of patients | Low (n=77) | High (n=83) | P-valuea |
| Age, years | 0.188 | |||
| <60 | 80 | 42 | 38 | |
| ≥60 | 80 | 35 | 45 | |
| Family history | 0.472 | |||
| Absent | 147 | 72 | 75 | |
| Present | 13 | 5 | 8 | |
| Tumor grade | 0.776 | |||
| I–II | 125 | 62 | 63 | |
| III | 35 | 15 | 20 | |
| TNM stage | 0.006 | |||
| I | 39 | 28 | 11 | |
| II | 55 | 32 | 23 | |
| III | 45 | 13 | 32 | |
| IV | 21 | 4 | 17 | |
| Lymph node metastasis | 0.017 | |||
| Negative | 78 | 27 | 51 | |
| Positive | 82 | 50 | 32 | |
| ER status | 0.696 | |||
| Negative | 87 | 43 | 44 | |
| Positive | 73 | 34 | 39 | |
| PR status | 0.574 | |||
| Negative | 20 | 8 | 12 | |
| Positive | 140 | 69 | 71 | |
| HER2 status | 0.726 | |||
| Negative | 47 | 22 | 25 | |
| Positive | 113 | 55 | 58 | |
Pearson's χ2 test. TNM, tumor-node-metastasis; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor-2; lncRNA, long non-coding RNA; HEIH, high expression in hepatocellular carcinoma
RT-qPCR
Total RNA was extracted from the tissues and cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA (0.2-0.5 µg) was reverse transcribed into cDNA using the PrimeScript RT Reagent kit (cat. no. RR037B; Takara Biotechnology Co., Ltd.) at 37°C for 15 min. qPCR was subsequently performed using SYBR Premix Ex Taq™ (cat. no. RR420B; Takara Biotechnology Co., Ltd.) on an ABI 7300 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for the qPCR: Initial incubation at 95°C for 15 sec; followed by 40 cycles of denaturation at 95°C for 5 sec and annellation at 60°C for 31 sec. The primer sequences used were as follows: HEIH forward 5′-CCTCTTGTGCCCCTTTCT-3′ and reverse, 5′-AGGTCTCATGGCTTCTCG-3′; and β-actin forward, 5′-GGGAAATCGTGCGTGACATTAAG-3′ and reverse, 5′-TGTGTTGGCGTACAGGTCTTTG-3′. Expression levels were quantified using the 2−ΔΔCq method (10) and normalized to the β-actin loading control.
Cell culture
MCF-10A (normal human mammary epithelial cell line) and four breast cancer cell lines, including MCF-7, SK-BR-3, MDA-MB-231 and MDA-MB-468, were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. These cell lines were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in the presence of 5% CO2.
HEIH small interfering (si)RNA transfection
Briefly, 50 nM HEIH siRNA or negative control (NC) siRNA (Guangzhou RiboBio Co., Ltd.) sequences were synthesized and transfected into 1×105 MDA-MB-231 cells/well seeded into 12-well plates. Transfection was achieved using Lipofectamine™ 2000 (Guangzhou RiboBio Co., Ltd.). The transfection was performed for 24 h at 37°C, and then used for subsequent experiments. The transfection efficiency was determined by measuring the HEIH expression via RT-qPCR. The siRNA segment sequences were obtained by a previous study (9).
Cell viability assay
Cell viability was confirmed by the CellTiter 96® AQueous One Solution Cell Proliferation assay kit (MTS; Promega Corporation). The transfected cells were seeded in 96-well plates at a concentration of 5×104 cells in each well. Following incubation for 24 h, 20 µl reagent was added into each well and the cells were subsequently incubated at 37°C for 2 h. The cell viability was determined by the OD value at 490 nm. The detailed protocol was performed following the manufacturer's instructions as described previously (11).
5-Ethynyl-20-deoxyuridine (EdU) incorporation assay
In addition to the cell viability assay, the EdU incorporation assay kit (Guangzhou RiboBio Co., Ltd.) was used to assess the proliferative activity of the cells. Briefly, the transfected cells were seeded in 96-well plates at a density of 1×104 cells/well, following incubation for 24 h at 37°C. A total of 100 µl medium containing EdU solution (50 µM) was added into each well. The cells were incubated for 3 h and paraformaldehyde (4%) with Triton X-100 (0.5%) was added to the cells. Moreover, the cell nuclei were stained with DAPI for 15 min at room temperature. The ratio of EdU positive cells (green cells) to total DAPI positive cells (blue cells) corresponded to proliferative activity.
Western blot analysis
Total protein was extracted using NP-40 lysis buffer (Beyotime Institute of Biotechnology). Total protein was quantified using a BCA protein quantification kit (cat. no. P0012S; Beyotime Institute of Biotechnology) and 20 µg protein/lane was resolved via 10% SDS-PAGE and electroblotted onto nitrocellulose membranes. Membranes were blocked with 5% non-fat milk solution at room temperature for 1.5 h, followed by overnight incubation with primary antibodies at 4°C. The blots were subsequently incubated with horseradish peroxidase-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (1:5,000; Abcam; cat. nos. ab205718 and ab205719) at room temperature for 1 h. The following primary antibodies (all purchased from Abcam) were used: Bcl-2 (1:2,000; cat. no. ab692), Bax (1:2,000; cat. no. ab32503), E-cadherin (1:1,000; cat. no. ab40772), N-cadherin (1:1,000; cat. no. ab18203), Vimentin (1:1,000; cat. no. ab92547), matrix metallopeptidase 9 (MMP-9; 1:1,000; cat. no. ab137867) and β-actin (1:2,000; cat. no. ab8227).
Caspase-3 activity assay
A Caspase-3 Activity kit (Beyotime Institute of Biotechnology; cat. no. C1168M) was used to determine caspase-3 activity. Total cellular protein was obtained using a lysis buffer. A total of 40 µg protein was diluted to a 50-µl final solution that was subsequently mixed with 75 µl caspase-3 substrate and incubated for 3 h. The hydrolysis of Ac-DEVD-pNA resulted in caspase-3 released free pNA (yellow formazan product) that was detected at 405 nm. Caspase-3 activity was expressed as the fold of enzyme activity compared with that of the synchronized cells.
Flow cytometry assay
The apoptotic assays were performed using an Annexin V-FITC/PI Apoptosis Detection kit (Miltenyi Biotec GmbH), according to the manufacturer's protocols. The apoptotic rate was calculated using both early and late apoptotic cells. Briefly, 1×106 cells were washed using 500 µl binding buffer, centrifuged at 560 × g, and stained with 10 µl Annexin V-FITC solution at room temperature for 30 min. A total of 5 µl PI solution was added to each sample and incubated at room temperature for 15 min. The apoptotic rate was evaluated using a BD FACSCalibur flow cytometer (BD Biosciences) and ModFit LT software version 2.0 (Verity Software House, Inc.). At least 10,000 events were analyzed for each sample. The analysis was repeated in three cell samples.
Transwell assay
MDA-MB-231 cells (5×104) were seeded in triplicate onto a 24-well plate with Transwell Boyden chambers (BD Biosciences) coated with Matrigel (37°C for 30 min) or without Matrigel for the invasion and migration assay, respectively. The upper chamber contained serum-free DMEM, and the lower chamber contained DMEM supplemented with 10% FBS as a chemoattractant. Cells were incubated for 36 h at 37°C, fixed with 4% paraformaldehyde at room temperature for 15 min and then stained with 0.5% crystal violet for 5 min at room temperature. Stained cells were visualized in five randomly selected fields using a light IX71 inverted research microscope (magnification, ×200; Olympus Corporation).
Scratch assay
MDA-MB-231 cells (5×104) were grown to 90–100% confluence in 6-well culture plates. A scratch on the cell layer was created using a 200 µl pipette tip. Cells were cultured in serum-free DMEM, and photographed immediately (0 h) after scratching and 24 h after scratching. The scratch closure was monitored using a light IX71 inverted research microscope (magnification, ×200; Olympus Corporation). An ocular ruler was used to verify the scratch sizes.
Statistical analysis
A paired Wilcoxon signed-rank test was employed to evaluate significant differences of HEIH expression in breast cancer tissues compared with adjacent normal tissues. Associations between HEIH expression and clinical features were performed by a Pearson's χ2 test. The optimal cut-off value of HEIH expression in tumor/normal tissues was determined by a receiver operating characteristic (ROC) curve analysis. The survival curve was evaluated by the Kaplan-Meier method and the log-rank test was used to compare differences between groups. The prognostic value of HEIH expression was assessed based on the high-HEIH vs. low-HEIH expression categorical definition. Hazard ratios (HRs) and 95% confidence intervals (95% CIs) were calculated with the Cox proportional hazard regression model and the data were adjusted for the clinicopathological prognostic factors. All the statistical tests were two-sided and considered significant when the P-value was <0.05. The data in Table I are presented as number (percentage). The cell experiment data were analyzed by one-way ANOVA with a Dunnett's post hoc test for multiple comparisons (≥3 groups). An unpaired Student's t-test was used to analyze the statistical differences between two groups, except for Fig. 1A, which was analyzed using a paired Student's t-test. Data are presented as the mean ± SEM. P<0.05 was considered to indicate a statistically significant difference.
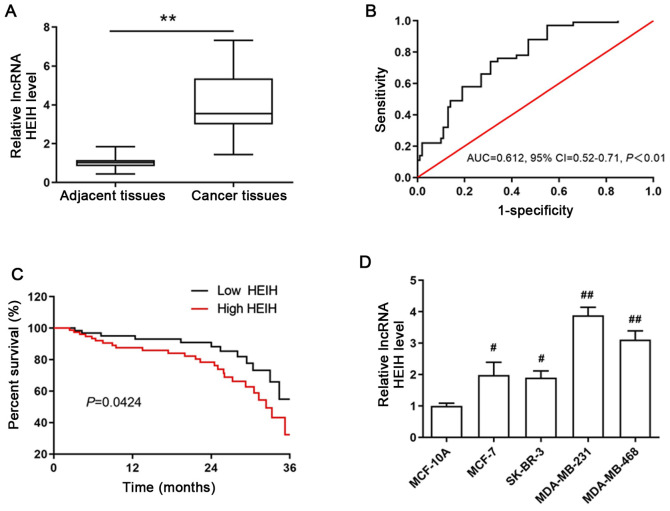
Figure 1.
Expression of HEIH in breast cancer and its association with clinical variables. (A) HEIH expression in breast cancer tissues and paired adjacent normal tissues. (B) The receiver operating characteristic curve for the 3-year overall survival was used to determine the optimal cut-off value of HEIH expression. (C) Kaplan-Meier 3-year overall survival curves were plotted in order to assess survival according to HEIH expression. (D) HEIH expression levels in the four breast cancer cell lines (MCF-7, SK-BR-3, MDA-MB-231 and MDA-MB-468) and in the normal human mammary epithelial cell line MCF-10A. The data are presented as the mean ± SEM. (A and B) n=160 patients and (C) n=3 experimental repeats. **P<0.01 vs. adjacent tissues; #P<0.05, ##P<0.01 vs. MCF-10A cells. HEIH, high expression in hepatocellular carcinoma; AUC, area under curve; CI, confidence interval; lncRNA, long non-coding RNA.
Results
Upregulation of HEIH expression is positively associated with malignancy status and poor disease prognosis
To determine the aberrant expression of HEIH in breast cancer, the expression levels of HEIH in cancer tissues and paired adjacent normal tissues from 160 patients with breast cancer were measured by RT-qPCR. HEIH was upregulated in breast cancer tissues (Fig. 1A). The area under the ROC was 0.612 (95% CI, 0.52-0.71; P<0.01; Fig. 1B). In order to explore the association of HEIH expression with the clinical characteristics of patients with breast cancer, they were divided into high or low expression groups according to the optimal cut-off value (0.43) of fold-expression change of HEIH. The results of the Pearson's χ2 analysis between HEIH expression and patient clinical characteristics indicated that high HEIH expression levels were associated with TNM stage and lymph node metastasis, whereas this type of association was not noted for patient age, family history, tumor grade, ER, PR and HER2 status (Table I).
In addition, the 3-year overall survival of patients was estimated using Kaplan-Meier analysis by the log-rank test. The data indicated that HEIH expression was negatively associated with 3-year overall survival and that high HEIH expression levels were positively associated with a poor prognosis of patients with breast cancer (log rank=4.118, P=0.0424; Fig. 1C). This finding was also confirmed by univariate analysis for the significant factors described in Table II. HEIH expression was an independent prognostic factor of the 3-year overall survival in patients with breast cancer (HR, 4.97, 95% CI, 3.19-9.66; P<0.001). Taken together, these results suggested that HEIH expression is positively associated with malignancy status and poor prognosis.
Table II.
Univariate and multivariate analysis of clinical characteristics of overall survival using Cox regression model.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Characteristics | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age | 1.51 (0.88-2.60) | 0.212 | 1.34 (0.70-2.25) | 0.375 |
| Family history | 1.34 (0.51-3.44) | 0.513 | ||
| Tumor grade | 1.89 (1.03-3.44) | 0.032 | 1.70 (0.93-3.11) | 0.082 |
| TNM stage | 2.42 (1.41-4.20) | 0.004 | 2.31 (1.34-4.02) | 0.006 |
| Lymph node metastasis | 1.98 (1.11-3.62) | 0.017 | 1.23 (0.37-3.86) | 0.672 |
| ER status | 0.74 (0.36-1.19) | 0.323 | ||
| PR status | 0.77 (0.38-1.21) | 0.513 | ||
| HER2 status | 0.81 (0.42-1.39) | 0.472 | ||
| HEIH expression | 5.19 (3.21-9.84) | <0.001 | 4.97 (3.19-9.66) | <0.001 |
HR, hazard ratio; CI, confidence interval; TNM, tumor-node-metastasis; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor-2; HEIH, high expression in hepatocellular carcinoma.
Proliferation of MDA-MB-231 cells can be inhibited by HEIH knockdown
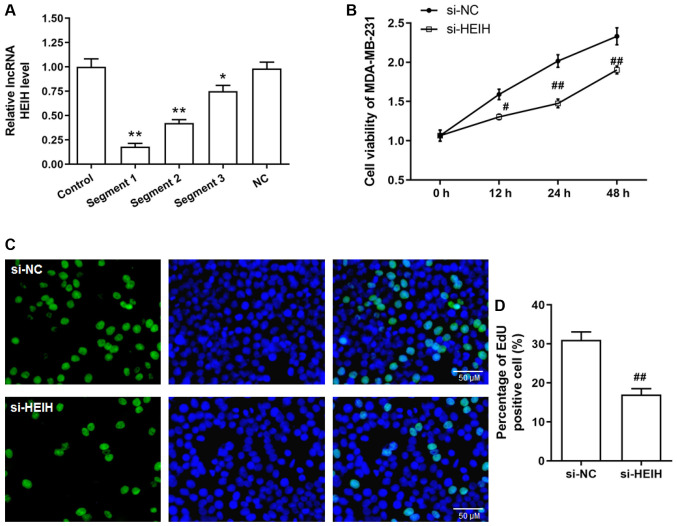
The aforementioned results allowed the determination of HEIH expression in breast cancer cells. The expression levels of HEIH were upregulated in four breast cancer cell lines (MCF-7, SK-BR-3, MDA-MB-231 and MDA-MB-468), which was most notable in MDA-MB-231 cells (Fig. 1D). Therefore, a functional study was performed in MDA-MB-231 cells. Initially, siRNA sequences were transfected into MDA-MB-231 cells in order to achieve HEIH knockdown. Among the three siRNA sequences tested, the first sequence indicated the maximum interference efficiency and was used for the following studies (Fig. 2A). Following transfection of the siRNAs into cells, cell viability and EdU-positive cell percentage were decreased in MDA-MB-231 cells, suggesting that HEIH knockdown inhibited the proliferation of breast cancer cells.
Figure 2.
Effect of HEIH knockdown on the proliferation of MDA-MB-231 cells. (A) The expression levels of HEIH in MDA-MB-231 cells following transfection of the cells with HEIH siRNA. (B) Cell viability was determined by the MTS assay. (C and D) Proliferative ability was evaluated by an EdU incorporation assay in MDA-MB-231 cells following transfection of the cells with HEIH siRNA. Data are presented as the mean ± SEM (n=3). *P<0.05, **P<0.01 vs. control; #P<0.05, ##P<0.01 vs. si-NC. HEIH, high expression in hepatocellular carcinoma; siRNA, small interfering RNA; NC, negative control; lncRNA, long non-coding RNA.
Apoptosis of MDA-MB-231 cells is promoted by HEIH knockdown
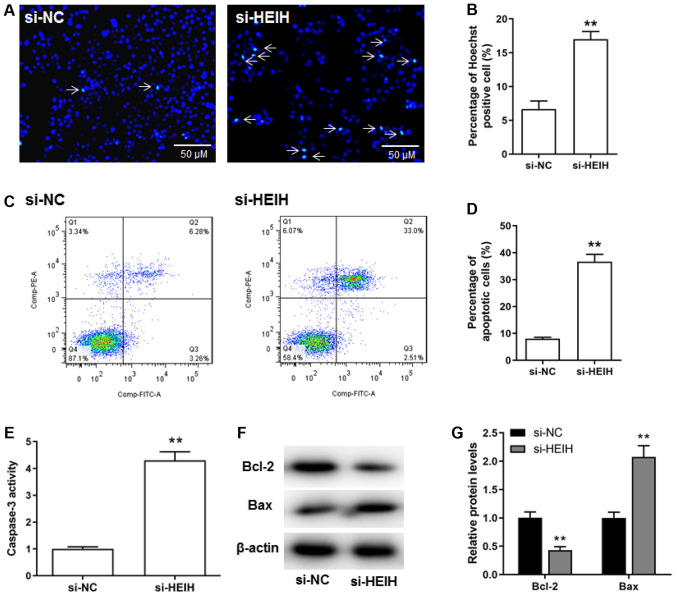
The effects of HEIH knockdown on breast cancer cell apoptosis were investigated. The percentage of Hoechst positive cells was increased following HEIH knockdown (Fig. 3A and B). The results of flow cytometry confirmed that HEIH knockdown increased the number of apoptotic cells (Fig. 3C and D). Moreover, upregulation of Bax protein expression and caspase-3 activity, as well as downregulation of Bcl-2 protein expression could be observed following HEIH knockdown in MDA-MB-231 cells (Fig. 3E-G). Taken collectively, the data suggested that HEIH knockdown promoted the apoptosis of breast cancer cells.
Figure 3.
Effect of HEIH knockdown on the induction of apoptosis of MDA-MB-231 cells. (A and B) Hoechst staining was used to detect apoptotic cells. (C and D) Flow cytometry was performed to determine the percentage of apoptotic cells. (E) Caspase-3 activity was assessed by a Caspase-3 Activity kit. (F and G) The expression levels of apoptotic proteins (Bcl-2 and Bax) were measured by western blotting following transfection with HEIH siRNA in MDA-MB-231 cells. Data are presented as the mean ± SEM (n=3). **P<0.01 vs. si-NC. HEIH, high expression in hepatocellular carcinoma; siRNA, small interfering RNA; NC, negative control.
MDA-MB-231 cell metastasis can be inhibited by HEIH knockdown
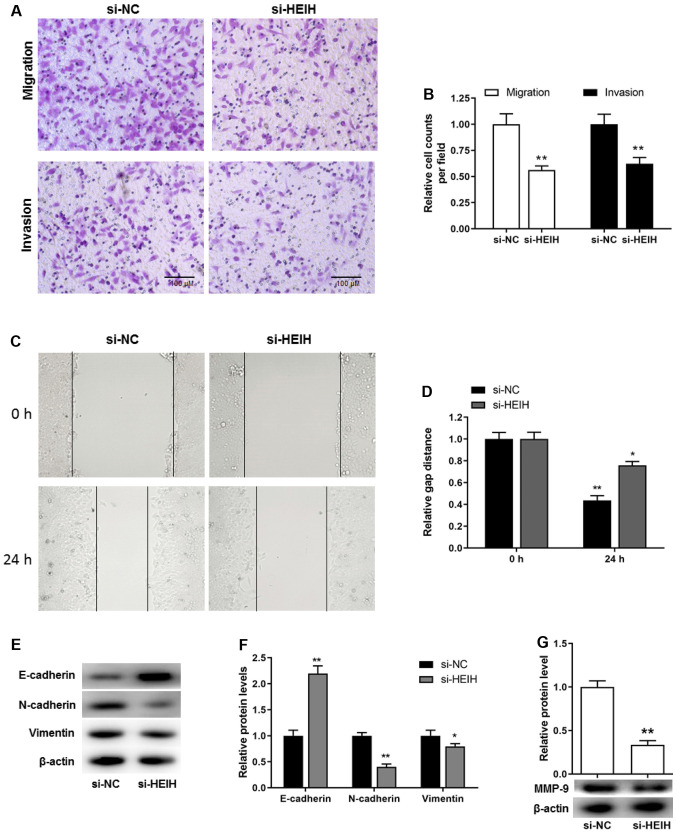
In addition to the proliferative potential and apoptosis resistance, cell metastasis is considered a key pathological feature during cancer development. Transwell assays, including migration and invasion assays, were used to investigate cell motility. The cell number, which corresponded with the cells passing through the membrane, was decreased following HEIH knockdown in MDA-MB-231 cells. This was determined by migration and invasion assays (Fig. 4A and B). In order to exclude that these effects were attributed to decreased proliferation and/or increased apoptosis, a scratch assay was performed to further investigate cell motility. The results demonstrated that the distance covering the wound in si-HEIH cells was higher than that noted in si-NC cells, suggesting that HEIH knockdown decreased cell motility (Fig. 4C and D). Furthermore, considering the key role of the epithelial-mesenchymal transition (EMT) in the development of metastasis, the expression levels of EMT-related proteins were determined following HEIH knockdown. HEIH knockdown upregulated E-cadherin protein expression, whereas it downregulated the expression of N-cadherin, Vimentin and MMP-9 proteins (Fig. 4E-G). These results indicated that HEIH knockdown inhibited metastasis of breast cancer cells.
Figure 4.
Effect of HEIH knockdown on MDA-MB-231 cell metastasis. (A and B) A Transwell assay was used to investigate cell migration and invasion. (C and D) Cell migration was further detected by the scratch test (magnification, ×200). (E-G) E-cadherin, N-cadherin, Vimentin and MMP-9 protein expression levels were measured by western blotting following transfection of MDA-MB-231 cells with HEIH siRNA. Data are presented as the mean ± SEM (n=3). *P<0.05, **P<0.01 vs. si-NC. HEIH, high expression in hepatocellular carcinoma; siRNA, small interfering RNA; NC, negative control; MMP-9, matrix metallopeptidase 9.
Discussion
As an intermediate between DNA and protein, RNA transmits genetic information that is translated into various biological processes. However, these intermediate RNAs (mRNAs) compose <3% of human DNA, as demonstrated from the development of human genome sequencing and the discovery of ncRNAs. Emerging evidence suggests that ncRNAs are key RNA molecules that play prominent roles in cell biology (12). According to their size, ncRNAs can be classified into small ncRNAs (<200 nt in length) and lncRNAs (≥200 nt in length) (4). The microRNAs are considered one of the well-known small ncRNAs that have been reported to regulate carcinogenesis (13–15). The investigation of the expression levels of ncRNAs can be used for diagnostic and prognostic purposes in cancer. In recent years, the investigation of the role of lncRNAs in cancer has become a research hot spot due to their functional relevance in physiological and pathological processes involved in this disease (4). An increasing number of lncRNAs are aberrantly expressed in breast cancer (16–18). Previously, Yang et al (19) sequenced >1,300 lncRNAs that were aberrantly expressed in breast cancer. Notably, the lncRNA with the highest upregulation was identified as AFAP1-AS1 (19). Our previous study further investigated the functional role of AFAP1-AS1 in breast cancer and found its diagnostic and prognostic value for this disease (20). Similarly, in the present study it was reported that the expression levels of HEIH were upregulated in breast cancer tissues. Moreover, high HEIH expression levels were shown to be positively associated with the malignancy status, including TNM stage and lymph node metastasis, which was also indicative of poor disease prognosis. However, assessing patient survival from the first three years may not be enough to support the aforementioned conclusion. The follow-up of patients with breast cancer from the present study is still on-going, and future studies would assess overall survival over a longer time period.
It has been previously reported that lncRNA HEIH is upregulated in hepatocellular carcinoma, indicating poor disease outcome of patients with hepatocellular carcinoma (5,6). Moreover, previous studies have proposed that it can promote cell cycle progression of hepatocellular carcinoma cells (5,6). Upregulation of HEIH was further observed in colorectal cancer (7), melanoma (8) and non-small cell lung cancer (9). The expression levels of HEIH are significantly increased in colorectal cancer tissues, and HEIH expression is positively associated with malignancy status and poor prognosis in patients with colorectal cancer. Furthermore, upregulation of HEIH has been reported to upregulate colorectal cancer cell proliferation and decrease apoptosis in vitro, whereas it further accelerates colorectal cancer tumor growth in vivo (7). Moreover, HEIH has been demonstrated to be highly expressed in melanoma tissues and cell lines and is associated with advanced clinical stage and poor disease outcome in patients with melanoma. Knockdown of HEIH inhibits melanoma cell proliferation, migration and invasion (8). In non-small cell lung cancer, it was revealed that HEIH accelerates cell proliferation and metastasis (9). These studies collectively suggest that HEIH serves as an oncogene in multiple cancer types. The present study demonstrated that HEIH expression was upregulated in breast cancer cell lines, and HEIH knockdown inhibited proliferation and induced apoptosis of breast cancer cells, suggesting that it acted as an oncogene in breast cancer.
Among the four breast cancer cell lines examined, upregulation of HEIH in MDA-MB-231 and MDA-MB-468 cells was higher than that noted in other types of breast cancer cell lines (MCF-7 and SK-BR-3). The highest expression was found in MDA-MB-231 cells. MDA-MB-231 and MDA-MB-468 cells belong to triple-negative breast cancer (TNBC) cell lines. It is well known that TNBC is an aggressive form of breast cancer that contains cells that overexpress HER2. TNBC cells do not express ER or PR and are consequently resistant to cancer therapy (21). The development of targeted therapy (hormonal agents and trastuzumab) has made significant improvements in the outcome of other subtypes of breast cancer, including ER-positive/HER2 overexpressing tumors. However, current treatments for TNBC do not contain specific tumor-targeting therapeutic agents (22). Moreover, TNBC easily recurs and its metastasis leads to a poor prognosis during treatment (23). Therefore, the present study further explored the role of HEIH in the metastatic activity of MDA-MB-231 cells. Although, it was demonstrated that high HEIH expression was not associated with ER, PR and HER2 status, HEIH knockdown inhibited the invasion and migration of MDA-MB-231 cells, suggesting the contribution of HEIH in TNBC metastasis.
In conclusion, in the present study, the upregulation of HEIH expression was confirmed in breast cancer tissues. This lncRNA was positively associated with the malignancy status and poor prognosis of patients with breast cancer. Furthermore, HEIH knockdown induced apoptosis and suppressed proliferation and metastatic activity in vitro. These findings suggested the oncogenic role of HEIH in breast cancer, which may be used as a novel diagnostic and prognostic indicator for this disease.
Acknowledgements
Not applicable.
Funding
The present work was supported by the Scientific Research Project of Gansu Provincial Health Commission (grant no. GSWSKY-2019-02) and Gansu Provincial Natural Scientific Foundation (grant no. 18JR3RA352).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
CC and LW designed the study; CG, QR, FGD, QP and YCN performed the experiments; DCM, QP and YCN performed the data analysis; CC and LW wrote the manuscript; and CC, CG, QR and LW revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All patients provided signed informed consent prior to surgery. Clinicopathological follow-up information of patients with breast cancer was available. All clinical investigations in the present study were approved by the Human Ethics Committee of the First Hospital of Lanzhou University (Lanzhou, China). The study protocols were conducted according to the principles described in the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gardezi SJS, Elazab A, Lei B, Wang T. Breast cancer detection and diagnosis using mammographic data: Systematic review. J Med Internet Res. 2019;21:2869. doi: 10.2196/14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merino Bonilla JA, Torres Tabanera M, Ros Mendoza LH. Breast cancer in the 21st century: From early detection to new therapies. Radiologia. 2017;59:368–379. doi: 10.1016/j.rx.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Kumar M, DeVaux RS, Herschkowitz JI. Molecular and cellular changes in breast cancer and new roles of lncRNAs in breast cancer initiation and progression. Prog Mol Biol Transl Sci. 2016;144:563–586. doi: 10.1016/bs.pmbts.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 6.Ma Y, Cao D, Li G, Hu J, Liu X, Liu J. Silence of lncRNA HEIH suppressed liver cancer cell growth and metastasis through miR-199a-3p/mTOR axis. J Cell Biochem. 2019;120:17757–17766. doi: 10.1002/jcb.29041. [DOI] [PubMed] [Google Scholar]
- 7.Cui C, Zhai D, Cai L, Duan Q, Xie L, Yu J. Long noncoding RNA HEIH promotes colorectal cancer tumorigenesis via counteracting miR-939mediated transcriptional repression of Bcl-xL. Cancer Res Treat. 2018;50:992–1008. doi: 10.4143/crt.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao H, Xing G, Wang Y, Luo Z, Liu G, Meng H. Long noncoding RNA HEIH promotes melanoma cell proliferation, migration and invasion via inhibition of miR-200b/a/429. Biosci Rep. 2017;37:BSR20170682. doi: 10.1042/BSR20170682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia K, Chen F, Xu L. Long noncoding RNA HEIH promotes the proliferation and metastasis of non-small cell lung cancer. J Cell Biochem. 2019;120:3529–3538. doi: 10.1002/jcb.27629. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Li WQ, Li XH, Wu YH, Du J, Wang AP, Li D, Li YJ. Role of eukaryotic translation initiation factors 3a in hypoxia-induced right ventricular remodeling of rats. Life Sci. 2016;144:61–68. doi: 10.1016/j.lfs.2015.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Ling H, Vincent K, Pichler M, Fodde R, Berindan-Neagoe I, Slack FJ, Calin GA. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–5011. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling H, Krassnig L, Bullock MD, Pichler M. MicroRNAs in testicular cancer diagnosis and prognosis. Urol Clin North Am. 2016;43:127–134. doi: 10.1016/j.ucl.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 14.Thomas J, Ohtsuka M, Pichler M, Ling H. MicroRNAs: Clinical relevance in colorectal cancer. Int J Mol Sci. 2015;16:28063–28076. doi: 10.3390/ijms161226080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Troppan K, Wenzl K, Deutsch A, Ling H, Neumeister P, Pichler M. MicroRNAs in diffuse large B-cell lymphoma: Implications for pathogenesis, diagnosis, prognosis and therapy. Anticancer Res. 2014;34:557–564. [PubMed] [Google Scholar]
- 16.Youness RA, Gad MZ. Long non-coding RNAs: Functional regulatory players in breast cancer. Noncoding RNA Res. 2019;4:36–44. doi: 10.1016/j.ncrna.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu N, Wang F, Lv M, Cheng L. Microarray expression profile analysis of long non-coding RNAs in human breast cancer: A study of Chinese women. Biomed Pharmacother. 2015;69:221–227. doi: 10.1016/j.biopha.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Shen X, Xie B, Ma Z, Yu W, Wang W, Xu D, Yan X, Chen B, Yu L, Li J, et al. Identification of novel long non-coding RNAs in triple-negative breast cancer. Oncotarget. 2015;6:21730–21739. doi: 10.18632/oncotarget.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Lyu S, Dong S, Liu Y, Zhang X, Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. OncoTargets Ther. 2016;9:761–772. doi: 10.2147/OTT.S97664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma D, Chen C, Wu J, Wang H, Wu D. Up-regulated lncRNA AFAP1-AS1 indicates a poor prognosis and promotes carcinogenesis of breast cancer. Breast Cancer. 2019;26:74–83. doi: 10.1007/s12282-018-0891-3. [DOI] [PubMed] [Google Scholar]
- 21.Park SY, Choi JH, Nam JS. Targeting cancer stem cells in triple-negative breast cancer. Cancers (Basel) 2019;11:965. doi: 10.3390/cancers11070965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCann KE, Hurvitz SA, McAndrew N. Advances in targeted therapies for triple-negative breast cancer. Drugs. 2019;79:1217–1230. doi: 10.1007/s40265-019-01155-4. [DOI] [PubMed] [Google Scholar]
- 23.Saleh R, Taha RZ, Sasidharan Nair V, Alajez NM, Elkord E. PD-L1 blockade by atezolizumab downregulates signaling pathways associated with tumor growth, metastasis, and hypoxia in human triple negative breast cancer. Cancers (Basel) 2019;11:1050. doi: 10.3390/cancers11081050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.