Abstract
We sequenced 29 carbapenemase-producing Klebsiella pneumoniae isolates from a neonatal intensive care unit in Ghana. Twenty-eight isolates were sequence type 17 with blaOXA-181 and differed by 0–32 single-nucleotide polymorphisms. Improved surveillance and infection control are needed to characterize and curb the spread of multidrug-resistant organisms in sub-Saharan Africa.
Keywords: neonatal intensive care unit, Ghana, Klebsiella pneumoniae, whole-genome sequencing, bloodstream infection, carbapenemase-producing, bacteria, carbapenem, carbapenemase, antimicrobial resistance
Carbapenems are antimicrobial drugs of last resort for infections caused by multidrug-resistant gram-negative bacteria. Therefore, the global spread of carbapenemase-producing Enterobacteriaceae, which are resistant to carbapenems, is troubling (1,2). Because of the high number of deaths associated with infections caused by these bacteria, the World Health Organization classifies Enterobacteriaceae as priority organisms for which new antimicrobial drugs are urgently needed (3).
Oxacillinase (OXA)-48–like carbapenemases are among the most common carbapenemases in Enterobacterales; of the OXA-48–like enzymes, OXA-181 is the second most common type (2). OXA-48 Klebsiella pneumoniae is considered endemic to North Africa and the Middle East; OXA-181 Klebsiella pneumoniae is endemic to the Indian subcontinent. However, nosocomial outbreaks of OXA-181 have occurred in sub-Saharan Africa (2). We describe the epidemiology and clonal spread of OXA-181–producing Klebsiella pneumoniae in a neonatal intensive care unit (NICU) in Ghana. The Institutional Review Board of the Korle-Bu Teaching Hospital granted ethics approval (no. IRB/0025/2017) for this study.
The Study
We whole-genome sequenced 29 carbapenemase-producing K. pneumoniae isolates: 18 from neonatal carriage (isolates from swabs of neonates) (4), 3 from the NICU environment (cots and trolley handles, incubator doors, tables), and 8 from neonatal bloodstream infections. These samples were isolated from the NICU of Korle-Bu Teaching Hospital (Accra, Ghana) from September 2017 through February 2019 (5) (Table; Appendix).
Table. Characteristics of neonates and carbapenemase-producing Klebsiella pneumoniae isolates from Korle-Bu Teaching Hospital, Ghana, 2017–2019*.
| Patient | Sample | Sex | MOD | DOS | Birthweight, kg | Cubicle | Shared space | Date of isolation | ST | Capsular serotype | Prior AB use | AB use after BSI | DOA, d | Vital status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KP007 | Carriage | F | CS | 13 | 1.4 | I | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP010 | Carriage | F | CS | 21 | 1.1 | II | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP011 | Carriage | M | SVD | 18 | 0.9 | I | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Dead |
| KP020 | Carriage | F | CS | 43 | 0.8 | II | No | 2017 Sep 21 | 48 | KL62 | AMK, CXC, MEM, CIP, VA | NA | NA | Alive |
| KP025 | Carriage | F | SVD | 15 | 1.2 | III | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP034 | Carriage | M | CS | 34 | 1.4 | III | Yes | 2017 Sep 21 | 17 | KL25 | AMK, CXC, CIP, MEM | NA | NA | Alive |
| KP035 | Carriage | F | CS | 30 | 1.4 | III | Yes | 2017 Sep 21 | 17 | KL25 | No | NA | NA | Alive |
| KP036 | Carriage | F | SVD | 36 | 1.3 | III | Yes | 2017 Sep 21 | 17 | KL25 | No | NA | NA | Alive |
| KP037 | Carriage | M | SVD | 12 | 2.1 | III | Yes | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP045 | Carriage | F | SVD | 11 | 1.7 | III | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | NA |
| KP047 | Carriage | F | SVD | 4 | 3.9 | III | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP052 | Carriage | M | CS | 12 | 3 | III | Yes | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP055 | Carriage | M | CS | 24 | 1.5 | III | Yes | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP056 | Carriage | M | SVD | 17 | 3.2 | III | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| K058 | Carriage | M | SVD | 33 | 1 | I | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| K221 | Carriage | F | SVD | 2 | 1.6 | III | No | 2017 Sep 21 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP233 | Carriage | M | SVD | 12 | 3.2 | III | No | 2018 Jan 19 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP242 | Carriage | M | SVD | 2 | 3.7 | III | No | 2018 Jan 19 | 17 | KL25 | AMK, CXC | NA | NA | Alive |
| KP0033 | Blood | M | SVD | 20 | 2.4 | I | NA | 2017 Oct 5 | 17 | KL25 | AMK, CXC, CAZ | MEM, CIP | 10 | Dead |
| KP0455 | Blood | M | SVD | 10 | 3.6 | II | NA | 2018 Jan 5 | 17 | KL25 | AMK, CXC | NA | 12 | Dead |
| KP0457 | Blood | F | SVD | 9 | 1.3 | II | NA | 2018 Jan 5 | 17 | KL25 | AMK, CXC | CIP | 69 | Alive |
| KP0879 | Blood | M | SVD | 13 | 4.4 | III | NA | 2018 Mar 19 | 17 | KL25 | MEM | MEM | NA | NA |
| KP2326 | Blood | F | A | 7 | 1.1 | I | NA | 2018 Dec 29 | 17 | KL25 | AMK, CXC | NA | 9 | Dead |
| KP2201 | Blood | M | CS | 10 | 1.3 | I | NA | 2018 Nov 24 | 17 | KL25 | AMK, CXC | MEM, CIP | 42 | Alive |
| KP2557 | Blood | M | SVD | 18 | 2.5 | I | NA | 2019 Feb 6 | 17 | KL25 | AMK, CXC | MEM | NA | Alive |
| KP2455 | Blood | F | CS | 10 | NA | III | NA | 2019 Jan 20 | 17 | KL25 | AMK, CXC | CIP | 27 | Alive |
| KP026 | Cot | II | 2017 Sep 21 | 17 | KL25 | |||||||||
| KP040 | Cot | III | 2017 Sep 21 | 17 | KL25 | |||||||||
| KP090 | Trolley handle | III | 2017 Sep 21 | 17 | KL25 |
*Blank spaces indicate not applicable. AB, antimicrobial drug; AMK, amikacin; BSI, bloodstream infections; CAZ, ceftazidime; CIP, ciprofloxacin; CS, cesarean section; CXC, cloxacillin; DOA, duration of admission; DOS, duration of stay before specimen collection; MOD, mode of delivery; MEM, meropenem; NA, not available; ST, multilocus sequence type.
Twenty-eight of the 29 isolates were sequence type (ST) 17 and capsular type KL25. We excluded 1 isolate from further analysis that was ST48 and KL64. Core-genome phylogeny showed a close genetic relationship of all ST17 isolates (0–32 single-nucleotide polymorphism [SNP] differences; median 5 SNP differences), suggesting a localized outbreak (Figure 1). We estimated that the most recent common ancestor of the outbreak emerged in April 2017 (year 2017.3; 95% highest posterior density interval 2017.0–2017.6) with an estimated mean substitution rate of 2.1 × 10–6 SNPs/site/year (9.9 SNPs/year) (Appendix Figure).
Figure 1.
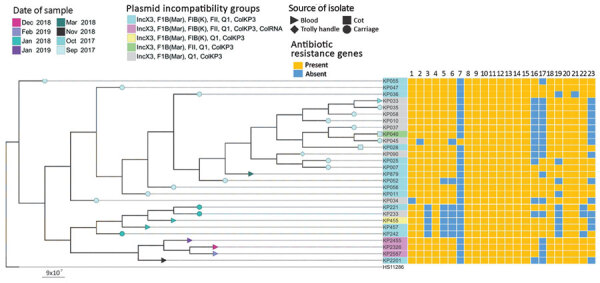
Phylogenetic tree of 28 carbapenemase-producing Klebsiella pneumoniae isolates and their acquired resistance genes from the neonatal intensive care unit at Korle-Bu Teaching Hospital, Accra, Ghana, 2017–2019. The tree was produced by analysis of single-nucleotide polymorphisms (SNPs) of core genomes. Maximum genetic distance was between isolates KP2201 and KP026, which differed by 32 SNPs. Tree used genome of K. pneumoniae reference strain HS11286 as outgroup. Lane 1, rmtB; lane 2, aph(3")-lb; lane 3, aph(3')-la; lane 4, aph(6)-ld; lane 5, aac(3)-lld; lane 6, aadA2; lane 7, aadA2b; lane 8, blaOXA-181; lane 9, blaTEM-1B; lane 10, blaSHV-94; lane 11, blaCTX-M-15; lane 12, qnrS; lane 13, oqxA; lane 14, oqxB; lane 15, fosA; lane 16, mph(A); lane 17, catA2; lane 18, sul2; lane 19, sul1; lane 20, tetA; lane 21, tetG; lane 22, dfrA12; lane 23, drfA14. Scale bar indicates substitutions per site.
All isolates were resistant to amoxicillin/clavulanic acid, gentamicin, amikacin, cefuroxime, ceftriaxone, ceftazidime, tazobactam/piperacillin, and ciprofloxacin. The isolates were susceptible to colistin and had MICs of <1 μg/mL. All outbreak isolates harbored the carbapenemase blaOXA-181 and extended-spectrum β-lactamase blaCTX-M-15 in addition to other β-lactamases (blaTEM-1B, blaSHV-94). We also found several genes encoding resistance to other antimicrobial drugs: aminoglycosides (rmtB, aph(3¢')-Ib, aph(3¢)-Ia, aph(6)-Id, aac(3)-IId, aadA2, aadA2b); fluoroquinolones (qnrS, oqxA, oqxB); fosfomycin (fosA); macrolide (mph (A)); phenicols (catA2); sulphonamides (sul2, sul1); tetracyclines (tetA, tetG); and trimethoprim (dfrA12, dfrA14) (Figure 1).
All isolates contained 4 common plasmid incompatibility (Inc) groups (IncX3, IncF1B (Mar), IncQ1, IncColKP3). Eighteen isolates also contained incompatibility groups IncFIB (K) and IncFII, and 3 contained additional IncColRNA (Figures 1, 2). Further analysis revealed that recently recovered isolates had more plasmid Inc groups than did older isolates (Figure 1). The accessory genome of the isolates showed large variation in gene content (Figure 2). These data illustrate that this variation existed at the time of the first sampling in September 2017, when the isolates formed 3 distinct clusters (Figure 2). The clustering is associated with differences in plasmid content of the isolates and represents the uptake or loss of 205 genes. On the basis of the phylogeny and metadata, we hypothesize that 4 major evolutionary events caused changes in Inc groups and the ancestor of the cluster of isolates with Inc groups IncX3, IncFIB, IncQ1 and ColKP3 (Figure 2).
Figure 2.
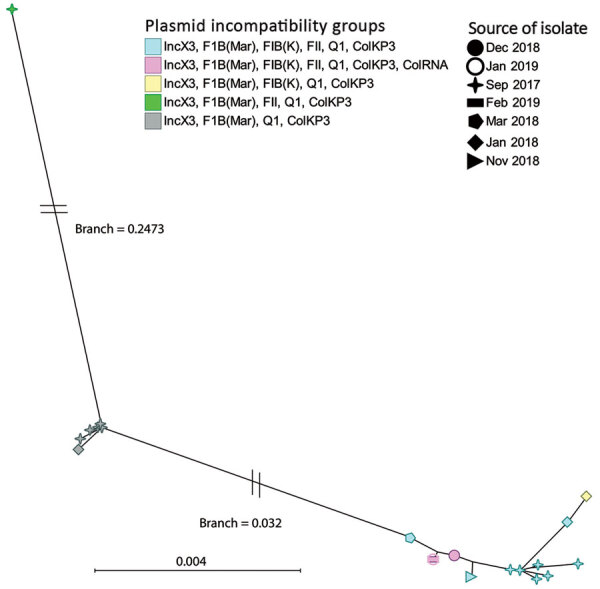
Binary rational tree illustrating genetic diversity (presence–absence of genes) of the accessory genome of carbapenemase-producing Klebsiella pneumoniae isolates from the neonatal intensive care unit at Korle-Bu Teaching Hospital, Accra, Ghana, 2017–2019. Different shapes represent different dates of organism isolation. Blue and green shapes evolved from the gray; pink and yellow evolved from the blue. Scale bar indicates genetic differences per site.
A study in South Africa identified a fully closed plasmid carrying blaOXA-181 (6). Using the short-read sequencing applied in this study, we cannot determine whether blaOXA-181 is carried on a plasmid or located in the chromosome. Mapping of raw reads toward the fully closed plasmid reveals complete coverage across the whole plasmid for 24 of the 28 isolates; the remaining 4 most recent isolates had reads covering the whole plasmid (except for 4 genes). This finding might indicate these isolates have a similar plasmid containing blaOXA-181, although we cannot rule out that these reads might belong to other related plasmids and not the previously reported plasmid (6).
Conclusions
We identified an outbreak of ST17 K. pneumoniae carrying blaOXA-181 in a NICU in Ghana. Outbreak isolates were resistant to all antimicrobial drugs commonly used to treat neonatal infections (although it was susceptible to colistin). Similar outbreaks of ST17 OXA-181–producing K. pneumoniae have been documented in South Africa (7), further confirming the spread of this type of resistance into nonendemic regions (2). Time-based phylogenetic analysis showed the outbreak isolates share a recent ancestor (approximately April 2017). This finding suggests that the outbreak strain had been introduced recently into the NICU or that the outbreak strain had limited genetic diversity because of a recent bottleneck or selective sweep in the outbreak strain population.
K. pneumoniae is an entry point of antimicrobial resistance into the family Enterobacteriaceae (8). Thus, carbapenemase-producing K. pneumoniae in the NICU might transmit resistance to other Enterobacteriaceae species. Other studies have associated blaOXA-181 with the insertion sequence element ISEcp1, which can spread cephalosporinases and extended-spectrum β-lactamases (9). In our study, all isolates possessed the IncX3 plasmid. This plasmid is self-transmissible and associated with worldwide dissemination of New Delhi metallo-β-lactamases 1 and 5 (10,11). Recent studies from countries in Africa have found blaOXA-181 carried on the IncX3 plasmid in Enterobacteriaceae species, including K. pneumoniae (2,6,7).
In Europe, the spread of carbapenem-resistant K. pneumoniae has been driven by 4 carbapenemase-positive clonal lineages that are often transmitted in hospitals (8). The isolates from the NICU were genetically diverse, especially in the plasmid content of the accessory genome. This diversity indicates the genome evolved rapidly, similar to isolates from an outbreak of K. pneumoniae in Beijing, China. In the outbreak in China, the isolates underwent rapid genotypic evolution mainly through rearrangement (including the gain and loss of genes) in the accessory genome (12). Antimicrobial pressure in hospitals might lead to adaptation and resistance transmission of K. pneumoniae in the hospital environment (8).
From our data, we infer the background transmission of carbapenemase-producing K. pneumoniae in the NICU before its detection. Neonatal carriage or environmental contamination by carbapenemase-producing K. pneumoniae might have started or maintained the outbreak. Improved surveillance of multidrug-resistant organisms, buttressed with improved infection prevention and control activities, are required to detect and control outbreaks in low-resource settings.
Additional information about methods of sampling Klebsiella pneumoniae isolates and analyzing their phylogenetic relationships.
Acknowledgments
The study falls under the HAI-Ghana Project funded by the Danish Ministry of Foreign Affairs (grant no. 16-PO1-GHA). M.B. and R.L.M. are supported by the Danish National Research Foundation (grant no. 126). K.L.N. is supported by Mica-foundation.
Biography
Dr. Labi is a doctoral student at the Centre for Medical Parasitology in the Department of Immunology and Microbiology at the University of Copenhagen. His research interests are antimicrobial resistance and stewardship and healthcare-associated infections.
Footnotes
Suggested citation for this article: Labi AK, Nielsen KL, Marvig RL, Bjerrum S, Enweronu-Laryea C, Bennedbæk M, et al. Oxacillinase-181 carbapenemase-producing Klebsiella pneumoniae in neonatal intensive care unit, Ghana, 2017–2019. Emerg Infect Dis. 2020 Sep [date cited]. https://doi.org/10.3201/eid2609.200562
References
- 1.Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pitout JDD, Peirano G, Kock MM, Strydom K-A, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33:e00102–19. 10.1128/CMR.00102-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. ; WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–27. 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 4.Labi A-K, Bjerrum S, Enweronu-Laryea CC, Ayibor PK, Nielsen KL, Marvig RL, et al. High carriage rates of multidrug-resistant gram-negative bacteria in neonatal intensive care units from Ghana. Open Forum Infect Dis. 2020;7:ofaa109. [DOI] [PMC free article] [PubMed]
- 5.Clinicaltrials.gov. Neonatal sepsis at neonatal intensive care units in Ghana [cited 2019 Nov 22]. https://clinicaltrials.gov/ct2/show/NCT03755635
- 6.Lowe M, Kock MM, Coetzee J, Hoosien E, Peirano G, Strydom K-A, et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014-2016. Emerg Infect Dis. 2019;25:739–47. 10.3201/eid2504.181482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strydom KA, Chen L, Kock MM, Stoltz AC, Peirano G, Nobrega DB, et al. Klebsiella pneumoniae ST307 with OXA-181: threat of a high-risk clone and promiscuous plasmid in a resource-constrained healthcare setting. J Antimicrob Chemother. 2020;75:896–902. 10.1093/jac/dkz550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al. ; EuSCAPE Working Group; ESGEM Study Group. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–29. 10.1038/s41564-019-0492-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Potron A, Nordmann P, Lafeuille E, Al Maskari Z, Al Rashdi F, Poirel L. Characterization of OXA-181, a carbapenem-hydrolyzing class D β-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2011;55:4896–9. 10.1128/AAC.00481-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnevend A, Al Baloushi A, Ghazawi A, Hashmey R, Girgis S, Hamadeh MB, et al. Emergence and spread of NDM-1 producer Enterobacteriaceae with contribution of IncX3 plasmids in the United Arab Emirates. J Med Microbiol. 2013;62:1044–50. 10.1099/jmm.0.059014-0 [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Fang L, Fu Y, Du X, Shen Y, Yu Y. Dissemination of NDM-1-producing Enterobacteriaceae mediated by the IncX3-type plasmid. PLoS One. 2015;10:e0129454. 10.1371/journal.pone.0129454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dorp L, Wang Q, Shaw LP, Acman M, Brynildsrud OB, Eldholm V, et al. Rapid phenotypic evolution in multidrug-resistant Klebsiella pneumoniae hospital outbreak strains. Microb Genom. 2019;5:e000263. 10.1099/mgen.0.000263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information about methods of sampling Klebsiella pneumoniae isolates and analyzing their phylogenetic relationships.


