Abstract
Magnetic Resonance Imaging (MRI) can be used to assess anatomical structure, and its sensitivity to a variety of tissue properties enables superb contrast between tissues as well as the ability to characterize these tissues. However, despite vast potential for quantitative and functional evaluation, MRI is typically used qualitatively, in which the underlying tissue properties are not measured, and thus the brightness of each pixel is not quantitatively meaningful. Positron Emission Tomography (PET) is an inherently quantitative imaging modality that interrogates functional activity within a tissue, probed by a molecule of interest coupled with an appropriate tracer. These modalities can complement one another to provide clinical information regarding both structure and function, but there are still technical and practical hurdles in the way of the integrated use of both modalities. Recent advances in MRI have moved the field in an increasingly quantitative direction, which is complementary to PET, and could also potentially help solve some of the challenges in PET/MR. Magnetic Resonance Fingerprinting (MRF) is a recently described MRI-based technique which can efficiently and simultaneously quantitatively map several tissue properties in a single exam. Here, the basic principles behind the quantitative approach of MRF are laid out, and the potential implications for combined PET/MR are discussed.
Keywords: magnetic resonance fingerprinting, magnetic resonance imaging, positron emission tomography, quantitative imaging
I. Introduction
MEDICAL imaging enables non-invasive assessment of structure and function within the body, aiding in clinical decision-making. Computed tomography (CT) and magnetic resonance imaging (MRI) have been ranked by physicians as the most important innovations in modern medicine [1]. MRI provides excellent soft tissue contrast due to its ability to probe a variety of tissue characteristics. In a traditional scan, data acquisition parameters are selected to highlight a specific tissue property, such as T1, T2, diffusion, susceptibility, etc. The pixel intensity values in these images reflect a “weighting” by the property of interest, where tissues appear bright or dark relative to one another, based on the interplay of the particular value of the tissue property in that pixel with the specific MRI scan parameters. Such images are typically used to qualitatively assess the relationships between different regions in the image (i.e. hyper- or hypo-intense regions relative to surrounding tissue).
As shown by [2], while imaging has become a cornerstone in modern medicine, the use of qualitative images instead of quantitative maps of tissue properties can lead to subjective interpretations and contributes to uncertainty in diagnosis. Radiologist uncertainty, expressed often in “wishy-washy” reports, or “hedges,” remains the most common complaint about radiology. Conversely, the potential advantage of quantitative tissue property maps is that they could be used to objectively classify tissues and compare images over time and across patients, and remove some of the uncertainty that is pervasive in medical imaging. Quantitative imaging biomarkers could enable clinicians to make more objective diagnoses and accurately stage and monitor disease. Accurate quantitative property mapping also contributes toward the development of rapid, comprehensive diagnostic MRI exams. Different properties can provide complementary tissue information, so multiple quantitative properties may be combined to improve the diagnostic sensitivity and specificity. Furthermore, comprehensive MRI exams may reduce the need for biopsies because the quantitative metrics reflect the composition and function of the tissue.
While MRI can theoretically be used to measure a wide array of physical and physiological tissue properties, actual quantitative measurement of tissue properties is seldom performed, due to inherent inefficiencies in quantitative mapping experiments (multiple weighted images are needed for a single map). Approximations can be made to increase the speed of quantitative mapping, but these short-cuts can decrease the accuracy and precision of the results, reducing the utility of these measurements. Moreover, the lack of normative and clinical data makes it challenging for physicians to interpret these tissue property maps.
MR Fingerprinting (MRF) is a quantitative imaging platform that can be applied to map any property to which MRI is sensitive. MRF has been demonstrated to be a robust and rapid method for mapping T1 and T2, as well as other quantitative tissue properties, in multiple applications.
While MRI is an incredibly powerful tool for medical imaging, other modalities provide important and complementary information. Positron emission tomography (PET) is an imaging modality that involves injecting a radioactive isotope, or tracer, into a patient and measuring the gamma photons emitted as a product of the positron annihilation process to create images of activity in the body. The tracer fluorodeoxyglucose (FDG) is most commonly used due to the increased uptake in metabolically active tissue. Multiple other tracers are available and under development [3]–[5], though a discussion of these molecules is beyond the scope of this paper. Standardized uptake value (SUV) is a semi-quantitative tissue property derived from PET images, and represents the radioactivity in a voxel normalized to the whole-body concentration of radioactivity. SUV is not truly quantitative due to inconsistencies in calculation and a variety of error sources [6]. However, using each patient as their own reference for SUV is still valuable for identifying lesions and tracking the response to treatment [7]. The metabolic information obtained with PET images is complementary to that from MRI images, and the use of the two modalities together allows a more complete assessment of tissue characteristics.
Despite the strengths of PET, this modality is seldom used alone. Due to the relatively low spatial resolution, a hybrid structural imaging protocol typically accompanies a PET exam. Integrated PET/CT scanners are widely available and commonly used in clinical applications. Integrated PET/MR imaging has been developed in recent years, and is being explored in the clinical setting.
Integrated PET/MR may enable physicians to better characterize tissues by combining the metabolic information provided by PET and the structural and functional information of MRI. However, the use of the two modalities together poses technical and clinical challenges that provide an opportunity for development in image acquisition and reconstruction technologies, attenuation correction, and in clinical integration of data from both modalities. For instance, MR images can be used for attenuation correction without the cost of additional radiation, but those methods typically rely on a limited set of standard attenuation coefficient values for broad tissue classes. It has been proposed that quantitative MRI methods, such as mapping, may improve the accuracy of attenuation coefficient calculations [8]. MRI can also be used to quantify motion during the PET exam [9], presenting opportunities for motion correction and more accurate PET images. Joint reconstruction of PET and MR images has been proposed [10], which takes advantage of underlying anatomical information in both modalities, particularly the soft tissue contrast available in MR images. Additionally, integrating PET and quantitative MRI data shows promising clinical utility [11], [12]. MRF allows simultaneous measurements of multiple properties efficiently and accurately, and thus may provide a pathway to addressing some of these challenges.
In this paper, we discuss the clinical and scientific implications of quantitative MRI, specifically MRF, and the future potential of combining MRF with PET/MR. In Section II we review the concept of MRF, and its utility in quantitative imaging. In Section III we describe recent developments in PET/MR and opportunities to integrate MRF. In Section IV we discuss some of the challenges to quantitative imaging and bringing these methodologies to clinical practice.
II. Quantitative MRI and Magnetic Resonance Fingerprinting
In recent years, there has been an increasing recognition that quantitative mapping of particular tissue properties can be clinically powerful. Diffusion coefficient [13] and tensor [14] mapping in numerous applications throughout the body [15]–[17] remain the most common clinically-used MR-based quantitative imaging techniques, but there have been critical technical and clinical application advances in fat fraction mapping [18], [19], MR elastography [20], quantitative susceptibility mapping [21], temperature mapping [22], and perfusion mapping [23], among others. This growing body of work is indicative of a new approach to MRI, where quantitative tissue property information can be used to inform clinical decisions.
While T1 and T2 relaxation times are properties that are used to generate contrast in almost all MR images, these tissue properties are rarely mapped clinically, for reasons described below. However, the clinical utility of quantitative mapping of relaxation times is becoming increasingly evident. For example, cardiac T1 and T2 values have been shown to be useful in characterizing myocarditis, myocardial scar, and edema [24]–[28]. Relaxation time maps, particularly T1, can be acquired in a time-efficient manner using methods such as MOLLI [29] or SASHA [30]. mapping is commonly used in the liver to quantify iron content, and multi-echo gradient echo techniques allow for rapid, breath-held acquisitions [31]. In musculoskeletal imaging, T1ρ mapping is a valuable tool for characterizing cartilage [32].
Quantitative MRI is challenging due to inefficiencies and inaccuracy. Typically, multiple high quality images are needed in order to calculate a single quantitative property map. Attempts at reducing the duration of time-expensive quantitative methods in turn results in reduced accuracy and precision. MRF is a quantitative imaging method that grew out of an attempt at addressing some of the difficulties associated with relaxation time mapping [33], [34].
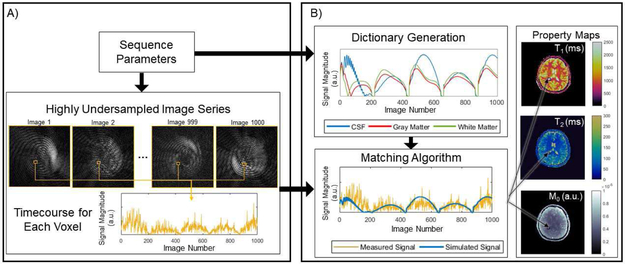
MRF is a technique for simultaneous measurement of multiple tissue properties using MRI [34]. The original experiment demonstrated that accurate measurements of relaxation time (T1), signal decay time (T2), and static field (B0) inhomogeneity could be performed simultaneously in a single rapid scan. The MRF data acquisition and image reconstruction approach is fundamentally different from traditional quantitative MRI techniques. The MRF experiment can be separated into two stages. First in the data acquisition stage, MR signals, termed “fingerprints”, are acquired using a rapid MRI pulse sequence with varying sequence parameters. Second, in the pattern matching stage, the fingerprint from each pixel is matched to atoms of a dictionary to determine the appropriate combination of tissue properties to assign to that pixel. A diagram of the basic structure of an MRF experiment is shown in Fig. 1.
Fig. 1.
Basic components of an MRF experiment. An MRF sequence is designed to quantify a set of properties, such as T1, T2, and M0. The MRF sequence yields highly undersampled images, used to construct the timecourse, or fingerprint, for each voxel. The value at a single voxel for all images in the series are concatenated to form the timecourse for that voxel. Sequence parameters are also used to generate dictionary atoms. The time courses from the undersampled images and the dictionary atoms are compared using a matching algorithm, which provides tissue property values for every pixel in the image. Variations of the elements in (A) are discussed in Section II-A and those in (B) are discussed in Section II-B.
A. Data Acquisition
The goal of MRF is not to generate weighted images, but solely to measure tissue property values at every voxel. In MRF, the acquisition of time-consuming high quality “weighted” images which are typically required by quantitative MRI techniques is replaced by the rapid collection of low quality images. This approach to data acquisition enables more frequent sampling of the information rich, rapidly evolving signal which contains information about T1 and T2 relaxation. In order to distinguish fingerprints that arise from different combinations of tissue properties, the magnetization must be kept out of the “steady state” so that each additional data point collected provides new information, making signals from different combinations of tissue properties unique. To this end, the MRI pulse sequence parameters are varied as data are collected.
The original MRF implementation used a balanced steady state free precession pulse sequence (bSSFP), along with varying repetition times (TR) and flip angles (FA) [34]. However, many different data acquisition schemes have been suggested for MRF. Fast imaging with steady-state precession (FISP or SSFP), proposed by Jiang et al. [35] is advantageous for MRF because it is insensitive to B0 inhomogeneity, but still sensitive to T1 and T2. In [36], Assländer et al. used a pseudo steady-state free precession (pSSFP), which restores the spin echolike behavior of SSFP sequences in MRF to yield accurate property maps that are also robust to B0 inhomogeneity. Amthor et al. [37] proposed reducing the relaxation intervals between MRF repetitions to force the spins into a “stationary state”, reducing the scan time. To avoid errors due to motion, Hamilton et al. [38] used a FISP sequence for cardiac MRF, but divided the MRF experiment into blocks that fit within the diastolic phase of a heartbeat. Furthermore, Hamilton et al. [38] used small FA values (<15°) to minimize the effect of inhomogeneous fields and included both T1 and T2 preparation pulses to improve sensitivity to those properties. Beyond these FA and timing modifications for cardiac MRF, Cohen et al. [39] and Sommer et al. [40] investigated methods for optimizing the pseudo-random FA and TR schedules in an MRF experiment. Cloos et al. [41] took a different approach by playing alternating, heterogeneous fields with the transmit RF coil, leveraging their encoding capabilities to generate unique fingerprints.
The use of a spiral readout trajectory is common in MRF, due to its potential for efficient k-space coverage and incoherent aliasing artifacts when undersampled. The need for noise-like artifacts stems from the assumption that such artifacts do not interfere with the pattern matching approach used to assign tissue properties to each pixel. However, nearly any trajectory can be used for MRF data collection, as long as the sampling pattern avoids coherent aliasing artifacts. Buonincontri et al. [42] have used a 3D Cartesian trajectory for data collection, with undersampling in ky-kz based on a Gaussian distribution, using an SSFP sequence. An echo planar imaging (EPI) readout and spoiled gradient echo sequence was proposed by Rieger et al. [43], which is faster than the undersampled Cartesian readout but not as efficient as the spiral trajectory. Cloos et al. [41] used a radial sampling pattern in which, with an undersampling factor of 50, a comparable scan time and spatial resolution to the spiral acquisition in [34] is achievable. Other trajectories, such as the rosette trajectory, have been proposed for the mapping of tissue properties beyond T1 and T2 due to the specialized properties of their point spread functions [44].
MRF can also be used to collect tissue property maps in 3D, but as the coverage increases, so does scan time. To accelerate volumetric imaging, undersampling can also be performed in the slice direction. Ma et al. [45] achieved whole brain coverage with spatial resolution of 1.2 mm × 1.2 mm × 3 mm in 4.6 minutes using a slice-interleaved acquisition corresponding to an acceleration factor of 3 in the slice direction. Rieger et al. [46] used a slice-interleaved EPI readout, allowing for an acceleration factor of four in the slice direction.
Advanced imaging techniques have also been integrated into MRF experiments. Liao et al. [47] incorporated Cartesian GRAPPA and a sliding window reconstruction into their MRF experiment to accurately measure property maps using only 420 time points, with an acceleration factor of 3 in the slice direction. This approach enabled mapping with a 1.0 mm isotropic resolution and full-brain coverage in 7.5 minutes. Simultaneous multi-slice (SMS) imaging has also been demonstrated to accelerate acquisition time, making volumetric coverage more feasible. Ye et al. [48] and Jiang et al. [49] both used a multiband factor of 2 to speed up data collection for brain MRF. Incorporating slice-GRAPPA with the SMS-MRF proposed in [48] also allowed acceleration to a multiband factor of 3 [50].
The MRF framework has been adapted to measure tissue properties beyond those proposed in the original implementation, specifically relaxation times T1 and T2, proton density, and off-resonance frequency. The spoiled gradient echo sequence used in EPI-MRF by Rieger et al. [43], [46] allows for quantification of instead of T2, which is useful in measuring iron content and blood oxygenation level. Christen et al. [51] introduced MR vascular fingerprinting (MRvF), where they defined their fingerprints as the ratio of the MR signals acquired before and after administering an iron-based contrast agent. The tissue properties measured in this embodiment of MRF were cerebral blood volume, mean vessel radius, and blood oxygen saturation, which allowed the authors to characterize the microvasculature in the brain. They also extended this work to include water diffusion quantification, and demonstrated that this approach could be used to assess diffusion under pathological conditions in rats [52]. Su et al. [53] and Wright et al. [54] have both demonstrated how to modify an arterial spin labeling (ASL) experiment to leverage an MRF-like approach for perfusion mapping. MRF-ASL is capable of simultaneously quantifying several hemodynamic properties, including cerebral blood volume and perfusion, in addition to T1. Anderson et al. [55] used MRF to quantify the local concentration of two contrast agents based on their individual, known relaxation rates. Wang et al. [56] developed an MRF protocol to quantify the chemical exchange rate between phosphocreatine (PCr) and adenosine triphosphate (ATP) via creatine kinase, which provides metabolic information about tissues, particularly muscle. Their MRF experiment alternated excitation of the PCr and ATP, and they quantified the rate of ATP synthesis, the concentration ratio of PCr to ATP, as well as the T1, T2, and resonant frequency for each species. Recently, Cohen et al. [57] and Zhou et al. [58] proposed MRF methods for quantitative chemical exchange saturation transfer (CEST) imaging. The method of Cohen et al. [57] can be used to quantify the exchange rate, exchangeable proton volume fraction, and water relaxation times, and has been demonstrated in phantoms and rat brain tissue.
B. Pattern Matching
Instead of relying on curve fitting to calculate maps of tissue properties from the series of weighted images, tissue properties are extracted directly from the highly accelerated MRF data using pattern matching, as in [59]. After acquiring the MRF data, the signal timecourse, or “fingerprint”, for each voxel is compared to the atoms of a dictionary to determine the tissue properties for that voxel. To calculate these fingerprints, the acquired k-space data are first transformed into the image domain. As noted above, and shown in Fig. 1(A), the resultant images are highly undersampled and not valuable for providing standard anatomical information. In order to generate the fingerprint for each voxel, the complex-valued image intensities are concatenated over time. An example of such a fingerprint can be seen in Fig. 1.
In order to extract the tissue property values from these fingerprints using pattern matching, a dictionary of all possible fingerprints is required. The dictionary describes the signal behavior, or fingerprint, for a specific combination of tissue properties given the application of a particular MRI pulse sequence. In the original MRF experiment, T1, T2, and off-resonance frequency were included as factors which influence the signal behavior, and the Bloch equations were used to calculate the fingerprints for combinations of these properties. A total of 563,784 fingerprints were generated, forming the dictionary used in the work [34]. While Bloch equation simulations are often used to create MRF dictionaries, the Bloch-McConnell equations and the extended phase graph formalism [35] have also been employed.
The acquired fingerprints and the dictionary are then compared to identify the tissue property combination to assign to each voxel. In the original MRF implementation, the vector dot product was used to determine the similarity between the acquired fingerprint for a given voxel and the simulated fingerprints in the dictionary. The dictionary atom that yielded the largest dot product was determined to be the best match, and the tissue properties used to simulate that dictionary fingerprint were assigned to that voxel. This process was repeated for each voxel in the image, and the result is the set of property maps for each of the modeled tissue properties.
As described in the previous section, the MRF framework has been adapted to quantify a number of properties beyond T1, T2, and off-resonance frequency. These approaches require different simulations to create the dictionary. For example, ASL MRF requires perfusion modeling [53], [54] and CEST MRF uses 2- and 3-pool exchange models [57].
In most cases, the MRF dictionary is only generated once for a specific pulse sequence and range of tissue properties. There are instances where the dictionary may need to be calculated for each experiment. These include cardiac MRF [38], where the sequence timing depends on the subject’s heartrate, and MRvF [51], where the signal depends on the dosage of contrast.
As the number of properties to be measured in an MRF experiment grows, so does the size of the dictionary. This can be problematic for both storing the dictionary as well as the time it takes to pattern match. McGivney et al. [60] developed a dictionary compression method in the time domain using the singular value decomposition (SVD). They demonstrated that the dictionary can be compressed to 10%-20% of the original length in the time domain, which offers an approximately 3-5 times reduction in matching speed without decreasing SNR or accuracy in the property maps. Another method to reduce matching time is fast group matching [61], which uses correlations in the dictionary to remove unlikely matches from the search process. Deep learning has been applied to MRF in the work by Cohen et al. [62], where a neural network was trained on simulated MRF data and subsequently used to reconstruct property maps with several orders of magnitude reduction in computation time.
An additional factor that affects the dictionary size is the range and intervals of the tissue properties for which the fingerprints must be modeled. The dictionary should include a representative set of the properties which could reasonably be expected in the tissue to avoid biasing the resulting tissue property maps. The range of values should be large, and it is desirable to use small steps to improve the accuracy of the property maps. However, a large range in the properties along with fine intervals between the tissue property values may be impractical and possibly have diminished returns. One recently proposed solution to this issue is a low rank approximation method by Yang et al. [63], which uses polynomial fitting in the randomized SVD space of an MRF dictionary to interpolate property values from a coarse dictionary. This approach reduces the size of the dictionary and mitigates the problems associated with both small and large step sizes.
An assumption underlying the MRF experiment is that, despite the amount of noise and artifact present in the raw images, the pattern matching can discern the underlying signal timecourse and find the appropriate dictionary entry. This assumption must be carefully tested in simulation, phantom, and in vivo experiments prior to use. The accuracy and precision of the tissue property maps can be improved by optimizing both the pulse sequence design and the tissue property extraction algorithms. It is imperative that the pulse sequence yields signal timecourses with minimal spatio-temporal correlations which are free from coherent artifacts, and that the variation in sequence parameters creates sufficiently different fingerprints for different tissues. Several modifications to the pattern matching step can be useful in reducing the incoherent artifacts in the fingerprints. As the noise-like artifacts are reduced, the amount of data required for accurate property mapping, and thus the scan time, is also reduced. Some of these approaches, including work by Doneva et al. [64], Assländer et al. [65], and Zhao et al. [66], involve a low rank approximation of the MRF data which decreases the aliasing “noise”. Iterative approaches include multi-scale reconstruction by Pierre et al. [67], a maximum likelihood reconstruction by Zhao et al. [68], and accelerated iterative reconstruction for MRF (AIR-MRF) by Cline et al. [69]. A simpler mathematical approach is the sliding window reconstruction described by Cao et al. [70], which effectively offers the same result of reduced aliasing without the need for advanced reconstruction algorithms.
In the dictionary simulation process, confounding factors including the slice profile, inhomogeneity, and intra-voxel dephasing are often ignored. The assumption that these factors do not contribute significantly to the signal timecourse may be valid for some pulse sequences, but not others. Several groups have addressed the impact of these potential confounders on the accuracy of MRF results in the brain [71], [72] and the heart [73]. These confounding factors can be estimated as part of the MRF dictionary, as in [42], [74], or measured separately, as in [75], [76]. Furthermore, it is typically assumed that a voxel will contain only one type of tissue and therefore only one set of tissue properties must be simulated in the dictionary for each pixel. However, a dictionary may misrepresent voxels exhibiting partial volume effects, where the signal timecourse may be better described by a combination of two or more dictionary entries (or an even more complex timecourse). McGivney et al. [77] developed a method for estimating multiple tissue components in a single voxel.
An important aspect of MRF, as with any quantitative imaging tool, is the repeatability of the method. While authors present accuracy measurements for novel MRF protocols in their respective work, the repeatability of MRF is an open research topic [78] and is discussed further in Section IV.
C. Clinical Applications
The original MRF experiment was demonstrated in the brain, and this work demonstrated good agreement between the measured T1 and T2 values for gray matter and white matter and those reported in the literature [34]. While initial work on the clinical applications of MRF relaxometry has been most focused on the brain and prostate [43], [47], [52], [53], [77], [79]–[81], application of MRF to other body regions, such as cardiac, abdomen, breast, and musculoskeletal imaging, is feasible [38], [41], [76], [82].
1). Neuroimaging:
Badve et al. [79] applied the original MRF protocol described in [34] to 56 normal volunteers. They noted regional tissue property differences that correlated with age and gender. Badve et al. [80] also examined T1 and T2 values in 31 patients with intra-axial brain tumors. They found significant differences in the mean T2 values of solid tumor regions of lower grade gliomas and metastases (means of 172 ± 53 ms and 105 ± 27 ms, respectively; P = 0.004) and in the mean T1 values of peritumoral white matter around lower grade gliomas and glioblastomas (means of 1066 ± 218 ms, and 1578 ± 331 ms, respectively; P = 0.004). They also found that the mean T2 values of solid tumor gave the best separation between glioblastomas and metastases, with an area under the receiver operating characteristic curve (AUC) of 0.86 (95% CI, 0.69 −1.00; P < 0.0001). This study provides evidence for using T1 and T2 values to differentiate between disease states, and MRF is an efficient method for measuring these tissue properties simultaneously. Work by McGivney et al. [77] demonstrated the ability to separate different tissue components from single voxels within a brain tumor and in the peritumoral region. This study provides an extra layer of information about the composition of a tumor.
In the EPI MRF work by Rieger et al. [46], the authors tested their method on a patient with multiple sclerosis (MS). The patient’s MS lesion was visible on both T1 and maps, with higher values than the surrounding tissue.
Liao et al. [83] studied the impact of MRF T1 and T2 maps on the diagnostic rate of hippocampal sclerosis (HS) in patients with mesial temporal lobe epilepsy (MSLE). An MR protocol including fluid-attenuated inversion recovery (FLAIR), T1- and T2-weighted imaging, and two-dimensional MRF was performed in 33 patients with MSLE and 30 healthy participants. Both the T1 and T2 values in HS lesions in patients were higher than those of normal hippocampal tissue of healthy participants (T1: 1361 msec ± 85 vs 1249 msec ± 59, respectively; T2: 135 msec ± 15 vs 104 msec ± 9, respectively; P < .0001). The authors found that the diagnosis rate of HS using MRF was 96.9%, as compared to 69.7% with conventional MRI methods. The multiparametric MRF maps are more sensitive to the subtle tissue changes in the HS lesions, and could be a valuable clinical tool in diagnosing and treating epilepsy. Ma et al. [84] also used MRF to detect epileptic lesions. In four of 15 total subjects, MRF T1 and T2 maps showed additional findings as compared to traditional weighted MR images. These additional findings were highly concordant with patients’ EEG results, suggesting that MRF may be a better tool for detecting epileptic lesions.
Diseases related to blood flow may benefit from the rapid tissue characterization possible with MRF. Lemasson et al. [52] used the MRvF approach to measure blood volume fraction, vessel diameter, and blood oxygenation simultaneously in stroke and brain tumor mouse models. They found that MRvF could robustly distinguish between healthy and pathological brain tissue. One notable result is the distinctly different vascular fingerprint of one tumor model as compared to two others. Su et al. [53] applied their ASL-MRF protocol to three patients with Moyamoya disease. The patients showed longer bolus arrival times in stenotic tissue as compared to normal tissue (1310 ± 79 ms, and 933 ± 236 ms, respectively; P = 0.028), which could not be detected with traditional ASL.
Future MRF work in the brain will require larger volumes of coverage in a clinically acceptable amount of time. The 3D methods described in Section II-A show that this is technically feasible, but patient data has yet to be published.
2). Prostate:
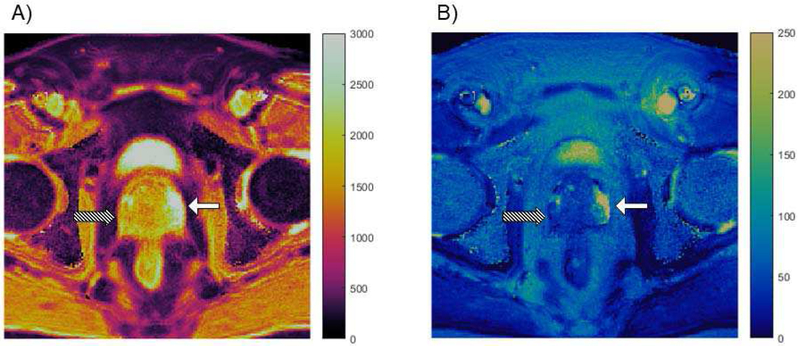
Multi-parametric MRI is a valuable tool in diagnosing prostate cancer, so it is a logical application for MRF. Fig. 2 shows an example of MRF property maps in which prostate cancer is clearly distinguished from normal tissue. Yu et al. [81] developed a prostate MRI exam consisting of MRF for T1 and T2 measurement in addition to conventional apparent diffusion coefficient (ADC) mapping. The protocol was evaluated in 140 patients suspected of having prostate cancer; 109 lesions were analyzed. A number of significant results were reported. In the differentiation between cancer and normal-appearing peripheral zone (NPZ), T1, T2, and ADC from cancerous lesions (means of 1628 ms ± 344, 73 ms ± 27, and 0.773 × 10−3 mm2/s ± 0.331, respectively) were significantly lower than those from NPZ (means of 2247 ms ± 450, 169 ms ± 61, and 1.711 × 10−3 mm2/s ± 0.269) (P < 0.0001 for each). In the differentiation between prostatitis and NPZ, T1, T2, and ADC in prostatitis (means of 1707 ms ± 377, 79 ms ± 37, and 0.911 × 10−3 mm2/s ± 0.239) were significantly lower than those in NPZ (P < 0.0005 for each). Furthermore, ADC and T2 produced the best separation between high- or intermediate-grade tumors and low-grade tumors (AUC = 0.83). The proposed combined MRF and ADC protocol took an average of 12 minutes, as compared to 21 minutes for the standard clinical protocol. The results of this study show the potential for multi-parametric MR protocols to objectively characterize tissues and exemplifies the efficiency of MRF as a quantitative tool. There is significant ongoing work to extend this same approach to characterization of transition zone lesions, to verify and further develop relationships with targeted biopsy and against prostatectomy specimens.
Fig. 2.
Property maps from a prostate MRF study. T1 map (A) and T2 map (B) are both in units of milliseconds. High grade prostate cancer in the peripheral zone (Gleason score = 8), indicated by the striped arrow, has much lower T1 and T2 values as compared to the normal peripheral zone, indicated by the solid white arrow.
3). Cardiac:
Cardiac MRF presents the challenge of imaging a moving object. Hamilton et al. [38] demonstrated a cardiac-gated MRF sequence in normal volunteers. The T1 and T2 values acquired with the cardiac MRF sequence were in good agreement with those acquired with traditional methods, but the MRF experiment only required 16 heartbeats for both T1 and T2 measurements. The comparative methods required 17 heartbeats for T1 mapping and 9 heartbeats for T2 mapping. Cardiac MRF was also demonstrated in heart transplant recipients by Coristine et al. [85], with good correlation to conventional property mapping techniques. The MRF technique could potentially be an efficient, non-invasive method for assessing graft health. MRF is a promising tool for mapping relaxation times in the heart, but, as with most applications, will benefit from volumetric coverage.
4). Abdomen:
Abdominal MRF presents similar challenges to cardiac MRF with respect to motion. Subjects may be instructed to hold their breath to eliminate respiratory motion, though patients have difficulty complying with breath-holds and the length of breath-holding also constrains exam time. Given the large field-of-view required for abdominal imaging, spatial resolution may suffer due to these limits. The presence of fat can also lead to inaccurate measurements. An abdominal MRF exam with an in-plane spatial resolution of 1.9 mm was demonstrated by Chen et al. [76] in eight asymptomatic subjects and six patients with 20 focal liver lesions. The MRF sequence measures T1 and T2 simultaneously in a 19 second breath-hold. The measured T1 and T2 values in metastatic carcinoma (1673 ms ± 331 and 43 ms ± 13) were significantly different from those in the surrounding liver parenchyma in patients (840 ms ± 113 and 28 ms ± 3; P < 0.0001 and P < 0.01, respectively). Measured T1 and T2 values in metastatic carcinoma were also significantly different from those in hepatic parenchyma in asymptomatic subjects (745 ms ± 65 and 31 ms ± 6, P < 0.0001 and P = 0.021, respectively). As with the prostate work, this clinical data suggests that quantitative values can be used to objectively characterize tissue.
5). Breast:
Previous studies have shown the value of T2 relaxation times in diagnosing and predicting outcomes for breast cancer. For example, Manton et al. [86] performed a prospective study on 16 women undergoing neoadjuvant chemotherapy for locally advanced breast cancer to study the ability to predict tumor response using quantitative MRI and MR spectroscopy methods. They found that a decrease in measured T2 early in the treatment period predicted treatment response in 69% of cases with 100% specificity and positive predictive value. Furthermore, the study by Tan et al. [87] showed the product of lesion T2 and tumor volume after two treatment cycles of neoadjuvant chemotherapy is a good predictor of treatment response (positive predictive value = 95.5%, negative predictive value = 84.6%). MRF is a more efficient method for collecting T2 maps, with the option to simultaneously measure additional tissue properties. Recently, Chen et al. [82] developed a three-dimensional acquisition to quantify T1 and T2 values in the breast with a spatial resolution of 1.6 × 1.6 × 3 mm3. They applied the technique to 15 healthy females and 14 females with breast cancer. Among the participants with invasive ductal carcinoma, T2 relaxation times were significantly higher than those in healthy breast tissue (68 ms ± 13 and 46 ms ± 7, respectively; P < 0.001). These results, in addition to the previous studies, support the use of MRF to measure relaxation times to aid in following breast cancer.
6). Alternate Quantitative Approaches:
Certainly, MRF is not the only MR experiment to yield quantitative results. Furthermore, T1 and T2 alone may not provide the complex tissue quantification required for all applications. The rich literature in quantitative MR includes, but is not limited to, fields such as fat fraction measurements, flow and perfusion, diffusion, elastography, and multiple additional approaches to relaxometry. A review of these methods is beyond the scope of this manuscript, but these methods can also be used in addition to or in combination with an MRF approach for quantitative analysis.
III. PET/MR
As described above, MRI can provide a multitude of information which can be used to characterize tissue. However, PET imaging offers complementary tissue information that is also valuable in a number of clinical applications. As quantitative MRI has emerged as a clinically viable approach to collecting MRI data, the combination of quantitative MRI and PET has been explored. While quantitative MRI information can be collected in many different ways, MRF is attractive for this purpose due to its ability to collect several different types of information in a single rapid scan. The following section describes how the combination of quantitative MRI, and specifically MRF, can be used to facilitate improved PET/MR.
A. Technical Challenges and Opportunities for PET/MR with MRF
A major challenge in PET/MR is accurate attenuation correction. Unlike CT, an MRI acquisition does not readily provide information needed for attenuation correction of the PET data. MRI does, however, provide the soft tissue contrast required to segment different tissue types. Attenuation correction in PET/MR is typically performed by segmenting the MR images and assigning attenuation coefficient values to different tissues [8]. One tissue that is particularly problematic to identify with MRI is bone, but ultra-short echo time (UTE) and zero echo time (ZTE) imaging can aid in its identification [88]. However, such an approach can be problematic, as the signal intensities from weighted images cannot be used to unambiguously assign tissue labels and attenuation coefficients, leading to errors in the PET reconstruction. In regions of insufficient attenuation information, PET data themselves can also be used in a joint estimation of attenuation and activity, as suggested by Rezaei et al. [89]. MR images can also be used to inform the reconstruction of PET images as an anatomical prior [90], [91] or in a joint reconstruction [10]. In the joint reconstruction proposed by Knoll et al. [10], mutual structural information is shared between the two modalities to improve reconstruction accuracy.
Quantitative MRI approaches can be used for more accurate tissue segmentation, as objective properties are available for tissue classification instead of relative gray scale image intensities. Relaxometry methods, including MRF, can readily provide differentiation between soft tissue types [92]. Fat-water separation, for which there has also been a proposed MRF experiment [93], can be used in separating fat from other soft tissues. The incorporation of a UTE or ZTE portion in the MRF scan, or simply using a second traditional scan, could provide the information needed to differentiate bone from air, yielding a comprehensive exam to accurately identify four different tissue types – fat, soft tissue, air, and bone. It has been demonstrated that T2* maps, which have been calculated using MRF, can be used for continuous attenuation coefficient calculations [8], allowing for more than four attenuation coefficient values and thereby improving attenuation correction accuracy. MRF is thus an ideal MRI acquisition for rapid and accurate tissue segmentation, which will lead to more accurate attenuation correction and thus more accurate PET images.
MRI has the potential to enhance PET exams in ways beyond attenuation correction. Simultaneous acquisition of PET and MR data enables motion correction in the PET image. Numerous motion measurement and correction techniques exist in MRI for cardiac motion, respiratory motion, and bulk patient motion. If motion is tracked throughout the entire PET exam, the PET counts can be retrospectively corrected to improve the effective resolution of the images. One approach to motion correction involves estimating or modeling motion fields to correct the PET data [9]. Motion correction has proven useful for small lesions in regions that experience a lot of motion, such as the lungs [94]. Furthermore, simultaneous data acquisition results in automatically co-registered multimodality images, which is a much more challenging problem when PET and MR are performed as separate exams.
One can envision a comprehensive MRI sequence for PET reconstruction during the PET exam that would provide motion information as well as tissue characteristics. An MRF sequence would be an excellent candidate. The simultaneous T1 and T2 mapping would provide information for tissue segmentation and attenuation correction, and may also serve a clinical purpose. Such an MRF sequence would require the addition of a navigator for motion correction and, ideally, the ability to identify bone and other tissue composition.
B. Clinical Opportunities for PET/MR with MRF
Given the increasing number and types of quantitative MR technologies in use clinically, there is an opportunity to combine these quantitative data streams from MR with both laboratory testing and other modalities, with PET chief amongst them. As noted above, PET has inherent advantages in quantitative analysis and in providing information that often MRI alone cannot. Integrated PET/MR with MRF has the potential to provide a rich quantitative space for sophisticated tissue characterization.
An important research topic that needs to be explored is the optimal combination of quantitative PET and MRI protocols to yield synergistic results. The individual exams, including the PET tracer and MRI sequences, must be designed to answer specific clinical questions. Moreover, the time of the exam must be limited, as with standard MRI exams, for efficiency purposes.
A recent review of current clinical PET/MR applications can be found in [95], but we will examine a few that could benefit from an MRF approach here.
1). Neuroimaging:
FDG PET has become an invaluable tool in assessing primary brain tumors, but whether there is increased value in an integrated PET/MR exam is undecided. In a recent study by Hojjati et al. [96] comparing PET/MR, DSC perfusion MRI, and PET/CT in differentiating radiation necrosis from tumor recurrence in 24 glioblastoma multiforme patients, the authors found that PET/MR with perfusion MRI provides the best diagnostic utility (AUC = 1.0). In a study by Yoon et al. [97], however, FDG-PET and multi-parametric MRI showed high concordance rates in differentiating high and low grade gliomas but little to no value added in using the two in combination. MR vascular fingerprinting [51], [52], MRF relaxometry with proven neurological applications and utilization [77], [79], [80], [83], and MRF-ASL [53], [54] all hold promise for combination with PET for this important application.
PET/MR also has the potential to be a valuable tool for dementia patients. PET imaging is sensitive to neurodegenerative processes, whereas MRI exams can provide information related to atrophy, iron deposition, and diffusion [98]. The modalities provide complementary information to potentially improve the differential diagnosis of dementias. As MRF may be used to provide the required T1, T2, T2*, and ADC measurements in a single rapid scan, this approach would be ideal for collecting PET/MR data in these patients.
2). Prostate:
The role of MRF in characterizing prostate cancer aggressiveness and differentiation from mimics has been discussed above. With the development of prostate-specific PET tracers, there is potential for improvement with the combined PET/MR exam. In a study by Eiber et al. [11], 66 patients underwent simultaneous 68Ga-PSMA HBED-CC PET/MR prior to prostatectomy. The sensitivity of PET/MR in detecting prostate cancer was significantly higher than that of the multi-parametric MRI exam (95% CI for difference in sensitivities, 21–45; p < 0.001). Furthermore, in localization of prostate cancer, the AUC was significantly higher for PET/MR than for multi-parametric MRI (Δ0.147; 95% CI, 0.081–0.213; p < 0.001). A study by Park et al. [99] used dualtime-point 68Ga-PSMA-11 PET/MR imaging to study 33 men with intermediate- or high-risk prostate cancer. The authors reported that cancer was identified in all patients using the PET images, but only in 29 patients using the multi-parametric MR images. They also suggested that the combined PET/MR imaging allowed for better localization of the cancer due to the higher spatial resolution of the MR images. Combining prostate MRF with PET imaging could possibly improve sensitivity, specificity, and localization of prostate cancer.
3). Cardiac:
PET imaging has been validated for the accurate assessment of myocardial viability and myocardial perfusion [100]. Cardiac T1 and T2 values, which can be mapped using MRF [38], have been shown to be associated with inflammation and myocarditis [24], [25], myocardial scar [26], [101], [102], fibrosis [27], [103], and edema [28], [104]. MRF property maps would contribute high-resolution structural and functional information to the PET/MR exam, yielding richer diagnostic information. In two myocarditis case studies [105], [106], FDG-PET was used to identify regions of active inflammation, increasing the sensitivity of PET/MR to inflammatory process. Some of the benefits of an integrated PET/MR exam include the motion correction and high resolution imaging provided by MRI. For example, an efficient free-breathing acquisition and reconstruction method by Munoz et al. [107] yields improved image sharpness in the myocardium and coronary arteries. The two modalities offer complementary information, in terms of structure versus function as well as anatomical location. The combination of MR angiography and cardiac MRF with structural MR images and PET perfusion information holds the potential for a very powerful cardiac imaging tool.
4). Abdomen:
PET/MR imaging in the liver has shown to be particularly useful in diagnosing liver metastases. Brendle et al. [12] investigated the diagnosis of metastatic colorectal cancer lesions, and found that using PET/MR with diffusion-weighted MRI and ADC mapping was more accurate than the individual modalities themselves. In particular, the highest diagnostic accuracy of the combined PET/MR exam was recorded when detecting liver metastases. PET/MR with diffusion weighted MRI and ADC mapping was superior to PET/CT, and the addition of PET to the MRI exam increased specificity. Kirchner et al. [108] examined the value of PET/MR in detecting and characterizing solid tumors and metastases in the liver. They reported that an exam consisting of FDG-PET and MRI with liver-specific contrast phase yielded the highest accuracy for classifying lesions as malignant or benign. However, they did not find a significant difference in the diagnostic confidence of the PET/MR exam as compared to MRI with liver-specific contrast phase alone. Quantitative MRI could potentially contribute complementary information to better characterize and diagnose liver lesions. Kong et al. [109] obtained maximum SUV values and ADC values in 41 patients with hepatic tumors and found SUV to be negatively correlated with ADC. They also noted that different tumor types had different metabolic and diffusion characteristics, possibly reflecting the underlying tumor biology. It is also possible that T1 and T2 values, which have been measured using MRF and shown to increase in metastatic lesions [76], could provide additional information about tumor composition. An obvious advantage of an integrated PET/MR exam in the abdomen is motion correction and registration. If accurate motion estimation were integrated into the MRF acquisition, then both the PET images and property maps would be motion-corrected and co-registered.
5). Response to Treatment:
Evaluation of tumor response to treatment using MRI typically involves Response Evaluation Criteria in Solid Tumors (RECIST) [110], which tallies the number of lesions and their sizes to assess response. Morphological assessment has limitations: it does not necessarily reflect the composition of the tumor, and waiting for notable size changes may cost valuable time. Both PET and quantitative MRI methods show promise for assessing tumor response. In a study of 378 patients with cervical cancer [111], post-therapy metabolic response, as measured by FDG-PET, predicted progression-free survival just three months after chemoradiation. MRI studies in mice have shown that T1 may be an indicator for tumor response to chemotherapy [112], [113]. Integrated PET/MR exams may provide comprehensive, quantitative metrics sufficient for assessing response to treatment before size-related changes for RECIST assessment are evident. Earlier response evaluation would allow for earlier treatment adaptations. Furthermore, the quantitative metrics that reflect the metabolic activity and composition of the tumor may be valuable in initial treatment planning.
Wang et al. [114] performed FDG-PET/MRI exams on 13 patients with pancreatic ductal adenocarcinoma before and four weeks after chemotherapy began. They were classified as responders or nonresponders according to RECIST using CT exams 8-12 weeks after treatment initiation. Results showed that at the four-week mark, responders had a higher mean reduction in tumor size than nonresponders, but the difference was not statistically significant (20.87% ± 17.86% and 3.4% ± 3.4%, respectively; p = 0.11). However, the responders did have significant differences compared to nonresponders in the reduction of metabolic tumor volume (MTV) (87.7% ± 13.6% and 47.8% ± 21.2%, respectively; p = 0.003), reduction of total lesion glycolysis (TLG) (89.5% ± 11.1% and 50.9% ± 24.5%, respectively; p = 0.006), and increase in mean ADC values (40.2% ± 12.1% and 7.5% ± 15.8%, respectively; p = 0.004). Responders also had a greater reduction in maximum SUV as compared to non-responders, although only approaching significance (46.6% ± 25.8% and 18.0% ± 19.7%, respectively; p = 0.066). These results are mirrored in a study by Sarabhai et al. [115], which included 8 patients with cervical cancer. From integrated FDG-PET/MR exams before and 2-6 weeks after the end of treatment, ADC values, perfusion parameters from DCE-MRI, and metabolic PET parameters were measured. Seven of the eight patients were classified as responders and demonstrated an increase in ADC values, reduction in SUV, and decrease in certain perfusion parameters. One interesting finding was one patient was mis-identified as a partial responder according to RECIST, but using PET data was correctly classified as a complete responder as confirmed with histopathology.
Quantitative imaging has the potential to improve prediction and classification of response to treatment over traditional morphological assessments. MRF will likely be beneficial by providing an efficient method for accurately quantifying a number of tissue properties to characterize tumors within a richer quantitative space.
IV. Open Needs in Quantitative Imaging
There are numerous opportunities for MRF and PET imaging to work synergistically in both the technical aspects and clinical applications of integrated PET/MR. Quantitative MRI could potentially improve attenuation correction and improve tissue characterization for identifying and monitoring disease. While there is certainly evidence to support the use of quantitative MRI, and MRF in particular, outside of PET/MR, there still exist major hurdles before it will be widely accepted in practice.
A major need, and a place of considerable ongoing effort, is multi-scanner standardization, and in the future, multivendor standardization. One important and on-going challenge to quantitative MRI is that the values measured may differ significantly when using different MRI scanners (from different vendors, with different software, hardware, and/or postprocessing tools. The inability to replicate these measurements over time and on different MRI scanners precludes the use of these quantitative metrics as biomarkers for disease. Two major proponents of quantitative imaging standardization are the Quantitative Imaging Biomarker Alliance®(QIBA®) and the National Institute of Standards and Technology (NIST). Both of these organizations have developed phantoms for multi-scanner, multi-site comparisons of DCE-MRI, T1, T2, and diffusion, among other metrics [116], [117]. Numerous other companies have been involved in developing phantoms for quantitative MRI methods, such as fat fraction, , and diffusion tensors [118]–[122]. Accuracy and repeatability are integral to the success of quantitative imaging to study disease across populations and over time.
In the same vein as standardization, precision and accuracy of individual modalities is important for integrated modalities such as PET/MR. Quantitative PET imaging hinges on accurate attenuation correction. The quantitative maps that can be generated with MRF could be used to improve MRI-based attenuation correction approaches, thereby improving the accuracy of quantitative PET measurements.
Another open need in quantitative imaging is the determination of best practices for comprehensive exams which maximize value for clinicians and patients. With the growing development of quantitative imaging, protocols will need to be edited based on the reason for the imaging study. As we have described in the Clinical Applications sections, certain properties are only useful in a select number of situations. Further research must be done to balance the amount of valuable quantitative information with imaging exam time. Efficiency, accuracy, and added-value must all be considered.
V. Summary
Quantitative imaging protocols are powerful clinical tools that can reduce subjectivity and uncertainty in medical imaging. MRF is a one such tool for quantitative MRI, attractive because of its flexibility, efficiency, and the potential to quantify multiple interesting properties simultaneously. PET/MR is a hybrid imaging modality that already combines the metabolic information from PET with an abundance of structural and functional information from MRI. The potential to combine quantitative MR approaches such as MRF with PET for improved image reconstruction informed by the MR acquisition, more accurate attenuation correction, and improved clinical tissue characterization are areas of ongoing investigation and hold significant promise for future development.
Acknowledgment
The authors thank Dr. Ananya Panda and Dr. Debra McGivney for their assistance with the figures, and Dr. Katherine Wright for manuscript revisions.
Contributor Information
Kathleen M. Ropella-Panagis, Department of Radiology, Case Western Reserve University, Cleveland, OH 44106 USA.
Nicole Seiberlich, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH 44106 USA..
Vikas Gulani, Department of Radiology, Case Western Reserve University, Cleveland, OH 44106 USA..
References
- [1].Fuchs VR and Sox HC, “Physicians’ views of the relative importance of thirty medical innovations: A survey of leading general internists provides a useful consensus on the relative importance of innovations to their patients,” Health Aff., vol. 20, pp. 30–42, 2001. [DOI] [PubMed] [Google Scholar]
- [2].Kransdorf MJ and Murphey MD, “Radiologic evaluation of soft-tissue masses: A current perspective,” Am. J. Roentgenol, vol. 175, no. 3, pp. 575–587, 2000. [DOI] [PubMed] [Google Scholar]
- [3].Fendler WP, Czernin J, Herrmann K, and Beyer T, “Variations in PET/MRI operations: Results from an international survey among 39 active sites,” J. Nucl. Med, vol. 57, no. 12, pp. 2016–2021, Dec. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Afshar-Oromieh A, Zechmann CM, Malcher A, Eder M, Eisenhut M, Linhart HG, Holland-Letz T, Hadaschik BA, Giesel FL, Debus J, and Haberkorn U, “Comparison of PET imaging with a 68Ga-labelled PSMA ligand and 18F-choline-based PET/CT for the diagnosis of recurrent prostate cancer,” Eur. J. Nucl. Med. Mol. Imaging, vol. 41, no. 1, pp. 11–20, Jan. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Mojtahedi A, Thamake S, Tworowska I, Ranganathan D, and Delpassand ES, “The value of (68)Ga-DOTATATE PET/CT in diagnosis and management of neuroendocrine tumors compared to current FDA approved imaging modalities: a review of literature,” Am. J. Nucl. Med. Mol. Imaging, vol. 4, no. 5, pp. 426–34, 2014. [PMC free article] [PubMed] [Google Scholar]
- [6].Kinahan PE and Fletcher JW, “Positron emission tomography-computed tomography standardized uptake values in clinical practice and assessing response to therapy,” Semin. Ultrasound. CT. MR, vol. 31, no. 6, pp. 496–505, Dec. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Keyes JW, “SUV: standard uptake or silly useless value?,” J. Nucl. Med, vol. 36, no. 10, pp. 1836–9, Oct. 1995. [PubMed] [Google Scholar]
- [8].Chen Y and An H, “Attenuation correction of PET/MR imaging,” Magn. Reson. Imaging Clin. N. Am, vol. 25, no. 2, pp. 245–255, May 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fayad H, Lamare F, Merlin T, and Visvikis D, “Motion correction using anatomical information in PET/CT and PET/MR hybrid imaging,” Q. J. Nucl. Med. Mol. Imaging, vol. 60, pp. 12–24, 2016. [PubMed] [Google Scholar]
- [10].Knoll F, Holler M, Koesters T, Otazo R, Bredies K, and Sodickson DK, “Joint MR-PET reconstruction using a multi-channel image regularizer,” IEEE Trans. Med. Imaging, vol. 36, no. 1, pp. 1–16, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Eiber M, Weirich G, Holzapfel K, Souvatzoglou M, Haller B, Rauscher I, Beer AJ, Wester H-J, Gschwend J, Schwaiger M, and Maurer T, “Simultaneous 68Ga-PSMA HBED-CC PET/MRI improves the localization of primary prostate cancer,” Eur. Urol, vol. 70, no. 5, pp. 829–836, Nov. 2016. [DOI] [PubMed] [Google Scholar]
- [12].Brendle C, Schwenzer NF, Rempp H, Schmidt H, Pfannenberg C, la Fougère C, Nikolaou K, and Schraml C, “Assessment of metastatic colorectal cancer with hybrid imaging: comparison of reading performance using different combinations of anatomical and functional imaging techniques in PET/MRI and PET/CT in a short case series,” Eur. J. Nucl. Med. Mol. Imaging, vol. 43, no. 1, pp. 123–132, Jan. 2016. [DOI] [PubMed] [Google Scholar]
- [13].Le Bihan D and Breton E, “Imagerie de diffusion in vivo par résonance magnétique nucléaire,” C R Acad Sci Pari, vol. 301, pp. 1109–1112, 1985. [Google Scholar]
- [14].Basser PJ, Mattiello J, and Lebihan D, “MR diffusion tensor spectroscopy and imaging,” Biophys. J, vol. 66, pp. 259–267, 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Le Bihan D, Mangin J-F, Poupon C, Clark CA, Pappata S, Molko N, and Chabriat H, “Diffusion tensor imaging: Concepts and applications,” J. Magn. Reson. Imaging, vol. 13, no. 4, pp. 534–546, Apr. 2001. [DOI] [PubMed] [Google Scholar]
- [16].Charles-Edwards EM and deSouza NM, “Diffusion-weighted magnetic resonance imaging and its application to cancer,” Cancer Imaging, vol. 6, no. 1, pp. 135–143, Sep. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Le Bihan D and Johansen-Berg H, “Diffusion MRI at 25: Exploring brain tissue structure and function,” Neuroimage, vol. 61, no. 2, pp. 324–341, Jun. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Reeder S and Sirlin M, “Quantification of liver fat with magnetic resonance imaging,” Magn. Reson. Imaging Clin. J, vol. 18, pp. 1–34, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu H, Shimakawa A, Hines CDG, McKenzie CA, Hamilton G, Sirlin CB, Brittain JH, and Reeder SB, “Combination of complex-based and magnitude-based multiecho water-fat separation for accurate quantification of fat-fraction,” Magn. Reson. Med, vol. 66, no. 1, pp. 199–206, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mariappan YK, Glaser KJ, and Ehman RL, “Magnetic resonance elastography: A review,” Clin. Anat, vol. 23, no. 5, pp. 497–511, Jul. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Y and Liu T, “Quantitative susceptibility mapping (QSM): Decoding MRI data for a tissue magnetic biomarker,” Magn. Reson. Med, vol. 73, no. 1, pp. 82–101, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rieke V and Butts Pauly K, “MR thermometry,” J. Magn. Reson. Imaging, vol. 27, no. 2, pp. 376–90, Feb. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jahng G-H, Li K-L, Ostergaard L, and Calamante F, “Perfusion magnetic resonance imaging: a comprehensive update on principles and techniques,” Korean J. Radiol, vol. 15, no. 5, pp. 554–77, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hinojar R, Nagel E, and Puntmann VO, “T1 mapping in myocarditis – headway to a new era for cardiovascular magnetic resonance,” Expert Rev. Cardiovasc. Ther, vol. 13, no. 8, pp. 871–874, Aug. 2015. [DOI] [PubMed] [Google Scholar]
- [25].Thavendiranathan P, Walls M, Giri S, Verhaert D, Rajagopalan S, Moore S, Simonetti OP, and Raman SV, “Improved detection of myocardial involvement in acute inflammatory cardiomyopathies using T2 mapping,” Circ. Cardiovasc. Imaging, vol. 5, no. 1, pp. 102–110, Jan. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Okur A, Kantarci M, Kizrak Y, Yildiz S, Pirimoglu B, Karaca L, Ogul H, and Sevimli S, “Quantitative evaluation of ischemic myocardial scar tissue by unenhanced T1 mapping using 3.0 Tesla MR scanner,” Diagnostic Interv. Radiol, vol. 20, no. 5, pp. 407–413, Aug. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bull S, White SK, Piechnik SK, Flett AS, Ferreira VM, Loudon M, Francis JM, Karamitsos TD, Prendergast BD, Robson MD, Neubauer S, Moon JC, and Myerson SG, “Human noncontrast T1 values and correlation with histology in diffuse fibrosis,” Heart, vol. 99, no. 13, pp. 932–937, Jul. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Park CH, Choi E-Y, Kwon HM, Hong BK, Lee BK, Yoon YW, Min P-K, Greiser A, Paek MY, Yu W, Sung YM, Hwang SH, Hong YJ, and Kim TH, “Quantitative T2 mapping for detecting myocardial edema after reperfusion of myocardial infarction: validation and comparison with T2-weighted images,” Int. J. Cardiovasc. Imaging, vol. 29, no. S1, pp. 65–72, Jun. 2013. [DOI] [PubMed] [Google Scholar]
- [29].Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, and Ridgway JP, “Modified Look-Locker Inversion Recovery (MOLLI) for high-resolution T 1 mapping of the heart,” Magn. Reson. Med, vol. 52, pp. 141–146, 2004. [DOI] [PubMed] [Google Scholar]
- [30].Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, and Thompson RB, “Saturation recovery single-shot acquisition (SASHA) for myocardial T1 mapping,” Magn. Reson. Med, vol. 71, pp. 2082–2095, 2014. [DOI] [PubMed] [Google Scholar]
- [31].Hernando D, Levin YS, Sirlin CB, and Reeder SB, “Quantification of liver iron with MRI: state of the art and remaining challenges,” J. Magn. Reson. Imaging, vol. 40, no. 5, pp. 1003–1021, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wang L and Regatte RR, “T1ρ MRI of human musculoskeletal system,” J. Magn. Reson. Imaging, vol. 41, no. 3, pp. 586–600, Mar. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Warntjes JBM, Leinhard OD, West J, and Lundberg P, “Rapid magnetic resonance quantification on the brain: Optimization for clinical usage,” Magn. Reson. Med, vol. 60, no. 2, pp. 320–329, Aug. 2008. [DOI] [PubMed] [Google Scholar]
- [34].Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, and Griswold MA, “Magnetic resonance fingerprinting,” Nature, vol. 495, no. 7440, pp. 187–192, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jiang Y, Ma D, Seiberlich N, Gulani V, and Griswold MA, “MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout,” Magn. Reson. Med, vol. 74, pp. 1621–1631, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Asslander J, Glaser SJ, and Hennig J, “Pseudo steady-state free Precession for MR-Fingerprinting,” Magn. Reson. Med, vol. 77, pp. 1151–1161, 2017. [DOI] [PubMed] [Google Scholar]
- [37].Amthor T, Doneva M, Koken P, Sommer K, Meineke J, and Börnert P, “Magnetic resonance fingerprinting with short relaxation intervals,” Magn. Reson. Imaging, vol. 41, pp. 22–28, 2017. [DOI] [PubMed] [Google Scholar]
- [38].Hamilton JI, Jiang Y, Chen Y, Ma D, Lo W-C, Griswold M, and Seiberlich N, “MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density,” Magn. Reson. Med, vol. 77, no. 4, pp. 1446–1458, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cohen O and Rosen MS, “Algorithm comparison for schedule optimization in MR fingerprinting,” Magn. Reson. Imaging, vol. 41, pp. 15–21, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sommer K, Amthor T, Doneva M, Koken P, Meineke J, and Börnert P, “Towards predicting the encoding capability of MR fingerprinting sequences,” Magn. Reson. Imaging, vol. 41, pp. 7–14, 2017. [DOI] [PubMed] [Google Scholar]
- [41].Cloos MA, Knoll F, Zhao T, Block KT, Bruno M, Wiggins GC, and Sodickson DK, “Multiparametric imaging with heterogeneous radiofrequency fields,” Nat. Commun, vol. 7, p. 12445, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Buonincontri G and Sawiak SJ, “MR fingerprinting with simultaneous B1 estimation,” Magn Reson Med, vol. 76, pp. 1127–1135, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rieger B, Zimmer F, Zapp J, Weingärtner S, and Schad LR, “Magnetic resonance fingerprinting using echo-planar imaging: Joint quantification of T1 and T2* relaxation times,” Magn. Reson. Med, vol. 78, no. 5, pp. 1724–1733, 2017. [DOI] [PubMed] [Google Scholar]
- [44].Liu Y, Hamilton J, Griswold M, and Seiberlich N, “Simultaneous quantification of T1, T2, and off-resonance using FISP-MRF with a rosette trajectory and readout segmentation,” Proc. Intl. Soc. Mag. Reson. Med. 26, p. 2895, 2018. [Google Scholar]
- [45].Ma D, Jiang Y, Chen Y, McGivney D, Mehta B, Gulani V, and Griswold M, “Fast 3D magnetic resonance fingerprinting for a whole-brain coverage,” Magn. Reson. Med, vol. 79, no. 4, pp. 2190–2197, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Rieger B, Akçakaya M, Pariente JC, Llufriu S, Martinez-Heras E, Weingärtner S, and Schad LR, “Time efficient whole-brain coverage with MR Fingerprinting using slice-interleaved echo-planar-imaging,” Sci. Rep, vol. 8, p. 6667, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liao C, Bilgic B, Manhard MK, Zhao B, Cao X, Zhong J, Wald LL, and Setsompop K, “3D MR fingerprinting with accelerated stack-of-spirals and hybrid sliding-window and GRAPPA reconstruction,” Neuroimage, vol. 162, pp. 13–22, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ye H, Ma D, Jiang Y, Cauley SF, Du Y, Wald LL, Griswold MA, and Setsompop K, “Accelerating magnetic resonance fingerprinting (MRF) using t-blipped simultaneous multislice (SMS) acquisition,” Magn. Reson. Med, vol. 75, pp. 2078–2085, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jiang Y, Ma D, Bhat H, Ye H, Cauley SF, Wald LL, Setsompop K, and Griswold MA, “Use of pattern recognition for unaliasing simultaneously acquired slices in simultaneous multislice MR fingerprinting,” Magn. Reson. Med, vol. 78, pp. 1870–1876, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ye H, Cauley SF, Gagoski B, Bilgic B, Ma D, Jiang Y, Du YP, Griswold MA, Wald LL, and Setsompop K, “Simultaneous multislice magnetic resonance fingerprinting (SMS-MRF) with direct-spiral slice-GRAPPA (ds-SG) reconstruction,” Magn. Reson. Med, vol. 77, pp. 1966–1974, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Christen T, Pannetier NA, Ni WW, Qiu D, Moseley ME, Schuff N, and Zaharchuk G, “MR vascular fingerprinting: A new approach to compute cerebral blood volume, mean vessel radius, and oxygenation maps in the human brain,” Neuroimage, vol. 89, pp. 262–270, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lemasson B, Pannetier N, Coquery N, Boisserand LSB, Collomb N, Schuff N, Moseley M, Zaharchuk G, Barbier EL, and Christen T, “MR vascular fingerprinting in stroke and brain tumors models,” Sci. Rep, vol. 6, p. 37071, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Su P, Mao D, Liu P, Li Y, Pinho MC, Welch BG, and Lu H, “Multiparametric estimation of brain hemodynamics with MR fingerprinting ASL,” Magn. Reson. Med, vol. 78, no. 5, pp. 1812–1823, Nov. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wright KL, Jiang Y, Ma D, Noll DC, Griswold MA, Gulani V, and Hernandez-Garcia L, “Estimation of perfusion properties with MR fingerprinting arterial spin labeling,” Magn. Reson. Imaging, vol. 50, pp. 68–77, Jul. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Anderson CE, Donnola SB, Jiang Y, Batesole J, Darrah R, Drumm ML, Brady-Kalnay SM, Steinmetz NF, Yu X, Griswold MA, and Flask CA, “Dual contrast-magnetic resonance fingerprinting (DC-MRF): A platform for simultaneous quantification of multiple MRI contrast agents,” Sci. Rep, vol. 7, p. 8431, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang CY, Liu Y, Huang S, Griswold MA, Seiberlich N, Yu X, Correspondence X, and Yu D, “31P magnetic resonance fingerprinting for rapid quantification of creatine kinase reaction rate in vivo,” NMR Biomed, vol. 30, p. e3786, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Cohen O, Huang S, McMahon MT, Rosen MS, and Farrar CT, “Rapid and quantitative chemical exchange saturation transfer (CEST) imaging with magnetic resonance fingerprinting (MRF),” Magn. Reson. Med, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhou Z, Han P, Zhou B, Christodoulou AG, Shaw JL, Deng Z, and Li D, “Chemical exchange saturation transfer fingerprinting for exchange rate quantification,” Magn. Reson. Med, no. December 2017, pp. 1352–1363, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Doneva M, Börnert P, Eggers H, Stehning C, Sénégas J, and Mertins A, “Compressed sensing reconstruction for magnetic resonance parameter mapping,” Magn. Reson. Med, vol. 64, no. 4, pp. 1114–1120, 2010. [DOI] [PubMed] [Google Scholar]
- [60].McGivney DF, Pierre E, Ma D, Jiang Y, Saybasili H, Gulani V, and Griswold MA, “SVD compression for magnetic resonance fingerprinting in the time domain,” IEEE Trans. Med. Imaging, vol. 33, no. 12, pp. 2311–2322, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Cauley SF, Setsompop K, Ma D, Jiang Y, Ye H, Adalsteinsson E, Griswold MA, and Wald LL, “Fast group matching for MR fingerprinting reconstruction,” Magn. Reson. Med, vol. 74, no. 2, pp. 523–528, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Cohen O, Zhu B, Rosen MS, and Martinos AA, “MR fingerprinting Deep RecOnstruction NEtwork (DRONE),” Magn. Reson. Med, vol. 80, pp. 885–894, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yang M, Ma D, Jiang Y, Hamilton J, Seiberlich N, Griswold MA, and McGivney D, “Low rank approximation methods for MR fingerprinting with large scale dictionaries,” Magn. Reson. Med, vol. 79, no. 4, pp. 2392–2400, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Doneva M, Amthor T, Koken P, Sommer K, and Börnert P, “Matrix completion-based reconstruction for undersampled magnetic resonance fingerprinting data,” Magn. Reson. Imaging, vol. 41, pp. 41–52, 2017. [DOI] [PubMed] [Google Scholar]
- [65].Asslander J, Cloos MA, Knoll F, Sodickson DK, Urgen Hennig J, and Lattanzi R, “Low rank alternating direction method of multipliers reconstruction for MR fingerprinting,” Magn. Reson. Med, vol. 79, pp. 83–96, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zhao B, Setsompop K, Adalsteinsson E, Gagoski B, Ye H, Ma D, Jiang Y, Ellen Grant P, Griswold MA, and Wald LL, “Improved magnetic resonance fingerprinting reconstruction with low-rank and subspace modeling,” Magn. Reson. Med, vol. 79, no. 2, pp. 933–942, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Pierre EY, Ma D, Chen Y, Badve C, and Griswold MA, “Multiscale reconstruction for MR fingerprinting,” Magn. Reson. Med, vol. 75, no. 6, pp. 2481–2492, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Zhao B, Setsompop K, Ye H, Cauley SF, and Wald LL, “Maximum likelihood reconstruction for magnetic resonance fingerprinting,” IEEE Trans. Med. Imaging, vol. 35, no. 8, pp. 1812–1823, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Cline CC, Chen X, Mailhe B, Wang Q, Pfeuffer J, Nittka M, Griswold MA, Speier P, and Nadar MS, “AIR-MRF: Accelerated iterative reconstruction for magnetic resonance fingerprinting,” Magn. Reson. Imaging, vol. 41, pp. 29–40, 2017. [DOI] [PubMed] [Google Scholar]
- [70].Cao X, Liao C, Wang Z, Chen Y, Ye H, He H, and Zhong J, “Robust sliding-window reconstruction for accelerating the acquisition of MR fingerprinting,” Magn. Reson. Med, vol. 78, pp. 1579–1588, 2017. [DOI] [PubMed] [Google Scholar]
- [71].Chiu S-C, Lin T-M, Lin J-M, Chung H-W, Ko C-W, Büchert M, and Bock M, “Effects of RF pulse profile and intra-voxel phase dispersion on MR fingerprinting with balanced SSFP readout,” Magn. Reson. Imaging, vol. 41, pp. 80–86, 2017. [DOI] [PubMed] [Google Scholar]
- [72].Hong T, Han D, Kim M-O, and Kim D-H, “RF slice profile effects in magnetic resonance fingerprinting,” Magn. Reson. Imaging, vol. 41, pp. 73–79, 2017. [DOI] [PubMed] [Google Scholar]
- [73].Hamilton JI, Jiang Y, Ma D, Lo W-C, Gulani V, Griswold M, and Seiberlich N, “Investigating and reducing the effects of confounding factors for robust T1 and T2 mapping with cardiac MR fingerprinting,” Magn. Reson. Imaging, vol. 53, pp. 40–51, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Buonincontri G, Schulte RF, Cosottini M, and Tosetti M, “Spiral MR fingerprinting at 7 T with simultaneous B1 estimation,” Magn. Reson. Imaging, vol. 41, pp. 1–6, 2017. [DOI] [PubMed] [Google Scholar]
- [75].Ma D, Coppo S, Chen Y, McGivney DF, Jiang Y, Pahwa S, Gulani V, and Griswold MA, “Slice profile and B1 corrections in 2D magnetic resonance fingerprinting,” Magn. Reson. Med, vol. 78, no. 5, pp. 1781–1789, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chen Y, Jiang Y, Pahwa S, Ma D, Lu L, Twieg MD, Wright KL, Seiberlich N, Griswold MA, and Gulani V, “MR fingerprinting for rapid quantitative abdominal imaging,” Radiology, vol. 279, pp. 278–286, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mcgivney D, Deshmane A, Jiang Y, Ma D,. Badve C, Sloan A, Gulani V, and Griswold M, “Bayesian estimation of multicomponent relaxation parameters in magnetic resonance fingerprinting,” Magn. Reson. Med, vol. 80, pp. 159–170, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Jiang Y, Ma D, Keenan KE, Stupic KF, Gulani V, and Griswold MA, “Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom,” Magn. Reson. Med, vol. 78, pp. 1452–1457, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Badve C, Yu A, Rogers M, Ma D, Liu Y, Schluchter M, Sunshine J, Griswold M, and Gulani V, “Simultaneous T 1 and T 2 brain relaxometry in asymptomatic volunteers using magnetic resonance fingerprinting,” Tomography, vol. 1, no. 2, pp. 136–144, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Badve C, Yu A, Dastmalchian S, Rogers M, Ma D, Jiang Y, Margevicius S, Pahwa S, Lu Z, Schluchter M, Sunshine J, Griswold M, Sloan A, and Gulani V, “MR fingerprinting of adult brain tumors: Initial experience,” Am. J. Neuroradiol, vol. 38, no. 3, pp. 495–499, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yu AC, Badve C, Ponsky LE, Pahwa S, Dastmalchian S, Rogers M, Jiang Y, Margevicius S, Schluchter M, Tabayoyong W, Abouassaly R, Mcgivney D, Griswold MA, and Gulani V, “Development of a combined MR fingerprinting and diffusion examination for prostate cancer,” Radiology, vol. 283, pp. 729–738, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chen Y, Panda A, Pahwa S, Hamilton JI, Dastmalchian S, McGivney DF, Ma D, Batesole J, Seiberlich N, Griswold MA, Plecha D, and Gulani V, “Three-dimensional MR fingerprinting for quantitative breast imaging,” Radiology, vol. In Press, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liao C, Wang K, Cao X, Li Y, Wu D, Ye H, Ding Q, He H, and Zhong J, “Detection of lesions in mesial temporal lobe epilepsy by using MR fingerprinting,” Radiology, vol. 288, pp. 804–812, 2018. [DOI] [PubMed] [Google Scholar]
- [84].Ma D, Jones SE, Deshmane A, Sakaie K, Pierre E, Larvie M, McGivney D, Blumcke I, Krishnan B, Lowe M, Gulani V, Najm I, Griswold M, and Wang I, “Development of high-resolution 3D MR fingerprinting for detection and characterization of epileptic lesions,” J. Magn. Reson. Imaging, vol. In Press, 2018. [DOI] [PubMed] [Google Scholar]
- [85].Coristine AJ, Hamilton J, van Heeswijk RB, Hullin R, and Seiberlich N, “Cardiac magnetic resonance fingerprinting in heart transplant recipients,” in Proceedings of the 26th Annual ISMRM, 2018, p. 675. [Google Scholar]
- [86].Manton DJ, Chaturvedi A, Hubbard A, Lind MJ, Lowry M, Maraveyas A, Pickles MD, Tozer DJ, and Turnbull LW, “Neoadjuvant chemotherapy in breast cancer: early response prediction with quantitative MR imaging and spectroscopy,” Br. J. Cancer, vol. 94, no. 3, pp. 427–35, Feb. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Tan PC, Pickles MD, Lowry M, Manton DJ, and Turnbull LW, “Lesion T2 relaxation times and volumes predict the response of malignant breast lesions to neoadjuvant chemotherapy,” Magn. Reson. Imaging, vol. 26, no. 1, pp. 26–34, Jan. 2008. [DOI] [PubMed] [Google Scholar]
- [88].Sekine T, Ter Voert EEGW, Warnock G, Buck A, Huellner M, Veit-Haibach P, and Delso G, “Clinical evaluation of zero-echo-time attenuation correction for brain 18F-FDG PET/MRI: Comparison with atlas attenuation correction,” J. Nucl. Med, vol. 57, no. 12, pp. 1927–1932, Dec. 2016. [DOI] [PubMed] [Google Scholar]
- [89].Rezaei A, Deroose CM, Vahle T, Boada F, and Nuyts J, “Joint reconstruction of activity and attenuation in time-of-flight PET: A quantitative analysis,” J. Nucl. Med, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Ehrhardt MJ, Markiewicz P, Liljeroth M, Barnes A, Kolehmainen V, Duncan JS, Pizarro L, Atkinson D, Hutton BF, Ourselin S, Thielemans K, and Arridge SR, “PET reconstruction with an anatomical MRI prior using parallel level sets,” IEEE Trans. Med. Imaging, vol. 35, no. 9, pp. 2189–2199, Sep. 2016. [DOI] [PubMed] [Google Scholar]
- [91].Schramm G, Holler M, Rezaei A, Vunckx K, Knoll F, Bredies K, Boada F, and Nuyts J, “Evaluation of parallel level sets and Bowsher’s method as segmentation-free anatomical priors for time-of-flight PET reconstruction,” IEEE Trans. Med. Imaging, vol. 37, no. 2, pp. 590–603, Feb. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bojorquez JZ, Bricq S, Acquitter C, Brunotte F, Walker PM, and Lalande A, “What are normal relaxation times of tissues at 3 T?,” Magn. Reson. Imaging, vol. 35, pp. 69–80, Jan. 2017. [DOI] [PubMed] [Google Scholar]
- [93].Koolstra K, Webb A, Koken P, Nehrke K, and Bornert P, “Water-Fat separation in spiral magnetic resonance fingerprinting using conjugate phase reconstruction,” in Proceedings of the 26th Annual ISMRM, 2018, p. 681. [Google Scholar]
- [94].Boada FE, Koesters T, Block KT, and Chandarana H, “Improved detection of small pulmonary nodules through simultaneous MR/PET imaging,” PET Cliniscs, vol. 13, pp. 89–95, 2018. [DOI] [PubMed] [Google Scholar]
- [95].Broski SM, Goenka AH, Kemp BJ, and Johnson GB, “Clinical PET/MRI: 2018 update,” Am. J. Roentgenol, vol. 211, pp. 295–313, 2018. [DOI] [PubMed] [Google Scholar]
- [96].Hojjati M, Badve C, Garg V, Tatsuoka C, Rogers L, Sloan A, Faulhaber P, Ros PR, and Wolansky LJ, “Role of FDG-PET/MRI, FDG-PET/CT, and dynamic susceptibility contrast perfusion MRI in differentiating radiation necrosis from tumor recurrence in glioblastomas,” J. Neuroimaging, vol. 28, pp. 118–125, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Yoon JH, Kim J, Kang WJ, Sohn C-H, Choi SH, Yun TJ, Eun Y, Song YS, and Chang K-H, “Grading of cerebral glioma with multiparametric MR imaging and 18F-FDG-PET: Concordance and accuracy,” Eur. Radiol, vol. 24, no. 2, pp. 380–389, Feb. 2014. [DOI] [PubMed] [Google Scholar]
- [98].Barthel H, Schroeter ML, Hoffmann K-T, and Sabri O, “PET/MR in dementia and other neurodegenerative diseases,” Semin. Nucl. Med, vol. 45, no. 3, pp. 224–233, May 2015. [DOI] [PubMed] [Google Scholar]
- [99].Park SY, Zacharias C, Harrison C, Fan RE, Kunder C, Hatami N, Giesel F, Ghanouni P, Daniel B, Loening AM, Sonn GA, and Iagaru A, “Gallium 68 PSMA-11 PET/MR imaging in patients with intermediate- or high-risk prostate cancer,” Radiology, vol. 288, no. 2, pp. 495–505, Aug. 2018. [DOI] [PubMed] [Google Scholar]
- [100].Ratib O and Nkoulou R, “Potential applications of PET/MR imaging in cardiology,” J. Nucl. Med, vol. 55, p. 40S–46S, Jun. 2014. [DOI] [PubMed] [Google Scholar]
- [101].Goldfarb JW, Arnold S, and Han J, “Recent myocardial infarction: Assessment with unenhanced T1-weighted MR imaging,” Radiology, vol. 245, no. 1, pp. 245–250, Oct. 2007. [DOI] [PubMed] [Google Scholar]
- [102].Dall’Armellina E, Piechnik SK, Ferreira VM, Le Si Q, Robson MD, Francis JM, Cuculi F, Kharbanda RK, Banning AP, Choudhury RP, Karamitsos TD, and Neubauer S, “Cardiovascular magnetic resonance by non contrast T1-mapping allows assessment of severity of injury in acute myocardial infarction,” J. Cardiovasc. Magn. Reson, vol. 14, no. 1, p. 15, Feb. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Karamitsos TD, Piechnik SK, Banypersad SM, Fontana M, Ntusi NB, Ferreira VM, Whelan CJ, Myerson SG, Robson MD, Hawkins PN, Neubauer S, and Moon JC, “Noncontrast T1 mapping for the diagnosis of cardiac amyloidosis,” JACC Cardiovasc. Imaging, vol. 6, no. 4, pp. 488–497, Apr. 2013. [DOI] [PubMed] [Google Scholar]
- [104].Giri S, Chung Y-C, Merchant A, Mihai G, Rajagopalan S, Raman SV, and Simonetti OP, “T2 quantification for improved detection of myocardial edema,” J. Cardiovasc. Magn. Reson, vol. 11, no. 1, p. 56, Dec. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].von Olshausen G, Hyafil F, Langwieser N, Laugwitz K-L, Schwaiger M, and Ibrahim T, “Detection of acute inflammatory myocarditis in Epstein Barr virus infection using hybrid 18F-fluoro-deoxyglucose-positron emission tomography/magnetic resonance imaging,” Circulation, vol. 130, no. 11, pp. 925–926, Sep. 2014. [DOI] [PubMed] [Google Scholar]
- [106].Piriou N, Sassier J, Pallardy A, Serfaty J-M, and Trochu J-N, “Utility of cardiac FDG-PET imaging coupled to magnetic resonance for the management of an acute myocarditis with non-informative endomyocardial biopsy,” Eur. Hear J. - Cardiovasc. Imaging, vol. 16, no. 5, pp. 574–574, May 2015. [DOI] [PubMed] [Google Scholar]
- [107].Munoz C, Neji R, Cruz G, Mallia A, Jeljeli S, Reader AJ, Botnar RM, and Prieto C, “Motion-corrected simultaneous cardiac positron emission tomography and coronary MR angiography with high acquisition efficiency,” Magn. Reson. Med, vol. 79, pp. 339–350, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kirchner J, Sawicki LM, Deuschl C, Grüneisen J, Beiderwellen K, Lauenstein TC, Herrmann K, Forsting M, Heusch P, and Umutlu L, “18 F-FDG PET/MR imaging in patients with suspected liver lesions: Value of liver-specific contrast agent Gadobenate dimeglumine,” PLoS One, vol. 12, no. 7, p. e0180349, Jul. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Kong E, Chun KA, and Cho IH, “Quantitative assessment of simultaneous F-18 FDG PET/MRI in patients with various types of hepatic tumors: Correlation between glucose metabolism and apparent diffusion coefficient,” PLoS One, vol. 12, no. 7, p. e0180184, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, and Gwyther SG, “New guidelines to evaluate the response to treatment in solid tumors,” JNCI J. Natl. Cancer Inst, vol. 92, no. 3, pp. 205–216, Feb. 2000. [DOI] [PubMed] [Google Scholar]
- [111].Schwarz JK, Grigsby PW, Dehdashti F, and Delbeke D, “The role of 18F-FDG PET in assessing therapy response in cancer of the cervix and ovaries,” J. Nucl. Med, vol. 50, no. Suppl 1, p. 64S–73S, May 2009. [DOI] [PubMed] [Google Scholar]
- [112].Jamin Y, Tucker ER, Poon E, Popov S, Vaughan L, Boult JKR, Webber H, Hallsworth A, Baker LCJ, Jones C, Koh D-M, Pearson ADJ, Chesler L, and Robinson SP, “Evaluation of clinically translatable MR imaging biomarkers of therapeutic response in the TH-MYCN transgenic mouse model of neuroblastoma,” Radiology, vol. 266, no. 1, pp. 130–140, Jan. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Weidensteiner C, Allegrini PR, Sticker-Jantscheff M, Romanet V, Ferretti S, and McSheehy PM, “Tumour T1 changes in vivo are highly predictive of response to chemotherapy and reflect the number of viable tumour cells – a preclinical MR study in mice,” BMC Cancer, vol. 14, no. 1, p. 88, Dec. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Wang ZJ, Behr S, Consunji MV, Yeh BM, Gao K, Ko AH, Cinar P, Tempero MA, and Collisson EA, “Early Response Assessment in Pancreatic Ductal Adenocarcinoma Through Integrated PET/MRI,” Am. J. Roentgenol, 2018. [DOI] [PubMed] [Google Scholar]
- [115].Sarabhai T, Tschischka A, Stebner V, Nensa F, Wetter A, Kimmig R, Forsting M, Herrmann K, Umutlu L, and Grueneisen J, “Simultaneous multiparametric PET/MRI for the assessment of therapeutic response to chemotherapy or concurrent chemoradiotherapy of cervical cancer patients: Preliminary results,” Clin. Imaging, vol. 49, pp. 163–168, May 2018. [DOI] [PubMed] [Google Scholar]
- [116].Bosca R, Ashton E, Zahlmann G, Jackson E, and Editors, “RSNA Quantitative Imaging Biomarker Alliance (QIBA) DCE-MRI phantom: goal, design, and initial results,” in Proceedings of the 98th Scientific Assembly and Annual Meeting of RSNA, 2012. [Google Scholar]
- [117].National Institute of Standards and Technology, “Quantitative MRI.” [Online]. Available: https://www.nist.gov/programs-projects/quantitative-mri. [Accessed: 06-Aug-2018].
- [118].Pullens P, Roebroeck A, and Goebel R, “Ground truth hardware phantoms for validation of diffusion-weighted MRI applications,” J. Magn. Reson. Imaging, vol. 32, no. 2, pp. 482–488, Jul. 2010. [DOI] [PubMed] [Google Scholar]
- [119].Hubbard PL, Zhou F-L, Eichhorn SJ, and Parker GJM, “Biomimetic phantom for the validation of diffusion magnetic resonance imaging,” Magn. Reson. Med, vol. 73, no. 1, pp. 299–305, Jan. 2015. [DOI] [PubMed] [Google Scholar]
- [120].Lichenstein SD, Bishop JH, Verstynen TD, and Yeh F-C, “Diffusion capillary phantom vs. human data: Outcomes for reconstruction methods depend on evaluation medium,” Front. Neurosci, vol. 10, p. 407, Sep. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Guise C, Fernandes MM, Nóbrega JM, Pathak S, Schneider W, and Fangueiro R, “Hollow polypropylene yarns as a biomimetic brain phantom for the validation of high-definition fiber tractography imaging,” ACS Appl. Mater. Interfaces, vol. 8, no. 44, pp. 29960–29967, Nov. 2016. [DOI] [PubMed] [Google Scholar]
- [122].Calimetrix, “Calimetrix.” [Online]. [Accessed: 07-Sep-2018]. Available: https://www.calimetrix.com/shop/.