Abstract
Plant sucrose transporters are required for phloem loading, and therefore are essential for plant growth and development. In common beans (Phaseolus vulgaris) there are only two sucrose transporters functionally characterized. Through a previous RNA‐seq study, we identified a putative sucrose transporter in common bean, which we hypothesize to function in import of sucrose into plant cells. In silico analysis revealed that PvSUT1.1 is a putative sucrose‐proton co‐transporter distinct from other characterized sucrose transporters in common bean indicating that this is a previously undescribed transporter protein in beans. Further analysis revealed that PvSUT1.1 shares high protein sequence homology to the phloem loader Arabidopsis SUC2; both have 12 transmembrane domains, a typical characteristic of plant sucrose transporters. Heterologous expression in yeast further showed PvSUT1.1 to be functional and it imported sucrose into yeast cells with a Km of 0.7 mM sucrose. Import of sucrose through PvSUT1.1 is also pH‐dependent with highest uptake at pH 4.0, and activity is lost in the presence of the uncoupler carbonyl cyanide 3‐chlorophenylhydrazone. Consistent with identification of PvSUT1.1 as a Type I transporter, PvSUT1.1 also transports esculin. Finally, PvSUT1.1 showed expression in multiple tissues and the protein was localized to the plasma membrane. The results show that PvSUT1.1 is a sucrose transporter that is probably involved in the uptake of sucrose into source and sink cells. The potential role of PvSUT1.1 in leaf phloem loading of sucrose in common beans and its importance in heat tolerance of reproductive tissues are further discussed.
Keywords: Esculin uptake, phloem loading, sucrose transporter, sucrose uptake, transporter regulation, uptake kinetics
1. INTRODUCTION
Leaves capture atmospheric CO2 and ultimately convert it to starch for transient storage, and sucrose for long distance transport to developing sink tissues. In most plant species, carbon is transported throughout the plant as sucrose. After its synthesis in the mesophyll cells, sucrose moves toward the vascular bundle symplasmically via the plasmodesmata. In apoplastic phloem loading species, sucrose reaches the phloem parenchyma cells where it is unloaded into the cell wall space. Unloading from the parenchyma cells to the apoplast was hypothesized to be carried out by sucrose exporter proteins. Recently these proteins were identified as SWEET11 and SWEET12 in Arabidopsis (Chen et al., 2012). From the apoplast, sucrose is taken up into the phloem companion cells by sucrose transporter proteins such as AtSUC2 in Arabidopsis and ZmSUT1 in maize for allocation to sink tissues (Baker et al., 2016; Gottwald, Krysan, Young, Evert, & Sussman, 2000; Slewinski, Meeley, & Braun, 2009; Srivastava, Ganesan, Ismail, & Ayre, 2008; Stadler & Sauer, 1996). Export and delivery of sucrose to reproductive sink tissues is critical for sink formation, growth, and development (Kühn, 2003; Riesmeier, Willmitzer, & Frommer, 1994). Since most of the food for the world's population depends on sink tissues (seed, fruit, and tuber), it is critical to fully understand the process of sucrose allocation to these tissues.
Plant sucrose transporters belong to the major facilitator superfamily (MFS), also called the uniporter‐symporter‐antiporter family (Goswitz & Brooker, 1995; Marger & Saier, 1993; Pao, Paulsen, & Saier, 1998). MFS transporters, depending on the subfamily, can adopt either inward open, outward open, inward facing occluded, or outward facing occluded conformational states where the transition involves both rigid‐body rotations, as in the rocker switch model for sucrose transporters, and structural changes in individual transmembrane helices (Quistgaard, Löw, Guettou, & Nordlund, 2016).
The SUC/SUT subfamily of MFS have been extensively studied in many plant species having different subgroups/types (Doidy, Vidal, & Lemoine, 2019; Reinders, Sivitz, & Ward, 2012). Evolutionary analysis of sucrose transporters from fission yeast, charophyte algae, red algae, non‐vascular plants, early vascular land plants, and angiosperms show three transporter types (Reinders et al., 2012). Type I sucrose transporters are only found in eudicots, they are localized to the plasma membrane and are able to transport coumarin glycosides (for example esculin) in addition to sucrose. In contrast, Type II and III sucrose transporters only transport sucrose and noncoumarin glycosides (Gora, Reinders, & Ward, 2012; Reinders, Sivitz, Starker, Gantt, & Ward, 2008; Reinders et al., 2012; Sivitz et al., 2007). Type II sucrose transporters localize to the plasma membrane while Type III transporters are found in the vacuole membrane (tonoplast).
Sucrose transporters have been studied intensively in Arabidopsis. Specific expression patterns and localization to either the plasma membrane or tonoplast indicate specific, nonredundant functions for each transporter in the family (Endler et al., 2006; Kühn, 2003; Sivitz et al., 2007; Truernit & Sauer, 1995). For example, AtSUC1 is expressed in trichomes, pollen grains, and roots and is essential for normal pollen function (Sivitz et al., 2007; Sivitz, Reinders, & Ward, 2008). AtSUC2 is important for phloem loading of sucrose (Srivastava et al., 2008) while AtSUT4/SUC4 is localized to the tonoplast and transports sucrose from the vacuole into the cytoplasm (Endler et al., 2006; Schulz et al., 2011). In legumes, particularly in common bean (Phaseolus vulgaris), there are very few sucrose transporters functionally characterized to date. PvSUT1 along with a putative sucrose facilitator PvSUF1, which were isolated from the seed coat, were shown to allow growth of yeast on sucrose (Zhou, Qu, Dibley, Offler, & Patrick, 2007). Recently, Soltani, Weraduwage, Sharkey, and Lowry (2019) reported RNA‐seq analysis showing that expression of putative sucrose transporter Phvul.007G088200 is significantly reduced in leaves when exposed to elevated temperature. This gene is homologous to AtSUC2 and was named PvSUT2 (Soltani et al., 2019). In a more recent phylogenetic analysis of legume sucrose transporters, however, this gene was named PvSUT1.1, as a member of the Type I clade (Doidy et al., 2019). Common bean, like other plants, encodes a Type II sucrose transporter and thus to avoid confusion Phvul.007G088200 will be referred to as PvSUT1.1 in this study. PvSUT1.1 is grouped within the SUT1.1 subclade of the legume Type I sucrose transporter while PvSUF1 and PvSUT1 analyzed by Zhou et al. (2007) are from the SUT1.4 and SUT1.5 subclades and thus renamed PvSUT1.4 and PvSUT1.5 respectively (Doidy et al., 2019). At present, the function of PvSUT1.1 in sucrose uptake has not been demonstrated.
In this study we functionally characterized a sucrose transporter, PvSUT1.1, recently identified in common bean and implicated in high temperature resilience during common bean reproduction (Soltani et al., 2019). Because no functional study has reported that PvSUT1.1 is a sucrose transporter, we hypothesized that PvSUT1.1 is functioning in sucrose import into the cells. Predictive, biochemical, and molecular evidence reveal that PvSUT1.1 is a high affinity sucrose‐proton co‐transporter expressed in multiple tissues and is possibly involved in phloem loading of sucrose. Furthermore, the effect of elevated temperature on expression of PvSUT1.1 and the role of sucrose phloem loading in heat tolerance is discussed.
2. MATERIALS AND METHODS
2.1. In silico analysis
Protein sequences were obtained from TAIR (www.arabidopsis.org; AtSUC2), Phytozome (www.phytozome.jgi.die.gov; GmSUT1; PvSUT1/PvSUT1.5, PvSUT1.1[Phvul.007G088200]), and Cool Season Food Legume Crop Database Resources (www.coolseasonfoodlegume.org) and aligned using Clustal Omega (Sievers et al., 2011) prior to generation of the phylogenetic tree. The phylogenetic tree was made using MEGA‐X software (Kumar, Stecher, Li, Knyaz, & Tamura, 2018) using the JTT Maximum Likelihood method with bootstrapping with 1,000 replications (Jones, Taylor, & Thornton, 1992). Clustal Omega (Sievers et al., 2011) was used to generate the protein identity matrix. Prediction of the number of transmembrane domains was carried out using the PvSUT1.1 peptide sequence and TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Three‐dimensional structure of PvSUT1.1 was predicted using PHYRE2 (Kelley, Mezulis, Yates, Wass, & Sternberg, 2015).
2.2. Cloning, and esculin and sucrose uptake
The coding sequence of PvSUT1.1 was obtained from Phytozome and was synthesized (Bio Basic Inc., Ontario, Canada) with PstI restriction sites on each end and cloned into yeast expression vector pDR196 (Rentsch et al., 1995; Gora et al., 2012). Yeast strain SEY6210 [MATα leu2‐3, 112 ura3‐52 his3‐Δ200 trp1‐Δ901 lys2‐Δ801 suc2‐Δ9] (Robinson, Klionsky, Banta, & Emr, 1988) was transformed with this construct and cells were grown on SD/‐URA plates (Takara Bio USA, Inc.) for 2 days at 28°C. Six independent transformants were then spread on fresh SD/‐URA plates and grown overnight (approximately 16 hr) at 28°C to test for esculin (a glycoside of coumarin that is diagnostic for Type I sucrose transporters) uptake, which was performed according to Gora et al. (2012) with modifications. Briefly, patched out yeast cells were scraped from the SD/‐URA plates and then resuspended in 2 ml of SD/‐URA in glass test tubes and grown overnight at 28°C with shaking. The following day, 1 ml of yeast culture was transferred into 1.5 ml tubes and centrifuged for five minutes at 1500 g and the supernatant was aspirated. The pellet was resuspended in 1 mM esculin prepared in 25 mM phosphate (Na2HPO4) buffer at pH 4.0 (or varying pH levels in Figure 4), then vortexed for 20 s and then incubated at 28°C for 1 hr with agitation. After incubation, the cells were pelleted at 1500 g for five min and the supernatant was aspirated. The cells were washed by resuspending the cells in 800 µl of 25 mM phosphate buffer with the same pH as the esculin solution. Yeast cells were then pelleted by centrifugation at 1500 g for five min. Finally, the cells were resuspended in 200 µl 25 mM phosphate buffer by gentle pipetting and then transferred to a black microtiter plate. Fluorescence was read using a Filter Max F5 Multi Mode Microplate Reader (Molecular Devices) at 360 nm (with bandwidth of 35 nm) excitation and 465 nm (with bandwidth of 35 nm) emission. A 50 µl cell suspension from the fluorescence reading was transferred and mixed with 150 µl of water in a clear microtiter plate and OD600 was measured. Relative fluorescence was determined by calculating the fluorescence per unit OD600. Yeast cells transformed with either potato (pDR196::StSUT1) or rice (pDR196::OsSUT1) sucrose transporters were used as positive and negative controls respectively (Gora et al., 2012).
FIGURE 4.
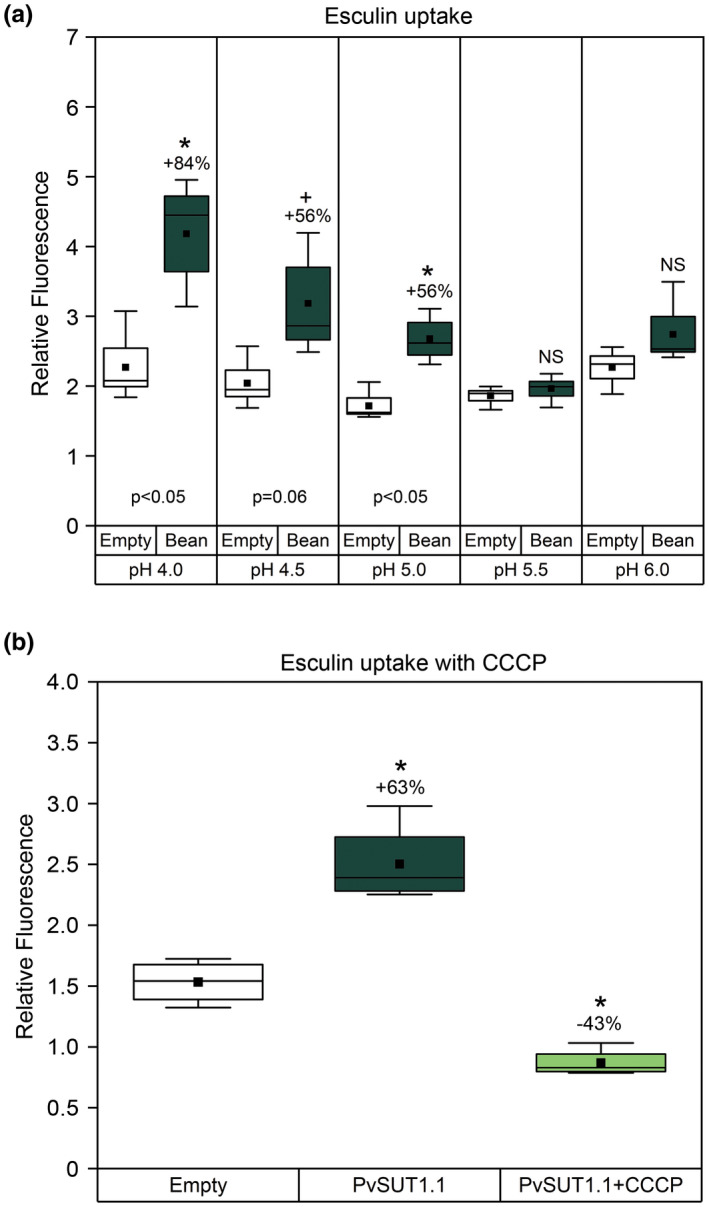
pH dependent Esculin uptake in yeast SEY6210 cells. The cells were transformed with either empty pDR196 (Empty) or with pDR196::PvSUT1.1 (Bean) and incubated in 1 mM esculin solution with either (A) varying pH levels or (B) with the protonophore CCCP (100M) for one hour at 28°C. Box plots represent relative fluorescence values normalized for cell density (n = 3‐4). Solid black squares represent the mean while the horizontal line inside the box represent the median. Numbers above the boxes indicate percent increase compared to the empty vector control. Asterisk represent significant difference at α = 0.05 level while plus symbol (+) represent difference at α=0.10 level. NS = no significant difference versus empty vector control
For sucrose transport kinetics analysis, the procedure described above was followed with slight modification. The yeast cells were incubated with 150 µL of sucrose (unlabeled sucrose + [14C]‐sucrose) instead of esculin, and then incubated for 1 hr. After incubation, the cells were pelleted and the supernatant was aspirated. The cells were then washed twice with cold 25 mM phosphate buffer, pH 4.0. After pelleting of the cells, 1 ml of distilled water was added to the tube and the cells were resuspended followed by measurements of OD600. Radioactivity was measured by liquid scintillation counting. Data for kinetic analysis were analyzed by using OroginPro 2018b (OriginLab Corp.) where nonlinear curve fitting was performed using the Michaelis‐Menten equation.
2.3. Subcellular localization
The PvSUT1.1 coding sequence without the stop codon was synthesized as described above but instead of restriction sites, the Gateway attL1 and attL2 sites were added on the N and C‐termini respectively. The coding sequence was ultimately cloned into pUC52 plasmid. An LR Gateway recombination reaction was then done between pUC57::PvSUT1.1 and the empty expression vector pEarleyGate100 (Earley et al., 2006), which contains an mGFP coding sequence downstream of the reaction site. After the LR reaction, the resulting vector constituted a C‐terminal fusion of PvSUT1.1 with mGFP (PvSUT1.1::mGFP) under the control of CaMV 35S promoter. The destination vector containing PvSUT1.1::mGFP was then transformed into Agrobacterium tumefaciens GV3101 strain. Young Nicotiana benthamiana leaves were then infiltrated with Agrobacterium harboring the destination vector. A confocal microscope (Olympus FluoViewTM FV1000) was used to take images of leaf epidermal cells two days after infiltration. As plasma membrane control, Arabidopsis PIP2A (At3g53420; Yoo et al., 2016) fused to mCherry (Shaner et al., 2004) was used.
2.4. Plant material, RNA extraction, and RT‐PCR
Common bean (P. vulgaris L. cv Sacramento) was grown in a greenhouse with a day/night temperature of 27°C/20°C with 16‐hr photoperiod and 400–500 µmol/m2 s‐1 light. The plants were fertilized weekly with half‐strength Hoagland's solution. For heat treatment, half of the plants from the control greenhouse were transferred to an adjacent greenhouse with 32°C/27°C upon the appearance of the first flower bud around 25 days after sowing. After 7 days of heat treatment, source leaves were collected from control and heat‐treated plants and immediately frozen in liquid nitrogen. The leaves from control and heat‐treated plants were then ground into a fine powder for total RNA extraction. Total RNA was extracted from plant tissues using TRIzol reagent (InvitrogenTM, Waltham, MA; Chomczynski, 1993) and reverse transcription polymerase chain reaction (RT‐PCR) was performed. cDNA was synthesized using oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (InvitrogenTM; Santiago & Tegeder, 2016). A subsequent polymerase chain reaction (PCR) was performed with 24 cycles using PvSUT1.1‐specific primers and common bean Actin (PvACTIN) gene primer pair was used to control for equal amounts of RNA. The primer sequence used to amplify PvSUT1.1 are Fwd: 5′‐ATGGAACCTCTCTCTGCCA‐3′, Rev: 5′‐CAGGAGCTGGACGTAGG‐3′ and for PvACTIN, Fwd: 5′‐TGCATACGTTGGTGATGAGG‐3′ and Rev: 5′‐AGCCTTGGGGTTAAGAGGAG‐3′.
2.5. Statistical analysis
Data are presented as mean ± standard error of at least four biological replications. Each experiment was carried out at least twice to test for reproducibility of data. For data with more than two groups, one‐way ANOVA and Tukey mean separation test was performed. Student's t‐test was used to determine significant differences between just two groups of data. Statistical analysis was done using OroginPro 2018b (OriginLab Corp.).
2.6. Accession numbers
Sequence data of PvSUT1.1 (Phvul.007G088200) CDS can be found in the EMBL/GenBank under the following accession number: XM_007143572.1.
3. RESULTS
3.1. Phaseolus vulgaris SUT1.1 has high sequence similarity to Arabidopsis and legume sucrose transporters (SUT)
An RNA‐Seq experiment using common bean leaves previously showed that the gene Phvul.007G088200 is homologous to Arabidopsis SUC2 and is downregulated under heat stress conditions (Soltani et al., 2019). Upon comparison with other legume sucrose transporters, analysis of the amino acid sequence alignment showed that this putative sucrose transporter protein is 94% identical with the soybean (Glycine max) sucrose transporter GmSUT1 (Figure 1; Aldape, Elmer, Chao, & Grimes, 2003), 86% identical with Vicia faba SUT1 (Weber, Borisjuk, Sauer, & Wobus, 1997) and with pea (Pisum sativum) PsSUT1 (Tegeder, Wang, Frommer, Offler, & Patrick, 1999). Further sequence comparison with another common bean sucrose transporter, PvSUT1 (Zhou et al., 2007), revealed that Phvul.007G088200 is about 68% identical with this transporter protein. Finally, between the common bean transporter proteins, Phvul.007G088200 showed higher amino acid sequence similarity with Arabidopsis SUC2 (69%; Figure 1). In a recent phylogenetic study, Phvul.007G088200 was named PvSUT1.1 (Doidy et al., 2019) and from here will be referred to as such. Analysis of the P. vulgaris and Arabidopsis peptide sequences further revealed many conserved regions shared within bean and between bean and Arabidopsis transporters (Figure S1).
FIGURE 1.
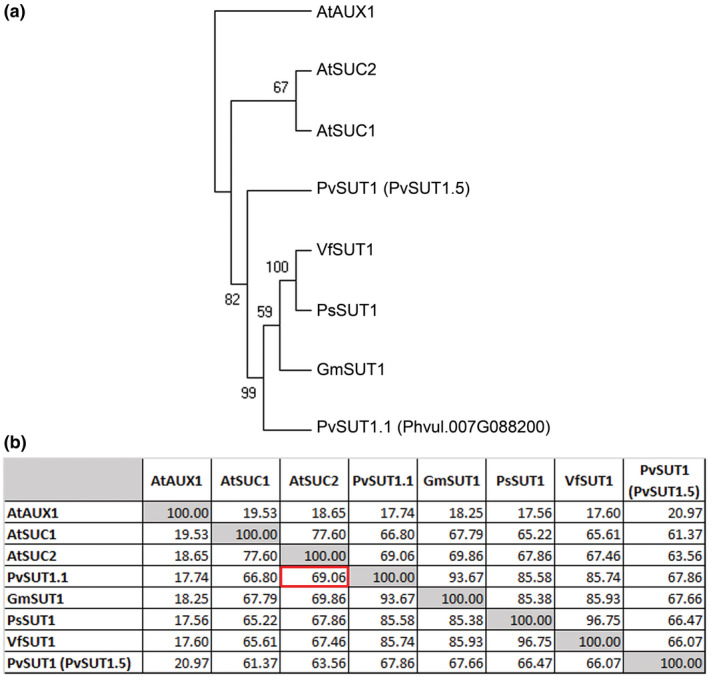
Protein sequence analysis of PvSUT1.1. (A) Rooted phylogenetic analysis of sucrose transporter proteins from Phaseolus vulgaris, Arabidopsis thaliana, Glycine max, Pisum sativum, and Vicia faba. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The Arabidopsis thaliana auxin transporter AUX1 was used as the root. (B) Protein sequence similarity (in percentage) between sucrose transporters
In silico prediction of the 3D structure of PvSUT1.1 revealed that it has 12 hydrophobic transmembrane domains (Figure 2a). These transmembrane regions form a basket‐like structure similar to the Arabidopsis SUC2 (Figure 2b). Rotation of the molecule to show the top and bottom views revealed an opening where sucrose molecules can potentially enter and be moved through the protein. Taken together, these in silico analysis results show close similarity of PvSUT1.1 to other legume and Arabidopsis sucrose transporter proteins. Furthermore, these data suggest that the putative transporter PvSUT1.1 functions as a sucrose transporter.
FIGURE 2.
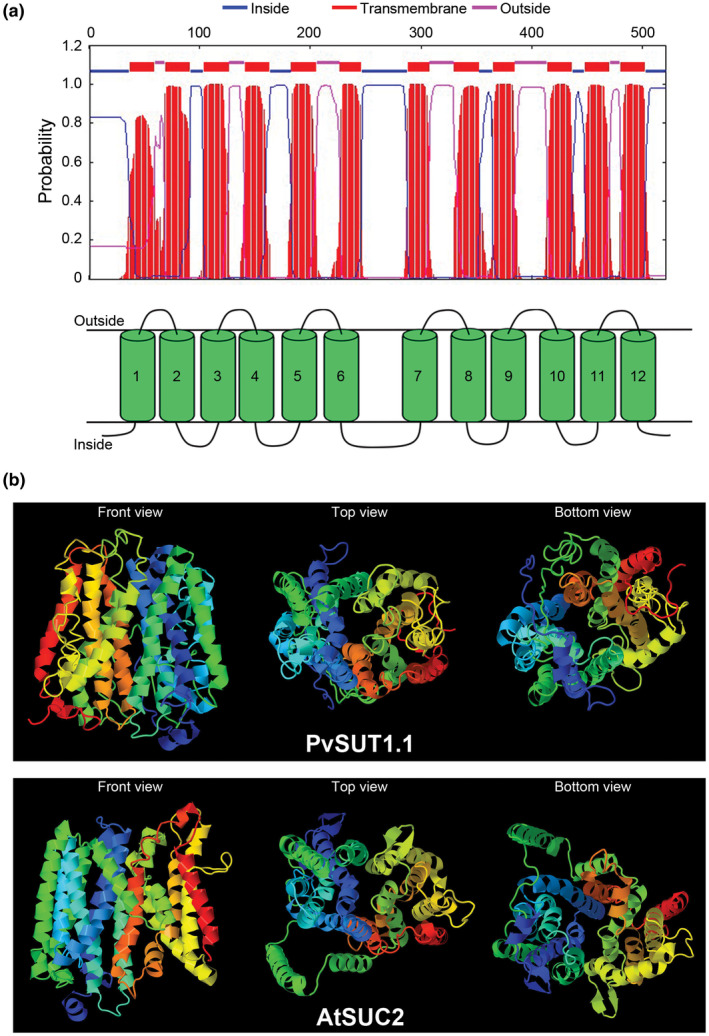
In silico prediction analysis of PvSUT1.1. (A) Transmembrane domain prediction of PvSUT1.1 showing twelve hydrophobic regions. (B) Predicted structure of putative common bean sucrose transporter (PvSUT1.1; top panel) in comparison with the phloem localized Arabidopsis sucrose transporter (AtSUC2; bottom panel). The front, top, and bottom views of the predicted protein structure is shown in each panel
3.2. PvSUT1.1 is a pH‐dependent Type I sucrose transporter
To test whether PvSUT1.1 functions similarly to other Type I sucrose transporters, an esculin uptake assay was performed using yeast cells. Yeast mutant SEY6210 that is lacking an endogenous invertase gene was transformed with PvSUT1.1 and then incubated with esculin. Yeast cells transformed with potato StSUT1 (Type I) and rice OsSUT1 (Type II) were used as positive and negative controls respectively. Type II SUTs are more selective for sucrose and do not transport esculin (Reinders et al., 2012; Sivitz et al., 2007). Results show that the PvSUT1.1‐transformed cells had increased relative fluorescence compared to the empty vector control. The positive control StSUT1‐transformed cells showed the highest fluorescence while no change in fluorescence was measured in the OsSUT1‐expressing yeast cells (Figure 3). These results indicate that PvSUT1.1 is functional in yeast cells and is able to transport esculin consistent with its identification as a Type I sucrose transporter. Transport of sucrose into cells by sucrose transporters is driven by 1:1 co‐transport with a proton (Bush, 1990; Carpaneto et al., 2005; Giaquinta, 1977; Komor, Rotter, & Tanner, 1977). When yeast cells expressing either empty vector or PvSUT1.1 were fed with esculin solutions of varying pH levels from 4.0 to 6.0, cells with PvSUT1.1 had increased fluorescence compared to the empty vector control, but only in pH 4.0 to 5.0. No change in fluorescence was detected in cells incubated in solutions with pH 5.5 or 6.0 (Figure 4a). The dependence of PvSUT1.1 on the proton gradient was further tested by adding a protonophore with the uptake solution (Figure 4b). The results show that yeast cells expressing PvSUT1.1 fed with 1mM esculin alone had increased relative fluorescence versus the empty vector control. In contrast, when cells were fed with the uptake solution (pH 4.0) plus the protonophore carbonyl cyanide m‐chlorophenyl hydrazone (CCCP), a significant reduction of relative fluorescence was observed showing that the import function of PvSUT1.1 was abolished by CCCP. This provides a conclusive evidence that solute transport with PvSUT1.1 is proton coupled.
FIGURE 3.
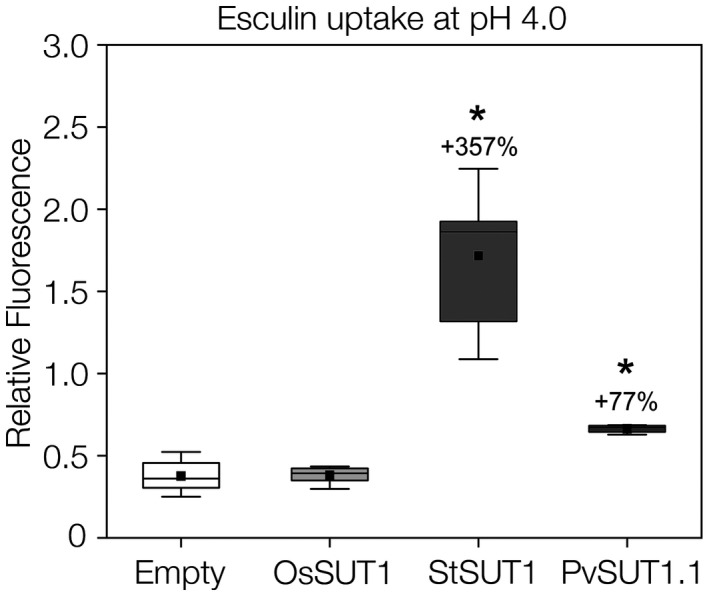
Esculin uptake assay using yeast SEY6210 cells. Potato (StSUT1; type I), rice (OsSUT1; type II), and common bean (PvSUT1.1) sucrose transporters were cloned into yeast expression vector pDR196 and then transformed into yeast cells. In addition to empty vector control, potato and rice transporters were used as positive and negative controls, respectively. Box plots represent relative fluorescence values normalized for cell density (n = 5‐6). Solid black squares represent the mean while the horizontal line inside the box represent the median. Numbers above the boxes indicate percent increase compared to the empty vector control. Asterisk represent significant difference at α = 0.05 level
The ability of yeast cells expressing PvSUT1.1 to transport sucrose was next tested using [14C]‐sucrose (Figure 5). When incubated with increasing concentrations of sucrose, the uptake rate for PvSUT1.1‐expressing cells was higher than the empty vector control showing that PvSUT1.1 is able to import sucrose into yeast cells (Figure 5a). Since the empty vector shows a linear uptake, these values were subtracted from those of the PvSUT1.1‐expressing yeast cells and the final uptake kinetics of the transporter is shown in Figure 5b. The kinetic analysis of [14C]‐sucrose uptake by SEY6210 cells expressing PvSUT1.1 revealed a Km for sucrose of 0.720 mM (Figure 5b). All together, these uptake experiment results show that PvSUT1.1 functions as a high affinity proton coupled transporter with an optimal uptake at pH 4.0 in a yeast system.
FIGURE 5.
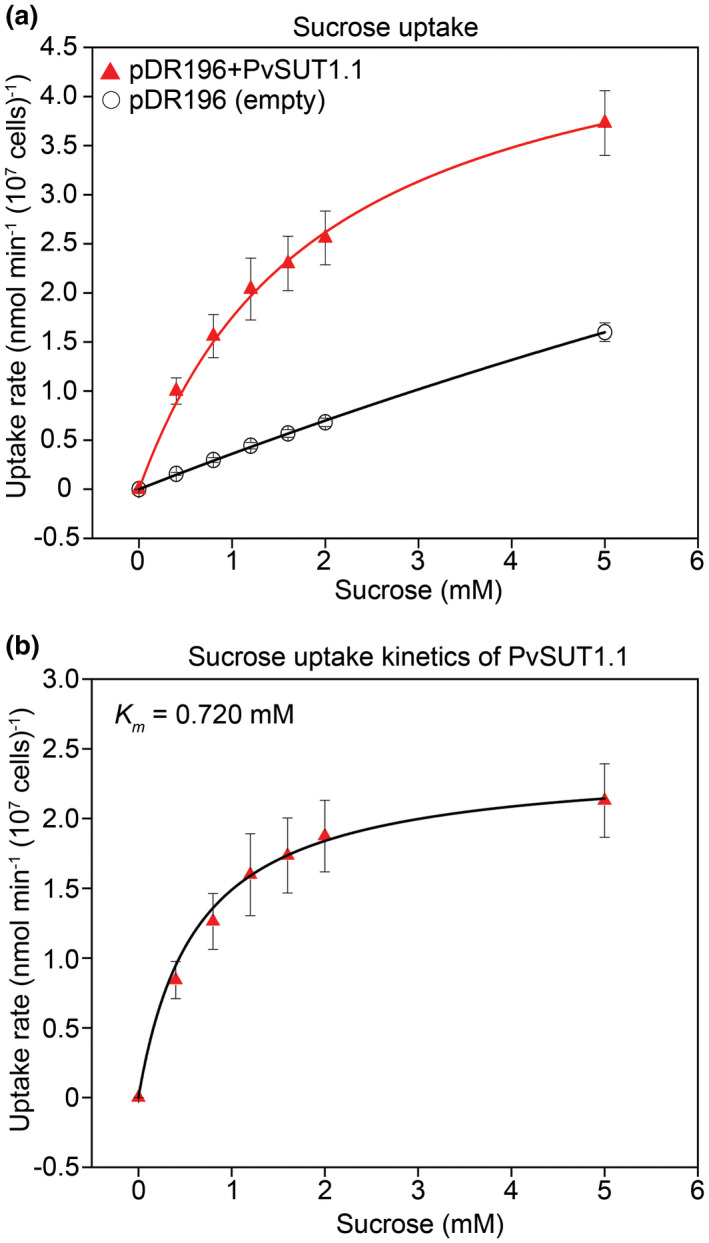
Analysis of sucrose uptake in yeast cells fed with varying total sucrose concentrations. (A) [14C]‐sucrose uptake by yeast strain SEY6210 expressing either empty pDR196 or pDR196+PvSUT1.1 (n = 4). (B) Sucrose uptake kinetics of PvSUT1.1 where a nonlinear regression fit using the Michaelis‐Menten equation is shown. Uptake duration was one hour at 28°C with n = 4 per sucrose concentration. The Km shown here represents the average Km of three independent experiments using independent transformants
3.3. PvSUT1.1 is expressed in multiple tissues and its expression is regulated by temperature
The gene structure of PvSUT1.1 was predicted to have four exons with a transcript that is 2039 bp long with 1566 bp coding sequence (Figure 6a). Using PvSUT1.1‐specific primers, organ‐specific expression of PvSUT1.1 was determined using RT‐PCR. Results show that PvSUT1.1 is expressed in all aerial organs tested but shows stronger expression in stem, flowers, and pods (Figure 6b). Further analysis of PvSUT1.1 expression revealed that its expression decreased by up to 50% due to elevated temperature (Figure 6c).
FIGURE 6.
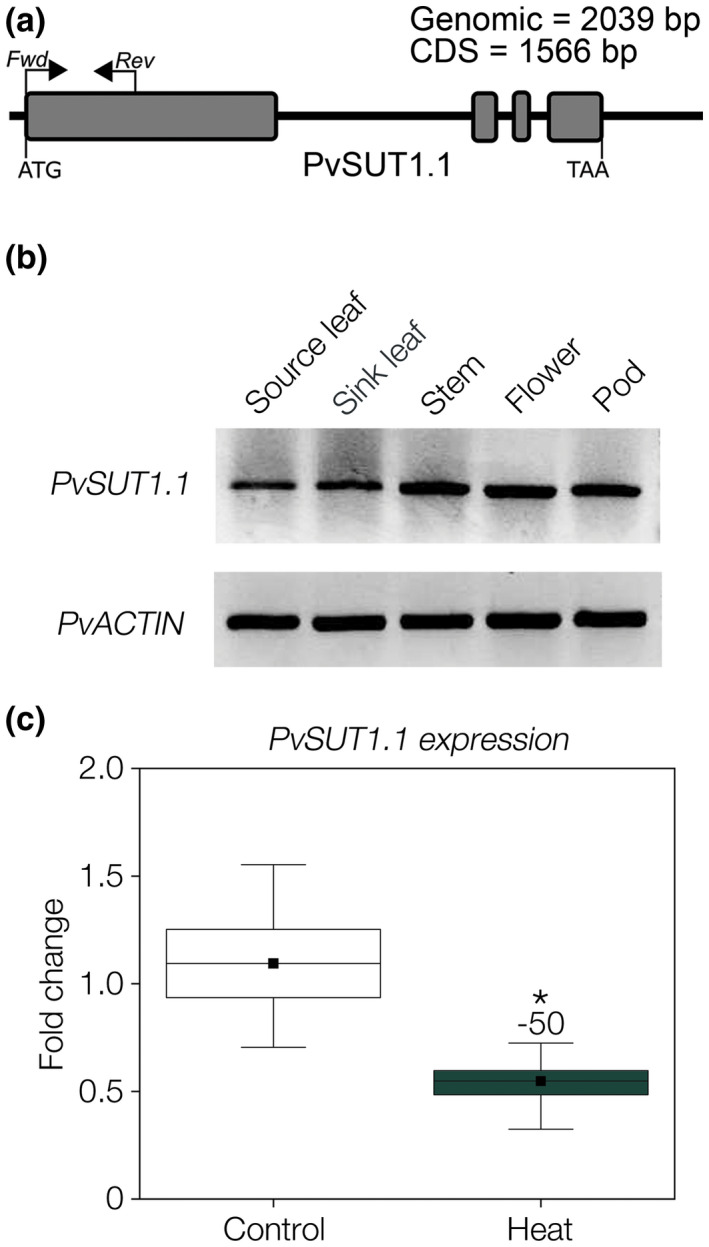
Analysis of PvSUT1.1 expression in common bean. (A) Gene model of PvSUT1.1 showing the primers used for RT‐PCR and qRT‐PCR. (B) Tissue expression analysis of PvSUT1.1 using RT‐PCR. PvACTIN was used as internal control for equal amounts of RNA. (C) Expression of PvSUT1.1 in source leaves exposed to ambient or elevated temperature (32°C) using qRT‐PCR (n=9)
3.4. PvSUT1.1 is targeted to the plasma membrane
To determine the subcellular localization of PvSUT1.1, the transporter fused with a Green Fluorescent Protein (GFP) was transiently expressed in Nicotiana benthamiana leaf epidermal cells (Figure 7). Visualization using a confocal microscope revealed a strong expression of the fluorescent fusion protein in the periphery of the cells. By using a protein known to be targeted to the plasma membrane (AtPIP2A; Yoo et al., 2016), it was resolved that PvSUT1.1 co‐localizes with the AtPIP2A control, thus confirming that PvSUT1.1 is targeted to the plasma membrane. It is also apparent that GFP seems to also be present in the nuclei and the cytosol, which is where free GFP is usually found (Brandizzi et al., 2003). However, the plasma membrane control mCherry is also present in these subcellular compartments indicating an error or defect in the secretory pathway and not on the actual localization of PvSUT1.1. All together, these results suggest that like AtSUC2, PvSUT1.1 might be involved in sucrose import in source and sink cells.
FIGURE 7.
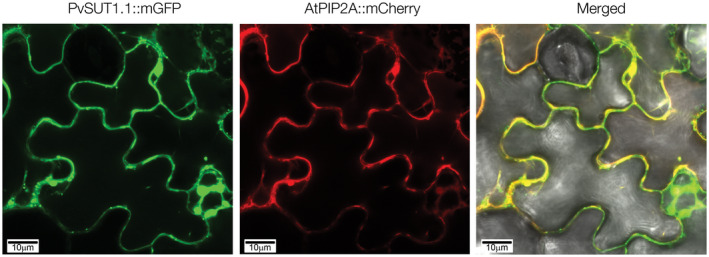
Subcellular localization of PvSUT1.1 in Nicotiana benthamiana leaves. PvSUT1.1::mGFP and AtPIP2A::mCherry (plasma membrane control) were co‐infiltrated and transiently expressed using the Agrobacterium infiltration method. The left and middle images show localization of PvSUT1.1 and AtPIP2A, respectively. The image on the right shows co‐localization of PvSUT1.1 and AtPIP2A to the plasma membrane. Bar = 10 µm
4. DISCUSSION
4.1. PvSUT1.1 shares common features with known sucrose transporters
In a recent study, the Fabaceae family is divided into three clades (SUT1, SUT2, and SUT4 clades; Doidy et al., 2019). Common bean has eight SUT members that were scattered in these three groups. Within the SUT1 clade, there are six subclades (SUT1.1 to SUT1.6) in which P. vulgaris has five paralogous SUT1 genes assigned to three of these six subclades (Doidy et al., 2019). Furthermore, some of these genes were previously indicated to be sucrose facilitators (SUF), for example, PvSUF1, which under the new nomenclature is renamed PvSUT1.4 (Doidy et al., 2019; Zhou et al., 2007). Among these P. vulgaris SUT1 paralogs, two sucrose transport proteins, PvSUT1.4 (PvSUF1) and PvSUT1.5 (PvSUT1), have been functionally characterized in yeast (Zhou et al., 2007). To date, no other member of the Phaseolus PvSUT1 clade has been functionally characterized. In this study, we functionally analyzed a putative sucrose transporter PvSUT1.1 through uptake experiments using 14C‐labeled sucrose fed to yeast cells; we determined that PvSUT1.1 is a sucrose transporter based on several lines of evidence.
Three key findings made during the present study provide evidence to support that PvSUT1.1 is a sucrose transporter. First, was the revelation of high protein sequence similarity between PvSUT1.1 and other legume and Arabidopsis Type I sucrose transporters. In this study PvSUT1.1, shared about 60 to 90% protein similarity with known sucrose transporters in legume and Arabidopsis. This is in agreement with previous studies where homologous sucrose transporters were shown to be 50 to 80% identical with each other (Aoki, Hirose, Scofield, Whitfield, & Furbank, 2003; Payyavula, Tay, Tsai, & Harding, 2011). Sucrose transporters are grouped into three types: Type I, Type II, and Type III (Reinders et al., 2012). Type I SUTs are lacking in nonvascular land plants and monocots and only dicots have this type (Lalonde & Frommer, 2012; Reinders et al., 2012). Recently, common bean was shown to have five Type I SUT genes, presumably each having nonredundant function in plants (Doidy et al., 2019). The specialization within Type I sucrose transporters is best seen in Arabidopsis where there are several Type I sucrose transporters with varying specialized functions (Reinders et al., 2012). For example, AtSUC1 functions in sucrose signaling in vegetative tissues as well as for normal gametophyte function (Sivitz et al., 2008), AtSUC2 in phloem loading (Srivastava et al., 2008), and AtSUC5 in sucrose nutrition of filial tissues during the early stage of embryo development (Baud et al., 2005). While much of the work on sucrose transporters was done with Arabidopsis, studies of legume Type I sucrose transporters exist. Some examples of these include Pisum sativum SUT1 expressed in seed coat and cotyledon epidermal transfer cells of garden pea (Tegeder et al., 1999), Vicia faba SUT1 in seed embryo epidermal cells (Weber et al., 1997), Glycine max GmSUT1 expressed in developing cotyledons (Aldape et al., 2003), and P. vulgaris PvSUT1.5 in vascular and parenchyma cells of the seed coat (Zhou et al., 2007). For common bean, there has been only one study of sucrose transporter function (Zhou et al., 2007). It is reasonable to assume that each of the common bean Type I sucrose transporters has specific functions based on the specificity of Arabidopsis homologous sucrose transporters and that PvSUT1.1 presumably functions in leaf phloem loading due to its high similarity with AtSUC2 at the protein level.
The second observation supporting the idea that PvSUT1.1 is a sucrose transporter is its ability to form a basket‐shaped structure and 12 transmembrane domains. One of the unifying characteristics of sucrose transporters is the presence of twelve α‐helix transmembrane segments and these structures form a basket‐shaped protein and operate by a “rocker‐switch” mechanism (Hirose, Imaizumi, Scofield, Furbank, & Ohsugi, 1997; Karpowich & Wang, 2008; Locher, Bass, & Rees, 2003; Shiratake, 2007; Williams, Lemoine, & Sauer, 2000). The commonality of these 12 transmembrane helices among sucrose transporters in plants was hypothesized to arise from duplication and fusion of a gene that coded for a six‐α‐helix transmembrane protein (Lemoine, 2000). In fact, this is suggested by the large cytoplasmic loop in between transmembrane domain number six and seven (Gottwald et al., 2000; Figure S2a). Within this basket‐like structure is a sucrose binding site but information about this site was lacking. However, it was recently found that when the Arg188 residue in Arabidopsis (AtSUC1, Type I), rice (OsSUT1, Type II), and lotus (LjSUT4, Type III) sucrose transporters were replaced with lysine, the substitution resulted in loss of transport activity showing that the arginine residue is essential for the transporter function (Sun & Ward, 2012). In Arabidopsis SUC2 and common bean SUT1.5 and SUT1.1, this arginine residue is conserved and highlights its important role in transport of sucrose (Figure S2b). The presence of this arginine residue in PvSUT1.1 (Arg164) further supports the idea that PvSUT1.1 is a sucrose transporter protein.
A third key finding is that yeast cells expressing PvSUT1.1 were able to import sucrose. Early characterization of sucrose transporters was done using yeast or Xenopus oocytes (Chandran, Reinders, & Ward, 2003; Meyer et al., 2000; Reinders et al., 2008; Sauer et al., 2004; Sauer & Stolz, 1994). In our study, analysis of PvSUT1.1 in yeast showed transport kinetics similar to the Arabidopsis sucrose phloem loader AtSUC2 having a Km of 0.7 mM (Sauer & Stolz, 1994). It is interesting to note that when expressed in Xenopus oocytes, the AtSUC2 Km is 1.4 mM (Chandran et al., 2003). The same can be said with known sucrose transporters in other plants such as carrot (0.5 mM), tomato (1 mM), and potato (1 mM) (cf. Kühn, 2003). In fact, members of the SUT1 clade show high affinity for sucrose and have a Km between 0.5 and 2 mM (cf. Lalonde, Wipf, & Frommer, 2004). One other characteristic of most plant sucrose transporters is their requirement for low pH to function where one sucrose molecule is moved across the membrane along with one proton (Boorer, Loo, Frommer, & Wright, 1996; Giaquinta, 1977; Sivitz, Reinders, & Ward, 2005; Slone & Buckhout, 1991; Zhou, Theodoulou, Sauer, Sanders, & Miller, 1997). For PvSUT1.1, this held true where the function of the transporter is negatively affected when either the solution pH was increased or when a proton uncoupler (CCCP) was introduced in the uptake solution. While most plant sucrose transporters have an acidic pH optimum, an exception was previously reported where Arabidopsis SUC9 function was shown to be pH independent (Sivitz et al., 2007). An active transport mechanism is needed to import sucrose into the phloem cells since sucrose concentration in the phloem is greater than in the apoplast and can exceed 1 osmolal (Ayre & Turgeon, 2018). Furthermore, previous measurements of sucrose concentration in the phloem sieve tube of Ricinus communis hypocotyls was determined to range between 150 and 300 mM (Kallarackal, Orlich, Schobert, & Komor, 1989; Verscht et al., 1998). In sugar beet source leaves, however, the concentration is in the micromolar range and the apoplastic sucrose concentration (70 µM) was about half of what was measured inside the cells suggesting that sucrose import across the membrane is against a concentration gradient and requires an active import mechanism such as co‐transport with a proton (Bush, 1990; Carpaneto et al., 2005; Fondy & Geiger, 1977; Giaquinta, 1977; Komor et al., 1977). Another requirement for phloem loading of sucrose is that the active transporter must be localized to the plasma membrane where it can transport sucrose across the lipid bilayer. The present study revealed that PvSUT1.1 is localized in the plasma membrane, which supports the idea that it functions in retrieval of sucrose from the apoplast space. Taken together, our results demonstrate that PvSUT1.1 is a Type I sucrose transporter located in the plasma membrane and expressed in multiple plant tissues, and that its function is pH dependent and is involved in import of sucrose into the cell.
4.2. PvSUT1.1 is regulated by temperature and could be involved in leaf sucrose phloem loading
In a recent bean leaf transcriptome study by Soltani et al. (2019), PvSUT1.1 mRNA was found to be reduced at high temperature, consistent with our observation. In a previous study, expression of sucrose transporters from several plant species were shown to respond to environmental cues including temperature, CO2, light, water limitation. In Arabidopsis and maize, expression of the phloem loaders SUC2 and SUT1, respectively, were shown to be downregulated by heat stress (Liesche et al., 2011; Xu, Chen, Yunjuan, Chen, & Liesche, 2018). At the protein level, sucrose transporters were also recently reported to be regulated by phosphorylation and ubiquitination (Xu et al., 2020). Coupled with the reduction of Arabidopsis sucrose transporter expression functioning in leaf phloem loading at elevated temperature, our results support the hypothesis of Soltani et al. (2019) that reduced expression of PvSUT1.1 could result in reduced leaf export of sucrose in common beans. The ability of the plant to export sucrose from leaves is important for thermotolerance (Basu and Minhas, 1991) and based on our expression analysis, this could explain why common beans are generally susceptible to elevated temperature. At present no sucrose transporter functioning in source leaf sucrose phloem loading in P. vulgaris had been identified and demonstrated in vivo. While functional analysis and specific cell and tissue localization in plants are still needed, based on data presented above and since members of the Fabaceae SUT1.1 subclade are presumably involved in sucrose import into the phloem, our results support the hypothesis that PvSUT1.1 functions as a source leaf sucrose phloem loader (Doidy et al., 2019) and is involved in heat tolerance of reproductive tissues. Further physiological analysis is needed to test this hypothesis.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
J.P.S. conceived, designed, and performed the experiments; J.P.S., J.M.W., and T.D.S. analyzed the data and wrote the article.
Supporting information
Fig S1‐S2
ACKNOWLEDGEMENT
The authors thank Professor Mechthild Tegeder (Washington State University) for providing the construct for plasma membrane control AtPIP2A that was used for the localization experiment in this study. The authors thank Dr. Sarathi Weraduwage (Michigan State University) for proofreading the manuscript and providing constructive comments. Finally, the authors thank the MSU Plant Resilience Institute for funding this project and Michigan AgBioResearch for partial salary support for TDS.
Santiago JP, Ward JM, Sharkey TD. Phaseolus vulgaris SUT1.1 is a high affinity sucrose‐proton co‐transporter. Plant Direct. 2020;4:1–11. 10.1002/pld3.260
Contributor Information
James P. Santiago, Email: santjpm@gmail.com.
Thomas D. Sharkey, Email: tsharkey@msu.edu.
REFERENCES
- Aldape, M. J. , Elmer, A. M. , Chao, W. S. , & Grimes, H. D. (2003). Identification and characterization of a sucrose transporter isolated from the developing cotyledons of soybean. Archives of Biochemistry and Biophysics, 409, 243–250. 10.1016/S0003-9861(02)00631-8 [DOI] [PubMed] [Google Scholar]
- Aoki, N. , Hirose, T. , Scofield, G. N. , Whitfield, P. R. , & Furbank, R. T. (2003). The sucrose transporter gene family in rice. Plant and Cell Physiology, 44, 223–232. 10.1093/pcp/pcg030 [DOI] [PubMed] [Google Scholar]
- Ayre, B. G. , & Turgeon, R. (2018). Export of photosynthates from the leaf In Adams W., & Terashima I. (Eds.), The leaf: A platform for performing photosynthesis. Advances in photosynthesis and respiration (Including bioenergy and related processes) (pp. 55–79). Heidelberg: Springer. [Google Scholar]
- Baker, R. F. , Leach, K. A. , Boyer, N. R. , Swyers, M. J. , Benitez‐Alfonso, Y. , Skopelitis, T. , … Braun, D. M. (2016). Sucrose transporter ZmSUT1 expression and localization uncover new insights into sucrose phloem loading. Plant Physiology, 172, 1876–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, P. S. , & Minhas, J. S. (1991). Heat tolerance and assimilate transport in different potato genotypes. Journal of Experimental Botany, 42, 861–866. [Google Scholar]
- Baud, S. , Wuillème, S. , Lemoine, R. , Kronenberger, J. , Caboche, M. , Lepiniec, L. , & Rochat, C. (2005). The AtSUC5 sucrose transporter specifically expressed in the endosperm is involved in early seed development in Arabidopsis. Plant Journal, 43, 824–836. 10.1111/j.1365-313X.2005.02496.x [DOI] [PubMed] [Google Scholar]
- Boorer, K. J. , Loo, D. D. F. , Frommer, W. B. , & Wright, E. M. (1996). Transport mechanism of the cloned potato H+/sucrose cotransporter StSUT1. Journal of Biological Chemistry, 271, 25139–25144. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F. , Hanton, S. , DaSilva, L. L. P. , Boevink, P. , Evans, D. , Oparka, K. , … Hawes, C. (2003). ER quality control can lead to retrograde transport from the ER lumen to the cytosol and the nucleoplasm in plants. Plant Journal, 34, 269–281. 10.1046/j.1365-313X.2003.01728.x [DOI] [PubMed] [Google Scholar]
- Bush, D. R. (1990). Electrogenicity, pH‐dependence, and stoichiometry of the proton‐sucrose symport. Plant Physiology, 93, 1590–1596. 10.1104/pp.93.4.1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpaneto, A. , Geiger, D. , Bamberg, E. , Sauer, N. , Fromm, J. , & Hedrich, R. (2005). Phloem‐localized, proton‐coupled sucrose carrier ZmSUT1 mediates sucrose efflux under the control of the sucrose gradient and the proton motive force. Journal of Biological Chemistry, 280, 21437–21443. 10.1074/jbc.M501785200 [DOI] [PubMed] [Google Scholar]
- Chandran, D. , Reinders, A. , & Ward, J. M. (2003). Substrate specificity of the Arabidopsis thaliana sucrose transporter AtSUC2. Journal Biological Chemistry, 278, 44320–44325. [DOI] [PubMed] [Google Scholar]
- Chen, L. Q. , Qu, X. Q. , Hou, B. H. , Sosso, D. , Osorio, S. , Fernie, A. R. , & Frommer, W. B. (2012). Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science, 335, 207–211. 10.1126/science.1213351 [DOI] [PubMed] [Google Scholar]
- Chomczynski, P. (1993). A reagent for the single‐step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques, 15, 532–534. [PubMed] [Google Scholar]
- Doidy, J. , Vidal, U. , & Lemoine, R. (2019). Sugar transporters in Fabaceae, featuring SUT, MST and SWEET families of the model plant Medicago truncatula and the agricultural crop Pisum sativum . PLoS One, 14, e0223173 10.1371/journal.pone.0223173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley, K. W. , Haag, J. R. , Pontes, O. , Opper, K. , Juehne, T. , Song, K. , & Pikaard, C. S. (2006). Gateway‐compatible vectors for plant functional genomics and proteomics. Plant Journal, 45, 616–629. 10.1111/j.1365-313X.2005.02617.x [DOI] [PubMed] [Google Scholar]
- Endler, A. , Meyer, S. , Schelbert, S. , Schneider, T. , Weschke, W. , Peters, S. W. , … Schmidt, U. G. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidpsis mesophyll cells by a tonoplast proteomic approach. Plant Physiology, 141, 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondy, B. R. , & Geiger, D. R. (1977). Sugar selectivity and other characteristics of phloem loading in Beta vulgaris L. Plant Physiology, 59, 953–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaquinta, R. (1977). Phloem loading of sucrose. pH dependence and selectivity. Plant Physiology, 59, 750–755. 10.1104/pp.59.4.750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gora, P. J. , Reinders, A. , & Ward, J. M. (2012). A novel fluorescent assay for sucrose transporters. Plant Methods, 8, 13 10.1186/1746-4811-8-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswitz, V. C. , & Brooker, R. J. (1995). Structural features of the uniporter/symporter/antiporter superfamily. Protein Science, 4, 534–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald, J. R. , Krysan, P. J. , Young, J. C. , Evert, R. F. , & Sussman, M. R. (2000). Genetic evidence for the in planta role of phloem‐specific plasma membrane sucrose transporters. Proceedings of the National Academy of Sciences of the United States of America, 97, 13979–13984. 10.1073/pnas.250473797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose, T. , Imaizumi, N. , Scofield, G. N. , Furbank, R. T. , & Ohsugi, R. (1997). cDNA cloning and tissue specific expression of a gene for sucrose transporter from rice (Oryza sativa L.). Plant and Cell Physiology, 38, 1389–1396. 10.1093/oxfordjournals.pcp.a029134 [DOI] [PubMed] [Google Scholar]
- Jones, D. T. , Taylor, W. R. , & Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences, 8, 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Kallarackal, J. , Orlich, G. , Schobert, C. , & Komor, E. (1989). Sucrose transport into the phloem of Ricinus communis L. seedlings as measured by the analysis of sieve‐tube sap. Planta, 177, 327–335. 10.1007/BF00403590 [DOI] [PubMed] [Google Scholar]
- Karpowich, N. K. , & Wang, D.‐N. (2008). Symmetric transporters for asymmetric transport. Science, 321, 781–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, L. A. , Mezulis, S. , Yates, C. M. , Wass, M. N. , & Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10, 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor, E. , Rotter, M. , & Tanner, W. (1977). A proton‐cotransport system in a higher plant: Sucrose transport in Ricinus communis . Plant Science Letters, 9, 153–162. 10.1016/0304-4211(77)90093-1 [DOI] [Google Scholar]
- Kühn, C. (2003). A comparison of the sucrose transporter systems of different plant species. Plant Biology, 5, 215–232. 10.1055/s-2003-40798 [DOI] [Google Scholar]
- Kumar, S. , Stecher, G. , Li, M. , Knyaz, C. , & Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde, S. , & Frommer, W. B. (2012). SUT sucrose and MST monosaccharide transporter inventory of the Selaginella genome. Frontiers in Plant Science, 3, 24 10.3389/fpls.2012.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde, S. , Wipf, D. , & Frommer, W. B. (2004). Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annual Review of Plant Biology, 55, 341–372. 10.1146/annurev.arplant.55.031903.141758 [DOI] [PubMed] [Google Scholar]
- Lemoine, R. (2000). Sucrose transporters in plants: Update on function and structure. Biochimica Et Biophysica Acta, 1465(1‐2), 246–262. 10.1016/S0005-2736(00)00142-5 [DOI] [PubMed] [Google Scholar]
- Liesche, J. , Krügel, U. , He, H. , Chincinska, I. , Hackel, A. , & Kühn, C. (2011). Sucrose transporter regulation at the transcriptional, post‐transcriptional, and post‐translational level. Journal of Plant Physiology, 168, 1426–1433. 10.1016/j.jplph.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Locher, K. P. , Bass, R. B. , & Rees, D. C. (2003). Breaching the barrier. Science, 301, 603–604. [DOI] [PubMed] [Google Scholar]
- Marger, M. D. , & Saier, M. H. Jr (1993). A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends in Biochemical Science, 18, 13–20. 10.1016/0968-0004(93)90081-W [DOI] [PubMed] [Google Scholar]
- Meyer, S. , Melzer, M. , Truernit, E. , Hümmer, C. , Besenbeck, R. , Stadler, R. , & Sauer, N. (2000). AtSUC3, a new gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in carpel cell layer. Plant Journal, 24, 869–882. [DOI] [PubMed] [Google Scholar]
- Pao, S. S. , Paulsen, I. T. , & Saier, M. H. Jr (1998). Major Facilitator Superfamily. Microbiology and Molecular Biology Reviews, 62, 1–34. 10.1128/MMBR.62.1.1-34.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payyavula, R. S. , Tay, K. H. C. , Tsai, C.‐J. , & Harding, S. A. (2011). The sucrose transporter family in Populus: The importance of a tonoplast PtaSUT4 to biomass and carbon partitioning. Plant Journal, 65, 757–770. 10.1111/j.1365-313X.2010.04463.x [DOI] [PubMed] [Google Scholar]
- Quistgaard, E. M. , Löw, C. , Guettou, F. , & Nordlund, P. (2016). Understanding transport by the major facilitator superfamily (MFS): Structures pave the way. Nature Reviews Molecular Cell Biology, 17, 123 10.1038/nrm.2015.25 [DOI] [PubMed] [Google Scholar]
- Rentsch, D. , Laloi, M. , Rouhara, I. , Schmelzer, E. , Delrot, S. , & Frommer, W. B. (1995). NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Letters, 370, 264–268. [DOI] [PubMed] [Google Scholar]
- Reinders, A. , Sivitz, A. B. , Starker, C. G. , Gantt, J. S. , & Ward, J. M. (2008). Functional analysis of LjSUT4, a vacuolar sucrose transporter from Lostus japonicus . Plant Molecular Biology, 68, 289. [DOI] [PubMed] [Google Scholar]
- Reinders, A. , Sivitz, A. B. , & Ward, J. M. (2012). Evolution of plant sucrose uptake transporters (SUTs). Frontiers in Plant Science, 3, 22 10.3389/fpls.2012.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesmeier, J. W. , Willmitzer, L. , & Frommer, W. B. (1994). Evidence for an essential role of sucrose transporter in phloem loading and assimilate partitioning. EMBO Journal, 13, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. S. , Klionsky, D. J. , Banta, L. M. , & Emr, S. D. (1988). Protein sorting in Saccharomyces cerevisiae: Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Molecular and Cellular Biology, 8, 4936–4948. 10.1128/MCB.8.11.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago, J. P. , & Tegeder, M. (2016). Connecting source with sink: The role of Arabidopsis AAP8 in phloem loading of amino acids. Plant Physiology, 171, 508–521. 10.1104/pp.16.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, N. , Ludwig, A. , Knoblauch, A. , Rothe, P. , Gahrtz, M. , & Klebl, F. (2004). AtSUC8 and AtSUC9 encode functional sucrose transporters but the closely related AtSUC6 and AtSUC7 genes encode aberrant proteins in different Arabidopsis ecotypes. Plant Journal, 40, 120–130. 10.1111/j.1365-313X.2004.02196.x [DOI] [PubMed] [Google Scholar]
- Sauer, N. , & Stolz, J. (1994). SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker’s yeast and identification of the histidine‐tagged protein. Plant Journal, 6, 67–77. 10.1046/j.1365-313X.1994.6010067.x [DOI] [PubMed] [Google Scholar]
- Schulz, A. , Beyhl, D. , Marten, I. , Wormit, A. , Neuhaus, E. , Poschet, G. , … Hedrich, R. (2011). Proton‐driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant Journal, 68, 129–136. 10.1111/j.1365-313X.2011.04672.x [DOI] [PubMed] [Google Scholar]
- Shaner, N. C. , Campbell, R. E. , Steinbach, P. A. , Giepmans, B. N. , Palmer, A. E. , & Tsien, R. Y. (2004). Improved monomeric red, orange, and yellow fluorescent proteins derived from Discosoma sp. Red Fluorescent Protein. Nature Biotechnology, 22, 1567–1572. 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shiratake, K. (2007). Genetics of sucrose transporter in plants. Genes, Genomes, Genomics, 1, 73–80. [Google Scholar]
- Sievers, F. , Wilm, A. , Dineen, D. , Gibson, T. J. , Karplus, K. , Li, W. , … Higgins, D. G. (2011). Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology, 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz, A. B. , Reinders, A. , Johnson, M. E. , Krentz, A. D. , Grof, C. P. L. , Perroux, J. M. , & Ward, J. M. (2007). Arabidopsis sucrose transporter AtSUC9. High affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiology, 143, 188–198. 10.1104/pp.106.089003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz, A. B. , Reinders, A. , & Ward, J. M. (2005). Analysis of the transport activity of barley sucrose transporter HvSUT1. Plant and Cell Physiology, 46, 1666–1673. 10.1093/pcp/pci182 [DOI] [PubMed] [Google Scholar]
- Sivitz, A. B. , Reinders, A. , & Ward, J. M. (2008). Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose‐induced anthocyanin accumulation. Plant Physiology, 147, 92–100. 10.1104/pp.108.118992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski, T. L. , Meeley, R. , & Braun, D. M. (2009). Sucrose transporter1 functions in phloem loading in maize leaves. Journal of Experimental Botany, 60, 881–892. 10.1093/jxb/ern335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slone, J. H. , & Buckhout, T. J. (1991). Sucrose‐dependent H+ transport in plasma‐membrane vesicles isolated from sugar beet leaves (Beta vulgaris L.). Planta, 183, 584–589. [DOI] [PubMed] [Google Scholar]
- Soltani, A. , Weraduwage, S. , Sharkey, T. D. , & Lowry, D. B. (2019). Elevated temperatures cause loss of seed set in common bean (Phaseolus vulgaris L.) potentially through disruption of source‐sink relationships. BMC Genomics, 20, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, A. C. , Ganesan, S. , Ismail, I. O. , & Ayre, B. G. (2008). Functional characterization of the Arabidopsis AtSUC2 sucrose/H+ symporter by tissue‐specific complementation reveals an essential role in phloem loading but not in long‐distance transport. Plant Physiology, 148, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R. , & Sauer, N. (1996). The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Botanica Acta, 109, 299–306. [Google Scholar]
- Sun, Y. , & Ward, J. M. (2012). Arg188 in rice sucrose transporter OsSUT1 is critical for substrate transport. BMC Biochemistry, 13, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder, M. , Wang, X.‐D. , Frommer, W. B. , Offler, C. E. , & Patrick, J. W. (1999). Sucrose transport into developing seeds of Pisum sativum L. Plant Journal, 18, 151–161. 10.1046/j.1365-313X.1999.00439.x [DOI] [PubMed] [Google Scholar]
- Truernit, E. , & Sauer, N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose‐H+ symporter gene directs expression of β‐glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta, 196, 564–570. [DOI] [PubMed] [Google Scholar]
- Verscht, J. , Kalusche, B. , Köhler, J. , Köckenberger, W. , Metzler, A. , Haase, A. , & Komor, E. (1998). The kinetics of sucrose concentration in the phloem of individual vascular bindles of the Ricinus communis seedling measured by nuclear magnetic resonance microimaging. Planta, 205, 132–139. [Google Scholar]
- Weber, H. , Borisjuk, L. , Sauer, N. , & Wobus, U. (1997). A role for sugar transporters during seed development: Molecular characterization of a hexose and a sucrose carrier in Faba bean seeds. The Plant Cell, 9, 895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L. E. , Lemoine, R. , & Sauer, N. (2000). Sugar transporters in higher plants – a diversity of roles and complex regulation. Trends in Plant Science, 5, 283–290. 10.1016/S1360-1385(00)01681-2 [DOI] [PubMed] [Google Scholar]
- Xu, Q. , Chen, S. , Yunjuan, R. , Chen, S. , & Liesche, J. (2018). Regulation of sucrose transporters and phloem loading in response to environmental cues. Plant Physiology, 176, 930–945. 10.1104/pp.17.01088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q. , Yin, S. , Ma, Y. , Song, M. , Song, Y. , Mu, S. , … Liesche, J. (2020). Carbon export from leaves is controlled via ubiquitination and phosphorylation of sucrose transporter SUC2. Proceedings of the National Academy of Sciences of the United States of America, 117(11), 6223–6230. 10.1073/pnas.1912754117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, Y.‐J. , Lee, H. K. , Han, W. , Kim, D. H. , Lee, M. H. , & Jeon, J. … Kim, J. S. et al (2016). Interactions between transmembrane helices with monomers of the aquaporin AtPIP2;1 play a crucial role in tetramer formation. Molecular Plant, 9, 1004–1017. [DOI] [PubMed] [Google Scholar]
- Zhou, J. J. , Theodoulou, F. , Sauer, N. , Sanders, D. , & Miller, A. J. (1997). A kinetic model with ordered cytoplasmic dissociation for SUC1, and Arabidopsis H+/sucrose cotransporter expressed in Xenopus oocytes. Journal of Membrane Biology, 159, 113–125. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. , Qu, H. , Dibley, K. E. , Offler, C. E. , & Patrick, J. W. (2007). A suite of sucrose transporters expressed in coats of developing legume seeds include novel pH‐independent facilitators. Plant Journal, 49, 750–764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1‐S2


