Abstract
Cancer is instigated by mutator phenotypes, including deficient mismatch repair and p53-associated chromosomal instability. More recently, a distinct class of cancers was identified with unusually high mutational loads due to heterozygous amino acid substitutions (most commonly P286R) in the proofreading domain of DNA polymerase ε, the leading strand replicase encoded by POLE. Immunotherapy has revolutionized cancer treatment, but new model systems are needed to recapitulate high mutational burdens characterizing human cancers and permit study of mechanisms underlying clinical responses. Here, we show that activation of a conditional LSL-PoleP286R allele in endometrium is sufficient to elicit in all animals endometrial cancers closely resembling their human counterparts, including very high mutational burden. Diverse investigations uncovered potentially novel aspects of Pole-driven tumorigenesis, including secondary p53 mutations associated with tetraploidy, and cooperation with defective mismatch repair through inactivation of Msh2. Most significantly, there were robust antitumor immune responses with increased T cell infiltrates, accelerated tumor growth following T cell depletion, and unfailing clinical regression following immune checkpoint therapy. This model predicts that human POLE-driven cancers will prove consistently responsive to immune checkpoint blockade. Furthermore, this is a robust and efficient approach to recapitulate in mice the high mutational burdens and immune responses characterizing human cancers.
Keywords: Genetics, Oncology
Keywords: Cancer immunotherapy, DNA repair, Mouse models
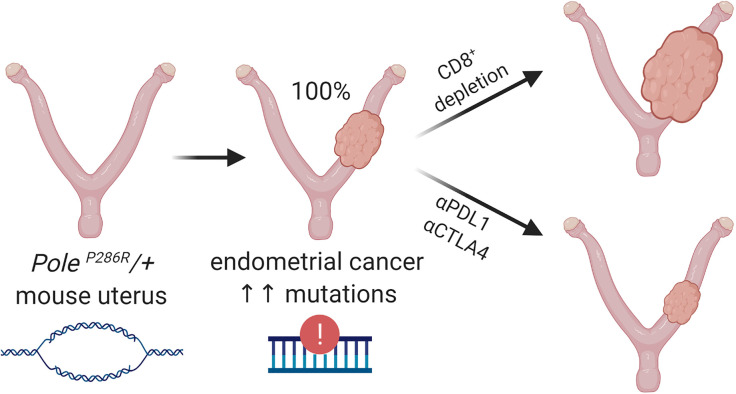
A mouse model of endothelial cancer based on polymerase ε-driven ultramutation recapitulates high human tumor burden and can be used to assess potential therapies.
Introduction
DNA mutations are the fundamental drivers of cancer (1). Accordingly, a central hallmark of cancer is an incidence of mutations more numerous than can be explained on the basis of the intrinsic mutation rate of normal (nonmalignant) cells (2, 3). In the last decade, systematic characterization of cancer genomes has underscored the high incidence of mutations in most cancers — especially carcinomas — and the underlying mutator mechanisms that initiate cancers and support subsequent diversification. These “mutator phenotypes” reflect the complexity of pathways that ensure high DNA replication fidelity and repair DNA damage sustained from mutagens, such as ionizing ultraviolet radiation and environmental toxicants, as well as the mutagenic potential of normal cell-intrinsic metabolic processes (3, 4). In many if not most cancers, the acquisition of a mutator phenotype is the initial instigating event driving tumorigenesis. For example, defective mismatch repair (dMMR) is common in endometrial and gastrointestinal carcinomas, and experimental evidence (genetic, genomic, mouse models, etc.) points to dMMR as the initial cancer-driving event (5–7).
The prevalence of somatic mutations (base substitution rate) varies dramatically across and within individual cancer types, ranging from less than 0.01/megabase to more than 500/megabase (Mb). Most carcinomas have base substitution rates of at least 1/Mb. Cancer types with the highest averages (5–12/Mb) include lung, colorectal, and endometrial carcinomas. A mutation rate 10/Mb or higher (hypermutation) is associated with dMMR. More recently, cancers with base substitution rates of at least 100/Mb (ultramutation) have been attributed to somatically acquired POLE missense mutations leading to single amino acid substitutions in the proofreading (exonuclease) domain, most commonly P286R (8). POLE encodes DNA polymerase ε, which replicates the leading strand during normal DNA synthesis (9). The incidence of POLE-driven ultramutation is highest in endometrial and colorectal cancers (~5%–10%), but POLE mutations also occur in sporadic sarcomas, hematopoietic malignancies, glioblastomas, and diverse carcinomas (10). P286R interferes with DNA binding and produces a hyperactive polymerase that introduces numerous errors during DNA synthesis (11, 12), leading to an error rate much higher than what results from inactivation of the exonuclease domain (13–15). POLEP286R and the rarer ultramutating amino acid substitutions (such as V411L) are genetically dominant, with retention of 1 WT allele (13). Analogous mutations leading to recurring single amino acid substitutions also occur in the lagging strand polymerase δ (encoded by POLD); however, these are less common and exceedingly rare in endometrial cancers, for unclear reasons (8, 9, 14, 16).
Intriguingly, some POLE-driven ultramutant cancers also exhibit dMMR. The majority of errors POLE mutants produce are presumed to be corrected by MMR (17), leading to the prediction that dMMR and POLE mutations should cooperate. On the other hand, excessive mutation rate might decrease cell fitness and lead to extinction (18). Children with inherited biallelic mismatch repair deficiency have very early onset of central nervous system and hematologic cancers with massive accumulation of mutations (>250/Mb), greater than all childhood and most adult cancers. All such cancers analyzed (10/10 cases) also harbored a somatically acquired mutation in POLE (7/10) or POLD (3/10) that appeared to be the initiating event (19). Some POLE-ultramutated adult colorectal and endometrial cancers are also dMMR, and specific mutational signatures have been ascribed to tumors harboring these combined defects (10, 20, 21). However, such cases may occur less frequently than predicted based on the incidence of POLE mutations and dMMR. Thus, whether POLE mutations cooperate with or antagonize dMMR in adult cancers is unclear, and defined genetic model systems are needed to investigate such interactions (22).
Immune checkpoint blockade (by monoclonal antibodies against programmed cell death 1/programmed cell death ligand 1 [PD1/PDL1] and cytotoxic T lymphocyte–associated protein 4 [CTLA-4]) result in long-term responses and even cures of otherwise untreatable malignancies. However, objective responses occur in a minority of patients, prompting concerted efforts to uncover mechanisms of blockade and resistance (23). There is a linear relationship between base substitutions and amino acid changes producing neoantigens that evoke immune responses by tumor-infiltrating CD8+ T lymphocytes (24, 25). In 2017, the FDA approved pembrolizumab as the first “tissue-agnostic” anticancer therapy for dMMR tumors irrespective of anatomic location or other histopathologic/molecular parameters (26, 27). There have not been systematic studies of immune checkpoint blockade in POLE-mutant tumors either in patients or in model systems, although there are isolated case reports of treatment responses (28, 29).
With this background in mind, it is notable that genetically engineered mouse cancer models have dramatically lower average mutational frequencies than human cancers. Egfr-, Kras-, or Myc-driven models of human lung cancer exhibit fewer than 0.1 mutations per megabase, several logs lower than human lung adenocarcinoma (30), and thus do not recapitulate mutational loads defining human cancers. That such models have not proved useful for testing immune checkpoint therapies has been attributed to mutational burdens too low to model human tumor immunology (31). Alternative strategies are needed to optimize mouse models with respect to mutational load, now known to define many aspects of tumor biology, clinical behavior, and treatment responses (30, 32, 33). In this study, we hypothesized that a new kind of mouse model of human cancer could be developed based on ultramutation driven by conditional PoleP286R expression and that such a model would be of broad investigational utility.
Results
Conditional PoleP286R expression provokes endometrial cancers with 100% penetrance.
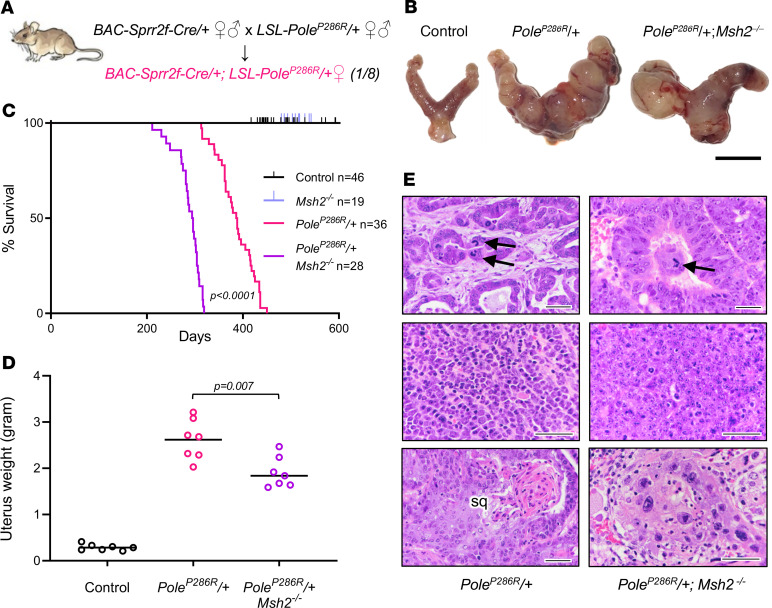
Heterozygous mice harboring BAC-Sprr2f-Cre and the conditional LSL-PoleP286R alleles were interbred. The functionality of LSL-PoleP286R, including Cre-mediated induction of PoleP286R expression equal to that of the WT Pole allele, was validated in a systemic knockin model, which elicited malignancies across many cell lineages (22). BAC-Sprr2f-Cre is a BAC-transgenic line with Cre inserted into the Sprr2f locus, which is expressed exclusively in the endometrial epithelial cells that give rise to all endometrial carcinomas. BAC-Sprr2f-Cre induces Cre-mediated recombination only in endometrium and is naturally estrogen inducible because of estrogen response elements in Sprr2f regulatory regions (34). Cre-mediated recombination by BAC-Sprr2f-Cre begins at 5 weeks (puberty onset), and is approximately 50% efficient within endometrial epithelial cells, leading to efficient (but mosaic) recombination (35). This single-generation breeding scheme is simpler and more efficient than for most mouse models, which can require multiple alleles and generations and are inefficient in yielding experimental animals. In this breeding scheme, one-fourth of the progeny are the desired genotype, and one-eighth are of the desired genotype and sex. Siblings not inheriting BAC-Sprr2f-Cre were used as controls (Figure 1A).
Figure 1. PoleP286R is a fully penetrant monoallelic driver of endometrial cancer and cooperates with Msh2 deficiency.
(A) Diagram showing single-generation breeding scheme for conditional PoleP286R/+ endometrial knockin mice. Genotypes shown are germline (determined by tail PCR). (B) Gross photographs of uteri. Images placed on white background. Scale bar: 1 cm. (C) Survival analyses, P value per log-rank test. (D) Uterus weight at necropsy as metric for uterine tumor burden; n = 7 per group, P value per 2-tailed Mann-Whitney U test. (E) Histologic features of uterine primary cancers, H&E-stained sections. Top panels show architecturally well-differentiated endometrioid adenocarcinomas with atypical mitoses (arrows). Middle panels show more poorly differentiated adenocarcinomas without gland formation. Bottom panels show cases with squamous differentiation (sq) or striking nuclear atypia (right panel) as described for human POLEP286R cancers. Scale bars: 50 μm.
First, we generated a cohort of transheterozygous BAC-Sprr2f-Cre/+LSL-PoleP286R/+ mice (abbreviated PoleP286R/+) to study age-related cancer onset. Whereas no deaths occurred in controls up to 600 days of age, the first death in PoleP286R mice occurred at 313 days (45 weeks), and all mice were dead within a short time span, by 450 days (64 weeks) (P < 0.0001, log-rank test) (Figure 1, B–D). The cause of death was always an aggressive endometrial cancer that replaced normal uterine tissues and metastasized to adjacent organs (ovary, bladder, kidney) or more distant sites. Malignancies of nonendometrial origin were not found. Histologically, the tumors invaded through the entire myometrial (uterine smooth muscle) layer. The tumors were histologically surprisingly homogenous, with most tumors appearing as well-differentiated endometrioid adenocarcinomas forming glands resembling normal endometrium. Nuclear atypia ranged from moderate to severe and atypical mitoses were characteristic (Figure 1E). However, some tumors were poorly differentiated (Figure 1E). About 20% of tumors exhibited squamous differentiation, seen in a similar percentage of human endometrial adenocarcinomas (36). Some cancers exhibited striking nuclear atypia (giant nuclei), implying abnormal ploidy (Figure 1E). These features (well-differentiated tumors with paradoxically high nuclear grade and giant nuclei) closely resembled human P286R endometrial cancers (see Supplemental Figure 1A for examples; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.138829DS1) (see also refs. 37, 38). However, some sporadic tumors exhibited distinctive patterns or histotypes, such as being highly invasive but lacking distinct gland formation or with exophytic, clear cell, or spindle cell components consistent with carcinosarcoma (Supplemental Figure 1, B–E). This also resembles the histologic distribution of human POLE-driven endometrial cancers, which are usually endometrioid but can be of the clear cell or carcinosarcoma subtypes (39–41). The aggressive and infiltrative nature of the mouse cancers was further evidenced by spread to adjacent structures such as ureters, frequent lymphovascular invasion, and metastases to more distant abdominal organs such as the pancreas and spleen (Supplemental Figure 1, F–I). Thus in summary, murine P286R/+ endometrial cancers closely resemble human counterparts histotypically and in clinical behavior.
dMMR induction through Msh2 inactivation accelerates PoleP286R-driven tumor progression.
To study genetic interactions with dMMR, we employed an Msh2L floxed allele used to investigate dMMR in colorectal cancer progression in vivo (5). Additional cohorts of mice were generated with BAC-Sprr2f-Cre to inactivate both alleles of Msh2 by itself or in combination with PoleP286R. Msh2 and Msh6 proteins form dimers that bind to DNA and upon detection of base-base mismatches recruit Mlh1/Pms2 dimers to excise mismatches on the newly synthesized strand (42). Inactivating mutations (point mutations or deletions) of either Msh2 or Msh6 resulting in dMMR destabilize Msh2/Msh6 dimers with degradation of both proteins (Supplemental Figure 2A). This destabilization is the basis of immunohistochemistry (IHC) as the principal assay in clinical practice to screen for dMMR defects (to identify Lynch syndrome or tumors likely to respond to pembrolizumab) (6, 43). In all n = 21 PoleP286R/+ tumors examined, there was retention of Msh2, Msh6, and Mlh1 in all cells, suggesting that spontaneous dMMR does not occur frequently in PoleP286R/+ murine endometrial cancers, consistent with human data (Supplemental Figure 2B) (21). All BAC-Sprr2f-Cre/+LSL-PoleP286R/+Msh2L/L mice (abbreviated PoleP286R/+Msh2–/–) harbored distinct multifocal clones of Msh2 and Msh6 loss, as expected, and all invasive primary or metastatic cancer cells at later time points were Msh2/Msh6 deficient (Supplemental Figure 2C).
In contrast to PoleP286R/+ mice, BAC-Sprr2f-CreMsh2L/L mice (abbreviated Msh2–/–) did not harbor cancers, and no deaths occurred up to 600 days of age (Figure 1C), showing that PoleP286R is a much more potent mutator allele and effective cancer driver than dMMR/Msh2 loss, at least in endometrium. Interestingly, however, dMMR and PoleP286R showed clear genetic cooperation, with a significant leftward shift of the survival curve (P < 0.0001, log-rank test). The cause of death was more aggressive endometrial cancers (Figure 1B), as evidenced by the decreased survival and extensive tumor spread found in this cohort. At death, uterine weight (a metric for primary tumor burden) was significantly decreased in PoleP286R/+Msh2–/– mice, consistent with more rapid spread from the primary site resulting in earlier deaths (Figure 1D). Histologic spectra were similar in PoleP286R/+ versus PoleP286R/+Msh2–/– mice, except that striking nuclear enlargement/atypia was more common in PoleP286R/+Msh2–/– mice (12/16 versus 6/21 for PoleP286R/+ alone, P = 0.0081 per Fisher exact test) (Figure 1E). Thus, the data showed that (a) most PoleP286R tumors do not spontaneously undergo dMMR and (b) dMMR and PoleP286R cooperate in tumor progression.
PoleP286R/+ tumors exhibit a propensity for tetraploidization.
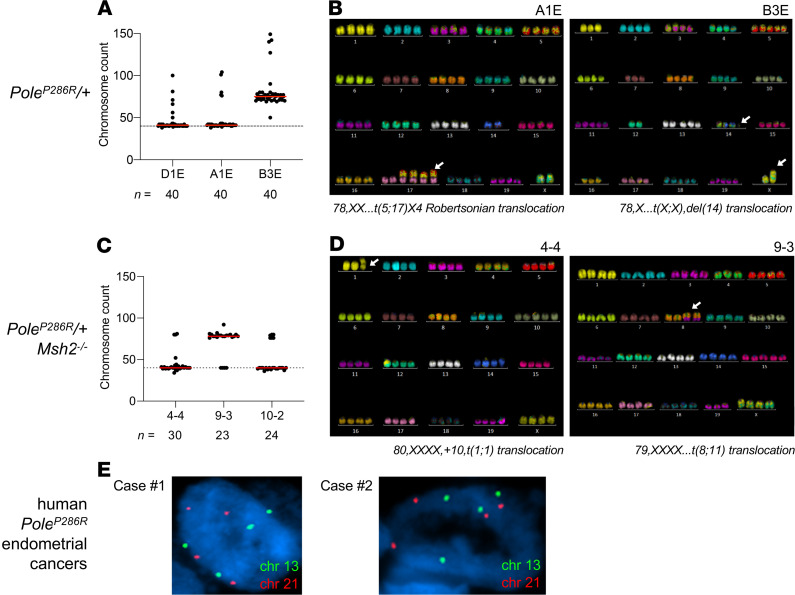
Nuclear atypia and enlargement imply abnormal karyotypes, suggesting that although PoleP286R is a pure base substitution mutator, tumor progression may be associated with additional, and possibly adaptive, layers of genomic instability. To explore this hypothesis, cell lines were established from n = 3 PoleP286R/+ and n = 3 PoleP286R/+Msh2–/– tumors. All 6 cell lines showed some tetraploid cells, and 1 cell line for each genotype was essentially tetraploid. Spectral karyotyping (SKY) of these cell lines confirmed the presence of tetraploid cells and showed a few chromosome-level aberrations such as fusions or translocations (Figure 2, A–D). These results indicate that PoleP286R/+ cancers exhibit a tendency toward tetraploidization. Tetraploidization, which occurs in some cancers, may be an adaptive response to buffer against high mutational loads (44). To explore whether this might also occur in human tumors, tissue sections from n = 6 POLEP286R endometrial cancers were subjected to DNA fluorescence in situ hybridization (FISH) with enumeration probes for chromosomes X, 8, 13, 18, and 21. In all cases, a substantial proportion of nuclei had 4 signals consistent with tetraploidy (Figure 2E), suggesting that tetraploidy is shared by human and murine POLE cancers, in agreement with prior studies (45).
Figure 2. SKY of mouse cancer cell lines.
(A) Chromosome counts of individual cells in PoleP286R/+ cell lines (n = 3). The normal diploid chromosomal complement (n = 40) is shown by a dotted line. All cell lines show some cells in tetraploid range, with 1 line (B3E) being predominantly tetraploid. (B) Representative SKY images of tetraploid cells from 2 lines as shown, showing only rare chromosomal translocations. The white arrows point to the translocations indicated below each image. (C) Chromosome counts of individual cells in PoleP286R/+Msh2–/– cell lines (n = 3). The normal diploid chromosomal complement (n = 40) is shown by a dotted line. All lines show some cells in tetraploid range, with 1 line (9-3) being predominantly tetraploid. (D) Representative SKY images of tetraploid cells from 2 lines as shown, showing only rare chromosomal translocations. The white arrows point to the translocations indicated below each image. (E) Ploidy analysis by interphase FISH of tissue sections. Representative images of single nuclei from 2 human PoleP286R/+ cases (confirmed by Sanger sequencing of exon 9) showing tetraploidy, with slides counterstained with DAPI.
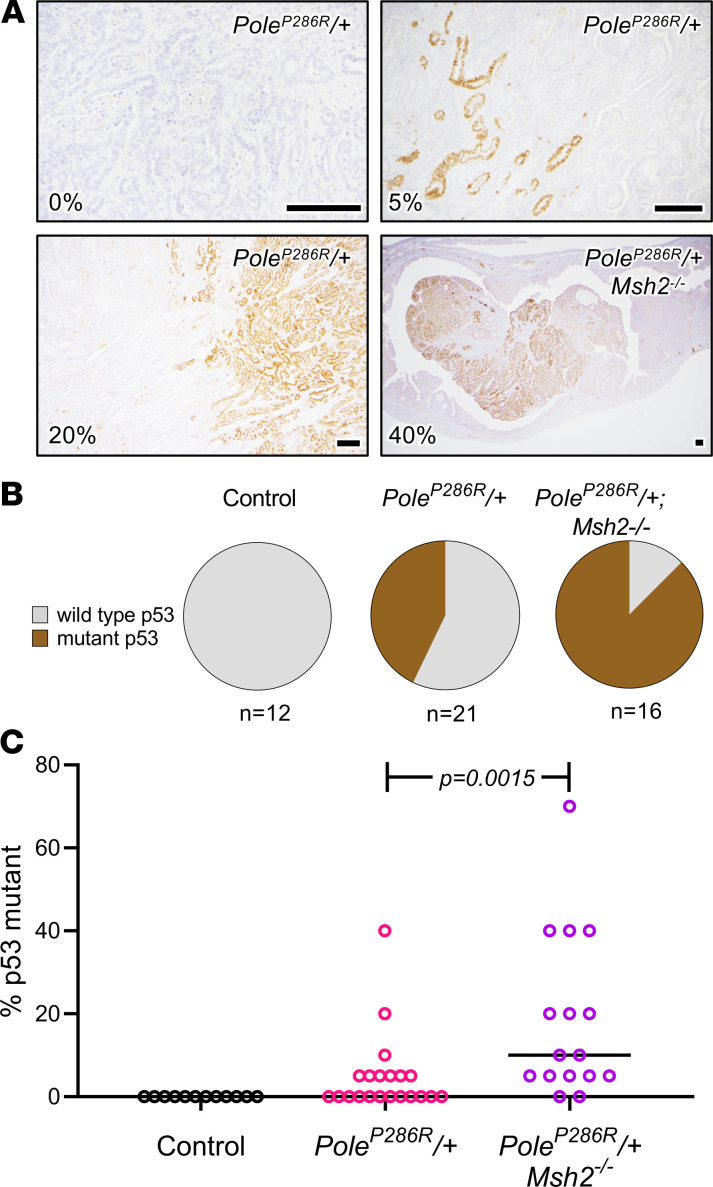
Tetraploidy in cancers is associated with p53 mutations, which favor cell survival in the context of aneuploidy and polyploidy (46, 47). Furthermore, p53 mutations are common in endometrial cancers, including POLE-mutant cancers (21). p53 mutations stabilize p53 protein through functional inactivation of the Mdm2 degradation loop, making IHC a reliable surrogate of such mutations (48–50). In n = 12 control uteri, no p53-mutant clones (evidenced by p53-mutant patterns; i.e., overexpression) were observed. In contrast, 43% (9/21) of PoleP286R tumors contained p53-mutant clones, versus 88% (14/16) in PoleP286R/+Msh2–/– tumors, P = 0.0031 per Fisher exact test (Figure 3, A and B). Furthermore, the percentage of p53-overexpressing tumor cells was higher in PoleP286R/+Msh2–/– versus PoleP286R/+ tumors (P = 0.0082, unpaired t test) (Figure 3C). These results demonstrate that p53 mutations are common but relatively late events in PoleP286R tumor progression, consistent with human POLE endometrial cancers, and show that p53 mutations occurred more frequently and presumably earlier in PoleP286R/+Msh2–/– tumors, perhaps as an adaptation to increased mutation rate.
Figure 3. Trp53 mutations are common late events in PoleP286R-driven endometrial cancers.
(A) Presence of Trp53-mutant clones per immunohistochemistry (IHC). Fields are selected at varying magnifications to best show each mutant clone. The percentage of tumor cells expressing p53 (5%, 10%, 20%, etc.) for the entire tissue section (not just the field shown) was semiquantitatively estimated and shown in the lower left-hand corner for each panel. The upper left panel shows an example of a tumor with no mutant clone. Scale bars: 200 μm. (B) Pie charts summarizing analysis as in A, for all tumors. Tumors were scored on presence or absence of Trp53-mutant clones by IHC. (C) Percentage of p53-positive cells in each tumor shown in B, P value per 2-tailed Mann-Whitney U test, showing that p53-expressing clones are significantly larger in PoleP286R/+Msh2–/– tumors.
PoleP286R/+ endometrial cancers harbor very high base substitution rates, in the range of human ultramutated tumors.
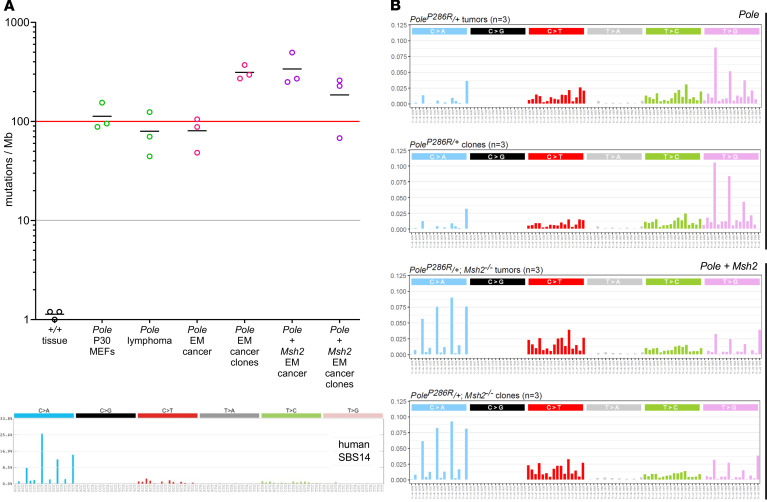
To define base substitution rates, n = 3 primary tumors and cell lines (total of 12 samples from the 2 mutant cohorts) were subjected to whole-genome sequencing (WGS) at an average depth of 40 times, in the general range of The Cancer Genome Atlas (TCGA) studies and permitting comparison to prior murine and human studies (22). PoleP286R/+ endometrial cancers exhibited base substitution rates of 48–105/Mb, far greater than previous genetically engineered mouse models of cancer and in the range of human ultramutant cancers. Cell lines exhibited a modest increase in base substitution rates of about 2–3 times relative to the primary tumors, perhaps in part because of clonal purification enhancing detection, although such relatively small differences could also be due to random variation. PoleP286R/+Msh2–/– tumors also exhibited very high base substitution rates that appeared modestly elevated as compared with PoleP286R/+ alone; cell lines exhibited similar base substitution rates as tumors (Figure 4A). Next, trinucleotide contexts for base substitutions were evaluated. All PoleP286R/+ samples exhibited virtually superimposable signatures, with few C>G and T>A substitutions, and a preponderance of T>G substitutions, especially with a T at the third position. All PoleP286R/+Msh2–/– samples also exhibited virtually superimposable signatures with each other, with a shift to C>A substitutions, especially when the third position was a T. This signature closely resembled the recently described SBS14 signature for rare human cancers harboring simultaneous POLE mutations and dMMR (20).
Figure 4. PoleP286R cancers have very high base substitution rates and specific mutational signatures, per WGS.
(A) Samples (n = 3 from different mice) include control (+/+) tissue, PoleP286R/+ mouse embryo fibroblasts at passage (P) 30 (22), T cell lymphomas from PoleP286R/+ global knockin mice (22), endometrial cancers (EM), or cell lines derived from single-cell clones. Gray and red lines correspond to human hypermutant and ultramutant thresholds. (B) Trinucleotide signatures for PoleP286R/+ tumors and cloned cell lines; each graph is derived from n = 3 independent samples, with the published SBS14 human signature for POLE + dMMR cancers shown for comparison (20).
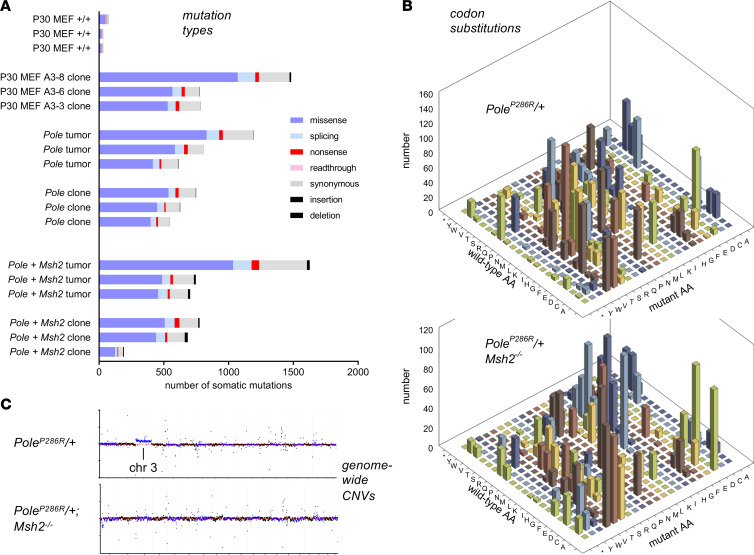
Next, we analyzed predicted coding impacts. As expected, PoleP286R/+ samples had a strong predominance of missense mutations, with a number of nonsense and splicing mutations, occasional readthrough mutations due to conversion of a terminal stop codon, and infrequent indels. PoleP286R/+Msh2–/– samples, in contrast, exhibited an elevation of the indel rate (Figure 5A and Supplemental Table 1) as a consequence of microsatellite instability, where expansion of microsatellite repeats leads to indels (20). The favored trinucleotide contexts described above resulted in highly skewed and nonrandom codon substitution tables, which were distinctive in PoleP286R/+ versus PoleP286R/+Msh2–/– samples (Figure 5B and Supplemental Table 1). Consistent with SKY results, read mapping to visualize genome-wide copy number variations (CNVs) revealed only modest alterations, with occasional chromosomes exhibiting copy number alterations relative to the whole genome (Figure 5C), and such CNVs were fewer than (for general comparison) in immortalized WT mouse embryo fibroblasts (Supplemental Figure 3).
Figure 5. PoleP286R endometrial cancers have a high rate of missense and nonsense mutations, while Msh2 deficiency adds to indel rate and alters codon substitutions.
(A) Mutations assigned into different categories using SnpEff with GENCODE M16 as reference. The x axis represents the number of somatic mutations in each sample. (B) Codon substitution matrix for nonsynonymous mutations across all samples (n = 6 per genotype). (C) Copy number plots from WGS data (representative individual samples). For each sample, the y axis represents normalized depth, log2(depth/median depth across genome). The x axis represents position among the 19 autosomes as illustrated (see Supplemental Figure 3). The analysis shows that PoleP286R does not result in large-scale chromosomal instability.
PoleP286R/+ endometrial cancers recruit T cells that serve to restrict tumor progression.
Amino acid substitutions create neoantigens that stimulate T cell–mediated antitumor immune responses (24). Concordantly, PoleP286R/+ endometrial cancers harbored numerous infiltrating CD3+ T cells (Figure 6A). PoleP286R could itself be immunogenic, particularly because polymerase ε is a housekeeping enzyme expressed in all cells, and P286R is believed to be the initiating tumor event shared by all tumor cells in a P286R-driven malignancy. However, no 8- to 11-mer peptides spanning P286R are predicted by NetMHC to bind mouse MHC (51), suggesting that PoleP286R does not create an effectively immunogenic neoepitope and that it is more likely the subsequent accumulation of amino acid substitutions that could invoke an antitumor response.
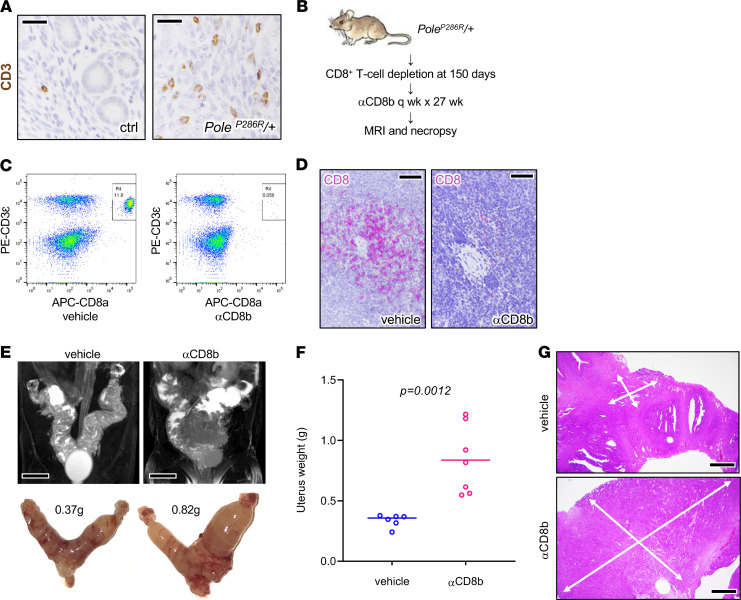
Figure 6. Acceleration of tumor progression following T cell depletion.
(A) CD3 IHC highlighting T cells, normal uterus versus tumor, with slides counterstained with hematoxylin. Scale bars: 25 μm. (B) Schematic summarizing CD8+ T cell depletion protocol. (C) αCD8 antibody resulted in near total CD8+ T cell depletion (inset boxes) in peripheral blood sampled 6 days after the first CD8 antibody injection. x axis, allophycocyanin-stained CD8; y axis, phycoerythrin-stained CD3ε. (D) IHC of CD8+ T cells in splenic tissue shows systemic tissue depletion. Tissue obtained 3 days after 27 weeks of treatment. Scale bars: 40 μm. (E–G) Mice euthanized after 27-week interval as shown in C. (E) MRI and matching gross images of uteri, from representative control and experimentally treated mice. Scale bars: 0.5 cm. (F) Uterus weights in control (n = 6) versus treated mice (n = 7), P value per 2-tailed Mann-Whitney U test. (G) Tumor burden per weight measurements likely underestimates overall tumor extent. White arrows show histologic extent of tumors. Scale bars: 250 μm.
T cells were systemically depleted in PoleP286R/+ mice by injection of an anti-CD8 (αCD8) antibody starting at 150 days of age, with mice followed serially by MRI to aid in determination of an appropriate time point for necropsy (Figure 6B). Flow cytometric analysis and tissue immunostains (CD8) confirmed CD8+ T cell depletion in peripheral blood (Figure 6C) and in tissues (Figure 6D). At necropsy, MRI and uterine weights showed significantly increased tumor burden in animals treated with αCD8 versus vehicle (P = 0.0014, unpaired t test) (Figure 6, E and F). However, while uterine weight is useful as an objective and easily measurable parameter, it likely underestimates increases in tumor burden; for example, residual normal uterine tissues make up a significant percentage of tumorous uteri. Consistent with this, histologic examination consistently revealed larger areas of tumor infiltration in the CD8+ T cell–depleted uteri (Figure 6G). These findings suggest that PoleP286R results in immunogenic responses that limit tumor progression.
PoleP286R/+ and PoleP286R/+Msh2–/– tumors are highly responsive to immune checkpoint blockade.
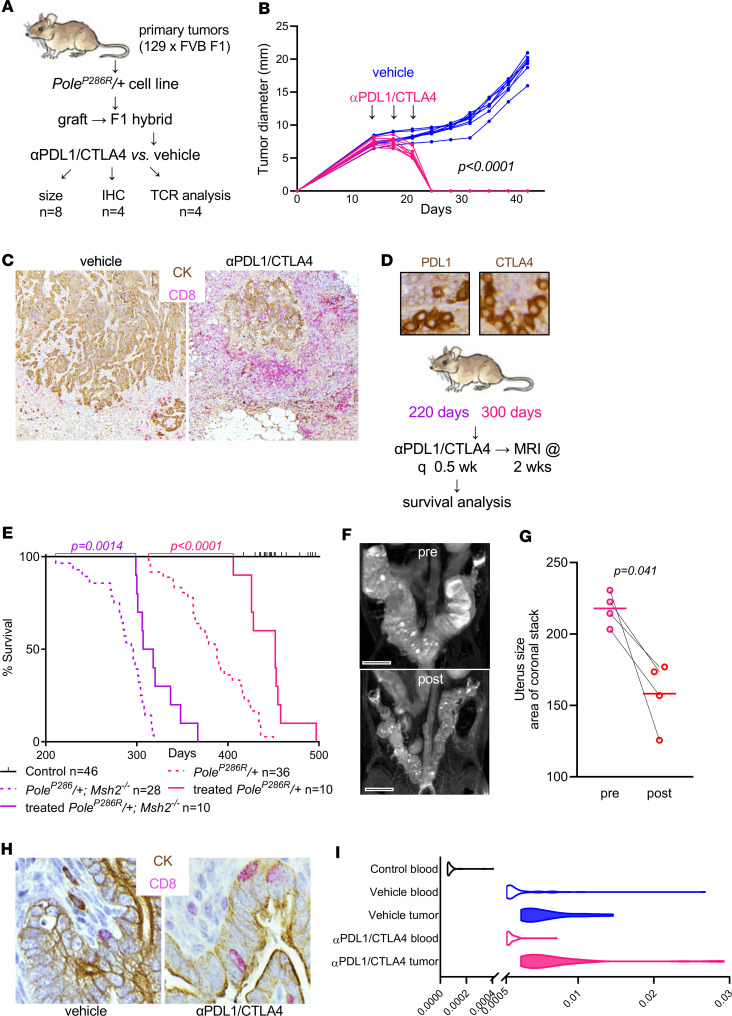
First, we established an F1 hybrid syngeneic graft model. The LSL-PoleP286R allele was generated in and maintained in a pure 129S6/SvEvTac background, whereas the BAC-Sprr2f-Cre allele was maintained (and extensively backcrossed) in an FVB background. Thus, experimental PoleP286R/+ mice were F1 hybrids comprising 50% each of the 2 backgrounds. Their tumors are thus syngeneic and should be engraftable into F1 hybrid mice generated by interbreeding WT animals of the 2 strains (Figure 7A). PoleP286R/+ endometrial cancer cell line B3E (Figure 2B) was engrafted into F1 hybrid mice, with tumors showing continual growth following successful engraftment (Figure 7B). The cell line was selected at random and was subsequently determined to be tetraploid (Figure 2B); the impact of ploidy on this experiment was not further investigated. Strikingly, tumor-engrafted animals subjected to just 3 injections of αPDL1/CTLA-4 combined therapy over 10 days showed complete regression of the tumors with lack of regrowth following cessation of treatment (P < 0.0001, Figure 7B). IHC double-labeling against pan-cytokeratin (CK) and CD8 showed that regression was accompanied by massive infiltration of CD8+ T cells (Figure 7C). Therefore, tumors were immunogenic and highly responsive to immune checkpoint blockade.
Figure 7. PoleP286R/+ and PoleP286R/+Msh2–/– tumors are highly responsive to immune checkpoint blockade.
(A) Schematic summarizing combined αPDL1/CTLA-4 treatment of syngeneically engrafted F1 hybrid mice. (B) Subcutaneous tumor graft measurements per Vernier caliper twice a week; n = 8 grafts for vehicle and treated, P value per 2-way ANOVA grouped analysis. (C) Double-labeling of grafts by IHC against CD8 to highlight T cells and pan-cytokeratin (CK) to highlight tumor cells. (D) PDL1 and CTLA-4 expression in lymphocytes in representative PoleP286R/+ tumors and schematic of αPDL1/CTLA-4 treatment of live mice. (E) Survival analysis following treatment of PoleP286R/+ and PoleP286R/+Msh2–/– tumors, P value per log-rank test. (F) Z-stack MRI of PoleP286R/+ mouse uterus before treatment and 2 weeks posttreatment. Scale bars: 0.5 cm. (G) Tumor response per MRI Z-stacks; n = 4 matched (pre/post) pairs, P value by paired t test. (H) IHC of CD8 and CK highlights increased numbers of tumor-infiltrating lymphocytes within malignant gland epithelium. (I) Distribution of the 50 most frequent TCRs in peripheral blood and tumors of 129 × FVB F1 engrafted mice; n = 4 per group; x axis, frequency.
Primary tumors expressed PDL1 and CTLA-4 in infiltrating lymphocytes (Figure 7D). To further test combined αPDL1/CTLA-4 blockade, an increasingly common therapeutic combination (52), treatment responses were measured in live PoleP286R/+ and PoleP286R/+Msh2–/– mice. PoleP286R/+ and PoleP286R/+Msh2–/– treatments were initiated at 300 days and 220 days, respectively, because of the accelerated mortality of the latter (Figure 7D). Survival analysis showed significant clinical benefit with statistically significant survival extension in both cohorts (Figure 7E). Pre- and posttreatment MRIs showed significant responses 2 weeks after initiation of therapy (P = 0.041 per paired t test, Figure 7, F and G). Tumors showed increased numbers of infiltrating CD8+ T cells, especially within malignant gland epithelium (Figure 7H). Holistic T cell receptor (TCR) sequence analysis showed that mice syngeneically engrafted with a PoleP286R/+ cancer cell line and treated with αPDL1/CTLA-4 had significantly increased TCR clonal expansion in both peripheral blood and tumor tissues. Tumor-grafted mice treated with vehicle showed significantly increased TCR expansion in tumor tissues but not in peripheral blood (Supplemental Figure 4, A and B). The expansion of the 50 most represented TCR rearrangements were analyzed, and tumors in treated mice had the highest expansion of TCRs (Figure 7I and Supplemental Figure 4C), indicating that immune checkpoint therapy resulted in larger changes in TCR repertoires associated with tumor diminution. The frequency of 1 TCR in blood samples from mice receiving vehicle was unusually high (>0.02, Supplemental Figure 4C), suggesting that this TCR might be related to a dominant T cell–responding clone. The significantly extended survival in both PoleP286R/+ and PoleP286R/+Msh2–/– mice was thus likely related to functional TCR repertoire expansion suppressing tumor development. These results demonstrate the mouse cancer models with PoleP286R-driven ultramutation are robust models for further investigations into the biology of Pole-driven immunogenicity and mechanisms of responsiveness versus nonresponsiveness to immune checkpoint blockade.
Discussion
In this study, we present a potentially novel and efficient conditional, tissue-specific approach using an LSL-PoleP286R allele to generate a specific cancer mouse model with a far higher mutational burden than previously feasible in live genetically engineered animal models. PoleP286R proved genetically dominant, as has been observed in human cancers. We documented a 100% incidence of aggressive and fatal endometrial cancers, even when the allele was only heterozygous. That cancers could be generated with a single monoallelic driver and in only 1 generation stands in contrast to previous mouse models of cancer, which have typically required multiple alleles and complex breeding schema. Murine PoleP286R endometrial cancers closely resembled their human counterparts in terms of histology and clinical behavior. We demonstrated that PoleP286R-driven endometrial cancers have high mutational burdens in the range of human ultramutant cancers and were sensitive to immune checkpoint blockade, providing a model with robust responses to immunotherapy. This work provides a new approach for modeling cancer that may overcome current limitations of mouse models, namely very low mutational load and consequently limited tumor heterogeneity — which are not representative of any human tumor (30–32).
The initial TCGA study of endometrial cancer reported that POLE-mutant endometrial cancers have an exceptionally good prognosis (16). POLE mutations are present in diverse endometrial cancer histologic subtypes, including some associated with poor outcome, such as clear cell carcinoma and carcinosarcoma. This suggests that POLE testing of tumors (e.g., by cancer gene panel) could be useful to identify patients who could forego additional treatments associated with substantial morbidity, such as surgical staging/lymph node dissection or adjuvant chemotherapy/radiotherapy (21), much as dMMR testing has become standard practice. Meta-analysis of 23 studies of dMMR and clinical outcome found no significant association between MMR status and survival in the setting of endometrial cancer (53), but dMMR testing is standard for all new endometrial cancer cases to (a) screen for Lynch syndrome and (b) identify patients who are candidates for immune checkpoint blockade (pembrolizumab) (54, 55). That POLE-ultramutated cancers will also prove responsive to immunotherapy has been suggested by isolated case reports of exceptional responders (56), but large clinical trials have not yet been conducted. Such trials will be complicated by (a) the need for prospective identification of POLE-mutant cancers by cancer gene panel (not yet routine), (b) the relative rarity of such cancers, and (c) the even smaller subset with advanced disease at the time of diagnosis. Thus, our preclinical model is useful in that it provides compelling in vivo evidence that ultramutant POLE-mutant endometrial cancers (and by extension, POLE-driven malignancies at other anatomic sites) will also prove consistently sensitive to immune checkpoint blockade.
Recent studies of patient cohorts have challenged the idea that POLE endometrial cancers have an invariably good prognosis. For example, in 1 single-institution study of n = 23 POLE endometrial cancers identified by cancer gene panel (MSK-IMPACT), 17% (4/23) were of advanced stage with extrauterine disease at the time of diagnosis, including 2 cases that were stage IV (distant metastasis). After a median follow-up of 30 months, 17% (4/23) of patients developed recurrences, of which 3 were distant metastases, including 2 brain metastases, and 1 patient died after 33 months (57). A separate large, multi-institutional study of POLE cancers by the NRG Oncology/Gynecologic Oncology Group found improved outcomes for the POLE group, but the differences were not statistically significant (58). Although additional patient studies are needed to better define clinical outcomes, these later studies found that a significant proportion of POLE cancers metastasize, and such patients should benefit from targeted therapeutic approaches.
In our BAC-Sprr2f-Cre models, tumors were aggressive, with metastatic disease present in 100% of animals. This apparently more aggressive clinical course in mice relative to women likely reflects the nature of the model. In women, a single endometrial epithelial cell spontaneously acquires a POLEP286R mutation, giving rise to a single somatic clone that eventually becomes malignant. Whereas some endometrial cancers can show heterogeneity with respect to drivers such as TP53 (see below), all studies to date suggest that POLEP286R and other POLE ultramutator alleles are present throughout the tumor and thus are the initial driver. In contrast, in our models, the PoleP286R mutation is induced in hundreds, and probably thousands, of independent clones. It seems very likely that such multiclonality provides greater opportunities for tumor evolution and escape from immune surveillance or other tumor-suppressive mechanisms normally restraining ultramutation-driven carcinogenesis. While such enforced induction in many cells is likely necessary for a robust, high-penetrance animal model, it may be interesting to study tumor progression with other cell type–specific Cre drivers or with methods permitting P286R induction in fewer cells.
In systematic analyses of PoleP286R/+ endometrial cancers, we found no evidence for spontaneous dMMR. There was not even focal loss of dMMR factor expression in any primary endometrial cancer, and all PoleP286R/+ samples subjected to WGS showed superimposable trinucleotide signatures readily distinguishable from the combined Pole + dMMR signature (Figure 4B). At the same time, we observed definitive cooperation with respect to overall tumor progression and survival in a defined genetic model where both defects were provoked simultaneously. These findings demonstrate that while Pole + dMMR can cooperate in tumor progression, such cooperation is not obligate. Pole-driven ultramutation is sufficient to drive tumor initiation and progression even in the context of proficient mismatch repair, although in a minority of cancers, both defects coexist and undoubtedly cooperate. POLE mutations may be secondary events that further accelerate the progression of initially dMMR cancers, as suggested by the secondary acquisition of mutations in children with constitutional dMMR (19) and the observation that some endometrial cancers in Lynch syndrome patients have POLE mutations (59). Thus, our results, combined with the available literature, suggest that strong POLE mutations occur secondarily in the context of dMMR-driven cancers, but perhaps not vice versa.
We documented definitive p53-mutant patterns in nearly half of PoleP286R/+ and the majority of PoleP286R/+Msh2–/– endometrial cancers. The patterns and sizes of p53-mutant clones indicated that p53 mutations were acquired late in tumor progression. These results demonstrate substantial selective pressure for p53 inactivation during PoleP286R-driven tumor progression and suggest that such selective pressure increases as a function of mutational load. This likely explains the higher incidence and larger p53-mutant clone sizes in PoleP286R/+Msh2–/– tumors. Similar processes may occur in human POLE cancers because most POLE-mutant, immunohistochemically p53-abnormal tumors show incomplete (i.e., subclonal) loss of p53 staining, again showing that the acquisition of p53 mutations is a late event (21). Whereas mutant p53 immunostaining patterns usually signify poor prognosis, “double-classifier” endometrial cancers harboring POLE mutations and mutant p53 immunostaining have a good prognosis similar to POLE-alone cancers (21). Our work provides further evidence that p53 and Pole mutations are functionally intertwined and should be viewed as a characteristic feature of Pole-driven carcinogenesis.
Mouse PoleP286R endometrial cancers exhibited striking nuclear atypia and giant nuclei, as described for human POLE endometrial cancers (37, 39). These findings initially seemed paradoxical because POLE is well established as a single base substitution mutator (60, 61), whereas nuclear atypia and giant nuclei imply aneuploidy or polyploidy. Our subsequent investigations suggest that tetraploidization is yet another distinctive feature of POLE-driven tumorigenesis shared by the mouse and human counterparts (45). We propose that tetraploidy, which occurs in diverse cancers, is particularly adaptive in ultramutant tumors because polyploidization provides additional copies of loci to permit “genetic buffering” against phenotypic variation, which is likely extreme in the context of ultramutation (62, 63). Tetraploidization would also promote even further genetic diversity and could contribute to the apparent base substitution rate as determined by sequencing. Tetraploid p53 WT cells fail to propagate in culture, whereas p53-null cells can be passaged, demonstrating that p53 is a key checkpoint suppressing tetraploidization and that p53 loss favors cell survival in this context (47). Tetraploidization in turn can promote further chromosome-level instability (64). Thus, we propose that POLE tumors, though initially driven by a pure single base substitution mutator phenotype, acquire additional layers of genome instability through the acquisition of p53 mutations, polyploidization, and modest chromosome-level instability, as documented by our SKY results.
In addition to models of Pole-driven neoplasia, our results suggest that the LSL-PoleP286R allele could be useful for other studies of cancer. For example, incorporation of the allele into genetically engineered mouse models (e.g., Kras-driven lung cancers) could be used in an experimental system to formally investigate the contribution of high mutational burden into diverse aspects of tumor progression, including immune surveillance and how the PoleP286R and PoleP286RMsh2–/– models eventually become resistant to immune checkpoint blockade. Thus, this approach may facilitate the development of additional experimental models of the immune landscape of cancer.
Methods
Mouse husbandry and survival analysis.
Mice were housed in a pathogen-free animal facility in microisolator cages and fed ad libitum on standard chow. Only females were used, with ages as described for each observation. All experiments used littermate controls. The LSL-PoleP286R allele was generated in and maintained in a pure 129S6/SvEvTac background; the BAC-Sprr2f-Cre allele was in an FVB background (backcrossed for 12 generations). Survival analyses were conducted on experimental and control animals selected at weaning.
Tissue processing and IHC.
Fixation, sectioning, antigen retrieval, blocking, and secondary detection for the following primary antibody dilutions in 2% BSA were performed as previously described (65): CD3 (Abcam, ab5690 rabbit polyclonal Ab, 1:2000 dilution), p53 (Cell Signaling Technology, 2524, mouse mAb, 1:2000 dilution), MSH2 (Cell Signaling Technology, 2017, rabbit mAb, 1:500 dilution), MSH6 (Abcam, ab92471, rabbit mAb, 1:500 dilution), MLH1 (Abcam, ab92312, rabbit mAb, 1:500 dilution), CD8 (Cell Signaling Technology, 98941S, rabbit mAb, 1:500 dilution), CK (Cell Signaling Technology, 98941S, rabbit mAb, 1:500 dilution), PDL1 (Cell Signaling Technology, 29122, mouse mAb, 1:400 dilution), and CTLA-4 (Santa Cruz Biotechnology, sc-376016, mouse mAb, 1:5000 dilution) (48). Antigen retrieval for all antibodies was performed in parboiling 10 mM Na citrate with 0.05% Tween-20, pH 6.0, for 12 minutes. Expression of p53 was assessed by a pathologist evaluating the entirety of a tumor present on a single immunostained slide. Strong nuclear expression in excess of background in more than 100 cells in close proximity was the criterion for designating a p53-positive clone.
Cell line derivation.
Tumor fragments were excised from uteri under a dissection microscope, chopped to fine pieces with a scalpel in cold 0.25% Trypsin-EDTA solution (25200-114, Thermo Fisher Scientific), moved to 37°C for 15 minutes, and then triturated 20 times with a transfer pipette. Cells were pelleted by centrifugation and resuspended in DMEM (Thermo Fisher Scientific 10566-016) with 10% FBS and 1× penicillin-streptomycin and then grown in this medium under standard tissue culture conditions. Cells were passaged 4 times before initiation of experiments, and epithelial character was confirmed by phase-contrast microscopy. For WGS studies only, cells were subcloned by flow sorting of single cells into 96-well plates as previously described (22).
SKY of mouse cell lines and interphase FISH of human cell lines.
Chromosome spreads were prepared by synchronizing cells with 100 ng/mL colcemid (KaryoMAX, Thermo Fisher Scientific) for 4 hours and harvesting by trypsinization. Cell pellets were gently resuspended in prewarmed 75 mM KCl solution and incubated at 37°C for 6 minutes. Cells were then fixed with ice-cold methanol/acetic acid (3:1) and dropped onto slides. For SKY, multicolor DNA FISH probes for mouse chromosomes (MetaSystems) were used per the manufacturer’s protocol. Briefly, slides were denatured in 0.07N NaOH at room temperature for 1 minute, and FISH probes were denatured at 75°C for 5 minutes. FISH probes were then applied to chromosome spreads, sealed with a coverslip, and incubated in a humidified chamber at 37°C for 1–2 days. Following hybridization, slides were washed with 0.4× SSC at 72°C for 2 minutes and 2× SSC, with 0.05% Tween-20, at room temperature for 1 minute. Slides were gently rinsed in water, air-dried, and DAPI counterstained. Images were acquired using a Zeiss Axio Imager Z2 equipped with a Metafer Slide Scanning System and analyzed using Isis (MetaSystems) software.
Interphase FISH was performed at the UT Southwestern Molecular Cytogenetics Clinical Laboratory on 4-μm-thick tissue sections using the AneuVysion kit (Abbott) with DAPI counterstaining per the manufacturer’s instructions. Cases of endometrial cancer with POLE mutations were identified by Sanger sequencing of exons 9 and 13, using DNA prepared from formalin-fixed, paraffin-embedded tissue sections.
DNA and library preparation for WGS.
DNA was extracted with the QIAamp DNA Mini Kit (QIAGEN, 51306) with concentrations per Qubit fluorometer (Invitrogen, Thermo Fisher Scientific). Sample integrity was confirmed by agarose gel electrophoresis. For preparation of libraries, 1.5 μg DNA was fragmented by Covaris ultrasonicator, then analyzed by gel electrophoresis. The fragmented DNA was combined with End-Repair Mix and incubated at 20°C for 30 minutes. The end-repaired DNA was purified with QIAquick PCR Purification Kit (QIAGEN) followed by addition of A-Tailing mix (Illumina) and incubated at 3°C for 30 minutes. This was combined with the purified Adenylate 3′ ends DNA, adapter and ligation mix, and the ligation reaction was incubated at 20°C for 15 minutes. Adapter-ligated DNA was run on 2% agarose gel to recover target fragments. DNA was gel-purified with QIAquick Gel Extraction Kit (QIAGEN). Several rounds of PCR amplification with PCR Primer Cocktail and Master Mix (both from Illumina) were performed to enrich Adapter-ligated DNA fragments. PCR products were run on a 2% agarose gel to recover the target fragments, followed by gel purification with QIAquick Gel Extraction Kit (QIAGEN). The final library was analyzed by determination of average molecule length per Agilent 2100 bioanalyzer (Agilent DNA 1000 Reagents) and quantification performed by real-time PCR by TaqMan assay. Qualified libraries were loaded onto the HiSeq X Ten sequencer (Illumina) for paired-end sequencing with read lengths of 100–150 bp.
Variant calling.
Reads were mapped to the mouse reference genome (GRCm38) using BWA 0.7.17 (66). Duplicated reads were marked using Picard, and base quality score recalibration was applied using GATK 4.0 (67). SNP and indel discovery were performed using samtools (68). A mutation was considered as existing in a sample only if the alternative allele frequency was more than 0.1. A mutation was considered somatic when it was not a known variant from the Mouse Genomes Project, including several FVB and 129 substrains (69), and the mutation was identified in only 1 sample but not in any other samples. All mutations were annotated using SnpEff (70), based on GENCODE M16 annotation (71).
Depth analyses.
Average depth in each window (1 Mbp) was estimated using samtools. Raw depth was normalized by dividing the median depth across the genome followed by a log2 transformation.
The pipelines for read mapping, quality control, mutation calling, and annotation were as described (22). Base substitution rate was calculated for each sample as the number of mutations identified divided by the number of genomic positions covered by at least 20 reads. Trinucleotide signature was generated using a self-written script according to the mutational signatures observed in PCAWG (20).
T cell depletion studies.
αCD8b antibody (Bio X Cell, BE0223) at 200 μg per mouse per week was administered by intraperitoneal (IP) injection. CD8+ T cell depletion in peripheral blood was confirmed by flow cytometry analysis at 6 days and 24 weeks after initial injection. Mice were euthanized after a 27-week treatment interval with final MRI performed a few days before euthanasia.
Immune checkpoint blockade therapy.
F1 syngeneic mouse hosts were treated with FTY720 (20 μg/mouse) (MilliporeSigma, SML0700) at 3 days and 1 day before tumor cell grafting and 1 day and 4 days after tumor cell grafting by IP injection. For syngeneic graft studies, 1 million PoleP286R/+ endometrial carcinoma cells were subcutaneously injected into the right flank. Immune checkpoint blockade was started 14 days after tumor cell engrafting. Two hundred micrograms each αPDL1 (Bio X Cell, BE0101, clone 10F.9G2) and αCTLA-4 (Bio X Cell, BE0164, clone 9D9) antibodies were administered by IP injection for 3.5 days × 3 doses. Tumor sizes were measured with calipers twice a week. For treatment of live PoleP286R/+ and PoleP286R/+Msh2–/– mice, combined αPDL1/CTLA-4 was also given at 200 μg each antibody per mouse, twice a week by IP injection.
MRI and data analysis.
MRI was conducted with a 7-T small-animal system (Bruker BioSpin Corp.) with a 40-mm (I.D.) radio frequency (RF) coil. Animals were anesthetized with 1%–2% isoflurane (AErrane, Baxter Healthcare Corporation) mixed in 100% O2 and placed prone with respiratory sensor, headfirst with abdomen centered with respect to the center of the RF coil. Low-resolution multislice gradient echo imaging, serving as the localizer, was first performed on the abdominal region to confirm location and orientation of the uterus. For volume measurements of tumorous uteri, axial and coronal T2-weighted multislice images encompassing an entire uterus were obtained with a fat suppression fast spin-echo sequence. Acquisition parameters for axial images were 4000-ms repetition time, 40-ms effective echo time, 32 × 32 mm field of view, 256 × 256 matrix, 1-mm slice thickness, 31 slices, gapless, 8 excitations, fat suppression, and scan time of 16 minutes and 10 seconds, and those for coronal images were 3000-ms repetition time, 40-ms effective echo time, 48 × 32 mm field of view, 384 × 256 matrix, 1-mm slice thickness, 19 slices, gapless, 8 excitations, fat suppression, and scan time of 9 minutes and 36 seconds. Volumes were calculated with image processing software (ImageJ, version 1.40g; NIH), as described previously (72).
TCR repertoire analyses.
DNA from all blood and tumor tissue samples was extracted with the QIAamp DNA Mini Kit (QIAGEN, 51306). DNA samples were sent to Adaptive Biotechnologies for deep TCR-β profiling resolution analysis. TCR diversity and clonality were analyzed per Adaptive Biotechnologies’ ImmunoSEQ assays guide (73).
Data and materials availability.
Sequence data have been deposited in the NCBI’s Sequence Read Archive database under accession number PRJNA613918.
Statistics.
Data are presented as mean ± SEM unless otherwise indicated. To determine P values, 2-tailed Student’s t tests or Fisher exact tests were performed (unless otherwise indicated). P < 0.05 was considered statistically significant. For survival curves, Kaplan-Meier analysis was used, with statistical comparison among curves performed with the log-rank test. The above statistical analyses were performed with GraphPad Prism (version 8). No statistical method was used to predetermine sample sizes. For treatment studies, mice were randomly assigned to treatment/no-treatment cohorts by alternating assignment to the 2 pools. Where possible (i.e., assessment of mutant clones and their size following IHC of tumors of differing genotypes), analysis was performed by a pathologist blinded to the genotype. Some histopathologic assessments could not be randomized.
Study approval.
All animal studies were approved by the UT Southwestern IACUC and in adherence to the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). Analysis of human samples was approved with a waiver of consent under a UT Southwestern Institutional Review Board protocol.
Author contributions
HDL, CL, YXF, and DHC conceived this study. ICC and DHC designed and generated the alleles. HDL, CL, QH, and JZ performed experiments. BL, YXF, PL, EGM, SZ, EAA, MA, GML, SSS, and CL helped design and interpret experiments. DHC conducted the IHC and histologic characterization of tumors. PK performed interphase FISH on human tumors and interpreted results. SZ and XW conducted MRI analyses. HZ designed and conducted computational and data analyses. DHC and HDL wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
This work was supported by NIH/National Cancer Institute grants R01CA211339 to DHC and R01CA237405 to DHC and YXF.
Version 1. 07/23/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(14):e138829.https://doi.org/10.1172/jci.insight.138829.
Contributor Information
Hao-Dong Li, Email: Hao-Dong.Li@utsouthwestern.edu.
Changzheng Lu, Email: Changzheng.Lu@UTSouthwestern.edu.
He Zhang, Email: zhangheqd@gmail.com.
Qing Hu, Email: qing.hu@utsouthwestern.edu.
Junqiu Zhang, Email: junqiu.zhang@utsouthwestern.edu.
Mitzi Aguilar, Email: mitzi.aguilar@utsouthwestern.edu.
Shanrong Zhang, Email: Shanrong.Zhang@jax.org.
Xiaojing Wang, Email: Xiaojing.Wang@UTSouthwestern.edu.
Guo-Min Li, Email: Guo-Min.Li@UTSouthwestern.edu.
Bo Li, Email: bo.li@utsouthwestern.edu.
Prasad Koduru, Email: prasad.koduru@utsouthwestern.edu.
Peter Ly, Email: peter.ly@utsouthwestern.edu.
Yang-Xin Fu, Email: yang-xin.fu@utsouthwestern.edu.
Diego H. Castrillon, Email: diego.castrillon@utsouthwestern.edu.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34(9):2311–2321. [PubMed] [Google Scholar]
- 3.Loeb LA. Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer. 2011;11(6):450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5(7):704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kucherlapati MH, et al. An Msh2 conditional knockout mouse for studying intestinal cancer and testing anticancer agents. Gastroenterology. 2010;138(3):993–1002.e1. doi: 10.1053/j.gastro.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong S, Hui P, Buza N. Frequent loss of mutation-specific mismatch repair protein expression in nonneoplastic endometrium of Lynch syndrome patients. Mod Pathol. 2020;33(6):1172–1181. doi: 10.1038/s41379-020-0455-x. [DOI] [PubMed] [Google Scholar]
- 7.Germano G, Amirouchene-Angelozzi N, Rospo G, Bardelli A. The clinical impact of the genomic landscape of mismatch repair-deficient cancers. Cancer Discov. 2018;8(12):1518–1528. doi: 10.1158/2159-8290.CD-18-0150. [DOI] [PubMed] [Google Scholar]
- 8.Rayner E, et al. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16(2):71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 9.Lujan SA, Williams JS, Kunkel TA. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016;26(9):640–654. doi: 10.1016/j.tcb.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell BB, et al. Comprehensive analysis of hypermutation in human cancer. Cell. 2017;171(5):1042–1056.e10. doi: 10.1016/j.cell.2017.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xing X, et al. A recurrent cancer-associated substitution in DNA polymerase ε produces a hyperactive enzyme. Nat Commun. 2019;10(1):374. doi: 10.1038/s41467-018-08145-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkash V, Kulkarni Y, Ter Beek J, Shcherbakova PV, Kamerlin SCL, Johansson E. Structural consequence of the most frequently recurring cancer-associated substitution in DNA polymerase ε. Nat Commun. 2019;10(1):373. doi: 10.1038/s41467-018-08114-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barbari SR, Kane DP, Moore EA, Shcherbakova PV. Functional analysis of cancer-associated DNA polymerase epsilon variants in Saccharomyces cerevisiae. G3 (Bethesda) 2018;8(3):1019–1029. doi: 10.1534/g3.118.200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbari SR, Shcherbakova PV. Replicative DNA polymerase defects in human cancers: Consequences, mechanisms, and implications for therapy. DNA Repair (Amst) 2017;56:16–25. doi: 10.1016/j.dnarep.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kane DP, Shcherbakova PV. A common cancer-associated DNA polymerase ε mutation causes an exceptionally strong mutator phenotype, indicating fidelity defects distinct from loss of proofreading. Cancer Res. 2014;74(7):1895–1901. doi: 10.1158/0008-5472.CAN-13-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497(7447):67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodel KP, et al. Explosive mutation accumulation triggered by heterozygous human Pol ε proofreading-deficiency is driven by suppression of mismatch repair. Elife. 2018;7:e32692. doi: 10.7554/eLife.32692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herr AJ, Kennedy SR, Knowels GM, Schultz EM, Preston BD. DNA replication error-induced extinction of diploid yeast. Genetics. 2014;196(3):677–691. doi: 10.1534/genetics.113.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shlien A, et al. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47(3):257–262. doi: 10.1038/ng.3202. [DOI] [PubMed] [Google Scholar]
- 20.Alexandrov LB, et al. The repertoire of mutational signatures in human cancer. Nature. 2020;578(7793):94–101. doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.León-Castillo A, et al. Interpretation of somatic POLE mutations in endometrial carcinoma. J Pathol. 2020;250(3):323–335. doi: 10.1002/path.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li HD, et al. Polymerase-mediated ultramutagenesis in mice produces diverse cancers with high mutational load. J Clin Invest. 2018;128(9):4179–4191. doi: 10.1172/JCI122095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 24.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal R, et al. Genetic diversity of tumors with mismatch repair deficiency influences anti-PD-1 immunotherapy response. Science. 2019;364(6439):485–491. doi: 10.1126/science.aau0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.American Association for Cancer Research. First tissue-agnostic drug approval issued. Cancer Discov. 2017;7(7):656. doi: 10.1158/2159-8290.CD-NB2017-078. [DOI] [PubMed] [Google Scholar]
- 27.Bruegl AS, Kernberg A, Broaddus RR. Importance of PCR-based tumor testing in the evaluation of Lynch syndrome-associated endometrial cancer. Adv Anat Pathol. 2017;24(6):372–378. doi: 10.1097/PAP.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johanns TM, et al. Immunogenomics of hypermutated glioblastoma: a patient with germline POLE deficiency treated with checkpoint blockade immunotherapy. Cancer Discov. 2016;6(11):1230–1236. doi: 10.1158/2159-8290.CD-16-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santin AD, et al. Regression of chemotherapy-resistant polymerase ε (POLE) ultra-mutated and MSH6 hyper-mutated endometrial tumors with nivolumab. Clin Cancer Res. 2016;22(23):5682–5687. doi: 10.1158/1078-0432.CCR-16-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McFadden DG, et al. Mutational landscape of EGFR-, MYC-, and Kras-driven genetically engineered mouse models of lung adenocarcinoma. Proc Natl Acad Sci U S A. 2016;113(42):E6409–E6417. doi: 10.1073/pnas.1613601113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedognetti D, et al. Toward a comprehensive view of cancer immune responsiveness: a synopsis from the SITC workshop. J Immunother Cancer. 2019;7(1):131. doi: 10.1186/s40425-019-0602-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McFadden DG, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell. 2014;156(6):1298–1311. doi: 10.1016/j.cell.2014.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Westcott PM, et al. The mutational landscapes of genetic and chemical models of Kras-driven lung cancer. Nature. 2015;517(7535):489–492. doi: 10.1038/nature13898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Contreras CM, et al. Lkb1 inactivation is sufficient to drive endometrial cancers that are aggressive yet highly responsive to mTOR inhibitor monotherapy. Dis Model Mech. 2010;3(3-4):181–193. doi: 10.1242/dmm.004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuevas IC, et al. Fbxw7 is a driver of uterine carcinosarcoma by promoting epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2019;116(51):25880–25890. doi: 10.1073/pnas.1911310116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zaino RJ, Kurman RJ. Squamous differentiation in carcinoma of the endometrium: a critical appraisal of adenoacanthoma and adenosquamous carcinoma. Semin Diagn Pathol. 1988;5(2):154–171. [PubMed] [Google Scholar]
- 37.Hussein YR, et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod Pathol. 2015;28(4):505–514. doi: 10.1038/modpathol.2014.143. [DOI] [PubMed] [Google Scholar]
- 38.Van Gool IC, et al. Blinded histopathological characterisation of POLE exonuclease domain-mutant endometrial cancers: sheep in wolf’s clothing. Histopathology. 2018;72(2):248–258. doi: 10.1111/his.13338. [DOI] [PubMed] [Google Scholar]
- 39.Bakhsh S, et al. Histopathological features of endometrial carcinomas associated with POLE mutations: implications for decisions about adjuvant therapy. Histopathology. 2016;68(6):916–924. doi: 10.1111/his.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gotoh O, et al. Clinically relevant molecular subtypes and genomic alteration-independent differentiation in gynecologic carcinosarcoma. Nat Commun. 2019;10(1):4965. doi: 10.1038/s41467-019-12985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leskela S, et al. Molecular basis of tumor heterogeneity in endometrial carcinosarcoma. Cancers (Basel) 2019;11(7):E964. doi: 10.3390/cancers11070964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groothuizen FS, Sixma TK. The conserved molecular machinery in DNA mismatch repair enzyme structures. DNA Repair (Amst) 2016;38:14–23. doi: 10.1016/j.dnarep.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 43.Lucas E, et al. Mismatch repair protein expression in endometrioid intraepithelial neoplasia/atypical hyperplasia: should we screen for Lynch syndrome in precancerous lesions? Int J Gynecol Pathol. 2019;38(6):533–542. doi: 10.1097/PGP.0000000000000557. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka K, Goto H, Nishimura Y, Kasahara K, Mizoguchi A, Inagaki M. Tetraploidy in cancer and its possible link to aging. Cancer Sci. 2018;109(9):2632–2640. doi: 10.1111/cas.13717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proctor L, et al. Assessment of DNA Ploidy in the ProMisE molecular subgroups of endometrial cancer. Gynecol Oncol. 2017;146(3):596–602. doi: 10.1016/j.ygyno.2017.06.020. [DOI] [PubMed] [Google Scholar]
- 46.Davaadelger B, Shen H, Maki CG. Novel roles for p53 in the genesis and targeting of tetraploid cancer cells. PLoS One. 2014;9(11):e110844. doi: 10.1371/journal.pone.0110844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 48.Nucci MR, et al. Biomarkers in diagnostic obstetric and gynecologic pathology: a review. Adv Anat Pathol. 2003;10(2):55–68. doi: 10.1097/00125480-200303000-00001. [DOI] [PubMed] [Google Scholar]
- 49.Yemelyanova A, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Mod Pathol. 2011;24(9):1248–1253. doi: 10.1038/modpathol.2011.85. [DOI] [PubMed] [Google Scholar]
- 50.Singh N, et al. p53 immunohistochemistry is an accurate surrogate for TP53 mutational analysis in endometrial carcinoma biopsies. J Pathol. 2020;250(3):336–345. doi: 10.1002/path.5375. [DOI] [PubMed] [Google Scholar]
- 51.Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics. 2016;32(4):511–517. doi: 10.1093/bioinformatics/btv639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14(8):561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 53.Diaz-Padilla I, et al. Mismatch repair status and clinical outcome in endometrial cancer: a systematic review and meta-analysis. Crit Rev Oncol Hematol. 2013;88(1):154–167. doi: 10.1016/j.critrevonc.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Lucas E, et al. Mismatch repair protein expression in endometrioid intraepithelial neoplasia/atypical hyperplasia: should we screen for lynch syndrome in precancerous lesions? Int J Gynecol Pathol. 2019;38(6):533–542. doi: 10.1097/PGP.0000000000000557. [DOI] [PubMed] [Google Scholar]
- 55.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehnert JM, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126(6):2334–2340. doi: 10.1172/JCI84940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stasenko M, et al. Clinical outcomes of patients with POLE mutated endometrioid endometrial cancer. Gynecol Oncol. 2020;156(1):194–202. doi: 10.1016/j.ygyno.2019.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cosgrove CM, et al. An NRG Oncology/GOG study of molecular classification for risk prediction in endometrioid endometrial cancer. Gynecol Oncol. 2018;148(1):174–180. doi: 10.1016/j.ygyno.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jansen AM, et al. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet. 2016;24(7):1089–1092. doi: 10.1038/ejhg.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bell DW, Ellenson LH. Molecular genetics of endometrial carcinoma. Annu Rev Pathol. 2019;14:339–367. doi: 10.1146/annurev-pathol-020117-043609. [DOI] [PubMed] [Google Scholar]
- 61.Henninger EE, Pursell ZF. DNA polymerase ε and its roles in genome stability. IUBMB Life. 2014;66(5):339–351. doi: 10.1002/iub.1276. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi KH. Multiple modes of canalization: Links between genetic, environmental canalizations and developmental stability, and their trait-specificity. Semin Cell Dev Biol. 2019;88:14–20. doi: 10.1016/j.semcdb.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 63.Kuznetsova AY, et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle. 2015;14(17):2810–2820. doi: 10.1080/15384101.2015.1068482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121(pt 23):3859–3866. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 65.Nakada Y, et al. The LKB1 tumor suppressor as a biomarker in mouse and human tissues. PLoS One. 2013;8(9):e73449. doi: 10.1371/journal.pone.0073449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Adams DJ, Doran AG, Lilue J, Keane TM. The Mouse Genomes Project: a repository of inbred laboratory mouse strain genomes. Mamm Genome. 2015;26(9-10):403–412. doi: 10.1007/s00335-015-9579-6. [DOI] [PubMed] [Google Scholar]
- 70. doi: 10.4161/fly.19695. Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6(2):80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wright JC, et al. Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow. Nat Commun. 2016;7:11778. doi: 10.1038/ncomms11778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kurman RJ, Carcangiu ML, Young RH, Herrington CS. WHO Classification of Tumours of Female Reproductive Organs. International Agency for Research on Cancer; 2014. [Google Scholar]
- 73. Adaptive Biotechnologies. Analyzer Manual. Adaptive Biotechnologies website. https://clients.adaptivebiotech.com/assets/downloads/immunoSEQ_AnalyzerManual.pdf Accessed June 25, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.