Abstract
Understanding the protein-secretion dynamics from single, specific tissues is critical toward the advancement of disease detection and treatments. However, such secretion dynamics remain difficult to measure in vivo due to the uncontrolled contributions from other tissue populations. Here, we describe an integrated platform designed for the reliable, near real-time measurements of cytokines secreted from an in vitro single-tissue model. In our setup, we grow 3D biomimetic tissues to discretize cytokine source, and we separate them from a magnetic microbead-based biosensing system using a Transwell insert. This design integrates physiochemically controlled biological activity, high-sensitivity protein detection (LOD < 20 pg mL−1), and rapid protein diffusion to enable non-invasive, near real-time measurements. To showcase the specificity and sensitivity of the system, we use our setup to probe the inflammatory process related to the protein Interleukine 6 (IL-6) and to the Tumor Necrosis Factor (TNF-α). We show that our setup can monitor the time-dependence profile of IL-6 and TNF-α secretion that results from the electrical and chemical stimulation of 3D skeletal muscle tissues. We demonstrate a novel and affordable methodology for discretizing the secretion kinetics of specific tissues for advancing metabolic-disorder studies and drug-screening applications.
Keywords: Microphysiological tissues; Tissue engineering; Electrochemical, biosensors; Magnetic particles; Skeletal muscle; Electric stimulation
Highlights
-
•
Animal models and 2D cells systems not always accurate.
-
•
3D skeletal muscle microphysiological systems (MPS) exhibit architecture that represents native tissue's characteristics.
-
•
Reproducible and viable for at least 4 weeks under physiological conditions.
-
•
Our MPS tissues allow personalized medicine for high content screening for repeated dose efficacy testing, and toxicology.
1. Introduction
Detecting changes in protein secretion with spatiotemporal accuracy remains a fundamental challenge toward understanding the role of proteins in regulating biological processes. For instance, to understand its role in immune and inflammatory responses, (Rothenberg, 2007) protein expressions of cytokine are routinely studied from blood serums using methods such as RNA sequencing, protein microarrays, enzyme-linked immunosorbent assay (ELISA) or mass spectroscopy (MS) (Mukherjee and Mani, 2013). However, because most in vivo tissues of interest are composed of a variety of cells, profiling cytokine expressions by specific cells directly from bulk serum samples remains impractical.
Recent studies approach this challenge in two ways, focusing either on maximizing detection sensitivity, or on tailoring the ideal functional-tissue models. The first approach targets real-time analysis and quantification of secreted cytokines using ultrasensitive detection method or imaged-based high-resolution spatiotemporal techniques (Shirasaki et al., 2015; Juan-Colás et al., 2018; Saxena et al., 2018). Here however, because the techniques involved rely on single-cell secretion, this approach cannot probe cytokine secretion by functional tissues.
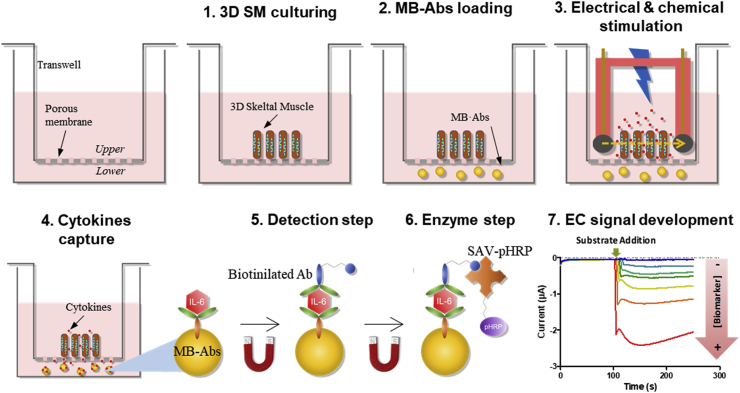
The second approach tailors specific, desired functional tissue samples that can be probed by established techniques such as ELISA or MS. Using this approach, cytokine secretions have been analyzed from in vivo muscles, in vitro contractile myofiber sheets, (Furrer et al., 2017; Takahashi et al., 2018) or even adipose and immune cells co-cultures without unwanted interactions from unknown cells (Kongsuphol et al., 2019). However, because techniques such as ELISA and MS lack temporal resolution, this approach cannot profile cytokine secretion at timescales relevant to biological processes (Zhang et al., 2017; Bruls et al., 2009; Esteban-Fernández de Ávila et al., 2013; Skardal et al., 2017). Here we present an integrated platform designed for reliable, near real-time profiling of cytokine secretions by combining the advantages of both aforementioned approaches: we maximize both the chemical and the time resolution of our detection system, and we tailor tissue samples that are ideal for our measurement. Fig. 1 illustrates our integration concept, which grows the desired 3D skeletal muscle (SM, blue oblongs) sample tissues on the top surface of the porous membrane (dashed lines) of a Transwell insert, in proximity of antibody-bioconjugated magnetic microbead (MB·abs) detectors, located underneath the membrane. Upon electrical and chemical stimulation applied to the SM in the Transwell insert, any secreted cytokine would rapidly diffuse through the porous membrane to be captured by the nearby microbeads. This concept enables our custom-grown in vitro 3D SM tissues (Ortega et al., 1039) to provide the ideal sample for our measurements, while exploiting the magnetic beads’ high area to volume ratio and its proximity to the sample maximize chemical and temporal resolutions (20 and 10 pg mL−1, near real-time, respectively).
Fig. 1.
Schematic illustration of the immunosensor measurement procedure. Firstly, 3D SM is fabricated on the Transwell membrane, then MB·Abs are loaded in the lower compartment. Secreted cytokines, under the chemical and electrical effect, are captured by the MB·Abs. Detection step is followed by the addition of a biotinylated antibody and SAV-pHRP. Finally, MB·Abs are resuspended in enzymatic buffer and captured on the surface of the SPCEs. After addition of the corresponding substrate, EC signal is obtained. The intensity of the current is directly related with the concentration of secreted cytokines.
In this article, we test the validity of our concept by profiling inflammatory process related cytokines: Interleukine 6 (IL-6) and Tumor Necrosis Factor (TNF-α), induced by chemical and electrical stimulation of 3D engineered skeletal muscle tissues. We demonstrate that our integrated system represents a simple and feasible tool profiling the secretion kinetics of specific proteins from specific in vitro tissue models. Our results provide a new strategy toward understanding the role of proteins in both diseased and healthy states and in drug-screening assays.
2. Experimental section
2.1. Materials
Electrochemical measurements were carried out using a Portable multipotentiostat μSTAT 8000P (Metrohm Dropsense AG Herisau, Switzerland)). Screen-printed carbon electrodes (SPCEs), were integrated by a Ag/AgCl pseudo-reference electrode and a carbon counter electrode (DRP-8X110, 2.56 mm Ø) originally used as working electrodes (Metrohm Dropsense AG Herisau, Switzerland). Round bottom non-treated plates were purchased from Nirco (Barberà del Vallés, Spain). Polystyrene MaxiSorp microtiter plates used to perform ELISA assays were purchased from Nunc (Roskilde, Denmark). Washing steps were carried out using a 405 TS microplate washer (BioTeck Instruments, Winooski, USA). The electrochemical data obtained was analyzed using DropView 8400 software (Metrohm Dropsense AG Herisau, Switzerland). Absorbances were read on Infinite® 200 PRO multiplate reader (TEKAN, Männedorf, Switzerland). Digital light processing (DLP) projector-based stereo-lithography 3D printer (Solus DLP model, Reify3D, CA, USA) was used to fabricate the device for the electrical stimulation. Crosslinking of composite hydrogel were performed by a UVP Crosslinker (CL-1000, Analytik Jena, Germany). Assessment of electrical stimulation was done using grade 1 graphite Rods (Ted Pella, Ca, USA), connected to a multifunctional wave generator (WF1947/WF1948, NF corporation, Japan).
2.1.1. Chemicals and immunochemicals
Recombinant mouse Interleukin 6 (ref. 200-02-100 μg) and recombinant mouse TNF-α (ref. 200-31-100 μg) were purchased from CliniSciences S.L. (Nanterre, France). Purified rat anti-mouse IL-6 (ref. 554400, clone MP5-20F3, capture antibody), biotin rat anti-mouse IL-6 (ref. 554402, clone MP5-32C11, detection antibody), purified rat anti-mouse TNF-α (ref. 551225, clone G281-2626, capture antibody), and biotin rat anti-mouse TNF-α (ref. 554415, clone MP6-XT3, detection antibody) were purchased from BD Biosciences (Barcelona, Spain). Streptavidin-poly Horseradish Peroxidase (SAV-pHRP), MF20 Alexa Fluor 488, DAPI, Tosyl-activated magnetic beads MB Dynabeads® MyOne™, Dulbecco's modified Eagle's medium (DMEM high glucose, L- glutamine, GIBCO, 11965092)), trypsin/EDTA, horse serum (HS, 26050088), fetal bovine serum (FBS, 16000044), penicillin/streptomycin (P/S, 15140122), 3.5 kDa MWCO membranes (68035) were purchased from Thermo Fisher (21140, Massachusetts, USA). Bovine serum albumin (BSA, A4737), Dexamethasone (Dex, D4901), Caffeine (C0750) and Lipopolysaccharides from E. Coli O55:B5 (LPS, L6529), Gelatin Type A from porcine skin (G2500), methacrylate anhydride (276685), 10% formalin solution (HT5011), Triton (X-100), 24 mm Transwell with 0.4 μm Pore Polyester Membrane Insert (CLS3450), Sodium carboxymethylcellulose (CMC, 419273), N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, E7750), N-hydroxysuccinimide (NHS, 130672), aminoethylmethacrylate (479659), Trypsin/EDTA (T4049) were purchased from Sigma-Aldrich (Sigma Aldrich Co. St Louis. MO, USA). Tris-buffered saline was purchased from Canvax (TBS, BR0042, Biotech, Spain). Rhodamine-Phalloidin 480 was purchased from Cytoskeleton (PHDR1, USA). The Bradford solution (BIO-RAD protein assay cat No. 500-0006) was purchased from BIO-RAD laboratories GmbH (Munich, Germany). Acetone was purchased from Panreac (161007, Barcelona, Spain). Polydimethylsiloxane (PDMS) was purchased from Dow Corning (SILPOT 184, Toray, Japan). 6–8 kDa MWCO membranes were purchased from Spectrumlabs (08-700-142, San Francisco, USA). Lithium Phenyl (2,4,6-trimethylbenzoyl)phosphinate (i.e., LAP) was purchased from TCI EUROPE N.V. (Belgium). Murine C2C12 skeletal myoblasts was purchased from American Type Culture Collection (CRL-1772, ATCC, Virginia, USA). Phosphate buffer saline (PBS) was 0.01 mol L−1 phosphate buffer, 0.14 mol L−1 in NaCl and 0.003 mol L−1 in KCl saline solution at pH 7,5. PBST was PBS with 0.05% (v/v) Tween 20. PBST-BSA was PBST with 1% (w/v) BSA. Coating buffer is 0.05 M carbonate- bicarbonate buffer, pH 9.6. Citrate buffer was a 0.04 M solution of sodium citrate, pH 5,5. The substrate solution for optical measurement and used also in the amperometric immunosensor was 0.01% TMB (3,3′,5,5′-tetramethylbenzidine) and 0.004% H2O2 in citrate buffer. For amperometric measurements, citrate buffer-KCl was prepared with citrate buffer that contains 0.1 mol L−1 KCl. Borate buffer was 0.1 M boric acid (pH 9.5).
2.2. Preparation and optimization of the MB·Abs-based immunosensing platform
The antibody was coupled covalently to the magnetic beads (Supplementary information) according to the supplier protocol but with slight modifications, and the final concentration of the functionalized microbeads (MB·Abs) was 8 mg mL−1. Prior to performing the detection of the secreted cytokines, the immunosensing platform was characterized and optimized (in terms of concentration) by checkboard titration. The incubation time for cytokines detection by MB·Abs was set at 60 min. Subsequent incubation steps for biotinylated antibody and SAV-pHRP bioconjugate were set at 30 min. Interleukin 6 and TNF-α solutions were prepared at 4 ng mL−1 in differentiation media as standard solutions during amperometric optimization measurements. The effects of different parameters, including the MB·Abs quantity, biotinylated antibody, and enzyme tracer concentrations, were analyzed. The effect of each parameter was evaluated individually, and optimized values were used in subsequent experiments. Initially, MB·Abs quantity was optimized in the range of 0.1 μg–5 μg. Other parameters including biotinylated antibody and SAV-pHRP concentrations were maintained constant at 1 μg mL−1 in both cases.
To optimize detection and enzymatic steps, MB·Abs concentration was set, and concentrations of both biotinylated antibodies and SAV-pHRP were evaluated in the range of 0.0625–1 μg mL−1, following the same procedure as described before.
Related to selectivity studies, cross-reactivity was evaluated for either IL-6 and TNF-α including a negative control (zero point). On the other hand, to evaluate accuracy in our experiments, different blind spiked samples were prepared in differentiation media and measured directly by our optimized immunoassays. Accuracy for both immunoassays was evaluated by establishing a linear regression between spiked and measured values. All experiments were performed in triplicate.
2.3. Cell culture
Murine C2C12 myoblast cells were cultured under a 5% CO2 atmosphere at 37 °C in growth medium, high glucose DMEM, containing L-Glut, 10% (v/v) FBS, 1% (v/v) P/S. When ∼70% confluency was reached, the cells were detached by using trypsin/EDTA and then subcultured or used in the experiment. High glucose DMEM, containing L-Glut, 2% (v/v) HS, and 1% (v/v) P/S was used as differentiation medium (DM) to promote C2C12 differentiation towards myotubes.
2.4. Synthesis and preparation of prepolymer solutions
Gelatin-methacryloyl (GelMA) and carboxymethyl cellulose-methacrylate (CMCMA) were synthesized according with our previous reported protocol (Supporting Information) (García-lizarribar et al., 2018). Prepolymer solutions were prepared by dissolving polymers and photoinitiator lithium phenyl (2,4,6-trimethylbenzoyl)phosphinate (LAP) in growth medium to obtain final concentrations of 5% (w/v) GelMA, 1% (w/v) CMCMA, and 0.1% (w/v) LAP. Prepolymer solutions were placed at 65 °C for 3 h to obtain homogeneous solutions.
2.5. Fabrication of the 3D SM engineered tissue (3SM)
Polymer solution was mixed with a C2C12 cell suspension to reach the final cell density of 25·106 cell mL−1. 3D skeletal muscle tissues were fabricated by photo-molding technique as we described previously (Ortega et al., 1039). Briefly, 20 μL of the cell-laden prepolymer were poured on the Transwell insert (24 mm) and a microstructured PDMS stamp was placed onto the prepolymer. The PDMS microgrooves were filled and cell-laden prepolymer was exposed for 24 s under a UV light source (40 W, UVP Crosslinker) (Fig. S2, Supplementary information). After carefully removing the stamp we obtained a 3D cell-microstructured hydrogel anchored on the Transwell membrane. Once 3SM was fabricated, a new cell medium was added on the upper part (1 mL) and the lower part (2 mL) and changed every 2 days. The growth medium was replaced at day 6 with differentiation media (DM). The DM was changed every 2 days until day 14, when the drug assays or electrical assays were carried out.
2.6. Immunostaining
The tissues were fixed in a 10% formalin solution at 15 days after fabrication. Then hydrogels were washed with TBS, cells were permeabilized with 0.1% Triton X- 100 in TBS for 15 min and blocked with a blocking buffer consisting of 0.3% Triton X- 100 and 3% donkey serum (brand) in TBS for 2 h. Afterwards, tissues were washed with TBS and incubated in 100 nM Rhodamine-Phalloidin 480 solution overnight to stain filamentous actin (F-actin). An additional overnight staining for Myosin Heavy Chain (MHC) was performed by incubating in a solution of 5 μg/mL MF20 Alexa Fluor 488 in blocking buffer. After washing with TBS, nuclei were counterstained with DAPI 1 μM for 15 min.
2.7. Immunosensor measurement procedure
Transwell permeable supports were used as a platform to develop the immunosensing measurement procedure, design and operation of the integrated platform is depicted in Fig. 1. 3SM were maintained in culture for up to 14 days and, just before electrical or chemical stimulation, MB·Abs (5 μg for IL-6 and 10 μg for TNF-α) were loaded in the lower side of the Transwell, where capture step was performed. Samples were incubated under soft stirring to avoid bead aggregation, inside an incubator at 37 °C and 5% CO2 atmosphere. During the experiments, upper part and lower part was filled with 1 mL and 2 mL of cell medium, respectively. Secreted cytokines, as a result of either electrical or chemical stimulation, diffused trough the Transwell membrane and were captured by the specific antibodies immobilized onto the surface of the beads. MB·Abs were replaced every hour to obtain a continuous monitoring of the secreted cytokines. After this time, MB·Abs were washed (3X) and resuspended in biotinylated antibody 0.125 μg mL−1 or 0.5 μg mL−1 (in the case of IL-6 or TNF-α respectively), prepared in PBST-BSA buffer, and leave it under stirring for 30 min to avoid bead aggregation. Then, MB·Abs were washed again and resuspended in SAV-pHRP 0.50 μg mL−1 (for both cytokines) prepared in PBST-BSA buffer, for 30 min under the same conditions described before. Afterwards, MB·Abs were washed and resuspended in 100 μL of citrate buffer-KCl and captured onto the surface of the working electrode using a custom-made PMMA cell with magnets located under the SPCEs (Fig. S3 Supporting Information). Finally, amperometric measurements were carried out by applying a constant potential of −0.2 V vs Ag pseudo-reference electrode. After current stabilization (100 s), 20 μL of substrate solution prepared in citrate buffer-KCl, was added to each well. Electrochemical signals were recorded once the current was again stabilized (250 s).
2.8. Drug stimulation assays
2D Drug stimulation screening assays. Caffeine, Dexamethasone (Dex) and Lipopolysaccharide (LPS) were tested in skeletal myotubes cultured in 2D (well plate configuration) using differentiation media for 7 days, to evaluate the effect of each drug in the cytokine's secretion at different times. Myotubes cultured in 2D were incubated for 0.5, 1, 2, 16, 24 and 48 h, in DM supplemented by the drug. Caffeine was diluted in DM to a concentration of 100 μM. Dex, was suspended in absolute ethanol at a concentration of 10 mM and diluted further in DM to obtain a final concentration of 100 μM. LPS was diluted in DM to a concentration of 10 μg mL−1. Three replicas for each condition were analyzed. The screening of the cytokine levels was carried out by ELISA (See supporting information).
3SM drug stimulation assay. Mature 3SM at day 14 were incubated in DM supplemented by LPS at different times: 0.5, 1, 2, 4, 8 and 24 h. MB·Abs were placed 1 hour before drug treatment in the lower compartment, and after drug treatment the immunoassay was performed as described in section 2.8.
2.9. Electrical pulse stimulation (EPS) assays
In order to stimulate 3SM on the Transwell membrane, a pair of graphite electrodes (3 mm diameter), separated at 1 cm, were used. A custom-made device was designed and fabricated using a DLP projector-based stereo-lithography 3D printer to hold the electrodes and fixed on well plate's lid. Copper wires were assembled to graphite rods and located out of the lid through the device, to permit the connection to a wave generator (Supplementary information). Under this configuration, electrical stimulation on Transwells was carried out in a high reproducible way, allowing the microscope imaging of the tissue. Mature 3SM were subjected to stimulations regimes with 0.25, 1 and 5 V peak-peak at 1 Hz of frequency and a duration of 2 ms during 1 h, to evaluate cell response upon electrical stimuli. To study time-dependent secretion of cytokines, samples were stimulated for seven hours in a continuous EPS, and on the other hand in cycles of 1 h stimulation with and 2 h of relaxing time (stimulation off) for a total of nine hours.
2.10. Data and statistical analysis
The calibration curves fittings and statistical analysis were performed using GraphPad Prims 5.03 (GraphPad Software Inc., San Diego, CA). Data is presented as the mean (⨱ ± SD). A t-test was used for statistical analysis of two sample comparison, and ANOVA one-way for statistical evaluation of one factor. A value of p < 0.05 was considered statistically significant.
3. Results and discussion
3.1. Immunoassay optimization and analytical features
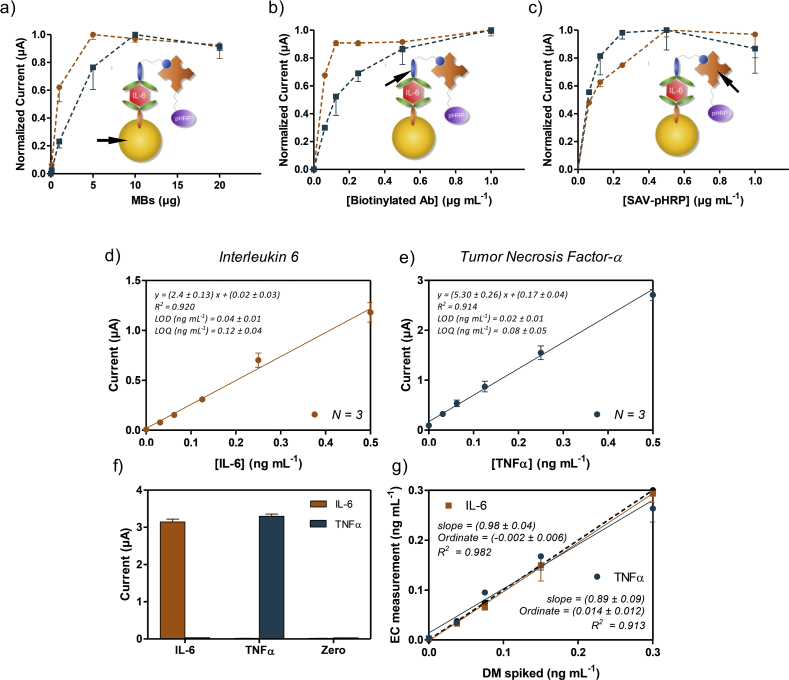
The conjugation efficiency (coupling of antibody to MB) supernatants after immobilization process were evaluated using Bradford assay. Our results indicate a coupling yield of 94 ± 8% for all replicates. The functionality of the bioconjugates by the electrochemical immunoassay, MB·Abs, biotinylated antibody, and SAV-pHRP concentracions were optimized by checkboard titration assay. Fig. 2a illustrates the effect of MB·Abs quantity on the normalized current obtained at −0.2 V applied potential, corresponding to IL 6 (orange circle) and TNF-α (blue squares) detection. The current reaches optimum signal at 5 μg (IL-6) and 10 μg (TNF-α) of MB·Abs, with non-significant enhancement of the signal using higher concentrations. These values of MB·Abs were selected for the optimization of the further steps.
Fig. 2.
Optimization of the electrochemical MB·Abs based immunoassay for IL-6 (orange circles) and TNF-α (blue squares) using 4 ng mL−1of each cytokine. Normalized current vs a) the number of microbeads in the 6 well Transwell set up, b) concentration of the biotinylated antibody, and c) SAV-pHRP concentration. Other parameters such as cytokine concentration and incubation time for each step were maintained fixed. Calibration curves for the detection of d) IL-6 and e) TNF-α respectively, in differentiation media and different analytical parameters such as limit of detection and quantification. f) Selectivity studies for both immunoassays, IL-6 and TNF-α at concentration of 4 ng mL−1. g) Accuracy studies performed using spiked samples prepared in differentiation media and used directly in the immunosensor without previous dilution. The data shown correspond to the average of at least two replicates. All data shown by mean ± SD. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2b shows the values of the maximum signal performance achieved at different biotinylated antibody concentrations. The current values for IL-6 reach saturation point at 0.125 μg mL−1 of biotinylated antibody, while for TNF-α we consider the saturation level above 0.5 μg mL−1, setting these values as optimal for further experiments.
Finally, the current response was tested at different concentrations of SAV-pHRP, from 0.0625 to 1 μg mL−1. As shown in Fig. 2c, in the case of IL-6, the signal response shows an increment of 50% μA at 0.5 μg mL−1, whereas in the case of TNF-α, this improvement takes place at a lower concentration of 0.25 μg mL−1, maintaining the same response at higher concentrations of the SAV-pHRP. For these reasons, concentration of 0.5 μg mL−1 of SAV-pHRP for both immunoassays were selected, simplifying the experimental procedure of the biosensor for both cytokines. These optimized values were used in later determination of calibrations curves, which represent the working conditions to obtained lowest limit of detection.
The dose-dependence of the magnetic bead-based immunosensors developed was studied for different concentrations of IL-6 and TNF-α in differentiation media. The assay was performed as is shown in Fig. 1, and described in Section 2, by assessing standard solutions of IL-6 and TNF-α (between 0 and 0.5 ng mL 1 in DM). As expected, the current signal recorded was directly proportional to the concentration of the cytokine in the sample in both cases. The limit of detection for IL-6 and TNF-α (Fig. 2d and e) were 40 ± 10 pg mL−1 and 20 ± 10 pg mL−1, respectively, which are better than ELISA tests.
Several works that have used magnetic beads to detect secreted biomarkers or cytokines have reported similar LOD's, about tens of pg mL−1. Riahi et al. succeeded on on-chip measurements with a LOD of 30 pg mL−1 by EC detection (Riahi et al., 2016). Other reports are based on the use of on-chip fluorescent bead-based immunoassay, (Cui et al., 2018) or by magnetic beads with the out-chip enzymatic step to monitor cytokine secretion from immune cells with a sensitivity of tens of pg mL−1 (Kongsuphol et al., 2016). Also, by a novel optofluidic nanobiosensor, Li et al., have reported a sensitivity of 39 pg mL−1, in the real-time detection of cytokine secretion using nanoplasmonics (Li et al., 2018). Thus, the limit of detection reached by the immunosensor described in this work is considered optimal for our purposes, taking into account the sensitivity reported by recent works, and the reported amount of secreted IL-6 (∼0.05 ng mL−1) without any stimulation in skeletal muscle 2D cultures (Nedachi et al., 2008; Farmawati et al., 2013). Our selectivity studies showed no cross reactivity between IL-6 and TNF-α and the feasibility of the system, while the assays used to study the accuracy in both cases showed a slope close to 1 (Fig. 3f and g), indicating that both immunosensors are high accurate in the working range of our interest.
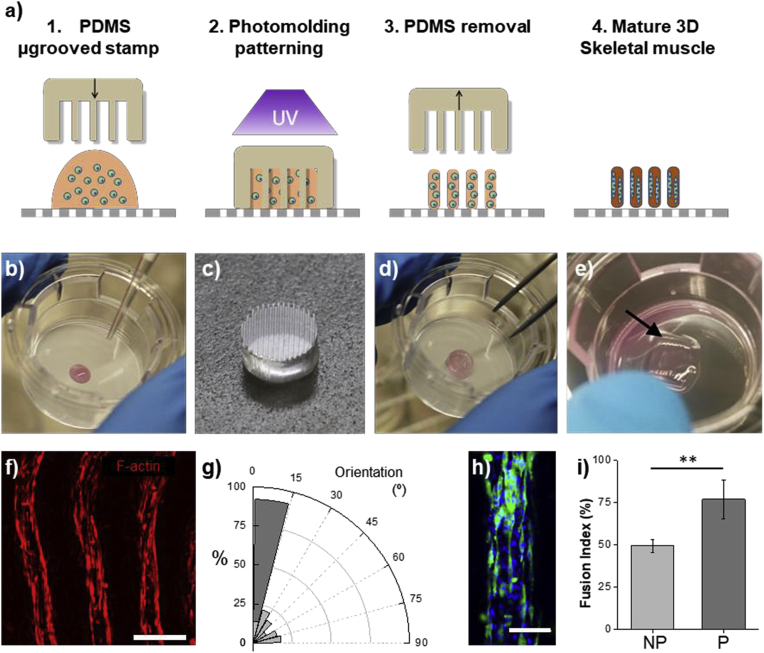
Fig. 3.
Fabrication of the 3D skeletal muscle in the integrated platform. a) Schematics of the photo mold patterning on the Transwell. 1. A drop of cell-laden solution is poured on the porous membrane of the Transwell, 2. A microstructured PDMS stamp is placed onto the prepolymer. The PDMS microgrooves were filled and cell-laden prepolymer is exposed under a UV light. 3. After removing the stamp we obtain a 3D cell-microstructured hydrogel anchored on the Transwell membrane. 4. After 14 days we obtain a mature 3D skeletal muscle. Pictures of the fabrication process: b) Cell-laden prepolymer drop onto a Transwell. c) Detail of the microgrooved stamp. d) PDMS microgrooves filled with the prepolymer cell-laden solution. e) Detail of the skeletal muscle tissue (arrow) on the Transwell, after fabrication. f) Confocal microscopy picture of 3SM at say 14, showing elongated cells and myotubes. F-actin in red. Scale bar = 500 μm. g) Normalized histogram (bin = 15°) depicting the distribution of the angles between cell cytoskeleton fibers inside non-patterned hydrogels (NP, light gray) and in the 3SM (dark gray). Inside the 3SM moytubes show high degree of alignment (>75%) following the pattern direction. h) Confocal image of the one structure of the mature 3SM depicting the cell, showing the presence of MHC positive myotubes (green). Nuclei in blue. Scale bar = 200 μm. i) Fusion index percentage (mean ± standard deviation, **p-value < 0.01) of the myotubes inside the non-patterned hydrogels (light gray) and in the 3SM (dark gray). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. 3D SM engineered tissue (3SM) and sensing system integration
To fabricate the 3D skeletal muscle constructs, muscle cells were encapsulated in a 5% w/v Gelatin methacryloyl (GelMA), and 1% w/v carboxy methyl cellulose methacrylate (CMCMA) polymer solution. Gelatin methacryloyl was combined with CMCMA to avoid cell degradation and improve stability, as we previously demonstrated (García-lizarribar et al., 2018). To control the 3D cellular organization of the encapsulated C2C12 cells, photo-mold patterning technique was applied using a microgrooved PDMS stamp and this method was directly carried out on the upper membrane of a Transwell (Fig. 3a–e). (Ortega et al., 1039)
The skeletal muscle constructs were differentiated from day 6 to day 14 and characterized by bright field microscopy during the skeletal muscle cells maturation. At day 14 constructs show aligned fibers (Fig. 3f), which compared to non-patterned samples show a high degree of alignment (Fig. 3g). The myotubes show a high content of myosin heavy chain positive stained cells (Fig. 3h), and a high fusion index compared to non-patterned samples (Fig. 3i). Thus, these results demonstrate the functional C2C12 differentiation towards myotubes and are in concordance with found by Ortega et al. (Ortega et al., 1039) We demonstrate that the reported fabrication method can be used on a permeable membrane without further modifications, and 3SM kept anchored on the membrane for at least 14 days supporting growth and skeletal muscle differentiation. In the present work, we exploit the advantages of Transwell insert membranes, to assess the invasive way of place MB·Abs and 3SM together. To demonstrate cytokine diffusion through the Transwell membrane (0.4 μm pore size), we placed IL-6 solution in the upper compartment. As expected, we found that IL-6 concentration increases with time in the lower compartment, indicating diffusion of cytokines through the porous membrane. Thus, permeable membranes, where 3SM are located, allow the diffusion of the secreted cytokines to the lower compartment where MB·Abs immediately capture the cytokines, but the membrane pore size prevents MB·Abs diffusion to the upper part. In this work, the integrated platform has been designed to monitor the secretion processes over time by the in situ capturing of the released cytokines. Several works have previously used complex chip arrays or devices to detect cytokines from immune cells by MB·Abs, which were located in separated compartments or chambers to avoid the cell-MB·Abs interaction (Riahi et al., 2016; Cui et al., 2018; Kongsuphol et al., 2016). In addition, previous work has reported complex systems based on fluorescence imaging or nanoplasmonic biosensors, which capture antibodies on the substrate for detecting cytokine secretion of single cells (Juan-Colás et al., 2018; Li et al., 2018; Nedachi et al., 2008; Farmawati et al., 2013; An et al., 2017). These examples have succeeded in “real-time” quantification of cytokine secretion but using single cells or cell solutions. Our platform incorporates the novelty of integrating a 3D tissue-like skeletal muscle instead of isolated cells.
3.3. Drug stimulation assays
To study the viability of the sensor and monitor the response of the skeletal muscle against chemical stimuli, IL-6 and TNF-α secretion were measured. To find the best chemical stimuli for promoting IL-6 secretion, 2D monolayers of differentiated myotubes were incubated with caffeine, which causes effects on muscle size and changes in signaling pathways, (Egawa et al., 2016) dexamethasone, which is used as a muscle atrophy inducer, (Shimizu et al., 2017) and LPS which stimulates the expression of IL-6 and TNF-α in both skeletal muscles in vivo and C2C12 myoblasts in vitro (Frost et al., 2002).
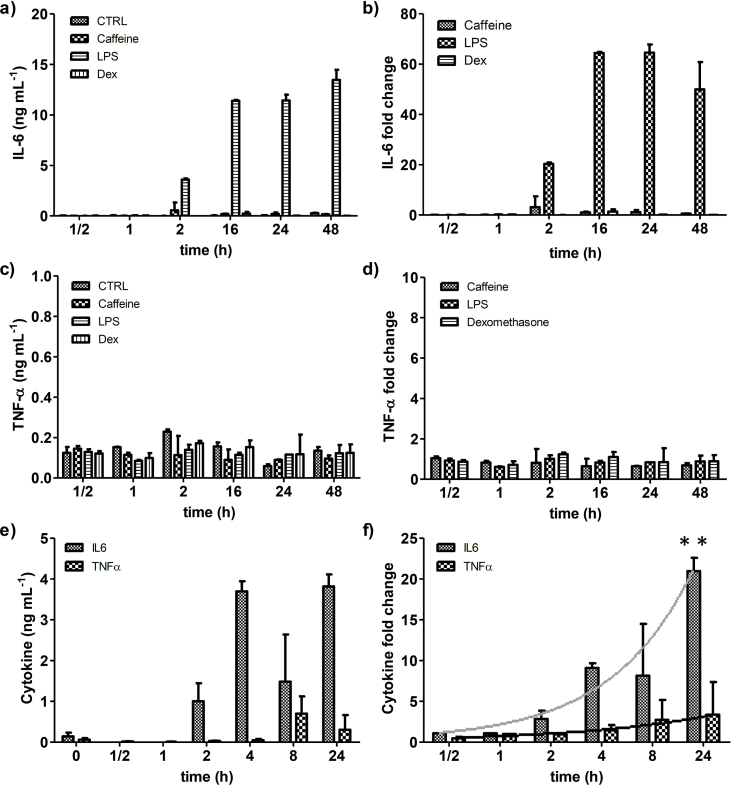
After we determined cytokine secretion by a in house ELISA, LPS treatment showed the highest secretion of IL-6 with no significant changes under caffeine and Dex treatment (Fig. 4a and b). The TNF-α values do not show significant changes under all the treatments (Fig. 4c and d). Thus, LPS was chosen to be tested in the integrated sensing system.
Fig. 4.
Monitoring of IL-6 and TNF-α secretion under the effect of chemical stimuli. Cytokine secretion by ELISA test of a) IL-6 concentration and b) normalized values by fold change (Treatment/Ctrol), and c) TNF-α concentration and d) normalized values by fold change of 2D differentiated myotubes under the effect of caffeine, Dex, and LPS. Continuous monitoring in integrated platform of e) IL6 secretion and TNF-α secretion concentration from 3D skeletal muscle constructs exposed to LPS (10 μg mL−1) for 24 h. f) Fold change of IL-6 and TNF-α secretion was calculated to assess responsiveness of 3SM to stimulation with LPS. Trend of cytokine secretion drawn as a gray line (IL-6) and a black line (TNF-α). ANOVA test **p-value < 0.01. Data are shown by mean ± SD, (n = 4–6).
The 3D skeletal muscle tissues were grown on the Transwell membrane and at day 14 and incubated with differentiation media containing LPS (10 μg mL−1). The IL-6 and TNF-α secretions were monitored in function of time for 24 hours. The IL-6 secretion showed an increasing profile along the incubation time as was found in 2D experiments. The TNF-α secretion showed an increasing but no statistically significant trend, and released less protein compared to IL-6 (Fig. 4e and f).
Our results prove that 3SM responds to LPS treatment, and that mature muscle cells secrete IL-6. On the other hand, we have demonstrated the capacity of the integrated system to monitor low concentrations of cytokine levels, immediately following chemical stimuli. After 24 h, we found that the IL-6 expression fold change (20.98 ± 1.61) was at least three times higher than the TNF-α fold change (3.37 ± 4.00); this correlates with the reported differences between IL-6 and TNF-α expression under LPS treatment in mouse (Frost et al., 2002).
3.4. Dependence between contraction/relaxation periods and IL-6 secretion from skeletal muscle
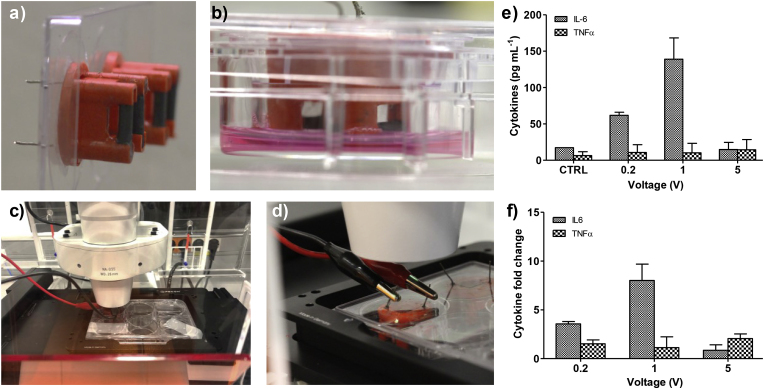
To electrically stimulate the 3SM, we designed and fabricated a device that fits in the Transwell insert containing a pair of graphite rods (Fig. 5a). A function generator was connected to the graphite rods by copper wires in the external part of the well plate's lid (Fig. 5b). The graphite rods are not in direct contact with the 3SM, enabling the imaging of the skeletal muscle tissue during the electrical stimulation (Fig. 5c). This compact configuration allows the routine use of culture-well plates inside the incubator.
Fig. 5.
a) Picture of the tailored device for the electrical stimulation. b) Lateral view of the device inside the Transwell in a well plate. c) integrated system coupled to a wave generator and located in a microscope. d) Detail of the connection between coper wires and wave generator. e) Effect of EPS regimes on the cytokine secretion, f) fold change in IL-6 and TNF-α secretion was calculated to assess responsiveness of 3SM to EPS regimes. Data are shown by mean ± SD. (n = 4–6) p < 0.05.
Previous works have reported that electrical pulse stimulation of in vitro skeletal muscle induced muscle contractions (Ortega et al., 1039; Nedachi et al., 2008; Marotta et al., 2004; Ramón-Azcón et al., 2013; Park et al., 2008; Evers-van Gogh et al., 2015; Banan Sadeghian et al., 2018). Other works have also reported that intense exercise by skeletal muscle increases IL-6 expression and secretion, but not of that of TNF-α (Banan Sadeghian et al., 2018; Bustamante et al., 2014). To obtain an appropriate stimulation regime with the tailored device, we optimized the stimulation regime to obtain high IL-6 secretion with a low TNF-α increase. The skeletal muscle constructs were electrically stimulated under three voltage regimes 0.25 V, 1 V, and 5 V. We found that electrical stimulations led to a significant increase of IL-6 secretion at 0.2 and 1 V, compared to the control, with a maximum level (0.14 ± 0.02 ng mL−1) by applying 1V (Fig. 5e). No significant changes in TNF-α secretion were found. Ours results suggest a successful EPS-evoked contraction of the 3SM, depicted by the increase of IL-6 expression, without effects on TNF-α release. Normalized values by fold change (EPS/Ctrol) (Fig. 5f) confirmed the highest IL 6/TNF-α signal at 1 V, which was fixed for the next experiments.
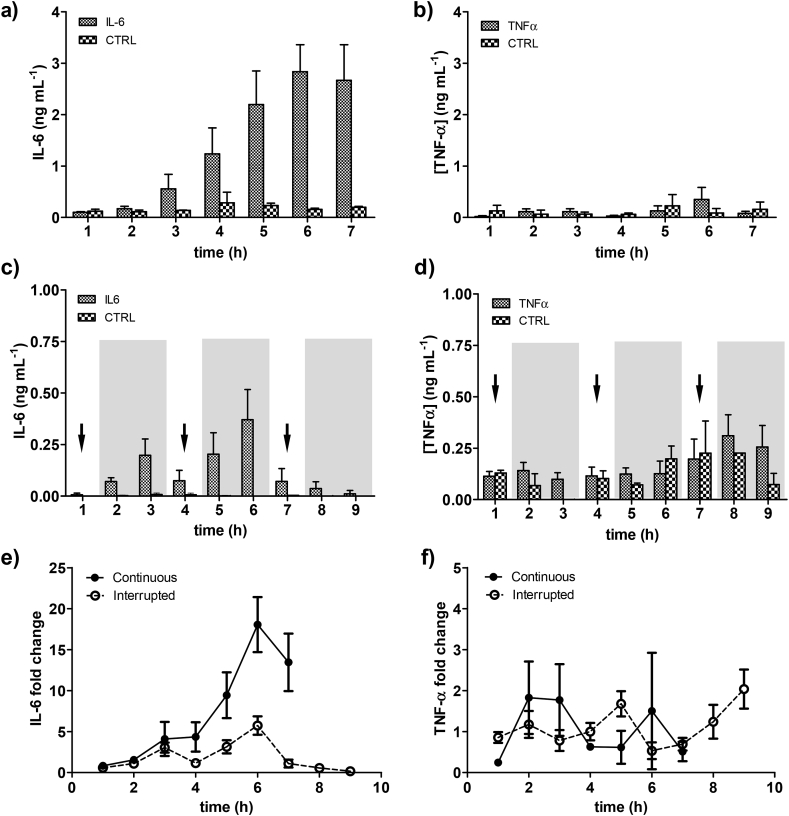
Once we fixed the EPS regime at 1 V, 1 Hz, and 0.002 s, we attempted to elucidate the time-dependent secretion of IL-6 from the 3D skeletal muscles tissues. First, we applied continuous EPS for 7 hours, to mimic endurance exercise and our system allows to detect changes in secreted IL-6 at low concentrations (0.5–3 ng mL−1). Fig. 6a shows a significant increase of IL-6 from 3 hours after starting EPS, while no significant increase in TNF-α release was observed (Fig. 6b). The effect of electrical stimulation on the IL-6 expression has been demonstrated previously in C2C12 myotubes, rat myotubes, with a peak after 2 hours of EPS, in agreement with our findings (Evers-van Gogh et al., 2015; Bustamante et al., 2014; Peake et al., 2015).
Fig. 6.
Monitoring of a) IL-6 secretion and b) TNF-α secretion under continuous EPS. Monitoring of c) IL6 secretion and d) TNF-α secretion under interrupted EPS. Arrows indicate EPS and gray background indicates relax period. Fold change increase in e) IL-6 and f) TNF-α secretion was calculated to assess responsiveness of 3SM to electrical stimulation. Results from continuous EPS (dots-line) and interrupted EPS (empty dots-dash line) show an impact on the IL-6 secretion but not on the TNF-α secretion. Data are shown by mean ± SD. (n = 4–6).
Next, we wanted to mimic interrupted exercise by alternating EPS with relaxed periods and use the biosensor platform to discretize when 3D skeletal muscle releases the cytokines. To this end, we applied stimulation periods of 1 h and relaxation periods of 2 h. First, in the case of IL-6 we observed lower secretion than in continuous EPS (Fig. 6c), while the TNF-α secretion kept similar, under 0.5 ng mL−1, as in the case of continuous EPS (Fig. 6d) without any pattern associated to the stimulation periods.
We also found an unexpected profile in the IL-6 secretion: a significant increase of secreted IL-6 took place during relaxation periods after 2 and 3 h, and after 5 and 6 h. To compare both regimes, we normalized the cytokine secretion by fold change calculation between stimulated and control samples (EPS/Ctrol). In the case of IL-6, we were able to measure differences based on the different EPS regimes. During the 3 first hours no differences are found in terms of fold change, but after that point, the endurance exercise (continuous EPS) induced higher IL-6 secretion, while alternate exercise produced to a different profile with lower IL-6 secretion levels and the presence of secretion peaks (Fig. 6e). As expected, the TNF-α profiles were similar with random tendencies along the time and low fold change (Fig. 6f). As we showed, the integrated platform allows to monitor the cytokine secretion of the 3D skeletal muscle tissue over the time to discretize the effect of different EPS regimes.
4. Conclusions
In this study, we developed an in vitro integrated platform to monitor the secretion of inflammatory processes related cytokines from single tissue-based models. When functionalized, magnetic beads were coupled with a 3D tissue engineered skeletal muscle, and chemical and electrical stimuli. First, we succeeded in the fabrication of the 3D tissue engineered skeletal muscle on a porous membrane of Transwells, what allowed the diffusion of the secreted cytokines to the lower compartment. Next, magnetic beads assay developed underneath the membrane, allowed amperometric measurements of IL-6 and TNF-α. Changes in the concentrations of IL-6 and TNF-α were measured clearly with limits of detection below of 40 and 20 pg mL−1, respectively. Through this configuration, secretion processes has been monitored by the in situ capturing of the released cytokines in a near real-time. Under proinflammatory drug exposure the expected changes in the cytokine levels were observed, and this effect was further monitored in the 3D tissue engineered skeletal muscle. In addition, this work showed different time-dependent profiles of IL-6 secretion, induced by the electrical stimulation-based contraction. These findings contribute to the better understanding roles of IL-6 in muscle related inflammation process. Therefore, this platform is an attractive candidate for further applications, since it does not require any complex modifications. Future work will involve study of other biomimetic tissues under a wide range of treatments or disorders, as in vitro test to high sensitivity metabolic studies.
CRediT authorship contribution statement
Alejandro Hernández-Albors: Conceptualization, Formal analysis, Writing - original draft, Writing - review & editing, Methodology, Investigation. Albert G. Castaño: Conceptualization, Methodology, Investigation, Writing - original draft. Xiomara Fernández-Garibay: Formal analysis, Writing - original draft, Conceptualization, Methodology, Investigation. María Alejandra Ortega: Conceptualization, Methodology, Investigation. Jordina Balaguer: Formal analysis, Investigation. Javier Ramón-Azcón: Conceptualization, Writing - review & editing, Methodology, Investigation, Supervision.
Acknowledgements
Funding for this project was provided by the Ramon y Cajal (RYC-2014-15022) fellowship and Severo Ochoa Program for Centers of Excellence (R&D 2016–2019) funded by the Ministerio de Economía, Industria y Competitividad; an ERC grant (ERC starting grant project – 714317 – DAMOC); the CERCA Programme/Generalitat de Catalunya (2014-SGR-1442 and 2014-SGR-1460); and the Fundación Bancaria "la Caixa"- Obra Social "la Caixa” (projecte IBEC-La Caixa Healthy Ageing). The authors thank Micro-Nano Technology Unit (U2 of the ICTS “NANBIOSIS”) from Institute of Microelectronics of Barcelona (IMB-CNM, Barcelona, Spain), for the design and fabrication of the several custom-made pieces used in this work. We thank Patrick Han at SayEdit.com for help with English editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biosx.2019.100025.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- An X., Sendra V.G., Liadi I., Ramesh B., Romain G., Haymaker C., Martinez-Paniagua M., Lu Y., Radvanyi L.G., Roysam B., Varadarajan N. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banan Sadeghian R., Ebrahimi M., Salehi S. J. Tissue Eng. Regen. Med. 2018;12:912–922. doi: 10.1002/term.2502. [DOI] [PubMed] [Google Scholar]
- Bruls D.M., Evers T.H., Kahlman J.A.H., van Lankvelt P.J.W., Ovsyanko M., Pelssers E.G.M., Schleipen J.J.H.B., de Theije F.K., Verschuren C.A., van der Wijk T., van Zon J.B.A., Dittmer W.U., Immink A.H.J., Nieuwenhuis J.H., Prins M.W.J. Lab Chip. 2009;9:3504. doi: 10.1039/b913960e. [DOI] [PubMed] [Google Scholar]
- Bustamante M., Fernández-Verdejo R., Jaimovich E., Buvinic S. Am. J. Physiol. Metab. 2014;306:E869–E882. doi: 10.1152/ajpendo.00450.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Liu Y., Hu D., Qian W., Tin C., Sun D., Chen W., Lam R.H.W. Lab Chip. 2018;18:522–531. doi: 10.1039/c7lc01183k. [DOI] [PubMed] [Google Scholar]
- Egawa T., Ohno Y., Goto A., Sugiura T., Ohira Y., Yoshioka T., Hayashi T., Goto K. J. Caffeine Res. 2016;6:88–96. [Google Scholar]
- Esteban-Fernández de Ávila B., Escamilla-Gómez V., Campuzano S., Pedrero M., Salvador J.-P., Marco M.-P., Pingarrón J.M. Sens. Actuators B Chem. 2013;188:212–220. [Google Scholar]
- Evers-van Gogh I.J.A., Alex S., Stienstra R., Brenkman A.B., Kersten S., Kalkhoven E. Sci. Rep. 2015;5:10944. doi: 10.1038/srep10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmawati A., Kitajima Y., Nedachi T., Sato M., Kanzaki M., Nagatomi R. Endocr. J. 2013:137–147. doi: 10.1507/endocrj.ej12-0316. [DOI] [PubMed] [Google Scholar]
- Frost R.A., Nystrom G.J., Lang C.H. Am. J. Physiol. Integr. Comp. Physiol. 2002;283:R698–R709. doi: 10.1152/ajpregu.00039.2002. [DOI] [PubMed] [Google Scholar]
- Furrer R., Eisele P.S., Schmidt A., Beer M., Handschin C. Sci. Rep. 2017;7:40789. doi: 10.1038/srep40789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-lizarribar A., Fernández-garibay X., Velasco-mallorquí F., Castaño A.G. J. Samitier J. Ramon-azcon. 2018;1800167:1–13. doi: 10.1002/mabi.201800167. [DOI] [PubMed] [Google Scholar]
- Juan-Colás J., Hitchcock I.S., Coles M., Johnson S., Krauss T.F. Proc. Natl. Acad. Sci. 2018;115:13204–13209. doi: 10.1073/pnas.1814977115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongsuphol P., Liu Y., Ramadan Q. Biomed. Microdevices. 2016;18:93. doi: 10.1007/s10544-016-0117-4. [DOI] [PubMed] [Google Scholar]
- Kongsuphol P., Gupta S., Liu Y., Bhuvanendran Nair Gourikutty S., Biswas S.K., Ramadan Q. Sci. Rep. 2019;9:4887. doi: 10.1038/s41598-019-41338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Soler M., Szydzik C., Khoshmanesh K., Schmidt J., Coukos G., Mitchell A., Altug H. Small. 2018;14:1800698. doi: 10.1002/smll.201800698. [DOI] [PubMed] [Google Scholar]
- Marotta M., Bragós R., Gómez-Foix A.M. Biotechniques. 2004;36:68–73. doi: 10.2144/04361ST01. [DOI] [PubMed] [Google Scholar]
- Mukherjee P., Mani S. Biochim. Biophys. Acta - Proteins Proteomics. 2013;1834:2226–2232. doi: 10.1016/j.bbapap.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedachi T., Fujita H., Kanzaki M. Am. J. Physiol. Metab. 2008;295:E1191–E1204. doi: 10.1152/ajpendo.90280.2008. [DOI] [PubMed] [Google Scholar]
- M. A. Ortega, X. Fernández-Garibay, A. G. Castaño, F. De Chiara, A. Hernández-Albors, J. Balaguer-Trias and J. Ramón-Azcón, Lab Chip, , DOI:10.1039/C9LC00285E. [DOI] [PubMed]
- Park H., Bhalla R., Saigal R., Radisic M., Watson N., Langer R., Vunjak-Novakovic G. J. Tissue Eng. Regen. Med. 2008;2:279–287. doi: 10.1002/term.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peake J.M., Della Gatta P., Suzuki K., Nieman D.C. Exerc. Immunol. Rev. 2015;21:8–25. [PubMed] [Google Scholar]
- Ramón-Azcón J., Ahadian S., Estili M., Liang X., Ostrovidov S., Kaji H., Shiku H., Ramalingam M., Nakajima K., Sakka Y., Khademhosseini A., Matsue T. Adv. Mater. 2013;25:4028–4034. doi: 10.1002/adma.201301300. [DOI] [PubMed] [Google Scholar]
- Riahi R., Shaegh S.A.M., Ghaderi M., Zhang Y.S., Shin S.R., Aleman J., Massa S., Kim D., Dokmeci M.R., Khademhosseini A. Sci. Rep. 2016;6:24598. doi: 10.1038/srep24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg E.V. Nat. Immunol. 2007;8:441–444. doi: 10.1038/ni1461. [DOI] [PubMed] [Google Scholar]
- Saxena A., Dagur P.K., Desai A., McCoy J.P. Front. Immunol. 2018 doi: 10.3389/fimmu.2018.02462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Genma R., Gotou Y., Nagasaka S., Honda H. Bioengineering. 2017;4:56. doi: 10.3390/bioengineering4020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki Y., Yamagishi M., Suzuki N., Izawa K., Nakahara A., Mizuno J., Shoji S., Heike T., Harada Y., Nishikomori R., Ohara O. Sci. Rep. 2015;4:4736. doi: 10.1038/srep04736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skardal A., Murphy S.V., Devarasetty M., Mead I., Kang H.-W., Seol Y.-J., Shrike Zhang Y., Shin S.-R., Zhao L., Aleman J., Hall A.R., Shupe T.D., Kleensang A., Dokmeci M.R., Jin Lee S., Jackson J.D., Yoo J.J., Hartung T., Khademhosseini A., Soker S., Bishop C.E., Atala A. Sci. Rep. 2017;7:8837. doi: 10.1038/s41598-017-08879-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Shimizu T., Okano T. Sci. Rep. 2018;8:13932. doi: 10.1038/s41598-018-32163-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.S., Aleman J., Shin S.R., Kilic T., Kim D., Mousavi Shaegh S.A., Massa S., Riahi R., Chae S., Hu N., Avci H., Zhang W., Silvestri A., Sanati Nezhad A., Manbohi A., De Ferrari F., Polini A., Calzone G., Shaikh N., Alerasool P., Budina E., Kang J., Bhise N., Ribas J., Pourmand A., Skardal A., Shupe T., Bishop C.E., Dokmeci M.R., Atala A., Khademhosseini A. Proc. Natl. Acad. Sci. 2017;114:E2293–E2302. doi: 10.1073/pnas.1612906114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.