Abstract
The magnitude of SARS-CoV-2 infection, the dynamic changes of immune parameters in patients with the novel coronavirus disease (COVID-19) and their correlation with the disease severity remain unclear. The clinical and laboratory results from 154 confirmed COVID-19 patients were collected. The SARS-CoV-2 RNA levels in patients were estimated using the Ct values of specific RT-PCR tests. The lymphocyte subsets and cytokine profiles in the peripheral blood were analyzed by flow cytometry and specific immunoassays. 154 confirmed COVID-19 patients were clinically examined up to 4 weeks after admission. The initial SARS-CoV-2 RNA Ct values at admission varied, but were comparable in the patient groups classified according to the age, gender, underlying diseases, and disease severity. Three days after admission, significant higher Ct values were found in severe cases. Significantly reduced counts of T cells and T cell subsets were found in patients with old age and underlying diseases at admission and were characteristic for the development of severe COVID-19. Severe COVID-19 developed preferentially in patients with underlying compromised immunity and was not associated with initial virus levels. Higher SARS-CoV-2 RNA levels in severe cases were apparently a result of impaired immune control associated with dysregulation of inflammation.
Electronic supplementary material
The online version of this article (10.1007/s00430-020-00693-z) contains supplementary material, which is available to authorized users.
Keywords: SARS-CoV-2, COVID-19, Severity, Viral load, Immune status
Introduction
A novel β-coronavirus, formally named as the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses, was identified as the cause of an outbreak that occurred in Wuhan in early December 2019. The epidemic of SARS-CoV-2 has now spread to over 210 countries, territories or areas with more than 7,000,000 confirmed cases worldwide [1]. The disease caused by SARS-CoV-2 has been recently named as coronavirus disease-2019 (COVID-19) by the World Health Organization. Patients with COVID-19 may develop mild or severe symptoms of acute respiratory infection. The mild and moderate cases show symptoms of fever, dry cough, fatigue, with or without abnormal chest CT findings, but with a good prognosis [2–4]. In contrast, some patients develop severe pneumonia, acute respiratory distress syndrome (ARDS) or multiple organ failure, with death rates ranging from 4.3 to 15% according to different study reports [2, 5–7]. Factors leading to severe COVID-19 have not been determined yet. Several viral and immunological parameters may provide clues for the underlying mechanisms. Among them, the viral loads may reflect the magnitude of SARS-CoV-2 infection in patients. Lymphopenia and inflammatory cytokine storm have been found to be associated with the disease severities in highly pathogenic severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) infections [8, 9]. In addition, decreased peripheral blood lymphocyte counts and elevated serum inflammatory cytokine levels have been reported in COVID-19 patients [5, 10]. However, a thorough investigation into the dynamic changes in different lymphocyte subsets, the kinetics of inflammatory cytokines changes in the peripheral blood of COVID-19 patients, and the relationships between these parameters and disease severity is still lacking.
In this study, a cohort of 154 clinically confirmed patients with positive SARS-CoV-2 detection by RT-PCR were enrolled and clinically treated and monitored for up to 4 weeks after admission. The clinical and laboratory results were collected and analyzed. Particularly, the viral loads in patients were assessed by using the Ct values of specific RT-PCR tests. The changes in lymphocyte subsets and cytokines profiles in the peripheral blood of COVID-19 patients were analyzed. The results of our analysis demonstrated that the initial SARS-CoV-2 RNA loads varied in patients, but were comparable in different patient groups stratified by age, gender, comorbidities and disease severity. In contrast, reduced counts of T cells and subsets were found in patients with old age and comorbidities at admission and were characteristic of severe COVID-19. Thus, severe COVID-19 developed preferentially in patients with compromised immunity, but was not associated with initial viral loads.
Materials and methods
Data collection
Written informed consents were obtained from all 154 patients upon admission into the Second People's Hospital, Fuyang, China. This retrospective single-center study was approved by the Ethics Committee of the Second People's Hospital, Fuyang (Number 2020020004). Medical record information including epidemiological, demographic, clinical manifestation, laboratory data, and outcome data was obtained for all patients.
The diagnosis of COVID-19 for patients was performed according to the Guidelines for the Diagnosis and Treatment of New Coronavirus Pneumonia (version 7) published by the National Health Commission of China. Mild and moderate cases met the following criteria: (1) epidemiological history, (2) fever or other respiratory symptoms, (3) frequently with typical CT image abnormities of viral pneumonia, and (4) positive RT-PCR result for SARS-CoV-2 RNA. Severe patients additionally met at least one of the following conditions: (1) shortness of breath, RR ≥ 30 times/min, (2) oxygen saturation (resting state) ≤ 93%, or (3) PaO2/FiO2 ≤ 300 mmHg.
Laboratory examination
Laboratory confirmation of SARS-CoV-2 infection was performed with real-time RT-PCR (Bojie Ltd, Shanghai, China) per Chinese CDC recommendation. The RT-PCR tests use two primer pairs and corresponding probes targeting the ORF1ab and N regions of the SARS-CoV-2 genome [11]. Sputum and/or throat-swab specimens were collected from all patients and the samples were maintained in viral-transport medium prior to laboratory testing. A test result was considered positive if the Ct values of both RT-PCR reactions were below 38 according to the guidelines published by the National Health Commission of China. Influenza A and B infections were excluded by real-time RT-PCR. Specific IgG and IgM for coxsackievirus, respiratory syncytial virus, parainfluenza virus, enterovirus, Chlamydia and Mycoplasma pneumonia were tested by chemiluminescence immunoassay. Specimens, including sputum or alveolar lavatory fluid, blood, urine, and feces, were cultured to identify pathogenic bacteria or fungi that may be associated with the SARS-CoV-2 infection. A lymphocyte test kit (Mindray Bio-medical Inc., Shenzhen, China) was used for lymphocyte subset analysis. Plasma cytokines (IL-6) were measured with Human Th1/2 Cytokine Kit II (Jinzhun Ltd., Shenzhen, China). All tests were performed according to product manuals.
Statistical analyses
Categorical variables were expressed in frequency or percentage, and significance was detected by Chi square or Fisher’s exact test. Continuous variables were expressed as mean ± standard deviation or mean ± standard error, and the significance was tested by t test. Nonparametric variables were expressed in median and quartile intervals or mean ± standard error, and significance was tested by Mann–Whitney U, ANOVA or Kruskal–Wallis test. P < 0.05 was considered statistically significant in all statistical analyses. Principal component analysis (PCA) was performed to identify the major contributing factors among clinical parameters to distinguish mild and severe cases of COVID-19 patients. The diagnostic values of selected parameters for differentiating mild and severe cases of COVID-19 patients were assessed by receiver operating characteristic (ROC) and the area under the ROC curve (AUC). SPSS statistical software (Macintosh version 26.0, IBM, Armonk, NY, USA) and R package were used for statistical analysis.
Results
Demographic and clinical characteristics of COVID-19 patients
A total of 154 patients were enrolled in this study and were divided into two groups according to the published guidelines, including 122 mild/moderate cases (79.2%, referred to as mild cases thereafter) and 32 severe/critical cases (20.7%, referred to as severe cases). Among 122 patients with mild symptoms, eight patients had no chest CT imaging abnormality. Among the 32 patients with severe symptoms, 4 patients required ICU care and ventilator-aided breathing.
The enrolled COVID-19 patients consisted of 86 males (55.8%) and 68 females (44.1%) (Table 1). The mean age of the patients was 42.4 years. The medium age of the severe symptom group (52.2 ± 14.0 years) was older than that of the mild group (39.9 ± 15.0 years). A total of 52 (33.8%) patients in both groups had baseline diseases, including hypertension (23 [14.9%]), diabetes (11 [7.1%]), chronic liver diseases (12 [7.8%]), coronary heart diseases (3 [1.9%]), malignant tumors (3 [1.3%]), cerebrovascular diseases (2 [1.3%], COPD (2 [1.3%]) and AIDS (1 [0.65%] (Table 1). Two severe group patients had mixed fungal and six bacterial infections, respectively. At admission, 31 (96.9%) patients of the severe group and 86.4% of mild group patients had fever, with a significant difference in body temperature (Table 1). The levels of the inflammatory markers ESR (P = 0.008), C-reactive protein (CRP, P < 0.001), serum amyloid A protein (SAA, P = 0.039), interleukin-6 (IL-6, P < 0.001), and other markers including albumin (P < 0.001), aspartate transaminase (P = 0.003), alanine transaminase (P = 0.043), lactate dehydrogenase (P < 0.001), fibrinogen (P = 0.004) and d-dimer (P < 0.001) in the peripheral blood of the severe group patients were significantly higher at admission compared to the mild group patients (Table 2). No significant differences in the numbers of red blood cells and platelets, or in the serum levels of procalcitonin, total bilirubin, creatinine, creatine kinase and PT were observed between the two groups (Table 2).
Table 1.
Baseline characteristics of patients infected with SARS-CoV-2
| Characteristics | All patients (n = 154) |
Mild type (n = 122) |
Severe type (n = 32) |
P value^ |
|---|---|---|---|---|
| Age, years | 154 (42.4) | 39.9 ± 14.97 | 52.2 ± 13.99 | < 0.001 |
| Sex | ||||
| Male | 86 (55.8) | 63 (51.6) | 23 (71.8) | |
| Female | 68 (44.1) | 59 (48.4) | 9 (28.1) | 0.041 |
| Comorbidities | ||||
| Hypertension | 23 (14.9) | 13 (10.7) | 10 (31.3) | 0.004 |
| Diabetes | 11 (7.1) | 3 (2.5) | 8 (25) | < 0.001* |
| Chronic liver disease | 12 (7.8) | 9 (7.4) | 3 (9.4) | 0.714* |
| Coronary heart disease | 3 (1.9) | 3 (2.5) | 0 | 1.000* |
| Malignant tumor | 3 (1.9) | 1 (0.8) | 2 (6.3) | 0.109* |
| Cerebrovascular disease | 2 (1.3) | 2 (1.6) | 0 | 1.000* |
| COPD | 2 (1.3) | 0 | 2 (6.3) | 0.042* |
| AIDS | 1 (0.6) | 1 (0.8) | 0 | 1.000* |
| Signs and symptoms | ||||
| Fever | 133 (86.4) | 102 (83.6) | 31 (96.9) | 0.052 |
| Maximum temperature, °C | 133 | 38 (37.7–38.5) | 38.6 (38.5–39) | < 0.001 |
| Fatigue | 92 (59.7) | 65 (53.2) | 27 (84.4) | 0.002 |
| Dry cough | 106 (68.8) | 80 (65.6) | 26 (81.3) | 0.204 |
| Expectoration | 60 (38.9) | 47 (38.5) | 13 (40.6) | 0.829 |
| Chills | 23 (14.9) | 15 (12.3) | 8 (25) | 0.073 |
| Chest tightness | 30 (19.5) | 13 (10.7) | 17 (53.1) | < 0.001 |
| Onset of symptom to hospital admission, median (IQR), days | 154 | 3 (2–7) | 7 (3.5–10) | 0.004 |
| Temperature on admission, °C | 154 | 36.9 (36.5–37.5) | 37 (36.6–37.7) | 0.287 |
| Respiratory frequency on admission, bpm | 153 | 20 (19–21) | 20 (19–22) | 0.679 |
| Pulse on admission, bpm | 153 | 85 (80–96) | 91 (75–101) | 0.838 |
| Systolic blood pressure on admission, median (IQR), mmHg | 153 | 127 (119–140) | 129 (116–141) | 0.839 |
| Diastolic pressure on admission, median (IQR), mmHg | 153 | 85 (75–92) | 84 (74–93) | 0.652 |
*Fisher's exact test
^P values indicate differences between mild and severe patients. A P value < 0.05 was considered as statistically significant
Table 2.
Laboratory findings of patients infected with SARS-CoV-2 on admission to hospital
| Items | Normal range | All patients (n = 154) |
Mild type (n = 122) |
Severe type (n = 32) | P value |
|---|---|---|---|---|---|
| Blood routine | |||||
| White blood cell (× 109/L) | 3.5–9.5 | 4.99 (3.87–6.37) | 4.99 (3.91–6.23) | 4.87 (3.52–6.91) | 0.796 |
| Neutrophis (× 109/L) | 1.8–6.3 | 3.47 (2.38–4.71) | 3.41 (2.38–4.58) | 3.63 (2.45–5.83) | 0.367 |
| Lymphocyte (× 109/L) | 1.10–3.20 | 1.11 (0.77–1.42) | 1.18 (0.89–1.50) | 0.76 (0.65–1.10) | < 0.001 |
| Red blood cell (× 1012/L) | 4.30–5.80 | 4.50 (4.20–4.80) | 4.50 (4.20–4.80) | 4.30 (4.10–4.80) | 0.251 |
| Hemoglobin (g/L) | 130–175 | 139.63 ± 15.90 | 140.37 ± 15.85 | 136.84 ± 16.08 | 0.802 |
| Platelet (× 109/L) | 125–350 | 179.5 (140.0–239.0) | 180.0 (146.0–240.0) | 165 (124.5–235.0) | 0.285 |
| Inflammatory biomarkers | |||||
| ESR (mm/h) | 0–20 | 22.0 (8.0–42.5) | 16.0 (7.0–39.5) | 31.0 (26.3–63.5) | 0.008 |
| C-reactive protein (mg/L) | < 6 | 15.4 (3.9–33.9) | 11.7 (2.7–25.2) | 34.0 (19.2–74.1) | < 0.001 |
| Serum amyloid A (mg/L) | < 10 | 84.2 (26.3–187.3) | 76.6 (15.9–176.7) | 132.3 (76.4–187.4) | 0.039 |
| Procalcitonin (ng/ml) | < 0.5 | 0.03 (0.02–0.07) | 0.03 (0.02–0.06) | 0.05 (0.02–0.09) | 0.187 |
| Interleukin-6 (pg/ml) | < 7 | 18.1 (6.1–36.1) | 12.9 (5.1–26.5) | 41.8 (23.6–80.2) | < 0.001 |
| Biochemical indicators | |||||
| Albumin (g/L) | 40–55 | 41.0 ± 4.14 | 41.9 ± 3.75 | 37.6 ± 3.79 | < 0.001 |
| Alanine transaminase (U/L) | 0–50 | 24.5 (15.0–37.0) | 23.0 (13.0–37.00 | 28.5 (20.0–47.5) | 0.043 |
| Aspartate transferase (U/L) | 15–40 | 26.0 (20.0–33.0) | 25.0 (19.0–33.0) | 29.5 (25.0–42.5) | 0.003 |
| Total bilirubin (μmol/L) | 0.0–26.00 | 10.9 (7.2–15.2) | 10.3 (7.0–14.8) | 12.7 (8.1–17.9) | 0.084 |
| Blood urea nitrogen (mmol/L) | 3.1–8.0 | 3.8 (3.1–4.6) | 3.7 (3.1–4.4) | 4.2 (3.2–5.4) | 0.037 |
| Creatinine (μmol/L) | 35–115 | 63.6 ± 16.71 | 63.3 ± 15.9 | 64.9 ± 19.6 | 0.635 |
| Glucose (mmol/L) | 3.89–6.11 | 6.2 (5.4–6.8) | 6.0 (5.4–6.8) | 6.5 (5.8–9.4) | 0.032 |
| Creatine kinase (U/L) | 50–310 | 62.0 (44.0–88.0) | 61.0 (46.3–88.0) | 63.0 (42.0–110.0) | 0.946 |
| Creatine kinase MB (U/L) | 0–24 | 9.0 (6.0–13.0) | 9.0 (6.0–12.0) | 9.5 (6.0–13.0) | 0.296 |
| Lactate dehydrogenase (U/L) | 120–250 | 234.0 (203.3–284.0) | 228.0 (196.8–264.3) | 336.5 (263.0–401.0) | < 0.001 |
| Potassium (μmol/L) | 3.50–5.30 | 3.7 (3.5–4.1) | 3.7 (3.5–4.1) | 3.7 (3.3–4.2) | 0.424 |
| Sodium (μmol/L) | 137.0–147.0 | 137.2 (135.2–138.9) | 137.7 (135.6–139.4) | 135.1 (132.5–137.7) | < 0.001 |
| Immune cell typing | |||||
| Lymphocyte (× 106/L) | 1530–3700 | 1061 (753–1469) | 1240 (812–1543) | 764 (585–987) | < 0.001 |
| Natural killer cell (× 106/L) | 84–724 | 147 (92–246) | 156 (92–258) | 130 (91–191) | 0.204 |
| B cell (× 106/L) | 80–616 | 124 (81–197) | 134 (87–212) | 106 (67–139) | 0.066 |
| T cell (× 106/L) | 723–2737 | 740 (507–1047) | 864 (598–1125) | 477 (337–675) | < 0.001 |
| CD4+T cell (× 106/L) | 404–1612 | 411 (278–587) | 462 (314–621) | 221 (185–407) | < 0.001 |
| CD8+T cell (× 106/L) | 220–1129 | 280 (173–433) | 326 (224–478) | 172 (119–269) | < 0.001 |
| CD4+/CD8+ | 0.71–2.78 | 1.37 (1.07–1.85) | 1.37 (1.09–1.83) | 1.36 (0.95–2.09) | 0.963 |
| Blood coagulation function | |||||
| PT (s) | 9.0–13.0 | 11.3 (10.8–12.3) | 11.3 (10.8–12.3) | 11.5 (10.9–12.4) | 0.835 |
| INR | 0.75–1.30 | 0.98 (0.95–1.07) | 0.98 (0.95–1.07) | 1.00 (0.95–1.08) | 0.927 |
| Fibrinogen (g/L) | 14.0–21.0 | 3.3 (2.5–4.1) | 3.1 (2.5–3.8) | 3.7 (3.3–4.9) | 0.004 |
| Thrombin time (s) | 14.0–21.0 | 14.6 (13.9–15.8) | 14.4 (13.6–15.4) | 15.4 (14.5–17.2) | < 0.001 |
| d-dimer (μg/L) | 0.00–0.55 | 0.28 (0.20–0.55) | 0.24 (0.19–0.40) | 0.50 (0.31–0.72) | < 0.001 |
^P values indicate differences between mild and severe patients. A P value < 0.05 was considered as statistically significant
The association of the severity of COVID-19 with comorbidities in our patient cohort was also analyzed (Fig. 1). Overall, patients with underlying diseases were more prone to developing severe COVID-19. However, patients with comorbidities such as chronic hepatitis B virus infection, liver cirrhosis, chronic respiratory diseases, HIV infection, and cancers (comorbidity group 1, CG-1) which are known to impair immunological functions were significantly more frequently affected compared to those with comorbidities unrelated to immune functions (comorbidity group 2, CG-2) (Fig. 1a), in the age-matched groups (Fig. 1b).
Fig. 1.
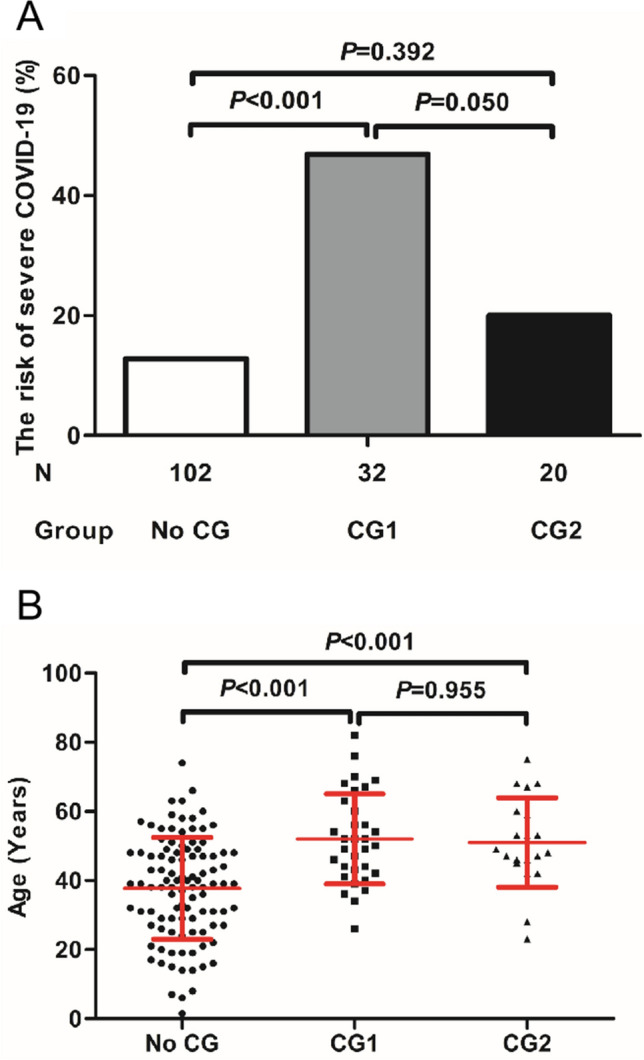
The association of the severity of COVID-19 with underlying diseases in the patient cohort. a The association of the severity of COVID-19 with comorbidities. The comorbidity group 1 (CG 1) includes chronic hepatitis B virus infection, liver cirrhosis, chronic respiratory diseases, HIV infection, and cancers which are known to impair immunological functions. The comorbidity group 2 (CG2) includes psychological diseases and fatty liver diseases which are primarily not related to host immune functions. The risk to develop severe COVID-19 was shown for patients without known comorbidities (No CG), CG1, and CG2. b The ranges of the ages of patients with and without comorbidities. The means ± SD are given in the graph. Results were tested for significance by Chi square test (a) or t test (b). A P value < 0.05 is considered as significant
Viral levels and the correlation with disease severities
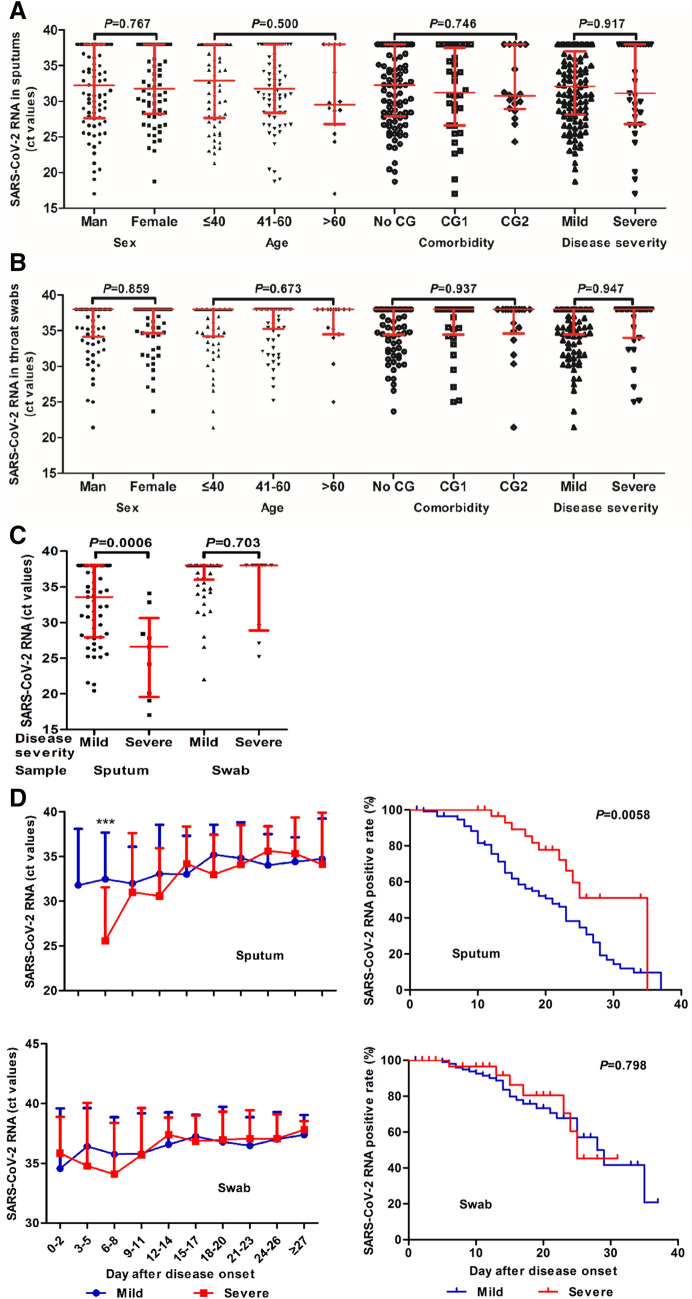
The Ct values of RT-PCR reactions performed were used to assess viral RNA levels in patient samples. An absolute quantification of viral RNA loads could not be obtained due to a lack of proved standard material. The amplification reaction for the N region regularly resulted in lower Ct values compared with that for the ORF1ab region, indicating a better sensitivity with N region amplification. Therefore, the Ct values of N-specific RT-PCR tests were analyzed to assess the dynamic changes in viral RNA levels (Fig. 2). For the detection of SARS-CoV-2 RNA, sputum or throat swab specimens were used. Overall, sputum specimens gave higher viral levels indicated by lower Ct values (Ct values up to 17) than throat swab specimens (Ct value up to 21), which was consistent with an early report [12]. Thus, the viral load data generated by using sputum or throat swab specimens were analyzed separately. We compared the measured SARS-CoV-2 RNA levels in sputum specimens from COVID-19 patients at admission among groups divided according to age, sex, underlying diseases and disease severity (Fig. 2a). The Ct values varied from > 38 up to 17 and no difference between patient groups was found. Consistently, the results obtained with throat swab samples showed no difference between patient groups (Fig. 2b). However, viral RNA levels in severe cases were found to be significantly higher in sputum specimens, but not in throat swab specimens between days 3 to 5 after disease onset compared to those in mild cases (Fig. 2c). Nevertheless, a number of mild cases had relatively high viral loads with a Ct value of up to 20. The viral RNA levels in the patients decreased and became undetectable (Ct > 38) over time (Fig. 2d). The mild cases became negative for viral RNAs with a median of 21 days after the disease onset, which was significantly earlier than severe cases with a mean of 35 days based on the test results using sputum specimens (Fig. 2e). No significant difference was found between the mild and severe cases at 28 and 25 days after disease onset if throat swab specimens were used (Fig. 2e).
Fig. 2.
Detection of SARS-CoV-2 RNA in patient samples by using a specific RT-PCR test. The Ct values of RT-PCR test for the SARS-CoV-2N region were used to assess viral RNA levels in patient samples. Sputum or throat swab specimens were used. a, b The measured SARS-CoV-2 RNAs levels in sputum (a) and throat swab (b) specimens from COVID-19 patients at admission were compared according to the age, sex, comorbidity, and the disease severity. c SARS-CoV-2 RNA levels in sputum and throat swab specimens from the mild and severe cases between the days 3 and 5 after disease onset. d Kinetics of SARS-CoV-2 RNA levels (Ct values) measured by RT PCR with sputum and throat-swap (swap) specimens. ***P < 0.001. e The Meier–Kaplan survival analysis for the detection of SARS-CoV RNAs based on the test results using sputum specimens and throat swab specimens. The median and quartile intervals (a–c) or means ± SD (d) are given in the graph. Results were tested for significance by t test, Mann–Whitney U or Kruskal–Wallis test (a–d), and log-rank (Mantel–Cox) test (e). A P value < 0.05 is considered as significant
Kinetic analysis of lymphocyte subsets in the peripheral blood of COVID-19 patients
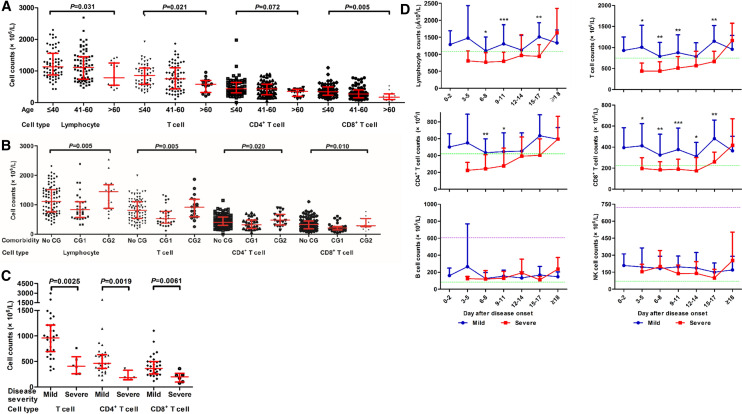
Lymphopenia was observed in 47.4% (73/154) of patients in our cohort, 40.2% (49/122) and 75% (24/32) in mild and severe cases at the onset of disease, respectively (Table 2). As shown in Table 2, the absolute counts of lymphocytes in the peripheral blood of patients subsequently developing severe COVID-19 was significantly lower at hospital admission and disease onset, respectively (Table 2, Fig. 3). Sustained lower lymphocyte counts in severe cases were observed, compared to those in mild cases (Fig. 3c). The absolute counts of total white blood cells (WBC) and neutrophils (NE) were initially comparable in both patient groups, but significantly increased only in severe cases at day 9 up to day 17 after disease onset (Fig. 3a, b). A significant difference in monocyte counts was observed between the two groups only at days 3–5, but not seen afterward (Table 2, Fig. 3d). The kinetic changes in different lymphocyte subsets in the peripheral blood of COVID-19 patients were monitored from disease onset up to 18 days. Flow cytometric analysis for CD3+ T cells, CD4+ and CD8+ T cell subsets, B cells and NK cells was performed (Fig. 4). Firstly, we analyzed initial lymphocyte counts and CD3+, CD4+ and CD8+ T cell counts at admission in patients by the age and presence of comorbidities (Fig. 4a, b). Clearly, patients of older age and with comorbidities of group 1 (CG1) had significantly lower counts for lymphocytes and T cell subsets. Consistent with the findings about lymphocytes, sustained significantly lower CD3+, CD8+ and CD4+ T cell counts were found in patients with severe disease compared to those of the mild cases during clinical observation (Fig. 4c). The lowest CD3+, CD4+ and CD8+ T cell counts in patients with severe COVID-19 were generally found 3–5 days after disease onset. However, they recovered and reached comparable levels to those in the mild cases after day 18 (Fig. 4d, Figure Suppl. 1). As severe COVID-19 cases had high SARS-CoV-2 RNA levels and low T, CD4+, and CD8+ cell counts, these parameters were tested for possible correlations. However, our analyses did not show any significant correlation between SARS-CoV-2 RNA level and T, CD4+or CD8+ T cell counts (Figure Suppl 2).
Fig. 3.
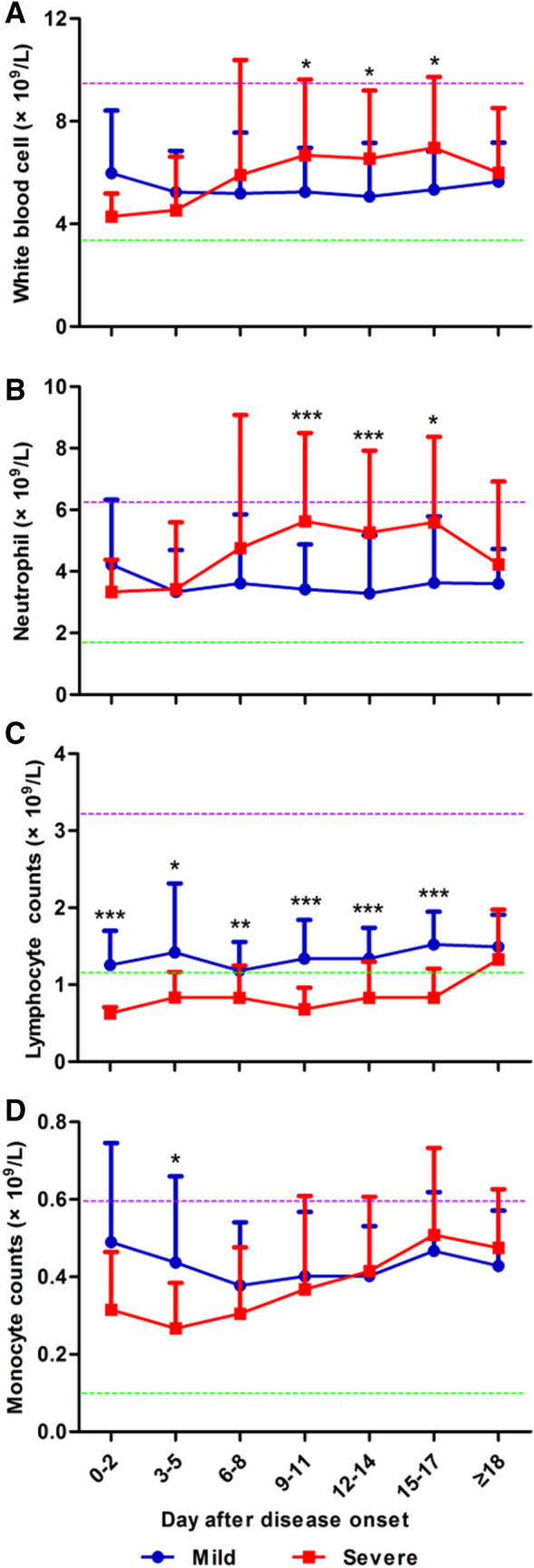
Kinetic analysis of cell counts of different populations of WBCs in COVID-19 patients. The absolute numbers of white blood cells (a), neutrophils (b), lymphocytes (c) and monocytes (d) in the peripheral blood of mild (blue line) and severe (red line) COVID-19 patients were analyzed at different time points after the disease onset. *P < 0.05; **P < 0.01; ***P < 0.001. The upper (purple line) and lower (green line) limits of normal values and the means ± SD are given in the graph. Results were tested for significance by t test or Mann–Whitney U test. A P value < 0.05 is considered as significant
Fig. 4.
Kinetic analysis of cell counts of different lymphocyte subsets in COVID-19 patients. The kinetic changes of different lymphocyte subsets in the peripheral blood of COVID-19 patients were measured from the disease onset up to 18 days later. Flow cytometric analysis to for CD3+ T cells, CD4+ and CD8+ T cell subsets, B cells and NK cells was performed. The initial lymphocyte counts and CD3+, CD4+ and CD8+ T cell counts at admission in patients according to age (a) and the presence of comorbidities (b). c The CD3+, CD8+ and CD4+ T cell counts in patients with mild and severe disease at 3–5 days after the disease onset. d The absolute numbers of CD3+ T cells, CD8+ T cells, CD4+ T cells, B cells and NK cells in the peripheral blood of mild (blue line) and severe (red line) COVID-19 patients were analyzed at different time points after the disease onset. *P < 0.05; **P < 0.01; ***P < 0.001. The median and quartile intervals (a–c) or means ± SD (d) are given in the graph. Results were tested for significance by two-way ANOVA (a), t test (b), Mann–Whitney U test (c) and t test or Mann–Whitney U test (d). A P value < 0.05 is considered as significant
No significant differences in B cell and NK cell counts were observed between the two groups during the whole disease course (Fig. 4d).
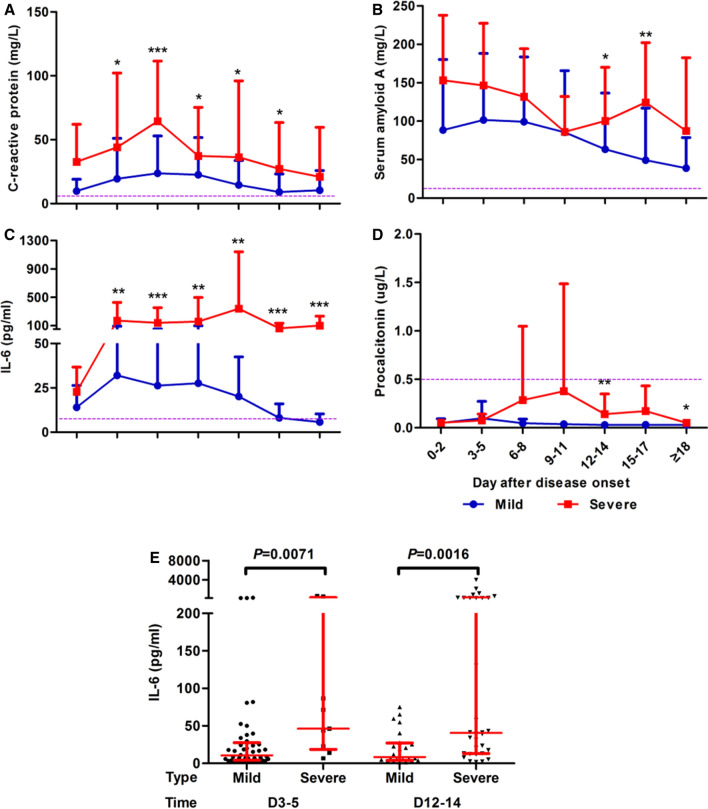
Kinetic analysis of serum inflammatory markers in COVID-19 patients
A previous study demonstrated a marked elevation of inflammatory cytokine levels such as IL-6 in the sera of COVID-19 patients [2]. Therefore, we further characterized the kinetic changes of inflammatory markers, including CRP, IL-6, SAA, and PCT in the sera of the patients. The serum levels of these markers in patients with mild disease showed minor fluctuations, while significant elevations of these markers were found severe cases (Fig. 5). CRP reached its peak in serum at 6–8 days after disease onset (Fig. 5a). IL-6 levels showed sustained increases in the severe group compared to the mild group over 18 days, even when some other markers returned to normal levels (Fig. 5b). The IL-6 levels in some patients with severe COVID-19 remained high as late as day 12–14 after disease onset, even though these patients had clinically recovered (Fig. 5e).
Fig. 5.
Kinetic analysis of serum levels of inflammatory markers in COVID-19 patients. The concentrations of C-reactive protein (a), IL-6 (b), serum amyloid A (c), procalcitonin (d), serum IL-6 concentrations at days 3–5 and 12–14 (e) in mild (blue line) and severe (red line) COVID-19 patients were analyzed at different time points after the disease onset. *P < 0.05; **P < 0.01; ***P < 0.001. The means ± SD (a–d) or median and quartile intervals (e) are given in the graph. Results were tested for significance by the Mann–Whitney U test. A P value < 0.05 is considered as significant
Significant increases in serum SAA and PCT levels were observed in the severe group after disease onset, but became only significantly different to those in the mild group at certain time points (Fig. 5c, d). The three examined markers CRP, SAA and PCT reached similar levels between the severe and mild patient groups at day 18 after disease onset (Fig. 5). A statistical analysis showed that SARS-CoV-2 RNA levels in patients did not correlate with the levels of serum inflammatory markers. No correlation was found between the initial levels of lymphocyte subsets in peripheral blood and serum inflammatory markers in the late period between days 12 and 14 after disease onset (not shown).
Factors associated with severe COVID-19
Next, we examined the possibility of using the above-mentioned parameters as prognostic factors for identifying severe cases in COVID-19 patients. PCA was firstly performed with R package “factoextra” to identify correlating variables for distinguishing severe patients from mild patients (Figure Suppl 3A). The lymphocyte and CD3+ T cell counts were found to be the two most important contributing variables among immunological markers. Previously, other factors such as neutrophil-to-CD8 + T cell ratio (N8R), neutrophil-to-lymphocyte ratio (NLR), neutrophil counts (NEC) and white blood cells counts (WBCC) had been proposed as potential prognostic factors for severe COVID-19 [13]. To assess the diagnostic value of these parameters, the receiver operating characteristic (ROC) curve and the area under ROC curve (AUC) were calculated by R package “pROC” (Figure Suppl 3B), and the AUC values for lymphocyte and CD3+ T cell counts were found to be 0.739 and 0.793, respectively. At the same time, the cutoff values were calculated from the ROC curves, with a value of 0.85 for lymphocyte counts (L) (specificity: 60.6%, sensitivity: 77.9%) and 665 for T cell counts (T) (665, 83.3%, 67.9%) (Figure Suppl 3B). Further, the ROC curves were calculated for other parameters: CD4+ T cell counts (CD4), CD8+ cell counts (CD8), and combined CD4+ and CD8+ T cell counts (CD4xCD8) with the AUC values 0.767 (cutoff: 269, specificity: 60.0%, sensitivity: 87.2%), 0.772 (236, 73.3%, 73.4%), and 0.800 (115,520, 90.0%, 62.4%), respectively (Figure Suppl 3B). Overall, the immune related parameters were found to have associations with the chance to develop severe COVID-19, but their diagnostic values need to be evaluated in larger cohorts.
Discussion
In this study, we analyzed the clinical features including SARS-CoV-2 RNA load and immunological characteristics of peripheral blood in a patient cohort with COVID-19 from Anhui Province, China. The major clinical features of this patient cohort are comparable with published data. The patients with severe COVID-19 were characteristic of older age and underlying diseases.
Our data revealed that the initial SARS-CoV-2 RNA levels in patients at admission did not differ with age, sex, comorbidities and disease severity at later time points. The viral loads in some mild cases were also high and comparable with those in severe cases. Thus, high viral load may be not the primary cause of severe COVID-19.
Recent studies reported that the lymphocyte counts were normal in COVID-19 patients with mild disease, and in contrast, the majority of patients with severe disease had lymphopenia [5, 13, 14]. Our study suggested that a number of patients, particularly those of old age and with underlying diseases might have initially low lymphocyte and T cell counts (Fig. 4). There was no correlation between initial SARS-CoV-2 RNA load and lymphocyte and T cell counts in patients at admission (Figure Suppl 2). A plausible explanation for these observations is that SARS-CoV-2 may replicate more efficiently in patients with compromised immunity and thus causes severe disease in these patients. Consistently, the viral loads in both mild and severe cases increased rapidly and reached peak levels within 3–5 days after disease onset. However, in severe cases, the viral loads were significantly higher than those in mild cases, especially at that time point. Strikingly, all patients requiring ICU treatment had high SARS-CoV-2 loads at days 3–5 after the onset of disease (Figure Suppl 4). Thus, accurate quantitative detection of SARS-CoV-2 loads may be an important parameter to guide the clinical handling of COVID patients.
Previous research on SARS-CoV and MERS-CoV infections has demonstrated the correlation between marked lymphopenia and the severity of the disease [15–19]. Altered antigen presenting cell (APC) function and impaired dendritic cell migration during SARS-CoV infection may result in insufficient T cell priming and thus contribute to decreased numbers of virus-specific T cells in the lungs [20, 21]. Moreover, delayed type I interferon response or inflammatory monocyte–macrophages derived pro-inflammatory cytokines could also sensitize T cells to undergo apoptosis [22]. In contrast, MERS-CoV was found to be able to infect large numbers of human immune cells, including dendritic cells, macrophages, and T cells [23–25]. MERS-CoV infection of T cells results in apoptosis mediated by a combination of extrinsic and intrinsic apoptosis pathways, which is believed to contribute to viral spread and severe immunopathology [25]. It remains unclear whether SARS-CoV-2 may induce T cell contraction by specific mechanisms. Yet, a correlation of initial SARS-Cov-2 RNA loads in patients with degrees of lymphopenia was not seen in our study.
Severe COVID-19 patients have significant and sustained higher levels of inflammatory markers including serum IL-6 concentrations than in mild cases, again suggesting that the magnitude of the cytokine storm is associated with disease severity. T cells are important for dampening overactive innate immune responses during viral infection [26, 27]. Thus, low T cell counts in patients with SARS-CoV-2 infection may result in aggravated inflammatory responses, while normal T cell levels and functions may maintain the immunological regulation and prevent overshooting inflammation. Early identification of risk factors for severe COVID-19 patients may facilitate appropriate supportive care and prompt access to intensive care unit if necessary. Some studies, such as one in a 61-patient cohort [13], reported that NLR was the most useful prognostic factor affecting the prognosis for severe COVID-19. However, the diagnostic value of such parameters to predict disease severity was not proven in our cohort involving a larger number of patients.
Finally, the differences in T cells, CRP, and IL6 in the patient groups classified by underlying disease were analyzed. The results showed that the differences between the No CG and CG1 groups were only compared in this study und significant at some points, as shown in Figure Suppl 5. A comparison of the No CG with CG2 did not show any significant difference. This is rather inconclusive due to the small sample size of CG2.
In conclusion, our analyses demonstrated that initial SARS-CoV-2 RNA load varied in patients, but were comparable in different patient groups classified according to age, gender, comorbidities and disease severity. In contrast, reduced counts of T cells and subsets were measured in patients with older age and comorbidities at admission and were characteristic of severe COVID-19. Thus, severe COVID-19 developed preferentially in patients with compromised immunity and was not associated with initial viral load. Our results highlighted the importance of close surveillance of SARS-CoV-2 infection for specific risk groups including elderly people and those with underlying diseases with impaired immunological functions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file1 The CD3+, CD4+ and CD8+ T cell counts in the patients with mild and severe disease at day 18 after disease onset. Flow cytometric analysis to for CD3+ T cells, CD4+ T and CD8+ T cell subsets was performed. The medians are given in the graph. Results were test for significance with the Mann Whitney U test. A p-value <0.05 is considered as significant (TIF 94 kb)
Supplementary file2 Lack of a significant correlation of SARS-CoV-2 RNA loads and lymphocyte, CD4+ T, and CD8+ T cell counts in COVID-19 patients. The correlation of SARS-CoV-2 RNA loads and T, CD4+ T, and CD8+ T cell counts in COVID-19 patients at days 3 to 5 after disease onset was tested. However, no statistical significant result was obtained. A p-value <0.05 is considered as significant (TIF 244 kb)
Supplementary file3 Prognostic factors of severe COVID-19. (A) Principal component analysis was performed by R package “factoextra” to identify correlated variables for distinguishing severe patients from mild COVID-19 patients. (B) ROC curve and AUC were calculated for the selected parameters by using R package “pROC”. NLR: neutrophil to lymphocyte ratio; N8R: neutrophil to CD8+ T cell ratio; WBC: white blood cells; NE: neutrophil; L: lymphocyte; T: CD3+ T cells; CD4: CD4+ T cells; CD8: CD8+ T cells; CD4_CD8: CD4+xCD8+T cell. A p-value <0.05 is considered as significant (TIF 308 kb)
Supplementary file4 Detection of SARS-CoV-2 RNA in patient samples by using a specific RT-PCR test. The Ct values of RT-PCR test for the SARS-CoV-2 N region were used to assess viral RNA loads in patient samples. SARS-CoV-2 RNA loads in sputum and throat-swab specimens from the mild to critical cases between the days 3 to 5 after disease onset. The patients were classified into 4 groups according to the Guidelines of the Diagnosis and Treatment of New Coronavirus Pneumonia (version 7) published by the National Health Commission of China. The median and quartile intervals are given in the graph; Results were tested for significance by two-way ANOVA. A p-value <0.05 is considered as significant (TIF 193 kb)
Supplementary file5 Kinetic analysis of cell counts of different lymphocyte subsets or inflammatory markersin COVID-19 patients with comorbidity. (A) The absolute numbers of lymphocyte counts, CD3+ T cells, CD4+ T cells and CD8+ T cells in the peripheral blood of No CG group (blue line) and CG1 group (red line) COVID-19 patients were analyzed at different time points after the disease onset. (B) The concentrations of C-reactive protein and IL-6 in No CG group (blue line) and CG1 group (red line) COVID-19 patients were analyzed at different time points after the disease onset.* p<0.05; ** p<0.01; *** p<0.001. The means ± SD are given in the graph Results were tested for significance by t-test or Mann Whitney U test. A p-value <0.05 is considered as significant (TIF 73 kb)
Acknowledgements
We thank Ms. Delia Cosgrove for language correction of this manuscript.
Abbreviations
- SARS-CoV-2
2019 Novel coronavirus
- IQR
Interquartile range
- ICU
Intensive care unit
- ARDS
Acute respiratory distress syndrome
- AIDS
Acquired immune deficiency syndrome
- AKI
Acute kidney injury
- COPD
Chronic obstructive pulmonary disease
- Bpm
Beats per minute
- ESR
Erythrocyte sedimentation rate
- PT
Plasma prothrombin time
- INR
International standard proportion of prothrombin time in plasma
Funding
This work was supported by the Natural Science Foundation of Anhui Province (Grant no. 1608085MH162) and a grant from an unrestricted grant of the Rudolf-Ackermann-Stiftung (Stiftung für Klinische Infektiologie) to O.W.
Data availability
All materials will be uploaded to the library of Anhui Medical University after publication.
Compliance with ethical standards
Conflict of interest
O.W. has received research grants for clinical studies, speaker’s fees, honoraria and travel expenses from Amgen, Astellas, Bristol-Myers Squibb, Chiesi, Hexal, Janssen-Cilag, MSD, Novartis, Roche, Pfizer, and Sanofi. The other authors disclose no conflicts of interest.
Ethics approval
This retrospective single-center study was approved by the Ethics Committee of the Second People's Hospital, Fuyang (Number 2020020004).
Consent to participate
Informed consent was obtained from all participants included in the study.
Consent for publication
All authors agreed the work to be published.
Code availability
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mingfeng Han, Mengyuan Xu, Yafei Zhang, Zhongping Liu and Shasha Li contributed equally to this work.
Contributor Information
Mengji Lu, Email: mengji.lu@uni-due.de.
Zhenhua Zhang, Email: zzh1974cn@163.com.
References
- 1.World Health Organization (2020) Coronavirus disease (COVID-2019) situation reports. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. Accessed 15 June 2020.
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, Yuan S, Kok K, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi H, Han X, Jiang N, Cao Y, Alwalid O, Gu J, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Yu Y, Xu J, Shu H, Xia J, Liu H, et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien JY, Hsueh PR, Cheng WC, Yu CJ, Yang PC. Temporal changes in cytokine/chemokine profiles and pulmonary involvement in severe acute respiratory syndrome. Respirology. 2006;11:715–722. doi: 10.1111/j.1400-1843.2006.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1101/2020.02.16.20023671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang D, Lin M, Wei L, Xie L, Zhu G, Dela Cruz CS, et al. Epidemiologic and clinical characteristics of novel coronavirus infections involving 13 patients outside Wuhan, China. JAMA. 2020;323:1092–1093. doi: 10.1001/jama.2020.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J, Zheng X, Tong Q, Li W, Wang B, Sutter K, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cui W, Fan Y, Wu W, Zhang F, Wang JY, Ni AP. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin Infect Dis. 2003;37:857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Qiu Z, Han Y, Wang Z, Fan H, Lu W, et al. Rapid loss of both CD4+ and CD8+ T lymphocyte subsets during the acute phase of severe acute respiratory syndrome. Chin Med J (Engl) 2003;116:985–987. doi: 10.1016/j.jinf.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Wong RS, Wu A, To KF, Lee N, Lam CW, Wong CK, et al. Haematological manifestations in patients with severe acute respiratory syndrome: retrospective analysis. BMJ. 2003;326:1358–1362. doi: 10.1136/bmj.326.7403.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li T, Qiu Z, Zhang L, Han Y, He W, Liu Z, et al. Significant changes of peripheral T lymphocyte subsets in patients with severe acute respiratory syndrome. J Infect Dis. 2004;189:648–651. doi: 10.1086/381535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Zhao J, Legge K, Perlman S. Age-related increases in PGD2 expression impair respiratory DC migration, resulting in diminished T cell responses upon respiratory virus infection in mice. J Clin Invest. 2011;121:4921–4930. doi: 10.1172/JCI59777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Zhao J, Van Rooijen N, Perlman S. Evasion by stealth: inefficient immune activation underlies poor T cell response and severe disease in SARS-CoV-infected mice. PLoS Pathog. 2009;5:e1000636. doi: 10.1371/journal.ppat.1000636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Channappanavar R, Perlman S. Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology. Semin Immunopathol. 2017;39:529–539. doi: 10.1007/s00281-017-0629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu H, Zhou J, Ho-Yin Wong B, Li C, Cheng Z, Lin X, et al. Productive replication of Middle East respiratory syndrome coronavirus in monocyte-derived dendritic cells modulates innate immune response. Virology. 2014;454–455:197–205. doi: 10.1016/j.virol.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou J, Chu H, Li C, Wong BH, Cheng Z, Poon VK, et al. Active replication of middle east respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J Infect Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu H, Zhou J, Wong BH, Li C, Chan JF, Cheng ZS, et al. Middle east respiratory syndrome coronavirus efficiently infects human primary T lymphocytes and activates the extrinsic and intrinsic apoptosis pathways. J Infect Dis. 2016;213:904–914. doi: 10.1093/infdis/jiv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim KD, Zhao J, Auh S, Yang X, Du P, Tang H, et al. Adaptive immune cells temper initial innate responses. Nat Med. 2007;13:1248–1252. doi: 10.1038/nm1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palm N, Medzhitov R. Not so fast: adaptive suppression of innate immunity. Nat Med. 2007;13:1142–1144. doi: 10.1038/nm1007-1142b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1 The CD3+, CD4+ and CD8+ T cell counts in the patients with mild and severe disease at day 18 after disease onset. Flow cytometric analysis to for CD3+ T cells, CD4+ T and CD8+ T cell subsets was performed. The medians are given in the graph. Results were test for significance with the Mann Whitney U test. A p-value <0.05 is considered as significant (TIF 94 kb)
Supplementary file2 Lack of a significant correlation of SARS-CoV-2 RNA loads and lymphocyte, CD4+ T, and CD8+ T cell counts in COVID-19 patients. The correlation of SARS-CoV-2 RNA loads and T, CD4+ T, and CD8+ T cell counts in COVID-19 patients at days 3 to 5 after disease onset was tested. However, no statistical significant result was obtained. A p-value <0.05 is considered as significant (TIF 244 kb)
Supplementary file3 Prognostic factors of severe COVID-19. (A) Principal component analysis was performed by R package “factoextra” to identify correlated variables for distinguishing severe patients from mild COVID-19 patients. (B) ROC curve and AUC were calculated for the selected parameters by using R package “pROC”. NLR: neutrophil to lymphocyte ratio; N8R: neutrophil to CD8+ T cell ratio; WBC: white blood cells; NE: neutrophil; L: lymphocyte; T: CD3+ T cells; CD4: CD4+ T cells; CD8: CD8+ T cells; CD4_CD8: CD4+xCD8+T cell. A p-value <0.05 is considered as significant (TIF 308 kb)
Supplementary file4 Detection of SARS-CoV-2 RNA in patient samples by using a specific RT-PCR test. The Ct values of RT-PCR test for the SARS-CoV-2 N region were used to assess viral RNA loads in patient samples. SARS-CoV-2 RNA loads in sputum and throat-swab specimens from the mild to critical cases between the days 3 to 5 after disease onset. The patients were classified into 4 groups according to the Guidelines of the Diagnosis and Treatment of New Coronavirus Pneumonia (version 7) published by the National Health Commission of China. The median and quartile intervals are given in the graph; Results were tested for significance by two-way ANOVA. A p-value <0.05 is considered as significant (TIF 193 kb)
Supplementary file5 Kinetic analysis of cell counts of different lymphocyte subsets or inflammatory markersin COVID-19 patients with comorbidity. (A) The absolute numbers of lymphocyte counts, CD3+ T cells, CD4+ T cells and CD8+ T cells in the peripheral blood of No CG group (blue line) and CG1 group (red line) COVID-19 patients were analyzed at different time points after the disease onset. (B) The concentrations of C-reactive protein and IL-6 in No CG group (blue line) and CG1 group (red line) COVID-19 patients were analyzed at different time points after the disease onset.* p<0.05; ** p<0.01; *** p<0.001. The means ± SD are given in the graph Results were tested for significance by t-test or Mann Whitney U test. A p-value <0.05 is considered as significant (TIF 73 kb)
Data Availability Statement
All materials will be uploaded to the library of Anhui Medical University after publication.