Summary
CD47 acts as a “Don’t eat me signal” that protects cells from phagocytosis by binding and activating its receptor SIPRA on macrophages. CD47 suppresses multiple different pro-engulfment “Eat Me” signals, including IgG, complement and calreticulin, on distinct target cells. This complexity has limited understanding of how the “Don’t eat me signal” is transduced biochemically. Here we utilized a reconstituted system with a defined set of signals to interrogate the mechanism of SIRPA activation and its downstream targets. CD47 ligation altered SIRPA localization, positioning SIRPA for activation at the phagocytic synapse. At the phagocytic synapse, SIRPA inhibited integrin activation to limit macrophage spreading across the surface of the engulfment target. Chemical reactivation of integrin bypassed CD47-mediated inhibition and rescued engulfment, similar to the effect of a CD47 function-blocking antibody. Thus, the CD47-SIRPA axis suppresses phagocytosis by inhibiting inside-out activation of integrin signaling in the macrophage, with implications to cancer immunotherapy applications.
Graphical Abstract
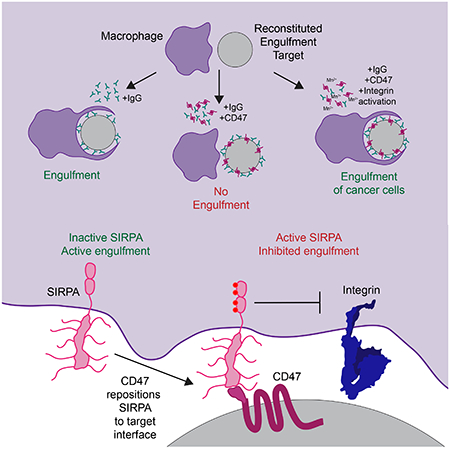
ETOC blurb
Morrissey et al use a reconstituted system to dissect the biochemical basis of the ‘Don’t Eat Me’ signal that is transmitted upon binding of CD47 on target cells its receptor SIRPA on macrophages. Steric exclusion of unligated SIRPA is required for phagocytosis. CD47 binding localized SIRPA to the phagocytic synapse and prevented integrin activation.
Introduction
The innate immune system is finely balanced to rapidly activate in response to pathogenic stimuli but remain quiescent in healthy tissue. Macrophages, key effectors of the innate immune system, measure activating and inhibitory signals to set a threshold for engulfment and cytokine secretion. The cell surface protein CD47 is a “Don’t Eat Me” signal that protects healthy cells from macrophage engulfment (Oldenborg et al., 2000). Hematopoietic cells lacking CD47 are rapidly engulfed by macrophages and trigger dendritic cell activation (Oldenborg et al., 2000; Yi et al., 2015). CD47 also functions in the nervous system, protecting active synapses from pruning by microglia (Lehrman et al., 2018). CD47 expression is often increased on cancer cells as a mechanism to evade immune detection (Chao et al., 2012; Jaiswal et al., 2009; Majeti et al., 2009; Oldenborg et al., 2001, 2000). CD47 function-blocking antibodies result in decreased cancer growth or tumor elimination (Advani et al., 2018; Chao et al., 2010a; Gholamin et al., 2017; Jaiswal et al., 2009; Willingham et al., 2012). Augmenting macrophage function by CD47 blockade may also be beneficial in other disease contexts such as atherosclerosis or viral infection (Cham et al., 2020; Kojima et al., 2016). Despite the therapeutic promise of manipulating CD47 signaling, there is limited insight into the mechanism whereby CD47 suppresses macrophage engulfment.
CD47 on the surface of target cells is recognized by SIRPA (Signal Regulatory Protein α) on macrophages or dendritic cells (Jiang et al., 1999; Liu et al., 2015; Okazawa et al., 2005; Oldenborg et al., 2000; Seiffert et al., 1999; Tseng et al., 2013; Yi et al., 2015). SIRPA is an inhibitory receptor containing multiple intracellular Immune Tyrosine-based Inhibitory Motifs (ITIMs; Kharitonenkov et al., 1997). Macrophages lacking SIRPA do not exhibit reduced phagocytosis of CD47-bearing targets, suggesting that SIRPA is the primary transducer of the CD47 signal (Okazawa et al., 2005; Oldenborg et al., 2000). Activation of SIRPA must be controlled with high fidelity to suppress engulfment of viable cells when CD47 is present while allowing for robust engulfment of targets lacking CD47. CD47 binding triggers SIRPA phosphorylation by Src family kinases (Barclay and Brown, 2006), but how CD47 binding is translated across the cell membrane to drive SIRPA phosphorylation is not known. Phosphorylated SIRPA recruits the phosphatases SHP-1 and SHP-2 (Fujioka et al., 1996; Noguchi et al., 1996; Okazawa et al., 2005; Oldenborg et al., 2001; Veillette et al., 1998), but the downstream targets of these phosphatases and their relationship to the engulfment process are not clear.
In vivo, CD47 suppresses multiple different pro-engulfment “Eat Me” signals, including IgG, complement and calreticulin (Chen et al., 2017; Gardai et al., 2005; Oldenborg et al., 2001). This complexity, in addition to substantial variation in target size, shape and concentration of “Eat Me” signals, can make a quantitative, biochemical understanding of receptor activation difficult. To overcome this problem, here we utilized a synthetic target cell-mimic with a defined set of signals to interrogate the mechanism of SIRPA activation and its downstream targets. We found that CD47 ligation altered SIRPA localization, positioning SIRPA for activation at the phagocytic synapse. At the phagocytic synapse, SIRPA inhibited integrin activation to limit macrophage spreading across the surface of the engulfment target. Directly activating integrin eliminated the effect of CD47 and rescued engulfment, similar to the effect of a CD47 function-blocking antibody. Thus, the CD47-SIRPA axis suppresses phagocytosis by inhibiting inside-out activation of integrin signaling in the macrophage, with implications to cancer immunotherapy applications.
Results
CD47 suppresses IgG and phosphatidylserine “Eat Me” signals
To study the mechanism of “Eat Me” and “Don’t Eat Me” signal integration during engulfment, we used a reconstituted engulfment target (Figure 1A). Silica beads were coated in a supported lipid bilayer to mimic the surface of a cancer cell. To activate engulfment, we introduced IgG, a well-defined “Eat Me” signal that synergizes with CD47 blockade to promote cancer cell clearance (Chao et al., 2010a; Freeman and Grinstein, 2014). IgG is recognized by the Fcγ Receptor family (FcR), which activates downstream signaling and engulfment (Freeman and Grinstein, 2014). To activate SIRPA, we incorporated the CD47 extracellular domain at a surface density selected to mimic the CD47 density on cancer cells (~600 molecules/μm2, Figure S1).
Figure 1: CD47-SIRPA suppresses IgG and PS dependent engulfment.
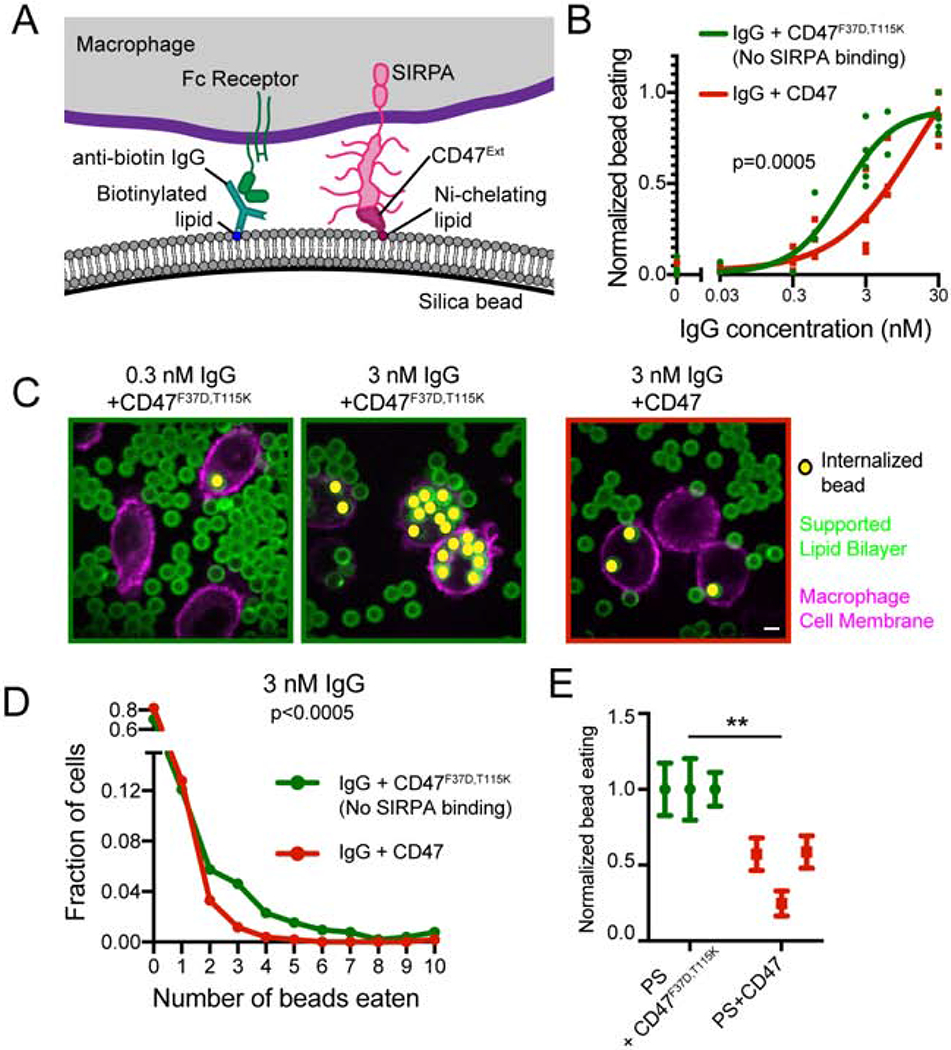
(A) Schematic shows the supported lipid bilayer system used in this study. Anti-biotin IgG is bound to biotinylated lipids. IgG is recognized by Fc Receptor on the macrophage. The extracellular domain of CD47-His10 is bound to Ni-NTA-conjugated lipids and recognized by SIRPA expressed by the macrophage. (B) Silica beads are coated with a supported lipid bilayer and incubated with the indicated concentration of IgG and either CD47 (red) or an inactive mutant CD47 (F37D, T115K; green). The functionalized beads were added to RAW264.7 macrophages and fixed after 30 min. The average number of beads per macrophage was assessed by confocal microscopy and normalized to the maximum average bead eating observed in that replicate. Each dot represents an independent replicate (n≥100 cells analyzed per experiment), and data are compared using a two-way ANOVA. For visualization, the data were fit using the “[Agonist] vs response – variable slope” model in Graphpad Prism. (C) Still images depict the assay described in (B). The supported lipid bilayers contain the fluorescently-labeled lipid atto390-DOPE (green) and the macrophages membranes are labeled with CellMask (magenta). Internalized beads are indicated with a yellow dot. (D) Graph depicts the fraction of cells engulfing the indicated number of beads (pooled data from three independent replicates included in (B)). Macrophages encountering CD47-conjugated beads (red) were less likely to engulf, and those that did engulfed fewer beads. CD47F37D,T115K, a mutant that cannot bind SIRPA, was used as a control (green). (E) Macrophages were incubated with beads coated with a supported lipid bilayer containing 10% phosphatidylserine and either CD47 or the inactive CD47F37D,T115K. Data were normalized to the maximum bead eating observed in that replicate. The complete, pooled data are shown in Supplementary Figure 1E. Dots and error bars denote the mean and standard error of independent replicates. *** indicates p<0.0005 and ** indicates p<0.005 by a Kruskal-Wallis test on the pooled data (D) or a Student’s T test on the means of individual replicates (E). Scale bar denotes 5 μm in this and all subsequent figures.
Using this system, we tested the effect of CD47 on engulfment across a titration of IgG densities (Figure S1). We mixed beads with the macrophage-like cell line RAW264.7 and measured the number of internalized beads by confocal microscopy. We found that CD47 suppressed engulfment at intermediate IgG densities, but did not appreciably affect engulfment of targets with high densities of bound IgG (Figure 1B–D). The presence of CD47 did not completely eliminate phagocytosis, but rather caused a quantitative decrease in the fraction of cells initiating engulfment and the number of beads engulfed per cell (Figure 1B, C, Figure S1). This suppression was dependent on CD47 binding as a mutated CD47 extracellular domain that is unable to bind to SIRPA (F37D, T115K; Hatherley et al., 2008) was unable to suppress engulfment.
We further examined whether CD47-SIRPA signaling could suppress engulfment of targets mimicking apoptotic corpses. A critical “Eat Me” signal from apoptotic corpses is phosphatidylserine, which becomes exposed on the outer leaflet of the plasma membrane during cell stress, apoptosis (Fadok et al., 1992; Poon et al., 2014), and on some cancer cells (Birge et al., 2016; Utsugi et al., 1991). We found that engulfment of beads containing 10% phosphatidylserine in the supported lipid bilayer was inhibited by the inclusion of CD47 on the bilayer (Figure 1E, Figure S1). Together, these data show that CD47-SIRPA signaling can block engulfment driven by IgG and phosphatidylserine. Moreover, bilayer-coated beads provide a well-defined and tunable platform for studying the integration of “Eat Me” and “Don’t Eat Me” signals during engulfment.
CD47 ligation relocalizes SIRPA to the phagocytic synapse
We next sought to determine the mechanism by which CD47 ligation regulates SIRPA activity. We first examined SIRPA localization during phagocytosis of IgG-coated beads. When not bound to CD47, SIRPA was segregated away from the phagocytic cup that enveloped IgG-coated beads (Figure 2A). Similarly, SIRPA was depleted at the center of the immunological synapse between a macrophage and a supported lipid bilayer containing phosphatidylserine (Figure S2). In contrast, in the presence of CD47, SIRPA remained at the phagocytic cup (Figure 2A). These data demonstrate that unligated SIRPA is excluded from the phagocytic synapse, whereas CD47-bound SIRPA remains at the phagocytic synapse.
Figure 2: Unligated SIRPA is excluded from the phagocytic synapse.
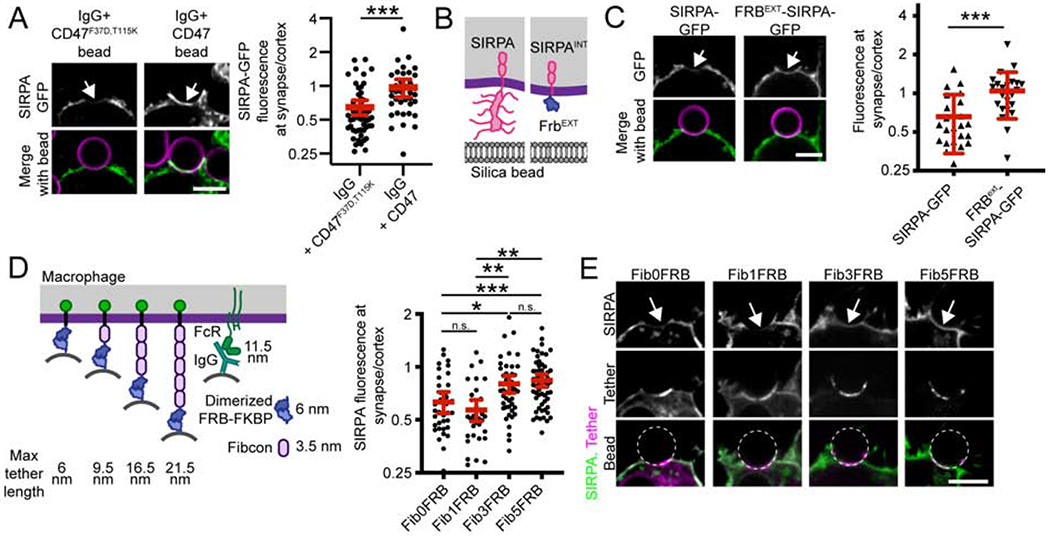
(A) SIRPA-GFP (top; green in merge) is depleted from the base of the phagocytic cup (arrow) when a macrophage engulfs a bead functionalized with IgG and CD47F37D, T115K, which cannot bind SIRPA (left; supported lipid bilayer, magenta). SIRPA is not depleted when CD47 is present (IgG+CD47, right). Graph depicts the ratio of SIRPA-GFP at the phagocytic cup/cell cortex for individual phagocytic cups. (B) A schematic shows full length SIRPA on the left and FRBext-SIRPA on the right. (C) SIRPA-GFP and FRBext-SIRPA-GFP are shown at cell-bead contacts (arrow). On the right, a graph depicts the ratio of GFP fluorescence at the synapse (arrow) compared to the cortex for the indicated SIRPA chimeras. (D) A schematic shows the synthetic tethers used to control macrophage-bead spacing. ‘Max tether length’ indicates the predicted height of these semi-rigid proteins if fully extended (Bakalar et al., 2018). Purified FKBP is attached to supported lipid-bilayer-coated beads, and the Fibcon repeat proteins with a N terminal FRB are expressed in the macrophage. In the presence of a rapamyacin analog, FKBP and FRB form a high affinity dimer tethering the bead to the macrophage. SIRPA-mCherry fluorescence was measured at the cell-bead synapse (arrow in E) and the cell cortex. The ratio of SIRPA fluorescence at the synapse compared to the cortex is graphed on the right. Representative images are shown in (E). Short tethers (Fib0FRB and Fib1FRB) drive a depletion of SIRPA at the cell-bead synapse, and long tethers (Fib3FRB and Fib5FRB) drive little or no depletion of SIRPA. In A, C, and D, dots represent individual cups, red lines show mean ± 95% confidence intervals, and data is pooled from three independent experiments. *** denotes p<0.0005, ** denotes p<0.005, * denotes p<0.05 and n.s. denotes p>0.05 as determined by a Student’s T test (A, C); or an Ordinary one-way ANOVA with Holm-Sidak multiple comparison test (D).
We next sought to address the mechanism of SIRPA segregation away from the phagocytic cup in the presence of IgG and absence of CD47. We hypothesized that exclusion of unligated SIRPA from the synapse could be driven by its heavily glycosylated extracellular domain, either by interactions with the surrounding glycocalyx or steric exclusion from the spatially restricted phagocytic synapse. We therefore created a SIRPA chimeric receptor where the extracellular domain was replaced with a small, inert protein domain (FRBEXT-SIRPA; Figure 2B). Unlike full-length SIRPA, FRBEXT-SIRPA was not segregated away from the cell-target synapse (Figure 2C). This result demonstrates that the extracellular domain of SIRPA is required for SIRPA exclusion from the phagocytic cup.
Exclusion of bulky phosphatases, like CD45, is driven by the steric constraints of a small distance between the macrophage and target membrane (Bakalar et al., 2018; Freeman et al., 2016). We hypothesized that the short membrane-membrane distance driven by FcR-IgG ligation (~11.5 nm, Lu et al., 2011) may be sufficient to exclude SIRPA. To test this, we generated a series of synthetic tethers varying in length (Figure 2D). In the macrophage, we expressed synthetic transmembrane proteins containing an intracellular GFP, and an extracellular domain with 0, 1, 3 or 5 repeats of a synthetic FNIII protein, Fibcon (Bakalar et al., 2018; Jacobs et al., 2012), plus half of an inducible dimerization system (Fib0FRB to Fib5FRB). The other half of the inducible dimerization system was attached to bilayer-coated beads (FKBP-His10). We then tethered beads to the macrophages in the absence of IgG or CD47 by adding a rapamycin analog to induce dimerization between the synthetic proteins (Spencer et al., 1993). We quantified SIRPA exclusion from the phagocytic synapse. We found that the tethers containing no Fibcon repeats (FRB-FKBP alone, ~6 nm) or one Fibcon repeat (Fib1FRB-FKBP, ~9.5 nm) drove SIRPA exclusion of a similar magnitude to FcR-IgG ligation (Figure 2D and E). The efficiency of SIRPA exclusion decreased with longer tether lengths (Fib3FRB-FKBP, 16.5 nm when fully extended; and Fib5FRB-FKBP, 21.5 nm). Together, these data suggest that SIRPA exclusion can be controlled by altering the height of the immunological synapse.
Targeting SIRPA to the phagocytic synapse suppresses engulfment
Receptor activation by Src family kinases at the phagocytic cup is favored due to exclusion of bulky phosphatases like CD45 (Freeman et al., 2016; Goodridge et al., 2011). SIRPA contains two immune tyrosine inhibitory motifs (ITIMs) that are phosphorylated by Src family kinases and essential for downstream signaling (Fujioka et al., 1996; Tsuda et al., 1998). We therefore hypothesized that positioning SIRPA at the phagocytic cup may drive ITIM phosphorylation and receptor activation. To distinguish between the effects of CD47 binding and synapse localization, we developed a chimeric SIRPA receptor that localized to the phagocytic synapse in the absence of CD47. We replaced the SIRPA extracellular domain with the IgG-binding extracellular domain of the FcγR3 α chain (Figure 3A; termed FcR3EXT-SIRPAINT). This receptor is driven into the synapse by IgG binding instead of CD47 (Figure 3A). Expression of this synapse-localized chimera suppressed engulfment of IgG-coated beads in the absence of CD47 (Figure 3B, Figure S3). As a control, we expressed a chimeric construct with the four tyrosines of the ITIM domains mutated to phenylalanines, prohibiting phosphorylation and activation of SIRPA (FcR3ext-SIRPA 4Fint). This construct did not affect engulfment, suggesting that the inhibitory effect of the SIRPA chimera is signaling dependent (Figure 3B). Thus, targeting SIRPA to the phagocytic cup is sufficient to inhibit engulfment, even in the absence of its natural ligand CD47.
Figure 3: Localizing SIRPA to the macrophage-target synapse suppresses engulfment.
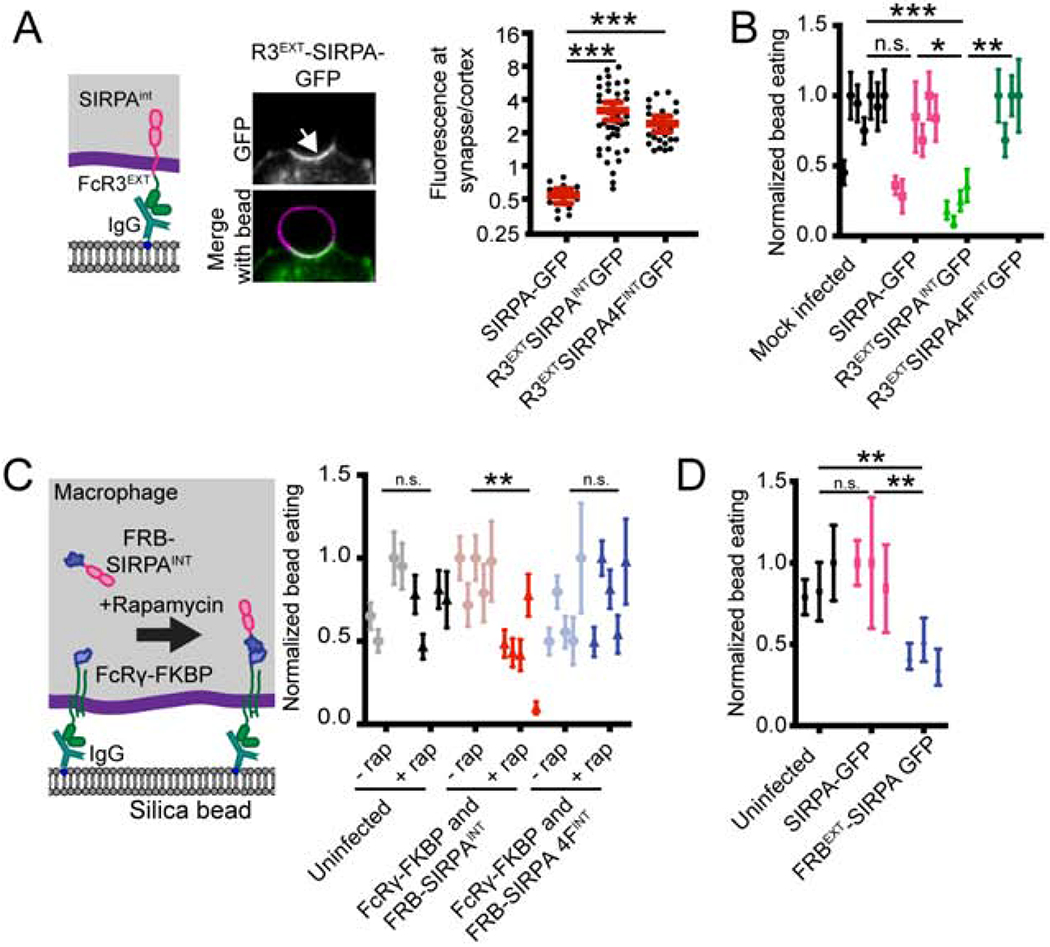
(A) A schematic of the chimeric synapse localized FcR3EXTSIRPAINT construct is shown on the left and representative images of its localization to the cell bead interface are shown in the center. On the right, a graph depicts the ratio of GFP fluorescence at the macrophage-bead synapse compared to the cortex for full length SIRPA, FcR3EXTSIRPAINT, and the chimeric SIRPA construct with the four tyrosines in the ITIM domains mutated to phenylalanine (FcR3EXTSIRPA 4FINT). (B) A graph depicts the average number of internalized IgG beads per macrophage expressing the indicated chimeric SIRPA constructs normalized to the maximum bead eating in that replicate. Mock infected macrophage or macrophages expressing membrane tethered GFP (GFP-CAAX) were used as a control.. (C) Schematic (left) shows a system for inducible recruitment of the SIRPA intracellular domain to the phagocytic cup. Recruiting SIRPA to the phagocytic cup suppresses engulfment compared to soluble SIRPA or compared to wild-type macrophages treated with a rapamycin analog (normalized to maximum bead eating in that replicate). Mutating the ITIM domains of SIRPA (SIRPA 4F) eliminated a rapamycin-induced effect. (D) The graph shows the number of beads engulfed by uninfected, SIRPA-GFP, or FRBEXT-SIRPA expressing macrophages normalized to the maximum observed eating in that replicate. In A, dots represent individual cups, red lines show mean ± 95% confidence intervals, and data is pooled from three independent experiments. In B, C, and D dots show the average from independent replicates with the error bars denoting SEM for that replicate. *** denotes p<0.0005, ** denotes p<0.005, * denotes p<0.05 and n.s. denotes p>0.05 as determined by an Ordinary one-way ANOVA with Holm-Sidak multiple comparison test.
As an alternative strategy to control the localization of SIRPA activity, we fused one half of the chemically inducible dimer to FcR (FcR γ chain-FKBP) and the second to a soluble SIRPA intracellular domain (FRB-SIRPAINT, Figure 3C). We then recruited the SIRPA intracellular domain to FcR by adding a rapamyacin analog to induce dimerization of FRB and FKBP. In the absence of rapamycin, cells efficiently engulfed IgG-coated beads (Figure 3C). In contrast, rapamycin-induced recruitment of the SIRPA intracellular domain to the FcR γ chain significantly suppressed engulfment (Figure 3C). When the ITIM domain of SIRPA was mutated, this construct no longer affected engulfment (Figure 3C).
We next returned to the extracellular domain truncation of SIRPA (FRBEXT-SIRPA), which was not excluded from the phagocytic synapse (Figure 2B,C), to determine if eliminating SIRPA exclusion is sufficient to suppress engulfment. FRBEXT-SIRPA constitutively suppressed engulfment (Figure 3D), demonstrating that exclusion of SIRPA is essential for efficient engulfment of targets presenting “Eat Me” signals. Taken together, these experiments show that exclusion of unligated SIRPA is essential for efficient phagocytosis and that CD47 activates SIRPA by positioning SIRPA at the phagocytic synapse.
CD47 does not suppress engulfment by altering Syk recruitment to IgG microclusters
We next sought to determine how activated SIRPA inhibits engulfment. Phosphorylated SIRPA recruits the phosphatases SHP-1 and SHP-2 via their phosphobinding SH2 domains but the downstream targets of SHP-1 and SHP-2 are not known (Fujioka et al., 1996; Noguchi et al., 1996; Okazawa et al., 2005; Oldenborg et al., 2001; Veillette et al., 1998). One potential target of SIRPA-bound SHP phosphatases is FcR itself. We used TIRF microscopy to examine the initial steps in the engulfment signaling cascade with high temporal and spatial resolution. When macrophages interacted with an IgG-bound supported lipid bilayer, the cells formed IgG microclusters that recruited Syk (Figure 4A; Lin et al., 2016). When we compared static images of macrophages that had landed on a bilayer containing IgG and CD47, or IgG and the inactive CD47F37D, T115K, we did not detect a significant difference in the fraction of cells forming IgG microclusters or the total area of the IgG microclusters under the cells. There was also no significant difference in the fraction of cells containing Syk microclusters or the amount of Syk-mCherry recruited to these clusters (Figure 4B,C). Further, we found that SIRPA did not co-localize with IgG clusters when macrophages landed on a bilayer containing IgG and CD47, suggesting that SIRPA is not positioned to dampen receptor activation (Figure 4D). Overall, this suggests that changes to FcR activation and Syk recruitment are unlikely to account for the effect of SIRPA, consistent with previous biochemical observations (Okazawa et al., 2005; Tsai and Discher, 2008).
Figure 4: CD47 does not affect IgG clustering and Syk recruitment.
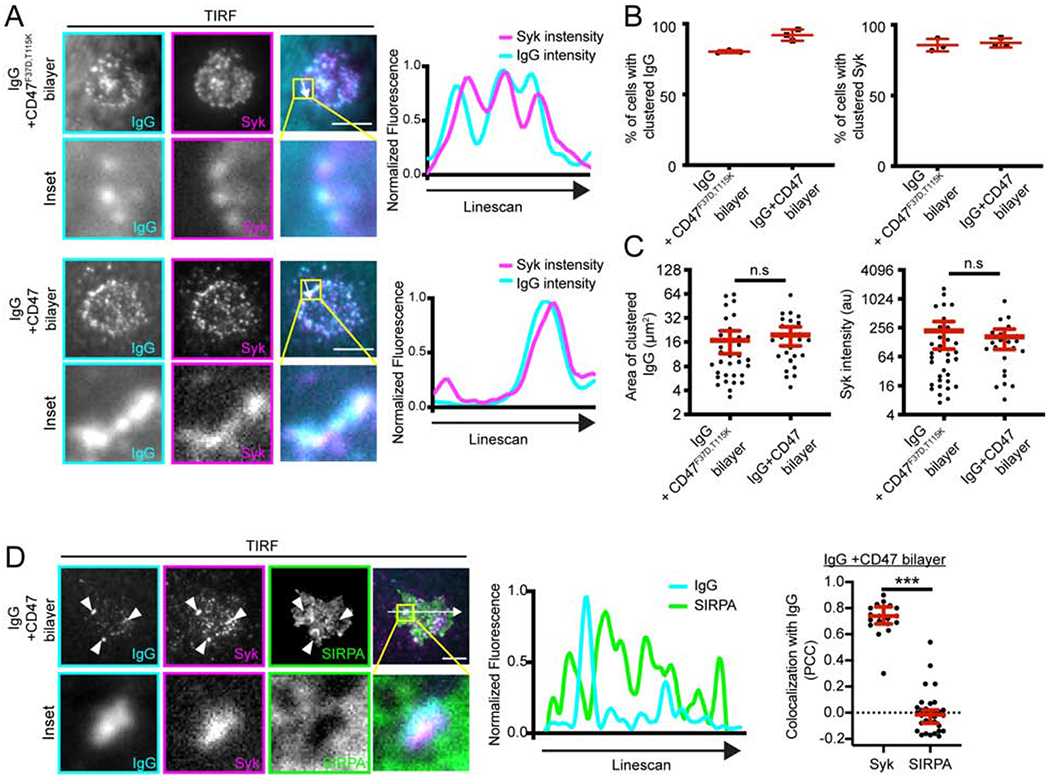
(A) TIRF microscopy shows that macrophages are able to form IgG microclusters (left; cyan in merged image) that recruit Syk (middle; magenta in merged image) when landing on bilayers containing IgG + CD47F37D,T115K (top) or IgG + CD47 (bottom). Inset shows the boxed region of the image above. The linescan shows the fluorescent intensity of AlexaFluor 647-IgG and Syk-mCherry at the indicated position (white arrow). Intensity was normalized so that 1 is the highest observed intensity and 0 is background. (B) The fraction of cells forming IgG (left) or Syk (right) was measured in static images of macrophages that had landed on the indicated supported lipid bilayer after 15 min. (C) Size of the IgG clusters (left), and the mean fluorescent intensity of Syk-mCherry colocalizing with these clusters (right) was measured at the same timepoint. (D) TIRF microscopy shows that, in the presence of CD47, SIRPA (green) does not co-localize with IgG clusters (cyan; arrowheads). Inset shows the boxed region in the above image. The linescan shows the fluorescent intensity of AlexaFluor 647-IgG and SIRPA-GFP at the position indicated by a white arrow. The Pearson’s Correlation Coefficient (PCC) for Syk and IgG, or SIRPA and IgG in individual cells is indicated in the graph on the right. In B, each dot represents the percent from an independent experiment (n≥ 20 cells per replicate) and the lines denote mean ± SD. In C and D, each dot represents an individual cell and data was pooled from three independent experiments. The red lines indicate the mean ± 95% confidence intervals. *** denotes p<0.0005, and n.s. denotes p>0.05 as determined by a Student’s T test.
CD47 prevents integrin activation
We next assessed the dynamics of cells landing on functionalized supported lipid bilayers. We found that cells on IgG-coated bilayers spread across the bilayer surface (Figure 5A, Movie S1). In contrast, macrophages encountering an IgG and CD47-containing bilayer exhibited reduced cell spreading (Figure 5A, Movie S2). TIRF imaging at a static timepoint revealed that fewer macrophages were interacting with the bilayer, and those interacting had a smaller footprint (Figure 5B). These data show that CD47 inhibits cell spreading across a target substrate.
Figure 5: CD47 prevents integrin activation.
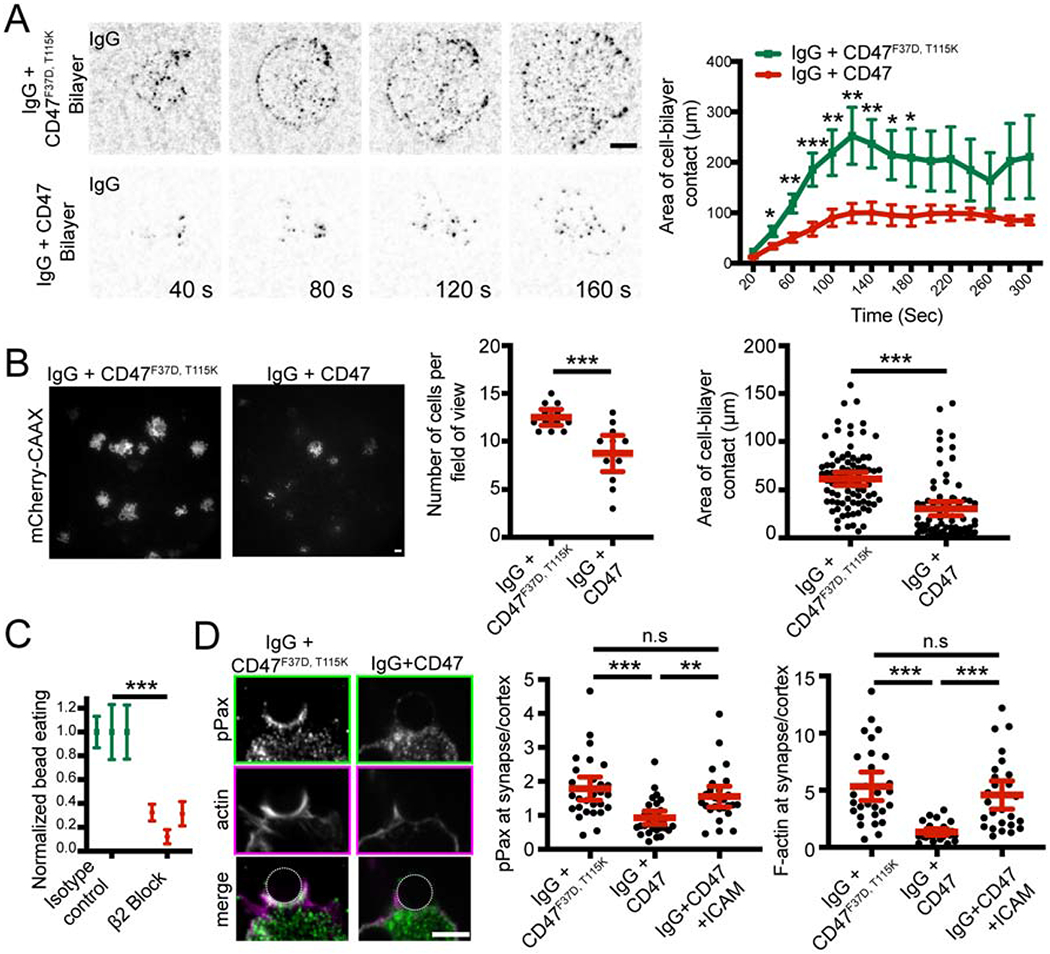
(A) Still images from a TIRF microscopy timelapse show that macrophages form IgG (black) microclusters as they spread across bilayers containing IgG and an inactive CD47F37D, T115K which cannot bind to SIRPA bilayer (top). Adding CD47 to the bilayer inhibits cell spreading (bottom; graphed on right, average area of contact from n≥11 cells ± SEM, pooled from three separate experiments). (B) TIRF images show the cell membrane (mCherry-CAAX; white) of macrophages engaging with an IgG and inactive CD47F37D, T115K (left), or IgG and CD47 (right) bilayer. Graphs depict the average number of cells seen contacting the bilayer after 10 min (center) and the average area of cell contact (right). Each dot represents an individual field of view (center) or cell (right) pooled from three independent experiments. (C) Blocking integrin activation using a function-blocking antibody (2E6) targeting the β2 integrin subunit decreased the efficiency of engulfment. Graph shows the number of beads engulfed normalized to the maximum observed eating in that replicate. Each data point represents an independent experiment and the error bars denote the SEM for that replicate. (D) Immunofluorescence images show phosphopaxillin (top; green in merge) and F-actin (center; magenta in merge; visualized with phalloidin) at the phagocytic cup of a bead containing IgG and inactive CD47F37D, T115K (left) or an IgG- and CD47-coated bead (right). Graphs show the ratio of phosphopaxilin (center) or actin (right) intensity at the phagocytic cup/cell cortex. Each dot represents an individual phagocytic cup; lines denote the mean ± 95% confidence intervals. *** denotes p<0.0005, ** denotes p<0.005, and * denotes p<0.05 as determined by Student’s T test (A, B, and C) or Ordinary one-way ANOVA with Tukey’s Multiple Comparison test (D).
Cell spreading is thought to involve activation of integrins and the actin cytoskeleton (Springer and Dustin, 2011). Inactive integrins exist in a low affinity, bent confirmation (Springer and Dustin, 2011). Upon activation, the extracellular domain extends into an open conformation that can bind many ligands with high affinity (Freeman and Grinstein, 2014; Springer and Dustin, 2011). FcR activation stimulates inside-out activation of integrins (Dupuy and Caron, 2008; Jones et al., 1998). Activated integrins can then promote engulfment, either by increasing adhesion to the target particle or by providing a platform for intracellular signaling and actin assembly (Dupuy and Caron, 2008; Wong et al., 2016). We found that inhibiting integrin with a β2 integrin function-blocking antibody (2E6) or Fab dramatically decreased the efficiency of IgG-mediated engulfment (Figure 5C and S4). We could also detect a role for αM integrin in engulfment, but not for β3 or αL (Figure S4). Thus, blocking αMβ2 integrin is sufficient to suppress engulfment.
Because integrin is required for cell spreading and engulfment (Springer and Dustin, 2011), we hypothesized that CD47-SIRPA signaling may inhibit engulfment by preventing inside-out activation of integrin. Supporting this hypothesis, a previous study identified phosphopaxillin, which is specifically recruited to sites of integrin activation, as one of a number of phosphoproteins affected by CD47 (Geiger et al., 2009; Tsai and Discher, 2008). Consistent with this, we found that the enrichment of phospho-paxillin at the interface of the macrophage with an IgG-coated bead was substantially diminished by the simultaneous presence of CD47 on the bead (Figure 5D). Together, these data indicate that CD47-SIRPA prevents integrin activation.
Activating integrin bypasses CD47-SIRPA inhibitory signaling
CD47-SIRPA has previously been reported to affect phosphorylation of multiple proteins, including paxillin and myosin (Tsai and Discher, 2008). We also found that SIRPA inhibited F-actin accumulation at the phagocytic cup (Figure 5D). It is not clear which of these pathways is a direct target of CD47 signaling and which is a secondary effect of altered upstream signaling. We hypothesized that if SIRPA signaling suppresses engulfment primarily by inhibiting integrin inside-out activation, then directly activating integrin might bypass SIRPA-mediated inhibition and permit bead engulfment (Figure 6A). Alternatively, if the target of CD47-SIRPA signaling is in a parallel pathway or downstream of integrin activation, then activating integrin should not rescue engulfment following SIRPA activation. To activate integrin, we treated macrophages with manganese, which locks integrin into a high-affinity open conformation (Dransfield et al., 1992). We found that macrophages treated with 1 mM manganese engulfed beads with a similar efficiency whether or not CD47 was conjugated to the supported lipid bilayer (Figure 6B). Importantly, manganese did not trigger bead engulfment on its own or dramatically enhance engulfment of IgG-coated beads in the absence of CD47 (Figure 6B,C), establishing that increasing integrin activation is not sufficient to trigger engulfment. Thus, a manganese-induced increase in engulfment was specific to beads coated with CD47 and IgG.
Figure 6: Bypassing inside out activation of integrin eliminates the effect of CD47.
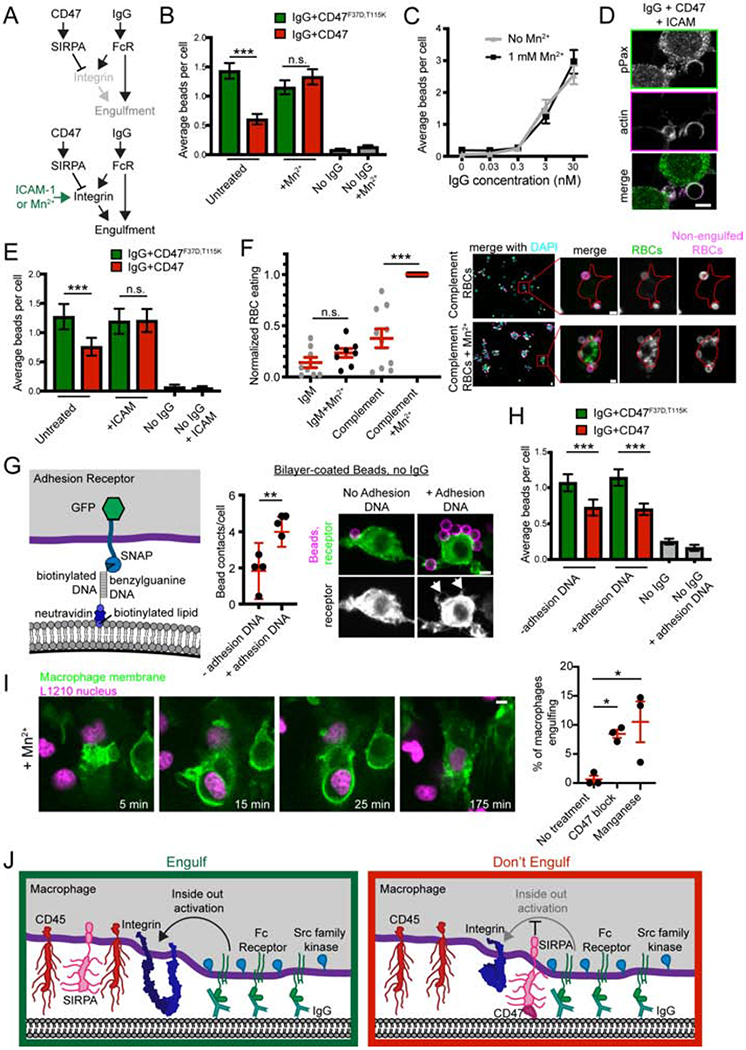
(A) The schematic shows a simplified signaling diagram. If CD47 and SIRPA act upstream of integrin, then providing an alternate means of integrin activation (Mn2+ or ICAM) should eliminate the effect of CD47. (B) Macrophages were treated with 1 mM Mn2+ and fed beads with IgG and either CD47 (red) or the non-signaling CD47F37D, T115K (green). Bars denote the average number of beads eaten from the pooled data of three independent replicates ± SEM. (C) Beads were incubated with the indicated concentration of IgG and added to macrophages. Treatment with Mn2+ did not dramatically enhance engulfment (black, compared to grey). Dots represent the average number of beads eaten ± SEM in one data set representative of three experiments. (D) Immunofluorescence shows that adding ICAM (10 nM coupling concentration) to IgG + CD47 beads rescues phosphopaxillin (top; green in merge, bottom) and actin (middle; magenta in merge) at the phagocytic cup. The quantification of this data is graphed in Figure 3D alongside the appropriate controls. (E) Beads were functionalized with IgG and either CD47 (red) or the non-signaling CD47F37D, T115K (green). Adding ICAM to the beads abrogated the effect of CD47 (center) but did not stimulate engulfment without IgG (right). (F) Complement-opsonized CD47+ mouse red blood cells (RBCs) were fed to control (grey) or 1 mM Mn2+ treated (black) macrophages. Unopsonized IgM treated RBCs were used as a negative control. Red blood cell internalization is graphed on the left. An anti-iC3b antibody was added after fixation, but before cell permeabilization to distinguish between engulfed (green) and non-engulfed (green and magenta) RBCs. On the right, representative images show increased adhesion and engulfment of RBCs (all RBCs, green; non-engulfed RBCs, magenta) in macrophages (nuclei stained with DAPI, cyan) treated with Mn2+. The inset (red box) highlights one example macrophage and is shown at higher magnification on the right. The macrophage cortex was determined through CellMask staining and is outlined in red. (G) Schematic shows the DNA-based adhesion system. Macrophages express a synthetic adhesion receptor containing an intracellular GFP, and an extracellular SNAP tag, which is conjugated to benzylguanine DNA. Graph depicts the mean number of bead contacts per cell, using beads functionalized only with neutravidin or with neutravidin and biotinylated ligand DNA (no IgG). Arrows point to cell membrane clinging to the adherent beads. (H) Beads were ligated to IgG, either CD47 (red) or the non-signaling CD47F37D, T115K (green), and biotinylated DNA to control adhesion. All cells express the adhesion receptor, which is conjugated to benzylguanine-DNA. Graph depicts the average number of beads engulfed per cell. (I) Bone marrow-derived macrophages expressing a membrane tethered GFP (green) were incubated with L1210 murine leukemia cells expressing H2B-mCherry (magenta). Treating with 100 mM manganese allowed for engulfment of whole cancer cells. These images correspond to frames from Movie S3. The percent of macrophages engulfing a cancer cell during an 8 hr timelapse is graphed on the right. Each dot represents an independent replicate, with red lines denoting mean ± SEM. (J) Model figure shows that in the absence of CD47 (left), SIRPA is segregated away from the phagocytic synapse and Fc Receptor binding triggers inside out activation of integrin. When CD47 is present (right), SIRPA localizes to the synapse and inhibits integrin activation. In F, G and I, each dot represents an independent replicate, with red lines denoting mean ± SEM. *** denotes p<0.0005, ** denotes p<0.005 and n.s. denotes p>0.05 as determined by a Kruskal-Wallis test (B, E, H), Student’s T test (F, G) or an Ordinary one way ANOVA with Holm-Sidak multiple comparison test (I).
As an alternative strategy to activate integrins, we incubated macrophages with beads containing a surplus of high affinity integrin ligand, ICAM-1 (Springer and Dustin, 2011). ICAM-1 was sufficient to activate integrin and recruit phophopaxillin even in the presence of CD47 (Figure 6D, 5D). Inclusion of high concentrations of ICAM-1 abrogated the inhibitory effect of CD47 on phagocytosis, but did not dramatically alter the engulfment efficiency of IgG coated beads in the absence of CD47 (Figure 6E). Despite the presence of CD47, ICAM-1-bound beads had similar levels of actin accumulation as beads lacking CD47 (Figure 6D, 5D). This demonstrates that activating integrins restores the ability of a macrophage to engulf targets in the presence of CD47. Together, these data suggest that inside-out activation of integrins may be a primary target for repression following CD47-SIRPA engagement.
CD47 has also been shown to suppress complement-mediated phagocytosis (Oldenborg et al., 2001). Because complement directly activates aMp2 integrin (Freeman and Grinstein, 2014), we sought to determine if preventing integrin activation could account for the suppressive effect of CD47 in complement-mediated phagocytosis. To address this, we examined whether manganese treatment could increase macrophage engulfment of complement-opsonized mouse red blood cells (RBCs), which present CD47 on their surface (Figure 6F; Oldenborg et al 2000; Yi et al., 2015). We found that activating integrin with 1 mM manganese dramatically increased engulfment of complement-opsonized RBCs but not control IgM-treated RBCs. This demonstrates that activating integrin enhances complement mediated engulfment, and is consistent with integrin activation bypassing the suppressive CD47 signal on complement-opsonized red blood cells.
We next sought to clarify whether integrin bypassed the CD47 signal by acting simply as a physical tether, or if intracellular integrin signaling was required. To distinguish between a tethering role and a signaling role for integrins, we created a DNA-based synthetic receptor that tethered the bilayer-coated beads to the macrophage but contained no intracellular signaling domains (Figure 6G). Unlike integrin activation, this inert tether was unable to bypass the suppressive CD47 signal (Figure 6H). This suggests that adhesion alone cannot overcome the effect of CD47 and that the intracellular signaling capabilities of integrin are essential.
Integrin activation drives cancer cell engulfment
Many cancer cells overexpress CD47 to evade the innate immune system despite increased expression of “Eat Me” signals such as calreticulin or phosphatidylserine (Birge et al., 2016; Chao et al., 2010b; Gardai et al., 2005; Utsugi et al., 1991). Blocking CD47 with a therapeutic antibody allows “Eat Me” signals to dominate, resulting in engulfment of whole cancer cells (Jaiswal et al., 2009; Majeti et al., 2009). We hypothesized that exogenous activation of integrin would bypass the CD47 signal on the surface of cancer cells, allowing for engulfment. To test this, we incubated bone marrow derived mouse macrophages expressing a membrane tethered GFP (GFP-CAAX) with a CD47-positive murine leukemia line, L1210, expressing nuclear H2B-mCherry (Chen et al., 2017). We then imaged macrophage-cancer cell interactions for 8 hours and found that activating integrins with 100 μM manganese increased the ability of macrophages to engulf cancer cells, reaching a similar efficiency as treatment with a CD47 function-blocking antibody (Figure 6I; Movie S3). Manganese did not directly affect cancer cell viability over the time course of this experiment (Figure S5). To confirm this result, we dyed L1210 cancer cells with CFSE. We then incubated these dyed cancer cells with primary bone marrow derived macrophages for 2 hours at a 2:1 cancer cell:macrophage ratio. We found that manganese increased whole cell engulfment in this assay as well (Figure S5). These data suggest that activating integrins bypasses the suppressive CD47 signal on the surface of cancer cells.
Discussion
CD47-SIRPA signaling suppresses engulfment, protecting viable cells and allowing cancer cells to evade the innate immune system (Jaiswal et al., 2009; Majeti et al., 2009; Oldenborg et al., 2000). Although CD47 blockade is a promising new target for cancer therapies (Advani et al., 2018; Gholamin et al., 2017; Willingham et al., 2012), there are many unresolved questions concerning CD47-SIRPA mechanism and potency. Using a reconstituted engulfment target, we quantitatively probed CD47-SIRPA signaling. By titrating CD47 and IgG, we found that CD47 dampened IgG-mediated phagocytosis but this suppressive effect could be overcome by a surplus of IgG. Mechanistically, we demonstrated that localizing SIRPA to the phagocytic synapse was sufficient to activate this inhibitory receptor. Once active, SIRPA suppressed engulfment by preventing integrin activation.
Our results demonstrate that SIRPA localization is a key determinant of its activity. In the absence of CD47, SIRPA is relegated to the phosphatase-rich zone outside the cell bead interface (Freeman et al., 2016; Goodridge et al., 2011). This localization prevents SIRPA activation. Conversely, CD47 binding retained SIRPA at the Src-kinase rich phagocytic cup, where it is activated and suppresses engulfment. Spatial segregation of Src-family kinase activity at the central phagocytic synapse and CD45 phosphatase activity at the periphery underlies the activation of many activating receptors (TCR, Fc Receptor, (Freeman et al., 2016; James and Vale, 2012). Our work expands this model, suggesting that exclusion of inhibitory receptors like SIRPA may be a pre-requisite for efficient engulfment. Further, these data suggest a new paradigm for regulating inhibitory receptors based on conditional recruitment to the immunological synapse.
SIRPA exclusion from the phagocytic synapse in the absence of CD47 prevents basal inhibition of engulfment and allows positive signaling to dominate. SIRPA may be sterically excluded from the phagocytic synapse based on the size of its bulky extracellular domain, as replacing the extracellular domain with a small, inert protein (FRB) allowed SIRPA to enter the phagocytic synapse. Could the bulky SIRPA extracellular domain be sterically excluded from the phagocytic synapse based on height alone? The FcR-IgG complex is ~11.5 nm tall (Lu et al., 2011), and our data demonstrate that both unligated SIRPA and CD47-bound SIRPA are excluded from these receptor-ligand clusters. Between IgG clusters, integrin forms a diffusion barrier in the phagocytic synapse that prevents bulky proteins from entering (Freeman et al., 2016). While extended integrin is quite tall, engaged integrin is tilted and has been shown to drive exclusion of the bulky transmembrane phosphatase CD45 (Freeman et al., 2016; Swaminathan et al., 2017). Although aglycosylated CD45 is larger than SIRPA (17 nm and 12 nm respectively), the size of both extracellular domains is increased by extensive glycosylation (Chang et al., 2016; Hatherley et al., 2008). Thus, steric exclusion may be sufficient to explain the depletion of SIRPA at the immunological synapse. To support this, we demonstrated that shortening the distance between the macrophage and its target increased SIRPA exclusion. How does CD47 binding alter SIRPA localization? Biophysical studies show that unligated proteins that are the same size or even slightly smaller than the height of a cell-cell synapse are excluded from the synapse (Schmid et al., 2016). Ligand binding is sufficient to drive synapse localization (Schmid et al., 2016). Thus, SIRPA may be sterically excluded unless CD47 ligation overcomes the energetic barrier preventing SIRPA from entering the immunological synapse. While our data demonstrates that SIRPA exclusion can be driven by altering the height of the immunological synapse, our studies do not rule out possible contribution of other exclusion mechanisms, such as lateral crowding or interactions with the surrounding glycocalyx.
After addressing the mechanism of SIRPA activation, we sought to identify the targets of CD47-SIRPA signaling. Previous work shows that SIRPA activation dramatically reduces global phosphotyrosine, including phosphorylation of mDia, paxillin, talin, alpha-actinin and non-muscle myosin IIA (Okazawa et al., 2005; Tsai and Discher, 2008). However, discerning between direct targets of SIRPA-bound phosphatases and indirect targets resulting from an upstream block in the engulfment signaling cascade has been challenging. Because blocking non-muscle myosin II decreases phagocytosis to a similar extent as CD47, myosin has been presumed to be the primary target of SIRPA, suggesting a model where CD47 inhibits “pulling” of the phagocytic target into the macrophage (Chao et al., 2012; Tsai and Discher, 2008). However, we demonstrate that the inhibitory effect of CD47-SIRPA can be eliminated by re-activating integrin, suggesting that the direct targets of SIRPA-bound SHP phosphatases are upstream of integrin activation. Instead of a pulling model, we propose that CD47 inhibits spreading of the macrophage around the phagocytic target. In our system, we found that blocking αMβ2 integrin had the largest effect on engulfment. However, instead of targeting a specific integrin subset directly, we hypothesize that SIRPA bound phosphatases deactivate an upstream step in the inside-out activation signaling pathway or an integrin regulator. SHP-2 has previously been shown to directly dephosphorylate Fak (Yu et al., 1998) and vinculin (Campbell et al., 2018), thus SHP-2 may act upon these key integrin regulators. However, given the broad specificity of SHP-1 and SHP-2, these phosphatases may dephosphorylate several targets at the phagocytic cup to suppress signaling.
We show that CD47-SIRPA prevents integrin activation, allowing macrophages to quickly discriminate between targets based on the presence of CD47. In addition to immediately inactivating integrin to prevent engulfment of a CD47-positive cell, SIRPA may also contribute to a long-term transcriptional down regulation of integrins (Liu et al., 2008). While this decrease in integrin expression does not explain how SIRPA prevents phagocytosis specifically of CD47-bound targets, it suggests that long-term exposure to activated SIRPA may decrease overall phagocytic capacity. Paradoxically, SIRPA may also be required for integrin-dependent cell migration, as fibroblasts lacking SIRPA have impaired motility (Alenghat et al., 2012; Inagaki et al., 2000; Motegi et al., 2003). In this context, SIRPA may promote integrin turnover to provide the dynamic interactions necessary for motility.
By suppressing integrin activation, CD47-SIRPA signaling may be able to suppress many different signaling pathways. CD47 has been reported to affect dendritic cell activation, cancer cell killing via a nibbling behavior (called trogocytosis), and complement-mediated engulfment (Caron et al., 2000; Matlung et al., 2018; Oldenborg et al., 2001; Tamada et al., 2004; Wu et al., 2018; Yi et al., 2015). These processes are triggered by diverse positive signaling receptors, but all require inside-out activation of integrin (Caron et al., 2000; Matlung et al., 2018; Oldenborg et al., 2001; Tamada et al., 2004; Wu et al., 2018; Yi et al., 2015). Targeting integrin, a common co-receptor, may explain how CD47-SIRPA signaling can regulate these diverse processes.
Finally, we found that integrin activation by manganese can drive engulfment of whole cancer cells by bone marrow-derived macrophages. As a cancer treatment, CD47 blockade synergizes with therapeutic antibodies, like rituximab (Advani et al., 2018; Chao et al., 2010a). Activating integrins with a small molecule agonist in combination with antibody therapeutics may have a similar synergistic effect as CD47 blockade. Small molecule agonists of the aM integrin subunit drive tumor regression in a macrophage-dependent manner (Panni et al., 2019; Schmid et al., 2018). Our data suggest that these small molecules may promote tumor regression partially by allowing macrophages to bypass the CD47 inhibitory signal.
Limitations of Study
To allow for high resolution imaging, this study was conducted ex vivo using mouse macrophage cell lines or bone marrow derived macrophages. Validating these findings in a model that more closely recapitulates the human tumor microenvironment would be an additional step forward. While we demonstrate that CD47-SIRPA prevents integrin activation, we have not shown whether this is through direct dephosphorylation of an integrin regulator by SIRPA-bound phosphatases, or through a less direct mechanism.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ron Vale (valer@janelia.hhmi.org).
Materials Availability
Plasmids generated in this study have been deposited to Addgene or can be obtained from the Lead Contact.
Data and Code Availability
All code used to acquire or analyze data in this paper is publicly available through μManager (micro-manager.org) or Fiji (fiji.sc). Complete imaging data sets are available from the Lead Contact.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
RAW264.7 macrophages were provided by the ATCC and certified mycoplasma-free. The cells were cultured in DMEM (Gibco, Catalog #11965–092) supplemented with 1 x Pen-Strep-Glutamine (Corning, Catalog #30–009 Cl), 1 mM sodium pyruvate (Gibco, Catalog #11360-070) and 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Catalog #S11150H). To keep variation to a minimum, cells were discarded after 20 passages. L1210 cells were also acquired from the ATCC.
J774A.1 macrophages were provided by the UCSF cell culture facility. J774A.1 and 293T cells were tested for mycoplasma using the Lonza MycoAlert Detection Kit (Lonza, Catalog# LT07-318) and control set (Lonza, Catalog #LT07-518).
Bone marrow-derived macrophages were generated from the hips and long bones of C57BL/6J mice as previously described (Weischenfeldt and Porse, 2008) except that purified 25 ng/ml M-CSF (Peprotech, Catalog # 315–02) was used.
METHOD DETAILS
Lentivirus production and infection
All constructs were expressed in RAW264.7 using lentiviral infection. Lentivirus was produced in HEK293T cells transfected with pMD2.G (a gift from Didier Tronon, Addgene plasmid # 12259 containing the VSV-G envelope protein), pCMV-dR8.91 (since replaced by second generation compatible pCMV-dR8.2, Addgene plasmid #8455), and a lentiviral backbone vector containing the construct of interest (derived from pHRSIN-CSGW, see STAR methods) using lipofectamine LTX (Invitrogen, Catalog # 15338–100). Constructs are described in detail in the Key Resources Table. The media was harvested 72 hr post-transfection, filtered through a 0.45 μm filter and concentrated using LentiX (Takara Biosciences). After addition of the concentrated virus, cells were centrifuged at 2000xg for 45 min at 37°C. Cells were analyzed a minimum of 60 hr later, and maintained for a maximum of one week.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-pPaxillin | Cell Signaling, Tech. | Cat#2541, RRID:AB_2174466 |
| AlexaFluor 647 anti-biotin IgG | Jackson Immuno Labs | Cat#200-602-211, RRID:AB 2339046 |
| CD18 monoclonal antibody 2E6 | ThermoFisher | Cat# MA1805, RRID:AB_223588 |
| Armenian hamster isotype control (for 2E6) | ThermoFisher | Cat#16-4888-81, RRID:AB 470171. |
| anti-mouse CD11a Antibody | BioLegend | 101109, RRID:AB_312782 |
| Rat IgG2a, κ Isotype Ctrl Antibody | BioLegend | 400515, RRID:AB_11147763 |
| anti-mouse/human CD11b Antibody | BioLegend | 101248, RRID:AB_2561479 |
| Rat IgG2b, κ Isotype Ctrl Antibody | BioLegend | 400643, RRID:AB_11147763 |
| Purified anti-mouse/rat CD61 Antibody | BioLegend | 104309, RRID:AB_313086 |
| Purified Armenian Hamster IgG Isotype Ctrl Antibody | BioLegend | 400969 |
| Anti mouse CD47 | BioLegend | 127517, RRID:AB_2571996 |
| Anti-mouse RBC. IgM fraction (polyclonal rabbit Igm) | MyBioSource | MBS524365 |
| APC anti-complement C3b/iC3b | BioLegend | 846106, RRID:AB_2632795 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Alexa Fluor 488 Phalloidin | Thermo/Molecular Probes | Cat# A12379 |
| Manganese | Sigma Aldrich | Cat# M8054 |
| POPS | Avanti | Cat# 840035 |
| Biotinyl Cap PE | Avanti | Cat# 870273 |
| POPC | Avanti | Cat# 850457 |
| Ni2+-DGS-NTA | Avanti | Cat# 790404 |
| PEG5000-PE | Avanti | Cat# 880230 |
| atto390 DOPE | ATTO-TEC GmbH | Cat# AD 390-161 |
| M-CSF | Peprotech | Cat# 315-02 |
| A/C heterodimerizer (rapamycin analog) | Takeda | Cat# 635056 |
| CFSE | ThermoFisher | Cat# C34554 |
| Alexa Fluor 488 NHS Ester (Succinimidyl Ester) | ThermoFisher | Cat# A20000 |
| Cell Mask orange plasma membrane stain | ThermoFisher | Cat # C10045 |
| PMA | Sigma-Aldrich | P1585 |
| Gelatin veronal buffer (GVB) | Millipore Sigma | G6514 |
| C5-deficient human serum | Sigma-Aldrich | C1163 |
| Critical Commercial Assays | ||
| Pierce Fab Preparation Kit | ThermoFisher | Cat# 44985 |
| Experimental Models: Cell Lines | ||
| HEK293T cells | UCSF Cell Culture Facility | |
| Hi Fives | ThermoFisher | Cat# BTI-TN-5B1-4 |
| Sf9 | ThermoFisher | Cat# 11496015 |
| Raw264.7 Macrophages | ATCC | Cat# ATCC® TIB-71™ |
| J774A.1 Macrophages | UCSF Cell Culture Facility | |
| Mouse CD1 RBCs | Innovative Research | IMSCD1RBC |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J | macrophages generated as described: PMID: 21356739 | |
| Oligonucleotides | ||
| Receptor DNA strand | this paper | Benzylguanine-5’- AATATGATGTATGTGG -3’, Oligonucleotide was ordered from IDT with a 5′ terminal amine. Conjugation to benzyl-guanine was performed as described (Farlow et al., 2013) |
| DNA ligand strand | IDT | Biotin-5’- TTTT-CCACATACATCATATT - 3’-Atto647 |
| Recombinant DNA | ||
| pHR-SIRPA-GFP | this paper | CDS: aa1-513 UniProtKB - P97797 (SHPS1_MOUSE), Linker: ADPPVAT, Fluorophore: mGFP |
| pHR-Syk mCherry | this paper | CDS: aa1-583 UniProtKB - P48025 (KSYK_MOUSE), Linker: ADPPVAT, Fluorophore: mCherry |
| pHR-Fcr3Ext-SIRPAint | this paper | CDS: Extracellular: aa1-215 UniProtKB P08508 (FCGR3_MOUSE) Linker: GSGSGSGGGGSS, TM and intracellular: aa374-513, UniProtKB - P97797 (SHPS1_MOUSE), Linker: ADPPVAT, Fluorophore: mGFP |
| pHR-Fcr3Ext-SIRPAint4F | this paper | as above, except the SIRPA intracellular domain contains 4 point mutations (Y440F, Y464F, Y481F, Y505F) |
| pHR-FRBext-SIRPA | this paper | Signal peptide: aa 1-20 Uniprot Q9UM88 (beta 2-microglobulin) Extracellular: FRB from Ariad Heterodimeriztion kit (see James and Vale, 2012), Linker: GSGSGSGGGGSS, TM and intracellular: aa374-513, UniProtKB - P97797 (SHPS1_MOUSE), Linker: ADPPVAT, Fluorophore: mGFP |
| pHR-Fc gamma p2a sirpa mch | this paper | FcR gamma chain: aa1-86 UniProtKB - P20491 (FCERG_MOUSE) Linker: GSGSGGGGSS , FKBP(F36V) from Ariad Heterodimerization kit (see James and Vale 2012) p2a; FRB(T2098L), SIRPAint: aa395-511 ProtKB - P97797 (SHPS1_MOUSE), Linker: ADPPVAT, Fluorophore: mCherry |
| pHR-Fc gamma p2a sirpa 4F mch | this paper | as above, except the SIRPA intracellular domain contains 4 point mutations (Y440F, Y464F, Y481F, Y505F) |
| ICAM-tagBFP-His10 | DOI: 10.7554/eLife.00778 | |
| pFastBAC-CD47ext-His10 | this paper | Signal peptide: from RPTPσ, CD47ext: aa40-182 (Uniprot Q61735), Linker: TS, Tag: His10 |
| pFastBAC-CD47(F37D, T115K)ext-His10 | this paper | As above, except F37D, T115K |
| pMD2.G lentiviral plasmid | D. Stainier, Max Planck; VSV-G envelope | Addgene 12259 |
| pCMV-dR8.91 | DOI: 10.1038/nature11220. | Current Addgene 8455 |
| pHRSIN-CSGW | DOI: 10.1038/nature11220. | |
| pHR-DNA adhesion receptor | this paper | Signal peptide: MQSGTHWRVLGLCLLSVGVWGQD Extracellular: HA tag, plus a linker (LPETGGGGGG), SNAPf (from the pSNAPf plasmid, New England Biolabs) Linker: GGSGGSGGS, TM and intracellular: aa 236-271 UniProtKB- P42081 (CD86TM), linker: SADASGG, Fluorophore: eGFP |
| pHR-FRBext-GFPint | this paper | Signal peptide: aa 1-20 Uniprot Q9UM88 (beta 2-microglobulin) Extracellular: FRB from Ariad Heterodimeriztion kit (see James and Vale, 2012), Linker: GGSGGSGGS TM: aa 236-271 UniProtKB- P42081 (CD86TM), intracellular: linker SADASGG, Fluorophore: mGFP |
| pHR-FRB-FIB1ext-GFPint | this paper | As above, except linker (TSSGS) and fibcon (LDAPTDLQVTNVTDTSITVSWTPPSATITGYRITYTPSNGPGEPKELTVPPSSTSVTITGLTPGVEYVVSVYALKDNQESPPLVGTQTT) inserted between FRB and extracellular linker |
| pHR-FRB-FIB3ext-GFPint | this paper | As above, except 3 fibcon repeats |
| pHR-FRB-FIB5ext-GFPint | this paper | As above, except 2 fibcon repeats and SG inserted before 3 fibcon repeats (5 fibcon repeats total) |
| pET28 - FKBP | doi.org/10.1073/pnas.1710358114 | |
| Software and Algorithms | ||
| ImageJ | NIH | |
| Illustrator | Adobe | CS6 |
| Photoshop | Adobe | CS6 |
| Fiji | https://fiji.sc/ | |
| Prism | GraphPad | 8 |
| Micromanager | DOI:10.14440/jbm.2014.36 | |
| Other | ||
| 5 um silica microspheres | Bangs | Cat# SS05N |
| Coverslips for TIRF chambers | Ibidi | Cat# 10812 |
| MatriPlate | Brooks | Cat# MGB096-1-2-LG-L |
Supported lipid bilayer assembly
SUV preparation
The following chloroform-suspended lipids were mixed and desiccated overnight to remove chloroform: 96.8% POPC (Avanti, Catalog # 850457), 2% Ni2+-DGS-NTA (Avanti, Catalog # 790404), 1% biotinyl cap PE (Avanti, Catalog # 870273), 0.1% PEG5000-PE (Avanti, Catalog # 880230, and 0.1% atto390-DOPE (ATTO-TEC GmbH, Catalog # AD 390–161). The lipid sheets were resuspended in PBS, pH7.2 (Gibco, Catalog # 20012050) and stored under argon. The lipids were broken into small unilamellar vesicles via several rounds of freeze-thaws. The mixture was cleared using ultracentrifugation (TLA120.1 rotor, 35,000 rpm / 53,227 x g, 35 min, 4°C). The lipids were then stored at 4°C under argon for up to two weeks.
Planar bilayer preparation for TIRF microscopy
Ibidi coverslips (catolog #10812) were RCA cleaned. Supported lipid bilayers were assembled in custom plasma cleaned PDMS (Dow Corning, catalog # 3097366-0516 and 3097358-1004) chambers at room temperature for 1 hour. Bilayers were blocked with 0.2% casein (Sigma, catalog # C5890) in PBS. Proteins were coupled to the bilayer for 45 min. Imaging was conducted in HEPES buffered saline (20 mM HEPEs, 135 mM NaCI, 4 mM KCl, 10 mM glucose, 1 mM CaCl2, 0.5 mM MgCl2). Bilayers were assessed for mobility by either photobleaching or monitoring the mobility of single particles.
Bead preparation
8.6 x 108 silica beads with a 5.02 μm diameter (10 μl of 10% solids, Bangs Labs, Catalog # SS05N) were washed three times with PBS, mixed with 1mM SUVs in PBS and incubated at room temperature for 0.5-2 hr with end-over-end mixing to allow for bilayer formation. Beads were then washed three times with PBS to remove excess SUVs and incubated in 100 μl of 0.2% casein (Sigma, catalog # C5890) in PBS for 15 min before protein coupling. Unless otherwise indicated, anti-biotin AlexaFluor647-IgG (Jackson ImmunoResearch Laboratories Catalog # 200-602-211, Lot # 137445) was added between 3 and 30 nM, always using the lowest IgG concentration that triggered engulfment. Purified CD47ext-His10 was added at 1 nm. Proteins were coupled to the bilayer for 1 hr at room temperature with end-over-end mixing.
Protein density estimation
Given the high affinity of His10 for Ni2+-DGS-NTA (0.6 nM (Hui and Vale, 2014)), and antibody-antigen interactions, we expect close to 100% coupling efficiency (Hui and Vale, 2014). Complete coupling would result in 600 molecules/μm2 CD47 and 300 molecules/μm2 IgG for the 3 nM coupling condition. This is well within the range of CD47 on the surface of a cancer cell (Figure S1). In addition, to estimate the amount of IgG bound to each bead, we compared the fluorescence of IgG on the bead surface to calibrated fluorescent beads (Quantum AlexaFluor 647, Bangs Lab) using confocal microscopy. Using this method, we measured 200-360 molecules/μm2 of IgG, which is consistent with the theoretical prediction of near-complete coupling.
Protein Purification
CD47ext-His10, CD47ext F37D, T115K-His10 (aa40-182; Uniprot Q61735) and ICAM-tagBFP-His10 (O’Donoghue et al., 2013) were expressed in SF9 or HiFive cells using the Bac-to-Bac baculovirus system as described previously (Hui and Vale, 2014). Briefly, the N-terminal extracellular domain of CD47 was cloned into a modified pFastBac HT A with an upstream signal peptide from chicken RPTPσ (Chang et al., 2016). Insect cell media containing secreted proteins was harvested 72 hr after infection with baculovirus. His10 proteins were purified by using Ni-NTA agarose (Qiagen, Catalog # 30230), followed by size exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare, Catalog # 17517501). The purification buffer was 30 mM HEPES pH 7.4, 150 mM NaCl, 2 mM MgCl2, 5% glycerol (CD47) or 150 mM NaCl, 50 mM HEPES pH 7.4, 5% glycerol, 2 mM TCEP (ICAM).
Microscopy and analysis
Images were acquired on a spinning disc confocal microscope (Nikon Ti-Eclipse inverted microscope with a Yokogawa spinning disk unit and an Andor iXon EM-CCD camera) equipped with a 40 × 0.95 NA air and a 100 × 1.49 NA oil immersion objective. The microscope was controlled using μManager (Edelstein et al., 2010). For TIRF imaging, images were acquired on the same microscope with a motorized TIRF arm, but using a Hamamatsu Flash 4.0 camera and the 100x 1.49 NA oil immersion objective. Data was analyzed in ImageJ (Rueden et al., 2017; Schindelin et al., 2012).
Quantification of engulfment
30,000 macrophages were plated in one well of a 96-well glass bottom MatriPlate (Brooks, Catalog # MGB096-1-2-LG-L) between 12 and 24 hr prior to the experiment. Unless otherwise noted, macrophages remained in culture media (DMEM with 10% heat inactivated serum) throughout the experiment. ~8 x 105 beads were added to well and engulfment was allowed to proceed for 30 min. Cells were fixed with 4% PFA and stained with CellMask (ThermoFisher, catalog # C10045) without membrane permeabilization to label cell boundaries. Images were acquired using the High Content Screening (HCS) Site Generator plugin in μManager (Edelstein et al., 2010). For Figure 1B–D, 2G-H, 4F-I, S3D, and S4, the analyzer was blinded during engulfment scoring using the position randomizer plug-in in μManager.
TIRF imaging
Macrophages were removed from their culture dish using 5% EDTA in PBS, two times washed and resuspended in the HEPES imaging buffer (20 mM HEPES, 135 mM NaCl, 4 mM KCl, 10 mM glucose, 1 mM CaCl2, 0.5 mM MgCl2) before being added to the TIRF chamber.
Quantification of IgG clusters and Syk recruitment
After 15 minutes of interacting with the bilayer, cells that had spread on the bilayer surface were selected for analysis. Otsu thresholding in ImageJ was used to select IgG clusters in an unbiased manner. This selection was used to generate an ROI that was then applied to the Syk-mCherry channel. The area of the ROI (area of IgG clusters) and the mean Syk intensity within that ROI were measured.
Pearson’s Correlation Coefficient
The region of cell-bilayer contact was manually selected in ImageJ and the Pearson’s correlation coefficient between AlexaFluor647-IgG and either SIRPA-GFP or Syk-mCherry for this ROI was measured using the Coloc2 plugin (Schindelin et al., 2012).
Quantification of cell-bilayer contact area
For 3A the area of the cell contacting the bilayer was traced in ImageJ beginning with the first frame where the cell can be detected. Only cells with mobile IgG clusters were included. For 3B, the number of macrophage-bilayer contacts and the area was quantified in still images of live cells between 10 and 15 min after cells were added to the bilayer. The macrophage membrane was labeled with mCherry-CAAX. All cells were included.
Phosphopaxillin staining
Macrophages were fixed in 4% PFA for 15 min, then permeabilized and blocked with 0.1% BSA in PBS with 0.5% Tween 20. The cells were incubated with the phosphopaxillin antibody at 1:50 dilution at 4° C overnight before incubating with Alexa Fluor 555 anti-rabbit secondary (21428), Alexa Fluor 488 phalloidin (A12379).
Quantification of synapse intensity of phosphoPaxillin, actin and SIRPA constructs
Phagocytic cups were selected for analysis based on the presence of clustered IgG at the cup base (SIRPA chimeras) or clear initiation of membrane extensions around the phagocytic target (actin, phosphopaxillin). The phagocytic cup and the cell cortex were traced with a line 3 pixels wide at the Z-slice with the clearest cross section of the cup. The average background intensity was measured in an adjacent region and subtracted from each measurement.
Integrin block and Fab generation
To disrupt integrin function, the blocking antibodies or isotype control indicated in the “Key Reagents” table were added to macrophages at 10 μg/ml 30 minutes before IgG-opsonized beads. To eliminate any effects of the Fc domain, we generated Fabs of the β2 blocking antibody and isotype control using the Pierce Fab separation kit (ThermoFisher 44985). In figures 3 and S4C, the antibodies and beads were added to macrophages in complete media containing heat inactivated serum. In figure S4B, macrophages were washed into serum-free HEPES imaging buffer (20 mM HEPES, 135 mM NaCl, 4 mM KCl, 10 mM glucose, 1 mM CaCl2, 0.5 mM MgCl2) prior to antibody treatment to eliminate any potential serum components that may serve as integrin ligands.
DNA tether experiments
For the DNA adhesion experiments, bilayers were assembled on silica beads, blocked with 0.2% casein, and coupled to IgG at a 3 nM concentration as described above. After 15 min of IgG coupling, 1 μg/ml neutravidin was added for 20 minutes. After washing out the neutravidin, 100 nM DNA ligand strand (5’ biotin, see Key Reagents table) and 1 nM CD47ext-His10 were added and coupled for 45 min. During this time, macrophages plated in a 96 well imaging plate expressing a DNA chimeric adhesion receptor (extracellular SNAP - CD86 transmembrane domain - intracellular EGFP) were incubated with 1 uM receptor DNA strand (5’ benzylyguanine, see Key Reagents table, Farlow et al., 2013) in 100 ul buffer for 10 minutes. Cells and beads were both washed 4 times. ~8 x 105 beads were added to well and engulfment was allowed to proceed for 30 min in HEPES imaging buffer. The DNA ligand and receptor strand sequences along with their modifications can be found in the key resources table.
Whole cell internalization assay
For timelapse imaging, 30,000 primary bone marrow derived macrophages infected with GFP-CAAX were plated in a 96-well glass bottom MatriPlate (Brooks, Catalog # MGB096-1-2-LG-L). 2 hours prior to imaging, cells were washed into serum-free, phenol free DMEM for imaging. Manganese (SigmaAldrich, M8054) was added at 100 μM 30 min prior to imaging. When indicated, CD47 function-blocking antibody clone miap301 (Biolegend, 127520) was used at 10 mg/ml. 100,000 H2B-mCherry expressing L1210 cells were added and the co-culture was imaged for 8 hr.
For the end-point analysis, 30,000 primary bone marrow derived macrophages were plated in a 96-well glass bottom MatriPlate. The following morning, the macrophages were serum starved for 2 hours. Then 60,000 L1210 dyed with CFSE (ThermoFisher, C34570) were added to the well. Engulfment was allowed to proceed for 4 hr, then cells were fixed and stained with DAPI to indicate nuclei and F4/80 to label macrophages. A blinded analyzer counted the fraction of F4/80+ cells containing CFSE+DAPI+ particles in each condition.
Mouse red blood cell internalization assay
For the mouse red blood cell internalization assay, 30,000 RAW264.7 macrophages (ATCC) were plated in one well of a 96-well glass bottom MatriPlate (Brooks, MGB096-1-2-LG-L) between 12 and 24 hr prior to the experiment. Mouse red blood cells (RBCs) (Innovative Research, 88R-M001) were washed into PBS and stained with CSFE (ThermoFisher, C34554) or Alexa Fluor 488 NHS Ester (ThermoFisher, A20000) for 1 hr at room temperature. RBCs were then opsonized with C3bi as previously described (Chow et al., 2004). Briefly, RBCs were incubated with anti-mouse IgM (MyBioSource, MBS524107) for 1 hour at 37C. A portion of RBCs were separated for IgM controls, and the remaining RBCs were incubated with C5 deficient serum (Sigma-Aldrich, C1163) for 1 hr at 37C. Macrophages were washed into serum-free HEPES imaging buffer and incubated with 150 ng/mL PMA and 1 mM Manganese or water. ~1 x 106 RBCs were added to each well and engulfment was allowed to proceed for 1 hour in incubator. Cells were washed with PBS, unengulfed RBCs were lysed with water for 2 min, and cells were fixed with 4% PFA. Remaining non-lysed, non-engulfed RBCs were stained with APC anti-iC3b antibody (Biolegend, 1:200 dilution, 846106) for 30 min. Macrophages were stained with CellMask orange (ThermoFisher, 1:5,000 dilution) and Hoechst (Invitrogen, H3570, 1:1,000 dilution) for 15 min. Images were acquired using the High Content Screening (HCS) Site Generator plugin in μManager (Edelstein et al., 2010). The analyzer was blinded during engulfment scoring using the position randomizer plug-in in μManager.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed in Prism 8 (GraphPad, Inc). The statistical test used is indicated in the relevant figure legend. Sample sizes were predetermined and indicated in the relevant figure legend. In general the analyzer was blinded during analysis using either manual renaming of the files or the data randomizer plugin in μManager. The details of each quantification method and blinding strategy are included in the Methods section.
Supplementary Material
Movie S3, related to Figure 6: An Mn2+-treated macrophage encounters an L1210 leukemia cell
A macrophage infected with GFP-CAAX encounters an L1210 leukemia cell labeled with H2B-mCherry. In the presence of 100 μM Mn2+ the macrophage is able to engulf the cancer cell. Images were acquired every 5 min for 140 min. The field of view is 53 μm by 53 μm.
Movie S1, related to Figure 5: Macrophage encounters IgG bound to a supported lipid bilayer.
TIRF imaging (see schematic in Figure S2) shows Alexa Fluor 647-IgG (black) in the supported lipid bilayer as a macrophage engages with an IgG-bound target. Frames were acquired every 20 sec and time is indicated in the top left. Scale bar denotes 5 μm.
Movie S2, related to Figure 5: Macrophage encounters IgG and CD47 bound to a supported lipid bilayer.
TIRF imaging shows Alexa Fluor 647-IgG (black) in the supported lipid bilayer as a macrophage engages with an IgG and CD47-bound target. Frames were acquired every 20 sec and time is indicated in the top left. Scale bar denotes 5 μm.
Highlights.
Steric constraints drive unligated SIRPA from the phagocytic synapse.
Localizing SIRPA to the phagocytic synapse is sufficient to suppress phagocytosis.
CD47 inhibits macrophage spreading and integrin activation.
Reactivating integrin allows for engulfment of CD47+ targets including cancer cells.
Acknowledgments
We thank R. Dong for providing the SNAP-DNA used in our DNA-based adhesion system, N. Stuurman for developing the image randomizer plug-in for blinding our analysis, as well as K. McKinley and O. Klein for providing mouse long bones as a source for hematopoietic stem cells. We thank members of the Vale lab for critical feedback on this manuscript. MAM was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number F32GM120990. This work was funded by the Howard Hughes Medical Institute to RDV.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
Ron Vale is a founder of Myeloid Therapeutics. Meghan Morrissey and Ron Vale have submitted a patent application related to this work.
References
- Advani R, Flinn I, Popplewell L, Forero A, Bartlett NL, Ghosh N, Kline J, Roschewski M, LaCasce A, Collins GP, et al. (2018). CD47 Blockade by Hu5F9-G4 and Rituximab in Non-Hodgkin’s Lymphoma. N. Engl. J. Med 379, 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat FJ, Baca QJ, Rubin NT, Pao LI, Matozaki T, Lowell CA, Golan DE, Neel BG, and Swanson KD (2012). Macrophages require Skap2 and Sirpa for integrin-stimulated cytoskeletal rearrangement. J. Cell Sci. 125, 5535–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakalar MH, Joffe AM, Schmid EM, Son S, Podolski M, and Correspondence DAF (2018). Size-Dependent Segregation Controls Macrophage Phagocytosis of Antibody-Opsonized Targets. Cell 174, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay AN, and Brown MH (2006). The SIRP family of receptors and immune regulation. Nat. Rev. Immunol 6, 457–464. [DOI] [PubMed] [Google Scholar]
- Birge RB, Boeltz S, Kumar S, Carlson J, Wanderley J, Calianese D, Barcinski M, Brekken RA, Huang X, Hutchins JT, et al. (2016). Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 23, 962–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell H, Heidema C, Pilarczyk DG, and DeMali KA (2018). SHP-2 is activated in response to force on E-cadherin and dephosphorylates vinculin Y822. J. Cell Sci. 131, jcs216648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E, Self AJ, and Hall A (2000). The GTPase Rap1 controls functional activation of macrophage integrin alphaMbeta2 by LPS and other inflammatory mediators. Curr. Biol 10, 974–978. [DOI] [PubMed] [Google Scholar]
- Cham LB, Bear L, Dulgeroff T, Caspi M, Weissman IL, Lang KS, and Correspondence KJH (2020). Immunotherapeutic Blockade of CD47 Inhibitory Signaling Enhances Innate and Adaptive Immune Responses to Viral Infection. CellReports 31, 107494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang VT, Fernandes RA, Ganzinger KA, Lee SF, Siebold C, McColl J, Jonsson P, Palayret M, Harlos K, Coles CH, et al. (2016). Initiation of T cell signaling by CD45 segregation at “close contacts”. Nat. Immunol 17, 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Alizadeh AA, Tang C, Myklebust JH, Varghese B, Gill S, Jan M, Cha AC, Chan CK, Tan BT, et al. (2010a). Anti-CD47 Antibody Synergizes with Rituximab to Promote Phagocytosis and Eradicate Non-Hodgkin Lymphoma. Cell 142, 699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Jaiswal S, Weissman-Tsukamoto R, Alizadeh AA, Gentles AJ, Volkmer J, Weiskopf K, Willingham SB, Raveh T, Park CY, et al. (2010b). Calreticulin Is the Dominant Pro-Phagocytic Signal on Multiple Human Cancers and Is Counterbalanced by CD47. Sci. Transl. Med 2, 63ra94–63ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Weissman IL, and Majeti R (2012). The CD47-SIRPa pathway in cancer immune evasion and potential therapeutic implications. Curr. Opin. Immunol 24, 225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhong M-C, Guo H, Davidson D, Mishel S, Lu Y, Rhee I, Perez-Quintero L-A, Zhang S, Cruz-Munoz M-E, et al. (2017). SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature 544, 493–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C-W, Downey GP, and Grinstein S (2004). Measurements of Phagocytosis and Phagosomal Maturation. Curr. Protoc. Cell Biol 22, 15.7.1–15.7.33. [DOI] [PubMed] [Google Scholar]
- Dheilly E, Moine V, Broyer L, Salgado-Pires S, Johnson Z, Papaioannou A, Cons L, Calloud S, Majocchi S, Nelson R, et al. (2017). Selective Blockade of the Ubiquitous Checkpoint Receptor CD47 Is Enabled by Dual-Targeting Bispecific Antibodies. Mol. Ther 25, 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dransfield I, Cabanas C, Craig A, and Hogg N (1992). Divalent Cation Regulation of the Function of the Leukocyte Integrin LFA1. [DOI] [PMC free article] [PubMed]
- Dupuy AG, and Caron E (2008). Integrin-dependent phagocytosis: spreading from microadhesion to new concepts. J. Cell Sci. 121, 1773–1783. [DOI] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, and Stuurman N (2010). Computer control of microscopes using manager. Curr. Protoc. Mol. Biol Chapter 14, Unit14 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, and Henson PM (1992). Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J. Immunol 148, 2207–2216. [PubMed] [Google Scholar]
- Farlow J, Seo D, Broaders KE, Taylor MJ, Gartner ZJ, and Jun YW (2013). Formation of targeted monovalent quantum dots by steric exclusion. Nat. Methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SA, and Grinstein S (2014). Phagocytosis: Receptors, signal integration, and the cytoskeleton. Immunol. Rev 262, 193–215. [DOI] [PubMed] [Google Scholar]
- Freeman SA, Goyette J, Furuya W, Woods EC, Bertozzi CR, Bergmeier W, Hinz B, Van Der Merwe PA, Das R, and Grinstein S (2016). Integrins Form an Expanding Diffusional Barrier that Coordinates Phagocytosis. Cell 164, 128–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y, Matozaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, Tsuda M, Takada T, and Kasuga M (1996). A novel membrane glycoprotein, SHPS-1, that binds the SH2-domain-containing protein tyrosine phosphatase SHP-2 in response to mitogens and cell adhesion. Mol. Cell. Biol 16, 6887–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg P-AA, Michalak M, and Henson PM (2005). Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123, 321–334. [DOI] [PubMed] [Google Scholar]
- Gardner B, Anstee DJ, Mawby WJ, Tanner MJ, and von dem Borne AE (1991). The abundance and organization of polypeptides associated with antigens of the Rh blood group system. Transfus. Med 1, 77–85. [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, and Bershadsky AD (2009). Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33. [DOI] [PubMed] [Google Scholar]
- Gholamin S, Mitra SS, Feroze AH, Liu J, Kahn SA, Zhang M, Esparza R, Richard C, Ramaswamy V, Remke M, et al. (2017). Disrupting the CD47-SIRPα anti-phagocytic axis by a humanized anti-CD47 antibody is an efficacious treatment for malignant pediatric brain tumors. Sci. Transl. Med 9, eaaf2968. [DOI] [PubMed] [Google Scholar]
- Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, Wolf AJ, Bose N, Chan ASH, Magee AS, Danielson ME, et al. (2011). Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse’. Nature 472, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatherley D, Graham SC, Turner J, Harlos K, Stuart DI, and Barclay AN (2008). Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with CD47. Mol. Cell 31, 266–277. [DOI] [PubMed] [Google Scholar]
- Hui E, and Vale RD (2014). In vitro membrane reconstitution of the T-cell receptor proximal signaling network. Nat. Struct. Mol. Biol 21, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki K, Yamao T, Noguchi T, Matozaki T, Fukunaga K, Takada T, Hosooka T, Akira S, and Kasuga M (2000). SHPS-1 regulates integrin-mediated cytoskeletal reorganization and cell motility. EMBO J. 19, 6721–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA, Diem MD, Luo J, Teplyakov A, Obmolova G, Malia T, Gilliland GL, and O’Neil KT (2012). Design of novel FN3 domains with high stability by a consensus sequence approach. Protein Eng. Des. Sel. 25, 107–117. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Jamieson CHM, Pang WW, Park CY, Chao MP, Majeti R, Traver D, van Rooijen N, and Weissman IL (2009). CD47 Is Upregulated on Circulating Hematopoietic Stem Cells and Leukemia Cells to Avoid Phagocytosis. Cell 138, 271–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JR, and Vale RD (2012). Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature 487, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Lagenaur CF, and Narayanan V (1999). Integrin-associated protein is a ligand for the P84 neural adhesion molecule. J. Biol. Chem 274, 559–562. [DOI] [PubMed] [Google Scholar]
- Jones SL, Knaus UG, Bokoch GM, and Brown EJ (1998). Two signaling mechanisms for activation of alphaM beta2 avidity in polymorphonuclear neutrophils. J. Biol. Chem 273, 10556–10566. [DOI] [PubMed] [Google Scholar]
- Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, and Ullrich A (1997). A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature 386, 181–186. [DOI] [PubMed] [Google Scholar]
- Kojima Y, Volkmer J-P, McKenna K, Civelek M, Lusis AJ, Miller CL, Direnzo D, Nanda V, Ye J, Connolly AJ, et al. (2016). CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature 536, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, Walker AJ, Heller MD, Umemori H, Chen C, et al. (2018). CD47 Protects Synapses from Excess Microglia-Mediated Pruning during Development. Neuron 100, 120–134.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Kurilova S, Scott BL, Bosworth E, Iverson BE, Bailey EM, and Hoppe AD (2016). TIRF imaging of Fc gamma receptor microclusters dynamics and signaling on macrophages during frustrated phagocytosis. BMC Immunol. 17, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DQ, Li LM, Guo YL, Bai R, Wang C, Bian Z, Zhang CY, and Zen K (2008). Signal regulatory protein α negatively regulates β 2 integrin-mediated monocyte adhesion, transendothelial migration and phagocytosis. PLoS One 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Pu Y, Cron K, Deng L, Kline J, Frazier WA, Xu H, Peng H, Fu Y-X, and Xu MM (2015). CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat. Med 21, 1209–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Ellsworth JL, Hamacher N, Oak SW, and Sun PD (2011). Crystal Structure of Fcy Receptor I and Its Implication in High Affinity γ-Immunoglobulin Binding. J. Biol. Chem 286, 40608–40613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD, van Rooijen N, and Weissman IL (2009). CD47 Is an Adverse Prognostic Factor and Therapeutic Antibody Target on Human Acute Myeloid Leukemia Stem Cells. Cell 138, 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlung HL, Babes L, Zhao XW, van Houdt M, Treffers LW, van Rees DJ, Franke K, Schornagel K, Verkuijlen P, Janssen H, et al. (2018). Neutrophils Kill Antibody-Opsonized Cancer Cells by Trogoptosis. Cell Rep. 23, 3946–3959.e6. [DOI] [PubMed] [Google Scholar]
- Michaels AD, Newhook TE, Adair SJ, Morioka S, Goudreau BJ, Nagdas S, Mullen MG, Persily JB, Bullock TNJ, Slingluffjr CL, et al. (2017). Cancer Therapy: Preclinical CD47 Blockade as an Adjuvant Immunotherapy for Resectable Pancreatic Cancer. [DOI] [PMC free article] [PubMed]
- Motegi S.-i., Okazawa H, Ohnishi H, Sato R, Kaneko Y, Kobayashi H, Tomizawa K, Ito T, Honma N, BQhring H, et al. (2003). Role of the CD47-SHPS-1 system in regulation of cell migration. EMBO J. 22, 2634–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouro-Chanteloup I, Delaunay J, Gane P, Nicolas V, Johansen M, Brown EJ, Peters LL, Van Kim C. Le, Cartron JP, and Colin Y (2003). Evidence that the red cell skeleton protein 4.2 interacts with the Rh membrane complex member CD47. Blood 101, 338–344. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Matozaki T, Fujioka Y, Yamao T, Tsuda M, Takada T, and Kasuga M (1996). Characterization of a 115-kDa protein that binds to SH-PTP2, a protein-tyrosine phosphatase with Src homology 2 domains, in Chinese hamster ovary cells. J. Biol. Chem 271, 27652–27658. [DOI] [PubMed] [Google Scholar]
- O’Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, and Groves JT (2013). Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. Elife 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa H, Motegi S.-i., Ohyama N, Ohnishi H, Tomizawa T, Kaneko Y, Oldenborg P-A, Ishikawa O, and Matozaki T (2005). Negative Regulation of Phagocytosis in Macrophages by the CD47-SHPS-1 System. J. Immunol 174, 2004–2011. [DOI] [PubMed] [Google Scholar]
- Oldenborg P-A, Gresham HD, and Lindberg FP (2001). Cd47-Signal Regulatory Protein α (Sirpa) Regulates Fcy and Complement Receptor-Mediated Phagocytosis. J. Exp. Med 193, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, and Lindberg FP (2000). Role of CD47 as a marker of self on red blood cells. Science 288, 2051–2054. [DOI] [PubMed] [Google Scholar]
- Panni RZ, Herndon JM, Zuo C, Hegde S, Hogg GD, Knolhoff BL, Breden MA, Li X, Krisnawan VE, Khan SQ, et al. (2019). Agonism of CD11b reprograms innate immunity to sensitize pancreatic cancer to immunotherapies. Sci. Transl. Med 11, eaau9240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon IKH, Lucas CD, Rossi AG, and Ravichandran KS (2014). Apoptotic cell clearance: basic biology and therapeutic potential. Nat. Rev. Immunol 14, 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, and Eliceiri KW (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid EM, Bakalar MH, Choudhuri K, Weichsel J, Ann HS, Geissler PL, Dustin ML, and Fletcher DA (2016). Size-dependent protein segregation at membrane interfaces. Nat. Phys 12, 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MC, Khan SQ, Kaneda MM, Pathria P, Shepard R, Louis TL, Anand S, Woo G, Leem C, Faridi MH, et al. (2018). Integrin CD11b activation drives antitumor innate immunity. Nat. Commun 9, 5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiffert M, Cant C, Chen Z, Rappold I, Brugger W, Kanz L, Brown EJ, Ullrich A, and BQhring HJ (1999). Human signal-regulatory protein is expressed on normal, but not on subsets of leukemic myeloid cells and mediates cellular adhesion involving its counterreceptor CD47. Blood 94, 3633–3643. [PubMed] [Google Scholar]
- Spencer DM, Wandless TJ, Schreiber SL, and Crabtree GR (1993). Controlling signal transduction with synthetic ligands. Science 262, 1019–1024. [DOI] [PubMed] [Google Scholar]
- Springer TA, and Dustin ML (2011). Integrin inside-out signaling and the immunological synapse. Curr. Opin. Cell Biol. 24, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Parthasarathy R, Sen S, Boder ET, and Discher DE (2006). Species-and cell type-specific interactions between CD47 and human SIRPα. Blood 107, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan V, Kalappurakkal JM, Mehta SB, Nordenfelt P, Moore TI, Koga N, Baker DA, Oldenbourg R, Tani T, Mayor S, et al. (2017). Actin retrograde flow actively aligns and orients ligand-engaged integrins in focal adhesions. Proc. Natl. Acad. Sci 114, 10648–10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamada M, Sheetz MP, and Sawada Y (2004). Activation of a Signaling Cascade by Cytoskeleton Stretch. Dev. Cell 7, 709–718. [DOI] [PubMed] [Google Scholar]
- Tsai RK, and Discher DE (2008). Inhibition of “self” engulfment through deactivation of myosin-II at the phagocytic synapse between human cells. J. Cell Biol. 180, 989–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng D, Volkmer J-PJ-P, Willingham SB, Contreras-Trujillo H, Fathman JW, Fernhoff NB, Seita J, Inlay MA, Weiskopf K, Miyanishi M, et al. (2013). Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc. Natl. Acad. Sci 110, 11103–11108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Matozaki T, Fukunaga K, Fujioka Y, Imamoto A, Noguchi T, Takada T, Yamao T, Takeda H, Ochi F, et al. (1998). Integrin-mediated tyrosine phosphorylation of SHPS-1 and its association with SHP-2. Roles of Fak and Src family kinases. J. Biol. Chem 273, 13223–13229. [DOI] [PubMed] [Google Scholar]
- Utsugi T, Schroit AJ, Connor J, Bucana CD, and Fidler IJ (1991). Elevated expression of phosphatidylserine in the outer membrane leaflet of human tumor cells and recognition by activated human blood monocytes. Cancer Res. 51, 3062–3066. [PubMed] [Google Scholar]
- Veillette A, Thibaudeaut E, and Latour S (1998). High expression of inhibitory receptor SHPS-1 and its association with protein-tyrosine phosphatase SHP-1 in macrophages. J. Biol. Chem 273, 22719–22728. [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J, and Porse B (2008). Bone Marrow-Derived Macrophages (BMM): Isolation and Applications. CSH Protoc. 2008, pdb.prot5080. [DOI] [PubMed] [Google Scholar]
- Willingham SB, Volkmer J-P, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, et al. (2012). The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci 109, 6662–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HS, Jaumouille V, Freeman SA, Doodnauth SA, Schlam D, Canton J, Mukovozov IM, Saric A, Grinstein S, and Robinson LA (2016). Chemokine Signaling Enhances CD36 Responsiveness toward Oxidized Low-Density Lipoproteins and Accelerates Foam Cell Formation. Cell Rep. 14, 2859–2871. [DOI] [PubMed] [Google Scholar]
- Wu J, Wu H, An J, Ballantyne CM, and Cyster JG (2018). Critical role of integrin CD11c in splenic dendritic cell capture of missing-self CD47 cells to induce adaptive immunity. Proc. Natl. Acad. Sci 201805542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi T, Li J, Chen H, Hu Y, Lowell CA, and Cyster Correspondence JG (2015). Splenic Dendritic Cells Survey Red Blood Cells for Missing Self-CD47 to Trigger Adaptive Immune Responses. Immunity 43, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DH, Qu CK, Henegariu O, Lu X, and Feng GS (1998). Protein-tyrosine phosphatase Shp-2 regulates cell spreading, migration, and focal adhesion. J. Biol. Chem 273, 21125–21131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S3, related to Figure 6: An Mn2+-treated macrophage encounters an L1210 leukemia cell
A macrophage infected with GFP-CAAX encounters an L1210 leukemia cell labeled with H2B-mCherry. In the presence of 100 μM Mn2+ the macrophage is able to engulf the cancer cell. Images were acquired every 5 min for 140 min. The field of view is 53 μm by 53 μm.
Movie S1, related to Figure 5: Macrophage encounters IgG bound to a supported lipid bilayer.
TIRF imaging (see schematic in Figure S2) shows Alexa Fluor 647-IgG (black) in the supported lipid bilayer as a macrophage engages with an IgG-bound target. Frames were acquired every 20 sec and time is indicated in the top left. Scale bar denotes 5 μm.
Movie S2, related to Figure 5: Macrophage encounters IgG and CD47 bound to a supported lipid bilayer.
TIRF imaging shows Alexa Fluor 647-IgG (black) in the supported lipid bilayer as a macrophage engages with an IgG and CD47-bound target. Frames were acquired every 20 sec and time is indicated in the top left. Scale bar denotes 5 μm.
Data Availability Statement
All code used to acquire or analyze data in this paper is publicly available through μManager (micro-manager.org) or Fiji (fiji.sc). Complete imaging data sets are available from the Lead Contact.


