Abstract
Hypertrophic scars (HSs) are a type of pathological scar which are induced by surgery, burn injuries or trauma during the healing process. Due to the high recurrence rates and strong invasive properties, HSs have become a major clinical issue. Resveratrol has been identified as a potential agent to suppress scar formation; however, the underlying mechanism of action remains unclear. Therefore, the present study aimed to investigate the effect of resveratrol on HS-derived fibroblasts (HSFBs) in vitro. MTT assay was performed to evaluate cell viability following the resveratrol treatment. Western blot and RT-qPCR analysis was used to identify the expression levels and the relationship among autophagic markers, miR-4654 and resveratrol treatment. Finally, GFP-LC3 stable HSFBs cells were generated to further assess the effect of resveratrol. The results revealed that resveratrol significantly induced cell death in a dose-dependent manner and induced autophagy by downregulating the expression levels of Rheb in HSFBs. Notably, microRNA-4654 (miR-4654) was significantly decreased in the HSFBs and re-upregulated by resveratrol treatment dose-dependently. Through the bioinformatic analysis and luciferase assay, miR-4654 was identified to directly target Rheb. Transfection studies showed that miR-4654 negative correlated with Rheb expression, suggesting that the autophagic process may be altered by the miR-4654/Rheb axis under the control of resveratrol. In conclusion, the results of the present study suggested that resveratrol may promote autophagy by upregulating miR-4654, which in turn may suppress Rheb expression via directly binding to the 3′-untranslated region of Rheb. These findings provided a novel insight into the development of potential therapeutic targets for HSs.
Keywords: hypertrophic scar, hypertrophic scar-derived fibroblasts, Rheb, miR-4654, autophagy
Introduction
Hypertrophic scars (HSs) comprise a type of pathological scar induced by surgery, burn injuries or trauma during the healing process (1). HSs most commonly occur in the outer layers of the skin and arthroses, resulting in damage to the individual appearance and severe dysfunction, including itchiness, susceptibility to infection, pain and disfigurement (2,3). It is well established that HSs are a type of tissue fibrosis caused by the accumulation of the extracellular matrix, exhibiting a robust inflammatory response and fibroblast proliferation (2,4). The therapeutic strategies for HSs include surgery, radiotherapy and combination therapy (1); however, the therapeutic efficacy of these treatments remains unsatisfactory. For example, previous studies investigated the efficacy of laser therapy combined with silicone gel sheeting and steroid injection, but found that there was no significant effect for HSs treatment (5,6). Furthermore, HSs have been reported to be regulated by a number of complicated regulatory mechanisms, including inflammation (7) and immune response (8). Nonetheless, to the best of our knowledge, the mechanisms behind the pathophysiological processes of HSs remain unknown (9). Therefore, it remains an urgent requirement to investigate the potential molecular events of HSs to identify novel therapeutic targets.
Autophagy is an evolutionarily highly conserved catabolic pathway that maintains the cellular energy balance through recycling cytoplasmic proteins and controlling the quality of organelles (10,11); it also provides efficient protection for cells under various stress conditions (12,13). Previous studies have reported that autophagy was involved in numerous types of disease, including cancer, lung disease and neurodegenerative diseases (14–17). In addition, the involvement of autophagy in the formation of HSs has been demonstrated under starvation stress (18). Thus, these findings provide reasoning for researchers to further investigate the relationship between HSs and autophagy.
MicroRNAs (miRNA/miR) are endogenous, conserved non-coding RNA molecules of 19–22 nucleotides in length (19). miRNAs serve as critical regulators of target genes through multiple mechanisms, including inhibiting translation, promoting mRNA degradation and repressing protein synthesis (20–22). In addition, an abundance of evidence has identified that miRNAs were involved in numerous metabolic reactions, including cell proliferation, differentiation, autophagy and apoptosis, by directly binding to the 3′-untranslated region (3′-UTR) of their target mRNAs (23–26). To date, a small number of studies have suggested a potential role between miRNAs and HSs; for example, the expression levels of miR-21 were reported to be upregulated in HS-derived fibroblasts (HSFBs) and inhibiting miR-21 expression significantly slowed the formation of HSs in vivo (27). Conversely, the expression levels of another miRNA, miR-137, were markedly downregulated in HSs, which induced the proliferation and metastasis of fibroblasts (28). Thus, as multiple miRNAs have been reported to be aberrantly expressed during HS formation, it deems worthy to investigate potential miRNAs candidates for HS therapy.
Resveratrol was discovered to be highly effective in the treatment of numerous types of tumor, including colon cancer, liver cancer and neuroendocrine tumors, as well as inflammatory reaction (29,30). Several previous studies have reported that resveratrol was involved in miRNA-induced autophagy during the treatment of multiple types of disease, such as chronic diabetic nephropathy (31), Alzheimer's disease (32) and cancer (33). For HSs, resveratrol has been approved as a potential agent to suppress scar formation (34). Interestingly, Zeng et al (35) identified that resveratrol significantly inhibited cell growth by inducing fibroblast apoptosis, whereas Bai et al (36) discovered that sirtuin 1 was upregulated by resveratrol, leading to autophagy during HSs treatment. Therefore, the present study hypothesized that resveratrol may inhibit the viability of hypertrophic scars by activating autophagy via the miRNAs. The results revealed that resveratrol induced autophagy by inhibiting the expression levels of Rheb. Notably, miR-4654 served as the ‘bridge’ between resveratrol and the GTP-binding protein Rheb. Taken together, the findings of the present study confirmed that Rheb was a target gene of miR-4654 and partially determined the novel mechanism of miR-4654-induced autophagy, thereby providing further insights into putative targets for HS therapy.
Materials and methods
Chemicals and cell culture
Resveratrol was purchased from Target Molecule Corp. Normal skin-derived fibroblasts (NSFBs) and HSFBs were kindly provided by Dr Li Min at Department of Dermatology, Gulou Hospital (Nanjing, China). 293T cells were obtained from the American Type Culture Collection. All cell lines were cultured in high glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% antibiotic-antimycotic (Thermo Fisher Scientific, Inc.), and maintained at 37°C in an atmosphere of 5% CO2. HSFBs were treated by resveratrol or transfected with miR-4654 mimic and inhibitor or Rheb vectors. HSFBs treated with mixed vehicle controls were used as control.
MTT assay
An MTT assay was used to determine cell viability. Briefly, HSFBs were seeded in the 96-well plate at a density of 106 cells per well. Then resveratrol was diluted to various concentrations (0, 1, 10 or 100 µmol/l) using PBS and incubated with the cells (1×106) for 0, 24, 48 or 72 h at 37°C. Following the incubation, 200 µl MTT medium was added/well and incubated with the cells at 37°C for a further 4 h. Following treatment with dimethyl sulfoxide (100 µl), the optical density was measured at a wavelength of 570 nm for each experimental group using a microplate reader (Thermo Fisher Scientific, Inc.).
Cell transfection
The miR-4654 mimics (5′-UGUGGGAUCUGGAGGCAUCUGG-3′), miR-4654 inhibitors (5′-AGAUGCCUCCAGAUCCCACAAA-3′), mimic-NC (5′-UUUGUACUACACAAAAGUACUG-3′) and inhibitor-NC (5′-CAGUCCUUUUGUGUAGUACAA−3′) were obtained from Shanghai GenePharma Co., Ltd. HSFBs were plated on the 6-well plate at a density of 106 cells pre well and transfected with miR-4654 mimics, miR-4654 inhibitors or the respective negative controls (NCs) (5 nM for all) for 72 h using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
According to the manufacturer's protocols, Rheb overexpression (OE) was accomplished using a Rheb OE vector (pcDNA3.1; Synthgene Biotech). The Rheb knockdown (KD) was accomplished using a short hairpin RNA targeting Rheb contained within a pcDNA3.1 vector. An empty pcDNA3.1 vector was used as the NC for the KD and OE vectors. HSFBs were plated in the 6-well plate (106 cells pre well) and transfected with these vectors at 5 nM using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cells were used for further study 72 h following transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 1 µg RNA was reverse transcribed into cDNA using AMV reverse transcriptase (RR019A, Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol, and a miR-4654 RT primer (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCAGAT-3′); U6 RT primer (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCATGCT-3′). The RT reaction conditions were as follows: 42°C for 30 min, 95°C for 5 min and 5°C for 5 min.
qPCR was subsequently performed using a SYBR Green qPCR mix (Vazyme Biotech Co., Ltd.), according to the manufacturer's protocol, on an ABI 7300 sequence detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for the qPCR (performed on a 96-well plates): Initial denaturation at 95°C for 10 min; followed by 40 cycles at 95°C for 30 sec, 56°C for 30 sec and 72°C for 1 min. The following primer sequences were used for the qPCR: miR-4654 forward, 5′-CGTGTGGGATCTGGAGGC−3′ and reverse, 5′-AGTGCAGGGTCCGAGGTATT-3′; U6 forward, 5′-CGGTCCAACGATACAGAGAAG−3′ and reverse, 5′-AGTGCAGGGTCCGAGGTATT-3′; Rheb forward, 5′-TGGGAATAAGAAAGACCTG-3′ and reverse, 5′-GAAGACTTGCCTTGTGAA−3′; and GAPDH forward, 5′-GATATTGTTGACATCAATGAC−3′ and reverse, 5′-TTGATTTTGGAGGGATCTCG-3′. The expression levels were quantified using the 2−ΔΔCq method (37), and GAPDH expression levels were used to normalize the relative abundance of Rheb, whereas U6 expression levels were used to normalize the relative abundance of miR-4654.
Detection of the target site of miR-4654 on the 3′-UTR of Rheb using a dual-luciferase reporter assay
The potential target sequence for miR-4654 on the 3′-UTR of Rheb was predicted using TargetScan (www.targetscan.org). Subsequently, 1×106 293T cells were plated into six-well plates and cultured for 12 h at 37°C and 5% CO2. The pGL3 luciferase reporter vector was obtained from Promega Corporation and the Rheb wild-type (WT) or mutant (MUT) 3′-UTR were cloned into the pGL3 plasmid to synthesize pGL3-Rheb-WT or pGL3-Rheb-MUT. To establish the 3′-UTR of mutant Rheb (Rheb-MUT), the binding sites were mutated via the site directed mutagenesis kit (NEB E0554, New England BioLabs, Inc.). 293T cells were co-transfected with 5 nM of miR-4564 mimics or mimic-NC and pGL3-Rheb-WT or pGL3-Rheb-MUT using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following incubation for 48 h at 37°C, the transfected cells were harvested by centrifugation (350 × g; 3 min; 20°C) and firefly luciferase activity was detected using a Dual-Luciferase Reporter Assay system (Promega Corporation). The data was normalized to Renilla luciferase activity.
Western blotting
Total protein was extracted from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific, Inc.). Total protein was quantified using a bicinchoninic acid assay (Thermo Fisher Scientific, Inc.) and 45 µg protein/lane was separated via 12% SDS-PAGE for 2.5 h at a voltage of 100 V. The separated proteins were subsequently transferred onto PVDF membranes for 1.5 h under a current of 320 mA and blocked with 5% non-fat milk for 2 h at 37°C. The membranes were then incubated with the following primary antibodies overnight at 4°C: Anti-Rheb rabbit polyclonal (ab92313, 1:2,000; Abcam), anti-microtubule-associated protein 1A/1B-light chain 3 (LC3) rabbit polyclonal (14600-1-ap, 1:1,000; ProteinTech Group, Inc.), anti-Beclin 1 rabbit monoclonal (ab210498, 1:2,000; Abcam) and anti-GAPDH mouse monoclonal (40004-1-lg, 1:10,000; ProteinTech Group, Inc.). GAPDH served as the internal loading control. Following the primary antibody incubation, the membranes were washed with PBS-Tween 20 (1%) and incubated with horseradish peroxidase-conjugated secondary antibodies: Anti-Rat IgG (HRP) (ab7097, 1:5,000, Abcam) or Anti-Mouse IgG (HRP) (ab97040, 1:5,000, Abcam) at room temperature for 2 h. Protein bands were visualized using an ECL reagent (Thermo Fisher Scientific, Inc.) and an ECL immunoblotting system (Tanon Science and Technology Co., Ltd.) and the expression levels were semi-quantified using ImageJ software (V1.8.0.112, National Institutes of Health).
Verification of the role of autophagy
HSFBs (1×106) were co-treated with 5 mM 3-Methyladenine (3-MA; cat. no. HY-19312; MedChemExpress) and 100 µmol/l resveratrol for 72 h at 37°C. Then, the cells were collected by centrifugation (350 × g; 3 min; room temperature) for further experiments.
Fluorescence assay
HSFBs were infected with adenoviruses expressing GFP-LC3B fusion protein [Umibio (Shanghai) Co., Ltd.] using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 72 h to obtain the stable GFP-LC3 cell line. Briefly, 1×106 cells were seeded upon glass confocal dishes and allowed to settle for 12 h at 37°C. miR-4654 mimics, miR-4654 inhibitors and the respective NCs were subsequently transfected into the stable cell line with or without Rheb OE plasmid or Rheb KD plasmid transfection. Subsequently, each group was treated with 100 µmol/l resveratrol or an equal volume of PBS. Following incubation for 72 h at 37°C, cells in each group were fixed by 4% paraformaldehyde for 10 min at room temperature. The cell nuclei were stained with DAPI (1:2,000; Abcam) for 5 min in the dark at room temperature. The images were observed at 200× magnification using a confocal microscope.
Statistical analysis
Statistical analysis was performed using GraphPad Prism v5 software (GraphPad Software, Inc.) and data are presented as the mean or percentage change ± SD from three independent experiments. Statistical differences between the two treatment groups were compared using a paired Student's t-test, whereas comparisons between >2 groups were performed using a one-way ANOVA and a Tukey's multiple comparisons test. P<0.05 was considered to indicate a statistically significant difference.
Results
Autophagy is triggered by resveratrol in a dose-dependent manner in HSFBs
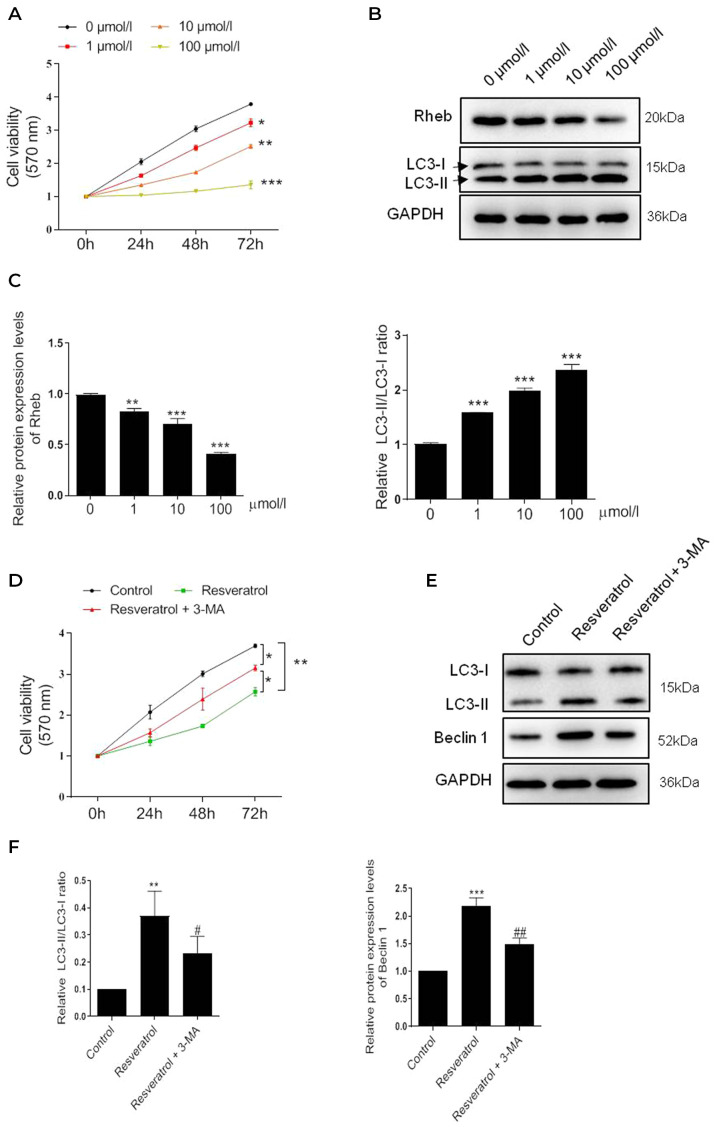
To identify the pharmacological effect of resveratrol, HSFBs were treated with resveratrol at different concentrations, and the cell viability was subsequently determined using a MTT assay, which revealed a significant dose-dependent decrease in cell viability following the treatment with resveratrol compared with the untreated cells (Fig. 1A). Notably, the most significant level of inhibition occurred following 100 µmol/l resveratrol treatment. Subsequently, changes in the expression levels of the autophagy-related protein marker, LC3, were investigated. The results revealed that the LC3-II/LC3-I ratio was significantly increased in a dose-dependent manner (Fig. 1B). Conversely, the expression levels of the upstream gene, Rheb, were significantly decreased dose-dependently. These findings suggested that resveratrol may induce autophagy in HSFBs.
Figure 1.
Resveratrol inhibits the viability of HSFBs through regulating Rheb and activating autophagy. (A) MTT assay was performed to determine the cell viability of HSFBs in each group at 72 h following the treatment with different concentrations of resveratrol. *P<0.05, **P<0.01, ***P<0.001 vs. 0 µmol/l group. (B) and (C) Western blotting was used to analyze the protein expression levels of Rheb and the LC3-II/I ratio in HSFBs following the treatment with different concentrations of resveratrol. **P<0.01, ***P<0.001 vs. 0 µmol/l group. (D) MTT assay was performed to detect the cell viability of HSFBs in each group at 72 h following the treatment with 100 µmol/l resveratrol with or without 5 mM 3-MA treatment. *P<0.05, **P<0.01. (E) and (F) Western blotting was used to analyze the protein expression levels of Rheb and LC3 in HSFBs following the treatment with resveratrol and 3-MA or resveratrol alone. **P<0.01, ***P<0.001 vs. the control group; #P<0.05, ##P<0.01 vs. the resveratrol group. Data are presented as the mean ± SD from three independent experiments. Control, the cell was treated with PBS; HSFBs, hypertrophic scar-derived fibroblasts; LC3, microtubule-associated protein 1A/1B-light chain 3; 3-MA, 3-Methyladenine.
To further confirm whether resveratrol inhibited the viability of HSFBs through activating autophagy, HSFBs were co-treated with the autophagy inhibitor 3-MA (5 mM) and 100 µmol/l resveratrol. Interestingly, 3-MA treatment partially reversed the inhibitory effect of resveratrol on cell viability (Fig. 1C). The western blotting results further revealed that 3-MA treatment markedly regulated the effect of resveratrol-induced autophagy; the LC3-II/LC3-I ratio and Beclin 1 expression levels were partially but notably downregulated in HSFBs treated with resveratrol and 3-MA compared with resveratrol alone (Fig. 1D). These results indicated that autophagy may be triggered by resveratrol in a dose-dependent manner to inhibit HSFB cell viability.
miR-4654 downregulates the protein expression levels of Rheb
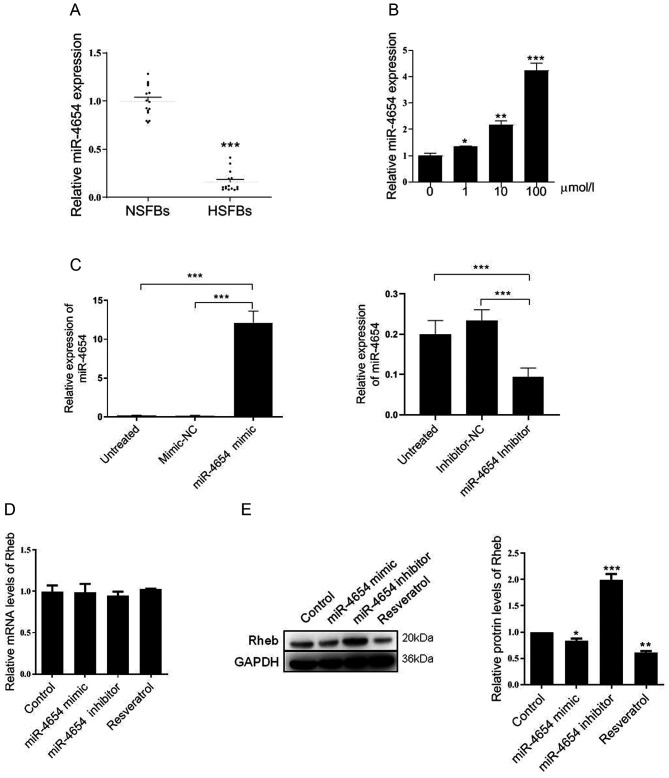
The subsequent experiments investigated the effect of separate controls and a mixed control on the expression levels of Rheb, cell viability and autophagy. The results revealed that the mimic-NC or inhibitor-NC transfections did not alter the protein or mRNA expression levels of Rheb compared with the untreated group (Fig. S1A and B). Similarly, following the transfection of the cells with the mixed control (mimic-NC + inhibitor-NC + PBS), the expression levels of Rheb were slightly downregulated; however, no statistical differences were recorded among the different groups (Fig. S1A and B). A similar trend in the cell viability response was observed in each group (Fig. S2A). Similarly, no significant differences were identified between the separate controls and mixed control transfections on the LC-3II/I ratio or Beclin 1 expression levels (Fig. S2B and C). In addition, the effect of the control transfections on the expression levels of LC3 were investigated using GFP-LC3 stable cells. Compared with the untreated group, each separated control group did not display significant changes in the LC3 signal intensity (Fig. S3). In fact, even in the mimic-NC + inhibitor-NC + vector + PBS group, the LC3 signal was similar compared to the untreated group. Taken together, the results identified that the mixed control presented a similar effect to the single controls, and had no effect on Rheb expression levels, HFSB viability or autophagy. Thus, the mixed controls were chosen for use in subsequent experiments.
The expression levels of miR-4654 were subsequently investigated in NSFBs and HSFBs from 14 independent repeated experiments. Compared with the NSFBs, the expression levels of miR-4654 in the HSFBs were significantly downregulated (Fig. 2A). Following resveratrol treatment, the expression levels of miR-4654 were significantly upregulated in a dose-dependent manner compared with the untreated HSFBs (Fig. 2B). Changes in miR-4654 expression in HSFBs transfected with miR-4654 mimic and inhibitor were assessed by RT-qPCR. The results revealed that the cells transfected with the miR-4654 mimic had significantly upregulated miR-4654 expression levels compared with the mimic-NC group. In addition, a 50% decrease was observed in the expression levels of miR-4654 following the transfection with the miR-4654 inhibitor compared with the inhibitor-NC group (Fig. 2C). Subsequently, the mRNA and protein expression levels of Rheb were analyzed in the presence of the miR-4654 mimic, miR-4653 inhibitor or resveratrol treatment; however, no significant differences were observed in the expression levels of Rheb mRNA between these groups (Fig. 2D). Interestingly, neither miR-4654 nor resveratrol were able to influence Rheb mRNA expression levels; however, resveratrol treatment significantly downregulated the protein expression levels of Rheb, while the inhibition of miR-4654 expression levels in HSFBs led to the significant upregulation of the protein expression levels of Rheb compared with the control group (Fig. 2E). These findings indicated that Rheb may be influenced by miR-4654 or resveratrol in the process of translation.
Figure 2.
Effect of miR-4654 on Rheb expression. (A) Relative expression levels of miR-4654 in NSFBs and HSFBs. N=14, ***P<0.001 vs. NFSBs. (B) RT-qPCR was performed to analyze the miR-4654 expression levels in HSFBs following the treatment with resveratrol at different concentrations. *P<0.05, **P<0.01, ***P<0.001 vs. 0 µmol/l group. (C) Transfection efficiency of miR-4654 mimic or miR-4654 inhibitor in HSFBs was determined using RT-qPCR. ***P<0.001. (D) RT-qPCR was performed to analyze the mRNA expression levels of Rheb in HSFBs transfected with miR-4654 mimic or miR-4654 inhibitor or treated with 100 µmol/l resveratrol. (E) Western blotting was used to analyze the protein expression levels of Rheb in HSFBs transfected with miR-4654 mimic or miR-4654 inhibitor, or treated with 100 µmol/l resveratrol. *P<0.05, **P<0.01, ***P<0.001 vs. the control group. Both the mRNA and protein expression levels from parts (D) and (E) were normalized to GAPDH. Data are presented as the mean ± SD; n=3. HSFBs, hypertrophic scar-derived fibroblasts; NSFBs, normal skin-derived fibroblasts; Control, mimic-NC + inhibitor-NC + PBS group; RT-qPCR, reverse transcription-quantitative PCR; miR, microRNA.
miR-4654 regulates the translation of Rheb through targeting its 3′-UTR region
To confirm the relationship between miR-4654 and Rheb, the potential binding sites of miR-4654 on the 3′-UTR of Rheb were investigated using bioinformatics analysis (Fig. 3A). To further confirm the hypothesis of the present study, a dual-luciferase reporter assay was performed by co-transfecting miR-4654 mimics or mimic-NC with the luciferase reporter vector containing the WT or MUT 3′-UTR of Rheb. The transfection of the miR-4654 mimic led to a significant reduction in the relative luciferase activity of Rheb-WT cells compared with the mimic-NC group; however, no significant differences were observed in the Rheb-MUT cells with miR-4654 treatment as compared with the mimic-NC group (Fig. 3B). These findings suggested that miR-4654 may target the 3′-UTR of WT Rheb and regulate the expression of Rheb negatively.
Figure 3.
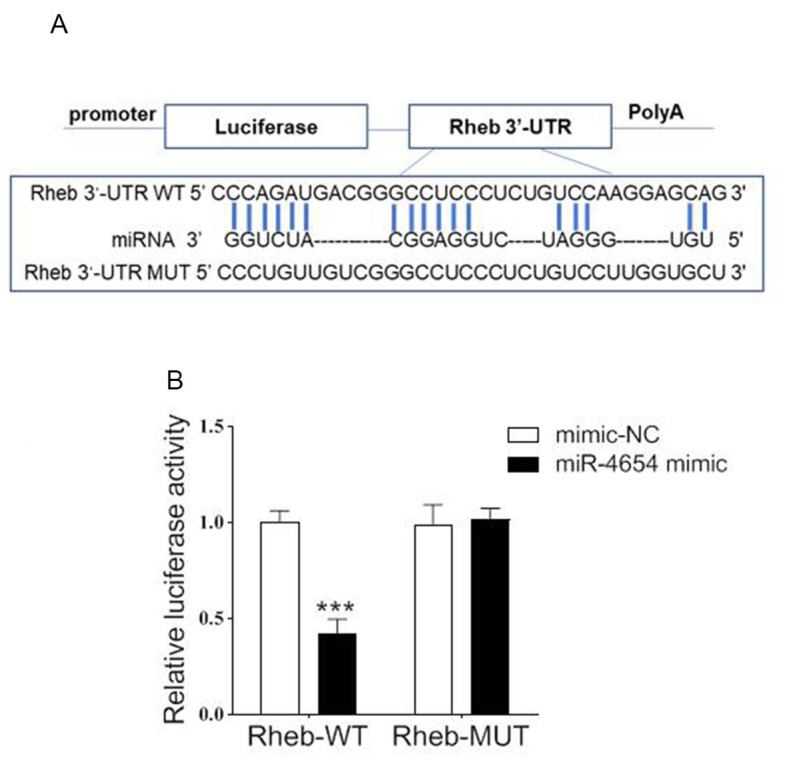
Rheb is a direct target gene of miR-4654. (A) Sequence analysis of binding sites between miR-4654 and the WT Rheb 3′-UTR according to TargetScan. The Rheb 3′-UTR MUT sequence is also shown. (B) Dual-luciferase reporter assay was performed in 293T cells co-transfected with the plasmid containing WT or MUT Rheb 3′-UTRs and miR-4654 mimic or mimic-NC. Data are presented as the mean ± SD. N=3, ***P<0.001 vs. mimic-NC. 3′-UTR, 3′-untranslated region; miR, microRNA; WT, wild-type; MUT, mutant.
miR-4654 enhances Rheb-induced autophagy in HSFBs
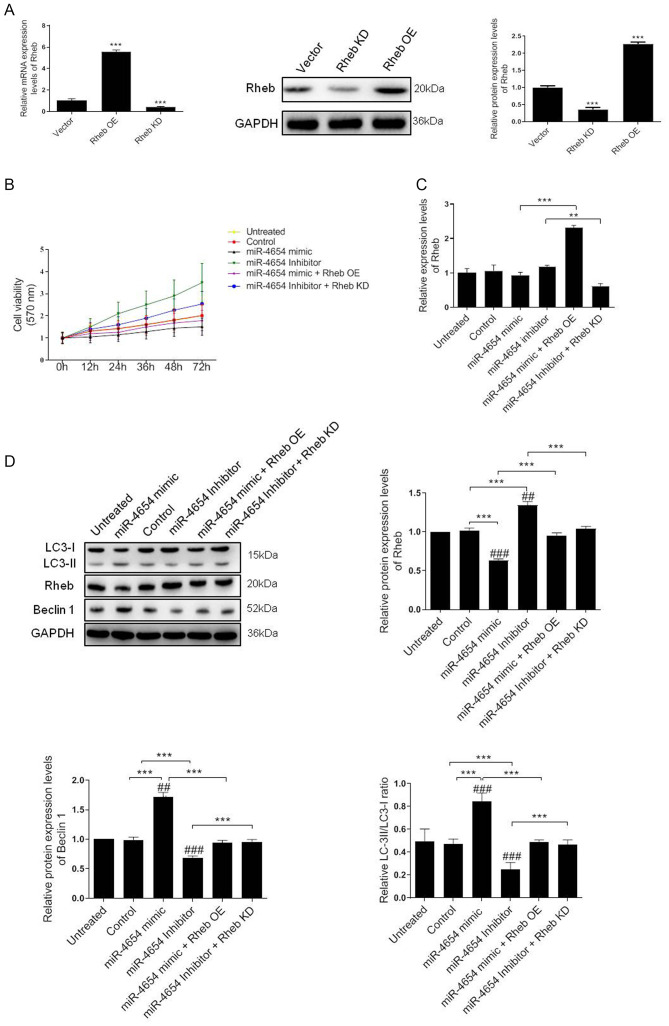
A previous study demonstrated the critical role of Rheb in the regulation of cell autophagy and proliferation processes (38). Therefore, the present study aimed to investigate whether the miRNA-dependent downregulation of Rheb affected these processes in HSFBs. A 5-fold upregulation in mRNA expression levels and a 2.4-fold upregulation in protein expression levels was identified in HSFBs transfected with the Rheb OE vector compared with the control vector. Conversely, the mRNA expression levels of Rheb were significantly downregulated in cells transfected with Rheb KD vector compared with the control vector and the western blotting results revealed a 60% reduction in Rheb expression levels following Rheb KD in the cells (Fig. 4A).
Figure 4.
Effect of miRNA-mediated Rheb downregulation on the viability and autophagy of HFSBs. (A) HFSBs were transfected with the Rheb OE plasmid, Rheb KD plasmid or the empty control vector. The mRNA and protein expression levels of Rheb were analyzed using RT-qPCR or western blotting, respectively. ***P<0.001 vs. vector group. (B) MTT assay was performed to determine the cell viability in each group at 72 h. (C) RT-qPCR analysis of Rheb expression levels in HFSBs in the different groups. **P<0.01, ***P<0.001. (D) Western blotting was used to determine the protein expression levels of Beclin 1, Rheb and LC3 in HFSBs in the different groups. ***P<0.001; ##P<0.01, ###P<0.001 vs. the untreated group. Data are presented as the mean ± SD from three independent experiments. HSFBs, hypertrophic scar-derived fibroblast; OE, overexpression; KD, knockdown; Control, mimic-NC + inhibitor-NC + vector treated group; LC3, microtubule-associated protein 1A/1B-light chain 3; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR.
Based on these results, the current study aimed to determine whether miR-4654 induced HSFB cell viability through a Rheb-dependent mechanism The MTT assay results revealed that the inhibition of miR-4654 expression levels increased the cell viability of HSFBs compared with the untreated group or control group. However, the cell viability was reduced following the transfection with the miR-4654 mimic. In addition, following the co-transfection with the Rheb KD vector prior to the miR-4654 inhibitor, the cell viability was re-increased compared with the miR-4654 inhibitor group (Fig. 4B). The mRNA expression levels of Rheb were subsequently investigated following the different transfections. The regulation of miR-4654 expression levels using the mimic or inhibitor did not significantly alter Rheb expression levels in the HSFBs compared with the control groups; however, a 2.3-fold increase and a 50% reduction was observed in Rheb expression levels in the Rheb OE + miR-4654 mimic and miR-4654 inhibitor + Rheb KD groups, respectively (Fig. 4C). Finally, the expression levels of autophagy-related protein markers, including LC3 and Beclin 1, were analyzed. The data revealed that the overexpression of miR-4654 significantly upregulated the expression levels of these proteins compared with the control group. Moreover, following the transfection with miR-4654 mimic, the expression levels of Rheb were significantly downregulated compared with the control group. In contrast, following the transfection of cells with the miR-4654 inhibitor, the expression levels of Rheb were significantly upregulated. Next, Rheb was overexpressed prior to the miRNA-mimic/inhibitor transfection; the results demonstrated that the transfection with Rheb OE partially reversed the effect of miR-4654 on autophagy. Similarly, the co-transfection of Rheb KD and the miR-4654 inhibitor into HSFBs also significantly reversed the effect of the miR-4654-inhibitor alone (Fig. 4D). These data suggested that Rheb may be one of the targets of miR-4654 responsible for autophagy and the viability of HSFBs.
Resveratrol induces HSFB autophagy through miR-4654/Rheb axis
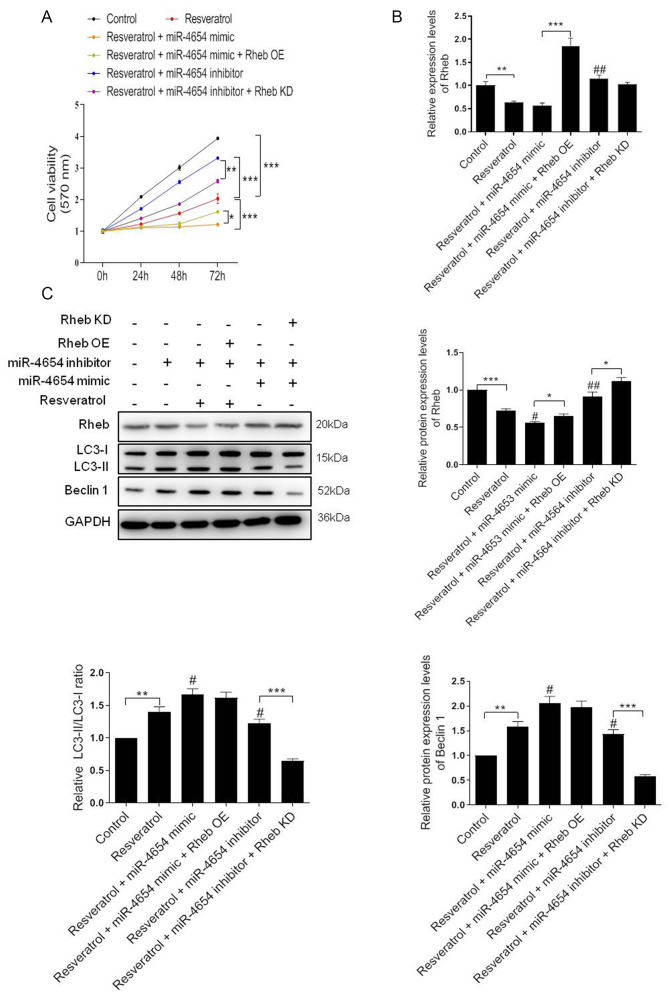
In light of the aforementioned findings, it was hypothesized that the effect of resveratrol on cell autophagy may be mediated by miRNA. As previously demonstrated, resveratrol treatment promoted the upregulation of miR-4654 expression levels in HSFBs. The subsequent results revealed that the cell viability was markedly reduced following resveratrol treatment compared with the control group, and this effect was exacerbated by the addition of the miR-4654 mimic (resveratrol + miR-4654 mimic group). Conversely, the inhibition of miR-4654 expression alongside resveratrol treatment effectively alleviated the effects of resveratrol treatment alone over cell viability. When Rheb was overexpressed before resveratrol and miR-4654 mimic treatment, the cell viability was re-increased significantly compared with the resveratrol + miR-4654 mimic group. The silencing of Rheb reversed the effect of resveratrol + miR-4654 inhibitor treatment (Fig. 5A). Moreover, RT-qPCR analysis revealed that neither resveratrol treatment alone or combined with miR-4654 mimics downregulated the expression level of Rheb compared with the control group (Fig. 5B). However, these effects were significantly reversed following the co-transfection with the Rheb OE plasmid (resveratrol + miR-4654 mimic + Rheb OE group; Fig. 5B). Similarly, the results obtained from the western blotting experiments also suggested that the downregulated expression levels of Rheb following resveratrol treatment in HSFBs were miR-4654 dependent. Rheb was markedly reduced following resveratrol treatment alone or combined with miR-4654 mimic compared with the control group. By contrast, the inhibition of miR-4654 expression alongside resveratrol treatment effectively increased the effects of resveratrol treatment alone. However, the expression level of Beclin 1 and the LC3-II/LC3-I ratio were both increased in the resveratrol group, and further increased in the resveratrol + miR-4654 mimic group. While the levels of such molecules were both downregulated when the cells were treated with miR-4654 inhibitor, which exhibited no difference compared with the resveratrol alone group. When the cells were co-transfected with the miR-4654 mimic and Rheb OE prior to resveratrol treatment, no change was observed in the expression level of Beclin 1 and the LC3-II/LC3-I ratio compared with the resveratrol + miR-4654 mimic group, while the protein level of Rheb was reupregulated significantly. Moreover, following the treatment of cells with the miR-4654 inhibitor and downregulating Rheb expression levels with the Rheb KD plasmid, resveratrol treatment significantly inhibited cell autophagy (Fig. 5C). These findings suggested that the increase in autophagy by resveratrol in HSFBs may be dependent on the miR-4654/Rheb axis.
Figure 5.
Resveratrol activates autophagy through the miR-4654/Rheb axis. (A) Cell viability was analyzed in HSFBs in the different groups using an MTT assay. *P<0.05, **P<0.01, ***P<0.001. (B) RT-qPCR analysis of the expression levels of Rheb in HSFBs in the different groups. **P<0.01, ***P<0.001; ##P<0.01 vs. the resveratrol group. (C) Western blotting was used to analyze the protein expression levels of Rheb, LC3 and Beclin 1 in HSFBs in the different groups. *P<0.05, **P<0.01, ***P<0.001; #P<0.05, ##P<0.01 vs. the resveratrol group. Data are presented as the mean ± SD. N=3. HSFBs, hypertrophic scar-derived fibroblasts; LC3, microtubule-associated protein 1A/1B-light chain 3; Control, mimic-NC + inhibitor-NC + PBS + vector group; OE, overexpression; KD, knockdown; miR, microRNA.
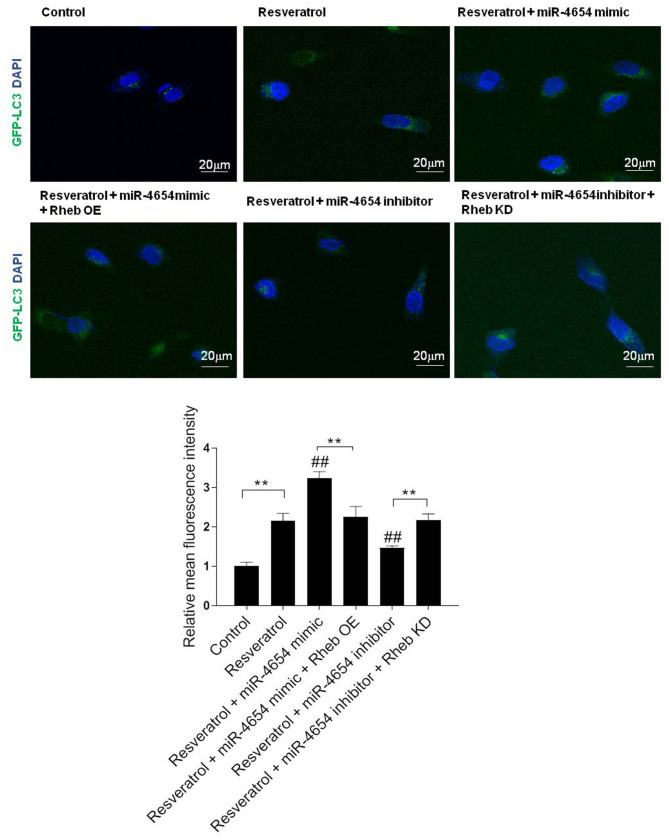
miR-4654 promotes the formation of autophagosomes in HSFBs following resveratrol treatment
To further confirm the role of miR-4654 in the activation of autophagy, the fluorescence intensity in the stable GFP-LC3 HSFBs cell line was analyzed in each group; miR-4654 mimics or inhibitors were transfected into stable GFP-LC3 cell lines. Following 72 h of transfection, DAPI was used to stain the cell nuclei and the cells were observed under confocal microscopy to determine the GFP-LC3 fluorescent intensities (Fig. 6). The results revealed that excessive autophagy was induced following resveratrol treatment, while the transfection with the miR-4654 inhibitor reversed the effect of resveratrol treatment. In addition, upon the co-transfection of the cells with the miR-4654 mimic and Rheb OE prior to resveratrol treatment, the increased GFP-LC3 fluorescent intensity in the resveratrol + miR-4654 mimic group was reduced. Moreover, the co-transfection of cells with the miR-4654 inhibitor and Rheb KD plasmid prior to resveratrol treatment re-increased the effect of resveratrol + miR-4654 inhibitor treatment.
Figure 6.
miR-4654 promotes the formation of autophagosomes in HSFBs following resveratrol treatment. HSFBs were infected with adenoviruses expressing GFP-LC3 fusion protein to obtain a stable GFP-LC3 cell line. Different treatments/transfections were applied in the stable cell line, including resveratrol treatment alone, resveratrol + miR-4654 mimic transfection, resveratrol + miR-4654 mimic + Rheb OE transfection, resveratrol + miR-4654 inhibitor transfection + Rheb KD, resveratrol + miR-4654 inhibitor transfection and the control group. After 72 h, DAPI was used to stain cells and the cells were observed at a magnification of ×200 using confocal microscopy. Scale bar=20 µm. The GFP-LC3 fluorescence intensity was normalized against the control group and determined using Image J software. Data are presented as the mean ± SD, n=3, **P<0.01; ##P<0.01 vs. the resveratrol group. LC3, microtubule-associated protein 1A/1B-light chain 3; OE, overexpression; KD, knockdown; Control, mimic-NC + inhibitor-NC + PBS + vector group; HSFBs, hypertrophic scar-derived fibroblasts; miR, microRNA.
Discussion
Autophagy serves a vital role in sustaining cellular metabolism; however, in certain cellular settings, autophagy is also known to induce cell death (39,40). Autophagy has also been identified to regulate the apoptotic response (41). It is also established that autophagy is an essential process for the maintenance of cellular homeostasis (42). Furthermore, the association between autophagy and HSs has been widely reported by an increasing number of studies; for example, Shi et al (43) compared the autophagic capacity of HSs and normal skin and discovered that the generation of LC3 is prevented in HSs, which was suggested to benefit HSs formation. In addition, Shi et al (44) demonstrated that WT p53-modulated autophagy and autophagic-induced fibroblast apoptosis suppresses the formation of HSs in a rabbit ear model. Moreover, Shi et al (45) reported that interleukin-10 inhibits autophagy in HSFBs under starvation stress to reduce HS formation. These findings suggested that autophagy may be involved in the proliferation and survival of HSFBs (46). The present study demonstrated that resveratrol could efficiently trigger HSFBs autophagy, as evidenced by the increased LC3-II/LC3-I ratio and upregulated expression level of Beclin 1, which are the markers of autophagy. The most marked effects were observed in the cells with 100 µmol/l resveratrol treatment for 72 h, in which the cell viability was seriously impaired. In addition, using the common inhibitor of the autophagic pathway 3-MA, it was found that inhibition of autophagy could significantly reverse the effect of resveratrol treatment. It was hypothesized that high level of autophagy in HSFBs might be essential to inhibit cell viability.
The present study also demonstrated that resveratrol treatment markedly reduced the expression levels of Rheb in a dose-dependent manner. Rheb has been identified as a critical regulator of autophagy during the disease state; a previous study has reported its ability to activate autophagy, thereby leading to its own inactivation via the mTOR complex 1 pathway (37). Another study suggested that reduction of Rheb can initiate autophagy in macrophages with increased Beclin1 and autophagy-related protein 7 (47). These finding suggest that inhibition of Rheb may be important to suppress HSFBs proliferation and induce autophagy, which was consistent with the present study.
An increasing number of miRNAs have been reported to serve as essential components of multiple pathophysiological and biomechanical processes (38,48,49). HSs have been discovered to be associated with abnormal changes in the expression level of multiple miRNAs, including the upregulation of miRNAs such as miR-30A-5p and miR-152, and the downregulation of miRNAs such as miR-143-5p and miR-4328 (50,51). In addition, miRNAs have been identified to affect the proliferation and apoptosis of fibroblasts, as well as extracellular matrix deposition (52,53). The roles of numerous miRNAs have been reported in both biological and clinical processes, although the roles for the majority of miRNAs have yet to be elucidated. The present study revealed that the expression levels of miR-4654 were significantly downregulated in HSFBs as compared to the NSFBs. While its level was significantly increased in the cells following the treatment with different concentrations of resveratrol. In addition, the expression levels of miR-4654 were closely associated with the degree of autophagy, which is consistent with the findings of a previous study (54). In the present study, overexpression of miR-4654 notably inhibited Rheb expression at the protein level and transfection of the miR-4654 inhibitor markedly increased the level of Rheb when compared with the control group. Subsequently, the current study further investigated the effect of miR-4654 and Rheb on the autophagic process. Results from bioinformatics analysis and a dual-luciferase reporter assay suggested that miR-4654 may directly inhibit the translation of Rheb by targeting its 3′-UTR region. Restoring the Rheb expression reversed the cell viability inhibition and autophagy initiation of HFSBs induced by miR-4654 overexpression. Similarly, suppressing the Rheb expression in Rheb-depleted cells inhibited the cell viability and re-enhanced cell autophagy as compared with the miR-4654 inhibitor group. Accordingly, these data indicated that resveratrol induced HFSBs autophagy might be regulated by upregulation of miR-4654 which in turn suppressed Rheb.
Finally, co-treatment with miR-4654 mimic further suppressed cell viability and enhanced cell autophagy in resveratrol-treated HFSBs. When the cells were treated with miR-4654 inhibitor prior to resveratrol administration, a higher cell viability and lower degree of autophagy were observed compared with the resveratrol treated group. By contrast, dysregulation of Rheb reversed the function of resveratrol and miR-4654 inhibitor treatment, and demonstrated no effect on the role of miR-4654 mimic treatment upon the resveratrol administration. The hypothesis of the present study was further verified by the fluorescence assay, which revealed that a higher number of autophagosomes were present following the transfection with the miR-4654 mimic and resveratrol treatment, whereas knocking down miR-4654 expression inhibited the fluorescence intensity of LC3 in HFSBs.
In conclusion, the findings of the present study suggested that resveratrol treatment may promote autophagy by upregulating miR-4654 expression levels, and thus downregulating the expression levels of the downstream gene, Rheb. However, the findings of the current study are limited due to the fact that the effect of resveratrol treatment on NSFBs was not investigated, which may limit the clinical impact of these results. However, despite this limitation, the findings still shed light on the molecular mechanism underlying the miR-4654-mediated activation of autophagy and the results suggested that miR-4654 may be a potential biomarker and therapeutic target for the treatment and diagnosis of HSs.
Supplementary Material
Acknowledgements
Not applicable.
Funding
The present study was supported by The National Natural Science Fund (grant no. 81774089), The Jiangsu Province, The Medical Innovation Team (grant no. CXTDA2017048) and Xuzhou Clinical Technology Diaphysis Training Program (grant no. 2019GG005).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
KP, BL, ZT, WY, LH, ZS, JZ, LC, RL, YL and QL made substantial contributions to the conception and design of the study, and acquired, analyzed and interpreted the data. LH, JZ, LC, RL, YL, and QL also contributed to drafting the manuscript and revising it critically for intellectual content. JD and CH made substantial contributions to the conception and design of the study, gave final approval of the version to be published and agreed to be accountable for the work in ensuring that questions related to the integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lee HJ, Jang YJ. Recent Understandings of biology, prophylaxis and treatment strategies for hypertrophic scars and keloids. Int J Mol Sci. 2018;19:3440. doi: 10.3390/ijms19030711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu Z, Ding J, Shankowsky HA, Tredget EE. The molecular mechanism of hypertrophic scar. J Cell Commun Signal. 2013;7:239–252. doi: 10.1007/s12079-013-0195-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang X, Zhang Y, Jiang BH, Zhang Q, Zhou RP, Zhang L, Wang C. Study on the role of Hsa-miR-31-5p in hypertrophic scar formation and the mechanism. Exp Cell Res. 2017;361:201–209. doi: 10.1016/j.yexcr.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- 5.Alster T. Laser scar revision: Comparison study of 585-nm pulsed dye laser with and without intralesional corticosteroids. Dermatol Surg. 2003;29:25–29. doi: 10.1046/j.1524-4725.2003.29024.x. [DOI] [PubMed] [Google Scholar]
- 6.Wittenberg GP, Fabian BG, Bogomilsky JL, Schultz LR, Rudner EJ, Chaffins ML, Saed GM, Burns RL, Fivenson DP. Prospective, single-blind, randomized, controlled study to assess the efficacy of the 585-nm flashlamp-pumped pulsed-dye laser and silicone gel sheeting in hypertrophic scar treatment. Arch Dermatol. 1999;135:1049–1055. doi: 10.1001/archderm.135.9.1049. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Hori K, Ding J, Huang Y, Kwan P, Ladak A, Tredget EE. Toll-like receptors expressed by dermal fibroblasts contribute to hypertrophic scarring. J Cell Physiol. 2011;226:1265–1273. doi: 10.1002/jcp.22454. [DOI] [PubMed] [Google Scholar]
- 8.Armour A, Scott PG, Tredget EE. Cellular and molecular pathology of HTS: Basis for treatment. Wound Repair Regen. 2007;15(Suppl 1):S6–S17. doi: 10.1111/j.1524-475X.2007.00219.x. [DOI] [PubMed] [Google Scholar]
- 9.Li P, He QY, Luo CQ. Overexpression of miR-200b inhibits the cell proliferation and promotes apoptosis of human hypertrophic scar fibroblasts in vitro. J Dermatol. 2014;41:903–911. doi: 10.1111/1346-8138.12600. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Wang J, Yang X. Functions of autophagy in pathological cardiac hypertrophy. Int J Biol Sci. 2015;11:672–678. doi: 10.7150/ijbs.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu H, Wang Y, Wang X, Li R, Yin D. MicroRNA-365 accelerates cardiac hypertrophy by inhibiting autophagy via the modulation of Skp2 expression. Biochem Biophys Res Commun. 2017;4:304–310. doi: 10.1016/j.bbrc.2017.01.108. [DOI] [PubMed] [Google Scholar]
- 12.Goutas A, Syrrou C, Papathanasiou I, Tsezou A, Trachana V. The autophagic response to oxidative stress in osteoarthritic chondrocytes is deregulated. Free Radical Biol Med. 2018;126:122–132. doi: 10.1016/j.freeradbiomed.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Camfield R, Gorski SM. The interplay between exosomes and autophagy-partners in crime. J Cell Sci. 2018;131:jcs215210. doi: 10.1242/jcs.215210. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Huang C, He Y, Sang Z, Liu G, Dai H. Knockdown of long non-coding RNA GAS5 increases miR-23a by targeting ATG3 involved in autophagy and cell viability. Cell Physiol Biochem. 2018;48:1723–1734. doi: 10.1159/000492300. [DOI] [PubMed] [Google Scholar]
- 15.Sun K, Deng W, Zhang S, Cai N, Jiao S, Song J, Wei L. Paradoxical roles of autophagy in different stages of tumorigenesis: Protector for normal or cancer cells. Cell Bio Sci. 2013;3:35. doi: 10.1186/2045-3701-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pabon MA, Ma KC, Choi AM. Autophagy and obesity-related lung disease. Am J Respir Cell Mol Biol. 2016;54:636–646. doi: 10.1165/rcmb.2016-0045PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo L, Qin ZH. Autophagy, aging, and longevity. Adv Exp Med Biol. 2019;1206:509–525. doi: 10.1007/978-981-15-0602-4_24. [DOI] [PubMed] [Google Scholar]
- 18.Lv L, Lin K, Gao W, He ZL, Gao ZM, Li ZF, Li JJ. Starvation induces autophagy of hypertrophic scar fibroblasts. Chin J Pathophysiology. 2013;29:330–333. [Google Scholar]
- 19.Xu J, Zhao J, Evan G, Xiao C, Cheng Y, Xiao J. Circulating microRNAs: Novel biomarkers for cardiovascular diseases. J Mol Med (Berl) 2012;90:865–875. doi: 10.1007/s00109-011-0840-5. [DOI] [PubMed] [Google Scholar]
- 20.Alevizos I, Illei GG. MicroRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol. 2010;6:391–398. doi: 10.1038/nrrheum.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Bei Y, Huang P, Zhou Q, Shi J, Sun Q, Zhong J, Li X, Kong X, Xiao J. Inhibition of miR-155 protects against LPS-induced cardiac dysfunction and apoptosis in mice. Mol Ther Nucleic Acids. 2016;5:e374. doi: 10.1038/mtna.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 23.Shi J, Bei Y, Kong X, Liu X, Lei Z, Xu T, Wang H, Xuan Q, Chen P, Xu J, et al. miR-17-3p contributes to exercise-induced cardiac growth and protects against myocardial ischemia-reperfusion injury. Theranostics. 2017;7:664–676. doi: 10.7150/thno.15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lv J, Yang L, Guo R, Shi Y, Zhang Z, Ye J. Ox-LDL-induced MicroRNA-155 promotes autophagy in human endothelial cells via repressing the Rheb/mTOR pathway. Cell Physiol Biochem. 2017;43:1436–1448. doi: 10.1159/000481875. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L, Zhuang L, Luo J, Chen H, Liu L, et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133–1143. doi: 10.1053/j.gastro.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 26.Li G, Zhou R, Zhang Q, Jiang B, Wu Q, Wang C. Fibroproliferative effect of microRNA-21 in hypertrophic scar derived fibroblasts. Exp Cell Res. 2015;339:360–366. doi: 10.1016/j.yexcr.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Guo B, Hui Q, Chang P, Tao K. miR-137 inhibits proliferation and metastasis of hypertrophic scar fibroblasts via targeting pleiotrophin. Cell Physiol Biochem. 2018;49:985–995. doi: 10.1159/000493236. [DOI] [PubMed] [Google Scholar]
- 28.Xu XH, Ding DF, Yong HJ, Dong CL, You N, Ye XL, Pan ML, Ma JH, You Q, Lu YB. Resveratrol transcriptionally regulates miRNA-18a-5p expression ameliorating diabetic nephropathy via increasing autophagy. Eur Rev Med Pharmacol Sci. 2017;21:4952–4965. [PubMed] [Google Scholar]
- 29.Han Y, Jo H, Cho JH, Dhanasekaran DN, Song YS. Resveratrol as a tumor-suppressive nutraceutical modulating tumor microenvironment and malignant behaviors of cancer. Int J Mol Sci. 2019;20:925. doi: 10.3390/ijms20040925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimenez-Gomez Y, Mattison JA, Pearson KJ, Martin-Montalvo A, Palacios HH, Sossong AM, Ward TM, Younts CM, Lewis K, Allard JS, et al. Resveratrol improves adipose insulin signaling and reduces the inflammatory response in adipose tissue of rhesus monkeys on high-fat, high-sugar diet. Cell Metab. 2013;18:533–545. doi: 10.1016/j.cmet.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kou X, Chen N. Resveratrol as a natural autophagy regulator for prevention and treatment of Alzheimer's disease. Nutrients. 2017;9:927. doi: 10.3390/nu9090927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu CG, Yang JM. Regulation of autophagy by a Beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;6:816–623. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehta M, Branford OA, Rolfe KJ. The evidence for natural therapeutics as potential anti-scarring agents in burn-related scarring. Burns Trauma. 2016;4:15. doi: 10.1186/s41038-016-0040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, Condorelli G, Sadoshima J. Rheb is a critical regulator of autophagy during myocardial ischemia: Pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–1146. doi: 10.1161/CIRCULATIONAHA.111.078212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng G, Zhong F, Li J, Luo S, Zhang P. Resveratrol-mediated reduction of collagen by inhibiting proliferation and producing apoptosis in human hypertrophic scar fibroblasts. Biosci Biotechnol Biochem. 2013;77:2389–2396. doi: 10.1271/bbb.130502. [DOI] [PubMed] [Google Scholar]
- 36.Bai XZ, Liu JQ, Yang LL, Fan L, He T, Su LL, Shi JH, Tang CW, Zheng Z, Hu DH. Identification of sirtuin 1 as a promising therapeutic target for hypertrophic scars. Br J Pharmacol. 2016;173:1589–1601. doi: 10.1111/bph.13460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Braunwald E. The war against heart failure: The Lancet lecture. Lancet. 2015;385:812–824. doi: 10.1016/S0140-6736(14)61889-4. [DOI] [PubMed] [Google Scholar]
- 39.Doherty J, Baehrecke EH. Life, death and autophagy. Nat Cell Biol. 2018;20:1110–1117. doi: 10.1038/s41556-018-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta. 2013;1833:3448–3459. doi: 10.1016/j.bbamcr.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Mizushima N, Komatsu M. Autophagy: Renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 43.Shi JH, Hu DH, Zhang ZF, Bai XZ, Wang HT, Zhu XX, Su YJ, Tang CW. Reduced expression of microtubule-associated protein 1 light chain 3 in hypertrophic scars. Arch Dermatol Res. 2012;304:209–215. doi: 10.1007/s00403-012-1204-x. [DOI] [PubMed] [Google Scholar]
- 44.Shi J, Xiao H, Li J, Zhang J, Li Y, Zhang J, Wang X, Bai X, Tao K, Hu D, Guan H. Wild-type p53-modulated autophagy and autophagic fibroblast apoptosis inhibit hypertrophic scar formation. Lab Invest. 2018;98:1423–1437. doi: 10.1038/s41374-018-0099-3. [DOI] [PubMed] [Google Scholar]
- 45.Shi J, Wang H, Guan H, Shi S, Li Y, Wu X, Li N, Yang C, Bai X, Cai W, et al. IL10 inhibits starvation-induced autophagy in hypertrophic scar fibroblasts via cross talk between the IL10-IL10R-STAT3 and IL10-AKT-mTOR pathways. Cell Death Dis. 2016;7:e2133. doi: 10.1038/cddis.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Felice B, Garbi C, Santoriello M, Santillo A, Wilson RR. Differential apoptosis markers in human keloids and hypertrophic scars fibroblasts. Mol Cell Biochem. 2009;327:191–201. doi: 10.1007/s11010-009-0057-x. [DOI] [PubMed] [Google Scholar]
- 47.Babuta M, Furi I, Bala S, Bukong TN, Lowe P, Catalano D, Calenda C, Kodys K, Szabo G. Dysregulated autophagy and lysosome function are linked to exosome production by Micro-RNA 155 in Alcoholic liver disease. Hepatology. 2019;70:2123–2141. doi: 10.1002/hep.30766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen LJ, Wei SY, Chiu JJ. Mechanical regulation of epigenetics in vascular biology and pathobiology. J Cell Mol Med. 2013;17:437–448. doi: 10.1111/jcmm.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Donaldson CJ, Lao KH, Zeng L. The salient role of microRNAs in atherogenesis. J Mol Cell Cardiol. 2018;122:98–113. doi: 10.1016/j.yjmcc.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 50.Wu ZY, Lu L, Liang J, Guo XR, Zhang PH, Luo SJ. Keloid microRNA expression analysis and the influence of miR-199a-5p on the proliferation of keloid fibroblasts. Genet Mol Res. 2014;13:2727–2738. doi: 10.4238/2014.April.14.2. [DOI] [PubMed] [Google Scholar]
- 51.Li C, Bai Y, Liu H, Zuo X, Yao H, Xu Y, Cao M. Comparative study of microRNA profiling in keloid fibroblast and annotation of differential expressed microRNAs. Acta Biochim Biophys Sin (Shanghai) 2013;45:692–699. doi: 10.1093/abbs/gmt057. [DOI] [PubMed] [Google Scholar]
- 52.Kozlova A, Pachera E, Maurer B, Jüngel A, Distler JHW, Kania G, Distler O. Regulation of fibroblast apoptosis and proliferation by MicroRNA-125b in systemic sclerosis. Arthritis Rheumatol. 2019;71:2068–2080. doi: 10.1002/art.41041. [DOI] [PubMed] [Google Scholar]
- 53.Zhang C, Zhang Y, Zhu H, Hu J, Xie Z. MiR-34a/miR-93 target c-Ski to modulate the proliferaton of rat cardiac fibroblasts and extracellular matrix deposition in vivo and in vitro. Cell Signal. 2018;46:145–153. doi: 10.1016/j.cellsig.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Jing Z, Han W, Sui X, Xie J, Pan H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015;356:332–338. doi: 10.1016/j.canlet.2014.09.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.