Abstract
Apelin is a well-established mediator of survival and mitogenic signaling through the apelin receptor (Aplnr) and has been implicated in various cancers; however, little is known regarding Elabela (ELA/APELA) signaling, also mediated by Aplnr, and its role and the role of the conversion of its precursor proELA into mature ELA in cancer are unknown. Here, we identified a function of mTORC1 signaling as an essential mediator of ELA that repressed kidney tumor cell growth, migration, and survival. Moreover, sunitinib and ELA showed a synergistic effect in repressing tumor growth and angiogenesis in mice. The use of site-directed mutagenesis and pharmacological experiments provided evidence that the alteration of the cleavage site of proELA by furin induced improved ELA antitumorigenic activity. Finally, a cohort of tumors and public data sets revealed that ELA was only repressed in the main human kidney cancer subtypes, namely clear cell, papillary, and chromophobe renal cell carcinoma. Aplnr was expressed by various kidney cells, whereas ELA was generally expressed by epithelial cells. Collectively, these results showed the tumor-suppressive role of mTORC1 signaling mediated by ELA and established the potential use of ELA or derivatives in kidney cancer treatment.
Keywords: Nephrology
Keywords: Calcium
ELA mediates the tumor-suppressive role of mTORC1 and has potential to be therapeutically exploited to treat kidney cancers.
Introduction
Renal cell carcinoma (RCC) is one of the most common malignancies worldwide, accounting for 140,000 cancer-related deaths and 330,000 new cases a year (1). Various environmental and daily life risk factors for RCC are well established, including hypertension, obesity, and several chronic kidney diseases. In adults, malignant kidney tumors mainly arise from the renal parenchyma and renal pelvis (2). RCC is classified into clear cell RCC (ccRCC), papillary RCC, and chromophobe RCC subtypes (2). Accounting for up to 75% of all RCCs, ccRCC is predominantly sporadic, and only 5% of ccRCC cases are associated with hereditary syndromes (von Hippel-Lindau disease) (2). Up to 50% of renal cancer patients develop metastatic disease, which remains incurable, and the median survival time is less than 28 months (3). Successful treatment procedures are currently limited because of the lack of thorough understanding of the basic molecular pathways involved in RCC carcinogenesis and lack of genetically pertinent animal models (4). Currently, signaling pathways that promote RCC metastatic progression are the subject of various studies, and tyrosine kinases have emerged as important determinants of RCC neoplasia (3–5).
The apelin peptides and their cognate G protein–coupled receptor (GPCR) known as the apelin receptor (Aplnr) play a key role in apoptosis, cell proliferation, angiogenesis, metabolic disorders, and various cancers (6, 7). After Aplnr activation, a wide range of signaling pathways mediate these biological processes. These signaling pathways include phosphorylation of protein kinase B (AKT), ERK1/2 (8), calcium mobilization (8, 9), cAMP (6), and NOS (10). Previously, 2 groups separately identified Elabela (ELA) (11, 12) as a new Aplnr ligand. Human ELA cDNA encodes a preproprotein of 54 aa residues. After the removal of the signal peptide, the proprotein (proELA) of 32aa (ELA32) is converted to mature peptide 11aa (ELA11) by the proprotein convertases (11–15). This well-conserved ELA gene (APELA) was initially associated with early cardiac development. Loss of APELA resulted in a phenotype similar to the deficient Aplnr gene, but different from the APLN gene, suggesting a functional link between ELA and Aplnr (8, 16). ELA is able to activate Aplnr in vitro, and activation of Aplnr by the apelin signaling pathway was shown to rescue ELA deficiency (11, 12). Hence, we could consider that ELA and Aplnr interaction might be involved in a series of yet-uncharacterized signals responsible for various physiopathological mechanisms, such as the recent report that ELA loss promotes preeclampsia and cardiovascular malformations (17). Herein, we report that the ELA gene and protein are systematically repressed in the main human kidney cancers. Activation of Aplnr by ELA and uncleaved ELA precursor peptide (mut ELA) induced inhibition of cell survival in an mTORC1-dependent manner. We also demonstrated that tumor cell proliferation and motility were repressed by ELA and more efficiently by mut ELA that selectively affected Aplnr affinity and internalization/recycling during its interaction with Aplnr. Our results highlighted the tumor suppressor features of ELA and mut ELA and identified the potentially new role of mTORC1 activation by ELA in these processes, suggesting the potential use of ELA and/or derivatives as a therapeutic approach in kidney cancers.
Results
Repressed expression of ELA in human kidney cancer patients.
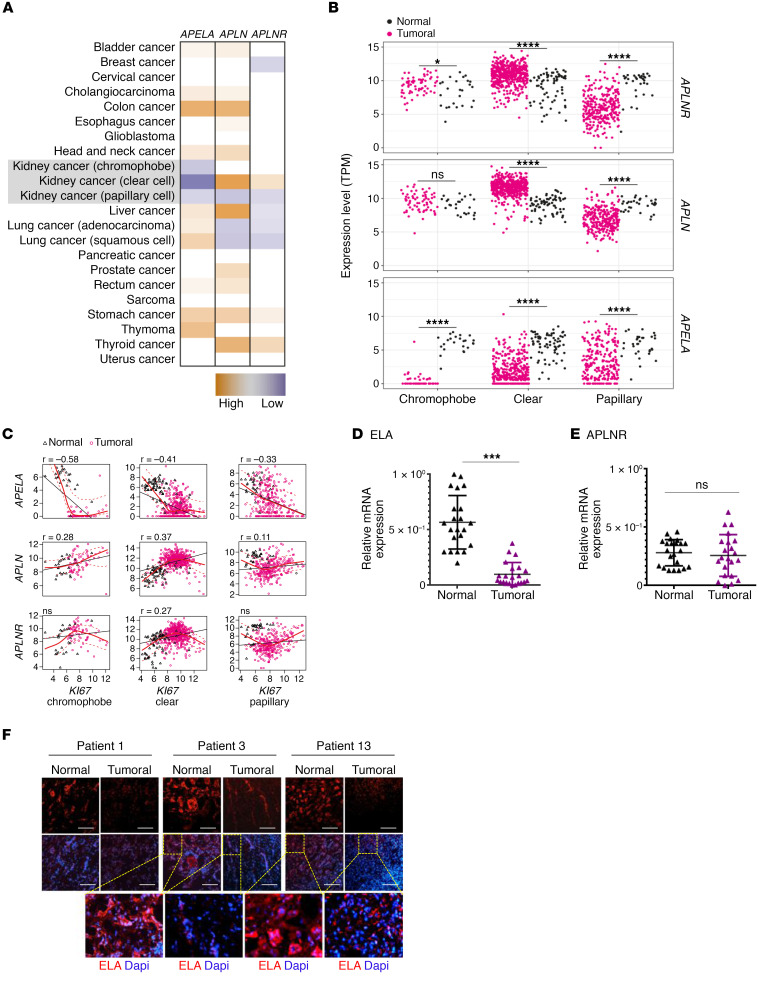
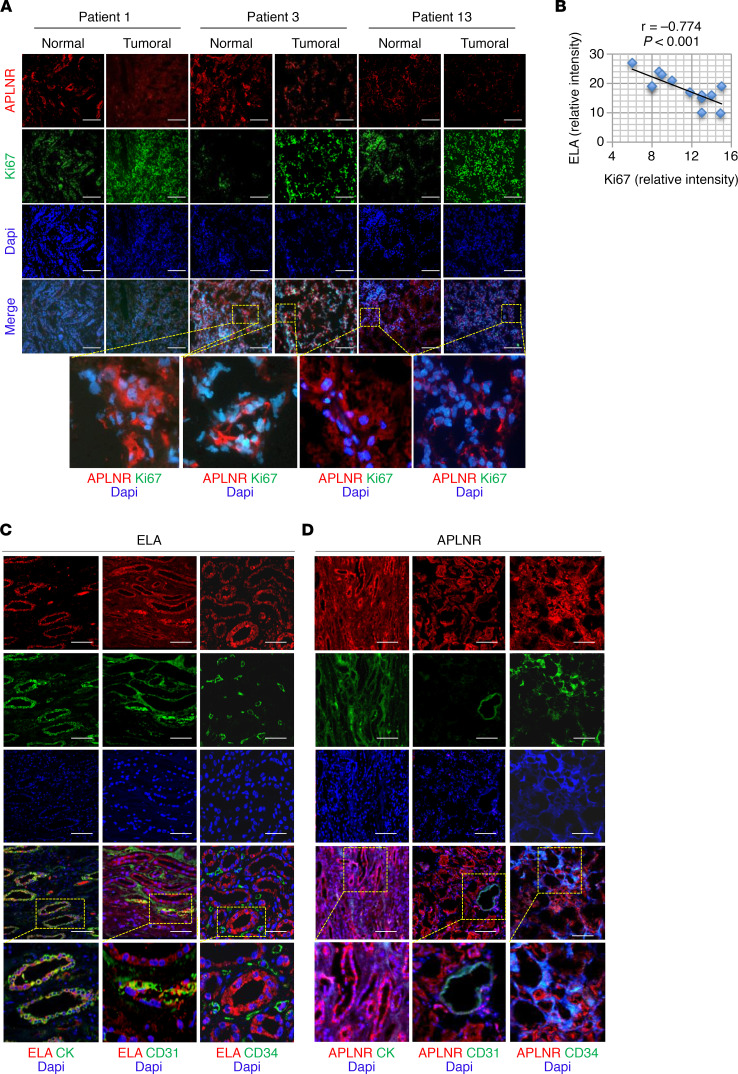
Using The Cancer Genome Atlas (TCGA) data sets, we analyzed ELA gene (APELA), APLN, and APLNR expression levels in various tumors and their corresponding normal tissues. We found that APELA expression was upregulated in colon, lung, stomach, and thymoma cancers but did not vary in the other types of cancer tissues analyzed except in kidney cancer tissues (Figure 1A). We found that APELA was systematically downregulated in all renal cancer types analyzed, including chromophobe RCC (n = 66), papillary RCC (n = 289), and ccRCC (n = 531), when compared with normal kidney tissues (n = 25, n = 32, and n = 72, respectively). APLN and APLNR expression were upregulated in ccRCC and downregulated in papillary RCC (Figure 1, A and B). A negative correlation between APELA and the marker of cellular proliferation KI67 expression was also noted in TCGA data set in all these renal cancer types (r = –0.58, r = –0.41, and r = –0.33 for chromophobe RCC, papillary RCC, and ccRCC, respectively) (Figure 1C). In contrast, the expression of APLN was positively correlated with KI67 expression in these kidney cancer subtypes (r = 0.28, r = 0.41, and r = 0.11, respectively (Figure 1C), and APLNR expression was positively correlated with KI67 only in ccRCC (r = 0.27). Similarly, analysis of tissues obtained from RCC patients with different cancer grades revealed that ELA mRNA expression was frequently strongly downregulated in renal cancer samples compared with normal tissues (Figure 1D). ELA was differentially repressed in more than 80% of the kidney cancer samples examined. Of the patients analyzed, up to 30% showed no ELA expression. Further analysis revealed that APLNR expression remained unchanged in 20%, repressed in approximately 45%, and increased in 20% of patients with tumors (Figure 1E). We also detected ELA (Figures 1F) and APLNR (Figure 2A) proteins in macroscopically normal kidney tissues. Quantitative analysis of the immunohistochemistry staining in RCC with matched normal kidney tissues indicated that the average of ELA protein staining was reduced (Figure 1F and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.129070DS1). These findings suggest that the expression of ELA was negatively correlated with human renal cancer. Indeed, when we analyzed and quantified the expression of Ki-67 protein (Figure 2B), we observed a negative correlation between ELA and Ki-67 expression (r = −0.774, P < 0.001). In some patients, the reduced expression of ELA was associated with reduced APLNR expression (Figure 1F, Figure 2A, and Supplemental Figure 2). Double-label immunostaining in human kidney sections revealed that ELA did not colocalize with endothelial cells and hematopoietic cells, but it specifically overlapped in the tubule epithelial cell area, which was detected using the anticytokeratin antibody (Figure 2C). In contrast, APLNR was expressed in all these cells (Figure 2D). Taken together, these data suggest that APELA may play a role in human renal cancer.
Figure 1. ELA is downregulated in renal cancers.
(A) TCGA data set analysis of ELA, APLN, and APLNR expression in various cancers and their corresponding normal tissues. (B) ELA was systematically repressed in the renal cancer subtypes chromophobe RCC (n = 66), papillary RCC (n = 289), and ccRCC (n = 531) compared with normal kidney tissues (n = 25, n = 32, and n = 72, respectively), as assessed by TCGA. (C) TCGA analysis identified a negative correlation between ELA and KI67 expression in all these renal cancer types. APLN expression was positively correlated with KI67 in all subtypes, and APLNR was positively correlated with KI67 only in ccRCC. (D) Relative expression of ELA mRNA level in 22 pairs of ccRCC tumors and their corresponding adjacent noncancerous tissues. (E) Relative expression of APLNR mRNA in the same samples is given for comparison. The relative amounts of mRNA were normalized against GAPDH mRNA and expressed relative to mRNA abundance in noncancerous tissue for each patient assigned 1 (mean ± SEM; n = 22). (F) Representative immunofluorescence images of renal tumor samples and adjacent healthy tissues from 3 patients (patients 1, 3, and 13) derived from (D–E) that were stained with anti-ELA and DAPI. Unpaired t tests were used to analyze the data. ***P < 0.001, ****P < 0.0001. Scale bar: 150 μm.
Figure 2. Expression of ELA by renal epithelial cells.
(A) Representative immunofluorescence images of renal tumor samples and adjacent healthy tissues from 3 patients derived from Figure 1, D and E, that were stained with anti-APLNR, anti-Ki67, and DAPI. (B) The correlation between ELA and Ki67 expression in tumor samples was determined by immunofluorescence following staining intensity analysis (Supplemental Figure 1) using Spearman’s correlation analysis. The test revealed a negative correlation between the expression of ELA and Ki67 (n = 11, r = –0.774, P < 0.001). (C) Representative confocal images of sections of human kidney tumors (n = 3) subjected to immunofluorescence double-labeling of ELA and cytokeratin (CK), CD31, or CD34. (D) Costaining of APLNR and CK, CD31, or CD34 in the same tumors. Scale bar: 100 μm. DAPI is in blue.
Inhibition of cell survival and the malignant phenotype by ELA and unprocessed ELA precursor (mut ELA).
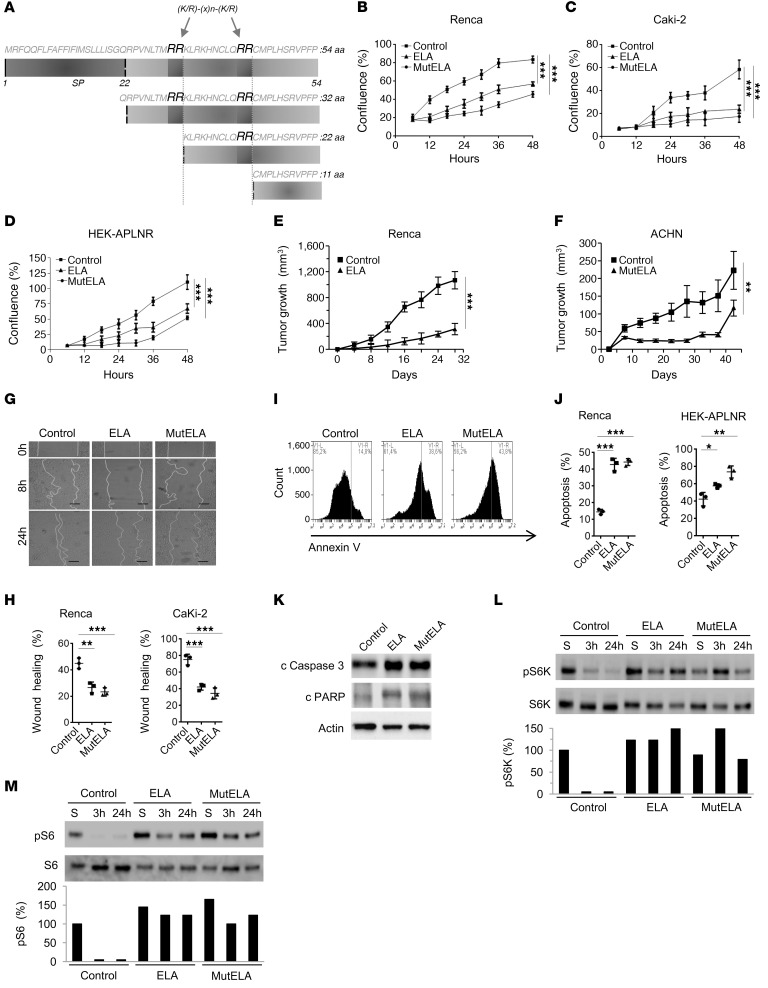
The ELA32 protein precursor (32 aa) is cleaved at 2 proprotein convertase–cleavage sites RX(K/R)RQ (18–20), namely R31/R32 and R42/R43, to generate mature ELA11 peptide (11 ELA32) (Figure 3A). Through GenBank databases, we found that these cleavage sites were conserved in all analyzed species (Supplemental Table 1), suggesting the biological importance of the proteolytic maturation of proELA (ELA32) by the proprotein convertases. To determine the role of ELA and proELA processing by these proteases on the malignant phenotype, lentiviral vectors were first used to deliver and stably express ELA and unprocessed proELA (mut ELA), in which the 2 cleavage sites were mutated to S31/S32 and S42/S43, respectively (Figure 3A), in the kidney cancer cells Renca and Caki-2. The proliferation of cells stably expressing ELA and mut ELA were analyzed using the IncuCyte Live Cell Analysis System (Figure 3, B and C). As illustrated, expression of ELA and mut ELA in these cells significantly decreased their confluence rate, suggesting the autocrine action of ELA and mut ELA in these cells. Interestingly, expression of ELA or mut ELA in Renca cells induced APLNR expression and repressed APLN expression (Supplemental Figure 3). The confluence rate of mut ELA–expressing cells was further inhibited compared with ELA-expressing cells and control cells (Figure 3, B and C). To confirm the autocrine effect of ELA on cell proliferation, we generated HEK293 cells stably overexpressing APLNR (HEK-APLNR). In agreement with our previous results, stable expression of ELA or mut ELA in HEK-APLNR cells repressed their confluence rate compared with controls (Figure 3D). To directly evaluate the effect of ELA on tumor cell growth in vivo, we first investigated whether secreted ELA by renal cells resulted in sustained tumor repression in a syngeneic mouse model developed with subcutaneously implanted murine Renca cells in BALB/c mice. Mice were inoculated subcutaneously with control cells and the same cells stably expressing ELA. As illustrated in Figure 3E, expression of ELA in Renca cells reduced their ability to induce tumor growth. Injection of Renca cells into the subcapsular space in the mouse kidney resulted in a decrease of tumor growth in mice bearing ELA tumors (Supplemental Figure 4). Similarly, using a human kidney cancer cell line, ACHN, as a second model, we found that stable expression of mut ELA in ACHN cells using lentiviral vectors inhibited their ability to mediate tumor growth in nude mice (Figure 3F). We next evaluated cell migration in a wound-healing assay and observed that while Renca and HEK-APLNR control cells closed the wound (~45% for Renca and ~80% for Caki-2 cells) within 24 hours, ELA- and mut ELA–expressing cells inefficiently closed the wound over the same time frame (~27% for Renca and ~42% for HEK-APLNR cells) (Figure 3, G and H). Next, we investigated whether ELA and mut ELA expression in cancer cells induced cell death. Apoptosis was examined in Renca and HEK-APLNR cells in the presence of serum or after serum starvation using an annexin V apoptosis detection kit. We observed that apoptosis levels were elevated in cells stably expressing ELA or mut ELA versus controls (Figure 3, I and J). Relatively greater effect was observed with mut ELA. Further experiments revealed that ELA and mut ELA expression in Renca cells increased the levels of cleaved caspase-3 and PARP (Figure 3K). This suggests the involvement of these apoptotic molecules in ELA- and mut ELA–induced cell death. Collectively, the database, the patient tissue analyses, and the in vivo results shown here were consistent with the potential tumor suppressor function of ELA in the kidney.
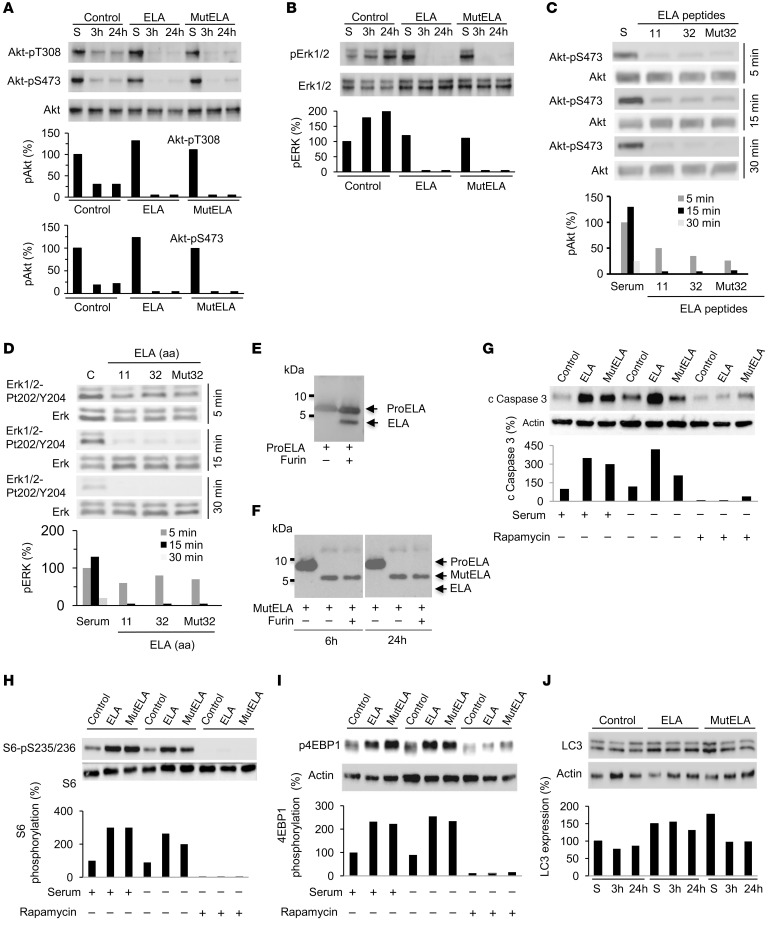
Figure 3. Repression of the malignant phenotype and increased activation of mTORC1 signaling by ELA.
(A) Primary structure of the 54 amino acids (ELA32) of human pre-proEla containing the signal peptide (SP) and ELA precursor (ELA32). The 2 proprotein convertase cleavage sites R31/R32 and R42/R43 that generate the ELA peptides are indicated. (B–D) Growth curves of control Renca (B), Caki-2 (C), and HEK-APLNR cells (D) or stably expressing ELA or mut ELA cDNA. Cells were plated at low confluence for time-lapse phase-contrast videomicroscopy using an IncuCyte microscope, and cell proliferation was monitored by automated confluence analysis at set intervals after plating (means, n = 6 wells per group, 3 independent experiments). (E) Tumor growth curves over time from representative experiment of subcutaneously injected syngeneic BALB/c mice with Renca cells or the same cells stably expressing ELA (n = 7 mice/group, 3 independent experiments). (F) Subcutaneously injected nude mice with the human ACHN cells or the same cells stably expressing mut ELA (n = 7 mice/group). (G) Cell migration was analyzed by scratch wound assay. Control Renca and Caki-2 cells or stably expressing ELA or mut ELA monolayers were subjected to scratch wounds and imaged after 8 hours and 24 hours (n = 6 wells per group, 3 independent experiments). Scale bar: 100 μm. (H) Quantification of wound closure after 24 hours of indicated control cells and the same cells stably expressing ELA or mut ELA. (I) FACS scatter plots of control cells and the same cells stably expressing ELA or mut ELA incubated for 24 hours in the absence of serum and stained with annexin V (n = 3 wells per group, 3 independent experiments). (J) Quantification of apoptosis in Renca and HEK-APLNR cells (annexin V–positive cells) under these conditions is shown. (K) Renca control cells expressing ELA or mut ELA were analyzed for cleaved proapoptotic protein caspase-3 (c caspase-3) and PARP by immunoblotting. (L and M) Western blot analysis of the activation of S6K-pT389 (L) and S6-pS235/236 (M) in control, ELA-expressing, and mut ELA–expressing cells starved for the indicated time period or in the presence of serum (S). Results in K, L, and M are representative of 3 independent experiments each. The mean ± SEM values are shown. One- or 2-way ANOVA with Tukey’s multiple comparisons test were used to analyze data in B, C, D, H, and J. Unpaired t tests were used to analyze the data in E and F. *P < 0.05, **P < 0.01, ***P < 0.001.
Regulation of AKT, ERK, and mTORC1 activation by ELA and mut ELA.
AKT, ERK, and mTOR kinases form a complex pathway, which regulates a diverse array of cellular functions, most prominently cell growth, migration, and survival. Therefore, we examined whether ELA- and mut ELA–expressing tumor cells display autocrine regulation of these pathways. Renca cells were first serum starved for short (3 hours) and long (24 hours) time periods, lysed, and analyzed by Western blot using phosphospecific antibodies. For comparison, after 24 hours of starvation, cells were stimulated with serum (S). Compared with control cells, we found that expression of ELA or mut ELA in Renca cells induced mTORC1 activation, as seen by the dramatic increase in the activation of mTORC1 substrate S6K (Figure 3L) and the S6K substrate S6 (Figure 3M). Indeed, mTORC1 was necessary for the phosphorylation of Thr389 in S6K, and as illustrated in Figure 3L, the phosphorylation of Thr389 was upregulated in ELA- and mut ELA–expressing cells. Analysis of NF-κB activation in these cells revealed that ELA and mut ELA had no effect on its activation (Supplemental Figure 5). In contrast, analysis of AKT (Thr308 and Ser473) and ERK activity under the same conditions revealed their repressed activity in ELA- and mut ELA–expressing cells (Figure 4, A and B). To further investigate the repressed ERK and AKT activation mediated by ELA and mut ELA, we analyzed ERK and AKT activation after treatment of HEK-APLNR cells with mature ELA11, ELA32, and mut ELA32 peptides during short time periods. We observed that although these peptides were able to induce rapid (5-minute) phosphorylation of AKT (Figure 4C) and ERK (Figure 4D), this activation was rapidly repressed and decreased over time. These results suggest that activation of APLNR by ELA peptides that activate mTORC1 was associated with rapid repression of AKT and ERK activation. In agreement with these results, treatment of control HEK cells and HEK-APLNR cells with ELA peptides revealed that although ELA11, ELA32, and mut ELA32 induced a weak effect on AKT and ERK activation in control HEK cells, overexpression of APLNR in these cells induced rapid (10-minute) ERK and AKT activation that was repressed over time in HEK-APLNR cells (3 hours) (Supplemental Figure 6). In contrast, increased activation of 4EBP-1 and S6K was observed at 3 hours in ELA peptide–treated HEK-APLNR cells compared with ELA peptide–treated control HEK cells (Supplemental Figure 6). To confirm that the mut ELA peptide is not cleaved by furin produced by HEK-APLNR cells during treatment, we performed an in vitro digestion assay. Incubation of ELA32 peptide with recombinant human furin (0.2 × 10−4 U) predominantly generated the mature ELA11 form as assessed by immunoblotting assay, suggesting that R31/R32 and R42/R43 sites were cleaved simultaneously (Figure 4E). In contrast, furin failed to cleave the mut ELA32 peptide (Figure 4F) at all the time points analyzed.
Figure 4. Repression of AKT and ERK activation by ELA and inhibition of ELA-induced mTORC1 signaling by rapamycin.
(A and B) Western blot analysis of the activation of AKT (A) and ERK (B) in control, ELA-expressing, and mut ELA–expressing cells starved for the indicated time period or in the presence of serum (S). (C and D) Control and Renca cells were serum starved and incubated with ELA11, ELA32, or mut ELA32 peptides for indicated time periods and the activation of AKT (C) and ERK (D) were analyzed by Western blot analysis. (E and F) Western blotting analysis using an anti-ELA antibody of WT proELA32 (E) and mut ELA (F) (cleavage sites R31/R32 and R42/R43 were replaced by S31/S32 and S42/S43) peptides incubated with furin (0.2 × 10−4 U) for 6 or 24 hours. (G) Western blot analysis of cleaved caspase-3 upon rapamycin treatment of control, ELA-expressing, and mut ELA–expressing cells for 24 hours in the absence or presence of serum. Graphs show quantification of cleaved caspase-3. (H and I) Western blot analysis of the activation of S6-pS235/236 (H) and 4EBP1 (I) in control, ELA-expressing, and mut ELA–expressing cells in the absence and presence of rapamycin and/or serum. (J) Western blot analysis of the levels of LC3 in control, ELA-expressing, and mut ELA–expressing cells. Bars denote the corresponding percentages of phosphorylated proteins (n = 3), expressed LC3 protein (n = 3), or cleaved caspase-3 (n = 3). All results shown are representative of at least 3 independent experiments.
Cell survival and autophagy inhibition by ELA and mut ELA.
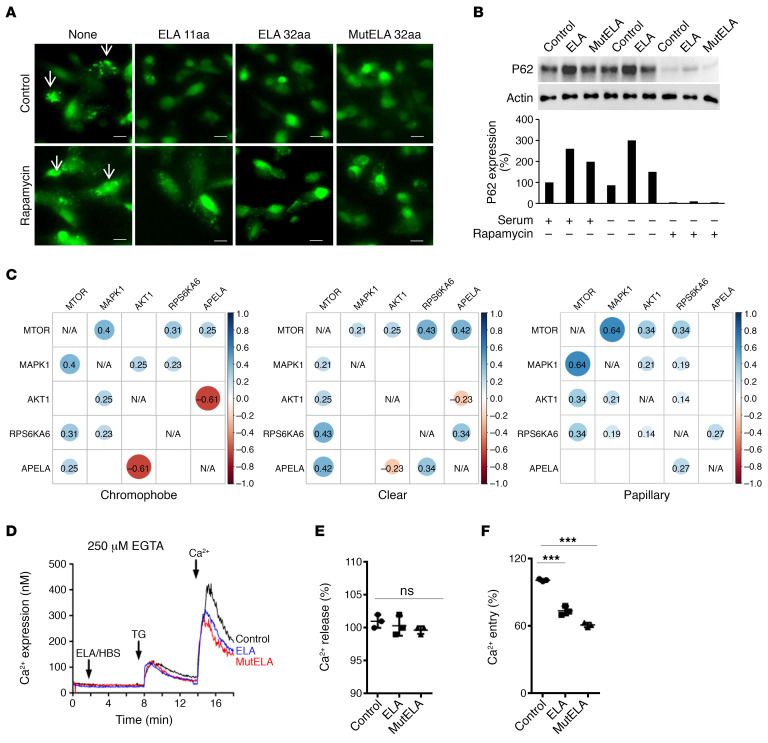
mTORC1 controls cell proliferation, survival, and metabolism by integrating a variety of signals from growth factors and nutrients (21). We investigated whether the activation of mTORC1 plays a mechanistic role in apoptosis induced by ELA and mut ELA. In agreement with this possibility, ELA and mut ELA expression in Renca cells induced caspase-3 cleavage, and rapamycin treatment of these cells was sufficient to inhibit caspase-3 cleavage (Figure 4G). Accordingly, we observed that although ELA and mut ELA induced the 2 mTORC1 targets, S6K and 4EBP1, in the absence or presence of serum, rapamycin inhibited S6K (Figure 4H) and 4EBP1 (Figure 4I) activation. We next explored whether apoptosis upon the activation of mTORC1 by ELA and mut ELA is linked to autophagy inhibition. For this purpose, we examined the levels of the autophagic marker LC3-II and the aggregation of a GFP-LC3 construct upon autophagosome formation. We first observed that there was no reduction in LC3 levels in ELA- and mut ELA–expressing cells compared with control cells during serum starvation, suggesting the inhibition of autophagy in these cells (Figure 4J). To confirm that autophagy acts downstream of ELA pathways, we next used osteosarcoma U2OS cells stably expressing GFP-LC3, a well-established cell model to study autophagy, particularly autophagosome formation (22). Thereby, incubation of the U2OS cells stably expressing a GFP-LC3 construct with ELA11, ELA32, and mut ELA32 peptides (100 nM) displayed a decrease in the number of GFP-LC3 aggregates compared with untreated cells (Figure 5A), showing an increase in autophagosome formation. These cells expressed APLNR but not ELA (Supplemental Figure 7). To clarify the role of mTORC1 on the blockage of autophagy by ELA and mut ELA, we inhibited the mTORC1 pathway using rapamycin. As shown in Figure 5B, mTORC1 inhibition by rapamycin repressed ELA- and mut ELA–mediated inhibition of autophagy flux, as determined by the increase in the amount of GFP-LC3 aggregates. In agreement with this finding, the autophagic protein sequestosome-1 (SQSTM1, p62) accumulated in ELA- and mut ELA–expressing Renca cells was inhibited by rapamycin (Figure 5B). Thus, the apoptotic cell death mediated by ELA- and mut ELA–induced activation of mTORC1 was linked to the inhibition of autophagy. These results indicate that mTORC1 activation by ELA and mut ELA was responsible for the inhibition of autophagy and cell death. The use of TCGA expression analysis to examine the correlation between APELA and mTOR interactants AKT, ERK, and S6K showed that the expression of APELA was positively correlated with mTOR expression in chromophobe RCC (r = 0.25, P = 0.016) and ccRCC (r = 0.42, P = 5.12 × 10–27). APELA and S6K expression positively correlated in papillary RCC (r = 0.27, P = 1.26 × 10–6) and ccRCC (r = 0.34, P = 1.27 × 10–17). In contrast, a negative correlation was noted between APELA and AKT in chromophobe RCC (r = –0.61, P = 1.1 × 10–10) and ccRCC (r = –0.23, P = 1.33 × 10–8) (Figure 5C). These results suggest that ELA is also involved in the regulation of the expression of mTORC1 network proteins.
Figure 5. Inhibition of mTOR-mediated autophagy blockade and calcium mobilization by ELA.
(A) GFP-LC3–expressing U2OS cells in the presence or absence of 100 nM ELA-11, ELA-32, or mut ELA-32 peptides and/or rapamycin for 24 hours. Autophagosome formation upon GFP-LC3 aggregation (white arrows) was determined using microscopy. Scale bar: 25 μm. (B) Western blot analysis of the levels of the autophagy protein P62 in control, ELA-expressing, and mut ELA–expressing cells in the absence (24 hours) or presence of serum and/or rapamycin. (C) TCGA data set analysis of correlated expression between APELA and mTOR interactants AKT, ERK, and S6K in indicated renal cancer subtypes compared with normal kidney tissues. (D) Traces of calcium mobilization in HEK/APLNR cells preincubated for 8 minutes in the absence or presence of 100 nM ELA-32 or mut ELA-32 peptides. (E and F) The corresponding percentages of Ca2+ release induced by thapsigargin (TG) (E) and Ca2+ entry (F) are represented as mean ± SEM (n = 3). Bars denote the corresponding percentage of accumulated P62 (n = 3). One-way ANOVA with Tukey’s multiple comparisons test was used to analyze the data. ***P < 0.001. All results shown are representative of at least 3 independent experiments.
Repression of thapsigargin-induced store-operated Ca2+ entry by ELA and mut ELA.
Store-operated Ca2+ entry (SOCE) is a mechanism for Ca2+ influx across the plasma membrane activated in response to depletion of intracellular Ca2+ stores, mostly in the ER. To evaluate the effect of ELA and mut ELA on Ca2+ mobilization, HEK-APLNR cells were incubated with ELA or mut ELA peptides (100 nM), and Ca2+ release and entry were measured. Cells were loaded with fura-2, preincubated with or without ELA32 and mut ELA32 peptides, and then stimulated with 1 μM thapsigargin to deplete the ER and activate SOCE. We observed that although the presence of ELA and mut ELA had no effect on Ca2+ release, these peptides inhibited Ca2+ entry into cells (Figure 5, D–F). These observations, together with the finding that ELA peptides were able to regulate mTORC1 activation, indicate that ELA and mut ELA play a key role in the repression of different pathways involved in the malignant phenotype (Figure 6).
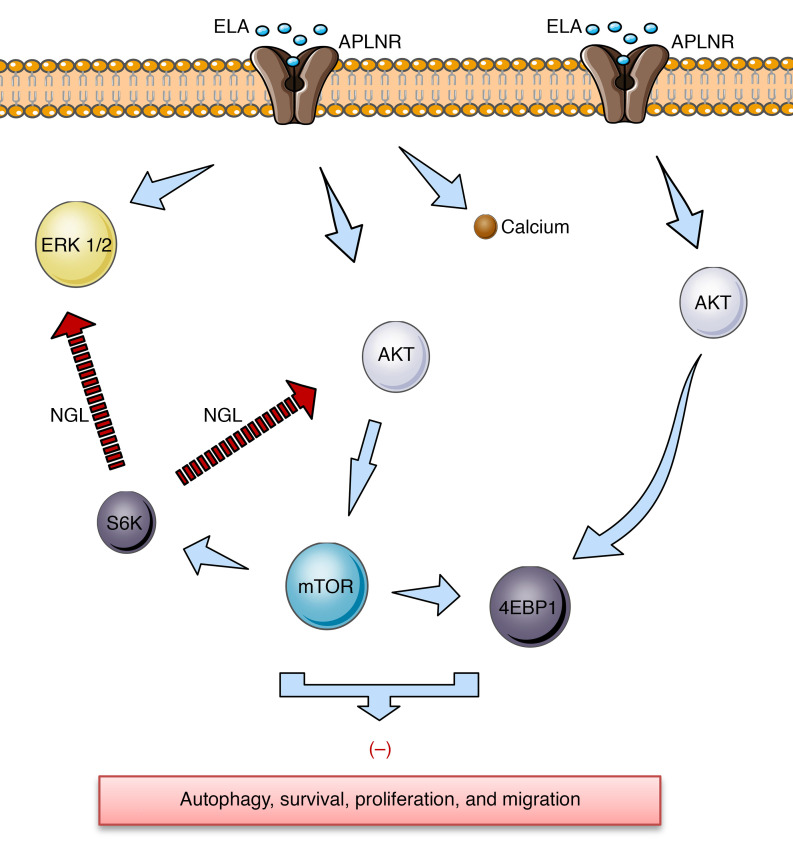
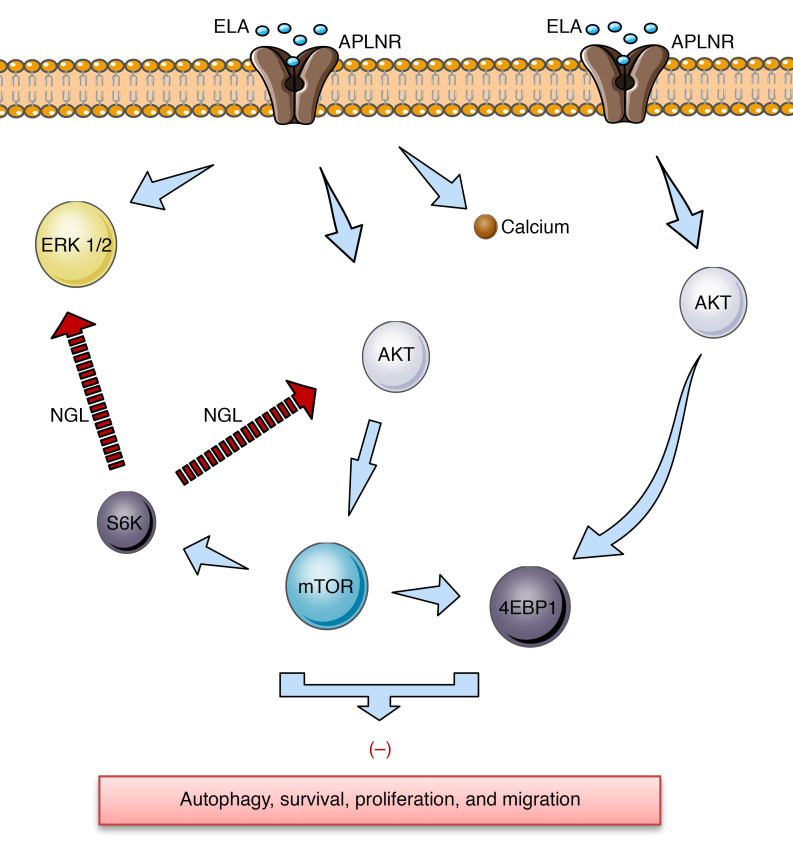
Figure 6. Schematic representation of proposed ELA-APLNR signaling in kidney cells.
ELA activates APLNR, which induces rapid and transit AKT and ERK activation that activates mTOR targets S6K and 4EBP1 and inhibits autophagy, cell survival, proliferation, and migration. Calcium (Ca2+) inhibition after ELA/APLNR interaction is also indicated. The negative feedback loop (NGL) from mTORC1 and S6K to AKT and ERK is shown.
Synergistic effect between sunitinib and mut ELA on tumor growth repression.
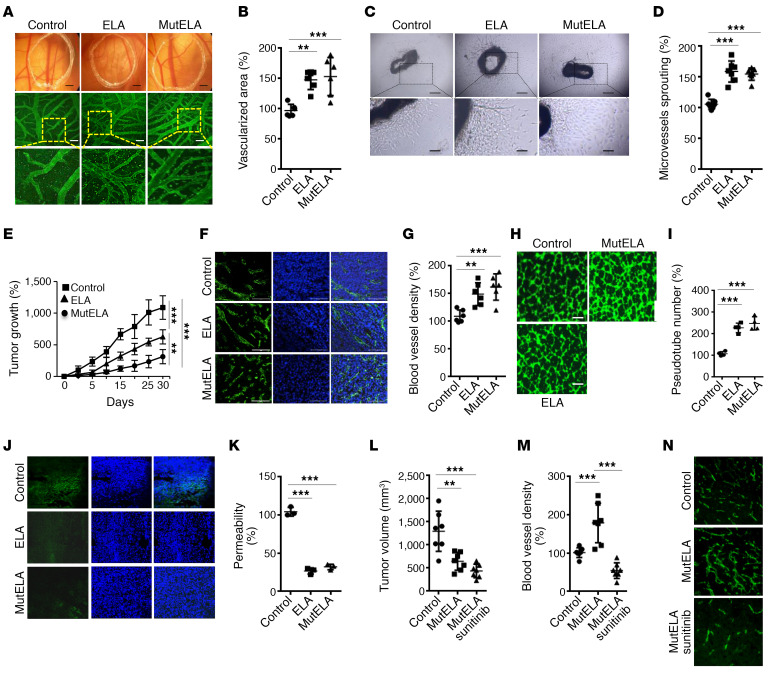
Previous studies have shown that ELA and APLNR interaction stimulates human umbilical vein endothelial cells (HUVECs) to form tube-like structures in vitro in a mechanism different from APLN and APLNR interaction (23). Thus, we sought to evaluate these findings and expand our analysis to further evaluate ELA and mut ELA in the context of tumor angiogenesis. We first used the chicken chorioallantoic membrane (CAM) assay to determine the effects of ELA32 and mut ELA32 peptides. As shown in Figure 7, A and B, the incorporation of ELA32 or mut ELA32 compared with vehicle alone resulted in induced growth of blood vessels. Similarly, using the aortic ring assay (Figure 7, C and D), we found that ELA32 and mut ELA32 peptides increased microvessel outgrowth compared with untreated aortic rings. Finally, to determine the direct effect of ELA and mut ELA on tumor angiogenesis, we compared the ability of ELA- and mut ELA–expressing cells to induce vessel formation after their subcutaneous implantation in mice. Although ELA- and mut ELA–expressing cells repressed tumor growth (with a potent effect observed in mut ELA–expressing cells) (Figure 7E), the developed tumors were characterized by an increase in the number of blood vessels, as assessed by CD31 staining quantification (Figure 7, F and G). To confirm that ELA and mut ELA derived from tumor cells inoculated in mice are directly involved in the observed increased tumor angiogenesis, we evaluated the ability of media derived from ELA- and mut ELA–expressing Renca cells to induce angiogenesis in vitro using the capillary-like tube formation assay. HUVECs were seeded and incubated with media derived from Renca control cells or cells expressing ELA or mut ELA for 18 hours. We observed that media derived from ELA- and mut ELA–expressing cells significantly induced HUVECs to form capillary-like tubes compared with control conditioned media (Figure 7, H and I). We next evaluated whether the observed antitumorigenic effect of ELA peptides is linked to tumor vessel normalization, a process previously reported to reduce abnormalities in the tumor microenvironment and ultimately tumor progression repression (24). The leakiness of blood vessels in control cells and ELA- and mut ELA–expressing ACHN cell–derived tumors was determined after FITC-dextran perfusion using a confocal microscope. The tumor vascular network in control tumors showed extensive leakiness. In contrast, vascular permeability to FITC-dextran was significantly reduced in ELA- and mut ELA–expressing cell-derived tumors (Figure 7J). To repress the ability of ELA-mediated tumor angiogenesis in mice, we next used the angiogenesis inhibitor sunitinib (Sutent). We found that administration of sunitinib in mice (40 mg/kg) inhibited more tumor growth (Figure 7L). The use of anti-CD31 immunostaining to determine blood vessel density within Renca tumors revealed that in contrast to mut ELA–derived tumors that displayed reduced tumor growth associated with high vessel density (Figure 7, L–N), in the presence of sunitinib the developed tumors exhibited markedly lower vessel densities and tumor growth. Therefore, the combination of mut ELA and sunitinib appeared to be highly effective.
Figure 7. Synergistic effect between mut ELA and sunitinib on tumor growth and angiogenesis repression.
(A) Capillaries of vehicle (control) and ELA peptide– and mut ELA peptide–treated CAMs (100 nM). (B) Quantification of vascularized area relative to control untreated CAMs assigned 100% (n = 6–8). Scale bar represents 2.5 mm for the upper panel and 200 μm for the lower panel. (C) Representative images of untreated (control) and ELA peptide– and mut ELA peptide–treated aortic rings (100 nM). Scale bar indicates 250 μm for the upper panel and 50 μm for the lower panel. (D) Quantification of aortic ring vascular sprout surface per aortic ring relative to control untreated aorta (100%) (n = 6–7). (E) Progression of subcutaneous tumors induced by Renca control cells and ELA- or mut ELA– expressing cells in syngeneic BALB/c mice (n = 7 per group). (F) Developed tumors shown in E were analyzed for angiogenesis using an anti-mouse CD31 monoclonal antibody. (G) Quantification of vascularized area relative to control tumors assigned 100% (n = 6). (H) Analyses of tube-like structure formation in the presence of media derived from Renca control cells and ELA- and mut ELA–expressing cells. (I) Quantification of tube formation via determining the number of cell cluster connections (tube number, n = 6). Scale bar: 200 μm. (J) Confocal images of sections from mice with developed ACHN tumors injected with FITC-dextran. (K) Quantification of vascular permeability to FITC-dextran relative to control tumors assigned 100% (n = 3). (L) Mice were injected subcutaneously with control Renca cells or the same cells stably expressing mut ELA (2 × 105 cells) (n = 7 per group). One week after tumor cell injection, vehicle or sunitinib was administered every 2 days by oral gavage. Tumor size (L) and angiogenesis (M and N) were determined at the end of the experiments. All results shown are representative of at least 3 independent experiments. The mean ± SEM values are shown. One- or 2-way ANOVA with Tukey’s multiple-comparisons test were used to analyze the data. **P < 0.01, ***P < 0.001.
mut ELA rapidly induces human APLNR recycling on the cell surface.
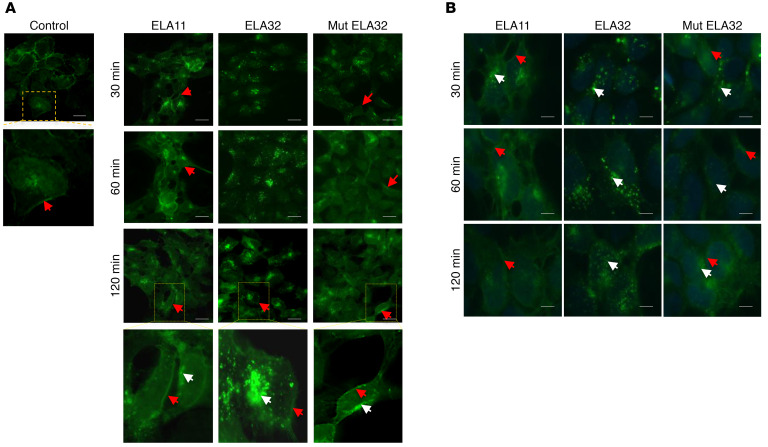
To investigate the importance of ELA precursor cleavage by the proprotein convertases during APLNR internalization, we used HEK-APLNR cells overexpressing GFP-APLNR fusion protein and examined APLNR intracellular localization in response to ELA11, ELA32, and mut ELA32 peptide treatment. In the absence of these peptides, APLNR localized mainly to the cell surface, as visualized by GFP staining (Figure 8A). Cell stimulation with ELA peptides for 30–120 minutes induced the formation of large intracellular vesicles clearly visible in the cytoplasm of ELA-32– and mut ELA32–treated cells. Only small vesicles were observed in ELA11-treated cells. In ELA32-treated cells, the majority of APLNR was internalized, compared with ELA11-treated cells and mut ELA32–treated cells (with reduced amount), which still showed APLNR on their cell surface (Figure 8A). To characterize the behavior of the APLNR vesicles after internalization, cells were treated with ELA peptides, and GFP location was analyzed at different time points after the peptide washout (Figure 8B). In cells treated with ELA11, the APLNR vesicles recycled rapidly and fused with the plasma membrane after 30 minutes of the washout. After 1 hour, the majority of the APLNR vesicles were localized to the cell membrane. In ELA32-treated cells, a larger part of the vesicles remained in the cytoplasm after 120 minutes of the washout. In ELA32 mut–treated cells, APLNR vesicles returned to the cell surface 1 hour after the washout; a lower amount was retained in the cytoplasm compared with ELA32-treated cells (Figure 8B). These findings suggest that the internalization of APLNR was less efficient with ELA11 as a ligand, and mut ELA32 recycled relatively faster than ELA32 to the cell surface (Figure 8B).
Figure 8. Effect of ELA peptides on APLNR recycling.
(A) HEK293 cells expressing APLNR-EGFP were analyzed under confocal microscopy after activation with ELA peptides (100 nM) at indicated time periods. Scale bar: 20 μm. White arrows indicate internalized receptors and red arrows indicate cell surface receptors. (B) HEK-APLNR cells were treated with ELA peptides and analyzed 30 minutes, 60 minutes, or 120 minutes after washout of the peptides. Scale bar: 50 μm. All experiments were repeated 3 times.
Docking computations of ELA11, ELA32, and mut ELA32 peptides against APLNR.
To explore the possible specific APLNR interaction with ELA11, ELA32, and mut ELA, docking simulations were performed using various in silico approaches. Among the different 3D structures of the peptide-receptor complex generated in silico, the most favorable one in terms of predicted binding energy values and structural analysis of the molecular interactions had the C-terminal end of the peptides inserted in the canonical ligand-binding pocket of the receptor (Figure 9, A and B). The overall orientation of these 3 peptides in this region of the receptor is similar to the experimental APLN mimetic peptide-receptor x-ray structure (25). However, for the N-terminal region of the ELA32 (or mut ELA32) peptide, 2 main positions were initially identified after docking computations. In the APLNR experimental structure cocrystallized with a peptide, 2 main grooves were observed and were referred to as groove 1 and groove 2 (Figure 9B) (25). The C-terminal region of the APLN-like peptide was found deeply inserted into site 1 and protruding outside the receptor, making several favorable interactions with site 2 (Figure 9, A and B). The docking results indicated equally favorable contacts for the entire N-terminal region of the ELA32 (or mut ELA32) peptide with the areas of groove 1 or with site 2 when the peptide was linear (i.e., the internal disulfide bond was not created). Thus, the N-terminal ELA32 (or mut ELA32) peptide residues were either in groove 1 for some models or into site 2 for some other predicted complexes. This result is understandable because there are several positively charged residues in this region of the ELA32 peptide and the receptor has many negatively charged amino acids in both regions. Moreover, the predicted interaction energy values were relatively similar on both sides. However, when the ELA32 disulfide bond was created and the peptide redocked with Surflex-Dock or the AutoDock suite, favorable interactions with both site 2 and with groove 1 were predicted. This orientation of the ELA32 and mut ELA32 peptides would seem reasonable because the peptide remains above the membrane while electrostatically interacting with polar phospholipid head groups (i.e., the positively charged regions of several positively charged residues of the ELA32 peptide could be at about 5–6 Å of the polar phospholipid heads).
Figure 9. Docking peptides into APLNR and PWR.
(A) ELA peptides were docked into the experimental APLNR structure. Colored spheres show the predicted position of the membrane. Two conserved disulfide bonds present in this family of receptors are shown in orange. The receptor (yellow) was shown to have 2 main grooves, 1 (red) and 2 (blue). Overall, the best predicted binding poses for the ELA peptide (blue tube) had the C-terminal region inserted in the canonical binding pocket region (site 1), while the N-terminal region seemed to fold back inside the groove 1 area. A disulfide bond in the ELA peptide is shown in magenta; this bond was not present in the apelin-like peptide cocrystallized with the APLNR receptor. ELA Arg residues that were mutated to serine in the present study to avoid cleavages by furin are shown in cyan. (B) View from the extracellular side. Proposed position of the ELA peptides at the surface of the receptor, top, ELA11, bottom, and mut ELA32. The receptor is shown as a solid surface and the peptide as a ribbon. Site 1 represents the canonical binding pocket in this family of receptors. The ELA11 peptide was essentially inserted into the receptor site 1; the longer ELA32 and mut ELA32 peptides protruded more outside the receptor and made several favorable interactions with residues of site 2 and folded back in the region of groove 1. One conserved disulfide bond in the receptor is shown (orange) to facilitate the reading of the figure. The mutated residues in ELA32 are shown in cyan. These residues point away from the receptor. (C) Schematic representation of APLNR with perpendicular p-polarized light (p-pol) to indicate vertical elongation of the receptor and parallel s-polarized light (s-pol) for horizontal elongation of the receptor on the cell membrane. (D–F) Binding curve for ELA11 (D), ELA32 (E), and mut ELA32 (F) interaction with APLNR in HEK-APLNR. D, E, and F are representative of 3 independent experiments, and graphs with mean ± SEM are shown in Supplemental Figure 7 (n = 3). Unpaired t tests were used to analyze the data. **P < 0.01, ***P < 0.001.
APLNR affinity and ELA peptide–induced conformational changes of APLNR.
To assess and compare the affinity and ligand-induced conformational changes of APLNR in response to ELA11, ELA32, and mut ELA32, we performed plasmon waveguide resonance (PWR). The PWR sensor was pretreated with polylysine to allow cell fragments to be captured by electrostatic interaction between polylysine-positive charges and the negative charges provided by the glycosaminoglycans in the cell membrane surface. Cell fragments were captured by pressing the polylysine-pretreated sensor against cells growing in 6-well plates. Mass gain after cell fragment deposition was monitored by PWR spectral changes that were reflected in shifts in the resonance angle to higher angles with both the p-polarized light (perpendicular to the sensor surface) and s-polarized light (parallel to the sensor surface) (Figure 9C). After cell membrane fragment capture, the ligand was added in an incremental fashion, and spectral changes were monitored for both polarizations with time. After ligand affinity analyses, the hyperbolic saturating curves revealed that ELA11 possessed the lowest affinity (Figure 9D) (highest KD of about 90 nM) for the cell membrane fragments compared with ELA32 (Figure 9E) and mut ELA32 (Figure 9F) affinity (KD of about 3 nM and 0.8 nM, respectively). Statistical analyses revealed that although the difference between ELA11 and ELA32 or mut ELA32 was highly significant, the difference between ELA32 and mut ELA32 was not significant (Supplemental Figure 8A). To investigate whether ELA32 and mut ELA32 could displace ELA11 separately, cell membrane fragments captured in the PWR sensor were first treated with 200 nM of ELA11, and then ELA32 or mut ELA32 was added at various concentrations. ELA32 and mut ELA32 were able to bind APLNR with a similar affinity despite the preincubation with ELA11 (Supplemental Figure 8B). These findings indicate that either the ELA32 and mut ELA32 binding site was independent of the ELA11 site (nonallosteric) or these peptides were able to bind to the same site, but because of the considerably higher affinity of ELA32 and mut ELA32 compared with ELA11, they displaced ELA11. By following the spectral changes induced by ligand binding with the 2 polarizations (p and s) (Figure 9C), information on the magnitude of the receptor conformational changes was obtained along the receptor axes. ELA11 induced very different receptor conformational changes in APLNR compared with ELA32 and mut ELA32 (Supplemental Figure 8C). ELA11 induced conformational changes that were of smaller magnitude for both polarizations and that were more isotropic (magnitude of the signal for p- and s- were similar) than those of ELA32 and mut ELA32. The latter induced similar conformation of the receptor (Supplemental Figure 8D). Preincubation of cell fragments with ELA11 did not alter the receptor conformational changes mediated by ELA32 and mut ELA32. The use of cell membranes derived from HEK cells lacking APLNR under the same conditions failed to induce any significant receptor conformational changes (Supplemental Figure 8E).
Discussion
Our studies identified ELA as an important positive regulator of mTORC1 that induced tumor repression. The repressed ELA expression level in the analyzed renal cancer patient specimens and public data sets supported the relevance of this mechanism in kidney cancers. TCGA RNA sequencing data sets revealed that ELA expression was systematically downregulated in the main kidney cancer subtypes and negatively correlated with Ki-67. We observed that in kidney cancer cells, ELA induced apoptosis and repressed cell proliferation and migration. These mechanisms involved the activation of downstream mTORC1 signal pathways and inhibition of autophagy, as demonstrated by ELA mediating the activation of 2 bona fide downstream targets of mTORC1, namely S6K and 4EBP1, and the accumulation of p62. In agreement with these findings, inhibition of mTORC1 by rapamycin was recently shown to promote cell survival upon amino acid starvation (26), elucidating, at least in part, the inefficacy of mTORC1 inhibitors observed in several treated patients. Indeed, aberrant activation of the mTORC1 pathway was found in up to 80% of human cancers, and repression of this pathway was initially proposed in various clinical studies as a pertinent strategy for cancer treatments (27). However, inhibition of mTORC1 activation by rapamycin analogs showed modest effects in a clinical setting (27). While targeting mTORC1 in patients seems to limit tumor growth, it increases resistance to apoptosis, leading to an enhanced resistance to cancer therapy. In addition, genetic inactivation of p62 was recently found to promote a more aggressive phenotype in tumor-bearing mice as a result of the activation of a wide range of genes involved in neoplasia (28). Increasing evidence suggests that the mTORC1/S6K axis activation mediates potent negative feedback loops that control upstream signaling through various tyrosine kinase receptors in both normal and cancer cells (29). Suppression of these feedback loops by inhibitors of mTORC1/S6K causes compensatory overactivation of upstream signaling nodes, including PI3K, AKT, and ERK, that potentially oppose the antiproliferative effects of these inhibitors, leading to drug resistance (29, 30). Therefore, removal of negative feedback by anticancer drugs is a major obstacle that limits the achievement of various targeted therapies (30). In our model, ELA induced AKT and ERK activation followed by their rapid repression, suggesting the implication of mTORC1/S6K activation in these processes. Similarly, calcium is considered as an activator of autophagy (31) and regulator of gene transcription involved in cell proliferation, migration, and resistance to apoptosis. AKT and ERK seemed to act downstream of SOCE, thereby reducing SOCE; ELA may also participate in the reduced activity of ERK and AKT through this pathway.
Although many proteins are processed by the proprotein convertases, the cleavage of some is not required for their functions (32). In contrast, the cleavage of other substrates was found to be critical for the mediation of their functions (19, 33–39). Our findings demonstrated that the blockade of proELA processing (mut ELA) seemed to increase the biological activity of ELA, probably because of its high stability and/or its affinity to APLNR. Structural analysis was performed on the previously reported crystal structure of the APLNR (25). This class of GPCR has 2 conserved disulfide bonds involving C19 and C281 and C102 and C181 in the extracellular region. The apelin-like peptide cocrystallized with the receptor was found to be deeply inserted into site 1 and to protrude outside the receptor in the area of site 2. The ELA11 peptide docked with favorable binding scores in an orientation similar to the apelin-like peptide. Furthermore, part of the N-terminal sequence of the ELA32 peptide fitted well in the site 2 region of the receptor, similar to the experimental apelin-receptor structure, whereas the longer ELA32 (or mut ELA32) N-terminal part was likely to fold back in the region of groove 1. With this orientation, the ELA peptides, like the APLN-like peptide, would be relatively distant from the N-glycosylated APLNR. Several additional hydrophobic and electrostatic contacts were noticed when comparing ELA32 and ELA11. This is consistent with the higher affinity of ELA32 versus ELA11 previously reported (40) and was further validated here using PWR analysis that also identified higher affinity of mut ELA32 for APLNR. The 2 furin sites of ELA32 (R31/R32 and R42/R43) did not seem to have strong interactions with the receptor and bent away from the protein molecular surface. Therefore, they could be cleaved by furin before and/or during ELA32-APLNR interaction. Although a substitution of arginine by serine residues in the mut ELA32 did not affect the affinity (i.e., the Arg in the WT peptide should not significantly contact the receptor as predicted here), it prevented the conversion of ELA32 to ELA11 that may promote a longer interaction of the unprocessed ELA32 (mut ELA) with APLNR. In addition, after APLNR activation by ELA peptides, mut ELA seemed to mediate rapid APLNR internalization and recycling at the cell surface, thereby quickly permitting available cell surface receptors for new activation.
Although reduced tumor size and tumor cell apoptosis were induced by ELA and mut ELA, there was an increase in tumor neovascularization in ELA and mut ELA tumors. The reasons for the opposing antitumorigenic and proangiogenic functions of ELA/mut ELA are not presently clear, but several mechanisms may be postulated. The ability of ELA to mediate vascular normalization, as revealed by the reduced vascular permeability observed in the ELA-expressing cell-derived tumors, may be a contributing factor. Indeed, attenuation of vessel permeability in tumors was reported to mediate the improvement in the tumor microenvironment and the delivery and efficacy of exogenously administered therapeutics (24). Accordingly, in our study, we also found that the antitumorigenic effect of ELA was more potent in the presence of sunitinib. The ability of ELA/mut ELA to repress AKT and ERK activation may be a second causal factor. Indeed, various components of the PI3K signaling pathway can exert either pro- or antiangiogenic effects, depending on the experimental conditions. One example is the role of endothelial AKT1, the main AKT isoform found in endothelial cells: deficiency of AKT1 in mice showed increased sensitivity to VEGF, and induced tumors grew more rapidly with increased neovascularization (41). This suggests that repression of endothelial AKT1 activity increases tumor angiogenesis as confirmed by the use of PI3K inhibitor (42). On the other hand, treatment of mice bearing tumors with rapamycin to repress AKT1 in endothelial cells resulted in normalization of vasculature and marked reduction in tumor angiogenesis (43, 44), demonstrating the irregularity in vascular response to AKT1 inhibition (45). Regardless, these studies, in addition to ours, strongly suggest that activation of mTORC1 and inhibition of the PI3K/AKT pathway, in addition to reduced vascular permeability by ELA, is a viable potential therapeutic approach. However, we also observed that targeting tumor angiogenesis using sunitinib constituted an adjuvant option to enhance the effect of ELA on tumor repression. In summary, our data point to a cell-autonomous tumor-suppressive function for ELA signaling through mTORC1 in synergy with other tumor suppressors to inhibit growth in the main renal cancer subtypes. Further studies should be aimed at resolving the question of whether the tumor suppressor function of ELA can be exploited to target specific or all kidney tumor subtypes.
Methods
Cell culture.
The BALB/c syngeneic kidney carcinoma Renca cell line, the HEK293 cells, the human renal carcinoma Caki-2 and ACHN cell lines (purchased from ATCC), and the human U2OS cells stably expressing GFP-LC3 (provided by Eyal Gottlieb, Cancer Research UK, Glasgow, United Kingdom) were grown at 37°C in 5% CO2 in DMEM (Life Technologies, Thermo Fisher Scientific) supplemented with 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin. All cell lines were determined to be free of Mycoplasma.
Constructs and cell infections.
Lentiviral vector construction and production were performed by the Vect’UB facility of the TMB-Core (Bordeaux University). The human ELA cDNA was subcloned into a multicistronic self-inactivating lentiviral vector containing a tdTomato reporter gene (pRRLsin-MND-hPGK-tdTomato-WPRE) under the control of the myeloproliferative sarcoma virus enhancer. Mutagenesis was carried out using PCR to generate the mut ELA cDNA (where the 2 cleavage sites of ELA precursor peptide, namely R31/R32 and R42/R43, were replaced by S31/S32 and S42/S43). The human APLNR cDNA was cloned into the pEGFP-N1 vector to generate the APLNR-pEGFP-N1 vector. All constructs were verified by sequencing. Renca, ACHN, and Caki-2 (5 × 104 cells/well) cell lines were seeded in 24-well plates with polybrene at 8 μg/mL. Lentiviruses coding for WT ELA, mut ELA, or only tdTomato were added to the medium at MOI 5 or MOI 10. Cell infection rates were observed 72 hours later using a fluorescence microscope. HEK293 cells were transfected with pEGFP-N1 empty vector or vector containing APLNR cDNA using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions. Stable HEK293 cells expressing APLNR-GFP (HEK-APLNR) were selected using G418 (Gibco, Thermo Fisher Scientific). In other experiments, HEK-APLNR cells stably expressing empty vector or containing ELA or mut ELA cDNA were generated using lentiviral vector construction, as described above.
Patient samples.
For frozen tissues, renal tumors (22 nonidentifiable ccRCC patients with different grades) and their corresponding normal tissues were obtained after resection. After surgery, tissue specimens were immediately transferred onto ice and snap-frozen in liquid nitrogen until used for further analysis.
Animal models.
Mice were housed in ventilated carousel racks and provided sterile food and drinking water. Control Renca or ACHN cells or the same cells stably expressing ELA or mut ELA were injected into the flank of female BALB/c and nude mice (Charles Rivers Laboratories), respectively. Tumor size was determined by collecting length and width measurements and calculating the tumor volume (mm3) as (tumor length × [tumor width]2) × 0.52. In other experiments, control Renca cells and ELA-expressing Renca cells were implanted into the subcapsular space of BALB/c mice. When mice were euthanized, tumors from each animal were collected and used for RNA and/or histopathological studies when required.
RNA extraction and real-time PCR.
Total RNA was extracted with an RNA isolation kit (MACHEREY-NAGEL) including DNase treatment (QIAGEN), according to the manufacturer’s instructions. The RNA quality for human samples was checked using the Agilent RNA 6000 Nano Kit according to the manufacturer’s instructions, and RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Thermo Fisher Scientific). The latter was used for real-time PCR in the presence of specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems, Thermo Fisher Scientific) (46). The quantitative PCR data were acquired with the StepOnePlus Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). The expression levels were normalized to mouse or human GAPDH.
Immunostaining and confocal microscopy.
Paraffin-embedded sections from kidney cancers and patients’ corresponding noncancerous tissues were deparaffinized and rehydrated. Antigens were retrieved by boiling sections in 10 mM citrate buffer (pH 6.0) for 20 minutes. Sections were blocked with 5% milk in PBS containing 0.5% Triton X-100, stained with unconjugated antibodies for 15 minutes at 4°C, and washed twice with PBS before incubation with fluorochrome-associated secondary antibodies. In other experiments, double-label immunostaining was performed. Frozen sections derived from mice injected with control Renca cells, ELA-expressing cells, or mut ELA–expressing cells were stained with unconjugated antibodies and fluorochrome-associated secondary antibodies. GFP-LC3–expressing U2OS cell line (2 × 105 cells) was grown on coverslips with the respective treatments for 72 hours. Thereafter, cells were fixed with 4% paraformaldehyde in PBS for 30 minutes at room temperature. All samples were mounted with ProLong mountant containing DAPI (Invitrogen, Thermo Fisher Scientific), and confocal immunofluorescence images were taken using the inverted microscope Nikon C2si Eclipse Ti-S with NIS-ElementsAR software (Nikon Instruments Europe B.V.).
Immunoblotting.
Western blot analysis was performed as previously described (47). Briefly, after their respective treatments, cells were lysed at 4°C with lysis buffer (20 mM HEPES at pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 6 mM β-octylglucoside, 10 μg/mL aprotinin, 20 μM leupeptin, 1 mM NaF, 1 mM DTT, and 100 μM sodium orthovanadate). In other experiments, ELA or mut ELA peptides were incubated alone or with recombinant furin for indicated time periods at 37°C in an enzymatic buffer of 25 mM Tris, 25 mM 2-(N-Morpholino)ethanesulfonic acid, and 2.5 mM CaCl2, pH 7.5 (46). Lysates or digested peptides were loaded on 10% SDS-PAGE gel and transferred by electroporation to nitrocellulose membrane. Membranes were incubated with indicated primary antibodies that were revealed by HRP-conjugated secondary antibodies (Amersham Pharmacia Biotech) and enhanced chemiluminescence (ECL Plus, Amersham Pharmacia Biotech) according to the manufacturer’s instructions. After imaging, band quantification was performed using ImageJ software (NIH) and protein levels were normalized to actin.
Vascular permeability assay.
Mice with developed tumors were injected with 0.5 mg of FITC-dextran in 200 μL of PBS via intravenous tail vein injection (7). Mice were euthanized 30 minutes after injection, and tumor tissues were submerged in 4% paraformaldehyde for 24 hours at 4°C. Samples were mounted with ProLong mountant containing DAPI and examined by confocal microscopy.
Calcium mobilization measurement.
HEK-APLNR cells were loaded with fura-2 and resuspended in HEPES-buffered saline. At the time of the experiment, 250 μM EGTA was added. Cells were preincubated for 8 minutes in the absence or presence of 100 nM of ELA or mut ELA and then stimulated with 1 μM thapsigargin to deplete ER Ca2+ stores. CaCl2 (1 mM) was added to the medium to initiate Ca2+ entry, and measurement used a Cary Eclipse Spectrophotometer (Varian Ltd.) with excitation wavelengths of 340 nm and 380 nm and emission at 505 nm.
Proliferation assay.
A proliferation assay for indicated cells was performed using the IncuCyte live-cell microscopy incubator (Essen Bioscience). After their respective treatments, cells (2 × 105) were detached with trypsin/EDTA and placed in the IncuCyte incubator, and phase-contrast images were taken at regular intervals over 48 hours. Results were calculated by the IncuCyte software and presented as confluence relative to time 0. Images were taken with a ×4 objective. Four images were taken from each well, and each condition had more than 6 wells.
Apoptosis.
Apoptosis was measured by means of flow cytometry after staining with annexin V using the Annexin V-FITC Apoptosis Detection Kit (Abcam), following the manufacturer’s instructions. Cells were analyzed by flow cytometry (BD Accuri C6) (46).
Tube-like formation assay.
HUVECs were seeded at a density of 25 × 103 cells per well onto Chamber Slides (Lab-Tek, Thermo Fisher Scientific) previously coated at 37°C with Geltrex (Gibco, Thermo Fisher Scientific) for 30 minutes to allow polymerization. For the tube formation assay, media derived from control cells, ELA-expressing cells, or mut ELA–expressing cells cultured in serum-free media for 24 hours were added directly after seeding the HUVECs. Tube formation was documented by Nikon epifluorescence microscopy.
Wound-healing assay.
Cells were seeded in 6-well plates and allowed to grow until they reached 90% confluence (2 × 106 cells per well of each cell line). The cell monolayer was scratched with a sterile micropipette tip, and the wound region was allowed to heal by cell migration. The area that remained clear of cells after 8 and 24 hours was quantified with ImageJ and compared with the area of the wound at time 0.
Aortic ring assay.
Male 6-week-old mice were anesthetized with isoflurane for 5 minutes before being killed. The descending aorta was isolated, cleared of adventitia, and placed in serum-free Opti-MEM (Gibco, Thermo Fisher Scientific) with 1× antibiotic/antimycotic (Gibco, Thermo Fisher Scientific, 100× stock solution). Aortas were sectioned into approximately 25 rings of 0.5-mm thickness and placed in fresh serum-free Opti-MEM with 1× antibiotic/antimycotic for 1 hour at 37°C. ELA and mut ELA were diluted to 100 nM in Opti-MEM and added to rings for overnight incubation at 37°C. Rings were washed in PBS and embedded in 2.5 mg/mL fibrin supplemented with 37.5 μL/mL of 1 mg/mL aprotinin/mL fibrinogen. EBM-2 medium (Lonza) supplemented with 0.2% FCS and 25 ng/mL VEGF-A was added to each well. Medium was changed on day 4, then every other day.
CAM angiogenesis assay.
Fertilized eggs were allowed to mature ex ovo. A small incision was made in the eggshell on day 3 of development, and 7 days later ELA and mut ELA peptides (100 nM) were added to the CAM tissue. On day 17, the CAMs were excised and fixed at room temperature for 30 minutes using 4% paraformaldehyde. Immunohistology in toto was performed for vessel staining. Images of CAM vessels were acquired using a Zeiss Axiophot epifluorescence microscope, and collected images were analyzed using ImageJ software.
Internalization assay.
HEK293A cells stably expressing human EGFP-APLNR fusion protein (HEK293-APLNR) were serum starved for 24 hours and treated for various time points with ELA11, ELA32, or mut ELA32 peptides at 100 nM. After formaldehyde treatment, cells were analyzed by fluorescence microscopy. For washout experiments, HEK293-APLNR cells were incubated with indicated peptides. Media were replaced and cells were imaged 30 minutes, 60 minutes, and 120 minutes after the washout. Confocal immunofluorescence images were taken using an inverted microscope (Nikon Instruments Europe B.V.).
TCGA database and data acquisition.
Expression data of ELA gene (APELA), APLN, APLNR, and other genes and the sample metadata were extracted from TGCA Pan-Cancer project through Xena (http://xena.ucsc.edu/). In brief, we used the RNA-Seq data quantified at the gene level with RSEM (48), normalized and log2(x + 1) transformed. Types of cancer (“primary disease”) without normal samples (“solid tissue normal”) were excluded. Expression of APELA, APLN, and APLNR was compared between normal and tumoral tissues in various cancers with unpaired t tests. To calculate Pearson correlations with associated P values, we analyzed the expression data between pairs of genes separately per each primary disease.
Docking computations and structural analyses.
ELA11, ELA32, and mut ELA peptides running from residues Q23 to P54 (or Q1 to P32 after removal of the signal peptide) as well as shorter versions of the peptide were generated in 3D with PEP-FOLD3 (49) using constraints from the APLNR receptor and with Surflex-Dock (50) and AutoDock (51) via the MTiOpenScreen server (52). Over 300 linear peptide structures were first generated in the presence of the receptor with PEP-FOLD3 using different parameters and peptide lengths. The best structures in terms of energetic parameters and after structural analysis of the molecular interactions were kept and compared with about 100 Surflex-Dock or AutoDock poses of the full-length peptide, ELA11, ELA32, and mut ELA, linear peptide, and peptide with the internal disulfide bond formed. The crystal structure of the apelin receptor in complex with a 17-aa apelin mimetic peptide was taken from the Protein Data Bank (53) (PDB entry 5VBL) (25). Some missing side chains and residues were rebuilt using the SWISS-MODEL server (54) and the resulting receptor structure energy minimized with MMTK (http://dirac.cnrs-orleans.fr/MMTK.html). Structural analysis was performed with Chimera and PyMOL (https://www.schrodinger.com). Prediction of the orientation of the receptor structure with respect to the lipid bilayer was done using the PPM server (55). Analysis of the protonation state for the titratable residues of the receptor was performed with the PCE server (56).
PWR.
PWR was used to follow receptor conformational changes upon ligand addition to HEK-APLNR cell membrane fragments overexpressing the apelin receptor immobilized in the sensor surface. PWR measurements were performed in a homemade instrument functioning at a fixed wavelength of 632 nm and variable incident angle with an angular resolution of about 0.5 millidegrees. The polarization angle of the incident light was placed at 45° to allow both p-polarized (parallel to the incident light and perpendicular to the sensor surface) and s-polarized (perpendicular to the incident light and parallel to the sensor surface) light resonances to be obtained within a single angular scan. The sensor consisted of a BK-7 prism coated with silver and silica to support waveguide. All measurements were performed at 22°C.
Cell fragment immobilization in the PWR sensor and ligand titration.
Adhesion of cell fragments on the PWR sensor (silica outer layer) was performed as previously reported (57). Briefly, the sensor silica surface was activated by plasma cleaner for 2 minutes (Diener) and incubated with polylysine solution (0.1 mg/mL) for 40 minutes. Cells grown to less than 50% confluence were covered with water to induce osmotic swelling, and the glass coverslip of the sensor was placed directly on top of cells. Pressure was applied for about 2 minutes on the glass slide or prism to induce cell rupture and capture of cell fragments. After that, they were removed by ripping off cell fragments containing the upper membrane specifically. The PWR cell sample (a Teflon block with a volume capacity of 250 μL) was placed in contact with the sensor containing the immobilized cell fragments and filled with PBS. After cell fragment deposition, there were positive shifts in the resonance minimum position that were correlated with the total mass gain occurring. We observed spectral shifts that correlated with those observed for the deposition of the lipid model membrane. In the case presented here, there were areas of the sensor covered with cell membranes and others uncovered; the PWR signal takes into account both covered and uncovered areas as the laser spot size is about 0.5 mm in diameter. At the same time, covered areas included both lipids and proteins and so possessed a higher mass than that of a pure lipid membrane. After stabilization of the signal (no changes in the resonance minimum position with time), the ligand was added in an incremental fashion to this chamber and spectral shifts followed with time. Ligand affinity to the receptor present in the cell membrane fragments was calculated by plotting the shifts in the resonance minimum position as a function of ligand concentration and fitting using a hyperbolic function that described ligand binding to a protein (single site) (GraphPad Prism). From one experiment to another, the amount in mass of cell membrane fragments and therefore receptor quantities varied, which was reflected in the magnitude of the ligand-induced spectral changes. Thus, the data were normalized relative to the spectral shifts observed due to cell fragment deposition.
Reagents and antibodies.
Information on all antibodies and reagents used in this study is in the Supplemental Methods.
Statistics.
Data are presented as the mean ± SEM. A 2-tailed t test and 1- or 2-way ANOVA with Tukey’s multiple comparisons test were used to analyze the data. The Pearson coefficient was calculated to determine the correlation between the normally distributed ELA and protein expressions in human tumors. The statistical significance level is illustrated with P values. A P value of less than 0.05 was considered statistically significant.
Study approval.
All tumor specimens collected were obtained from frozen and FFPE tissues with the approval of the relevant ethics committee (Bergonié Institute, Bordeaux, France). Informed written consent was obtained from all patients. All in vivo experiments were approved by the Animal Housing and Experiment Board of the University of Bordeaux and performed in compliance with the French guidelines for experimental animal studies.
Author contributions
AMK, GS, and SE conceived the study; AMK and GS designed the methodology; FS, CB, KH, JD, IA, MD, AM, ME, JLH, JJL, JAR, IS, MT, RVD, MN, BOV, and GS investigated; AMK, GS, and SE wrote the original draft; and AMK, GS, and SE reviewed and edited the draft.
Supplementary Material
Acknowledgments
This work was supported by INSERM, SIRIC Brio, Region Nouvelle Aquitaine, and La Ligue Contre le Cancer.
Version 1. 06/09/2020
In-Press Preview
Version 2. 07/23/2020
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2020, American Society for Clinical Investigation.
Reference information: JCI Insight. 2020;5(14):e129070.https://doi.org/10.1172/jci.insight.129070.
Contributor Information
Fabienne Soulet, Email: fabienne.soulet@u-bordeaux.fr.
Clement Bodineau, Email: clement.bodineau@u-bordeaux.fr.
Katarzyna B. Hooks, Email: katarzyna.hooks@gmail.com.
Jean Descarpentrie, Email: jean.descarpentrie@inserm.fr.
Marielle Dubreuil, Email: marielle.dubreuil@hotmail.fr.
Amandine Mouchard, Email: mouchardam@msn.com.
Malaurie Eugenie, Email: malaurieeugenie@gmail.com.
Jean-Luc Hoepffner, Email: dr.jlhoepffner@wanadoo.fr.
Juan A. Rosado, Email: jarosado@unex.es.
Isabelle Soubeyran, Email: soubeyran_i@bergonie.org.
Mercedes Tomé, Email: mercedes.tome.montesinos@gmail.com.
Raúl V. Durán, Email: raul.duran@inserm.fr.
Macha Nikolski, Email: macha.nikolski@u-bordeaux.fr.
Bruno O. Villoutreix, Email: bruno.villoutreix@inserm.fr.
Serge Evrard, Email: S.Evrard@bordeaux.unicancer.fr.
Geraldine Siegfried, Email: geraldine.siegfried@inserm.fr.
Abdel-Majid Khatib, Email: majid.khatib@inserm.fr.
References
- 1.Bray F, et al. Cancer incidence in five continents: inclusion criteria, highlights from volume X and the global status of cancer registration. Int J Cancer. 2015;137(9):2060–2071. doi: 10.1002/ijc.29670. [DOI] [PubMed] [Google Scholar]
- 2.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7(5):245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Motzer RJ, Hutson TE, McCann L, Deen K, Choueiri TK. Overall survival in renal-cell carcinoma with pazopanib versus sunitinib. N Engl J Med. 2014;370(18):1769–1770. doi: 10.1056/NEJMc1400731. [DOI] [PubMed] [Google Scholar]
- 4.Sivanand S, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Sci Transl Med. 2012;4(137):137ra75. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagstaff J, et al. Treatment patterns and clinical outcomes in patients with renal cell carcinoma in the UK: insights from the RECCORD registry. Ann Oncol. 2016;27(1):159–165. doi: 10.1093/annonc/mdv504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman NA, Dupré DJ, Rainey JK. The apelin receptor: physiology, pathology, cell signalling, and ligand modulation of a peptide-activated class A GPCR. Biochem Cell Biol. 2014;92(6):431–440. doi: 10.1139/bcb-2014-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uribesalgo I, et al. Apelin inhibition prevents resistance and metastasis associated with anti-angiogenic therapy. EMBO Mol Med. 2019;11(8):e9266. doi: 10.15252/emmm.201809266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamada J, et al. Evaluation of novel cyclic analogues of apelin. Int J Mol Med. 2008;22(4):547–552. [PubMed] [Google Scholar]
- 9.Adam F, et al. Apelin: an antithrombotic factor that inhibits platelet function. Blood. 2016;127(7):908–920. doi: 10.1182/blood-2014-05-578781. [DOI] [PubMed] [Google Scholar]
- 10.Tatemoto K, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun. 1998;251(2):471–476. doi: 10.1006/bbrc.1998.9489. [DOI] [PubMed] [Google Scholar]
- 11.Chng SC, Ho L, Tian J, Reversade B. ELABELA: a hormone essential for heart development signals via the apelin receptor. Dev Cell. 2013;27(6):672–680. doi: 10.1016/j.devcel.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Pauli A, et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. 2014;343(6172):1248636. doi: 10.1126/science.1248636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siegfried G, Descarpentrie J, Evrard S, Khatib AM. Proprotein convertases: key players in inflammation-related malignancies and metastasis. Cancer Lett. 2020;473:50–61. doi: 10.1016/j.canlet.2019.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scamuffa N, et al. Selective inhibition of proprotein convertases represses the metastatic potential of human colorectal tumor cells. J Clin Invest. 2008;118(1):352–363. doi: 10.1172/JCI32040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Z, et al. The proprotein convertase furin is a pro-oncogenic driver in KRAS and BRAF driven colorectal cancer. Oncogene. 2020;39(17):3571–3587. doi: 10.1038/s41388-020-1238-z. [DOI] [PubMed] [Google Scholar]
- 16.Zeng XX, Wilm TP, Sepich DS, Solnica-Krezel L. Apelin and its receptor control heart field formation during zebrafish gastrulation. Dev Cell. 2007;12(3):391–402. doi: 10.1016/j.devcel.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 17.Ho L, et al. ELABELA deficiency promotes preeclampsia and cardiovascular malformations in mice. Science. 2017;357(6352):707–713. doi: 10.1126/science.aam6607. [DOI] [PubMed] [Google Scholar]
- 18.Scamuffa N, Calvo F, Chrétien M, Seidah NG, Khatib AM. Proprotein convertases: lessons from knockouts. FASEB J. 2006;20(12):1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- 19.Ma J, Evrard S, Badiola I, Siegfried G, Khatib AM. Regulation of the proprotein convertases expression and activity during regenerative angiogenesis: role of hypoxia-inducible factor (HIF) Eur J Cell Biol. 2017;96(5):457–468. doi: 10.1016/j.ejcb.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Lalou C, et al. Inhibition of the proprotein convertases represses the invasiveness of human primary melanoma cells with altered p53, CDKN2A and N-Ras genes. PLoS One. 2010;5(4):e9992. doi: 10.1371/journal.pone.0009992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashoor R, Yafawi R, Jessen B, Lu S. The contribution of lysosomotropism to autophagy perturbation. PLoS One. 2013;8(11):e82481. doi: 10.1371/journal.pone.0082481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, et al. Elabela-apelin receptor signaling pathway is functional in mammalian systems. Sci Rep. 2015;5:8170. doi: 10.1038/srep08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goel S, et al. Normalization of the vasculature for treatment of cancer and other diseases. Physiol Rev. 2011;91(3):1071–1121. doi: 10.1152/physrev.00038.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, et al. Structural basis for apelin control of the human apelin receptor. Structure. 2017;25(6):858–866.e4. doi: 10.1016/j.str.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Villar VH, et al. mTORC1 inhibition in cancer cells protects from glutaminolysis-mediated apoptosis during nutrient limitation. Nat Commun. 2017;8:14124. doi: 10.1038/ncomms14124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chiarini F, Evangelisti C, McCubrey JA, Martelli AM. Current treatment strategies for inhibiting mTOR in cancer. Trends Pharmacol Sci. 2015;36(2):124–135. doi: 10.1016/j.tips.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, et al. Adipocyte p62/SQSTM1 suppresses tumorigenesis through opposite regulations of metabolism in adipose tissue and tumor. Cancer Cell. 2018;33(4):770–784.e6. doi: 10.1016/j.ccell.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Efeyan A, Sabatini DM. mTOR and cancer: many loops in one pathway. Curr Opin Cell Biol. 2010;22(2):169–176. doi: 10.1016/j.ceb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer Discov. 2012;2(4):311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Decuypere JP, et al. mTOR-controlled autophagy requires intracellular Ca(2+) signaling. PLoS One. 2013;8(4):e61020. doi: 10.1371/journal.pone.0061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creemers JW, Khatib AM. Knock-out mouse models of proprotein convertases: unique functions or redundancy? Front Biosci. 2008;13:4960–4971. doi: 10.2741/3055. [DOI] [PubMed] [Google Scholar]
- 33.Siegfried G, et al. Regulation of the stepwise proteolytic cleavage and secretion of PDGF-B by the proprotein convertases. Oncogene. 2005;24(46):6925–6935. doi: 10.1038/sj.onc.1208838. [DOI] [PubMed] [Google Scholar]
- 34.Siegfried G, et al. The secretory proprotein convertases furin, PC5, and PC7 activate VEGF-C to induce tumorigenesis. J Clin Invest. 2003;111(11):1723–1732. doi: 10.1172/JCI17220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomé M, et al. Inactivation of proprotein convertases in T cells inhibits PD-1 expression and creates a favorable immune microenvironment in colorectal cancer. Cancer Res. 2019;79(19):5008–5021. doi: 10.1158/0008-5472.CAN-19-0086. [DOI] [PubMed] [Google Scholar]
- 36.He Z, Khatib AM, Creemers JWM. Loss of the proprotein convertase furin in T cells represses mammary tumorigenesis in oncogene-driven triple negative breast cancer. Cancer Lett. 2020;484:40–49. doi: 10.1016/j.canlet.2020.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Lin H, et al. Proprotein convertase furin inhibits matrix metalloproteinase 13 in a TGFβ-dependent manner and limits osteoarthritis in mice. Sci Rep. 2018;8(1):10488. doi: 10.1038/s41598-018-28713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scamuffa N, Metrakos P, Calvo F, Khatib AM. Identification of the myosin heavy polypeptide 9 as a downstream effector of the proprotein convertases in the human colon carcinoma HT-29 cells. Methods Mol Biol. 2011;768:207–215. doi: 10.1007/978-1-61779-204-5_10. [DOI] [PubMed] [Google Scholar]
- 39.Basak A, et al. A novel enediynyl peptide inhibitor of furin that blocks processing of proPDGF-A, B and proVEGF-C. PLoS One. 2009;4(11):e7700. doi: 10.1371/journal.pone.0007700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang P, et al. Elabela/toddler is an endogenous agonist of the apelin APJ receptor in the adult cardiovascular system, and exogenous administration of the peptide compensates for the downregulation of its expression in pulmonary arterial hypertension. Circulation. 2017;135(12):1160–1173. doi: 10.1161/CIRCULATIONAHA.116.023218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen J, et al. Akt1 regulates pathological angiogenesis, vascular maturation and permeability in vivo. Nat Med. 2005;11(11):1188–1196. doi: 10.1038/nm1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sourbier C, et al. The phosphoinositide 3-kinase/Akt pathway: a new target in human renal cell carcinoma therapy. Cancer Res. 2006;66(10):5130–5142. doi: 10.1158/0008-5472.CAN-05-1469. [DOI] [PubMed] [Google Scholar]
- 43.Phung TL, et al. Endothelial Akt signaling is rate-limiting for rapamycin inhibition of mouse mammary tumor progression. Cancer Res. 2007;67(11):5070–5075. doi: 10.1158/0008-5472.CAN-06-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phung TL, et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. 2006;10(2):159–170. doi: 10.1016/j.ccr.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnell CR, et al. Effects of the dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor NVP-BEZ235 on the tumor vasculature: implications for clinical imaging. Cancer Res. 2008;68(16):6598–6607. doi: 10.1158/0008-5472.CAN-08-1044. [DOI] [PubMed] [Google Scholar]
- 46.Scamuffa N, et al. Prodomain of the proprotein convertase subtilisin/kexin Furin (ppFurin) protects from tumor progression and metastasis. Carcinogenesis. 2014;35(3):528–536. doi: 10.1093/carcin/bgt345. [DOI] [PubMed] [Google Scholar]
- 47.Sfaxi F, et al. Repression of liver colorectal metastasis by the serpin Spn4A a naturally occurring inhibitor of the constitutive secretory proprotein convertases. Oncotarget. 2014;5(12):4195–4210. doi: 10.18632/oncotarget.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016;44(W1):W449–W454. doi: 10.1093/nar/gkw329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cleves AE, Jain AN. Knowledge-guided docking: accurate prospective prediction of bound configurations of novel ligands using Surflex-Dock. J Comput Aided Mol Des. 2015;29(6):485–509. doi: 10.1007/s10822-015-9846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc. 2016;11(5):905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labbé CM, et al. MTiOpenScreen: a web server for structure-based virtual screening. Nucleic Acids Res. 2015;43(W1):W448–W454. doi: 10.1093/nar/gkv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40(Database issue):D370–D376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miteva MA, Tufféry P, Villoutreix BO. PCE: web tools to compute protein continuum electrostatics. Nucleic Acids Res. 2005;33(Web Server issue):W372–W375. doi: 10.1093/nar/gki365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alves ID, Lecomte S. Study of G-protein coupled receptor signaling in membrane environment by plasmon waveguide resonance. Acc Chem Res. 2019;52(4):1059–1067. doi: 10.1021/acs.accounts.9b00007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.