Abstract
Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) has been demonstrated to be oncogenic. The aim of the present study was to examine the effects of SNHG12 on the progression of endometrial cancer (EC). The expression levels of SNHG12 and microRNA (miR)-4429 were assessed in EC cell lines by reverse transcription-quantitative PCR. Plasmids, including SNHG12 short hairpin RNAs (shRNAs), shRNA negative control (NC), SNHG12 overexpression (OV), OV-NC, miR-4429 mimic and mimic-NC, were transfected into RL95-2 cells. Post-transfection, Cell Counting Kit-8, Transwell Matrigel and wound-healing assays were performed to assess cell proliferation, invasion and migration, respectively. Cell cycle phase distribution was assessed by flow cytometry. The protein expression levels of matrix metalloproteinase (MMP)2 and MMP9 were detected by western blotting. miR-4429 target genes were predicted by bioinformatics analysis using target prediction online tools; the findings of this analysis were verified using a dual-luciferase reporter system. Identified as a target of miR-4429, SNHG12 was overexpressed in EC cell lines with decreased expression of miR-4429. Further experiments demonstrated that SNHG12 silencing and overexpression of miR-4429 markedly suppressed proliferation, migration and invasion of RL95-2 cells, arrested cells in the G1 phase, and markedly downregulated the expression of MMP2 and MMP9. The opposite effects were observed in miR-4429 mimic-transfected RL95-2 cells after SNHG12 was overexpressed. The findings of the present study established the role of SNHG12 and miR-4429 in EC. Therefore, targeting the SNHG12/miR-4429 axis could serve as a potential future therapeutic target for treatment of EC.
Keywords: endometrial cancer, long noncoding RNA SNHG12, microRNA-4429, matrix metalloproteinases
Introduction
According to 2018 statistics by The World Cancer Research Fund, endometrial cancer (EC) is the 15th most common malignant tumor worldwide, the incidence rate of which is still on the rise (1). Patients with EC are often diagnosed when the disease is still confined to the uterus (2). Standard treatment consists of primary hysterectomy and bilateral salpingo-oophorectomy, often using minimally invasive approaches (laparoscopic or robotic). The 5-year overall survival rate ranges between 74 and 91% in patients with EC without metastatic disease (2), whereas patients with stage III or IV EC exhibit 5-year overall survival rates of 57–65 and 20–26%, respectively (3). Numerous patients with EC suffer from cancer recurrence and eventually succumb to the disease, and patients with EC relapse have poor prognosis (4). Unfortunately, effective treatments are still rare for patients with advanced and relapsed EC; therefore, there is an urgent requirement to develop new EC treatment strategies.
Increasing evidence has suggested that long non-coding RNAs (lncRNAs) and microRNAs (miRNAs/miRs) play important roles in the occurrence and development of several types of human cancer by affecting the malignant phenotypes of tumor cells, such as proliferation, invasion and apoptosis, including with breast, lung and ovarian cancer (5–8). In carcinoma cells, lncRNAs and miRNA can function as tumor suppressors or oncogenes. A recent study demonstrated that increased expression of the lncRNA HOX transcript antisense RNA may promote cell proliferation and inhibit apoptosis by binding to phosphatase and tensin homolog in EC, suggesting the important roles that lncRNAs serve in EC (9). Previous studies have demonstrated that small nucleolar RNA host gene 12 (SNHG12), a lncRNA transcribed from human chromosome 1 and located at the p35.3, is overexpressed in a few types of human cancer (10–12). SNHG12 has been reported to enhance the proliferative and migratory abilities of ovarian cancer cells by sponging miR-129 to upregulate SOX4 (13). It has also been suggested to play a pivotal role in the development of cervical cancer by sponging miR-125b (14). Recently, miRNAs have been demonstrated to be involved in the malignant behaviors of cancer by regulating cell growth and metastasis, including miR-4429 (15). lncRNA LINC00313 has been reported to modulate papillary thyroid cancer tumorigenesis by sponging miR-4429. In addition, miR-4429 can repress tumor progression and epithelial-mesenchymal transition by targeting cyclin-dependent kinase 6 (CDK6) in clear cell renal cell carcinoma, sensitize cervical cancer cells to irradiation by downregulating RAD51 recombinase (RAD51), and prevent gastric cancer progression by targeting methyltransferase like 3, thus suppressing m6A-induced stabilization of SEC62 homolog, preprotein translocation factor (16). However, it is not clear whether SNHG12 and miR-4429 act as oncogenes or anti-oncogenes in EC. Therefore, the present study aimed to examine the biological function and possible molecular mechanism of SNHG12 and miR-4429 in the tumorigenesis of EC.
Materials and methods
Cell culture
The human EC cell lines Ishikawa, KLE, RL95-2 and AN3 CA were purchased from Shanghai Cell Collection and the human normal endometrial stromal cell line ESC (BNCC340262) was obtained from BeNa Culture Collection. Cells were cultured in DMEM (Sigma-Aldrich; Merck KGaA) supplemented with 15% FBS (Sigma-Aldrich; Merck KGaA) at 37°C and 5% CO2.
Cell transfection
The sequences of short hairpin RNAs (shRNAs) targeting SNHG12 (shRNA-SNHG12-1 and shRNA-SNHG12-2) and their negative control (NC) sequence (shRNA-NC), were designed and synthesized by Shanghai GeneChem Co., Ltd, and cloned into the pSUPER-retro-puromycin plasmids (cat. no. VEC-pRT-0002; OligoEngine), according to the manufacturer's instructions. The SNHG12 overexpression plasmid (OV) was established by inserting the full-length human SNHG12 cDNA (Aksomics Inc.) into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.), whereas an empty vector served as the NC (OV-NC). miR-4429 mimics (cat. no. miR10018944-1-5) and corresponding NC mimics (mimic-NC; cat. no. miR1N0000002-1-5) were purchased from Guangzhou RiboBio Co., Ltd. After RL95-2 cells (1×106) were transfected with 50 nM plasmids, including shRNA-SNHG12-1, shRNA-SNHG12-2, shRNA-NC, OV-SNHG12, OV-NC, miR-4429 mimics and mimic-NC, using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C, the transfection efficiency was evaluated at 48 h after transfection using reverse transcription-quantitative PCR (RT-qPCR).
RT-qPCR
Total RNA was extracted from EC cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, cDNA was synthesized using a cDNA Synthesis kit (Takara Biotechnology Co., Ltd.). The reverse transcription conditions were as follows: 25°C for 10 min, 48°C for 30 min and a final step at 95°C for 5 min. RT-qPCR was conducted using SYBR-Green Master Mix (Takara Biotechnology Co., Ltd.) on an ABI 7500 fast PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with a thermocycling profile of initial denaturation for 10 min at 95°C, followed by 40 cycles at 95°C for 20 sec, 60°C for 30 sec and 72°C for 20 sec. The expression levels of SNHG12 and miR-4429 were normalized to the levels of GAPDH or U6, respectively. All reactions were repeated at least three times. The sequences of PCR primers are presented in Table I. Data were analyzed using the 2−ΔΔCq method (17) to calculate relative levels.
Table I.
Primer sequences for reverse transcription-quantitative PCR.
| Gene | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|
| SNHG12 | TCTGGTGATCGAGGACTTCC | ACCTCCTCAGTATCACACACT |
| GAPDH | CTCTGCTCCTCCTGTTCGAC | CGACCAAATCCGTTGACTCC |
| miR-4429 | GGCCAGGCAGTCTGAGTTG | GGGAGAAAAGCTGGGCTGAG |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
SNHG12, small nucleolar RNA host gene 12; miR-4429, microRNA-4429.
Cell Counting Kit-8 (CCK-8) assay
RL95-2 cells (2×103 cells/well) were seeded into a 96-well plate and incubated at 37°C in a humidified atmosphere containing 5% CO2. A total of 24, 48 and 72 h post-transfection, 10 µl CCK-8 solution (Dojindo Molecular Technologies, Inc.) was added to each well for 4 h. The absorbance (450 nm) of reduced water-soluble tetrazolium salt was evaluated using a microplate reader.
Wound-healing assay
RL95-2 cells (5×104 cells/well) were seeded in a 6-well plate and cultured for 24 h. When cells reached ~80% confluence, the cell monolayer was scratched with a 100-µl pipette tip to create an artificial homogenous wound, the cells were then washed twice with serum-free DMEM. The cells were subsequently cultured in serum-free DMEM for 72 h. The degree of scratch healing was observed with images captured in each group at 0 and 72 h using a light microscope (Olympus Corporation). The cell migration rate (%) was calculated using the following equation: [Initial distance (0 h)-final distance (72 h)]/Initial distance (0 h) ×100.
Transwell invasion assay
A Transwell Matrigel invasion assay was performed using a 24-well Transwell plate. Briefly, a 200-µl cell suspension (5×104 cells/well) in serum-free DMEM was added into the upper chamber, which was coated with Matrigel, and 600 µl DMEM supplemented with 10% FBS was added to the lower chamber. After 48 h, a cotton swab was used to wipe the cells above the Matrigel layer, whereas the cells below the membrane were fixed with 4% methanol for 10 min at room temperature and stained with 0.1% crystal violet solution for 15 min. The numbers of invasive cells in five random fields (magnification, ×200) were counted using a light microscope (Olympus Corporation).
Cell cycle assay
After transfection, RL95-2 cells were collected and fixed in precooled 70% ethanol in PBS for 12 h at 4°C. After washing with precooled PBS, the cells were resuspended in 1 mg/ml propidium iodide (PI; Sigma-Aldrich; Merck KGaA) and 0.5 mg/ml RNase A (Thermo Fisher Scientific, Inc.) in PBS for 25 min at 37°C in the dark. Finally, cell cycle analysis was conducted using the FACSCalibur flow cytometer and ModFit LT™ software (version 3.1; Verity Software House, Inc.).
Dual luciferase reporter assay
Using starBase version 3.0 (http://starbase.sysu.edu.cn/index.php), SNHG12 was discovered to be a target of miR-4229. For the dual-luciferase reporter assay, SNHG12 3′-untranslated region (UTR) sequences were amplified and cloned downstream of the luciferase reporter gene in pMIR-REPORT luciferase vectors (Thermo Fisher Scientific, Inc.). Cells (5×104 cells/well) were plated in 24-well plates and cultured until the cells were ~50% confluent. After 48 h of co-transfection with 50 nM miR-4429 mimics or mimic-NC and 30 ng/ml wild-type (WT) or mutant (MUT) firefly luciferase reporter plasmids at room temperature using Lipofectamine® 2000, cells were collected. The mutated binding site of the SNHG12 3′-UTR was created using a QuikChange Multi Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.). After 48 h of culture, the luciferase activities were measured with a dual-luciferase reporter system (Promega Corporation), and Renilla luciferase activity was used for normalization. All experiments were performed in triplicate.
Western blotting
After transfection, RL95-2 cells were lysed using a protein lysis buffer (RIPA; Beyotime Institute of Biotechnology) and the protein concentrations were measured using a micro bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). The proteins (40 µg) were separated by SDS-PAGE on 10% gels and transferred onto polyvinylidene fluoride membranes, then blocked with 5% non-fat milk in 20 mM TBS-0.1% Tween for 2 h at room temperature. The membranes were then incubated with primary antibodies, including anti-matrix metalloproteinase (MMP)2 (1:1,000; cat. no. ab92536; Abcam), anti-MMP9 (1:1,000; cat. no. ab137867; Abcam) and anti-GAPDH (1:10,000; cat. no. ab181603; Abcam) at 4°C overnight, followed by incubation with goat anti-rabbit horseradish peroxidase-conjugated immunoglobulin G secondary antibodies (1:5,000; cat. no. ab6721; Abcam) for 2 h at room temperature. Subsequently, membranes were visualized using an ECL kit (Invitrogen; Thermo Fisher Scientific, Inc.), and imaged with a gel imaging system (Bio-Rad Laboratories, Inc.). GAPDH protein levels were used for normalization. ImageJ software (version 1.47; National Institutes of Health) was used to analyze and semi-quantify the relative protein levels.
Statistical analysis
All experiments were performed at least three times. All data are presented as the mean ± SD. An unpaired Student's t-test and one-way analysis of variance (ANOVA) followed by a post hoc Tukey's test were used to analyze the statistical differences, as appropriate. The analyses were performed using SPSS 19.0 software. P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA SNHG12 is upregulated in EC cell lines, and SNHG12 silencing prevents proliferation, migration and invasion, and induces cell cycle arrest of RL95-2 cells
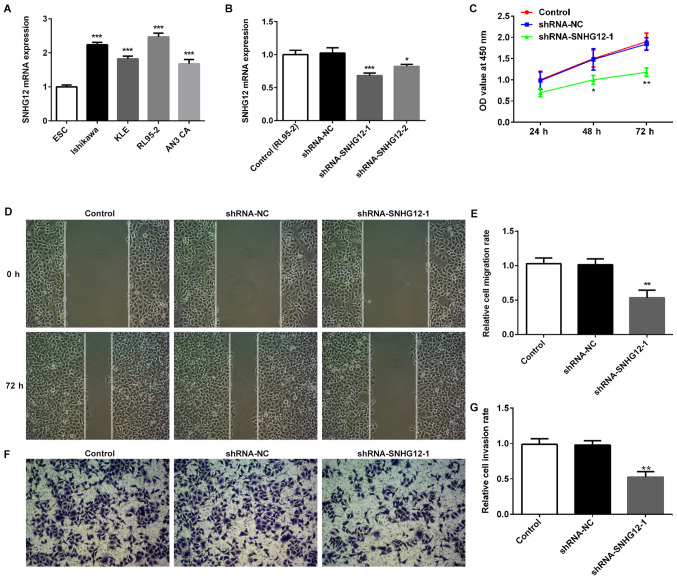
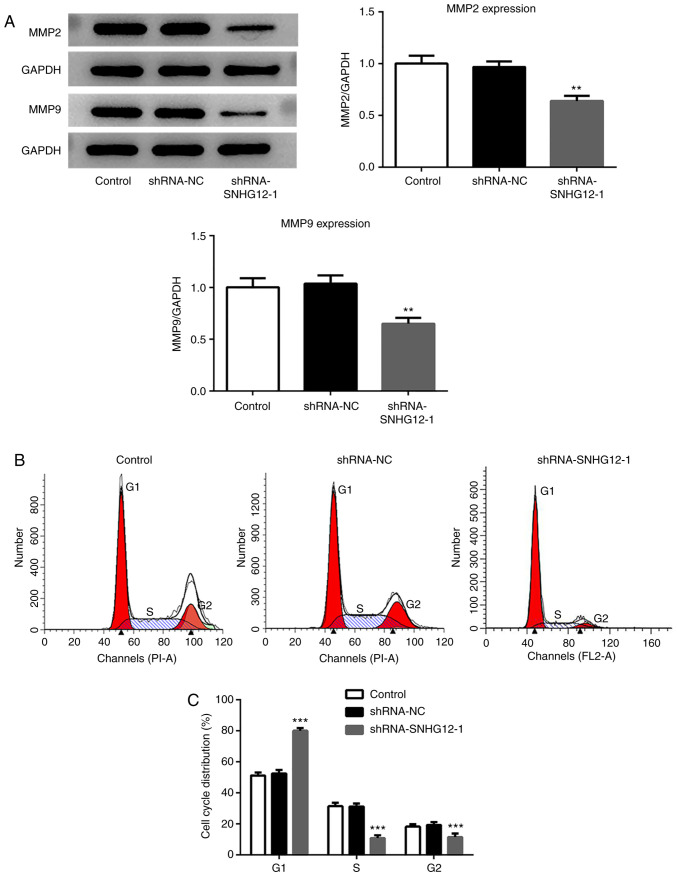
The expression of SNHG12 in EC cell lines was examined to determine whether SNHG12 was involved in EC. Results from RT-qPCR demonstrated that SNHG12 was higher in four EC cell lines, including Ishikawa, KLE, RL95-2 and AN3 CA cells, compared with the normal EC cell line, ESC (Fig. 1A). Subsequently, SNHG12 was silenced in RL95-2 cells, as that cell line possessed the highest SNHG12 mRNA levels, using shRNA-SNHG12-1 and shRNA-SNHG12-2 (Fig. 1B). As shRNA-SNHG12-1 had a greater inhibitory effect, shRNA-SNHG12-1 was selected for subsequent studies. Silencing SNHG12 significantly suppressed the proliferation of RL95-2 cells (Fig. 1C). Wound-healing assays demonstrated that SNHG12 depletion effectively prevented RL95-2 cells from entering the wounded area after 72 h, compared with the shRNA-NC and untransfected control groups, thus suggesting that shRNA-SNHG12-1 was able to inhibit cell migration (Fig. 1D and E). A Transwell Matrigel assay was also used to evaluate the invasion of RL95-2 cells transfected with shRNA-SNHG12-1. The present study demonstrated that silencing SNHG12 could significantly suppress invasion of RL95-2 cells (Fig. 1F and G). In addition, the effects of shRNA-SNHG12-1 treatment on the expression of proteins related to RL95-2 cell migration were assessed. The western blotting results demonstrated that the expression of MMP2 and MMP9 were significantly decreased in the shRNA-SNHG12-1 group when compared with the untransfected control and shRNA-NC groups (Fig. 2A).
Figure 1.
Effects of SNHG12 on EC cells. Reverse transcription-quantitative PCR was used to detect SNHG12 expression in (A) EC cell lines and ESC cells (***P<0.001 vs. ESC cells), and (B) in RL95-2 cells after transfection with shRNAs. (C) Cell Counting Kit-8 assay was performed to examine proliferation of RL95-2 cells transfected with shRNA-SNHG12-1. (D and E) Wound-healing and (F and G) Transwell assays were used to measure migration and invasion of RL95-2 cells in different treatment conditions, respectively. Magnification, ×200. Data are presented as the mean ± SD (n=3). *P<0.05, **P<0.01 and ***P<0.001 vs. control and shRNA-NC groups. EC, endometrial cancer; SNHG12, small nucleolar RNA host gene 12; shRNA, short hairpin RNA; NC, negative control; OD, optical density.
Figure 2.
Effects of SNHG12 on the expression of proteins related to migration and cell cycle progression of RL95-2 cells. (A) Western blot analysis of MMP2 and MMP9 in SNHG12-silenced RL95-2 cells. (B and C) Cell cycle distribution was assessed using flow cytometry. The percentage of cell numbers in G1, S and G2 phases was shown as plasmid the mean ± SD (n=3). **P<0.01 and ***P<0.001 vs. control and shRNA-NC groups. SNHG12, small nucleolar RNA host gene 12; MMP, matrix metalloproteinase; shRNA, short hairpin RNA; NC, negative control.
Flow cytometry was performed to determine the effects of SNHG12 on the progression of the RL95-2 cell cycle. As presented in Fig. 2B and C, shRNA-SNHG12-1 treatment led to a significant increase in the proportion of cells at the G1 phase, and significantly reduced arrest in the S and G2 phases compared with the untransfected control and shRNA-NC cells.
miR-4429 targets SNHG12 in RL95-2 cells
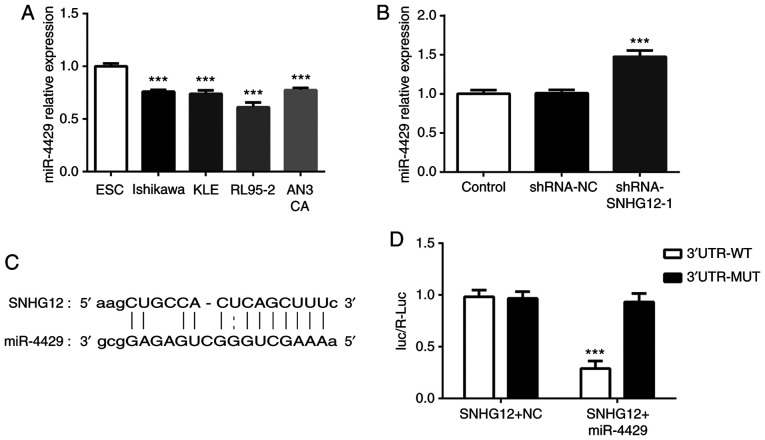
It was determined that miR-4429 was expressed at low levels in four EC cell lines compared with ESC cells (Fig. 3A). Notably, silencing SNHG12 markedly increased the level of miR-4429 in RL95-2 cells (Fig. 3B). The binding sequences of SNHG12 for miR-4429 are presented in Fig. 3C. It was observed in the luciferase reporter assay that miR-4429 significantly decreased the luciferase activity of SNHG12 WT but had no influence on that of SNHG12 MUT (Fig. 3D). These findings indicated that SNHG12 may negatively affect miR-4429 expression in EC cells.
Figure 3.
SNHG12 is a direct target of miR-4429. (A) RT-qPCR was used to examine miR-4429 expression in ESC cells and endometrial cancer cell lines. ***P<0.001 vs. ESC cells. (B) miR-4429 expression in SNHG12-silenced cells was detected by RT-qPCR. ***P<0.001 vs. control and shRNA-NC groups. (C) Putative binding site of miR-4429 in the 3′-UTR of SNHG12. (D) Analysis of luciferase activity. Relative luciferase activity was reduced when cells were co-transfected with miR-4429 mimic and pMIR-SNHG12-3′-UTR (WT). ***P<0.001 vs. mimic-NC group. Data are presented as the mean ± SD (n=3). SNHG12, small nucleolar RNA host gene 12; miR, microRNA; RT-qPCR, reverse transcription quantitative PCR; UTR, untranslated region; WT, wild-type; MUT, mutant.
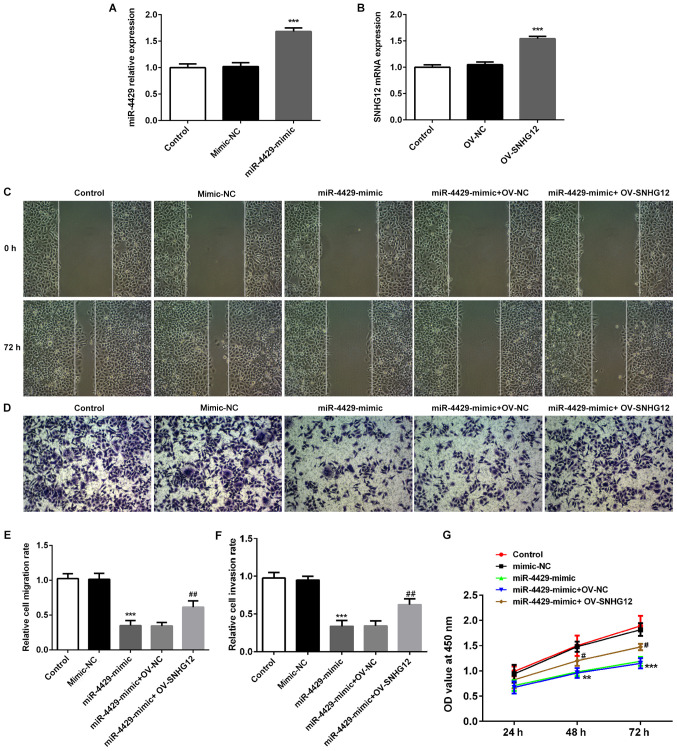
miR-4429 suppresses cell proliferation and induces cell cycle arrest at the S phase via SNHG12 in RL95-2 cells
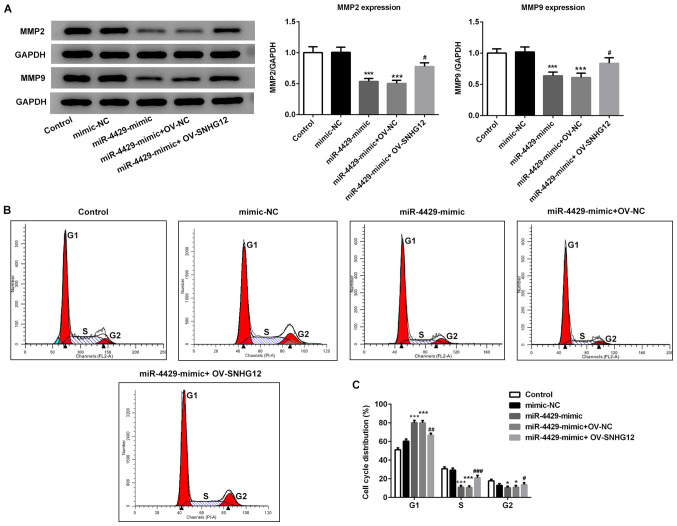
miR-4429 was overexpressed in RL95-2 cells by transfection with miR-4429 mimic (Fig. 4A) and overexpression of SNHG12 was also induced in RL95-2 cells using OV-SNHG12 plasmids (Fig. 4B). The rescue assays were performed to evaluate whether miR-4429 regulates EC procession by targeting SNHG12. As presented in Fig. 4C-F, the migration and invasion of RL95-2 cells were significantly decreased upon transection with the miR-4429 mimic; however, they were rescued by overexpression of SNHG12. Additionally, the miR-4429 mimic suppressed the proliferation of RL95-2 cells, which was then partially reversed by OV-SNHG12 (Fig. 4G). Furthermore, the expression levels of proteins related to migration in response to miR-4429 overexpression were detected by western blotting. As presented in Fig. 5A, the results demonstrated that transfection with miR-4429 mimic significantly decreased the expression of MMP2 and MMP9 compared with the control and mimic-NC groups. However, RL95-2 cells in the miR-4429 mimic + OV-SNHG12 group presented higher expression levels of MMP2 and MMP9 when compared with the cells in the miR-4429 mimic + OV-NC group. The flow cytometry data demonstrated that the miR-4429 mimic increased the number of cells at G1 phase, whereas the percentage of cells in the S phase were significantly decreased compared with the mimic-NC group (Fig. 5B and C). By contrast, OV-SNHG12 effectively reversed cell cycle arrest at G1 and increased the number of cells at S phase compared with the miR-4429 mimic + OV-NC group (Fig. 5B and C). Taken together, these findings suggested that miR-4429 suppressed EC progression and induced cell cycle arrest through the targeting of SNHG12.
Figure 4.
Overexpression of miR-4429 inhibits endometrial cancer migration, invasion and proliferation through targeting SNHG12 in RL95-2 cells. (A) Expression levels of miR-4429 in RL95-2 cells transfected with a miR-4429 mimic were assessed using RT-qPCR. ***P<0.001 vs. control and mimic-NC groups. (B) SNHG12 expression in RL95-2 cells transfected with OV-SNHG12 was detected using RT-qPCR. ***P<0.001 vs. control and OV-NC groups. Data are presented as the mean ± SD (n=3). (C-F) RL95-2 cells were transfected with mimic-NC, miR-4429 mimic, miR-4429 mimic + OV-NC and miR-4429 mimic + OV-SNHG12, respectively. The capacities of (C and E) migration and (D and F) invasion of RL95-2 cells were evaluated using wound healing and Transwell assays. Magnification, ×200. (G) Proliferation of RL95-2 cells was detected by Cell Counting Kit-8 assay. Data are represented as the mean ± SD (n=3). **P<0.01 and ***P<0.001 vs. control and mimic-NC groups; #P<0.05 and ##P<0.01 vs. miR-4429 mimic + OV-NC group. SNHG12, small nucleolar RNA host gene 12; miR, microRNA; RT-qPCR, reverse transcription quantitative PCR; NC, negative control; OV, overexpression plasmid; OD, optical density.
Figure 5.
Overexpression of miR-4429 inhibits migration-related proteins and induces cell cycle arrest of RL95-2 cells via SNHG12. RL95-2 cells were transfected with mimic-NC, miR-4429 mimic, miR-4429 mimic + OV-NC and miR-4429 mimic + OV-SNHG12, respectively. (A) Protein expression levels of MMP2 and MMP9 in RL95-2 cells were determined by western blotting. (B and C) Flow cytometry was performed to detect changes of cell cycle distribution in RL95-2 cells. Data are represented as mean ± SD (n=3). *P<0.05 and ***P<0.001 vs. mimic-NC group; #P<0.05, ##P<0.01 and ###P<0.001 vs. miR-4429 mimic + OV-NC group. SNHG12, small nucleolar RNA host gene 12; miR, microRNA; MMP, matrix metalloproteinase; NC, negative control; OV, overexpression plasmid.
Discussion
EC is the most common gynecologic malignancy. The main treatment for EC is total hysterectomy with bilateral salpingo-oophorectomy (18). However, the prognosis and survival rate are not satisfactory. Despite the roles that radiation and chemotherapy serve in EC treatment, and the promising therapies in drug development for the treatment of advanced/recurrent EC, no targeted therapies beyond hormonal therapy have been approved at present (19). Furthermore, early diagnosis is key to improving the probability of EC survival, which has a <20% survival rate for the advanced disease at 5 years but >90% survival rate for the early-stage disease (20). However, to the best of our knowledge, there are no validated biological markers for its early detection. Thus, there is an urgent need for scientifically validated therapy with predictive biomarkers. In the present study, it was demonstrated that SNHG12 was significantly upregulated in EC cells, which was negatively associated with miR-4429. Therefore, SNHG12 and miR-4429 may represent biomarkers that can be used for early diagnosis and targeted therapy of EC.
lncRNAs, miRNAs, piwiRNAs and other non-coding RNAs comprise ~90% of human genomes and can act as oncogenes or tumor suppressor genes (21,22). SNHG12, also known as LNC04080, is 1,867 bases in length and is involved in a number of human tumors, including non-small cell lung cancer, gastric cancer, triple-negative breast cancer, hepatocellular carcinoma, colorectal cancer, cervical cancer and ovarian cancer (23). Changes in the expression levels of SNHG12 have been reported to be associated with viability, proliferation, migration and invasion of tumor cells, thus affecting the prognosis and survival of patients with cancer (24). In the present study, SNHG12 silencing inhibited the proliferation, migration and invasion of EC cells, and induced cell cycle arrest in the G1 phase, which may enhance apoptosis. In addition, SNHG12 silencing markedly inhibited the expression of MMP2 and MMP9, which are key molecules in cancer invasion and metastasis (25). The present data identified a potential mechanism on how SNHG12 depletion may inhibit cell proliferation, invasion and migration.
SNHG12 also acts as a competitive endogenous RNA by harboring multiple miRNA-binding sites, thereby regulating their downstream targets by ‘sponging’ these miRNAs (23). Given that the interaction between lncRNAs and their mRNA targets has been predicted to rely on miRNAs, an increased density of miRNA response elements may increase the likelihood of lncRNAs to share miRNAs to communicate with and coregulate each other (26). The miRNAs that have been reported to be regulated by SNHG12 include miR-195 (27) and miR-101-3p (28), which have roles in the regulation of cell proliferation, and miR-424-5p and miR-125b, which regulate invasion and metastasis (14). Through the TargetScan Human prediction site, it was identified that miR-4429 potentially interacted with SNHG12. Previous studies have demonstrated that miR-4429 can be sponged by lncRNA LINC00313 to modulate papillary thyroid cancer tumorigenesis (16). miR-4429 was also demonstrated to sensitize cervical cancer cells to irradiation by targeting RAD51 (29) and inhibit the progression of clear cell renal cell carcinoma by targeting CDK6 (30). These findings demonstrated that miR-4429 may function as a tumor suppressor gene in several types of cancer. The present study demonstrated that miR-4429 mimic inhibited EC cell proliferation, migration and invasion, and at least partially inhibited cell cycle arrest. The present study also demonstrated the inhibitory effect of miR-4429 mimic on the proliferation of RL95-2 cells by enhancing the number of cells in G1 phase. Notably, the number of cells in G1 phase was increased after mimic-NC transfection compared with in the untransfected control group; it was suspected that the reason may be the mimic-NC plasmid affected the G1 phase of the RL95-2 cell cycle, whereas it had no significant effect on the S phase and G2 phase. However, in general, the miR-4429 mimic plasmid had a significant effect on all phases the cell cycle. All these suppressive effects of miR-4429 mimic on EC cells were attenuated by SNHG12 overexpression.
In summary, the present data demonstrated that silencing of SNHG12 was able to significantly suppress cell viability and cause cell cycle arrest, which was similar to the effects of the miR-4429 mimic. To the best of our knowledge, this is the first study to identify SNHG12 as a target molecule of miR-4429, which may serve as potential diagnostic and prognostic biomarkers, or targets for novel therapeutic strategies for EC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PYC and MXW designed the study, wrote the manuscript and performed the cell culture and molecular biology experiments; BZ, SYW and HYW performed the cell culture and molecular biology experiments and analyzed the data; and LW designed the study and critically revised the manuscript for intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Brooks RA, Fleming GF, Lastra RR, Lee NK, Moroney JW, Son CH, Tatebe K, Veneris JL. Current recommendations and recent progress in endometrial cancer. CA Cancer J Clin. 2019;69:2842–279. doi: 10.3322/caac.21561. [DOI] [PubMed] [Google Scholar]
- 2.Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387:1094–1108. doi: 10.1016/S0140-6736(15)00130-0. [DOI] [PubMed] [Google Scholar]
- 3.Van Nyen T, Moiola CP, Colas E, Annibali D, Amant F. Modeling endometrial cancer: Past, present, and future. Int J Mol Sci. 2018;19:2348. doi: 10.3390/ijms19082348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou M, Zhong L, Xu W, Sun Y, Zhang Z, Zhao H, Yang L, Sun J. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci Rep. 2016;6:31038. doi: 10.1038/srep31038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi H, Cheng L, Sun J. Relapse-related long non-coding RNA signature to improve prognosis prediction of lung adenocarcinoma. Oncotarget. 2016;7:29720–29738. doi: 10.18632/oncotarget.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao H, Zhong Z, Sun J. Comprehensive analysis of lncRNA expression profiles reveals a novel lncRNA signature to discriminate nonequivalent outcomes in patients with ovarian cancer. Oncotarget. 2016;7:32433–32448. doi: 10.18632/oncotarget.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Xue W, Lv J, Han P, Liu Y, Cui B. Identification of potential long non-coding RNA biomarkers associated with the progression of colon cancer. Oncotarget. 2017;8:75834–75843. doi: 10.18632/oncotarget.17924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang XH, Hu P, Xie YQ, Kang YJ, Li M. lncRNA HOTAIR promotes endometrial carcinoma cells proliferation by binding to PTEN via activating PI3k/Akt signaling pathway. Mol Cell Biol. 2019;39:e00251–e00219. doi: 10.1128/MCB.00251-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu ZB, Tang C, Jin X, Liu SH, Pi W. Increased expression of lncRNA SNHG12 predicts a poor prognosis of nasopharyngeal carcinoma and regulates cell proliferation and metastasis by modulating notch signal pathway. Cancer Biomark. 2018;23:603–613. doi: 10.3233/CBM-181873. [DOI] [PubMed] [Google Scholar]
- 11.Feng X, Dong X, Wu D, Zhao H, Xu C, Li H. Long noncoding RNA small nucleolar RNA host gene 12 promotes papillary thyroid carcinoma cell growth and invasion by targeting miR-16-5p. Histol Histopathol. 2020;35:217–224. doi: 10.14670/HH-18-155. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Zhou J, Wang S, Song Y, Zhou J, Ren F. Long non-coding RNA SNHG12 promotes proliferation and invasion of colorectal cancer cells by acting as a molecular sponge of microRNA-16. Exp Ther Med. 2019;18:1212–1220. doi: 10.3892/etm.2019.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun D, Fan XH. LncRNA SNHG12 accelerates the progression of ovarian cancer via absorbing miRNA-129 to upregulate SOX4. Eur Rev Med Pharmacol Sci. 2019;23:2345–2352. doi: 10.26355/eurrev_201903_17378. [DOI] [PubMed] [Google Scholar]
- 14.Jin XJ, Chen XJ, Zhang ZF, Hu WS, Ou RY, Li S, Xue JS, Chen LL, Hu Y, Zhu H. Long noncoding RNA SNHG12 promotes the progression of cervical cancer via modulating miR-125b/STAT3 axis. J Cell Physiol. 2019;234:6624–6632. doi: 10.1002/jcp.27403. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Chen R, Liu L. SP1-DLEU1-miR-4429 feedback loop promotes cell proliferative and anti-apoptotic abilities in human glioblastoma. Biosci Rep. 2019;39:BSR20190994. doi: 10.1042/BSR20190994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu WJ, Yin H, Hu JJ, Wei XZ. Long noncoding RNA LINC00313 modulates papillary thyroid cancer tumorigenesis via sponging miR-4429. Neoplasma. 2018;65:933–942. doi: 10.4149/neo_2018_180219N125. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Braun MM, Overbeek-Wager EA, Grumbo RJ. Diagnosis and management of endometrial cancer. Am Fam Physician. 2016;93:468–474. [PubMed] [Google Scholar]
- 19.Lee YC, Lheureux S, Oza AM. Treatment strategies for endometrial cancer: Current practice and perspective. Curr Opin Obstet Gynecol. 2017;29:47–58. doi: 10.1097/GCO.0000000000000338. [DOI] [PubMed] [Google Scholar]
- 20.Njoku K, Chiasserini D, Whetton AD, Crosbie EJ. Proteomic biomarkers for the detection of endometrial cancer. Cancers (Basel) 2019;11:1572. doi: 10.3390/cancers11101572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khandelwal A, Malhotra A, Jain M, Vasquez KM, Jain A. The emerging role of long non-coding RNA in gallbladder cancer pathogenesis. Biochimie. 2017;132:152–160. doi: 10.1016/j.biochi.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhotra A, Sharma U, Puhan S, Chandra Bandari N, Kharb A, Arifa PP, Thakur L, Prakash H, Vasquez KM, Jain A. Stabilization of miRNAs in esophageal cancer contributes to radioresistance and limits efficacy of therapy. Biochimie. 2019;156:148–157. doi: 10.1016/j.biochi.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamang S, Acharya V, Roy D, Sharma R, Aryaa A, Sharma U, Khandelwal A, Prakash H, Vasquez KM, Jain A. SNHG12: An LncRNA as a potential therapeutic target and biomarker for human cancer. Front Oncol. 2019;9:901. doi: 10.3389/fonc.2019.00901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan T, Ma W, Hong Z, Wu L, Chen X, Yuan Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36:11. doi: 10.1186/s13046-016-0486-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P, Hu MD, Deng XF, Li B. Genistein reinforces the inhibitory effect of cisplatin on liver cancer recurrence and metastasis after curative hepatectomy. Asian Pac J Cancer Prev. 2013;14:759–764. doi: 10.7314/APJCP.2013.14.2.759. [DOI] [PubMed] [Google Scholar]
- 26.Tan JY, Marques AC. miRNA-mediated crosstalk between transcripts: The missing ‘linc’? Bioessays. 2016;38:295–301. doi: 10.1002/bies.201500148. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang Z, Li Z, Zhang L, Liu Y. Inhibition of TDP43-mediated SNHG12-miR-195-SOX5 feedback loop impeded malignant biological behaviors of glioma cells. Mol Ther Nucleic Acids. 2018;10:142–158. doi: 10.1016/j.omtn.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Liu J, Chu L, Yang W, Liu H, Li C, Yang J. Long noncoding RNA SNHG12 facilitates the tumorigenesis of glioma through miR-101-3p/FOXP1 axis. Gene. 2018;676:315–321. doi: 10.1016/j.gene.2018.08.034. [DOI] [PubMed] [Google Scholar]
- 29.Sun H, Fan G, Deng C, Wu L. miR-4429 sensitized cervical cancer cells to irradiation by targeting RAD51. J Cell Physiol. 2020;235:185–193. doi: 10.1002/jcp.28957. [DOI] [PubMed] [Google Scholar]
- 30.Pan H, Hong Y, Yu B, Li L, Zhang X. miR-4429 inhibits tumor progression and epithelial-mesenchymal transition via targeting CDK6 in clear cell renal cell carcinoma. Cancer Biother Radiopharm. 2019;34:334–341. doi: 10.1089/cbr.2018.2697. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.