Abstract
Infantile hemangioma (IH) is one of the most common vascular tumors that occurs during childhood, but its pathogenesis is currently not completely understood. Even though lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) plays vital roles in tumorigenesis of malignant tumors, its roles in IH remain unclear. Therefore, we evaluate the function of lncRNA NEAT1 in IH. Reverse transcription-quantitative PCR indicated that IH tissues exhibited high expression levels of NEAT1 and hypoxia-inducible factor 1α (HIF1α), and low expression levels of the microRNA (miR)-33a-5p. Small interfering RNA-mediated depletion of NEAT1 suppressed hemangioma endothelial cell (HemEC) proliferation, migration and invasion. The data suggested that NEAT1 positively regulated HIF1α expression by sponging miR-33a-5p in HemECs. miR-33a-5p overexpression or HIF1α silencing also acted to suppress HemEC proliferation, migration and invasion. Furthermore, the results indicated that the NEAT1/miR-33a-5p/HIF1α axis regulated the NF-κB signaling pathway. Collectively, the results revealed that depletion of lncRNA NEAT1 suppressed the tumorigenesis of IH by competitively binding miR-33a-5p and thereby stimulating the HIF1α/NF-κB signaling pathway.
Keywords: nuclear paraspeckle assembly transcript 1, microRNA-33a-5p, hypoxia-inducible factor 1α, NF-κB signaling pathway, infantile hemangioma
Introduction
Infantile hemangioma (IH) is one of the most common vascular tumors that occurs during childhood (1). IH can lead to life-threatening disease owing to rapid growth and invasion of tumor cells. IHs consist of two phases: The proliferating phase and the involuting phase. The proliferating phase is characterized by dysregulated proliferation of the immature endothelial cells, whereas the involuting phase is characterized by the appearance of larger and fewer capillary-like vessels (2). Although several genes have been reported to be involved during the development of IH (3,4), the mechanism underlying IH progression is not yet completely understood.
Increasing evidence suggests that non-coding RNAs serve important roles during the progression of a number of diseases (5). Long non-coding RNAs (lncRNAs) are a type of non-coding RNA that are >200 nucleotides in length. Recently, a number of lncRNAs have been reported to have crucial roles during cancer cell proliferation, apoptosis and metastasis (6,7). Microarray and RNA-sequencing methods have previously been used to analyze lncRNA expression profiles during IH, which identified differentially expressed lncRNAs and microRNAs (miRNAs/miRs), and allowed for competing endogenous RNA (ceRNA) and lncRNA-mRNA co-expression networks to be constructed (8,9). Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is upregulated in IH tissues, and loss of MALAT1 expression has been shown to inhibit IH cell proliferation, migration and tube formation, and also promotes cell apoptosis by sponging miR-424 to inactivate the MEKK3/NF-κB signaling pathway (10). Furthermore, the lncRNA linc00152 is upregulated in IH tissues, and loss of lin00152 expression inhibits cell proliferation and induces cell apoptosis by inactivating the AKT/mTOR and NOTCH1 signaling pathways (11). The expression of linc00152 in hemangioma tissues was higher in the proliferating phase compared with the involuting phase. Depletion of linc00152 inhibited hemangioma tumor progression of by sponging miR-139-5p and by downregulating its target gene tumor protein D52 (12).
Nuclear paraspeckle assembly transcript 1 (NEAT1) is an oncogenic lncRNA that confers docetaxel resistance in prostate cancer cells by sponging miR-204-5p and miR-34a-5p (13). NEAT1 has been shown to enhance bladder cancer progression by negatively regulating miR-410 and positively regulating downstream HMGB1 (14). During endometrial cancer, NEAT1 promotes cell proliferation, migration and invasion by sponging miR-144-3p, leading to the activation of its target gene enhancer, zeste 2 polycomb repressive complex 2 subunit (15). However, the roles and mechanisms underlying NEAT1 during IH progression are not yet completely understood. Hypoxia-inducible factor 1α (HIF1α) is a crucial regulator during hypoxia-induced angiogenesis, but it is also involved in the growth of hemangiomas (16,17). Curcumin-mediated downregulation of HIF1α inhibits the proliferation of human hemangioma endothelial cells (HemECs) (18). Furthermore, by using starBase, NEAT1 was predicted as a ceRNA that regulates HIF1α by sponging miR-33a-5p (19). Therefore, the roles of the NEAT1/miR-33a-5p/HIF1α axis during the tumorigenesis of IH along with the downstream regulatory mechanisms were assessed in the present study.
Materials and methods
Tissue samples and cell line
Tissue specimens (n=12 patients/group), including normal subcutaneous tissues and infantile hemangioma tissues in the involuting stage (9 females and 3 males; median age, 7 months) and proliferating stage (10 females and 2 males; median age, 6 months), were obtained from patients at Kunming Children's Hospital (Kunming, China) between June 2016 and September 2018. Tissue samples were immediately frozen at −80°C until further analysis. The present study was approved by the Ethics Committee of Kunming Children's Hospital (approval no. 2018-005). Written informed consent was obtained from the parents/legal guardians of each patient.
HemECs were isolated from infant hemangioma tissues in the proliferating phase as previously described (20). HemECs were cultured in human endothelial-serum free medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) with 5% CO2 at 37°C.
Cell transfection
miR-33a-5p mimic, mimic negative control (NC) and small interfering (si)RNAs targeted against NEAT1 and HIF1α were purchased from Shanghai GenePharma Co., Ltd. The sequences were as follows: miR-33a-5p mimic, 5′-GTGCATTGTAGTTGCATTGCA-3′; mimic NC, 5′-TTCTCCGAACGTGTCACGTT-3′; NEAT1 si1, 5′-GCCATCAGCTTTGAATAAATT-3′; NEAT1 si2, 5′-TGGCTAGCTCAGGGCTTCAG-3′; HIF1α si1, 5′-GCAAGACGTTGTTTGAAATTT-3′; HIF1α si2, 5′-ACACACTGTGTCCAGTTAG-3′; NC siRNA, 5′-ACTGTTCTATGACTTGTCGTGAATA-3′. HemECs at 50–60% confluency were transfected with siRNA (50 nM) or miR-33a-5p mimic (50 nM) using Lipofectamine® 3000 (Gibco; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following incubation for 48 h at 37°C, reverse transcription-quantitative PCR (RT-qPCR) and western blotting were performed.
Cell proliferation
At 24 h post-transfection, HemECs (1×104 cells/well) were seeded into 96-well plates and cultured for 24, 72 or 120 h at 37°C. Subsequently, 10 µl Cell Counting Kit-8 (CCK-8) solution (Dojindo Molecular Technologies, Inc.) was added to each well and incubated at 37°C for 1 h, according to the manufacturer's protocol. The absorbance of each well was measured at a wavelength of 450 nm using a Bio-Rad 680 microplate reader (Bio-Rad Laboratories, Inc.). The relative proliferation was calculated by normalizing the absorbance of the NEAT1 si1- or si2-transfected cells to that of the NC group at the same time point.
Migration and invasion assays
To assess invasion, the Transwell membranes were coated with Matrigel® (BD Biosciences) for 1 h at 37°C. Subsequently, the membranes were hydrated in 100% FBS for 2 h at 37°C. A total of 1×105 cells in serum-free culture medium were seeded into the upper chamber. Culture medium with 20% FBS was plated into the lower chambers. Following incubation at 37°C for 36 h, the Transwell membranes were stained with 0.1% crystal violet solution for 10 min at room temperature. The average number of invasive cells per field was assessed by counting 10 random fields under a light microscope (magnification, ×40). To assess migration, the aforementioned protocol was performed; however, the Transwell membranes were not pre-coated with Matrigel®.
RT-qPCR
Total RNA was extracted from tissues and cells using the RNeasy Mini kit (Qiagen GmbH), according to the manufacturer's protocol. Total RNA was reverse-transcribed and miR-33a-5p expression was detected using the Hairpin-it™ qRT-PCR Primer Set for miR-33a-5p (Shanghai GenePharma Co., Ltd.). The other mRNAs were reverse-transcribed into cDNA using M-MLV reverse transcriptase (BioTeke Corporation) in the presence of oligo(dT) (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The expression levels of NEAT1, HIF1α and c-myc were determined by qPCR using the Power SYBR™ Green PCR master mix (Thermo Fisher Scientific, Inc.), according to the manufacturer's instruction. The PCR procedure was as follows: 95°C for 10 min; followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The following primer pairs were used for qPCR: NEAT1 forward, 5′-GTGGCTGTTGGAGTCGGTAT-3′, reverse 5′-TAACAAACCACGGTCCATGA-3′; c-myc, forward 5′-GGCTCCTGGCAAAAGGTCA-3′, reverse 5′-CTGCGTAGTTGTGCTGATGT-3′; HIF1α, forward 5′-GAACGTCGAAAAGAAAAGTCTCG-3′, reverse 5′-CCTTATCAAGATGCGAACTCACA-3′; GAPDH, forward 5′-CCAGGTGGTCTCCTCTGA-3′, reverse 5′-GCTGTAGCCAAATCGTTGT-3′; U6, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse 5′-CGCTTCACGAATTTGCGTGTCAT-3′. mRNA and miRNA expression levels were quantified using the 2−ΔΔCq method (21), and normalized to the internal reference genes GAPDH and U6, respectively.
Western blotting
Total protein was extracted using RIPA protein extraction buffer (Beyotime Institute of Biotechnology) and quantified using the BCA Protein assay kit (Beyotime Institute of Biotechnology). Equal amounts of proteins (7–10 µg) were separated by 10% SDS-PAGE and transferred to a PVDF membrane (EMD Millipore). After blocking with 5% skimmed milk for 1 h at room temperature, the membrane was incubated at 4°C overnight with the following primary antibodies: Anti-HIF1α (cat. no. ab51608; 1:1,000; Abcam), anti-NF-κB (p65; cat. no. ab16502; 1:1,000; Abcam), anti-phosphorylated (p)-NF-κB (p-p65; cat. no. ab86299; 1:1,000; Abcam) and anti-β-actin (cat. no. 66009-1-IG; 1:20,000; ProteinTech Group, Inc.). Subsequently, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5,000; cat. no. 5210-0174) and goat anti-mouse IgG (1:5,000; cat. no. 5210-0185; KPL, Inc.) secondary antibodies. Protein bands were visualized by Immobilon Enhanced Chemiluminescence (EMD Millipore). β-actin was used as the loading control. Optical density values of the protein bands were semi-quantified and analyzed using a gel image processing system Image Lab software (version 3.0; Bio-Rad Laboratories, Inc.).
Dual-luciferase reporter assay
A Dual-Luciferase Reporter assay system (Promega Corporation) was used to detect the binding between miR-33a-5p and lncRNA NEAT1 or 3′-untranslated region (UTR) of HIF1α according to the manufacturer's protocol. HemECs (5×105 cells/well) were added to a 6-well plate and cultured with 5% CO2 at 37°C. After 24 h, cells were co-transfected with the pGL3-NEAT1 WT, or pGL3-NEAT1 Mut (pGL3-HIF1α 3′UTR WT or pGL3-HIF1α 3′UTR Mut) and miR-33a-5p mimic/mimic NC using Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Following incubation for 48 h at 37°C, luciferase activities were measured. The firefly luciferase enzyme activity was normalized to Renilla luciferase enzyme activity.
StarBase database
The starBase v2.0 database (http://starbase.sysu.edu.cn/) is a powerful database to study non-coding RNAs, such as lncRNA, miRNA and circRNA. The starBase database was used to predict the binding between NEAT1 and miR-33a-5p, and between miR-33a-5p and the 3′UTR of HIF1α.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software (version 6; GraphPad Software, Inc.). Data are presented as the mean ± SD. Student's t-test and ANOVA (followed by Tukey's post hoc test for multiple comparisons) were used to analyze the differences. Correlations were analyzed by Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
NEAT1 and HIF1α expression levels are increased and miR-33a-5p expression is decreased in IH tissues
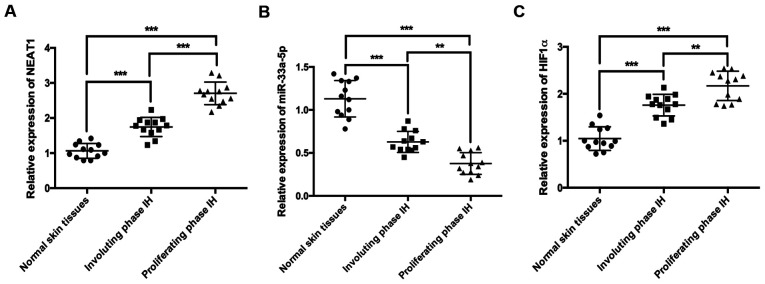
To investigate the associations between NEAT1, HIF1α and miR-33a-5p during IH, the expression levels of the three molecules were detected in normal skin, proliferating phase IH and involuting phase IH tissues by RT-qPCR. NEAT1 was significantly upregulated in IH tissues compared with normal skin tissues (Fig. 1A), and its expression was also significantly higher in proliferating phase IH tissues compared with involuting phase IH tissues (Fig. 1A). By contrast, the expression of miR-33a-5p was significantly lower in IH tissues compared with normal skin tissues (Fig. 1B), and its expression in proliferating phase IH tissues was significantly lower compared with involuting phase IH tissues (Fig. 1B). HIF1α expression was significantly increased in IH tissues compared with normal skin tissues (Fig. 1C), and the expression of HIF1α in proliferating phase IH tissues was also significantly higher compared with involuting phase tissues (Fig. 1C).
Figure 1.
NEAT1 and HIF1α are highly expressed, and miR-33a-5p expression is decreased in IH tissues. The expression levels of (A) NEAT1, (B) miR-33a-5p and (C) HIF1α were analyzed by reverse transcription-quantitative PCR. The data were analyzed by ANOVA followed by Tukey's post hoc test. **P<0.01 and ***P<0.001. HIF1α, hypoxia-inducible factor 1α; IH, infantile hemangioma; miR, microRNA; NEAT1, nuclear paraspeckle assembly transcript 1.
Depletion of NEAT1 mRNA suppresses HemEC proliferation, migration and invasion
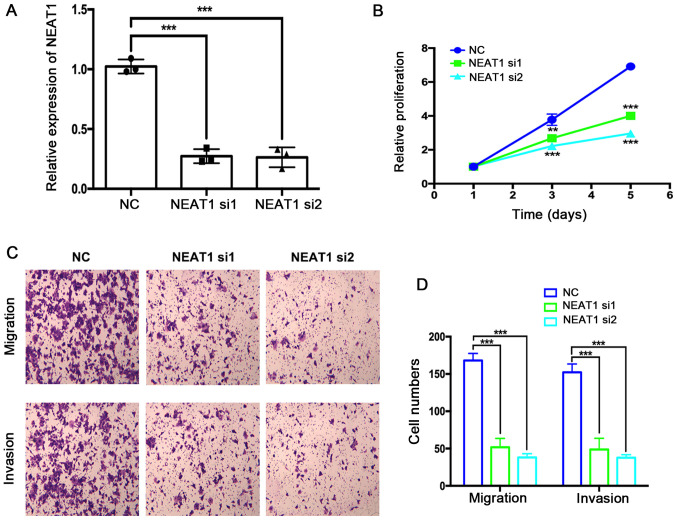
The effects of NEAT1 siRNA transfection on HemEC proliferation, migration and invasion were assessed using the CCK-8, Transwell and Matrigel assays, respectively. First, the efficiency of NEAT1 mRNA depletion by two NEAT1-targeting siRNAs was verified by RT-qPCR (Fig. 2A). NEAT1-depleted HemECs exhibited significantly decreased proliferation compared with the NC group (Fig. 2B). The results of the Transwell and Matrigel assays suggested that the migration and invasion of HemECs were also markedly suppressed by depletion of NEAT1 mRNA expression levels (Fig. 2C and D).
Figure 2.
NEAT1 depletion suppresses HemEC proliferation, migration and invasion. (A) The expression levels of NEAT1 were analyzed by reverse transcription-quantitative PCR following siRNA transfection. (B) The relative proliferation of HemECs were analyzed using a Cell Counting Kit-8 assay. (C and D) The migratory and invasive abilities of HemECs were analyzed using Transwell and Matrigel assays, respectively. The data were analyzed by ANOVA followed by Tukey's post hoc test. **P<0.01 and ***P<0.001 vs. NC group at the same time point or as indicated in figure. HemEC, hemangioma endothelial cells; NEAT1, nuclear paraspeckle assembly transcript 1; NC, negative control; si, small interfering RNA.
NEAT1 regulates HIF1α expression by sponging miR-33a-5p in HemECs
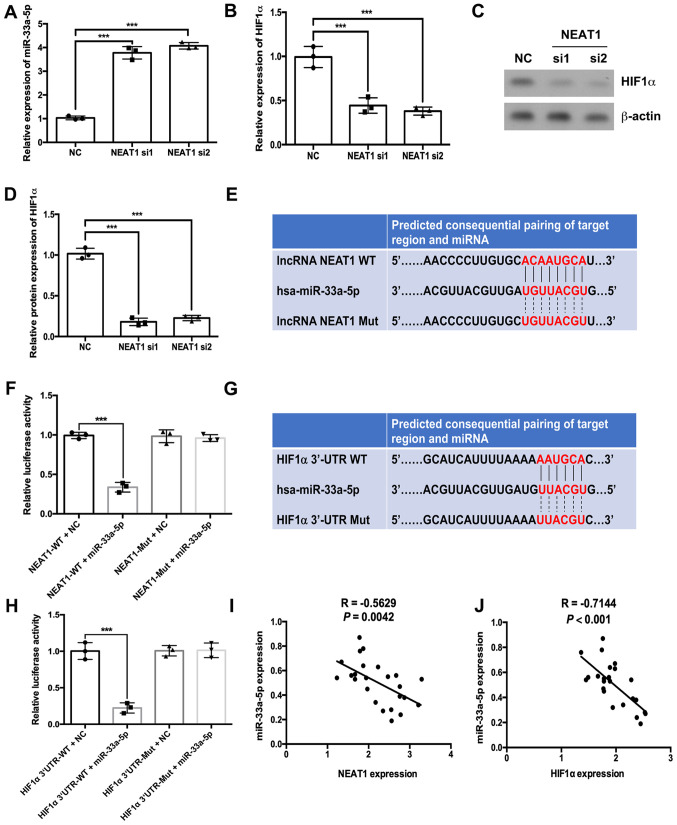
RT-qPCR results indicated that depletion of NEAT1 significantly increased the expression levels of miR-33a-5p and significantly decreased the mRNA expression of HIF1α (Fig. 3A and B, respectively). Western blotting results further demonstrated that depletion of NEAT1 decreased HIF1α protein expression levels (Fig. 3C and D). Subsequently, a dual-luciferase assay was performed to evaluate the associations between NEAT1, miR-33a-5p and HIF1α. Bioinformatics analysis conducted using the StarBase database suggested that miR-33a-5p could bind to NEAT1 (Fig. 3E). Results from the dual-luciferase assay suggested that the luciferase activities of wild-type NEAT1 were decreased by miR-33a-5p mimic compared with the miRNA NC group (Fig. 3F). However, in the mutated NEAT1 group, the luciferase activities were not significantly altered in the miR-33a-5p mimic group compared with the NC group (Fig. 3F). The starBase database also predicted binding sites between miR-33a-5p and the 3′UTR of HIF1α (Fig. 3G). The luciferase activity of the HIF1α 3′UTR-WT + miR-33a-5p group was lower compared with the HIF1α 3′UTR-WT + NC group (Fig. 3H). However, in the HIF1α 3′UTR-Mut group, the luciferase activities of the miR-33a-5p mimic group were not significantly different compared with the NC group (Fig. 3H). Notably, the expression of miR-33a-5p was negatively correlated with NEAT1 and HIF1α expressions in IH tissues (Fig. 3I and J, respectively).
Figure 3.
NEAT1 regulates HIF1α expression by sponging miR-33a-5p in HemECs. The expression levels of (A) HIF1α mRNA and (B) miR-33a-5p were analyzed by reverse transcription-quantitative PCR in NEAT1-siRNA transfected cells. (C and D) The protein expression level of HIF1α was analyzed by western blotting in NEAT1-siRNA transfected cells. (E) The predicted binding site between NEAT1 and miR-33a-5p. (F) A dual-luciferase activity assay was conducted to assess the binding affinity between NEAT1 and miR-33a-5p. (G) The predicted binding site between miR-33a-5p and the 3′UTR of HIF1α. (H) A dual-luciferase activity assay was conducted to assess the binding affinity between miR-33a-5p and the 3′UTR of HIF1α. Correlations were determined between (I) miR-33a-5p and NEAT1 or (J) miR-33a-5p and HIF1α expression levels. ***P<0.001. HemEC, hemangioma endothelial cells; HIF1α, hypoxia-inducible factor 1α; miR, microRNA; Mut, mutant; NC, negative control; NEAT1, nuclear paraspeckle assembly transcript 1; si, small interfering RNA; UTR, untranslated region.
Overexpression of miR-33a-5p or depletion of HIF1α suppresses HemEC proliferation, migration and invasion
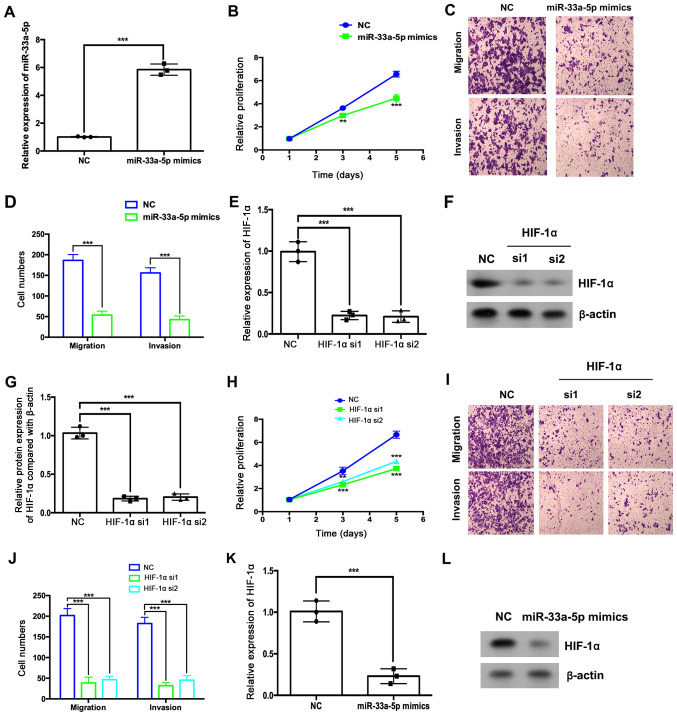
The effects of miR-33a-5p overexpression and HIF1α depletion on HemEC proliferation, migration and invasion were assessed. RT-qPCR results indicated that miR-33a-5p mimic transfection significantly increased the expression of miR-33a-5p in HemECs (Fig. 4A), which significantly inhibited HemEC proliferation, migration and invasion (Fig. 4B-D, respectively). HIF1α siRNA transfection resulted in decreased HIF1α mRNA and protein expression levels compared with the NC group (Fig. 3E-G). HIF1α depletion also significantly suppressed HemEC proliferation, migration and invasion (Fig. 4H-J). Furthermore, miR-33a-5p overexpression downregulated the mRNA and protein expression levels of HIF1α (Fig. 4K and L).
Figure 4.
miR-33a-5p overexpression or HIF1α depletion suppresses HemEC proliferation, migration and invasion. (A-C) HemECs were transfected with miR-33a-5p mimics or miRNA NC. (A) The expression levels of miR-33a-5p were analyzed by RT-qPCR. (B) The relative proliferation was analyzed using a CCK-8 assay. (C) Migratory and (D) invasive abilities were analyzed by Transwell and Matrigel assays, respectively. (F-J) HemECs were transfected with HIF1α or NC siRNAs. (E) The expression levels of HIF1α were analyzed by RT-qPCR. (F and G) The protein expression levels of HIF1α were analyzed by western blotting. (H) The relative proliferation of HemECs was analyzed using a CCK-8 assay. (I) Migratory and (J) invasive abilities of HemECs were analyzed by Transwell and Matrigel assays, respectively. HemECs were transfected with miR-33a-5p mimics or miRNA NC and the expression levels of HIF1α (K) mRNA were analyzed by RT-qPCR and (L) protein was analyzed by western blotting. **P<0.01 and ***P<0.001 vs. NC group at the same time point or as indicated in figure. CCK-8, Cell Counting Kit-8; HemEC, hemangioma endothelial cells; HIF1α, hypoxia-inducible factor 1α; miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative PCR; si, small interfering RNA.
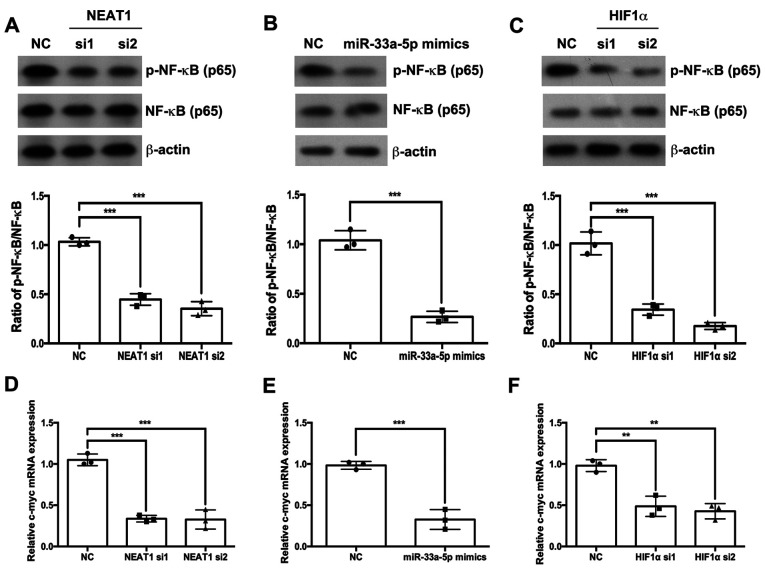
NEAT1/miR-33a-5p/HIF1α axis regulates the NF-κB signaling pathway
The effects of the NEAT1/miR-33a-5p/HIF1α axis on the NF-κB signaling pathway were also investigated. Western blotting results indicated that the depletion of NEAT1 or HIF1α (Fig. 5A and C, respectively) and the overexpression of miR-33a-5p (Fig. 5B) significantly inhibited the phosphorylation of p65 NF-κB compared with the respective NC groups. Similarly, the depletion of NEAT1 or HIF1α and the overexpression of miR-33a-5p decreased the mRNA expression levels of c-myc (Fig. 5D-F).
Figure 5.
NEAT1/miR-33a-5p/HIF1α axis regulates the NF-κB signaling pathway. The protein expression levels of p-NF-κB (p65) and NF-κB (p65) were analyzed by western blotting following transfection of HemEC cells with (A) NEAT1 siRNAs, (B) miR-33a-5p mimics or (C) HIF1α siRNA. The mRNA expression levels of c-myc were analyzed by reverse transcription-quantitative PCR following transfection of HemEC cells with (D) NEAT1 siRNAs, (E) miR-33a-5p mimics or (F) HIF1α siRNA. **P<0.01 and ***P<0.001. HIF1α, hypoxia-inducible factor 1α; miR, microRNA; NC, negative control; NEAT1, nuclear paraspeckle assembly transcript 1; si, small interfering RNA.
Discussion
A number of lncRNAs have been reported to display important roles during IH progression. For example, the lncRNA linc00342 enhances HemEC proliferation and suppresses HemEC apoptosis by sponging miR-3619-5p and increasing the expression of its target gene hepatoma-derived growth factor (22). Higher expression levels of lncRNA urothelial cancer-associated 1 (UCA1) have previously been detected in proliferating phase hemangioma samples compared with involuting phase hemangioma samples (23). UCA1 depletion decreased EOMA mouse hemangioendothelioma endothelial cell viability, migration and invasion, and also promoted EOMA cell apoptosis by regulating miR-200c (23). However, the roles and mechanisms underlying NEAT1 during IH are not yet completely understood.
The results of the present study indicated that the expression of NEAT1 was higher in IH tissues compared with normal skin tissues, in particular in proliferating phase samples compared with involuting phase samples. In vitro studies indicated that NEAT1 depletion inhibited HemEC proliferation, migration and invasion. The starBase database predicted that NEAT1 may function as a sponge for miR-33a-5p and further regulate downstream HIF1α. The results of the present study further suggested the binding between miR-33a-5p and NEAT1, as well as between miR-33a-5p and the 3′UTR of HIF1α.
miR-33a-5p expression is downregulated in prostate cancer tissues, and low expression levels are positively associated with poor prognosis and bone metastasis-free survival (24). miR-33a-5p, which was transcriptionally suppressed by zinc-finger E-box-binding homeobox 1 overexpression in prostate cancer, affected migration, invasion and the epithelial-to-mesenchymal transition of cancer cells (24). During colorectal cancer, miR-33a-5p is also downregulated, and its overexpression significantly suppressed colorectal cancer cell proliferation, colony formation, G1/S progression and migration (25). To the best of our knowledge, the present study suggested for the first time that miR-33a-5p was downregulated in IH tissues and inactivated by binding to an abundant amount of lncRNA NEAT1.
HIF1α plays important roles during the development of IH. HIF1α is significantly upregulated in proliferating IH tissues compared with involuting IH tissues, and an in vitro study indicated that hypoxia upregulated HIF1α expression in hemangioma stem cells (26). Curcumin-mediated HIF1α downregulation inhibits cell proliferation and induces infantile hemangioma endothelial cell apoptosis (18). The results of the present study suggested that HIF1α was highly expressed in IH tissues, and the expression levels of HIF1α in proliferating IH tissues were higher compared with involuting IH tissues. The results also further suggested that depletion of NEAT1 mRNA downregulated HIF1α, and this may be through sponging miR-33a-5p during IH, although further studies are needed to confirm this. A previous study reported that NEAT1 bound and inactivated miR-186-5p thereby upregulating HIF1α and promoting cell proliferation and invasion during osteosarcoma (21). Collectively, the aforementioned studies suggested that NEAT1 regulated HIF1α through different mechanisms in different types of cancer.
The NF-κB signaling pathway serves important roles during cancer progression (27,28). The expression levels of NF-κB (p65), p-NF-κB inhibitor-α (IκBα) and p-NF-κB inhibitor-β (IKKβ) were higher in proliferative hemangioma compared with involutional hemangioma (29). Propranolol treatment could decrease the expression of NF-κB (p65), and inhibit the phosphorylation of IκBα and IKKβ in vitro and in vivo during IH (30). Inactivating the NF-κB signaling pathway further decreased vascular endothelial growth factor A expression levels in IH-derived stem cells (31). The results of the present study indicated that the NF-κB signaling pathway could be regulated by lncRNA NEAT1 during IH.
The present study suggested that lncRNA NEAT1 depletion suppressed the tumorigenesis of infantile hemangioma by competitively binding miR-33a-5p to stimulate the HIF1α/NF-κB signaling pathway. Therefore, blocking the NEAT1/miR-33a-5p/HIF1α signaling pathway may represent a potential antitumor therapeutic strategy.
Acknowledgements
Not applicable.
Funding
The present study was funded by The Gathering Plan of Scientific and Technological Innovation Elements (grant no. 2017-1-S-16631).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
LY and HS designed the study. LY, HS, NZ, LX, ML, LL and YX performed the experiments. ZZ, LZ and YX analyzed the data. LY and HS wrote the paper.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Kunming Children's Hospital (approval no. 2018-005). Written informed consent was obtained from the parents/legal guardians of each patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Acevedo LM, Cheresh DA. Suppressing NFAT increases VEGF signaling in hemangiomas. Cancer Cell. 2008;14:3358–430. doi: 10.1016/j.ccr.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Leaute-Labreze C, Prey S, Ezzedine K. Infantile haemangioma: Part I. Pathophysiology, epidemiology, clinical features, life cycle and associated structural abnormalities. J Eur Acad Dermatol Venereol. 2011;25:1245–1253. doi: 10.1111/j.1468-3083.2011.04102.x. [DOI] [PubMed] [Google Scholar]
- 3.Yang L, Dai J, Li F, Cheng H, Yan D, Ruan Q. The expression and function of miR-424 in infantile skin hemangioma and its mechanism. Sci Rep. 2017;7:11846. doi: 10.1038/s41598-017-10674-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calicchio ML, Collins T, Kozakewich HP. Identification of signaling systems in proliferating and involuting phase infantile hemangiomas by genome-wide transcriptional profiling. Am J Pathol. 2009;174:1638–1649. doi: 10.2353/ajpath.2009.080517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu ZZ, Tian YF, Wu H, Ouyang SY, Kuang WL. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1α/VEGF axis. Neoplasma. 2020;67:111–118. doi: 10.4149/neo_2019_190121N61. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Qiu Q, Qian X, Yi J, Jiao Y, Yu M, Li X, Li J, Mi C, Zhang J, et al. Long noncoding RNA LCAT1 functions as a ceRNA to regulate RAC1 function by sponging miR-4715-5p in lung cancer. Mol Cancer. 2019;18:171. doi: 10.1186/s12943-019-1107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, Qiu J, He G, Liang Y, Wang L, Liu C, Pan H. LncRNA MALAT1 acts as a miR-125a-3p sponge to regulate FOXM1 expression and promote hepatocellular carcinoma progression. J Cancer. 2019;10:6649–6659. doi: 10.7150/jca.29213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Lv R, Zhang L, Xu G, Bi J, Gao F, Zhang J, Xue F, Wang F, Wu Y, et al. Long noncoding RNA expression profile of infantile hemangioma identified by microarray analysis. Tumour Biol. 2016 Oct 5; doi: 10.1007/s13277-016-5434-y. doi: 10.1007/s13277-016-5434-y (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 9.Li J, Li Q, Chen L, Gao Y, Zhou B, Li J. Competitive endogenous RNA networks: Integrated analysis of non-coding RNA and mRNA expression profiles in infantile hemangioma. Oncotarget. 2018;9:11948–11963. doi: 10.18632/oncotarget.23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li MM, Dong CX, Sun B, Lei HZ, Wang YL, Gong YB, Sun LL, Sun ZW. LncRNA-MALAT1 promotes tumorogenesis of infantile hemangioma by competitively binding miR-424 to stimulate MEKK3/NF-κB pathway. Life Sci. 2019;239:116946. doi: 10.1016/j.lfs.2019.116946. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Li M, Dong C, Ma Y, Xiao L, Zuo S, Gong Y, Ren T, Sun B. Linc00152 knockdown inactivates the Akt/mTOR and Notch1 pathways to exert its anti-hemangioma effect. Life Sci. 2019;223:22–28. doi: 10.1016/j.lfs.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Wang SJ, Li YJ, Gao B, Li XL, Li YT, He HY. Long non-coding RNA 00152 slicing represses the growth and aggressiveness of hemangioma cell by modulating miR-139-5p. Biomed Pharmacother. 2019;120:109385. doi: 10.1016/j.biopha.2019.109385. [DOI] [PubMed] [Google Scholar]
- 13.Jiang X, Guo S, Zhang Y, Zhao Y, Li X, Jia Y, Xu Y, Ma B. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell Signal. 2020;65:109422. doi: 10.1016/j.cellsig.2019.109422. [DOI] [PubMed] [Google Scholar]
- 14.Shan G, Tang T, Xia Y, Qian HJ. Long non-coding RNA NEAT1 promotes bladder progression through regulating miR-410 mediated HMGB1. Biomed Pharmacother. 2019;121:109248. doi: 10.1016/j.biopha.2019.109248. [DOI] [PubMed] [Google Scholar]
- 15.Wang W, Ge L, Xu XJ, Yang T, Yuan Y, Ma XL, Zhang XH. LncRNA NEAT1 promotes endometrial cancer cell proliferation, migration and invasion by regulating the miR-144-3p/EZH2 axis. Radiol Oncol. 2019;53:434–442. doi: 10.2478/raon-2019-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 17.Kleinman ME, Greives MR, Churgin SS, Blechman KM, Chang EI, Ceradini DJ, Tepper OM, Gurtner GC. Hypoxia-induced mediators of stem/progenitor cell trafficking are increased in children with hemangioma. Arterioscler Thromb Vasc Biol. 2007;27:2664–2670. doi: 10.1161/ATVBAHA.107.150284. [DOI] [PubMed] [Google Scholar]
- 18.Lou S, Wang Y, Yu Z, Guan K, Kan Q. Curcumin induces apoptosis and inhibits proliferation in infantile hemangioma endothelial cells via downregulation of MCL-1 and HIF-1α. Medicine (Baltimore) 2018;97:e9562. doi: 10.1097/MD.0000000000009562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42((Database Issue)):D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan ZA, Melero-Martin JM, Wu X, Paruchuri S, Boscolo E, Mulliken JB, Bischoff J. Endothelial progenitor cells from infantile hemangioma and umbilical cord blood display unique cellular responses to endostatin. Blood. 2006;108:915–921. doi: 10.1182/blood-2006-03-006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Kang Z, Dai Y, Zheng H, Wang Y. Long noncoding RNA LINC00342 promotes growth of infantile hemangioma by sponging miR-3619-5p from HDGF. Am J Physiol Heart Circ Physiol. 2019;317:H830–H839. doi: 10.1152/ajpheart.00188.2019. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Zhang C. Silence of long non-coding RNA UCA1 inhibits hemangioma cells growth, migration and invasion by up-regulation of miR-200c. Life Sci. 2019;226:33–46. doi: 10.1016/j.lfs.2019.03.038. [DOI] [PubMed] [Google Scholar]
- 24.Dai Y, Wu Z, Lang C, Zhang X, He S, Yang Q, Guo W, Lai Y, Du H, Peng X, Ren D. Copy number gain of ZEB1 mediates a double-negative feedback loop with miR-33a-5p that regulates EMT and bone metastasis of prostate cancer dependent on TGF-β signaling. Theranostics. 2019;9:6063–6079. doi: 10.7150/thno.36735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan Y, Zhang D, Lei T, Zhao C, Han J, Cui J, Wang Y. MicroRNA-33a-5p suppresses colorectal cancer cell growth by inhibiting MTHFD2. Clin Exp Pharmacol Physiol. 2019;46:928–936. doi: 10.1111/1440-1681.13125. [DOI] [PubMed] [Google Scholar]
- 26.Xia HF, Zhu JY, Wang JN, Ren JG, Cai Y, Wang FQ, Zhang W, Chen G, Zhao YF, Zhao JH. Association of ATF4 expression with tissue hypoxia and M2 macrophage infiltration in infantile hemangioma. J Histochem Cytochem. 2017;65:285–294. doi: 10.1369/0022155417694872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, De Menna M, Groenewoud A, Thalmann GN, Kruithof-de Julio M, Snaar-Jagalska BE. A NF-kB-Activin A signaling axis enhances prostate cancer metastasis. Oncogene. 2020;39:1634–1651. doi: 10.1038/s41388-019-1103-0. [DOI] [PubMed] [Google Scholar]
- 28.Xiu Y, Dong Q, Fu L, Bossler A, Tang X, Boyce B, Borcherding N, Leidinger M, Sardina JL, Xue HH, et al. Coactivation of NF-κB and Notch signaling is sufficient to induce B cell transformation and enables B-myeloid conversion. Blood. 2020;135:108–120. doi: 10.1182/blood.2019001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Xu W, Li S, Zhong Z, Li Y, Wang W, Sun C. Expression and significance of the proteins in TSP-1 and NF-κB signal pathways of infantile capillary hemangioma. Zhonghua Zheng Xing Wai Ke Za Zhi. 2016;32:441–446. (In Chinese) [PubMed] [Google Scholar]
- 30.Xu W, Li S, Yu F, Zhang Y, Yang X, An W, Wang W, Sun C. Role of Thrombospondin-1 and nuclear Factor-κB signaling pathways in antiangiogenesis of infantile hemangioma. Plast Reconstr Surg. 2018;142:310e–321e. doi: 10.1097/PRS.0000000000004684. [DOI] [PubMed] [Google Scholar]
- 31.Greenberger S, Adini I, Boscolo E, Mulliken JB, Bischoff J. Targeting NF-κB in infantile hemangioma-derived stem cells reduces VEGF-A expression. Angiogenesis. 2010;13:327–335. doi: 10.1007/s10456-010-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.