SUMMARY:
Background:
Body CT scans are frequently performed for a wide variety of clinical indications, but potentially valuable biometric information typically goes unused. We investigated the prognostic ability of automated CT-based body composition biomarkers derived from previously-developed deep-learning and feature-based algorithms for predicting major cardiovascular events and overall survival in an adult screening cohort, compared with clinical parameters.
Methods:
Mature and fully-automated CT-based algorithms with pre-defined metrics for quantifying aortic calcification, muscle density, visceral/subcutaneous fat, liver fat, and bone mineral density (BMD) were applied to a generally-healthy asymptomatic outpatient cohort of 9223 adults (mean age, 57.1 years; 5152 women) undergoing abdominal CT for routine colorectal cancer screening. Longitudinal clinical follow-up (median, 8.8 years; IQR, 5.1–11.6 years) documented subsequent major cardiovascular events or death in 19.7% (n=1831). Predictive ability of CT-based biomarkers was compared against the Framingham Risk Score (FRS) and body mass index (BMI).
Findings:
Significant differences were observed for all five automated CT-based body composition measures according to adverse events (p<0.001). Univariate 5-year AUROC (with 95% CI) for automated CT-based aortic calcification, muscle density, visceral/subcutaneous fat ratio, liver density, and vertebral density for predicting death were 0.743(0.705–0.780)/0.721(0.683–0.759)/0.661(0.625–0.697)/0.619 (0.582–0.656)/0.646(0.603–0.688), respectively, compared with 0.499(0.454–0.544) for BMI and 0.688(0.650–0.727) for FRS (p<0.05 for aortic calcification vs. FRS and BMI); all trends were similar for 2-year and 10-year ROC analyses. Univariate hazard ratios (with 95% CIs) for highest-risk quartile versus others for these same CT measures were 4.53(3.82–5.37) /3.58(3.02–4.23)/2.28(1.92–2.71)/1.82(1.52–2.17)/2.73(2.31–3.23), compared with 1.36(1.13–1.64) and 2.82(2.36–3.37) for BMI and FRS, respectively. Similar significant trends were observed for cardiovascular events. Multivariate combinations of CT biomarkers further improved prediction over clinical parameters (p<0.05 for AUROCs). For example, by combining aortic calcification, muscle density, and liver density, the 2-year AUROC for predicting overall survival was 0.811 (0.761–0.860).
Interpretation:
Fully-automated quantitative tissue biomarkers derived from CT scans can outperform established clinical parameters for pre-symptomatic risk stratification for future serious adverse events, and add opportunistic value to CT scans performed for other indications.
Introduction
There has been substantial and growing interest in applying artificial intelligence (AI) to medicine, using various machine- and deep-learning algorithms.1 Along with other “big data” challenges, diagnostic imaging has been identified as a logical early target.2–4 In particular, body computed tomography (CT) represents an ideal modality with vast potential, as these scans are widely performed and contain additional robust, objective volumetric data that is highly reproducible and consistent across patients. In fact, opportunistic use of CT data beyond the clinical indication has already shown value from a variety of manual and semi-automated approaches, most notably with incidental osteoporosis screening.5–7 Beyond bone mineral density (BMD) information, every abdominal CT scan contains additional rich body composition data that can be objectively measured, including vascular calcification, muscle mass and density, visceral and subcutaneous fat, and liver fat content.8–13 If properly leveraged, this additional opportunistic data could further augment the value of a CT scans for the benefit of patients by potentially providing risk stratification for future adverse events and overall mortality. Of note, a recent report has emphasized the relative lack of such prevention research among studies supported by the U.S. National Institutes of Health (NIH).14 Importantly, this body composition data is freely available on essentially any abdominal CT scan, regardless of the initial clinical indication for imaging.
We have previously developed, trained, tested, and validated fully-automated a number of algorithms for measuring body composition at abdominal CT, including quantification of aortic calcification, muscle density, visceral and subcutaneous fat, liver fat, and BMD.15–19 These CT-based biomarkers may hold potential for identifying those at increased risk for a variety of adverse clinical outcomes. With all of the artificial intelligence (AI) learning steps complete and the specific automated tool outputs already pre-selected, our next logical step was to apply these mature pre-defined “static” tools to an external cohort. To this end, we have access to a unique external screening cohort of generally healthy asymptomatic adults who underwent abdominal CT for the purpose of colorectal cancer prevention and screening, using CT colonography (CTC) technique.20 Through longitudinal follow-up, we have identified subsequent defined adverse events in this patient cohort, including heart attack, stroke, and death. The main purpose of this study was to investigate the prognostic ability of a fully-automated pre-defined panel of CT-based body composition biomarkers for pre-symptomatic prediction of future cardiovascular events and overall survival in a healthy adult screening cohort.
Methods
Patient cohort and CT protocol
This HIPAA-compliant investigation was approved by the Institutional Review Board at the University of Wisconsin and the Office of Human Subjects Research Protection at the NIH Clinical Center. The requirement for signed informed consent was waived. After exclusion of 82 individuals for inadequate follow-up (<1 year in the absence of an adverse event), the final study cohort consisted of 9223 generally healthy consecutive asymptomatic outpatient adults (mean age, 57.1 years; 5152 women, 4071 men), undergoing low-dose unenhanced abdominal CT for colorectal cancer screening (as part of routine health maintenance) between 2004 and 2016 at a single medical center. The low-dose non-contrast supine multi-detector CT scans utilized for this investigation were all performed at 120 kVp using a single vendor (GE Medical Systems), with modulated mA to achieve a noise index of 50, typically resulting in an effective dose of 2–3 mSv. The specific additional CTC-related techniques for bowel preparation and colonic distention have been previously described.20,21 and are beyond the scope of this investigation.
Automated CT Biomarkers
The deep learning and image processing algorithms utilized for this predictive trial were previously developed, trained, and tested at the NIH Clinical Center. These CT-based algorithms include automatically segmenting and quantifying the spine, aortic calcium, abdominal musculature, visceral and subcutaneous fat, and liver. The current study represents an external validation for these tools,22 which were all trained and tested on CT cohorts separate from the current study cohort. Both the preliminary works and this culminating predictive trial all made use of the high performance computing capabilities of the Biowulf system at the NIH. The specific AI methodology for these automated CT-based anatomic tissue segmentation and quantification tools have been previously described elsewhere15–19,23–29 (see Supplement for additional methodology details). Briefly, these tools fall into two main categories: a deep-learning group and a feature-based image-processing group. Deep-learning algorithms were utilized to segment and analyze the entire liver, the abdominal wall musculature, and calcified atherosclerotic aortic plaque. These models consisted of a modified 3D U-Net for segmentation of liver and muscle, and the Mask-RCNN algorithm for segmentation of aortic calcium. For bone and fat quantification, feature-based image processing algorithms were used, starting with fully-automated spine segmentation and labeling software to identify each vertebral level from T12–L5. This was followed by isolation of the anterior trabecular space of each vertebra for BMD, as well as the visceral and subcutaneous fat compartments at each level. Because these validated CT-based tools were utilized herein in a static manner whereby no additional “learning” was employed, the need for additional training, testing, or cross-validation is obviated.
Preliminary works utilizing our CT screening cohort were performed for each automated CT tool to establish normative values, success/failure rates, and to narrow down each tool to a single stable quantitative measure for each tissue composition, without additional learning or adjustment.15–19 Each tissue measure can be reported in a variety of ways. For example, CT attenuation numbers measured in Hounsfield units (HU) reflect mean tissue density. Tissue bulk can be expressed according to cross-sectional area at specified levels or by volume. The final selected static measure for each of five body composition areas (Fig 1) was chosen according to our preliminary investigations to optimize overall success, and included: 1) the visceral-to-subcutaneous (V/S) fat ratio at the L1 level, 2) mean muscle density (in HU) at the L3 level, 3) volumetric liver density (in HU), 4) aortic calcification between the L1–L4 vertebral levels, quantified by an Agatston score, and 5) trabecular BMD at the L1 level (in HU). The technical failure rates for these tools were all on the order of 1% or less. Figure 1 depicts visual correlates of the quantitative output for the automated CT tools. This final panel of biomarkers was derived from CT scans in this study cohort in a fully-automated fashion.
Figure 1A. Depiction of the fully-automated CT biomarkers tools utilized in this study.
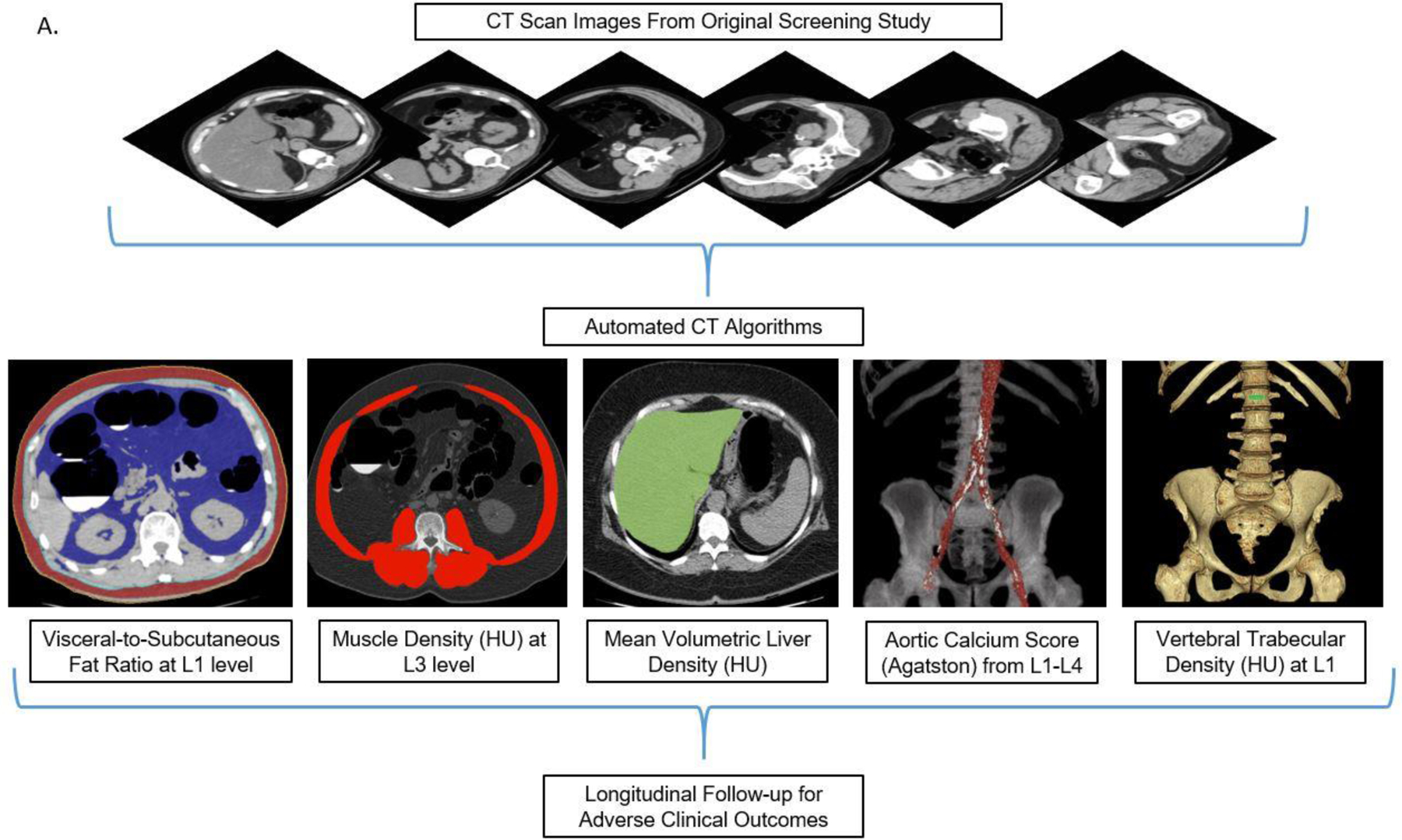
Schematic depiction (A) of the automated process for asses sing fat, muscle, liver, aortic calcium, and bone from original abdominal CT scan data. In practice, the visual tool outputs allow for slice-by-slice quality assurance for automated segmentation results in individual patients. CT biomarkers results were then correlated with subsequent adverse clinical outcomes. Importantly, these automated algorithms were pre-selected based on prior work and applied in a static fashion, without additional “learning” or adjustment.
Clinical Parameters and Adverse Outcomes
Beyond patient age and sex, the main clinical parameters we considered were body mass index (BMI), defined as weight (kg) divided by the square of height (m2) and the data inputs necessary for the Framingham Risk Score (FRS). The FRS for assessing risk for cardiovascular disease (CVD) is a well-established, validated multivariate algorithm combining the factors of age, sex, blood pressure, cholesterol, lipids, diabetic status, and smoking.30 Data points closest to the timing of the CT scan were included.
Adverse clinical outcomes were defined by either patient death or major cardiovascular events subsequent to CT scanning, including myocardial infarction (MI), cerebrovascular accident (CVA), or development of congestive heart failure (CHF) to reflect the endpoints considered by the FRS for CVD. We constructed a broad algorithmic EHR search for the relevant clinical data points and the defined clinical events.
Statistical Analysis
The analysis and modeling were developed specifically for this study, and have not been applied previously to other cohorts or scenarios. Summary statistics were compiled and compared for those patients with and without subsequent adverse events. To assess the association between the predictive measures and downstream adverse events, we utilized both an event-free survival analysis and logistic regression to compute receiver operating characteristic (ROC) curves. Relevant p-values were derived using two-sided t-tests for normally distributed variables, and the Wilcoxon rank sum test when the normality assumption did not hold. AUROC comparisons were made using DeLong’s method; p<0.05 was used to determine statistical significance. For the time-to-event survival analysis, Kaplan-Meier curves were generated by splitting predictor variables into quartiles. Cox proportional hazards models were used to derive concordance values and individual risk predictions. For ROC curve analysis, data sets were restricted to defined time intervals since time to event is not considered. Three arbitrary cutoffs included only patients having at least 2-year, 5-year, or 10-year follow-up, respectively, if they did not experience an event within those time frames. Area under the ROC curves (AUROC) with 95% confidence intervals were calculated. Univariate and multivariate analyses of CT biomarkers were performed. Age and sex were considered as potential confounders. Of note, FRS is already a multivariate predictor. Hazard ratios with 95% confidence intervals (CI) were computed for each CT biomarker, comparing the “highest-risk” quartile against the other three quartiles.
Results
The final study cohort consisted of 9223 generally healthy asymptomatic adults (mean age, 57.1 years; 5152 women, 4071 men), who underwent low-dose unenhanced abdominal CT. After final longitudinal clinical follow-up subsequent to CT scanning (median time interval, 8.8 years; IQR, 5.1–11.6 years), adverse clinical outcomes of interest, including major cardiovascular events (MI, CVS, or CHF) or death, were confirmed in 1831 (19.7%) patients. Of the 549 (5.9%) patients who died during the surveillance interval, the median time interval from CT scan to death was 6.1 years (mean, 6.2 years; IQR, 3.2–9.2 years). Median time to cardiovascular event was 4.4 years (mean, 5.0; IQR 2.0–7.8 years). Significant differences (p<0.001) were observed in all five automated CT-based measures (aortic calcification, muscle density, visceral/subcutaneous fat ratio, liver density, and L1 vertebral density) between those with and without an adverse event (Table 1). Summary data for clinical parameters according to adverse events are shown in Table S1.
Table 1.
Summary Data of CT Biomarkers According to Clinical Outcomes
| Summary Data of CT Biomarkers According to Clinical Outcomes | |||||
|---|---|---|---|---|---|
| CT Biomarker | Total Cohort (n=9223) | CV Event | Death | ||
| Yes (n=1831) | No (n=7392) | Yes (n=549) | No (n=8674) | ||
| AoCa (Ag) | |||||
| Mean | 699 | 1628 | 469 | 2471 | 587 |
| Median | 59 | 449 | 31 | 873 | 48 |
| IQR | 0–493 | 38–1945 | 0–313 | 131–3541 | 0–428 |
| Muscle HU | |||||
| Mean | 28.9 | 25.4 | 29.8 | 20.8 | 29.4 |
| Median | 31 | 27 | 31 | 22 | 31 |
| IQR | 22–38 | 17–35 | 23–38 | 12–31 | 23–38 |
| V/S Fat Ratio | |||||
| Mean | 0.91 | 1.13 | 0.58 | 1.22 | 0.89 |
| Median | 0.72 | 0.94 | 0.68 | 0.98 | 0.71 |
| IQR | 0.5–1.2 | 0.6–1.4 | 0.5–1.1 | 0.7–1.5 | 0.5–1.1 |
| Liver HU | |||||
| Mean | 55.4 | 54.1 | 55.7 | 53.6 | 55.5 |
| Median | 58 | 56 | 58 | 56 | 58 |
| IQR | 52–62 | 50–61 | 52–62 | 50–60 | 52–62 |
| L1 HU | |||||
| Mean | 171.2 | 159.0 | 174.2 | 150.9 | 172.4 |
| Median | 168 | 156 | 171 | 146 | 169 |
| IQR | 142–197 | 128–186 | 146–200 | 113–180 | 144–198 |
CV = cardiovascular; AoCa = aortic calcification; Ag = Agatston score; HU = Hounsfield units; V/S = visceral-tosubcutaneous; IQR = interquartile ratio; Subsequent CV events defined as acute MI, CVA, CHF, or death
All comparisons of CT biomarkers with versus without events were statistically significant (p<0.001)
The diagnostic performance of the clinical parameters (FRS and BMI) and the CT-based metabolic biomarkers for predicting overall survival is shown in Table 2 according to ROC curve and Cox proportional hazards analyses. For all data points (ie, 2-year, 5-year, and 10-year AUROC; and Cox model concordance), the automated CT-based univariate results for aortic calcification and muscle density were higher than the FRS, without including any demographic input data. For example, as shown in Figure 2a, the univariate 5-year AUROC values (with 95% CIs) for CT-based aortic calcification and muscle density were 0.743 (0.705–0.78sss0) and 0.721 (0.683–0.759), respectively, compared with 0.688 (0.650–0.727) for FRS (p<0.05 for aortic calcium vs. FRS). Automated CT-based fat, liver, and bone measures also performed fairly well as univariate measures, whereas BMI was a poor predictor, with 5-year AUROC of 0.499 (0.454–0,544) (Fig 2a). Similar performance trends were observed for prediction of downstream major cardiovascular events (Table S2). For example, all AUROC values for aortic calcification, whether alone or in combination with other CT-based automated measures, were significantly greater than for FRS (p<0.05). In general, multivariate combinations of CT biomarkers further improved prediction over clinical parameters (Tables 2 and S2). For example, combining the three CT-based quantitative biomarkers of aortic calcification, muscle density, and liver fat resulted in a 2-year AUROC of 0.811 (0.761–0.860) (Fig 2b).
Table 2.
Diagnostic Performance for Predicting Overall Survival
| Diagnostic Performance for Predicting Death | ||||
|---|---|---|---|---|
| 2-year AUROC (n=7849) | 5-year AUROC (n=6891) | 10-year AUROC (n=4029) | Cox PH Model Concordance | |
| Clinical Parameters | ||||
| FRS | 0.700 | 0.688 | 0.693 | 0.681 |
| BMI | 0.546 | 0.499 | 0.533 | 0.520 |
| Automated CT Biomarkers | ||||
| Univariate | ||||
| AoCa (Ag) | 0.746 | 0.743* | 0.746* | 0.735 |
| Muscle HU | 0.736 | 0.721 | 0.717 | 0.700 |
| V/S Fat Ratio | 0.685 | 0.661 | 0.656 | 0.648 |
| Liver HU | 0.644 | 0.619 | 0.628 | 0.602 |
| L1 HU | 0.627 | 0.646 | 0.640 | 0.637 |
| Multivariate** | ||||
| AoCa + Muscle | 0.780 | 0.768 | 0.768 | 0.772 |
| AoCa + Muscle + Liver | 0.811 | 0.782 | 0.777 | 0.778 |
| AoCa + Muscle + Liver + V/S | 0.817 | 0.789 | 0.780 | 0.780 |
| AoCa + FRS° | 0.774 | 0.744 | 0.746 | 0.733 |
| AoCa+Muscle+Liver+V/S+FRS° | 0.847 | 0.796 | 0.792 | 0.778 |
Cox PH model = Cox proportional hazards model; AUROC = area under the ROC curve; FRS = Framingham Risk Score; BMI = body mass index; AoCa = aortic calcification; Ag = Agatston score; HU = Hounsfield units; V/S = visceral-to-subcutaneous
p<0.05 compared with FRS performance
p<0.05 compared with FRS performance
No significant improvement compared with CT-based performance without FRS (p=0.509–0.965 for AoCa comparison and p=0.406–0.806 for AoCa+Muscle+Liver+V/S comparison)
Figure 2. ROC curves for predicting overall survival.
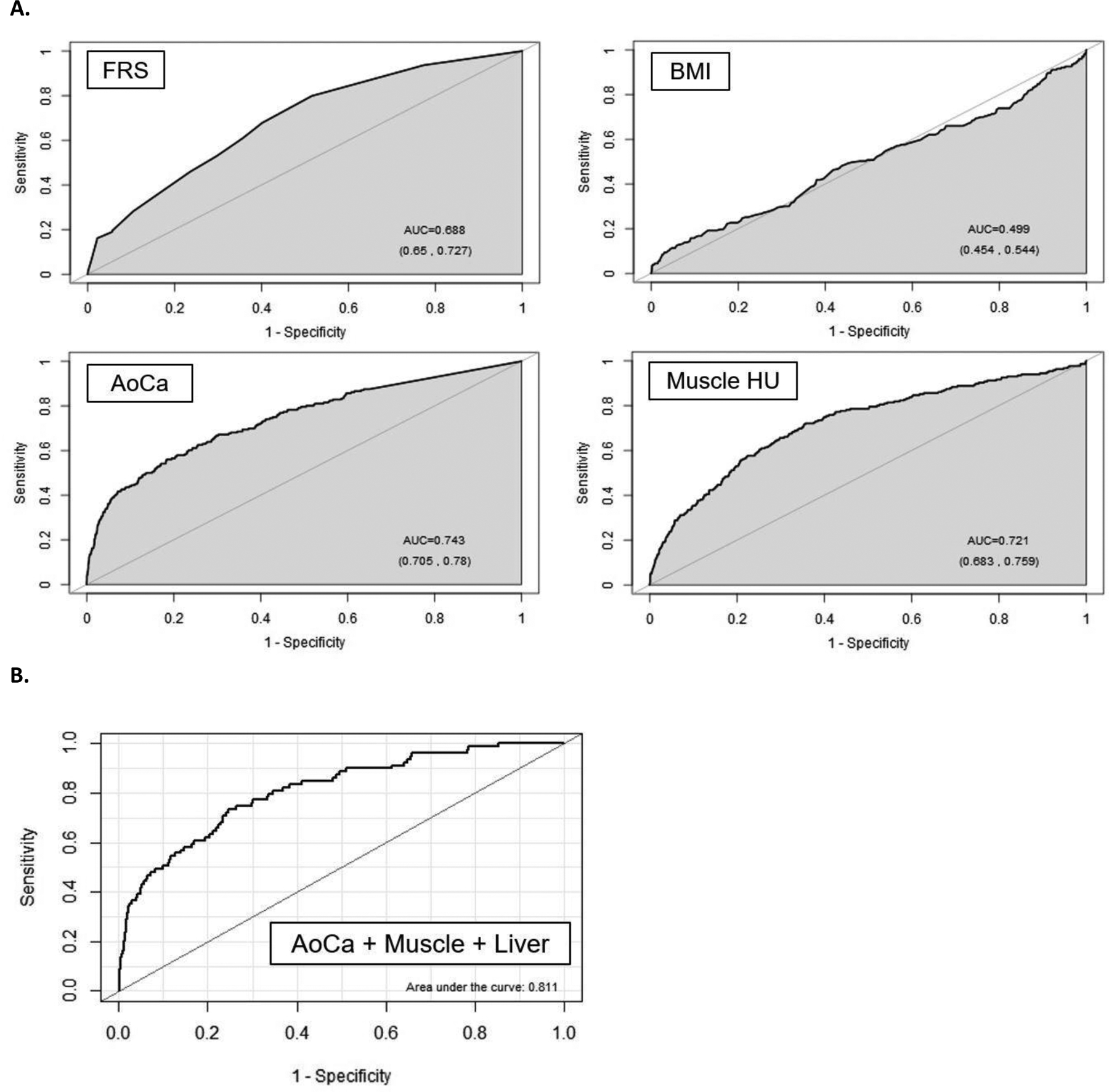
A. ROC curves for the clinical parameters of FRS and BMI, as well as univariate CT measures of aortic calcification and muscle density for predicting death over a 5-year time horizon. Without additional demographic or other input data, both CT parameters outperform the multivariate FRS according to area under the curve (AUC). In general, BMI was a poor predictor of outcomes at all time points in this study. The numbers in parentheses represent 95% confidence intervals).
B. Multivariate combination of CT-based biomarkers further improved prediction. In this example, CT-based aortic calcification, muscle density, and liver density for predicting overall survival over 2-year time horizon give an AUC or 0.811 (95% CI, 0.761–0.860).
When FRS was added to the CT-based aortic calcium score, there was no significant improvement of this automated CT measure alone for either CV events or overall survival, with p-values all 0.509 or greater for all AUROC comparisons (Tables 2 and S2). Similarly, adding FRS to CT-based multivariate combinations did not significantly improve performance either (Tables 2 and S2). Also of note, adding potential confounders of patient age and sex to the multivariate analysis provided only minor incremental benefit to the automated CT data alone (Fig S1b).
Kaplan-Meier time-to-death plots by quartile for the clinical parameters and univariate CT biomarkers are shown in Figure 3. Good separation between the “highest-risk” quartile versus the other three quartiles over time was observed for the automated aortic calcification score, with a similar trend for automated muscle density. Although quartile separation was less pronounced for the CT-based fat and liver measures, each is noticeably better then BMI. Univariate hazard ratios (with 95% CI) comparing the highest-risk quartile with the other three quartiles for CT-based aortic calcification, muscle density, visceral/subcutaneous fat ratio, liver density, and vertebral density were 4.53 (3.82–5.37), 3.58 (3.02–4.23), 2.28 (1.92–2.71), 1.82 (1.52–2.17), and 2.73 (2.31–3.23), respectively. Corresponding hazard ratios for BMI and FRS were 1.36 (1.13–1.64) and 2.82 (2.36–3.37), respectively. Similar time-to-event results were observed when cardiovascular events are included (Fig S1a); univariate hazard ratios ranged from 1.62–3.53 for the five metabolic CT markers (and 1.34 and 2.59 for BMI and FRS, respectively). When combining CT-based parameters in a multivariate fashion, further improvement in worst quartile separation was observed (Fig S1b).
Figure 3. Kaplan-Meier time-to-death plots by quartile for clinical parameter and univariate CT biomarkers.
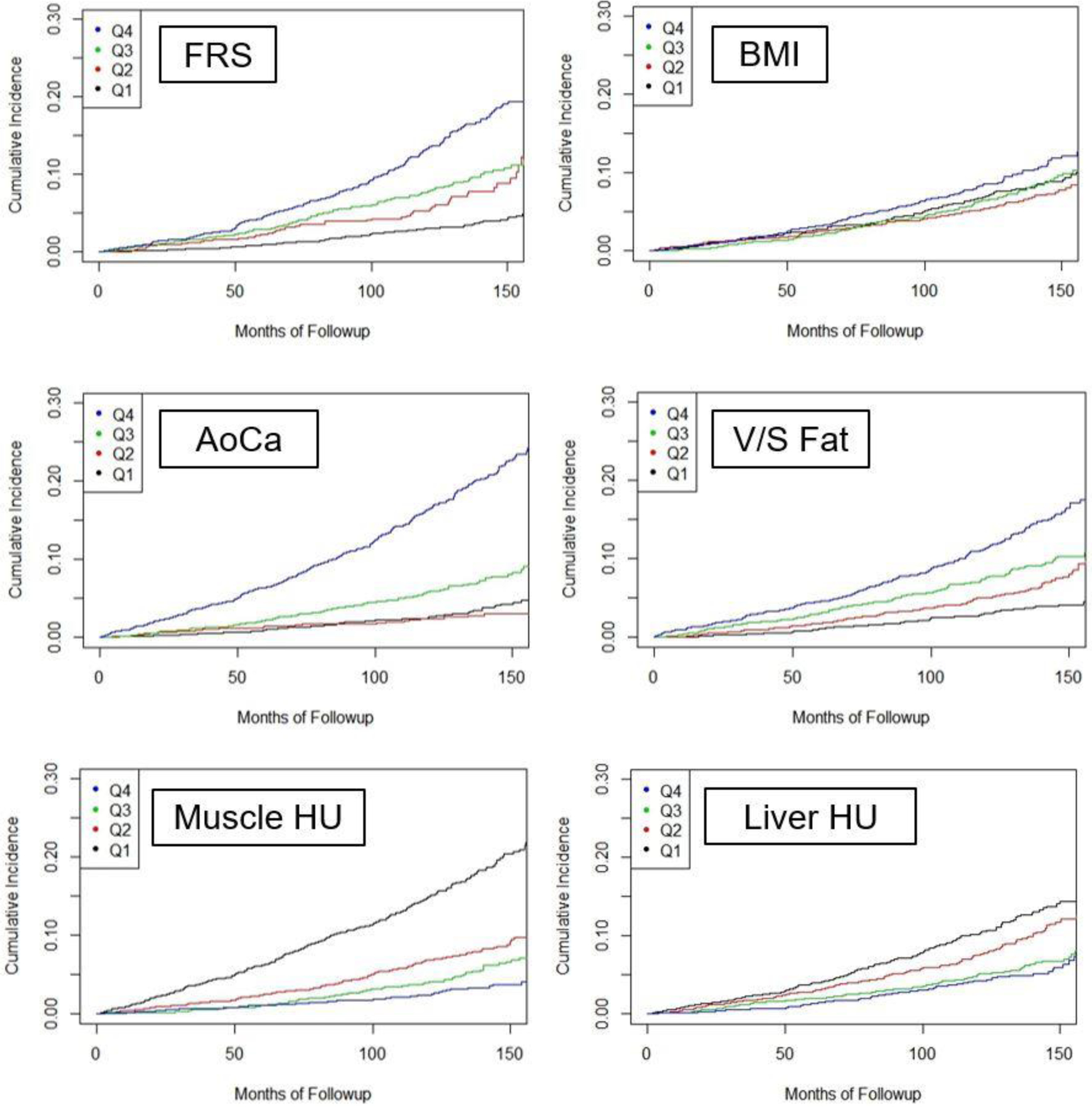
Good separation between the “worst” versus other quartiles over time was observed for the automated aortic calcium and muscle density values. Quartile separation was less pronounced for the CT-based fat and liver measures, but each is noticeably better than BMI. The L1 BMD results are not shown but separation of the worst quartile was comparable to muscle assessment.
Figure 1B demonstrates a case example that shows how predictive modeling derived from the quantitative CT data can be applied to an individual patient, similar to the multivariate FRS approach.
Figures 1B and 1C. Case example in an asymptomatic 57-year-old man undergoing CT for colorectal cancer screening.
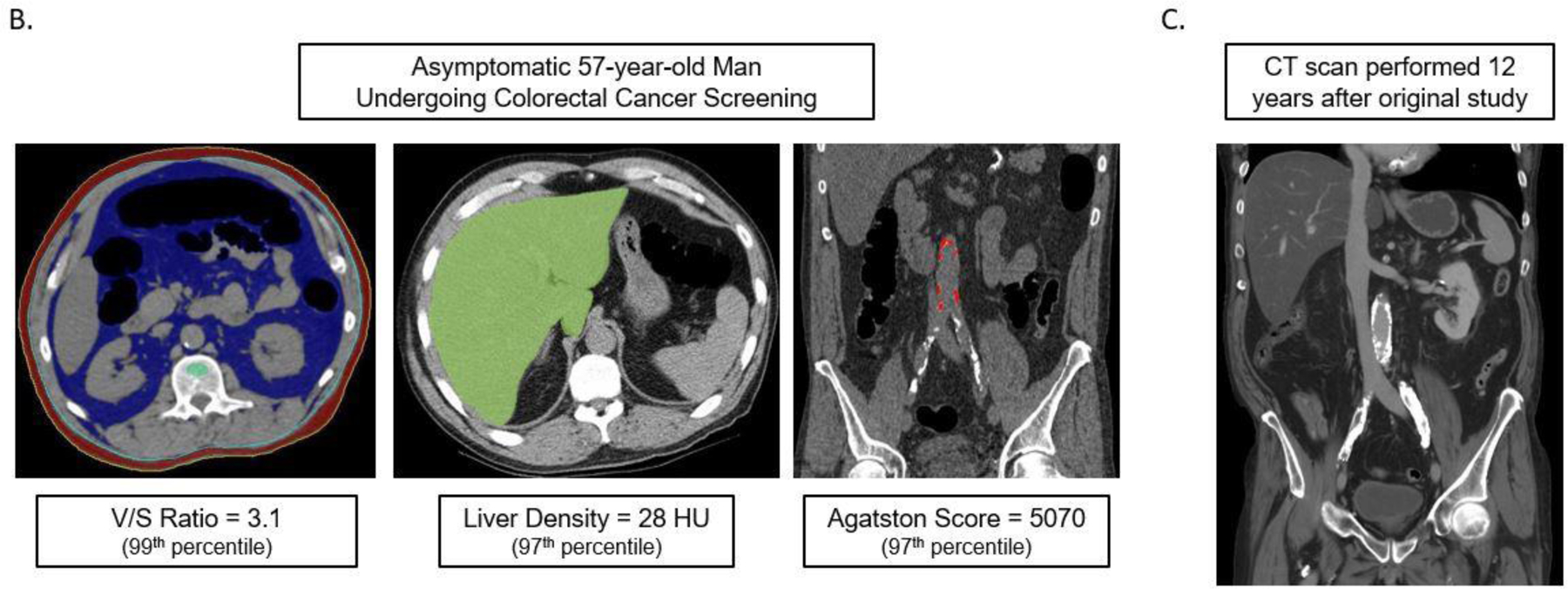
At the time of CT screening, he had a BMI of 27.3 and FRS of 5% (low risk). However, several CT-based metabolic markers were indicative of underlying disease (B), including a visceral-to-subcutaneous fat ratio of 3.1 (99th percentile), abdominal aortic Agatston score of 5070 (97th percentile), and steatotic liver density of 28 HU (97th percentile). Multivariate Cox model prediction based on these three CT-based results put risk of CV event within 2, 5, and 10 years at 19%, 40%, and 67%, respectively, and of death at 4%, 11%, and 27%, respectively. At longitudinal clinical follow-up, the patient suffered an acute MI three years after this initial CT and died 12 years after CT at the age of 64. Contrast-enhanced CT (C) performed seven months before death for minor trauma was interpreted as negative but does show significant progression of vascular calcification, visceral fat, and hepatic steatosis.
Discussion
This study demonstrates the potential value of harnessing the rich biometric tissue data embedded within all body CT scans that typically go unused in routine practice. Although such an opportunistic approach can be applied using manual or semi-automated measures, the maturation of robust fully-automated AI algorithms provides for a more efficient and objective means for high-volume population-based opportunistic screening. With over 80 million body CT scans performed each year in the U.S.,31 much of the focus has been placed on negative concerns about “incidentalomas” and radiation exposure.32,33 However, since most scans are performed on older adults, the opportunistic screening potential also becomes apparent. We applied these CT-based tools for assessing body composition to a generally healthy, outpatient screening cohort to start, which uniquely reflects our general adult population over 50, but this approach can also be applied to other cohorts, including those with symptoms or increased risk factors. We envision a (not-too-distant) future where this valuable prognostic CT information might be routinely captured and reported for the benefit of the patient, regardless of the clinical indication for imaging. The added value from these CT-based metabolic biomarkers requires no additional patient time or radiation exposure, and has the potential for improved individualized risk profiling.
We found that the AI panel of automated CT-based tissue biomarkers used in this study compared favorably with the FRS for pre-symptomatic prediction of future cardiovascular events and death. In fact, in terms of AUROCs and HRs, the univariate CT-based measures of aortic calcification alone significantly outperformed the multivariate FRS for major CV events and overall survival. Based on prior preliminary work that required manual case-by-case interaction for abdominal aortic calcium scoring in a smaller cohort,9 we expected the automated calcium tool to be valuable for cardiovascular risk profiling. BMI, which does not account for the relative anatomic distribution of fat,10 was a poor predictor of cardiovascular events and overall survival, whereas the CT-based visceral/subcutaneous fat ratio performed significantly better. Although BMI quartile separation was minimal, the slightly greater risk for death observed for the 1st and 4th quartiles (Figure 3) likely reflects the previously described U-shaped risk curve for this parameter.34 Liver density at non-contrast CT directly correlates with fat content,11,19 and reflects the high prevalence of hepatic steatosis, which has relevance for metabolic syndrome. Although its univariate performance was not stellar, liver density appears to have complementary value in terms of AUROC when combined with other CT biomarkers, such as aortic calcification and muscle density. In general, a multivariate combination of these CT-based biomarkers is likely the best way forward for optimized risk stratification. Furthermore, these CT biomarkers appear to be stronger predictors of future CV events compared with a panel of previously studied blood- and urine-based biomarkers reported by Wang et al.35
A recent publication by Vargas et al has emphasized the relative lack of prevention research that measures leading risk factors for death or disability as outcomes among studies supported by the U.S. National Institutes of Health (NIH).14 This study was intended in part to help address this research gap. While the current study focused only on the clinical outcomes of subsequent cardiovascular events and overall survival, these automated CT biomarkers have prognostic value for other “cardiometabolic” endpoints, such as osteoporotic fragility fractures and metabolic syndrome. We are only advocating for using this additional CT-based body composition data in an opportunistic fashion, and not as the sole reason for scanning. However, when coupled with an established indication such as CTC for colorectal cancer prevention, the concept of standalone population-based CT screening of asymptomatic adults could potentially be considered. In this scenario, the cumulative value of the screening CT data would need to clearly outweigh the potential harms, including cost and radiation exposure, and provide benefit beyond the more typical clinical means. Nonetheless, in current practice, this additional CT data is largely going unused in the many patients being scanned for a wide variety of established clinical indications. Automated CT measures of muscle, fat, and bone might also be valuable for opportunistic frailty monitoring in cancer patients, who often undergo repeated CT scanning for treatment response and surveillance.
The ever-expanding attention focused on the potential of AI in medicine is nearly ubiquitous, both in the medical literature and the lay press.1 The application of countless algorithms ranging from classic machine learning to more complex deep learning with convolutional neural networks is omnipresent. Along with a few other specific areas in medicine, medical image analysis represents a logical target for AI application.3,4 Despite predictions by some that “disruptive” AI technology is destined to soon displace the radiologist,36 the complexity of creating, training, and modifying the vast number of necessary algorithms argues instead for active engagement over replacement.3,4,37,38 Furthermore, AI is really nothing new in radiology, and co-authors of the current work have been involved in CT-based computer-aided detection (CAD) for many years.39 Although we believe that AI-based advances will ultimately enhance the practice of radiology, the recent hype has greatly outpaced true progress to date. The validated CT-based AI tools that we demonstrate herein represent the culmination of years of development, training, and testing. Although some of the processes are rooted in deep-learning algorithms, the output of these quantitative tools is straightforward and can be visually confirmed for quality assurance (ie, “explainable” AI), as opposed to the more “black box” feel of many other deep-learning AI solutions.
We acknowledge limitations to our investigation. All CT studies were performed with non-contrast technique; we are currently validating the use of these automated tools in a separate asymptomatic healthy cohort who underwent CT both without and with intravenous contrast. Risk stratification was based on analysis of the initial CTC examination in this screening cohort. A subset of more than 2000 patients underwent subsequent CT screening 5–10 years later, for which we plan to assess for interval changes in these automated measures that may offer additive value. Although our relatively unique CT screening cohort comprised of generally healthy outpatient adults was ideal for initial investigation, external validation in other screening populations with broader racial diversity is warranted, as our cohort was about 90% Caucasian. Application to new cohorts, including symptomatic patients at other centers, would also allow for further testing of the predictive models. This could be performed using a federated approach.40 One could argue that the FRS is outdated as a clinical comparator and used less often in the clinic. However, the FRS has served well for a number of previous trials, providing greater context as a common reference standard. Furthermore, the recent 2019 ACC/AHA guidelines state that the FRS may still be appropriate for use as an alternative risk prediction tool.41 It is conceivable that unforeseen confounders between the earlier testing/training cohorts and the current study group could exist with regard to measuring body composition by CT. Given the nature of the EHR search for adverse outcomes, it is possible that some definable events were not captured. However, our population tends to be quite stable. The quartile separation approach we have chosen likely does not reflect the optimal division of the data, but instead represents a starting point for further investigation, potentially with even larger cohorts. Finally, it is also important to consider the potential for possible unintended harm if subsequent intervention or inaction resulted from an incorrect classification of cardiovascular risk based on the CT-based body composition data.
In conclusion, we have shown that fully-automated quantitative tissue biomarkers derived from abdominal CT scans can outperform established clinical parameters for pre-symptomatic prediction of future cardiovascular events and overall survival. This approach leverages robust biometric data embedded in all such scans, and can add opportunistic value to abdominal CT scans performed for a wide variety of other indications.
Supplementary Material
Research in context.
Evidence before this study
There is a robust literature on how certain objective measures derived from abdominal CT scans can provide useful health information beyond the specific clinical indication for scanning. We and others have previously shown that manual “opportunistic” measures of aortic calcification, abdominal musculature, visceral fat, liver fat, and bone mineral density can help stratify patient risk in terms of future adverse cardiometabolic events, including death. In some cases, these manual CT-based measures outperformed established clinical predictive tools. We have also recently demonstrated that these CT-based biometric measures can all be fully automated using artificial intelligence (AI) techniques to allow for objective, large-scale investigation of larger patient cohorts.
Added value of this study
This is the first study to our knowledge to apply a battery of validated, fully-automated CT biomarkers to a large adult asymptomatic screening cohort with long-term clinical follow-up to assess their ability to predict future adverse clinical events, such as myocardial infarction, stroke, and death. Predictive ability of these CT biomarkers was compared with the well-established Framingham Risk Score (FRS). We found that the automated CT-based prediction was overall superior to the FRS. Some univariate CT measures outperformed the multivariate FRS, with further improvement in CT-based prediction when combining biomarkers. These CT biomarkers are typically ignored in current clinical practice, but this tissue-based information resides in all CT scans, regardless of clinical indication for imaging,
Implications of all the available evidence
Our study demonstrates the rich prognostic value that can be automatically derived from abdominal CT scans, incidental to the indication for imaging. Given the many millions of CT scans performed each year in many countries, harnessing this valuable data could identify many pre-symptomatic patients who are at high risk for future serious adverse events, potentially allowing for earlier intervention and prevention.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health Clinical Center. This study utilized the high performance computing capabilities of the NIH Biowulf cluster.
Declaration of Interests
R.M.S.: receives royalties from iCAD, Philips, PingAn, and ScanMed; receives research support from PingAn and NVIDIA.
P.J.P.: advisor/consultant for Zebra Medical Vision and Bracco Diagnostics; shareholder in Cellectar, Elucent, and SHINE.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data sharing
The numerical output of the automated CT-based tools may be shared upon request, subject to an internal review by P.J.P. and R.M.S. to ensure that participant privacy is protected, and subject to completion of a data sharing agreement, as well as approval from both the University of Wisconsin School of Medicine & Public Health and The NIH Clinical Center. Pending the aforementioned approvals, data sharing will be made in a secure setting, on a per-case-specific manner. Please submit such requests to P.J.P. (ppickhardt2@uwhealth.org).
References
- 1.Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. The New England journal of medicine 2019;380:1347–58. [DOI] [PubMed] [Google Scholar]
- 2.Wang SJ, Summers RM. Machine learning and radiology. Med Image Anal 2012;16:933–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choy G, Khalilzadeh O, Michalski M, et al. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018;288:318–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreyer KJ, Geis JR. When Machines Think: Radiology’s Next Frontier. Radiology 2017;285:713–8. [DOI] [PubMed] [Google Scholar]
- 5.Jang S, Graffy PM, Ziemlewicz TJ, Lee SJ, Summers RM, Pickhardt PJ. Opportunistic Osteoporosis Screening at Routine Abdominal and Thoracic CT: Normative L1 Trabecular Attenuation Values in More than 20 000 Adults. Radiology 2019;291:360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickhardt PJ, Pooler BD, Lauder T, del Rio AM, Bruce RJ, Binkley N. Opportunistic Screening for Osteoporosis Using Abdominal Computed Tomography Scans Obtained for Other Indications. Annals of Internal Medicine 2013;158:588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee SJ, Graffy PM, Zea RD, Ziemlewicz TJ, Pickhardt PJ. Future Osteoporotic Fracture Risk Related to Lumbar Vertebral Trabecular Attenuation Measured at Routine Body CT. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research 2018;33:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah RV, Yeri AS, Murthy VL, et al. Association of Multiorgan Computed Tomographic Phenomap With Adverse Cardiovascular Health Outcomes: The Framingham Heart Study. JAMA cardiology 2017;2:1236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connor SD, Graffy PM, Zea R, Pickhardt PJ. Does Nonenhanced CT-based Quantification of Abdominal Aortic Calcification Outperform the Framingham Risk Score in Predicting Cardiovascular Events in Asymptomatic Adults? Radiology 2019;290:108–15. [DOI] [PubMed] [Google Scholar]
- 10.Abraham TM, Pedley A, Massaro JM, Hoffmann U, Fox CS. Association between visceral and subcutaneous adipose depots and incident cardiovascular disease risk factors. Circulation 2015;132:1639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickhardt PJ, Graffy PM, Reeder SB, Hernando D, Li K. Quantification of Liver Fat Content With Unenhanced MDCT: Phantom and Clinical Correlation With MRI Proton Density Fat Fraction. AJR American journal of roentgenology 2018;211:W151–w7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JC, Caan BJ, Prado CM, et al. Body Composition and Cardiovascular Events in Patients With Colorectal Cancer: A Population-Based Retrospective Cohort Study. JAMA oncology 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derstine BA, Holcombe SA, Ross BE, Wang NC, Su GL, Wang SC. Skeletal muscle cutoff values for sarcopenia diagnosis using T10 to L5 measurements in a healthy US population. Sci Rep 2018;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vargas AJ, Schully SD, Villani J, Ganoza Caballero L, Murray DM. Assessment of Prevention Research Measuring Leading Risk Factors and Causes of Mortality and Disability Supported by the US National Institutes of Health. JAMA network open 2019;2:e1914718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SJ, Liu J, Yao J, Kanarek A, Summers RM, Pickhardt PJ. Fully automated segmentation and quantification of visceral and subcutaneous fat at abdominal CT: application to a longitudinal adult screening cohort. The British journal of radiology 2018;91:20170968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graffy PM, Liu J, O’Connor S, Summers RM, Pickhardt PJ. Automated segmentation and quantification of aortic calcification at abdominal CT: application of a deep learning-based algorithm to a longitudinal screening cohort. Abdom Radiol 2019. [DOI] [PubMed] [Google Scholar]
- 17.Pickhardt PJ, Lee SJ, Liu JM, et al. Population-based opportunistic osteoporosis screening: Validation of a fully automated CT tool for assessing longitudinal BMD changes. The British journal of radiology 2019;92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graffy PM, Liu J, Pickhardt PJ, Burns JE, Yao J, Summers RM. Deep learning-based muscle segmentation and quantification at abdominal CT: application to a longitudinal adult screening cohort for sarcopenia assessment. The British journal of radiology 2019:20190327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graffy PM, Sandfort V, Summers RM, Pickhardt PJ. Automated Liver Fat Quantification at Nonenhanced Abdominal CT for Population-based Steatosis Assessment. Radiology 2019:190512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DH, Pickhardt PJ, Taylor AJ, et al. CT colonography versus colonoscopy for the detection of advanced neoplasia. New England Journal of Medicine 2007;357:1403–12. [DOI] [PubMed] [Google Scholar]
- 21.Pickhardt PJ, Choi JR, Hwang I, et al. Computed tomographic virtual colonoscopy to screen for colorectal neoplasia in asymptomatic adults. New England Journal of Medicine 2003;349:2191–200. [DOI] [PubMed] [Google Scholar]
- 22.Luo W, Phung D, Tran T, et al. Guidelines for Developing and Reporting Machine Learning Predictive Models in Biomedical Research: A Multidisciplinary View. J Med Internet Res 2016;18:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns JE, Yao J, Chalhoub D, Chen JJ, Summers RM. A Machine Learning Algorithm to Estimate Sarcopenia on Abdominal CT. Acad Radiol 2019. [DOI] [PubMed] [Google Scholar]
- 24.Chellamuthu K, Liu J, Yao J, et al. Atherosclerotic Vascular Calcification Detection and Segmentation on Low Dose Computed Tomography Scans Using Convolutional Neural Networks. IEEE ISBI; Melbourne, Australia: 2017:388–91. [Google Scholar]
- 25.Liu J, Lu L, Yao J, Bagheri M, Summers RM. Pelvic artery calcification detection on CT scans using convolutional neural networks. In: Armato SG, Petrick NA, eds. SPIE Medical Imaging 2017:101341A. [Google Scholar]
- 26.Summers RM, Baecher N, Yao JH, et al. Feasibility of Simultaneous Computed Tomographic Colonography and Fully Automated Bone Mineral Densitometry in a Single Examination. J Comput Assist Tomogr 2011;35:212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao JH, O’Connor SD, Summers RM, Ieee. Automated spinal column extraction and partitioning. 2006. 3rd Ieee International Symposium on Biomedical Imaging: Macro to Nano, Vols 1–32006:390–3. [Google Scholar]
- 28.Liu JM, Yao JH, Bagheri M, Sandfort V, Summers RM, Ieee. A Semi-Supervised CNN Learning Method with Pseudo-class Labels for Atherosclerotic Vascular Calcification Detection 2019. Ieee 16th International Symposium on Biomedical Imaging. New York: Ieee; 2019:780–3. [Google Scholar]
- 29.Sandfort V, Yan K, Pickhardt PJ, Summers RM. Data augmentation using generative adversarial networks (CycleGAN) to improve generalizability in CT segmentation tasks. Sci Rep 2019;9:16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D’Agostino RB, Vasan RS, Pencina MJ, et al. General cardiovascular risk profile for use in primary care - The Framingham Heart Study. Circulation 2008;117:743–53. [DOI] [PubMed] [Google Scholar]
- 31.IMV 2018 CT Market Outook Report. Des Plains, IL: IMV Medical Information Division; 2018. [Google Scholar]
- 32.Berland LL, Silverman SG, Gore RM, et al. Managing incidental findings on abdominal CT: white paper of the ACR incidental findings committee. J Am Coll Radiol 2010;7:754–73. [DOI] [PubMed] [Google Scholar]
- 33.Brenner DJ, Hall EJ. Current concepts - Computed tomography - An increasing source of radiation exposure. New England Journal of Medicine 2007;357:2277–84. [DOI] [PubMed] [Google Scholar]
- 34.Zheng W, McLerran DF, Rolland B, et al. Association between body-mass index and risk of death in more than 1 million Asians. The New England journal of medicine 2011;364:719–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang TJ, Gona P, Larson MG, et al. Multiple biomarkers for the prediction of first major cardiovascular events and death. The New England journal of medicine 2006;355:2631–9. [DOI] [PubMed] [Google Scholar]
- 36.Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. New England Journal of Medicine 2016;375:1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohli M, Prevedello LM, Filice RW, Geis JR. Implementing Machine Learning in Radiology Practice and Research. Am J Roentgenol 2017;208:754–60. [DOI] [PubMed] [Google Scholar]
- 38.Langlotz CP, Allen B, Erickson BJ, et al. A Roadmap for Foundational Research on Artificial Intelligence in Medical Imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology 2019;291:781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Summers RM, Yao JH, Pickhardt PJ, et al. Computed tomographic virtual colonoscopy computer-aided polyp detection in a screening population. Gastroenterology 2005;129:1832–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Q, Liu Y, Chen TJ, Tong YX. Federated Machine Learning: Concept and Applications. ACM Trans Intell Syst Technol 2019;10:19. [Google Scholar]
- 41.Arnett DK, Blumenthal RS, Albert MA, et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease. J Am Coll Cardiol 2019;74:E177–U76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


