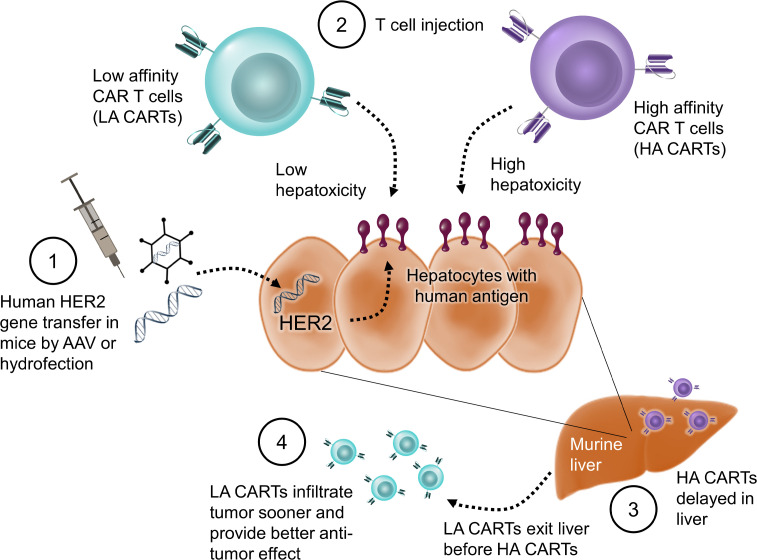
Abstract
Off-tumor targeting of human antigens is difficult to predict in preclinical animal studies and can lead to serious adverse effects in patients. To address this, we developed a mouse model with stable and tunable human Her2 (hHer2) expression on normal hepatic tissue and compared toxicity between affinity-tuned Her2 chimeric antigen receptor T cells (CARTs). In mice with hHer2-high livers, both the high-affinity (HA) and low-affinity (LA) CARTs caused lethal liver damage due to immunotoxicity. In mice with hHer2-low livers, LA-CARTs exhibited less liver damage and lower systemic levels of IFN-γ than HA-CARTs. We then compared affinity-tuned CARTs for their ability to control a hHer2-positive tumor xenograft in our model. Surprisingly, the LA-CARTs outperformed the HA-CARTs with superior antitumor efficacy in vivo. We hypothesized that this was due, in part, to T cell trafficking differences between LA and HA-CARTs and found that the LA-CARTs migrated out of the liver and infiltrated the tumor sooner than the HA-CARTs. These findings highlight the importance of T cell targeting in reducing toxicity of normal tissue and also in preventing off-tumor sequestration of CARTs, which reduces their therapeutic potency. Our model may be useful to evaluate various CARTs that have conditional expression of more than 1 single-chain variable fragment (scFv).
Keywords: Immunology, Therapeutics
Keywords: Cancer immunotherapy
Off-tumor toxicity of CAR T cells is assessed in a preclinical mouse model using gene transfer to express human antigen targets in healthy tissue.
Introduction
Chimeric antigen receptor–modified (CAR-modified) T cell therapy has emerged as an effective treatment for blood malignancies and is currently being developed to treat solid tumor cancers. A key aspect of the antitumor effectiveness of CAR T cell (CART) therapy is its ability to target and kill malignant cells by recognizing cell surface antigens. CART therapy can result in severe adverse effects due to on-target, off-tumor toxicity, which arises in patients who have target antigen expressed on both tumor and healthy tissue. This expression pattern is typical for the vast majority of target antigens used in CART therapies. One such antigen is Her2, which is an attractive target for CART therapy since it can be overexpressed 40- to 100-fold in tumors and, thus, has long been targeted therapeutically using monoclonal antibodies such as trastuzumab (Herceptin) (1). A Her2 CART therapy based on the trastuzumab sequence was used to treat a patient with colorectal cancer; unfortunately, off-tumor targeting of their cardiopulmonary system caused lethal toxicity (2). This adverse effect was not foreseen based on clinical studies of trastuzumab or in preclinical animal experiments.
Current FDA guidelines for preclinical animal testing of cellular therapies require the use of a relevant animal model capable of eliciting a biological response that would reasonably predict the expected human response (3). In many cases, animal models used for pharmacological testing of therapeutic human CARTs cannot adhere to this standard due to variability in cross-species reactivity to nonhuman target antigens. Therefore, preclinical animal studies have little chance of identifying potential adverse events in humans and often offer a false sense of safety (4). This limitation highlights the need for an animal model that expresses human targets in normal tissue to better predict off-tumor toxicity and improve the safety profile of immune therapies.
On-target, off-tumor toxicity can be reduced by using CAR targeting systems that can improve the recognition of tumor cells (5). One such strategy is to affinity-tune CARs so they detect tumor cells with a high density of surface antigens and do not react against normal cells that have low antigen densities (6). Recently, a low-affinity (LA) CD19 CART therapy was reported to exhibit promising therapeutic outcomes in patients with mostly low-burden relapsed/refractory pediatric B cell acute lymphoblastic leukemia (7). The activity of CARTs is dependent on the strength and duration of activation signals transmitted through the antigen receptor (8, 9) and by the antigen density on target cells (10, 11). Genetically tuning the affinity of a CAR can be done by altering the single-chain variable fragment (scFv) binding region via mutagenesis (12) or by recombining heavy and light chains (13). Recent in vivo studies have used tumors as surrogates for normal tissue to compare off-tumor toxicity between affinity-tuned CARTs (11, 14). Lower-affinity CARTs demonstrated improved safety by bypassing tumors with low-antigen expression while maintaining recognition and elimination of tumors with high-antigen expression. However, neither of these mouse models had human antigen expression on normal mouse tissue; therefore, on-target, off-tumor toxicity was not measured directly. Our first aim was to develop a mouse model that contains human antigen targets on both tumor and normal tissue. Our second aim was to assess the therapeutic index of affinity-tuned CARTs. Our study showed that the lower-affinity CARTs resulted in less off-tumor toxicity and improved tumor control. We concluded that this model is an effective preclinical tool to assess the consequences of tumor recognition by CARTs, permitting improved prediction of the safety and efficacy of CARTs compared with conventional tumor xenografts.
Results
Human tumor antigens can be expressed in murine livers using AAV8 gene delivery.
Human T cells respond differently than mouse T cells; thus, most preclinical studies use human CARTs in immune-deficient mice (e.g., NOD/scid/IL2rγ−/− [NSG] mice) (15). The antigen recognition portion of CARs is commonly derived from monoclonal antibodies that are highly specific for their cognate antigen but are usually not species cross-reactive between mice and humans. The absence of human targets in mice poses a problem for testing on-target, off-tumor toxicity of CARTs in preclinical mouse models. To address this limitation, we genetically introduced human tumor antigen targets into mouse livers using 2 different methods of gene transfer.
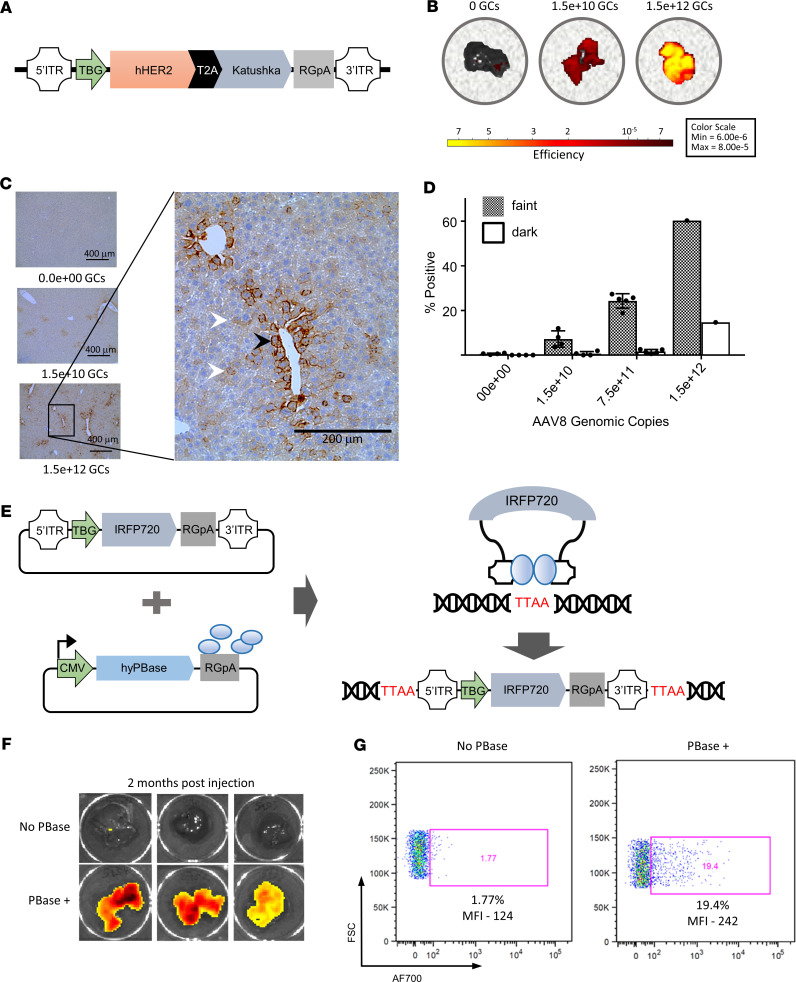
One method of gene transfer used adeno-associated virus serotype 8 (AAV8) that contained a truncated human Her2 gene (hHer2) and a fluorescent reporter (Katushka), which was then transduced into murine hepatocytes by i.v. tail vein injection (Figure 1A). The expression of the fluorescent reporter in murine hepatocytes was detected by ex vivo imaging of livers, which showed greater fluorescence intensity in the livers of mice that received a higher number of genomic copies (GCs) of AAV8 (Figure 1B). Similarly, the expression of the hHer2 antigen was detected by IHC and showed widespread expression with a more pronounced perivascular staining pattern (Figure 1C). Her2 staining of hepatocytes was quantified, and we observed a positive correlation with the number of GCs of AAV8 and the percentage of positive hepatocytes, thus demonstrating that the level of expression can be regulated through AAV8 dosing (Figure 1D). Our results confirm and extend previous in vivo studies that showed that transgene expression levels in hepatocytes directly correlate with AAV8 dosage (16).
Figure 1. Hepatic gene transfer of hHer2 by AAV8 delivery and transposase gene editing.
(A) Design of the AAV8 vector, which includes a truncated human Her2 gene (hHer2) and a fluorescent reporter gene (Katushka) that are expressed by the liver-specific thyroid hormone-binding globulin (TBG) promoter. Mice were i.v. injected with AAV8, and their livers were harvested 1 month later. (B) IVIS imaging of fluorescence in ex vivo livers harvested from mice that received either no genomic copies (GCs), 1.5 × 1010 GCs, or 1.5 × 1012 GCs of AAV8. (C) Immunohistological assessment of hHer2 expression in mice that received either 0, 1.5 × 1010, or 1.5 × 1012 GCs of AAV8. Darker Her2-stained cells have a perivascular pattern (black arrowhead) and are less frequent than the fainter Her2-stained cells (white arrowheads). Scale bars: 400 μm (left) and 200 μm (right). (D) Mean hepatocytes ± SEM were quantified for dark or faint Her2 staining by digitizing the IHC images using ImageScope and then analyzed using Aperio imaging software. Each group contained 4 mice except for 1.5 × 1012 GC, which had an n of 1. (E) Overview of gene editing using the piggyBac transposase system. The transposon vector contained a fluorescent reporter gene (IRFP720) that was expressed using the liver-specific TBG promoter. (F) Imaging of 6 mouse livers harvested 2 months after injection with the IRFP720 fluorescent reporter plasmid and either with or without the PiggyBac transposase DNA vector. (G) Detection of fluorescent reporter expression by flow cytometry in isolated mouse hepatocytes representative of the livers shown in F.
Transgene transposition by the piggyBac transposon system can create stable human antigen expression in murine livers.
We next wanted to explore the suitability of transgene transposition using a transposon system delivered to hepatocytes in vivo by hydrodynamic injection. The advantages to this system are that it requires no virus production, generates no viral antigens, has a large cargo delivery size that could accommodate multiple antigen transgenes, and genetically integrates the transgene into the cell genome for stable expression. The piggyBac (PB) and Sleeping Beauty (SB) transposons are movable genetic elements that can efficiently transpose vector DNA into mammalian genomes through a “cut-and-paste” mechanism that is effective in genetically modifying cells in vivo (Figure 1E, adapted from ref. 17). Although either the PB or SB system could be used in our model, we chose the PB system because it allows for a larger transgene cargo size (18, 19). We replaced the Katushka reporter from our AAV8 construct with the IRFP720 protein since near-infrared fluorescent proteins are preferable for deep-tissue imaging (20). Injection of nontransposon plasmid DNA resulted in transient episomal expression that was ill-suited for prolonged mouse toxicity studies (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.136012DS1). To test whether stable transgene expression could be achieved via the transposon system, mice were injected with 10 μg of the fluorescent reporter plasmid DNA pPB7-IRFP720 either with or without 10 μg of the PB transposase plasmid DNA pCMV-HypPBase (Figure 1E). The murine livers were harvested 8 weeks after injection, and IRFP720 expression was measured by IVIS imaging, which showed fluorescence in the mice that was dependent on injection with the PB transposase plasmids (Figure 1F). One liver from each group was enzymatically digested, and dissociated hepatocytes were isolated and then analyzed by flow cytometry to detect fluorescence in individual hepatocytes (Figure 1G). We observed more IRFP720 expression in the mouse that received both the transposon and transposase plasmids versus the control mouse that only received the transposon plasmid, as evidenced by the proportion of positive cells (20% versus 2%, respectively) and by the geometric mean fluorescence intensity (MFI; 242 vs. 124, respectively).
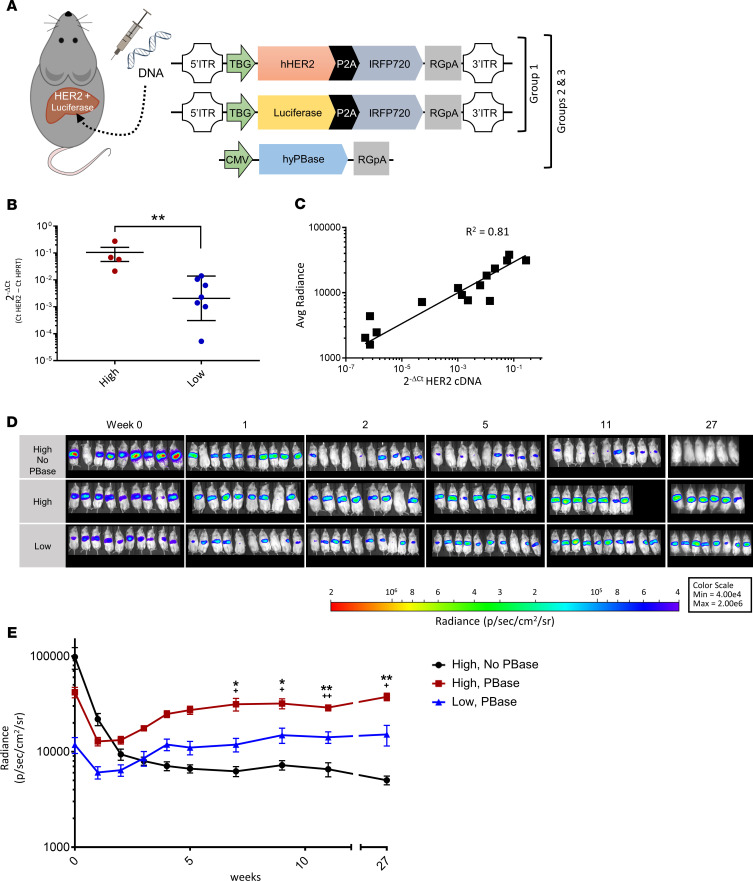
Next, we performed a longitudinal study of the PB gene editing system to evaluate the long-term kinetics of transgene expression (Figure 2A). The versatility of this system allowed us to customize the pattern of hepatic protein expression by using different combinations and concentrations of plasmids. We created 3 groups of mice that differed in their hydrodynamic tail vein injections of hHer2 transposon DNA, luciferase transposon DNA, and PB transposase DNA to ask if we could maintain stable expression of the transgenes at various levels. Group 1 received injections of luciferase transposon DNA and hHer2 transposon DNA but without transposase DNA. Group 2 received injections that combined luciferase transposon DNA, hHer2 transposon DNA, and the PB transposase DNA. Group 3 received the same plasmid combination as group 2 but at one-tenth the DNA concentration.
Figure 2. Murine expression of human antigen is stable and tunable using PiggyBac transposase gene transfer.
(A) Design of the luciferase and truncated human Her2 (hHer2) transposon vectors and the transposase vector used in the experiment. Group 1 mice were injected with 5 μg of the hHer2 transposon plasmid and 5 μg of the luciferase transposon plasmid, but no transposase plasmid. Group 2 mice were injected with 5 μg of the hHer2 transposon plasmid, 5 μg of the luciferase transposon plasmid, and 10 μg of the transposase plasmid, hyPBase. Group 3 mice were injected with the same plasmids as group 2 but at one-tenth the DNA concentration. (B) Comparison of hHer2 mRNA expression between mice that received higher versus lower concentrations of piggyBac transposon and transposase plasmids. hHer2 RNA was measured in murine livers using real-time PCR and normalized to mouse HPRT expression to calculate 2–ΔCt values. All data are shown as means ± SD (n = 4–7 mice per group). A 2-tailed Mann-Whitney U test of ΔCt values was used for statistical analysis. (C) Comparison of hHer2 DNA content and luciferase expression in the murine livers after hydrodynamic DNA injections (n = 15 mice). (D) In vivo imaging of luciferase expression in mice that either received piggyBac transposon but not transposase plasmid (group 1) or mice injected with either a higher or lower dose of piggyBac transposon and transposase plasmid (groups 2 and 3, respectively). (E) Mean ± SEM radiance over time with n = 9 mice per group. A 2-way repeated-measures ANOVA with Tukey’s multiple correction test was used for statistical analysis. Statistical significance for group 2 versus either group 1 (*) or group 3 (+) is denoted as */+P < 0.5 and **/++P < 0.01.
hHer2 mRNA expression from the harvested murine livers was evaluated by real-time PCR using TaqMan assays for hHer2, which was normalized to liver RNA using the mouse housekeeping gene HPRT (Figure 2B). Relative expression is shown as 2–ΔCt, where ΔCt is calculated as Ct HPRT minus Ct Her2 and was calculated for the mice that received all 3 of the plasmids (i.e., groups 2 and 3) and also for control mice that did not receive hHer2 transposon DNA, which were used to establish background Ct levels. This showed that a higher level of Her2 expression was achieved in the mice from group 2 versus mice from Group 3 and, thus, confirmed that we could regulate antigen levels by the amount of transfected DNA, which is consistent with previous studies (18). We observed a positive correlation in the livers between luciferase radiance and Her2 mRNA expression, suggesting that the Her2 and the luciferase transposon plasmids were transfected with equal efficiency and, thus, luciferase measurements were indicative of Her2 antigen levels (Figure 2C).
Weekly IVIS imaging of the murine liver was measured to determine longitudinal luciferase expression. This imaging revealed that Group 1 mice initially had the highest BLI levels (i.e., high concentration of the transposon DNA but no transposase DNA), but then bioluminescence was found to decrease dramatically during the first 2 weeks and then continued to decline during the following 6 months (Figure 2, D and E). The mice from groups 2 and 3, which received the additional transposase plasmid, showed a luciferase expression pattern that decreased markedly in the first week but then increased steadily before stabilizing approximately 1 month later (Figure 2, D and E). A comparison of groups 2 and 3 showed the group that received the higher concentration of DNA had a higher final average bioluminescent signal (i.e., transgene expression), which was consistent with the previous hHer2 mRNA expression results. Our findings demonstrate that either the PB transposon system or AAV8 can be used to effectively obtain stable human antigen expression in the mouse liver and at predetermined levels.
Human antigens in the murine liver can be targeted by CARTs.
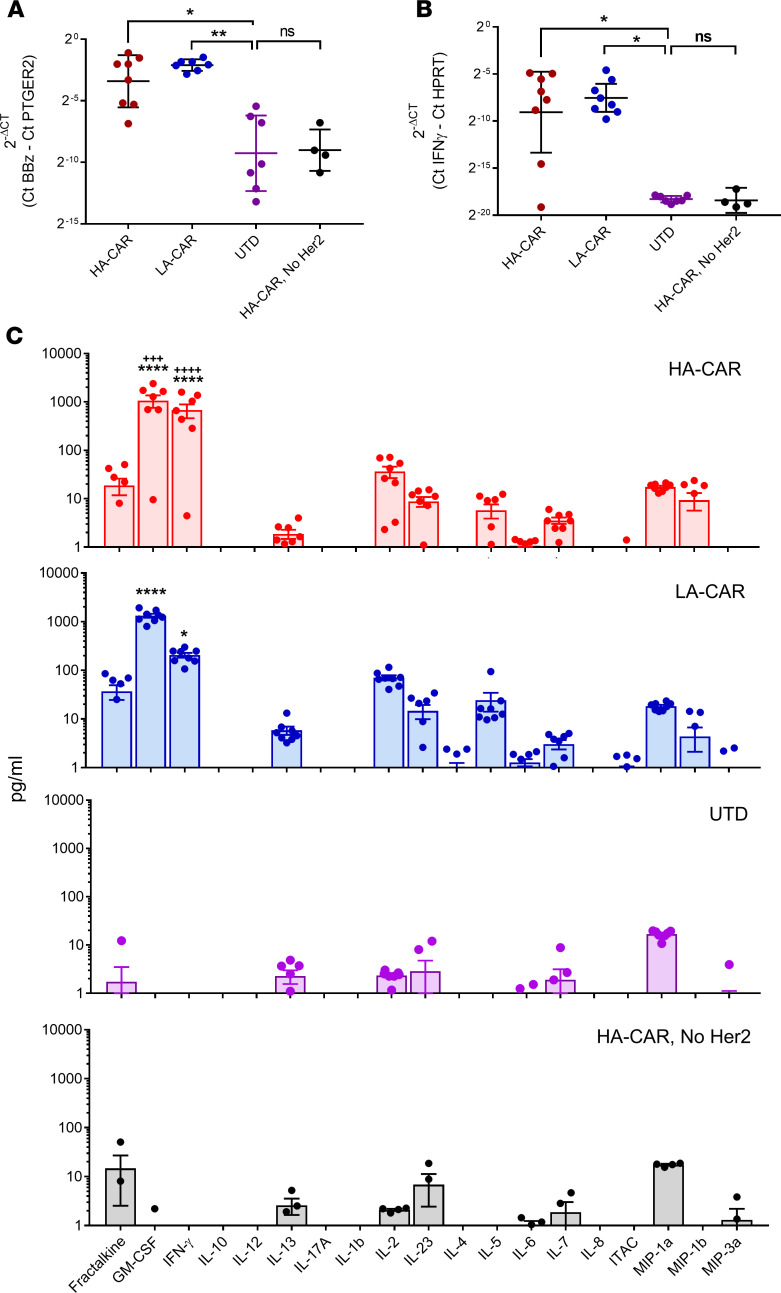
We next tested whether the hHer2 antigen that was expressed in the mouse livers could provoke on-target toxicity following infusion of anti–Her2-CARTs. Her2 CARTs with 2 different scFvs (4D5, high affinity [HA]; 4D5-5, low affinity [LA]) were i.v. infused into mice that had hepatic expression of hHer2 antigen, which was established using our PB transfection method. To detect a Her2-independent T cell response in the livers, we included 1 negative control group that expressed Her2 but received untransduced T cells and a second negative control group that received Her2-CARTs but in Her2-negative mice. The presence of CARTs in the murine livers were measured 1 week after injection using a CAR DNA TaqMan assay, which was normalized against a mouse PTGER2 TaqMan assay. Her2-CART (HA or LA) DNA was elevated only in mice that had hHer2 expression (Figure 3A). The mice that had CARTs present in their livers also had elevated hepatic expression of human IFN-γ mRNA, as measured using a TaqMan assay normalized against mouse HPRT mRNA (Figure 3B). The serum cytokine levels in the 4 groups revealed that only mice that were injected with Her2-CARTs and expressed hepatic hHer2 antigen had elevated human cytokines associated with T cell activation (i.e., IFN-γ, GM-CSF, IL-2, IL-5, and MIP-1b), which suggests antigen-dependent activation and no response to hydrodynamic injection (Figure 3C). Serum levels of IFN-γ and GM-CSF were significantly higher in the mice that received the HA versus LA CARTs, consistent with increased activation. To demonstrate that hepatic antigen expression delivered by AAV8 could also elicit an antigen-specific immune response, mice were injected with Her2-AAV8 and then infused with luciferase-expressing Her2 CARTs (Supplemental Figure 2). Mouse livers were analyzed by bioluminescent imaging and IHC, which showed an infiltration of Her2 CARTs. These results confirmed that the human antigen we delivered to the liver can act as a target for CARTs and that presence of this human antigen was sufficient for CART infiltration and activation. We concluded that either AAV8 or hydrodynamic tail vein injection of transposon DNA were suitable methods for promoting an antigen-dependent immune response in hepatocytes.
Figure 3. CARTs recognize cognate human antigen in mice.
hHer2 antigen was expressed in mouse hepatocytes following piggyBac gene transfer. Mice were then injected with 2.5 × 106 anti-hHer2 CART (either HA-CAR or LA-CAR), and livers were harvested 1 week later for analysis. Control groups included mice that had hepatic Her2 expression but untransduced T cells and mice that received Her2 CART but lacked hepatic Her2 expression due to empty transposon vector transfections. (A) Her2 CARTs were detected in mouse livers (n = 4–8) by performing real-time PCR assays for CAR DNA. 2–ΔCt values for CARTs were calculated using PCR assays that amplify the CAR intracellular signaling domain, 4-1BBz-CD3z, and were normalized to mouse PTGER2 genomic DNA content. (B) Expression of human IFN-γ mRNA from T cells was measured in murine livers (n = 4–8) using real-time PCR and normalized to mouse HPRT expression. Kruskal-Wallis test with Dunn’s multiple comparisons tests was used for statistical analysis of real-time PCR data. (C) Systemic cytokine release by T cells was detected in mouse serum (n = 4–8) by Luminex assay. A 2-way ANOVA with Tukey’s multiple comparison test was performed, and comparisons are shown between all groups and the untransduced (UTD) group (*) or between the HA-CAR and LA-CAR groups (+). Statistical significance is denoted as *P < 0.5, **P < 0.01,+++P < 0.001, and ****/++++P < 0.0001.
Off-tumor toxicity can be reduced using a lower-affinity CART.
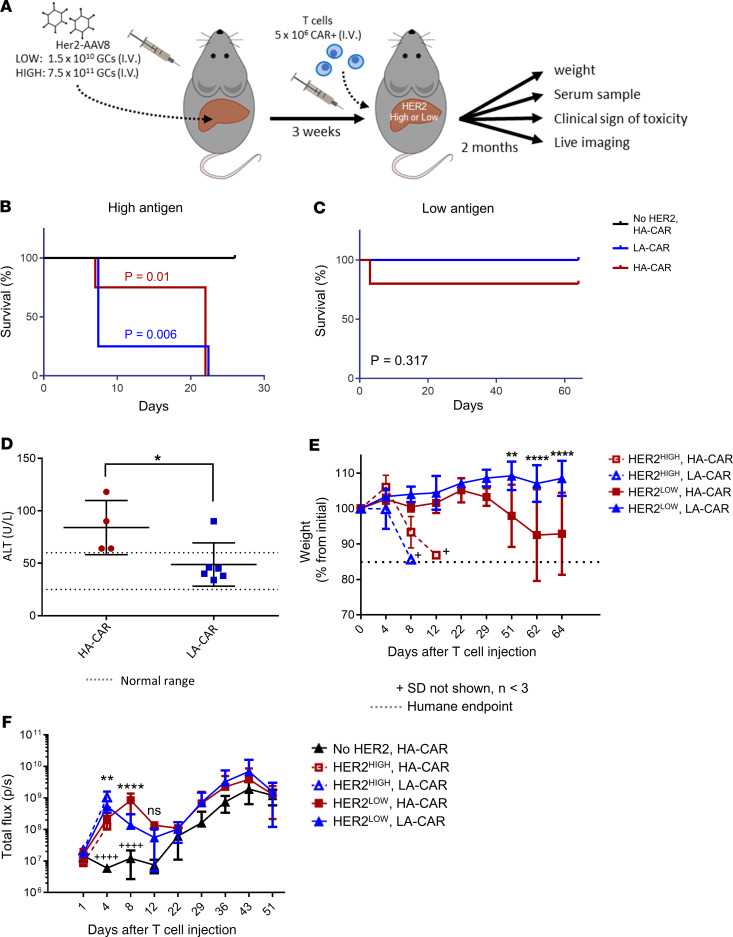
The amount of toxicity in Her2-expressing livers was compared between mice that received a HA versus a LA CART (4D5 and 4D5-5 scFv, respectively). We postulated that the HA CAR would cause more liver damage than its LA counterpart when hepatic Her2 expression was low, since the Her2 levels would be below the limit of detection for the LA CARTs. We also hypothesized that, when Her2 expression in the murine livers was high, hepatocytes would be recognized by both the HA and LA CARTs and, thus, the degree of liver damage would be equivalent. To test our assumption, we injected 1 group of mice with a high dose of Her2-AAV8 to create livers in mice with high antigen levels, while another group of mice received a comparatively low dose of Her2-AAV8 to generate low-antigen livers (Figure 4A). To determine if the AAV8 viral antigens alone would elicit T cell–mediated toxicity, a Her2-negative control group received a high dose of AAV8 that expressed GFP instead of Her2; then, these mice were injected with HA Her2-CARTs. As predicted, there was severe toxicity in mice with high antigen levels due to both the HA and LA CARTs, as observed by markedly increased mortality (Figure 4B) and postmortem analysis of liver pathology (Supplemental Table 1). In addition to liver damage, cytokine release as shown in Figure 3C may have also contributed to morbidity in these mice. In contrast, the HA CARTs were nonlethal in mice that lacked hepatic Her2 expression, excluding xenoreactivity as a cause of morbidity (Figure 4B).
Figure 4. CARTs cause lethal on-target, off-tumor toxicity in mice.
(A) Overview of the experimental design for comparing on-target liver toxicity between affinity-tuned Her2 CARTs. Two groups of mice received either 1.5 × 1010 or 7.5 × 1011 GCs of Her2-AAV8 and then were infused with either 5 × 106 high-affinity (HA) or low-affinity (LA) Her2-CARTs. A control group of mice received 4 × 1011 GCs of GFP-AAV8 (i.e., no Her2) and 5 × 106 HA CARTs. n = 6 mice per group are shown in each panel, unless stated otherwise. (B) Survival curves of mice that received the 7.5 × 1011 GCs of Her2-AAV8 and then CART injection. Statistical analysis was performed using a log-rank Mantel-Cox test. (C) Survival curves of mice that received the 1.5 × 1010 GCs of Her2-AAV8 and then CART injections. (D) Liver function profile as determined by serum ALT levels collected 25 days after T cell injection. Mean ALT ± SEM in mice (n = 4–6) that received 1.5 × 1010 GCs of Her2-AAV8 and either HA-CAR or LA-CAR. A 1-tailed unpaired 2-sample t test of ALT was used for statistical analysis. (E) Weight change shown by percent change from initial weight ± SD in mice that received either 7.5 × 1011 (dashed lines) or 1.5 × 1010 (solid lines) GCs of Her2-AAV8 and then either HA-CAR or LA-CAR. (F) Mean total flux ± SD for whole body bioluminescence imaging (BLI) of T cell luciferase. A 2-way repeated measures ANOVA with Bonferroni’s multiple comparison test was used for statistical analysis of weight change and BLI. Statistical significance for D–F is denoted as *P < 0.5, **P < 0.01, ***P < 0.001, and ****/++++P < 0.0001.
Toxicity was decreased in mice with low antigen levels, as seen by reduced mortality (Figure 4C) and liver pathology (Supplemental Table 1). The low-antigen mice had no significant difference in mortality between affinity-tuned CARTs (Figure 4C), but more liver damage was caused by the HA versus LA CARTs, according to elevated serum ALT levels (Figure 4D). The mean ALT from the mice that received HA CARTs is 84 U/L with a range of 64–118 U/L versus the negative control mice with a mean ALT of 46 U/L and a range of 34–66 U/L. The resulting fold change for the HA CART group is about double the normal values, which would be considered serious in humans. According to Hy’s law, drug-induced hepatocellular injury that is 3 times or greater above the upper limit of normal presents a high risk of fatal drug-induced liver injury. Toxicity effects were also apparent by mouse weight loss, which was profound in the high-antigen groups that received either the LA or HA CARTs and also occurred in the low-antigen mice that were infused with the HA but not LA CARTs (Figure 4E). Differences in abundance between the HA and LA CARTs were assessed by bioluminescence imaging (BLI) of their luciferase reporter gene (Figure 4F). In the Her2-negative control mice (i.e., AAV8-GFP group), the number of HA Her2 CARTs remained constant during the first 2 weeks after T cell injection, indicating an absence of antigen-dependent activation (Figure 4F). In the low-antigen mice, we initially observed an increase in abundance via luminescence for both the HA and LA CARTs, followed by a decrease after 4 days by the LA CARTs. In contrast, the HA CARTs continued to increase until day 8 and remained higher than the LA CARTs, until dropping to equivalent values by day 22. This suggests that the HA CARTs remained activated longer due to prolonged antigen recognition. All 3 groups had similar upward trends in T cell abundance starting at day 22, which was presumably due to xenogeneic graft-versus-host disease (GVHD). Overall, the LA CARs were better able to distinguish between low- and high-antigen density tissues in our safety model.
An increase in off-tumor targeting is associated with a delay in tumor CART infiltration and a decrease in antitumor efficacy.
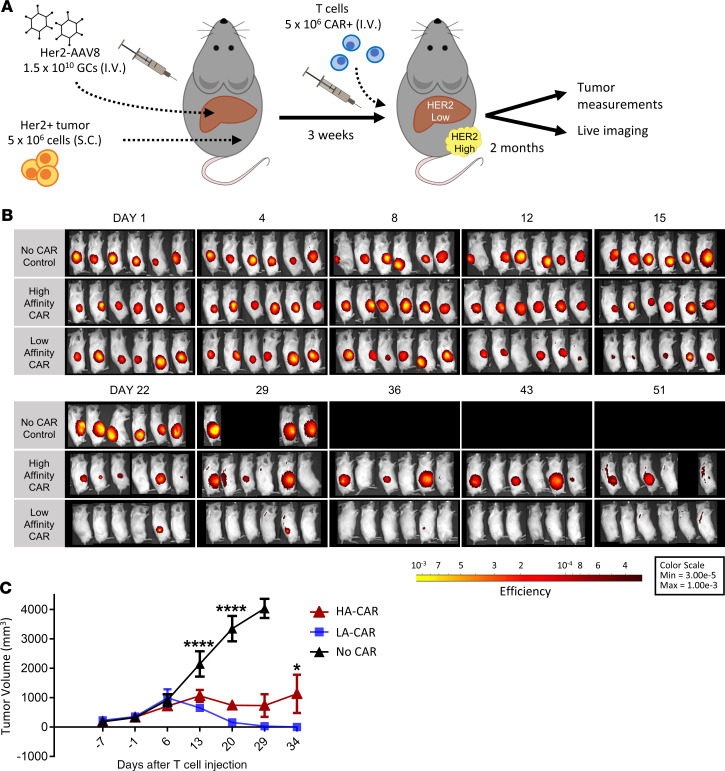
We next wanted to test our affinity-tuned CART treatments for their tumor control in a mouse model that simulated a common clinical scenario, in which antigen is overexpressed in a patient’s tumor but is also found at lower levels in some of their healthy tissue. To accomplish this, mice were engrafted with a high Her2–expressing tumor xenograft and additionally injected with a low dose of Her2-AAV8 to produce a low Her2–expressing liver (Figure 5A). The mice were then infused with either HA or LA CARTs. Surprisingly, the mice that received the LA CARTs showed significantly better antitumor efficacy than the ones that were treated with the HA CARTs (Figure 5, B and C). Tumor size was visualized by the expression of the fluorescent reporter IRFP720, which was congruent with our caliper measurements (Figure 5C and Supplemental Figure 3).
Figure 5. The low-affinity CAR has better tumor control than the high-affinity CAR when antigen is also expressed in normal tissue.
(A) Overview of the experimental design for comparing Her2+ tumor control between affinity-tuned Her2 CARTs. All mice received 1.5 × 1010 GCs of Her2-AAV8 and were implanted with 5 × 106 Her2+ SKOV3 tumor cells. Then, 3 groups were injected with either 5 × 106 high-affinity (HA) or low-affinity (LA) Her2-CARTs or no CAR control T cells. (B) The Her2+ tumor cells, SKOV3, were genetically modified to express the fluorescent reporter, IRFP720, for in vivo imaging. Tumor xenograft fluorescence is shown in a yellow-to-red spectrum. Lateral views of fluorescent tumor imaging. (C) Mean tumor volume ± SEM measured by calipers in n = 6 mice per group. A 2-way repeated measures ANOVA with Bonferroni’s multiple comparisons test was used for statistical analysis. Statistical significance is denoted as *P < 0.5 and ****P < 0.0001.
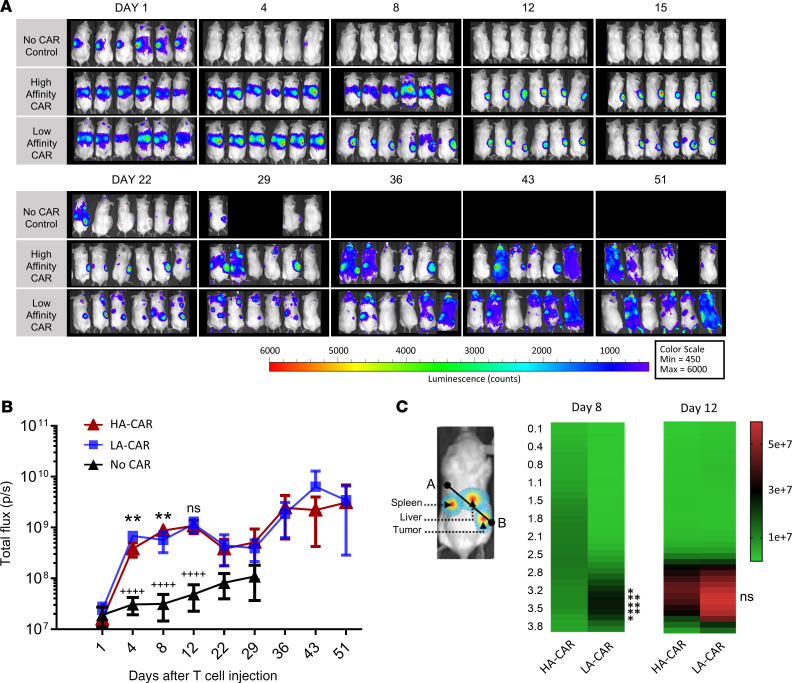
To investigate whether differences in tumor control between the 2 groups were due to differences in T cell abundance and/or trafficking, we observed luciferase-expressing T cells using in vivo imaging (Figure 6A). Slight differences in overall abundance between the HA and LA CARTs were seen by whole body BLI measurements in the first week and then became comparable throughout the rest of the experiment (Figure 6B). Migratory behavior between the T cell treatment groups was similar at day 1, in which the HA and LA CARTs were both seen in the liver while the T cells in the control group were observed in the spleen (Figure 6C). However, by day 8, there were striking differences in the trafficking of the CARTs as the HA CARTs remained in the mouse livers, whereas the LA CARTs had emigrated from the liver and homed to the tumor. By day 12, the HA CARTs had left the liver and infiltrated the tumor, while the LA CARTs had already caused measurable tumor regression. By day 43, the LA CART group had no measurable tumors by either fluorescent reporter expression or caliper readings (Figure 5, B and C). Conversely, the HA CART group at day 43 had detectable tumor in 4 of the 6 mice and less tumor infiltration of CARTs in those tumors compared with earlier time points. In tumor-bearing mice without hepatic antigen expression, no difference between affinity-tuned CARTs was observed in trafficking (Supplemental Figure 4) or tumor control (11). Thus, in our mouse model, the LA CARTs were better able to discriminate between low-antigen healthy tissue and high-antigen tumor tissue, which resulted in a better therapeutic outcome.
Figure 6. Low-affinity CARTs spend less time off -tumor than high-affinity CARTs.
In vivo CART kinetics were captured using IVIS imaging for n = 6 mice per group. (A) T cells were engineered to express a luciferase gene for in vivo luminescent imaging. The dorsal views of the mice that were kept in the same order as in Figure 5B, and luminescence intensity is shown in a blue-to-red spectrum. In addition to luciferase expression, the T cells contained either no CAR expression (negative control) or they were engineered to express a high-affinity (HA) or low-affinity (LA) Her2 CAR. (B) Whole body bioluminescent imaging (BLI) of T cell luciferase. Statistical significance for HA-CAR versus LA-CAR (*) or HA-CAR vs. No CAR (+) was compared by 2-way repeated measures ANOVA with a Tukey’s multiple comparison test. (C) Spatial luciferase expression was measured along a line that starts in the upper left thorax (point A) and ends in the lower right abdomen (point B). Luminescence from the spleen, liver, and tumor appear at the beginning (~0–1.5 cm), middle (~1–3 cm), and end (~2.5–4 cm) of the line, respectively. Mean luminescence along the line was compared between groups by 2-way repeated measures ANOVA with Bonferroni’s multiple comparisons test. Statistical significance is denoted as **P < 0.01 and ++++P < 0.0001.
Discussion
On-target, off-tumor toxicities from CART therapies can have serious clinical consequences that are difficult to predict in current animal models because of the specificity of therapeutic CARs for human antigens. Our study showed that the improved targeting by the LA CARTs resulted in less off-tumor toxicity and improved tumor control than the HA CARTs. However, the lower-affinity CARTs would be less effective at controlling tumors with low antigen expression than the higher-affinity CARTs. This was seen clinically with a LA Herceptin-based CAR, which showed a safer profile in sarcoma patients but had only modest clinical activity (21). This is an unavoidable consequence of enhancing the tumor specificity of CARTs via its antigen receptor. We concluded that our current off-tumor model is an effective preclinical tool to assess tumor recognition by CARTs that better predicts the therapeutic index of CARTs compared with tumor xenografts alone.
The liver is a suitable organ for testing CART toxicity because it tolerates damage well and it is well perfused. Hepatocytes have the added benefit of providing easy access to antigen, since they lack basement membranes (22). In addition, hepatocytes are convenient cells to express human antigens, as they have a high tropism for AAV8 (23) and they can be transfected in vivo by hydrodynamic gene delivery (24). Both methods for gene delivery were used in our study with equal effect, although there are advantages and disadvantages to each method. Liver-targeted gene transfer in mice using AAV8 has the advantages of being highly efficient for both dividing and nondividing cells, it is technically easy to administer via i.v. or i.p. injections, it provides stable expression in adult mice, and it has minimal toxicity and immunogenicity (25). However, limitations to using AAV8 include their small packaging capacity (<5 kb), the dilution of transgene expression in replicating cells, and the cost and time of viral production. In comparison, the hydrodynamics-based method coupled with the PB transposon system allows for efficient gene transfer, has a large packaging capacity (>200 kb), and forms genomic integration resulting in stable expression; furthermore, the transgene is delivered by naked plasmid DNA, so it does not introduce viral antigens or require virus production (26). The disadvantage to using the hydrodynamics-based method is the increased technical difficulty and physiological stress on the mouse compared with standard i.v. injections (27).
Immune tolerance is an important function of the liver to prevent an autoimmune response to food-derived and microbial antigens from the digestive system. As the HA CARTs spent more time in the livers compared with the LA CARTs, they had longer exposure to the liver microenvironment, which — in addition to sequestration — could potentially explain their decreased tumor clearing capacity. Hepatic immune suppression has been extensively reported in various animal models, including humans (28–30). Mechanistically, hepatic tolerization has been attributed to inhibitory ligand expression and immunosuppressive cells (31), T cell exhaustion (32, 33), elimination of CD8+ T by suicidal emperipolesis (34), and elimination of CD4+ T cells by enclysis (35). Our mouse model was devised to study off-tumor toxicity of human CARTs, but it may also serve as a useful tool to elucidate the mechanisms of liver tolerance.
Immune tolerance of autoreactive T cells is not unique to the liver and has been observed to a lesser extent in other organs such as the skin, the lung (36), immune privileged sites (37), and the gastrointestinal tract (38). It is unknown whether these or other tissues may contribute to on-target, off-tumor tolerance in immune cell therapies. At the very least, off-tumor targeting by immune cell therapies can cause immune cells to be sequestered and delayed from reaching their intended tumor targets, as was demonstrated in this study. This effect is in addition to the toxicity that on-target, off-tumor targeting can cause healthy tissue. Taken together, these effects highlight the need to reduce off-tumor targeting by developing better targeting strategies for CARTs (5) or by modifying the route of CART delivery to avoid damage to healthy tissue (39).
In summary, we have developed a mouse model to test on-target, off-tumor toxicity of CARTs that recognize human antigens in normal murine livers. Another use of our model would be to test various forms of CARTs that have conditional expression of 2 CAR molecules (40, 41), as our model would permit the assessment of on- and off-switching kinetics. In future experiments, gene editing of the liver can be used in conjunction with antigen gene delivery to further manipulate target hepatocytes and study effector cell interactions (42, 43).
Methods
Cell lines, primary human lymphocytes and primary mouse hepatocytes
The cell lines SKOV3 and HEK293T were obtained from ATCC, and tumor cell lines were regularly validated to be mycoplasma free. SKOV3 cells were authenticated by autosomal DNA profiling in 2018 by the University of Arizona Genetics Core (Tucson, Arizona, USA). The University of Pennsylvania Human Immunology Core provided the human primary CD3+ T cells from healthy donors. Primary lymphocytes were stimulated with Dynabeads coated with CD3 and CD28 stimulatory antibodies (Invitrogen) at a 1:3 cell/bead ratio as previously described (44).
Expanded T cells were cryopreserved on day 10 following activation in a solution of 90% FCS and 10% DMSO. Cells were cultured in R10 (RPMI 1640 media supplemented with 10% FCS, 100-U/mL penicillin, 100 μg/mL streptomycin sulfate, 10 mM HEPES; Invitrogen) in a 37°C and 5% CO2 incubator. Cells were transduced with lentiviral vectors containing CAR constructs approximately 24 hours following stimulation at a MOI of 3.
Primary mouse hepatocytes were dissociated following the nonperfusion method described previously (45), except that enzymatic digestion was done using the Liver Dissociation Kit (MACS Miltenyi Biotec), and liver homogenates were treated with the Debris Removal Solution (MACS Miltenyi Biotec) instead of Percoll. Hepatocytes were analyzed immediately by flow cytometry or cryopreserved in a solution of 90% FCS and 10% DMSO.
Her2 scFVs
We have previously described the Her2 CARs (11). The parental scFv has a HA with KD 0.58 nM, Ka 2.95 × 105 and Kd 1.71 × 10–4. The LA scFv has a KD of 1119 nM and dissociation rate constant (Ka) that is too rapid to measure.
Virus production
AAV8.
The AAV8 vector used to construct pENN.AAV.TBG.Her2/Neu-T2A-Katushka and a Her2-negative control AAV8 vector AAV8.TBG.PI.EGFP.WPRE.bGH (catalog AV-8-PV0146) were obtained from the Penn Vector Core. Customized AVV8 vector was manufactured by the Penn Vector Core. Supplemental Figure 5, A and B, shows schematic diagrams of the AAV8 vectors used in this study.
Lentivirus.
DNA for the LA or HA scFv (4D5-5 and 4D5, respectively) were linked to the CD8 transmembrane domain and 4-1BB and CD3ζ intracellular signaling domains. These were subcloned into pTRPE lentiviral vectors that coexpressed the click-beetle-red (CBR) luciferase gene. A CAR-negative lentiviral vector was used as a negative control and contained CBR and GFP. Supplemental Figure 5, C–E, shows schematic diagrams of the lentiviral vectors used in this study. The plasmids were transformed into Stbl3 chemically competent cells (Invitrogen), and plasmid DNA was isolated using a PureLink HiPure Plasmid Maxiprep kit (Invitrogen). Lentivirus was produced as described previously (46).
Flow cytometric analysis
Data were collected on either a LSRFortessa or LSR II (BD Biosciences) and analyzed using FlowJo software. Cell suspensions were stained with a fixable live/dead violet stain (L34955, Invitrogen) in PBS before surface antibody staining in FACS buffer. T cells were stained using the antibodies for ant–human CD4-BV510 (317444, BioLegend) and anti–human CD8-APC (17-0086-42, eBioscience). CAR positivity was assessed using either recombinant ErbB2/Her2-Fc Chimera (1129-ER-050, R&D Systems) with anti-Human IgG-Fc-DyLight 650 (ab98622, Abcam) or with biotinylated recombinant protein L (29997, Thermo Fisher Scientific) and anti–biotin-PE (12-9895-82, eBioscience). Primary mouse hepatocytes were gated using the gating schema described previously (47), and fluorescence from IRFP720 protein was detected in the AF100 spectrum (Supplemental Figure 6).
In vivo xenograft and gene transfer mouse studies
Immunodeficient NSG mice were purchased from The Jackson Laboratory and then bred, housed, and handled at the University of Pennsylvania in pathogen-free conditions according to institutional guidelines.
In vivo gene transfer of hepatocytes was performed using either tail vein injections of AAV8 or hydrodynamic tail vein injections of DNA. Mice received tail vein injections of AAV8 at the GCs shown in 200 μL of PBS. For hydrodynamic tail vein transfections of hepatocytes, mice received 1.5 mL of PBS in less than 10 seconds with the amount of DNA shown in each figure as described previously (48). Supplemental Figure 7 shows schematic diagrams of the PB vectors used in this study, which include the PB transposase vector pCMV-hyPBase that was obtained from the Wellcome Trust Sanger Institute (49) and the PB transposon plasmid PB007 SPB-007 that was obtained from Transposagen.
To establish xenografts of SKOV3 cells that expressed the fluorescent reporter IRFP720, adult male or female NSG mice were s.c. injected while under anesthesia with 5 × 106 SKOV3 cells resuspended in Matrigel (Corning) and diluted with equal parts PBS. Fluorescence imaging of the IRFP720 reporter gene in SKOV3 cells was done on an IVIS Spectrum imaging system (PerkinElmer). Bioluminescent imaging of T cells that expressed the CBR luciferase reporter gene was performed 10 minutes after i.p. injection of 123 mg/kg D-Lucifer (PerkinElmer) on an IVIS Spectrum imaging system (PerkinElmer).
Mice were euthanized if the tumor volume exceeded 4.2 cm3, if tumor ulceration exceeded 25% of the surface area of the tumor, or when they became moribund. Tumor volumes were calculated from caliper measurements using the formula V = 1⁄2 × L × W × W, where L is length and W is width.
Serum levels of cytokines and liver enzymes
Mouse peripheral blood was collected either by retro-orbital bleeding or by intracardiac puncture at the time of euthanasia. Blood samples were left to clot at room temperature for 30 minutes, centrifuged at 1,000g for 10 minutes at 4°C, and then the resulting serum was collected and stored at –80°C. Serum levels of mouse ALT were measured by ELISA at the University of Pennsylvania’s Veterinary Hospital, and human cytokines were measured on the Luminex panel (HSTCMAG28SPMX21, MilliporeSigma), which was performed by the University of Pennsylvania’s Human Immunology Core.
Nucleic acid isolation and quantitative PCR (qPCR) analysis
For nucleic acid isolation, fresh frozen liver samples were thawed and immediately homogenized using Lysing Matrix D beads in a Fastprep homogenizer. DNA and RNA was isolated from the liver homogenate using an AllPrep DNA/RNA/Protein Mini kit (QIAGEN) and quantified by NanoDrop.
For qPCR analysis, RNA was treated with exonuclease and DNAse to remove genomic DNA, plasmid DNA, or AAV8 DNA from the liver samples before cDNA synthesis. cDNA was generated from RNA using a High Capacity first-strand synthesis kit (Applied Biosystems). Either CART cDNA or genomic DNA were detected using a custom TaqMan assay with the following primer and probe set: forward primer, 5′ - CCAGAAGAAGAAGAAGGAGGATG - 3′; reverse primer, 5′ - GCTCGTTATAGAGCTGGTTCT - 3′; probe, 5′ - FAM-TGAGAGTGAAGTTCAGCAGGAGCG - 3′. Reverse transcriptase negative controls were used for quantification of cDNA, and only those samples that were negative were used in our analysis. The ABI gene expression assays for human IFN-γ, Hs00989291; human Erb2/Her2, Hs01001580; human HPRT1, Hs99999909; and mouse HPRT, Mm00446968, were used. HPRT was used to normalize gene expression, since it was found to be consistently expressed in the liver in the setting of inflammation and tissue repair (50, 51). DNA content was normalized against PTGER genomic DNA (gDNA) as described previously (52). All qPCR assays were performed using a ViiA 7 realtime PCR system (Applied Biosystems). Each assay was done twice in duplicate, and ΔCt was calculated by ΔCtsample – ΔCtreference (53).
IHC
Liver and tumor tissues were formalin fixed and then sent to the University of Pennsylvania’s Cancer Imaging Core for paraffin embedding and sectioning. Slides were immunostained with anti-Her2 (AMAb90627, MilliporeSigma) or anti-CD8 (RB9009-PO, Thermo Fisher Scientific) antibodies and counterstained with Hematoxylin, which was performed by the Pathology Core Laboratory at The Children’s Hospital of Philadelphia. For stain quantification, slides were digitally scanned using an Aperio Scanscope and analyzed using Aperio ImageScope software (Release 6).
Statistics
All growth curves, MFI, and engraftment plots were plotted using Prism 7 (GraphPad Software). For comparisons of 2 groups, 2-tailed unpaired t tests were used. One-way ANOVA with Tukey’s post hoc test was used for comparison of 3 or more groups in a single condition. Statistical analysis for tumor volume and weight change was performed using 2-way repeated-measures ANOVA. Kaplan-Meier survival data were analyzed using a log-rank (Mantel-Cox) test. Correlation was estimated by calculation of 2-tailed Pearson coefficients and significance. The statistical test used for each figure is described in the corresponding figure legend. Data were transformed when needed to normalize variance. *P < 0.05; **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Study approval
Animal studies were carried out under a University of Pennsylvania IACUC-approved protocol.
Author contributions
MC, JAF, and CHJ designed experiments. MC, CS, TD, and KG performed experiments. MC, CS, and EB analyzed data. JS and YZ provided technical expertise and provided reagents. MC wrote the manuscript. CS, JS, JAF, and CHJ edited the manuscript.
Supplementary Material
Acknowledgments
Grant support came, in part, by the Canadian Institute of Health Research and the Breast Cancer Society of Canada (MC) and by NIH grant U54-CA244711 to CHJ.
The authors would like to thank Shannon McGettigan and Kathleen Haines for assistance and expert technical assistance, Ashley O’Dacre for edits to the manuscript, Ling Zhao from the University of Pennsylvania’s Human Immunology Core for support with the Luminex cytokine assays, Celeste Simon from the University of Pennsylvania’s Cancer Histology Core for histology services, and Daniel Martinez from the Children’s Hospital of Pennsylvania’s Pathology Core for providing IHC services.
Version 1. 06/16/2020
In-Press Preview
Version 2. 07/23/2020
Electronic publication
Funding Statement
In partnership with Breast Cancer Society of Canada
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(14):e136012.https://doi.org/10.1172/jci.insight.136012.
Contributor Information
Mauro Castellarin, Email: mauroc@pennmedicine.upenn.edu.
Caroline Sands, Email: caroline.sands@pennmedicine.upenn.edu.
Tong Da, Email: tda@pennmedicine.upenn.edu.
John Scholler, Email: schoj@mail.med.upenn.edu.
Kathleen Graham, Email: grahamka@pennmedicine.upenn.edu.
Elizabeth Buza, Email: ebuza@vet.upenn.edu.
Joseph A. Fraietta, Email: jfrai@upenn.edu.
Yangbing Zhao, Email: Yangbing@upenn.edu.
Carl H. June, Email: cjune@upenn.edu.
References
- 1.Kallioniemi OP, et al. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc Natl Acad Sci USA. 1992;89(12):5321–5325. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA. Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther. 2010;18(4):843–851. doi: 10.1038/mt.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.U.S. Department of Health and Human Services, Food and Drug Administration, Center for Biologics Evaluation and Research. Guidance for human somatic cell therapy and gene therapy. Hum Gene Ther. 2001;12(3):303–314. doi: 10.1089/10430340150218431. [DOI] [PubMed] [Google Scholar]
- 4.Morgan RA. Human tumor xenografts: the good, the bad, and the ugly. Mol Ther. 2012;20(5):882–884. doi: 10.1038/mt.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellarin M, Watanabe K, June CH, Kloss CC, Posey AD. Driving cars to the clinic for solid tumors. Gene Ther. 2018;25(3):165–175. doi: 10.1038/s41434-018-0007-x. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y, et al. A herceptin-based chimeric antigen receptor with modified signaling domains leads to enhanced survival of transduced T lymphocytes and antitumor activity. J Immunol. 2009;183(9):5563–5574. doi: 10.4049/jimmunol.0900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghorashian S, et al. Enhanced CAR T cell expansion and prolonged persistence in pediatric patients with ALL treated with a low-affinity CD19 CAR. Nat Med. 2019;25(9):1408–1414. doi: 10.1038/s41591-019-0549-5. [DOI] [PubMed] [Google Scholar]
- 8.Gerardin J, Reddy NR, Lim WA. The Design Principles of Biochemical Timers: Circuits that Discriminate between Transient and Sustained Stimulation. Cell Syst. 2019;9(3):297–308.e2. doi: 10.1016/j.cels.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho JH, Collins JJ, Wong WW. Universal Chimeric Antigen Receptors for Multiplexed and Logical Control of T Cell Responses. Cell. 2018;173(6):1426–1438.e11. doi: 10.1016/j.cell.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majzner RG, et al. Tuning the Antigen Density Requirement for CAR T-cell Activity. Cancer Discov. 2020;10(5):702–723. doi: 10.1158/2159-8290.CD-19-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, et al. Affinity-Tuned ErbB2 or EGFR Chimeric Antigen Receptor T Cells Exhibit an Increased Therapeutic Index against Tumors in Mice. Cancer Res. 2015;75(17):3596–3607. doi: 10.1158/0008-5472.CAN-15-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carter P, et al. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89(10):4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drent E, et al. A Rational Strategy for Reducing On-Target Off-Tumor Effects of CD38-Chimeric Antigen Receptors by Affinity Optimization. Mol Ther. 2017;25(8):1946–1958. doi: 10.1016/j.ymthe.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caruso HG, et al. Tuning Sensitivity of CAR to EGFR Density Limits Recognition of Normal Tissue While Maintaining Potent Antitumor Activity. Cancer Res. 2015;75(17):3505–3518. doi: 10.1158/0008-5472.CAN-15-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172(5):2731–2738. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, et al. Systematic evaluation of AAV vectors for liver directed gene transfer in murine models. Mol Ther. 2010;18(1):118–125. doi: 10.1038/mt.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy A, Imre G, Mátés L. Transposon-based gene delivery vectors for gene therapy. Acta Biologica Szegediensis. 2015;59:247–260. [Google Scholar]
- 18.Nakanishi H, Higuchi Y, Kawakami S, Yamashita F, Hashida M. piggyBac transposon-mediated long-term gene expression in mice. Mol Ther. 2010;18(4):707–714. doi: 10.1038/mt.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Matteo M, et al. Hyperactive piggyBac transposons for sustained and robust liver-targeted gene therapy. Mol Ther. 2014;22(9):1614–1624. doi: 10.1038/mt.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods. 2013;10(8):751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed N, et al. Human Epidermal Growth Factor Receptor 2 (HER2) -Specific Chimeric Antigen Receptor-Modified T Cells for the Immunotherapy of HER2-Positive Sarcoma. J Clin Oncol. 2015;33(15):1688–1696. doi: 10.1200/JCO.2014.58.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crispe IN. Hepatic T cells and liver tolerance. Nat Rev Immunol. 2003;3(1):51–62. doi: 10.1038/nri981. [DOI] [PubMed] [Google Scholar]
- 23.Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16(6):1073–1080. doi: 10.1038/mt.2008.76. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6(7):1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 25.Nakai H, Fuess S, Storm TA, Muramatsu S, Nara Y, Kay MA. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J Virol. 2005;79(1):214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodard LE, Wilson MH. piggyBac-ing models and new therapeutic strategies. Trends Biotechnol. 2015;33(9):525–533. doi: 10.1016/j.tibtech.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang G, et al. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004;11(8):675–682. doi: 10.1038/sj.gt.3302210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calne RY, et al. Induction of immunological tolerance by porcine liver allografts. Nature. 1969;223(5205):472–476. doi: 10.1038/223472a0. [DOI] [PubMed] [Google Scholar]
- 30.LoDuca PA, Hoffman BE, Herzog RW. Hepatic gene transfer as a means of tolerance induction to transgene products. Curr Gene Ther. 2009;9(2):104–114. doi: 10.2174/156652309787909490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holz L, Rehermann B. T cell responses in hepatitis C virus infection: historical overview and goals for future research. Antiviral Res. 2015;114:96–105. doi: 10.1016/j.antiviral.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochel A, et al. Effective intrahepatic CD8+ T-cell immune responses are induced by low but not high numbers of antigen-expressing hepatocytes. Cell Mol Immunol. 2016;13(6):805–815. doi: 10.1038/cmi.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tay SS, et al. Antigen expression level threshold tunes the fate of CD8 T cells during primary hepatic immune responses. Proc Natl Acad Sci USA. 2014;111(25):E2540–E2549. doi: 10.1073/pnas.1406674111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benseler V, et al. Hepatocyte entry leads to degradation of autoreactive CD8 T cells. Proc Natl Acad Sci USA. 2011;108(40):16735–16740. doi: 10.1073/pnas.1112251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies SP, et al. Hepatocytes Delete Regulatory T Cells by Enclysis, a CD4+ T Cell Engulfment Process. Cell Rep. 2019;29(6):1610–1620.e4. doi: 10.1016/j.celrep.2019.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbari O, DeKruyff RH, Umetsu DT. Pulmonary dendritic cells producing IL-10 mediate tolerance induced by respiratory exposure to antigen. Nat Immunol. 2001;2(8):725–731. doi: 10.1038/90667. [DOI] [PubMed] [Google Scholar]
- 37.Griffith TS, Yu X, Herndon JM, Green DR, Ferguson TA. CD95-induced apoptosis of lymphocytes in an immune privileged site induces immunological tolerance. Immunity. 1996;5(1):7–16. doi: 10.1016/S1074-7613(00)80305-2. [DOI] [PubMed] [Google Scholar]
- 38.Steele L, Mayer L, Berin MC. Mucosal immunology of tolerance and allergy in the gastrointestinal tract. Immunol Res. 2012;54(1-3):75–82. doi: 10.1007/s12026-012-8308-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adusumilli PS, et al. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci Transl Med. 2014;6(261):261ra151. doi: 10.1126/scitranslmed.3010162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roybal KT, et al. Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors. Cell. 2016;167(2):419–432.e16. doi: 10.1016/j.cell.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma JS, et al. Versatile strategy for controlling the specificity and activity of engineered T cells. Proc Natl Acad Sci USA. 2016;113(4):E450–E458. doi: 10.1073/pnas.1524193113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang WC, et al. CRISPR/Cas9 Technology Targeting Fas Gene Protects Mice From Concanavalin-A Induced Fulminant Hepatic Failure. J Cell Biochem. 2017;118(3):530–536. doi: 10.1002/jcb.25722. [DOI] [PubMed] [Google Scholar]
- 43.von Knethen A, et al. Tolerizing CTL by Sustained Hepatic PD-L1 Expression Provides a New Therapy Approach in Mouse Sepsis. Theranostics. 2019;9(7):2003–2016. doi: 10.7150/thno.28057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barrett DM, et al. Treatment of advanced leukemia in mice with mRNA engineered T cells. Hum Gene Ther. 2011;22(12):1575–1586. doi: 10.1089/hum.2011.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gonçalves LA, Vigário AM, Penha-Gonçalves C. Improved isolation of murine hepatocytes for in vitro malaria liver stage studies. Malar J. 2007;6:169. doi: 10.1186/1475-2875-6-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milone MC, et al. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol Ther. 2009;17(8):1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zakrzewska KE, Samluk A, Wencel A, Dudek K, Pijanowska DG, Pluta KD. Liver tissue fragments obtained from males are the most promising source of human hepatocytes for cell-based therapies - Flow cytometric analysis of albumin expression. PLoS ONE. 2017;12(8):e0182846. doi: 10.1371/journal.pone.0182846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell JB, Podetz-Pedersen KM, Aronovich EL, Belur LR, McIvor RS, Hackett PB. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat Protoc. 2007;2(12):3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA. 2011;108(4):1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xing W, Deng M, Zhang J, Huang H, Dirsch O, Dahmen U. Quantitative evaluation and selection of reference genes in a rat model of extended liver resection. J Biomol Tech. 2009;20(2):109–115. [PMC free article] [PubMed] [Google Scholar]
- 51.Shi G, et al. Selection of reference genes for quantitative real-time reverse transcription-polymerase chain reaction in concanavalin A-induced hepatitis model. Anal Biochem. 2010;401(1):81–90. doi: 10.1016/j.ab.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 52.Alcoser SY, Kimmel DJ, Borgel SD, Carter JP, Dougherty KM, Hollingshead MG. Real-time PCR-based assay to quantify the relative amount of human and mouse tissue present in tumor xenografts. BMC Biotechnol. 2011;11:124. doi: 10.1186/1472-6750-11-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.