Abstract
Silver nanoparticles (AgNPs) are used in food packaging materials, dental care products and other consumer goods and can result in oral exposure. To determine whether AgNP coatings modulate transcriptional responses to AgNP exposure, we exposed mice orally to 20 nm citrate (cit)-coated AgNPs (cit-AgNPs) or polyvinylpyrrolidone (PVP)-coated AgNPs (PVP-AgNPs) at a 4 mg/kg dose for 7 consecutive days and analyzed changes in the expression of protein-coding genes and long noncoding RNAs (lncRNAs), a new class of regulatory RNAs, in the liver. We identified unique and common expression signatures of protein-coding and lncRNA genes, altered biological processes and signaling pathways, and coding-non-coding gene interactions for cit-AgNPs and PVP-AgNPs. Commonly regulated genes comprised only about 10 and 20 percent of all differentially expressed genes in PVP-AgNP and cit-AgNP exposed mice, respectively. Commonly regulated biological processes included glutathione metabolic process and cellular oxidant detoxification. Commonly regulated pathways included Keap-Nrf2, PPAR, MAPK and IL-6 signaling pathways. The coding-non-coding gene co-expression analysis revealed that protein-coding genes were co-expressed with a variable number of lncRNAs ranging from one to twenty three and may share functional roles with the protein-coding genes. PVP-AgNP exposure induced a more robust transcriptional response than cit-AgNP exposure characterized by more than two-fold higher number of differentially expressed both protein- coding and lncRNA genes. Our data demonstrate that the surface coating strongly modulates the spectrum and the number of differentially expressed genes after oral AgNP exposure. On the other hand, our data suggest that AgNP exposure can alter drug and chemical sensitivity, metabolic homeostasis and cancer risk irrespective of the coating type, warranting further investigations.
Keywords: Silver nanoparticles, transcriptomics, long noncoding RNA, mouse, in vivo
Graphical Abstract
1. Introduction
Due to rapid growth of nanotechnology, the human exposure levels to engineered nanoparticles (NPs) will increase (Royce et al., 2014; Yang & Westerhoff, 2014). Silver nanoparticles (AgNPs) are one of the most widely used NPs. AgNPs are incorporated into medical devises, food contact items, dental care products, dietary supplements, children’s items and other consumer products mainly due to their unique antibacterial and antimicrobial effects (Ge et al., 2014; PEN, 2018; Vance et al., 2015). The annual production of AgNPs is estimated to reach 800 tons by 2025 (Calderon-Jimenez et al., 2017). A number of studies examined the release of AgNPs from consumer products. These studies included quantification of silver (Ag) from food storage containers (Echegoyen & Nerin, 2013), toothbrushes (Mackevica et al., 2017), socks (Benn & Westerhoff, 2008; Geranio et al., 2009) and various children’s items (Quadros et al., 2013) into water, food, drink or sweat simulating solutions and verified that the released species included AgNPs and Ag ions (Ag+). The released levels varied from product to product with Ag concentrations ranging from 1.4 to 270,000 μg/g product in 500 ml of water (Yang & Westerhoff, 2014).
Since bare AgNPs aggregate in suspension (Shkilnyy et al., 2009), they are synthetized with surface coatings to enhance colloidal stability and extend shelf-life. Citrate (cit) and polyvinylpyrrolidone (PVP) are one of the most commonly used agents for AgNP stabilization (Huynh & Chen, 2011; Wang et al., 2014). Cit provides electrostatic stabilization, whereas PVP provides electrostatic and steric stabilization to AgNPs. Both agents impart a negative surface charge but cit is weakly bound and can be displaced easily with other molecules, while PVP is very tightly bound to the metal surface and confers high AgNP stability in various solvents (NanoComposix, 2018). Both cit and PVP have numerous applications including widespread use in pharmacy, cosmetics and food industry. However, which specific nanotechnology-enabled products contain AgNPs with cit or PVP coatings is unknown due to proprietary information.
Studies showed that protein absorption ability (Pang et al., 2016), protein corona composition (Shannahan et al., 2013), Ag+ release (Gliga et al., 2014; Wang et al., 2014) and intracellular uptake and localization (Gliga et al., 2014; Pang et al., 2016) were similar for cit-AgNPs and PVP-AgNPs, suggesting that protein corona, dissolution and/or cellular uptake of AgNPs are not altered by cit and PVP coatings. However, differences were noted in the magnitude of their induced adverse effects with cit-AgNPs generally being more potent than PVP-AgNPs (Anderson et al., 2015; Guo et al., 2016; Nallanthighal, Chan, Bharali, et al., 2017; Powers et al., 2011; Seiffert et al., 2015; Vecchio et al., 2014; Wang et al., 2014). For example, cit-AgNPs reduced survival and altered morphology of zebrafish embryos, while an equivalent dose of PVP-AgNPs was nontoxic (Powers et al., 2011). In rats exposed to AgNPs by intratracheal instillation, cit-AgNPs caused a greater increase in lung macrophages at 21 days post-exposure than PVP-AgNPs (Anderson et al., 2015). The adverse effects of AgNPs have been attributed to the released Ag+, NPs or a combination of both.
Previously, we observed that a sub-acute (7-days) oral exposure to 20 nm cit-AgNPs at a 4 mg/kg daily dose caused genotoxic effects (oxidative DNA damage, double stranded breaks and micronuclei) in mice, while PVP-AgNPs were not genotoxic at an equivalent dose (Nallanthighal, Chan, Bharali, et al., 2017). The molecular mechanisms leading to differential effects of cit-AgNPs and PVP-AgNPs are unclear. We hypothesized that cit-AgNPs and PVP-AgNPs induce different transcriptional responses and, therefore, analyzed global gene expression changes in the organ in which AgNPs are known to accumulate. While oral bioavailability of AgNPs is low (1 – 4%), the fraction that is absorbed from the gastrointestinal tract into the systemic circulation distributes to various organs with the highest levels found in the liver, kidney, spleen and lung (Park et al., 2011). Small size AgNPs (10 – 20 nm) are better absorbed than large size AgNPs (70 – 110 nm), particles larger than 320 nm are not absorbed through gastrointestinal tract (Boudreau et al., 2016; Park et al., 2010) and the effect of coating is unclear. In this study, we chose liver that has been described as the main organ for accumulation of AgNPs, coated or uncoated, following oral, intravenous or subcutaneous exposure (Bergin et al., 2016; Boudreau et al., 2016; Garcia et al., 2016; Lee et al., 2013; Loeschner et al., 2011; Pang et al., 2016; van der Zande et al., 2012) and as organ susceptible to AgNP-mediated damage (Kim et al., 2010; Ansar et al., 2017; Patlolla et al., 2015; Tiwari et al., 2011). In addition to protein-coding genes, we analyzed long non-coding RNAs (lncRNAs) that are transcripts longer than 200 nucleotides that are not translated into proteins. Since their discovery in the early 2000s, lncRNAs emerged as a new class of functional RNAs that act as regulators of transcriptional, post-transcriptional and epigenetic gene regulation of eukaryotic genomes during development and various disease states (Jarroux et al., 2017). It has been proposed that lncRNAs are novel biomarkers and regulators of toxicological responses to noxious chemicals (Bai et al., 2014; Gao et al., 2016; Li & Cui, 2018; Zhou et al., 2015) but lncRNA expression signatures have not been reported for AgNPs or other commonly used NPs. The current study has identified unique and common expression signatures of protein-coding and lncRNA genes, the altered biological processes and signaling pathways, and coding-non-coding gene interactions for cit-AgNPs and PVP-AgNPs in an in vivo system.
2. Materials and Methods
2.1. AgNPs and their characterization
20 nm spherical AgNPs coated with cit or PVP were obtained from nanoComposix (San Diego, CA). Particles were supplied as aqueous AgNP dispersions at Ag concentration of 1 mg/ml (Biopure™). AgNPs were characterized by the manufacturer by various analytic techniques that included transmission electron microscopy (TEM), dynamic light scattering (DLS), zeta potential measurements and UV-visible (UV-vis) spectroscopy. According to the manufacturer’s data sheets, cit-AgNPs (lot# KJW 1726) had a mean diameter of 21.3 ± 3.4 nm determined by TEM, a hydrodynamic diameter of 26.2 nm determined by DLS, zeta potential of −34.4 mV and a Surface Plasmon Resonance (SPR) peak was at 394 nm as determined by UV-vis spectroscopy. PVP-AgNPs (lot# JEA0252) had a mean 18.8 ± 2.9 nm TEM diameter, 39 nm hydrodynamic diameter, −32 mV zeta potential and the SRP peak was at 392 nm.
In-house, AgNPs were characterized by scanning transmission electron microscopy (STEM), single particle inductively coupled mass spectroscopy (sp-ICP-MS) and UV-vis spectroscopy. STEM was used to analyze the particle morphology, size and size distribution. Samples were prepared by drop casting a drop of undiluted nanoparticle dispersion on a 400 mesh formvar/carbon copper transmission electron microscope grid and air dried. STEM images were acquired on a FEI Titan 80 – 300 microscope operated at an accelerating voltage of 300 keV using the high angular annular dark field (HAADF) detector and/or bright field (BF) detector. STEM images were collected with a convergence angle of 15 mrad and a working distance of 185 mm for both HAADF and BF detectors. Images were obtained using a beam current of approximately 0.5 nA. For particle size distribution analyses, Fiji image processing program (open-source ImageJ software focused on biological-image analysis) was utilized to analyze the size of at least 450 particles from HAADF images. Sp-ICP-MS analysis was performed using a NexION 350X ICP-MS Spectrometer (PerkinElmer, Waltham, MA) and a Meinhard nebulizer, and operated at a radio frequency power of 1600 W. AgNPs were diluted in deionized water to an estimated concentration of 105 particles per ml. Ag was measured at m/z (molecule mass/number of elementary charges) values of 107 and 109. Data acquisition was performed using PerkinElmer Syngistix Nano Application Module software in the time-resolved analysis mode with a dwell time of 100 μs and an acquisition time of 100 s. The particle sizes were calculated from the measured Ag masses by assumption of a spherical shape of the particles. Particle size calibration was performed using AgNP standards from nanoComposix. AgNP spectral properties were analyzed by UV-visible spectroscopy on samples diluted 1:30 in deionized water with a NanoDrop1000 Spectrophotometer v3.8 (Thermo Fisher Scientific, Franklin, MA).
2.2. Mice and AgNP treatments
C57BL/6J pun/pun mice (Jackson Laboratory, Bar Harbor, ME), congenic to C57BL/6J strain, were bred in the virus-free animal facility at the University at Albany Cancer Research Center under standard conditions that include a 12 h light/dark cycle and a standard rodent diet and water ad libitum. All procedures were approved by the institutional animal use and care committee. Ten to 12 weeks-old mice, 4 mice per group (two males and two females), were exposed to cit-AgNPs or PVP-AgNPs by oral gavage at a daily dose of 4 mg of Ag/kg for 7 consecutive days as in our previous studies (Nallanthighal, Chan, Bharali, et al., 2017; Nallanthighal, Chan, Murray, et al., 2017), while control mice received water only. Allometric dose conversion of 4 mg/kg in a mouse yields 0.3 mg/kg in an average (70 kg) human (Reagan-Shaw et al., 2008) and would correspond to AgNP intake after taking AgNP dietary supplements for 6 weeks (Munger et al., 2014). Dosing was performed between noon and 2 PM. Mice were euthanized 24 h after the final dose and their livers were harvested, flash frozen in liquid nitrogen and stored in −80°C freezer until analyses.
2.3. Total RNA extraction
Total RNA was extracted with TRIzol® reagent (Thermo Fisher Scientific, Franklin, MA). RNA concentrations were quantified using a NanoDrop 1000 Spectrophotometer (ThermoScientific, Waltham, MA). Integrity of total RNA samples was evaluated by Agilent2100 Bioanalyzer (Agilent Technologies Inc. Santa Clara, CA). Samples with RNA integrity values above 8.0 were used for transcriptome analyses.
2.4. Clariom D assay and computational analysis
RNAs were processed for hybridization into the Clariom™ D for mouse array platform (Affymetrix) at the Center for Functional Genomics, University at Albany, Rensselaer, NY. Briefly total RNA (100 ng) was processed using the WT Plus Reagent kit (Affymetrix, Santa Clara, CA). Sense target cDNAs were generated using the standard Affymetrix WT protocol and hybridized to Affymetrix Mouse Clariom D arrays. Arrays were washed, stained and scanned on a GeneChip 3000 7G scanner using Affymetrix GeneChip Command Console Software (AGCC). Transcriptome Analysis Console Software (TAC v3.0.1.5) was used to identify differentially expressed genes. Briefly the CEL files were summarized using the SST-RMA algorithm in TAC and the normalized data were subjected to one-way ANOVA with a Benjamin Hochberg False Discovery Rate correction included (p<0.05). A 1.5-fold change was used to select entities that were statistically and differentially expressed between AgNP exposed and control mouse samples. The complete gene list was submitted to the Gene Expression Omnibus (series accession number GSE139560).
2.5. Gene ontology and pathway analysis
The differentially expressed protein-coding genes were subjected to gene ontology (GO) analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 to find overrepresentations of GO terms (Huang da et al., 2009a;2009b). The Mus musculus (house mouse) whole genome was used as background. Statistical enrichment was determined using default settings. Pathway analysis was conducted with the TAC software.
2.6. Construction of the lncRNA-mRNA co-expression network and lncRNA function prediction
The lncRNA-mRNA co-expression network was constructed based on the Pearson’s correlation coefficient values between differentially expressed lncRNA and mRNA pairs. The Pearson’s correlation coefficient was calculated using R studio software. RNAs with the Pearson’s correlation coefficient of ≥0.99 were considered significant. The lncRNA-mRNA co-expression network was constructed using the Cytoscape software (The Cytoscape consortium, SanDiego, CA). The lncRNA function prediction analysis was performed as described by Luo et al. (Luo et al., 2017). Briefly, lncRNA function can be predicted based on the function annotation of the co-expressed coding genes given that two co-expressed genes frequently show functional relationships. When the coding genes within the group, referred to as a module or subnetwork, of tightly connected lncRNAs and mRNAs in the co-expression network are enriched for at least one GO term, the lncRNA within the group can be assigned this term.
2.7. Real-time quantitative PCR
Total RNA was transcribed to cDNA with MultiScribe™ Reverse Transcriptase (Thermo Fisher Scientific, Franklin, MA). RNA was prepared from the same tissue samples that were used in Clariom D assay. Real-time quantitative PCR (qPCR) was performed using iTaq™ Universal SYBR® Green Supermix (Bio-Rad Laboratories, Hercules, CA) on the ABI7900 HT Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA). Quantification of gene expression levels was calculated using the 2−ΔΔCT method and presented relative to the expression of the housekeeping gene Gapdh. The primer sequences are provided below.
Gapdh (5’ TCGTCCCGTAGACAAAATGG, 3’ TTGAGGTCAATGAAGGGGTC)
Arrdc3 (5’ GGT GCT GGG AAA GGT AAA GAG T, 3’ TGA TTC CGT CCA CCG TAC TTT)
Nrf2 (5’ GTA AGA ATA AAG TCG CCG CCC, 3’ AGC CGC TTC AGT AGA TGG)
Foxo1 (5’ TTC TCT CGT CCC CAA CAT CT, 3’ TTG CTG TCC TGA AGT GTC TG)
Lurap1 (5’ GCC TGT GTA GTT TGC TGG AG, 3’ CAT CAT CTG CAA AAG TGT CCA)
Slc38a2 (5’ GAA AAG CCA TTA TGC CGA CG, 3’ GCA TAA GAA AGC CCA AGG ATT)
Lpin1 (5’ CCC TCA ACA CCA AAA AGT GAC, 3’ TGA AGA CTC GCT GTG AAT GG)
Ilr1 (5’ GAC TCC TGC TCT GGT TTT CTT, 3’ CAA ACT GTC CCT CCA AGA CCT)
Klf10 (5’ GCG ACT GGA AGT CTC ATT TCA AG, 3’ GGC TGT AAG GTG GCG TTA)
Gsta2 (5’ CCA GAG CCA TTC TCA ACT ACA TC, 3’ TTG GCT TCT CTT TGG TCT GG)
Gstm3 (5’ CCA TTT TCC CAA TCT GCC CTA C, 3’ TGC GGG TGT CCA TAA CTT)
Saa1 (5’ TGG TCT TCT GCT CCC TGC T, 3’ CCT TTG GGC AGC ATC ATA GT)
3. Results
3.1. Characterization of AgNPs
Cit-AgNPs and PVP-AgNPs were characterized by STEM, sp-ICP-MS and UV-vis spectroscopy. STEM analysis showed that particles were spherical in shape and had an average diameter of 18.3 ± 7.2 nm (Dv10, Dv50 and Dv90 of 12.4, 17.7 and 22.3 nm, respectively) for cit-AgNPs and 19.4 ± 4.2 nm (Dv10, Dv50 and Dv90 of 14.4, 19.9 and 24.2 nm, respectively) for PVP-AgNPs (Fig.1 A). Particle size distribution analysis by sp-ICP-MS indicated that the average diameter of AgNP dispersions in deionized water was 21.0 ± 2.5 nm for cit-AgNPs and 20.7 ± 3.3 nm for PVP-AgNPs (Fig. 1B). UV-vis spectra showed absorbance maxima at about 400 nm for both cit-AgNPs and PVP-AgNPs, depicting their characteristic size-dependent SPR wavelengths (Fig. 1C). Thus, both kinds of AgNPs had similar physicochemical characteristics and the results were consistent with the manufacturer’s AgNP characterization data (see Materials and Methods for manufacturer’s data).
Fig 1. AgNP characterization.
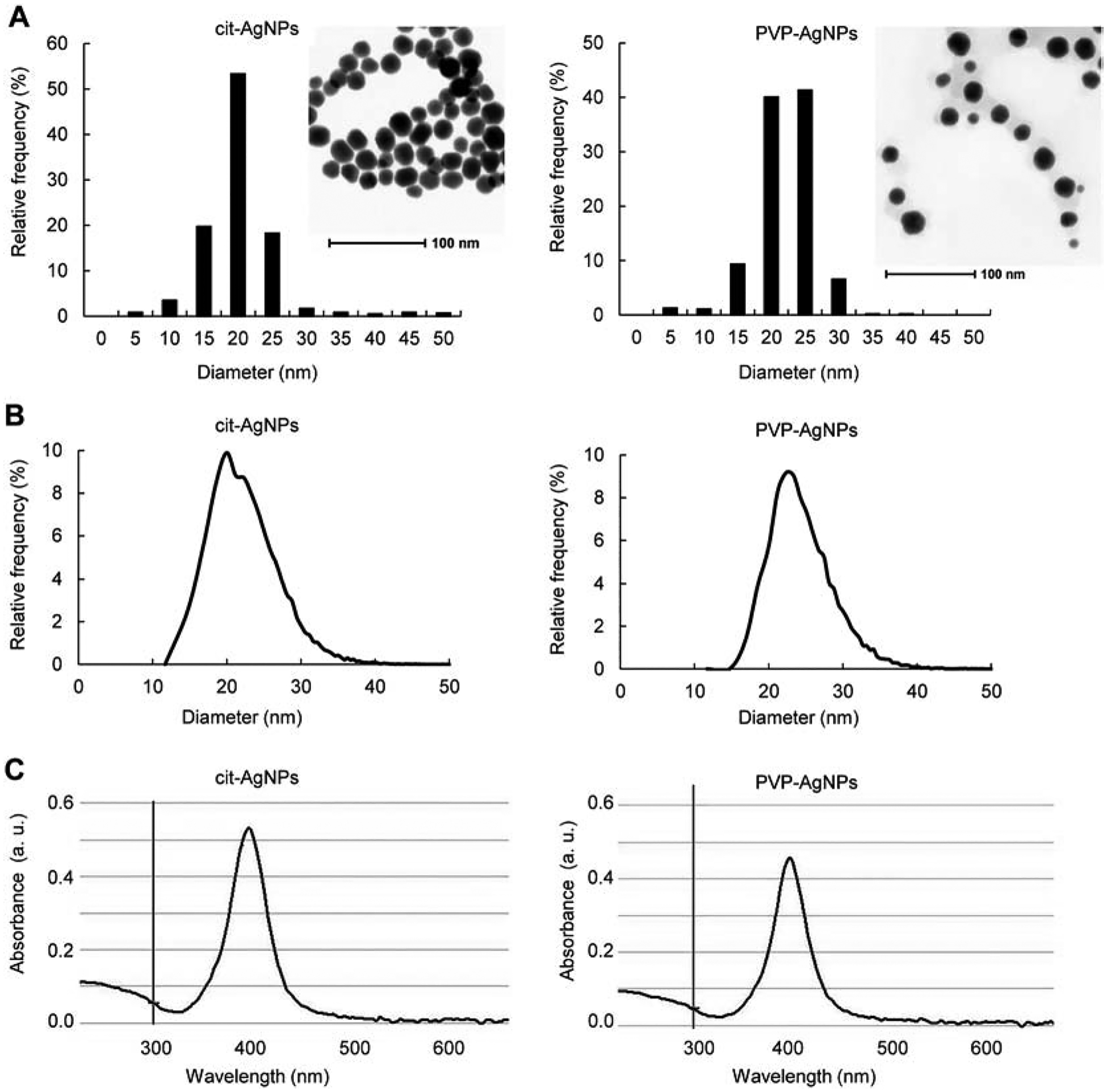
(A) AgNP morphology and size distribution by STEM. (B) AgNP size distribution by sp-ICP-MS. (C) UV-Vis absorbance spectra of AgNPs. Insets in panel A show representative STEM images of AgNPs.
3.2. mRNA expression profiles in AgNP exposed mice
A total of 140 differentially expressed mRNAs were identified in mice exposed to cit-AgNPs and 358 differentially expressed mRNAs were found in mice exposed to PVP-AgNPs (Fig. 2A). Approximately one third of mRNAs were upregulated and two thirds of mRNAs were downregulated in both exposure groups. Differentially expressed mRNAs with the highest up/down regulation in each group are shown in Table 1. Twenty four mRNAs were shared between cit-AgNP and PVP-AgNP exposed mice (Fig. 2A). This number represents 17% and 7% of all differentially expressed mRNAs in cit-AgNP and PVP-AgNP exposed mice, respectively. In summary, the mRNA expression profiles showed that the number of differentially expressed mRNAs was 2.5-times higher in PVP-AgNP than in cit-AgNP exposed mice and a small number of mRNAs were common between the groups, demonstrating that AgNP surface coatings alter transcriptional responses to AgNPs.
Fig 2. Differentially expressed mRNAs and lncRNAs in cit-AgNP and PVP-AgNP exposed mice.
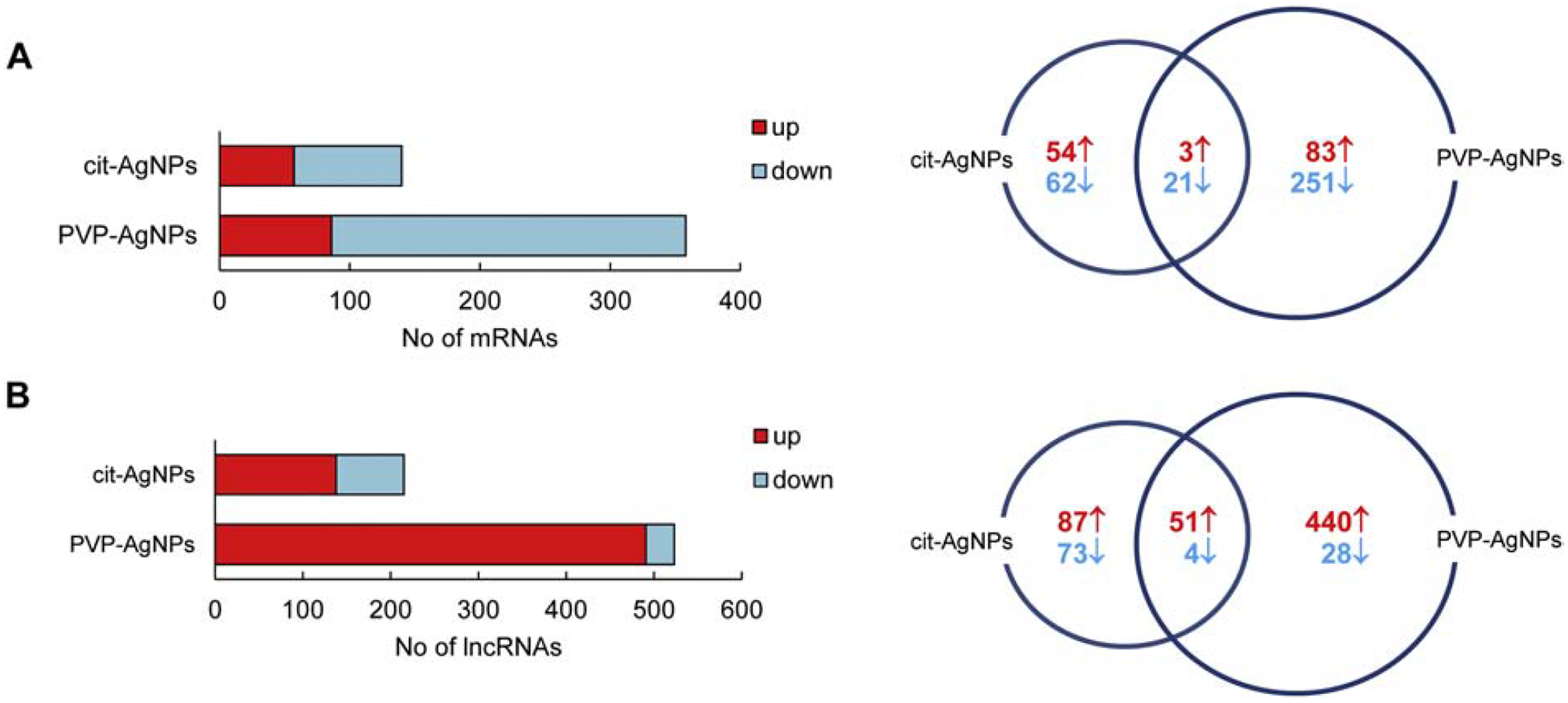
(A) Numbers of differentially expressed mRNAs. (B) Numbers of differentially expressed lncRNAs. The Vent diagrams represent shared and AgNP-specific numbers of genes upregulated (↑) or downregulated (↓) after AgNP exposure.
Table 1.
Top 10 up/down regulated mRNAs in cit-AgNP and PVP-AgNP exposed mice.
| Gene | Gene Description | Fold - Change | P value |
|---|---|---|---|
| Cit-AgNPs | |||
| Arrdc3 | arrestin domain containing 3 | −6.52 | 3.41E-04 |
| Serpina7 | serine (or cysteine) peptidase inhibitor, clade A, member 7 | −3.37 | 4.97E-02 |
| Id2 | inhibitor of DNA binding 2 | −3.05 | 1.26E-02 |
| Acaa1b | acetyl-Coenzyme A acyltransferase 1B | −2.9 | 3.62E-02 |
| Cyr61 | cysteine rich protein 61 | −2.87 | 3.44E-02 |
| Nr1d1 | nuclear receptor subfamily 1, group D, member 1 | −2.55 | 2.60E-02 |
| Gsta2 | glutathione S-transferase, alpha 2 (Yc2) | −2.45 | 3.55E-02 |
| Cited2 | Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 2 | −2.41 | 1.24E-03 |
| Fzd4 | frizzled homolog 4 | −2.35 | 1.76E-02 |
| Slc30a10 | solute carrier family 30, member 10 | −2.33 | 4.40E-02 |
| Arhgef26 | Rho guanine nucleotide exchange factor (GEF) 26 | 2.24 | 1.07E-03 |
| B3galt1 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 | 2.27 | 9.99E-03 |
| Grem2 | gremlin 2 homolog, cysteine knot superfamily | 2.29 | 1.58E-02 |
| Fam35a | family with sequence similarity 35, member A | 2.39 | 1.23E-02 |
| Pfkfb3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | 2.5 | 1.42E-02 |
| Slc38a2 | solute carrier family 38, member 2 | 2.53 | 3.20E-02 |
| Nop58 | NOP58 ribonucleoprotein | 2.57 | 2.45E-02 |
| II1r1 | interleukin 1 receptor, type I | 2.83 | 4.02E-03 |
| Zbtb16 | zinc finger and BTB domain containing 16 | 5.32 | 1.71E-03 |
| Lpin1 | lipin 1 | 12.36 | 1.16E-03 |
| PVP-AgNPs | |||
| Cyp26a1 | cytochrome P450, family 26, subfamily a, polypeptide 1 | −4.03 | 4.22E-02 |
| Arrdc3 | arrestin domain containing 3 | −3.87 | 2.20E-02 |
| Txnip | thioredoxin interacting protein | −3.49 | 1.88E-03 |
| Krt23 | keratin 23 | −3.48 | 3.18E-02 |
| Slc17a4 | solute carrier family 17 (sodium phosphate), member 4 | −3.17 | 4.74E-02 |
| Pklr | pyruvate kinase liver and red blood cell | −3.16 | 3.36E-02 |
| Tubb2a | tubulin, beta 2A class IIA | −3.13 | 3.75E-03 |
| Gstm3 | glutathione S-transferase, mu 3 | −2.97 | 2.51E-02 |
| Gsta1 | glutathione S-transferase, alpha 1 (Ya) | −2.97 | 3.76E-02 |
| Tuba1c | tubulin, alpha 1C | −2.89 | 6.80E-03 |
| Ccdc152 | coiled-coil domain containing 152 | 1.93 | 1.49E-02 |
| C4b | complement component 4B (Chido blood group) | 2.01 | 1.86E-02 |
| Lurap1l | leucine rich adaptor protein 1-like | 2.02 | 1.99E-02 |
| Pros1 | protein S (alpha) | 2.08 | 2.87E-02 |
| Slc38a2 | solute carrier family 38, member 2 | 2.28 | 3.98E-02 |
| Sftpa1 | surfactant associated protein A1 | 2.32 | 7.58E-03 |
| Slc41a2 | solute carrier family 41, member 2 | 2.76 | 3.78E-02 |
| Fgl1 | fibrinogen-like protein 1 | 6.99 | 3.59E-02 |
| Saa1 | serum amyloid A1 | 45.94 | 1.29E-02 |
| Saa2 | serum amyloid A2 | 81.32 | 1.66E-02 |
mRNAs are arranged from top down to top up regulated. Commonly regulated mRNAs are highlighted in bold.
3.3. lncRNA expression profiles in AgNP exposed mice
lncRNA expression profiling showed that there were a total of 215 differentially expressed lncRNAs in cit-AgNP exposed mice and 523 differentially expressed lncRNAs in PVP-AgNP exposed mice (Fig. 2B). About two thirds of lncRNAs were upregulated and about one third of lncRNAs were downregulated in cit-AgNP exposed mice. Most lncRNAs (94%) were upregulated in PVP-AgNP exposed mice. Differentially expressed lncRNAs with the highest up/down regulation in each treatment group are shown in Table 2. Fifty five differentially expressed lncRNAs were shared between cit-AgNP and PVP-AgNP exposed mice, which represents 25% and 10% of all differentially expressed lncRNAs in cit-AgNP and PVP-AgNP exposed mice, respectively. In summary, the number of differentially expressed lncRNAs was 2.4-times higher in PVP-AgNP than cit-AgNP exposed mice and the number of shared lncRNAs was relatively low, supporting that transcriptional responses to AgNPs are modulated by the surface coatings.
Table 2.
Top 10 up/down regulated lncRNAs in cit-AgNP and PVP-AgNP exposed mice.
| Probe Set ID | Transcript ID | Fold-Change | P value |
|---|---|---|---|
| Cit-AgNPs | |||
| TC0800002074.mm.1 | NONMMUT065295 | −4.83 | 4.98E-02 |
| TC0800000855.mm.1 | NONMMUT066192 | −3.24 | 3.66E-02 |
| TC0100002519.mm.1 | NONMMUT001378 | −3.18 | 2.92E-02 |
| TC1600000871.mm.1 | NONMMUT027374 | −3.09 | 2.73E-02 |
| TC1300002177.mm.1 | NONMMUT017922 | −2.86 | 2.79E-02 |
| TC0400000690.mm.1 | NONMMUT047558 | −2.67 | 4.68E-02 |
| TC0700001724.mm.1 | NONMMUT063452 | −2.2 | 2.98E-03 |
| TC0700004410.mm.1 | NONMMUT063950 | −2.11 | 3.38E-02 |
| TC0400002399.mm.1 | NONMMUT046698 | −2.09 | 4.46E-02 |
| TC1600000873.mm.1 | NONMMUT027376 | −2.0 | 1.95E-03 |
| TC0100003045.mm.1 | NONMMUT002543 | 2.41 | 1.52E-02 |
| TC0800002116.mm.1 | NONMMUT065392 | 2.59 | 2.13E-02 |
| TC0100003044.mm.1 | NONMMUT002542 | 2.67 | 3.28E-02 |
| TC0X00003076.mm.1 | NONMMUT074093 | 2.88 | 1.94E-02 |
| TC1700001758.mm.1 | NONMMUT028948 | 3.12 | 7.15E-03 |
| TC1800000231.mm.1 | NONMMUT031786 | 3.26 | 3.58E-03 |
| TC0100003043.mm.1 | NONMMUT002540 | 3.46 | 1.87E-02 |
| TC1700001760.mm.1 | NONMMUT028951 | 3.48 | 1.38E-02 |
| TC0900002242.mm.1 | NONMMUT069015 | 5.01 | 3.30E-04 |
| TC0400001628.mm.1 | NONMMUT049977 | 6.49 | 3.03E-03 |
| PVP-AgNPs | |||
| TC1700002700.mm.1 | NONMMUT031066 | −2.25 | 1.72E-02 |
| TC0700001724.mm.1 | NONMMUT063452 | −2.21 | 1.27E-02 |
| TC0800001918.mm.1 | NONMMUT064996 | −2.07 | 6.06E-03 |
| TC0700003768.mm.1 | NONMMUT062842 | −1.89 | 1.50E-02 |
| TC0400000131.mm.1 | NONMMUT046287 | −1.84 | 3.10E-02 |
| TC0700003794.mm.1 | NONMMUT062890 | −1.84 | 2.98E-02 |
| TC1100001277.mm.1 | NONMMUT011167 | −1.8 | 2.19E-02 |
| TC1200001297.mm.1 | NONMMUT016114 | −1.78 | 8.06E-03 |
| TC1800000765.mm.1 | NONMMUT032936 | −1.75 | 1.44E-02 |
| TC0600002110.mm.1 | NONMMUT056495 | −1.75 | 4.58E-02 |
| TC0100002692.mm.1 | NONMMUT001731 | 2.93 | 2.05E-02 |
| TC1900000088.mm.1 | NONMMUT033637 | 3.04 | 2.13E-02 |
| TC0800001107.mm.1 | NONMMUT066768 | 3.11 | 2.60E-02 |
| TC0800002116.mm.1 | NONMMUT065392 | 3.14 | 9.64E-03 |
| TC1400000183.mm.1 | NONMMUT019891 | 3.26 | 2.00E-02 |
| TC0100002415.mm.1 | NONMMUT001146 | 3.32 | 2.16E-02 |
| TC0100002416.mm.1 | NONMMUT001147 | 3.56 | 9.91E-03 |
| TC0800000487.mm.1 | NONMMUT065394 | 3.71 | 5.46E-03 |
| TC1900000087.mm.1 | NONMMUT033627 | 4.42 | 7.01E-03 |
| TC0800001306.mm.1 | NONMMUT067160 | 9.86 | 3.97E-02 |
lncRNAs are arranged from top down to top up regulated. Transcript IDs are from NONCODE database. Commonly regulated lncRNAs are highlighted in bold.
3.4. Comparison of mRNA and lncRNA regulation by AgNPs
The number of differentially expressed lncRNAs was 1.5-times higher than the number of differentially expressed mRNAs in both cit-AgNP and PVP-AgNP exposed mice. In addition, more lncRNAs were upregulated than downregulated (138 vs.77 and 491 vs. 32 lncRNAs in cit-AgNP and PVP-AgNP exposed mice, respectively), while more mRNAs were downregulated than upregulated (83 vs. 57 and 272 vs. 86 mRNAs in cit-AgNP and PVP-AgNP exposed mice, respectively) irrespective of the coating type. Thus, the pattern of mRNA and lncRNA regulation by AgNPs was coating-independent.
3.5. Gene ontology analysis
DAVID functional annotation analysis was performed to highlight the GO-term enrichment in the biological process category. The analysis revealed 29 GO terms associated with downregulated genes and 6 terms associated with upregulated genes in cit-AgNP exposed mice (Suppl. Table 1) and 60 GO terms associated with downregulated genes and 8 terms associated with upregulated genes in PVP-AgNP exposed mice (Suppl. Table 2). In cit-AgNP exposed mice, the top 5 enriched GO terms associated with downregulated genes were positive regulation of transcription, negative regulation of transcription, positive regulation of transcription from RNA pol II promotor, circadian rhythm and positive regulation of fatty acid biosynthetic process, while the top 5 enriched GO terms associated with upregulated genes were organ regeneration, cellular oxidant detoxification, fructose 2,6-biphosphate metabolic process, fructose metabolic process and cytokine-mediated signaling (Fig. 3A). The gene list in the most enriched GO term (positive regulation of transcription) consisted of transcription factors and transcriptional regulators (Suppl. Table 1). In PVP-AgNP exposed mice, the top 5 significantly enriched GO terms associated with downregulated genes were metabolic process, oxidation-reduction process, glutathione metabolic process, positive regulation of mononuclear cell migration and lipid metabolic process, while the top 5 enriched GO terms associated with upregulated genes were response to stilbenoid, complement activation, acute-phase response, cellular oxidant detoxification and cell chemotaxis (Fig. 3B). The gene list in the most enriched GO term (metabolic process) consisted mainly of Gst (glutathione-s-transferase), Aldh (aldehyde dehydrogenase), Acsl (long-chain fatty-acid-coenzyme A ligase) family genes and other metabolic enzymes (Suppl. Table 2).
Fig 3. GO term enrichment of differentially expressed genes in cit-AgNP and PVP-AgNP exposed mice.
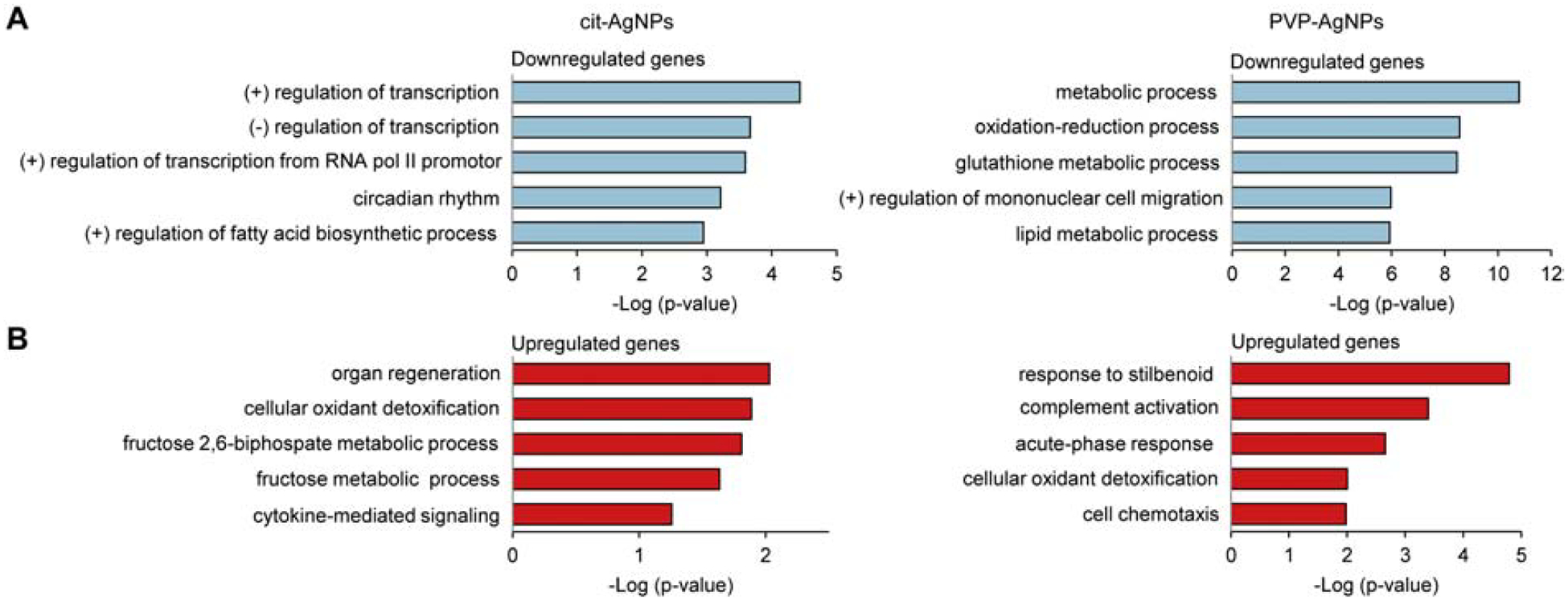
(A) Downregulated genes. (B) Upregulated genes. Top five significantly enriched GO terms in the biological process category are shown. The higher minus Log(p-value), the more significant association a particular GO term has with the gene list. (+), positive; (−), negative.
Glutathione metabolic process and cellular oxidant detoxification were common GO terms in cit-AgNP and PVP-AgNP exposed animals. Glutathione metabolic process was a common GO term among downregulated genes and included Gst genes that are involved in detoxification of xenobiotics (Daniel, 1993; Hayes & Pulford, 1995). Cellular oxidant detoxification was a common GO term among upregulated genes and included hemoglobin alpha chain genes Hba-a1 and Hba-a2 that, in addition to their classical roles in oxygen transport, play a role in protection against oxidative stress (X. Li et al., 2013; W. Liu et al., 2011).
3.6. Pathway analysis
TAC pathway analysis revealed 16 pathways significantly regulated by cit-AgNPs and 17 pathways significantly regulated PVP-AgNPs (Table 3). In cit-AgNP exposed mice, the most significantly regulated pathways were Keap-Nrf2 signaling, white fat cell differentiation, adipogenesis, PluriNetWork and interleukin-1 signaling. The genes involved in these pathways showed bi-directional (up or down) regulation by cit-AgNPs. The most significantly regulated pathways in PVP-AgNP exposed mice were tricarboxylic acid cycle (TCA), fatty acid biosynthesis, glycolysis and gluconeogenesis, electron transport chain and mitochondrial LC-fatty acid β-oxidation. All genes implicated in the pathways regulated by PVP-AgNPs were downregulated with the exception of the complement activation genes. Six pathways, PPAR, MAPK, interleukin-6, Keap-Nrf2, exercise-induced circadian regulation and glycolysis, were shared between cit-AgNP and PVP-AgNP exposed mice. The gene lists associated with these pathways included common as well as unique differentially expressed genes (Table 3).
Table 3.
Pathways altered by cit-AgNPs and PVP-AgNPs.
| Pathway | P value | Gene Count | Genes | |
|---|---|---|---|---|
| Upregulated | Downregulated | |||
| Cit-AgNPs | ||||
| Keap1-Nrf2* | 1.00E-06 | 4 | Cebpb | Gsta2, Gclc, Nfe2l2 |
| White fat cell differentiation | 1.70E-05 | 4 | Cebpb, Foxo1 | Nr2f2, Srebf1 |
| Adipogenesis genes | 5.00E-05 | 6 | Lpin1, Cebpb, Foxo1, Lifr | Irs 1, Srebf1 |
| PluriNetWork | 5.98E-04 | 7 | Lifr, Pim3, Rock2 | Nr2f2, Irs1, Zfp143, Ocln |
| IL-1 signaling | 8.36E-04 | 3 | II1r1, II1r2 | Irak2 |
| Focal Adhesion-PI3K-Akt-mTOR | 1.13E-03 | 7 | Pfkfb1, Pfkfb3, Foxo1 | Efna1, Lpar6, Srebf1, Irs 1 |
| ID signaling | 2.08E-03 | 3 | Srebf1, Irs1, Id2 | |
| PPAR signaling* | 7.51E-03 | 3 | Acsl4 | Acaa1a, Acaa1b |
| MAPK signaling* | 9.46E-03 | 4 | II1r2, II1r1 | Gstt2, Gsta2 |
| IL-6 signaling* | 1.25E-02 | 3 | Cebpb, Foxo1 | Erbb3 |
| IL-7 signaling | 1.93E-02 | 2 | Foxo1 | Irs 1 |
| ErbB signaling | 2.10E-02 | 2 | Nrg4 | Erbb3 |
| Exercise-induced circadian regulation* | 2.37E-02 | 2 | Cebpb | Nr1d1 |
| Glycolysis* | 2.55E-02 | 2 | Pfkfb 1 | Pfkfb3 |
| Lung fibrosis | 3.87E-02 | 2 | Cebpb | Nfe2l2 |
| Ptf1a related regulatory pathway | 4.74E-02 | 1 | Prox1 | |
| PVP-AgNPs | ||||
| TCA cycle | 1.52E-04 | 5 | Dlst, Pdk2, Mdh2, Sdhd, Idh3g | |
| Fatty acid biosynthesis | 4.44E-04 | 4 | Fasn, Acsl5, Echdc3, Acsl1 | |
| Glycolysis & gluconeogenesis | 1.46E-03 | 5 | Pklr, Got2, Mdh2, Pgam1, Eno1 | |
| Electron transport chain | 1.65E-03 | 7 | Ndufa10, Ndufb9, Slc25a5, Ndufa3, Sdhd, Uqcrc2, Ndufs8 | |
| Mitochondrial LC-fatty acid p-oxidation | 2.21E-03 | 3 | Cpt2, Ehhadh, Acsl1 | |
| MAPK signaling* | 2.26E-03 | 9 | Gstt1, Mapk3, Hspb1, Gsta4, Gstm2, Gstm3, Gstm4,Gsta1, Gsta2 | |
| Complement activation | 2.65E-03 | 3 | C4a, C4b, Hc | |
| Fatty acid beta oxidation | 2.68E-03 | 4 | Acsl1, Cpt2, Crat, Acsl5 | |
| Glutathione metabolism | 3.68E-03 | 3 | Gstm2, Gstt1, Gstm7 | |
| Glycolysis* | 1.04E-02 | 4 | Pklr, Eno1, Pgam1, Pdk2 | |
| PPAR signaling* | 1.22E-02 | 5 | Acsl1, Cpt2, Ehhadh, Acsl5, Pltp | |
| Oxidative phosphorylation | 1.51E-02 | 4 | Ndufa3, Ndufb9, Ndufa10, Ndufs8 | |
| Glycogen metabolism | 1.89E-02 | 3 | Pygl, Ugp2, Ppp2r1a | |
| Keap1-Nrf2* | 2.22E-02 | 2 | Nfe2l2,Gsta2 | |
| IL-6 signaling* | 2.54E-02 | 5 | Hsp90aa1, Erbb3, Ppp2r1a, Hspb1, Mapk3 | |
| Urea cycle & metabolism of amino groups | 4.34E-02 | 2 | Otc, Gamt | |
| Exercise-induced circadian regulation* | 4.85E-02 | 3 | Ugp2, Gstm3, Nr1d1 | |
Pathway analysis was performed using TAC.
common pathways modulated by cit-AgNPs and PVP-AgNPs.
3.7. lncRNA-mRNA co-expression analysis
The coding-non-coding gene co-expression network was constructed based on the correlation analysis between differentially expressed lncRNAs and mRNAs. A total of 31,450 lncRNA-mRNA pairs were obtained in cit-AgNP exposed mice and 207,901 lncRNA-mRNA pairs were obtained in PVP-AgNP exposed mice, with the Pearson’s correlation coefficient values ranging between −0.1 to 0.999. The lncRNA-mRNA pairs with the values greater than 0.99 and less than −0.99 were filtered and used to generate the co-expression networks (Fig. 4A, 4C). The input for the co-expression network for cit-AgNPs consisted of 182 positively correlated lncRNA-mRNA pairs and 162 negatively correlated lncRNA-mRNA pairs. The input for co-expression network for PVP-AgNPs consisted of 1127 positively correlated lncRNA-mRNA pairs and 1714 negatively correlated lncRNA-mRNA pairs. One coding gene was co-expressed with one or more than one lncRNAs and vice versa. For example, in cit-AgNP exposed mice, Gclc (glutamate-cysteine ligase catalytic subunit) was co-expressed with 5 lncRNAs including lncRNA NONMMUT017922 (Table 4).The lncRNA NONMMUT017922 was co-expressed with 8 coding genes including Gclc (Fig. 4B). The co-expressed coding genes were significantly enriched in the GO term monooxygenase activity, predicting that lncRNA NONMMUT017922 shares this function. In PVP-AgNP exposed mice, Sdhd (succinate dehydrogenase complex subunit D) was co-expressed with 21 lncRNAs including lncRNA NONMMUT001145 (Table 4) and lncRNA NONMMUT001145 was co-expressed with 10 coding genes including Sdhd (Fig.4 D). The 10 coding genes were significantly enriched in the GO terms positive regulation of binding and glycolytic process, which predicts that lncRNA NONMMUT017922 has a role in these processes. Table 4 shows more examples of the tightly connected coding genes and lncRNAs in the co-expression network and the pathways in which the coding gene is involved. The co-expressed lncRNAs may have functional roles in these pathways because neighboring genes in the co-expression networks are often related functionally (Luo et al., 2017; Nayak et al., 2009).
Fig 4. lncRNA and mRNA co-expression network in cit-AgNP and PVP-AgNP exposed mice.
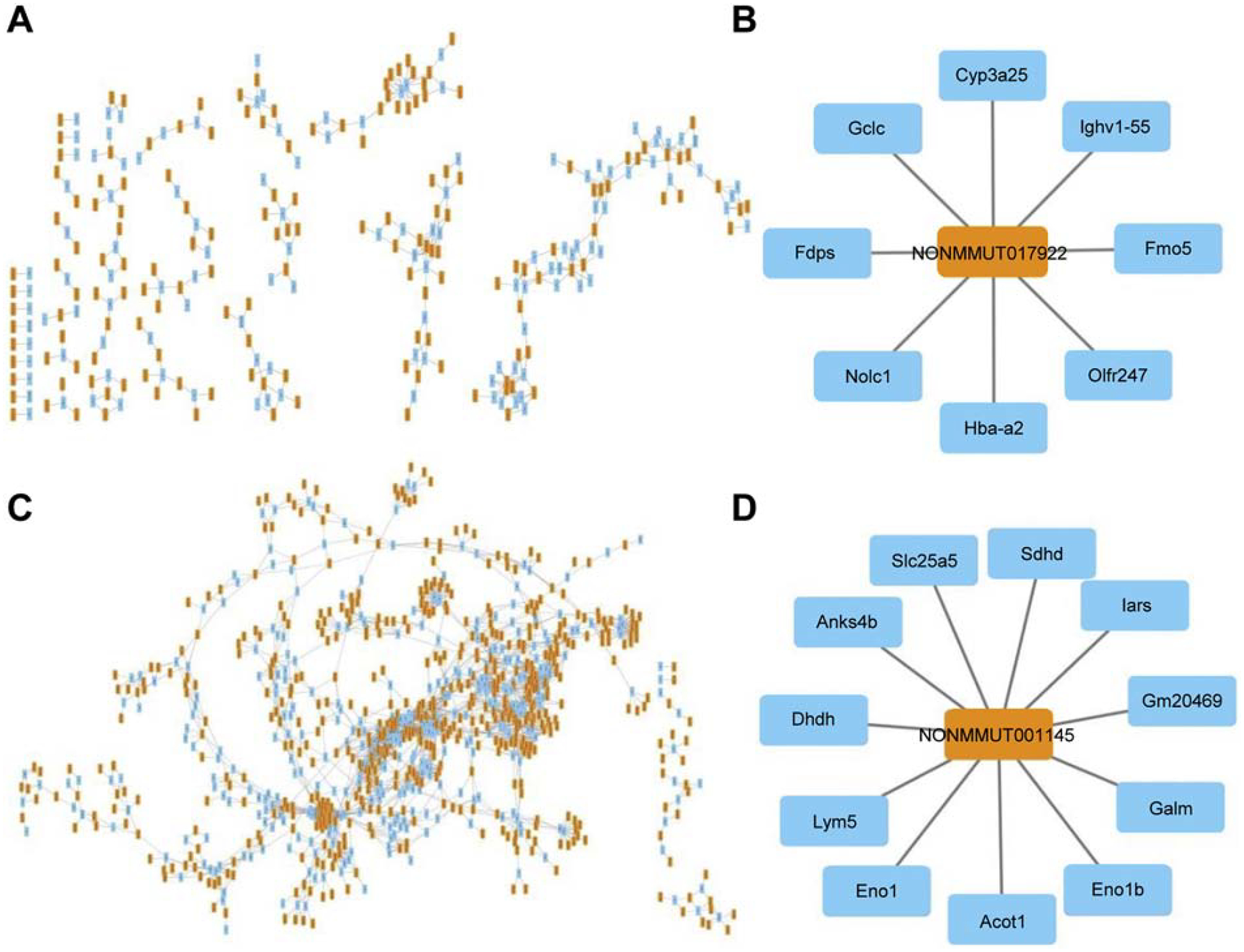
(A) lncRNA and mRNA co-expression network in cit-AgNP exposed mice. (B) A lncRNA and interacting mRNAs in cit-AgNP exposed mice. (C) lncRNA-mRNA co-expression network in PVP-AgNP exposed mice. (D) A lncRNA and interacting mRNAs in PVP-AgNP exposed mice. The co-expression network was constructed based on the correlation analysis between differentially expressed lncRNAs and mRNAs. Yellow rectangles indicate lncRNA and blue rectangle indicate mRNA.
Table 4.
mRNAs-lncRNAs co-expression.
| Gene | Gene description | Associated Pathway | No. of lncRNAs | ||
|---|---|---|---|---|---|
| Total No. | (−) correlation | (+) correlation | |||
| Cit-AgNPs | |||||
| Cebpb | CCAAT/enhancer-binding protein | Keap1-Nrf2 | 5 | 2 | 3 |
| Gclc | glutamate—cysteine ligase catalytic subunit | Keap1-Nrf2 | 5 | 2 | 3 |
| Gsta2 | glutathione S-transferase A2 | Keap1-Nrf2, MAPK | 2 | 1 | 1 |
| Gstt2 | glutathione S-Transferase theta 2 | MAPK | 3 | 1 | 2 |
| II1r1 | Interleukin 1 receptor, type I | MAPK, IL-1 signaling | 1 | 0 | 1 |
| Irak2 | Interleukin-1 receptor-associated kinase-like 2 | IL-1 signaling | 4 | 2 | 2 |
| Srebf1 | sterol regulatory element-binding transcription factor 1 | Focal Adhesion-PI3K-Akt-mTOR signaling | 3 | 2 | 1 |
| Pfkfb3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | Focal Adhesion-PI3K-Akt-mTOR signaling | 2 | 1 | 1 |
| Efna1 | ephrin A1 | Focal Adhesion-PI3K-Akt-mTOR signaling | 2 | 1 | 1 |
| Acaa1a | acetyl-Coenzyme A acyltransferase 1A | PPAR | 6 | 1 | 5 |
| Acaa1b | acetyl-Coenzyme A acyltransferase 1B | PPAR | 2 | 1 | 1 |
| PVP-AgNPs | |||||
| Dlst | dihydrolipoamide S-succinyltransferase | TCA cycle | 8 | 6 | 2 |
| Pdk2 | pyruvate dehydrogenase kinase, isoenzyme 2 | TCA cycle | 4 | 2 | 2 |
| Mdh2 | malate dehydrogenase 2, NAD (mitochondrial) | TCA cycle Glycolysis & Gluconeogenesis |
3 | 2 | 1 |
| Idh3g | isocitrate dehydrogenase 3 (NAD+), gamma | TCA cycle | 13 | 11 | 2 |
| Sdhd | succinate dehydrogenase complex, subunit D | TCA cycle Electron transport Chain |
21 | 5 | 16 |
| Ndufs8 | NADH dehydrogenase (ubiquinone) Fe-S protein 8 | Electron transport chain | 12 | 11 | 1 |
| Ndufb9 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 | Electron transport chain | 13 | 3 | 10 |
| Pklr | pyruvate kinase liver and red blood cell | Glycolysis & gluconeogenesis | 12 | 9 | 3 |
| Eno1 | enolase 1, alpha non-neuron | Glycolysis & gluconeogenesis | 16 | 3 | 13 |
| Gsta2 | glutathione S-transferase A2 | MAPK, Keap1-Nrf2 | 6 | 1 | 5 |
| Gstt1 | glutathione S-transferase theta 1 | MAPK | 10 | 9 | 1 |
| Mapk3 | mitogen-activated protein kinase 3 | MAPK, IL-6 signaling | 20 | 18 | 2 |
| Hspb1 | heat shock protein 1 | MAPK, IL-6 signaling | 9 | 7 | 2 |
| Cpt2 | carnitine palmitoyltransferase 2 | Fatty Acid Beta Oxidation, PPAR | 5 | 5 | 0 |
| Crat | carnitine acetyltransferase | Fatty Acid Beta Oxidation | 9 | 3 | 6 |
| Acsl5 | acyl-CoA synthetase long-chain family member 5 | Fatty Acid Beta Oxidation, PPAR | 4 | 1 | 3 |
| Ehhadh | enoyl-Coenzyme A hydratase/3-hydroxyacyl Coenzyme A dehydrogenase | PPAR | 4 | 1 | 3 |
| Pltp | phospholipid transfer protein | PPAR | 18 | 18 | 0 |
| Hsp90aa1 | heat shock protein 90, alpha, class A member 1 | IL-6 signaling | 16 | 15 | 1 |
| Ppp2r1a | protein phosphatase 2 regulatory subunit A (PR 65), alpha | IL-6 signaling | 23 | 18 | 5 |
Differentially expressed mRNA, the associated pathway and the number of total, negatively (−) and positively (+) correlated lncRNAs are shown. Pathway information is derived from TAC analysis.
3.8. Data validation by qPCR
Differentially expressed protein-coding genes that showed highest up/down regulation and/or were shared between cit-AgNP and PVP-AgNP exposure groups were selected for validation by qPCR. The gene list included Arrdc3, Gsta2, Gstm3, Foxo1, Ilr1, Klf10, Lpin1, Lurap1, Nrf2, Slc38a2 and Saa1. The up/down regulation was confirmed for most genes (Table 5).
Table 5.
Clariom D assay data validation with qPCR.
| Gene | Gene Description | Fold-change (Clariom D) | Fold-change (qPCR) |
|---|---|---|---|
| cit-AgNPs | |||
| Arrdc3 | arrestin domain containing 3 | −6.5 | −2.8 |
| Gsta2 | glutathione S-transferase A2 | −2.4 | −2.2 |
| Nfe2l2 | nuclear factor, erythroid derived 2, like 2 | −1.6 | −1.3 |
| Lurap1 | leucine rich adaptor protein 1-like | 2 | 1.7 |
| Foxo1 | forkhead box O1 | 2.2 | 1.8 |
| Slc38a2 | solute carrier family 38, member 2 | 2.5 | 2.2 |
| II1r1 | interleukin 1 receptor, type I | 2.8 | 2.4 |
| Lpin1 | lipin 1 | 12.4 | 5.6 |
| PVP-AgNPs | |||
| Arrdc3 | arrestin domain containing 3 | −3.9 | −2 |
| Gstm3 | glutathione S-transferase, mu 3 | −3 | −1.2 |
| Gsta2 | glutathione S-transferase A2 | −2.7 | −2.7 |
| Klf10 | Kruppel-like factor 10 | −2.4 | −1.6 |
| Nfe2l2 | nuclear factor, erythroid derived 2, like 2 | −1.7 | 1.1 |
| Lurap1 | leucine rich adaptor protein 1-like | 2 | 3.1 |
| Slc38a2 | solute carrier family 38, member 2 | 2.3 | 3.8 |
| Saa1 | serum amyloid A 1 | 46 | 33 |
4. Discussion
Using a systems biology approach we examined whether and how oral exposure to AgNPs alters expression of protein-coding and lncRNA genes in a target organ and whether nanoparticle coatings modulate the gene expression profiles. While the pattern of gene expression regulation was similar by cit-AgNPs and PVP-AgNPs with more protein-coding genes being downregulated than upregulated and vice versa for lncRNAs, the number of differentially expressed genes, the gene spectrum and the altered biological processes were markedly different.
Overall, PVP-AgNPs induced a more robust transcriptional response characterized by more than two-fold higher number of differentially expressed genes than cit-AgNPs. Commonly regulated genes comprised only about ten percent of all differentially expressed genes in PVP-AgNP exposed mice and about twenty percent of all differentially expressed genes in cit-AgNP exposed mice. Functional annotation analysis showed that few significantly altered biological processes were shared between the exposure groups and included downregulation of glutathione metabolic process and upregulation of cellular oxidant detoxification. Commonly regulated pathways comprised a somewhat greater proportion than commonly regulated genes. About one third of the pathways were shared between cit-AgNP and PVP-AgNP exposed mice and include Keap-Nrf2, PPAR, MAPK and IL-6 signaling pathways. However, all genes associated with these pathways were downregulated by PVP-AgNPs, while bidirectional regulation of gene expression was observed after cit-AgNP exposure. For example, Cebpb (CCAAT/enhancer-binding protein), which is associated with Keap-Nrf2, IL-6 and other pathways, was upregulated by cit-AgNPs but unaltered by PVP-AgNPs. Cebpb is known to regulate the expression of genes involved immune and inflammatory responses, liver regeneration, gluconeogenesis and adipogenesis (Noh et al., 2011; Roy et al., 2005; van der Krieken et al., 2015). Thus, these data demonstrate that the nature of the surface coating strongly influences transcriptional responses to AgNPs. The reasons for a more robust transcriptional response to PVP-AgNPs than cit-AgNPs are presently unknown. We postulate that oral exposure to PVP-AgNPs results in a higher AgNP accumulation in the liver than exposure to cit-AgNPs due to a less pronounced agglomeration in gastrointestinal juices altering their tissue distribution after absorption into blood. Using an in vitro gastrointestinal digestion model mimicking AgNP transformation in the gastrointestinal tract, we found that after a 3-step digestion in saliva, gastric and intestinal juices PVP-AgNPs and cit-AgNPs and with a primary size of 20 nm agglomerated and formed two populations: nanoscale agglomerates and large agglomerates (>1000 nm) (Nallanthighal, Chan, Bharali, et al., 2017). The mean diameter of the nanoscale agglomerates was 50 nm for PVP-AgNPs and 120 nm for cit-AgNPs. The sizes of these agglomerates suggest that both can be absorbed from the gastrointestinal tract into blood to a similar degree, given that NPs of 50 – 100 nm show maximal gastrointestinal absorption (Florence, 2005). However, NPs smaller than the liver fenestrae pores (~100 nm) are better taken up by the liver, while NPs larger than the liver fenestrae pores are better taken up by the spleen (Li & Huang, 2008). Since cit-AgNP agglomerates exceed the liver fenestrae pores, they are likely better taken up by the spleen than liver, while PVP-AgNPs agglomerates are likely better taken up by the liver. Consistent with our hypothesis, Lankveld et al showed that following intravenous exposure small AgNPs (20 nm) distributed mainly to liver followed by kidney and spleen, while large AgNPs (80 and 110 nm) distributed mainly to spleen followed by liver and lung (Lankveld et al., 2010). Likewise, highest Ag levels were found in the spleen after intravenous injection of 90.5 nm AgNP agglomerates (Xue et al., 2012).
Although the proportion of commonly regulated genes was small, these genes may serve as biomarkers of AgNP exposure given that they were modulated by AgNPs independently of the nature of the coating material. Candidate genes include the genes discussed below. We found that both cit-AgNPs and PVP-AgNPs downregulated glutathione metabolic process characterized by downregulation of Gst family genes. Gst genes encode multifunctional enzymes that play important roles in the detoxification of xenobiotics and oxidative stress response (Daniel, 1993; Hayes & Pulford, 1995). The levels of expression of Gst genes determine the sensitivity of cells to chemical carcinogens and other toxic chemicals (Hayes & Pulford, 1995). In addition to enzymatic functions, Gsts play a role in the modulation of signal transduction pathways involved in cell survival and apoptosis via protein-protein interactions, where they control the activity of the MAPK family members (Allocati et al., 2018). Metals including Ag+ have been shown to inhibit the activity of Gsts (Dobritzsch et al., 2019). One of the possible implications of AgNP-mediated downregulation of Gst genes in the liver is an increased susceptibility to carcinogens, drugs and noxious chemicals. Downregulation of Gst genes is an unexpected finding given that a broad spectrum of chemicals including oxidants induce the expression of Gst genes as a cellular defense mechanism (Daniel, 1993; Hayes & Pulford, 1995). On the other hand, Nair et al. observed that exposure to AgNPs for 24 and 48 h increased expression of Gst genes in C.riparius larvae, while exposure for 72 h decreased expression levels of the same genes (Nair & Choi, 2011). This suggests Gst genes are induced early after AgNP exposure and suppressed thereafter.
Arrdc3 (arrestin domain containing 3) was one of the top ten downregulated genes in both AgNP exposure groups. It encodes a member of the arrestin superfamily of proteins that mediate intracellular trafficking of cell-surface receptors, G protein-coupled receptors (GPCRs), and thereby regulate cell signaling (Kang et al., 2014; Shenoy & Lefkowitz, 2011). GPCRs are relevant drug targets in cancer, cardiovascular, chronic inflammatory and other human disorders (Mason et al., 2012; O’Hayre et al., 2014). In mammals, the α-arrestin family, a subgroup of the arrestin superfamily, consists of Arrdc1 to 5 and Txnip (thioredoxin interacting protein that is also called vitamin D3 upregulated protein 1 or thioredoxin-binding protein-2) (Kang et al., 2014). Arrdc3 is required for ubiquitination and degradation of the GPCR, β2-adrenergic receptor (β2-AR) (Nabhan et al., 2010), and for lysosomal sorting and degradation of the GPCR, protease-activated receptor-1 (PAR1) (Dores et al., 2015). Arrdc3 acts as a tumor suppressor by controlling PAR1 trafficking and signaling (Arakaki et al., 2018). In addition, Arrdc3 regulates metabolism and body mass (Patwari & Lee, 2012). These roles of Arrdc3 suggest that downregulation of Arrdc3 by AgNPs can have multiple effects such as heightening the risk of cancer and altering metabolism in the body. Txnip, another member of the α-arrestin family, was significantly downregulated in PVP-AgNP exposed mice and was downregulated by more than 1.5-fold but did not reach statistical significance in cit-AgNP exposed mice. Consistent with our data, decreased levels of Txnip protein was observed in cit-AgNP exposed bone marrow-derived mast cells isolated from mice of the same genetic background as used in our study (Johnson et al., 2018). Txnip is a multifunctional protein that was initially identified as an inhibitor of thioredoxin and thus promoter of oxidative stress (Junn et al., 2000). In addition, Txnip is known to inhibit cellular glucose uptake, adipogenesis and body weight gain and act as a tumor suppressor via direct interactions with thioredoxin and by independent mechanisms (Patwari & Lee, 2012). Given these functions, downregulation of Txnip by AgNPs may be viewed as an adaptive response to oxidative stress but contributing to metabolic syndrome and cancer.
With regards to upregulated genes, lpin1 (lipin 1) was the top upregulated (>10-fold) gene in cit-AgNP exposed mice. Studies in transgenic animal models showed that overexpression of Lpin1 results in obesity (Phan & Reue, 2005). Lpin1 functions as a phosphatidate phosphatase enzyme in triglyceride and phospholipid biosynthesis and as an inducible transcriptional coactivator in the regulation of lipid metabolism genes (Reue & Zhang, 2008). Through these functions, Lpin1 plays an important role in metabolic homeostasis in adipose tissue, liver, muscle and other cell types. These data warrant follow-up studies to understand whether the magnitude of AgNP-mediated upregulation of Lpin1 along with other adipogenesis genes is sufficient to significantly disrupt metabolic homeostasis in the relevant tissues. The top upregulated genes (>45-fold) in PVP-AgNP exposed mice, were Saa1 (serum amyloid A1) and Saa2 (serum amyloid A2). Saa1 and Saa2 encode isoforms of an acute-phase protein serum amyloid A that is highly expressed in response to inflammation and tissue injury (Gabay & Kushner, 1999). Moderately but chronically elevated serum amyloid A is associated with a wide variety of chronic pathological conditions including atherosclerosis, rheumatoid arthritis and cancer (Kisilevsky & Manley, 2012). Our observation that PVP-AgNPs upregulated serum amyloid A expression is in line with the findings that exposure to AgNPs results in increased plasma levels of other acute-phase proteins (complement component 3, c-reactive protein, fibrinogen and plasminogen-activator inhibitor 1) (Ansar et al., 2017; Ferdous et al., 2019; Meng et al., 2014).
We did not observe upregulation of metallothionein family genes involved in metal ion sequestration, indicating heavy metal stress caused by Ag+ released from AgNPs. Increased expression of metallothionein genes was reported in cultured cells (Sahu et al., 2015; van der Zande et al., 2016) and rat lung (C. Guo et al., 2018) following AgNP exposure. However, study results of the current and published studies cannot be directly compared because of markedly different study designs. Transformation of AgNPs in the gastrointestinal tract, blood and liver may be different than that in culture medium or lungs. Our previous study suggests that both cit-AgNPs and PVP-AgNPs agglomerate during their transit through gastrointestinal tract (Nallanthighal, Chan, Bharali, et al., 2017). Agglomeration reduces the dissolution rate of AgNPs to Ag+ because dissolution rates decrease with the increasing AgNP sizes (Wang et al., 2014). While low pH and high ionic strength in the acidic stomach favor AgNP dissolution, the released Ag+ precipitate with Cl− strongly limiting bioavailability of the free Ag+ (Liu et al., 2012). In addition, Ag+ forms complexes with proteins containing thiol groups and small molecules such as glutathione further limiting availability of Ag+. Glutathione concentrations are particularly high in the liver (Sies, 1999). Therefore, oral AgNP exposures at relatively low doses may not yield high levels of the free Ag+, which could explain the lack of upregulation of metallothionein genes in our study.
In addition to protein-coding genes, we profiled expression of lncRNAs that are considered new biomarkers of exposure and regulators of toxicological responses (Bai et al., 2014; Gao et al., 2016; Li & Cui, 2018; Zhou et al., 2015). Similar to protein-coding genes, the proportion of differentially expressed lncRNAs shared between the two AgNP exposure groups was small (10 – 25%). The coding-non-coding gene co-expression analysis revealed that coding genes were co-expressed with a variable number of lncRNAs ranging from one to twenty three. Because co-expressed genes often have similar functions, predictions can be made that the co-expressed lncRNAs and mRNAs have shared functions (Luo et al., 2017; Nayak et al., 2009). Co-expression networks have been used successfully in a number of studies to identify unknown protein-coding gene functions (Luo et al., 2017). Thus, based on the coding-non-coding gene co-expression network, a number of lncRNAs can have functional roles with protein coding genes involved in Keap-Nrf2, MAPK, IL-1, IL-6 and PPAR and other pathways regulated by AgNPs. However, the predicted lncRNA functions have to be validated by further studies utilizing overexpression, knockdown and other genetic approaches. To date, very few lncRNAs have been functionally validated and mainly in human cells. No studies examined global changes in lncRNA expression as a function of AgNP exposure. One study analyzed expression levels of select lncRNAs by qPCR in a human erythroid cell line K562 exposed to PVP-AgNPs at 40 μg/ml for 24 h (Gao et al., 2017). The authors chose lncRNAs with reported functions in erythroid cell survival and differentiation and found that several lncRNAs were modulated by AgNPs. The mouse homologs of these human lncRNAs are currently unknown. Since the study of lncRNAs is at the early stage of development, it is premature to conclude which lncRNAs that were differentially expressed in our study could serve as novel biomarkers and regulators of toxicological responses to AgNPs. However, our lncRNA profiling data can serve as the basis for future studies focusing on lncRNAs as biomarkers of AgNP exposure or regulators of their mediated effects.
5. Conclusions
The current study revealed protein-coding and lncRNA gene expression signatures of cit-AgNPs and PVP-AgNPs in the mouse liver following oral AgNP exposure. The surface coating had a strong effect on AgNP-mediated gene expression by altering the number and the spectrum of modulated genes, pathways and biological processes. However, data generated in this study also predict that exposure to AgNPs can alter drug and chemical sensitivity, metabolic homeostasis and the risk of cancer irrespective of the coating type, warranting further investigations of the health effects associated with AgNP exposure.
Supplementary Material
Suppl. Table 1. GO terms and associated genes in cit-AgNP exposed mice.
Suppl. Table 2. GO terms and associated genes in PVP-AgNP exposed mice.
Highlights.
Only 10–20% of genes were commonly regulated by cit-AgNPs and PVP-AgNPs.
Protein-coding genes were mainly downregulated and lncRNAs were upregulated.
PVP-AgNPs induced a more robust transcriptional response than cit-AgNPs.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences (ESO24123) and the Dominic Ferraioli Foundation to RR. The authors would like to acknowledge Krishna Nallani for his help with R studio software and Cytoscape software used in generating lncRNA-mRNA co-expression networks.
Abbreviations:
- AgNPs
silver nanoparticles
- cit
citrate
- PVP
polyvinylpyrrolidone
- lncRNA
long noncoding RNA
- SPR
Surface Plasmon Resonance
- sp-ICP-MS
single particle inductively coupled mass spectroscopy
- STEM
scanning transmission electron microscopy
- GO
gene ontology
- TAC
Transcriptome Analysis Console
- GPCRs
G protein-coupled receptors
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest
Authors have no conflict of interests.
References
- Allocati N, Masulli M, Di Ilio C,Federici L 2018. Glutathione transferases: substrates, inihibitors and pro-drugs in cancer and neurodegenerative diseases. Oncogenesis. 7 (1), 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DS, Silva RM, Lee D, Edwards PC, Sharmah A, Guo T, Pinkerton KE,Van Winkle LS 2015. Persistence of silver nanoparticles in the rat lung: Influence of dose, size, and chemical composition. Nanotoxicology. 9 (5), 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansar S, Alshehri SM, Abudawood M, Hamed SS,Ahamad T 2017. Antioxidant and hepatoprotective role of selenium against silver nanoparticles. Int J Nanomedicine. 12, 7789–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakaki AKS, Pan WA, Lin H,Trejo J 2018. The alpha-arrestin ARRDC3 suppresses breast carcinoma invasion by regulating G protein-coupled receptor lysosomal sorting and signaling. J Biol Chem. 293 (9), 3350–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai W, Yang J, Yang G, Niu P, Tian L,Gao A 2014. Long non-coding RNA NR_045623 and NR_028291 involved in benzene hematotoxicity in occupationally benzene-exposed workers. Exp Mol Pathol. 96 (3), 354–360. [DOI] [PubMed] [Google Scholar]
- Benn TM,Westerhoff P 2008. Nanoparticle silver released into water from commercially available sock fabrics. Environ Sci Technol. 42 (11), 4133–4139. [DOI] [PubMed] [Google Scholar]
- Bergin IL, Wilding LA, Morishita M, Walacavage K, Ault AP, Axson JL, Stark DI, Hashway SA, Capracotta SS, Leroueil PR, et al. 2016. Effects of particle size and coating on toxicologic parameters, fecal elimination kinetics and tissue distribution of acutely ingested silver nanoparticles in a mouse model. Nanotoxicology. 10 (3), 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau MD, Imam MS, Paredes AM, Bryant MS, Cunningham CK, Felton RP, Jones MY, Davis KJ,Olson GR 2016. Differential Effects of Silver Nanoparticles and Silver Ions on Tissue Accumulation, Distribution, and Toxicity in the Sprague Dawley Rat Following Daily Oral Gavage Administration for 13 Weeks. Toxicol Sci. 150 (1), 131–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Jimenez B, Johnson ME, Montoro Bustos AR, Murphy KE, Winchester MR,Vega Baudrit JR 2017. Silver Nanoparticles: Technological Advances, Societal Impacts, and Metrological Challenges. Front Chem. 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel V 1993. Glutathione S-transferases: gene structure and regulation of expression. Crit Rev Biochem Mol Biol. 28 (3), 173–207. [DOI] [PubMed] [Google Scholar]
- Dobritzsch D, Grancharov K, Hermsen C, Krauss GJ,Schaumloffel D 2019. Inhibitory effect of metals on animal and plant glutathione transferases. J Trace Elem Med Biol. 57, 48–56. [DOI] [PubMed] [Google Scholar]
- Dores MR, Lin H, J.G. N, Mendez F,Trejo J 2015. The alpha-arrestin ARRDC3 mediates ALIX ubiquitination and G protein-coupled receptor lysosomal sorting. Mol Biol Cell. 26 (25), 4660–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echegoyen Y,Nerin C 2013. Nanoparticle release from nano-silver antimicrobial food containers. Food Chem Toxicol. 62, 16–22. [DOI] [PubMed] [Google Scholar]
- Ferdous Z, Al-Salam S, Greish YE, Ali BH,Nemmar A 2019. Pulmonary exposure to silver nanoparticles impairs cardiovascular homeostasis: Effects of coating, dose and time. Toxicol Appl Pharmacol. 367, 36–50. [DOI] [PubMed] [Google Scholar]
- Florence AT 2005. Nanoparticle uptake by the oral route: Fulfilling its potential? Drug Discov Today Technol. 2 (1), 75–81. [DOI] [PubMed] [Google Scholar]
- Gabay C,Kushner I 1999. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 340 (6), 448–454. [DOI] [PubMed] [Google Scholar]
- Gao C, He Z, Li J, Li X, Bai Q, Zhang Z, Zhang X, Wang S, Xiao X, Wang F, et al. 2016. Specific long non-coding RNAs response to occupational PAHs exposure in coke oven workers. Toxicol Rep. 3, 160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Zhao B, Chen M, Liu Y, Xu M, Wang Z, Liu S,Zhang C 2017. Nrf-2-driven long noncoding RNA ODRUL contributes to modulating silver nanoparticle-induced effects on erythroid cells. Biomaterials. 130, 14–27. [DOI] [PubMed] [Google Scholar]
- Garcia T, Lafuente D, Blanco J, Sanchez DJ, Sirvent JJ, Domingo JL,Gomez M 2016. Oral subchronic exposure to silver nanoparticles in rats. Food Chem Toxicol. 92, 177–187. [DOI] [PubMed] [Google Scholar]
- Ge L, Li Q, Wang M, Ouyang J, Li X,Xing MM 2014. Nanosilver particles in medical applications: synthesis, performance, and toxicity. Int J Nanomedicine. 9, 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geranio L, Heuberger M,Nowack B 2009. The behavior of silver nanotextiles during washing. Environ Sci Technol. 43 (21), 8113–8118. [DOI] [PubMed] [Google Scholar]
- Gliga AR, Skoglund S, Wallinder IO, Fadeel B,Karlsson HL 2014. Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release. Part Fibre Toxicol. 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Buckley A, Marczylo T, Seiffert J, Romer I, Warren J, Hodgson A, Chung KF, Gant TW, Smith R, et al. 2018. The small airway epithelium as a target for the adverse pulmonary effects of silver nanoparticle inhalation. Nanotoxicology. 12 (6), 539–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Li Y, Yan J, Ingle T, Jones MY, Mei N, Boudreau MD, Cunningham CK, Abbas M, Paredes AM, et al. 2016. Size- and coating-dependent cytotoxicity and genotoxicity of silver nanoparticles evaluated using in vitro standard assays. Nanotoxicology. 10 (9), 1373–1384. [DOI] [PubMed] [Google Scholar]
- Hayes JD,Pulford DJ 1995. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 30 (6), 445–600. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT,Lempicki RA 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 (1), 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT,Lempicki RA 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4 (1), 44–57. [DOI] [PubMed] [Google Scholar]
- Huynh KA,Chen KL 2011. Aggregation kinetics of citrate and polyvinylpyrrolidone coated silver nanoparticles in monovalent and divalent electrolyte solutions. Environ Sci Technol. 45 (13), 5564–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarroux J, Morillon A,Pinskaya M 2017. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol. 1008, 1–46. [DOI] [PubMed] [Google Scholar]
- Johnson M, Alsaleh N, Mendoza RP, Persaud I, Bauer AK, Saba L,Brown JM 2018. Genomic and transcriptomic comparison of allergen and silver nanoparticle-induced mast cell degranulation reveals novel non-immunoglobulin E mediated mechanisms. PLoS One. 13 (3), e0193499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Han SH, Im JY, Yang Y, Cho EW, Um HD, Kim DK, Lee KW, Han PL, Rhee SG, et al. 2000. Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol. 164 (12), 6287–6295. [DOI] [PubMed] [Google Scholar]
- Kang DS, Tian X,Benovic JL 2014. Role of beta-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr Opin Cell Biol. 27, 63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Song MY, Park JD, Song KS, Ryu HR, Chung YH, Chang HK, Lee JH, Oh KH, Kelman BJ, et al. 2010. Subchronic oral toxicity of silver nanoparticles. Part Fibre Toxicol. 7, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisilevsky R,Manley PN 2012. Acute-phase serum amyloid A: perspectives on its physiological and pathological roles. Amyloid. 19 (1), 5–14. [DOI] [PubMed] [Google Scholar]
- Lankveld DP, Oomen AG, Krystek P, Neigh A, Troost-de Jong A, Noorlander CW, Van Eijkeren JC, Geertsma RE,De Jong WH 2010. The kinetics of the tissue distribution of silver nanoparticles of different sizes. Biomaterials. 31 (32), 8350–8361. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim P, Yoon J, Lee B, Choi K, Kil KH,Park K 2013. Serum kinetics, distribution and excretion of silver in rabbits following 28 days after a single intravenous injection of silver nanoparticles. Nanotoxicology. 7 (6), 1120–1130. [DOI] [PubMed] [Google Scholar]
- Li CY,Cui JY 2018. Regulation of protein-coding gene and long noncoding RNA pairs in liver of conventional and germ-free mice following oral PBDE exposure. PLoS One. 13 (8), e0201387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SD,Huang L 2008. Pharmacokinetics and biodistribution of nanoparticles. Mol Pharm. 5 (4), 496–504. [DOI] [PubMed] [Google Scholar]
- Li X, Wu Z, Wang Y, Mei Q, Fu X,Han W 2013. Characterization of adult alpha- and beta-globin elevated by hydrogen peroxide in cervical cancer cells that play a cytoprotective role against oxidative insults. PLoS One. 8 (1), e54342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang Z, Liu FD, Kane AB,Hurt RH 2012. Chemical transformations of nanosilver in biological environments. ACS Nano. 6 (11), 9887–9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Baker SS, Baker RD, Nowak NJ,Zhu L 2011. Upregulation of hemoglobin expression by oxidative stress in hepatocytes and its implication in nonalcoholic steatohepatitis. PLoS One. 6 (9), e24363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeschner K, Hadrup N, Qvortrup K, Larsen A, Gao X, Vogel U, Mortensen A, Lam HR,Larsen EH 2011. Distribution of silver in rats following 28 days of repeated oral exposure to silver nanoparticles or silver acetate. Part Fibre Toxicol. 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Bu D, Sun L, Fang S, Liu Z,Zhao Y 2017. Identification and function annotation of long intervening noncoding RNAs. Brief Bioinform. 18 (5), 789–797. [DOI] [PubMed] [Google Scholar]
- Mackevica A, Olsson ME,Hansen SF 2017. The release of silver nanoparticles from commercial toothbrushes. J Hazard Mater. 322 (Pt A), 270–275. [DOI] [PubMed] [Google Scholar]
- Mason JS, Bortolato A, Congreve M,Marshall FH 2012. New insights from structural biology into the druggability of G protein-coupled receptors. Trends Pharmacol Sci. 33 (5), 249–260. [DOI] [PubMed] [Google Scholar]
- Meng J, Ji Y, Liu J, Cheng X, Guo H, Zhang W, Wu X,Xu H 2014. Using gold nanorods core/silver shell nanostructures as model material to probe biodistribution and toxic effects of silver nanoparticles in mice. Nanotoxicology. 8 (6), 686–696. [DOI] [PubMed] [Google Scholar]
- Munger MA, Radwanski P, Hadlock GC, Stoddard G, Shaaban A, Falconer J, Grainger DW,Deering-Rice CE 2014. In vivo human time-exposure study of orally dosed commercial silver nanoparticles. Nanomedicine. 10 (1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Pan H,Lu Q 2010. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 11 (8), 605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair PM,Choi J 2011. Identification, characterization and expression profiles of Chironomus riparius glutathione S-transferase (GST) genes in response to cadmium and silver nanoparticles exposure. Aquat Toxicol. 101 (3–4), 550–560. [DOI] [PubMed] [Google Scholar]
- Nallanthighal S, Chan C, Bharali DJ, Mousa SA, Vasquez E,Reliene R 2017. Particle coatings but not silver ions mediate genotoxicity of ingested silver nanoparticles in a mouse model. NanoImpact. 5, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallanthighal S, Chan C, Murray TM, Mosier AP, Cady NC,Reliene R 2017. Differential effects of silver nanoparticles on DNA damage and DNA repair gene expression in Ogg1-deficient and wild type mice. Nanotoxicology. 11 (8), 996–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NanoComposix. 2018. https://nanocomposix.com/pages/nanoparticle-surfaces.
- Nayak RR, Kearns M, Spielman RS,Cheung VG 2009. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 19 (11), 1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh K, Kim YM, Kim YW,Kim SG 2011. Farnesoid X receptor activation by chenodeoxycholic acid induces detoxifying enzymes through AMP-activated protein kinase and extracellular signal-regulated kinase 1/2-mediated phosphorylation of CCAAT/enhancer binding protein beta. Drug Metab Dispos. 39 (8), 1451–1459. [DOI] [PubMed] [Google Scholar]
- O’Hayre M, Degese MS,Gutkind JS 2014. Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr Opin Cell Biol. 27, 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang C, Brunelli A, Zhu C, Hristozov D, Liu Y, Semenzin E, Wang W, Tao W, Liang J, Marcomini A, et al. 2016. Demonstrating approaches to chemically modify the surface of Ag nanoparticles in order to influence their cytotoxicity and biodistribution after single dose acute intravenous administration. Nanotoxicology. 10 (2), 129–139. [DOI] [PubMed] [Google Scholar]
- Park EJ, Bae E, Yi J, Kim Y, Choi K, Lee SH, Yoon J, Lee BC,Park K 2010. Repeated-dose toxicity and inflammatory responses in mice by oral administration of silver nanoparticles. Environ Toxicol Pharmacol. 30 (2), 162–168. [DOI] [PubMed] [Google Scholar]
- Park K, Park EJ, Chun IK, Choi K, Lee SH, Yoon J,Lee BC 2011. Bioavailability and toxicokinetics of citrate-coated silver nanoparticles in rats. Arch Pharm Res. 34 (1), 153–158. [DOI] [PubMed] [Google Scholar]
- Patlolla AK, Hackett D,Tchounwou PB 2015. Silver nanoparticle-induced oxidative stress-dependent toxicity in Sprague-Dawley rats. Molecular and Cellular Biochemistry. 399 (1–2), 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwari P,Lee RT 2012. An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab. 23 (5), 216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PEN, C.P.I. 2018. Consumer Products Inventory. The project on emerging nanotechnologies (PEN). http://www.nanotechproject.org/inventories/consumer/
- Phan J,Reue K 2005. Lipin, a lipodystrophy and obesity gene. Cell Metab. 1 (1), 73–83. [DOI] [PubMed] [Google Scholar]
- Powers CM, Slotkin TA, Seidler FJ, Badireddy AR,Padilla S 2011. Silver nanoparticles alter zebrafish development and larval behavior: distinct roles for particle size, coating and composition. Neurotoxicol Teratol. 33 (6), 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros ME, Pierson R.t., Tulve NS, Willis R, Rogers K, Thomas TA,Marr LC 2013. Release of silver from nanotechnology-based consumer products for children. Environ Sci Technol. 47 (15), 8894–8901. [DOI] [PubMed] [Google Scholar]
- Reagan-Shaw S, Nihal M,Ahmad N 2008. Dose translation from animal to human studies revisited. FASEB J. 22 (3), 659–661. [DOI] [PubMed] [Google Scholar]
- Reue K,Zhang P 2008. The lipin protein family: dual roles in lipid biosynthesis and gene expression. FEBS Lett. 582 (1), 90–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SK, Shuman JD, Platanias LC, Shapiro PS, Reddy SP, Johnson PF,Kalvakolanu DV 2005. A role for mixed lineage kinases in regulating transcription factor CCAAT/enhancer-binding protein-{beta}-dependent gene expression in response to interferon-{gamma}. J Biol Chem. 280 (26), 24462–24471. [DOI] [PubMed] [Google Scholar]
- Royce SG, Mukherjee D, Cai T, Xu SS, Alexander JA, Mi Z, Calderon L, Mainelis G, Lee K, Lioy PJ, et al. 2014. Modeling Population Exposures to Silver Nanoparticles Present in Consumer Products. J Nanopart Res. 16 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu SC, Zheng J, Yourick JJ, Sprando RL,Gao X 2015. Toxicogenomic responses of human liver HepG2 cells to silver nanoparticles. J Appl Toxicol. 35 (10), 1160–1168. [DOI] [PubMed] [Google Scholar]
- Seiffert J, Hussain F, Wiegman C, Li F, Bey L, Baker W, Porter A, Ryan MP, Chang Y, Gow A, et al. 2015. Pulmonary toxicity of instilled silver nanoparticles: influence of size, coating and rat strain. PLoS One. 10 (3), e0119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannahan JH, Lai X, Ke PC, Podila R, Brown JM,Witzmann FA 2013. Silver nanoparticle protein corona composition in cell culture media. PLoS One. 8 (9), e74001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy SK,Lefkowitz RJ 2011. beta-Arrestin-mediated receptor trafficking and signal transduction. Trends Pharmacol Sci. 32 (9), 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkilnyy A, Souce M, Dubois P, Warmont F, Saboungi ML,Chourpa I 2009. Poly(ethylene glycol)-stabilized silver nanoparticles for bioanalytical applications of SERS spectroscopy. Analyst. 134 (9), 1868–1872. [DOI] [PubMed] [Google Scholar]
- Sies H 1999. Glutathione and its role in cellular functions. Free Radic Biol Med. 27 (9–10), 916–921. [DOI] [PubMed] [Google Scholar]
- Tiwari DK, Jin T,Behari J 2011. Dose-dependent in-vivo toxicity assessment of silver nanoparticle in Wistar rats. Toxicol Mech Methods. 21 (1), 13–24. [DOI] [PubMed] [Google Scholar]
- van der Krieken SE, Popeijus HE, Mensink RP,Plat J 2015. CCAAT/enhancer binding protein beta in relation to ER stress, inflammation, and metabolic disturbances. Biomed Res Int. 2015, 324815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zande M, Undas AK, Kramer E, Monopoli MP, Peters RJ, Garry D, Antunes Fernandes EC, Hendriksen PJ, Marvin HJ, Peijnenburg AA, et al. 2016. Different responses of Caco-2 and MCF-7 cells to silver nanoparticles are based on highly similar mechanisms of action. Nanotoxicology. 10 (10), 1431–1441. [DOI] [PubMed] [Google Scholar]
- van der Zande M, Vandebriel RJ, Van Doren E, Kramer E, Herrera Rivera Z, Serrano-Rojero CS, Gremmer ER, Mast J, Peters RJ, Hollman PC, et al. 2012. Distribution, elimination, and toxicity of silver nanoparticles and silver ions in rats after 28-day oral exposure. ACS Nano. 6 (8), 7427–7442. [DOI] [PubMed] [Google Scholar]
- Vance ME, Kuiken T, Vejerano EP, McGinnis SP, Hochella MF Jr., Rejeski D,Hull MS 2015. Nanotechnology in the real world: Redeveloping the nanomaterial consumer products inventory. Beilstein J Nanotechnol. 6, 1769–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio G, Fenech M, Pompa PP,Voelcker NH 2014. Lab-on-a-chip-based high-throughput screening of the genotoxicity of engineered nanomaterials. Small. 10 (13), 2721–2734. [DOI] [PubMed] [Google Scholar]
- Wang X, Ji Z, Chang CH, Zhang H, Wang M, Liao YP, Lin S, Meng H, Li R, Sun B, et al. 2014. Use of coated silver nanoparticles to understand the relationship of particle dissolution and bioavailability to cell and lung toxicological potential. Small. 10 (2), 385–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhang S, Huang Y, Zhang T, Liu X, Hu Y, Zhang Z,Tang M 2012. Acute toxic effects and gender-related biokinetics of silver nanoparticles following an intravenous injection in mice. J Appl Toxicol. 32 (11), 890–899. [DOI] [PubMed] [Google Scholar]
- Yang Y,Westerhoff P 2014. Presence in, and release of, nanomaterials from consumer products. Adv Exp Med Biol. 811, 1–17. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Liu H, Wang C, Lu Q, Huang Q, Zheng C,Lei Y 2015. Long non-coding RNAs as novel expression signatures modulate DNA damage and repair in cadmium toxicology. Sci Rep. 5, 15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suppl. Table 1. GO terms and associated genes in cit-AgNP exposed mice.
Suppl. Table 2. GO terms and associated genes in PVP-AgNP exposed mice.


