Abstract
The discovery of prolyl hydroxylase domain proteins (PHDs) as key enzymes in the hypoxia inducible factor (HIF) pathway has been followed by reports of a multitude of non-HIF substrates of PHD. Reporting in eLife, Cockman et al. (2019) find a surprising lack of detectable PHD activity toward any of them.
Hypoxia impacts many aspects of cellular metabolism, the best known being the shift from oxidative phosphorylation toward glycolysis. In metazoans, the main transcriptional response to hypoxia is orchestrated by hypoxia inducible factor (HIF), which consists of an α subunit (HIF-1α, HIF-2α) and a β subunit (HIF-β, also known as ARNT) (Kaelin and Ratcliffe, 2008; Semenza, 2012). HIF-α protein levels are exquisitely regulated by oxygen levels, the key event being oxygen-dependent prolyl hydroxylation by a family of three PHDs (also known as EGLNs) that targets HIF-α for degradation. Given the central role that PHDs play in oxygen sensing in the HIF pathway, it was naturally of great interest to determine whether the PHDs might have non-HIF substrates, thereby potentially enlarging the scope of the hypoxic response under PHD control. There were hints that this might turn out to be the case. For example, the collagen prolyl hydroxylases, homologous to the PHDs in their catalytic domain, were originally identified based on their capacity to prolyl hydroxylate collagen, but were subsequently found to have a number of non-collagen substrates (Gorres and Raines, 2010). In the HIF pathway, the asparaginyl hydroxylase, factor inhibiting HIF (FIH), hydroxylates a specific asparaginyl residue in the transactivation domain of HIF-α and thereby controls HIF-α transcriptional activity. FIH was subsequently found to have numerous non-HIF substrates (Cockman et al., 2009). Perhaps more broadly, identification of protein kinases in signal transduction pathways has often been followed by the identification of non-canonical substrates. Accordingly, it was of substantial interest when reports emerged describing non-HIF substrates of the PHDs. They encompass a broad range of proteins involved in signal transduction, metabolism, transcription, translation, cytoskeletal integrity, and cell division, among others.
Writing in eLife, Cockman et al. systematically examined the capacity of these proteins to serve as PHD substrates in vitro (Cockman et al., 2019). They employed recombinant PHDs and prepared substrates in two different ways. First, they obtained synthetic peptides encompassing the reported sites of prolyl hydroxylation; in the case of HIF-α, such peptides serve as good substrates for the PHDs. Second, they prepared full-length versions of the proteins by in vitro transcription and translation (IVTT) using HeLa cell extracts. Prolyl hydroxylation was assessed primarily by mass spectrometry. In addition, in some cases, IVTT-generated proteins were labeled with [3H] proline, and the PHD-catalyzed production of 4-hydroxy [3H] proline was assessed by radiochemical procedures. Of note, under conditions in which the PHDs efficiently prolyl hydroxylate HIF-α, they could not detect hydroxylation of any of the >20 proteins that have been reported as substrates. All experiments were well controlled and meticulously conducted. The authors also describe the challenges of assigning prolyl hydroxylation events in the presence of potentially confounding neighboring oxidation events on residues such as methionine, as has been observed by others (Arsenault et al., 2015).
In addition to these results appearing to be at odds with numerous reports in the literature, the findings are surprising for a number of reasons. First, while the HIF-αs share a Leu-Xaa-Xaa-Leu-Ala-Pro (LXXLAP) motif at the site of prolyl hydroxylation (site of hydroxylation underlined), proteins bearing mutations at either of the leucines or at the alanine in this motif can still serve as substrates (Huang et al., 2002), suggesting that the LXXLAP motif is not a strict consensus. Second, the PHDs are presumably constitutively active under normoxic conditions and hence readily available to act on suitable substrates. In line with this, FIH hydroxylates numerous non-HIF substrates under steady-state, normoxic conditions in cell culture (Cockman et al., 2009). Given these considerations, an intriguing question that arises is how specificity in this pathway is achieved at all; in other words, how are the PHDs able to selectively target proteins such as HIF-α when an obligatory consensus does not seem to exist? Cockman et al. suggest a number of factors, including conformational changes in the PHDs, enzyme:substrate contacts, and secondary structure requirements in the substrate.
There are caveats to the present study. As discussed by the authors, their studies were performed in vitro. It is conceivable that the reported proteins may be more efficiently hydroxylated by the PHDs within cells than in an in vitro setting, for any of a number of reasons. For example, there could be cell-type-specific scaffold/adaptor proteins that facilitate hydroxylation of particular proteins. Alternatively, there could be non-hydroxylation posttranslational modifications, perhaps not induced by the HeLa cell extract employed in their experiments, that could be necessary for efficient hydroxylation. It is also conceivable that prolyl hydroxylation levels on particular proteins, while below the limit of detection in their assays, might still be physiologically significant. For example, in some protein kinase pathways, phosphorylation of a small fraction of the total pool of protein kinase is sufficient to induce a physiologically important response. Because HIF-α is virtually undetectable under normoxic conditions in many cell lines and hydroxylation targets it for degradation, by necessity this reaction has to be efficient and quantitative. HIF-α thus sets a high bar as a PHD substrate, and it is conceivable that non-HIF substrates may not need to be as efficiently hydroxylated as HIF-α. Finally, while not directly addressed in the present study, it is also possible that the PHDs may have non-catalytic roles.
Diversity in the outputs in response to hypoxia can be achieved by any number of mechanisms (Figure 1). The study of Cockman et al. raises the possibility that diversity in the PHD pathway response may be achieved primarily through the large number of HIF target genes (Schödel et al., 2013) (Figure 1A), rather than a large number of PHD substrates (Figure 1B). Clearly, additional experimental work needs to be done to clarify this issue. It should also be recognized that diversity can be achieved at the level of oxygen sensors (Figure 1C). FIH is a 2-oxoglutarate-dependent dioxygenase that is one such protein. Other 2-oxoglutarate-dependent dioxygenases, such as the KDM6A and KDM5A histone demethylases, are others (Batie et al., 2019; Chakraborty et al., 2019). Thus, the search for non-HIF PHD substrates continues and is paralleled by the search for non-PHD oxygen sensors. Undoubtedly, these efforts will enlarge our understanding of the hypoxic response.
Figure 1. Models for the Hypoxic Response.
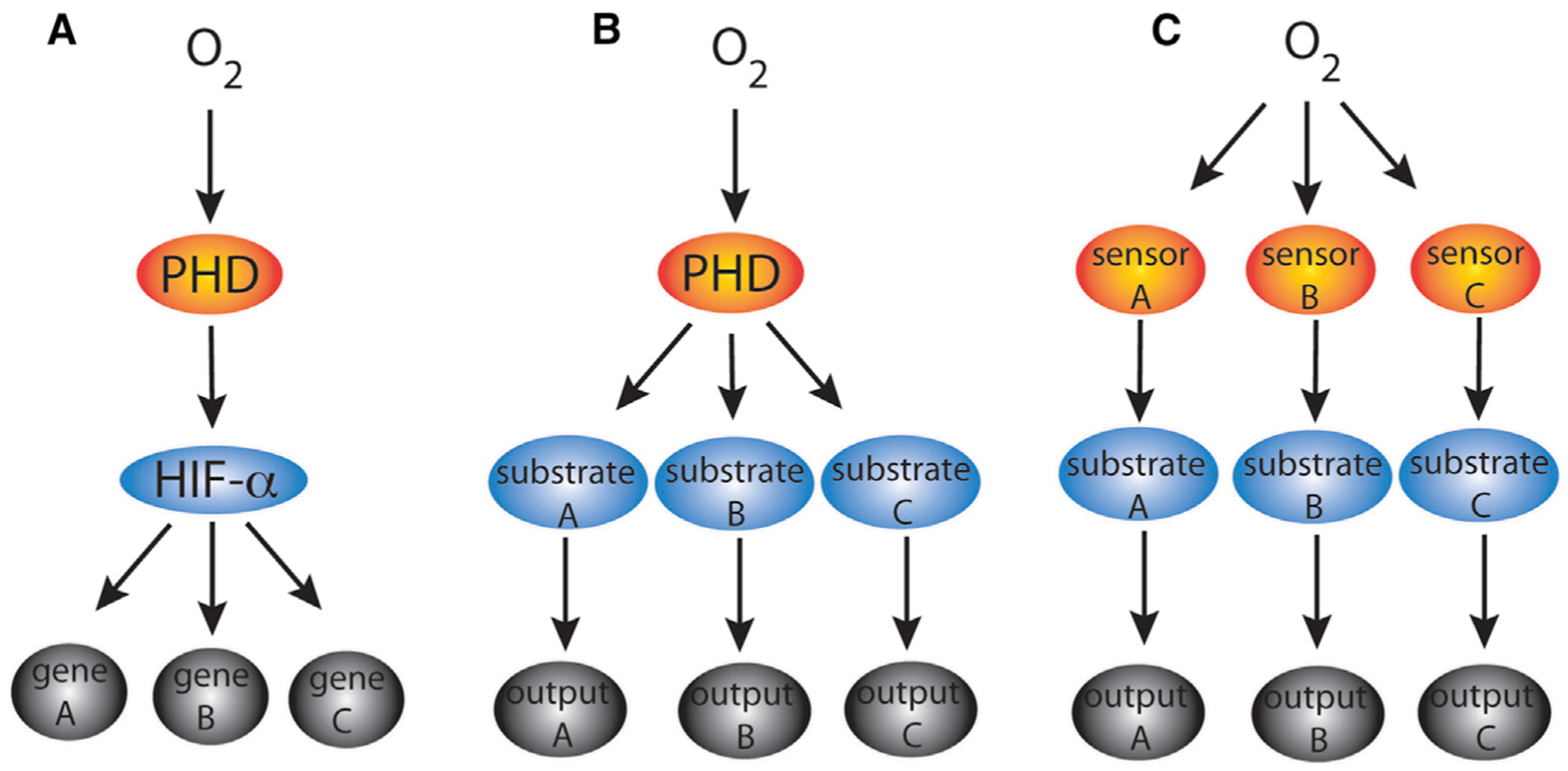
Generation of diversity of responses to hypoxia can be achieved by (A) multiple HIF gene targets, (B) multiple PHD substrates, and/or (C) multiple oxygen sensors.
ACKNOWLEDGMENTS
Work in the author’s laboratory is supported by grants R01-DK104796 and R33-HL120751 from the United States National Institutes of Health.
REFERENCES
- Arsenault PR, Heaton-Johnson KJ, Li LS, Song D, Ferreira VS, Patel N, Master SR, and Lee FS (2015). Identification of prolyl hydroxylation modifications in mammalian cell proteins. Proteomics 15, 1259–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batie M, Frost J, Frost M, Wilson JW, Schofield P, and Rocha S (2019). Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science 363, 1222–1226. [DOI] [PubMed] [Google Scholar]
- Chakraborty AA, Laukka T, Myllykoski M, Ringel AE, Booker MA, Tolstorukov MY, Meng YJ, Meier SR, Jennings RB, Creech AL, et al. (2019). Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockman ME, Lippl K, Tian Y-M, Pegg HB, Figg WD, Abboud MI, Heilig R, Fischer R, Myllyharju J, Schofield CJ, and Ratcliffe PJ (2019). Lack of activity of recombinant HIF prolyl hydroxylases (PHDs) on reported non-HIF substrates. Elife. Published online September 10, 2019. 10.7554/eLife.46490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockman ME, Webb JD, Kramer HB, Kessler BM, and Ratcliffe PJ (2009). Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates wide-spread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Mol. Cell. Proteomics 8, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorres KL, and Raines RT (2010). Prolyl 4-hydroxylase. Crit. Rev. Biochem. Mol. Biol 45, 106–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao Q, Mooney SM, and Lee FS (2002). Sequence determinants in hypoxia-inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J. Biol. Chem 277, 39792–39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr., and Ratcliffe PJ (2008). Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402. [DOI] [PubMed] [Google Scholar]
- Schödel J, Mole DR, and Ratcliffe PJ (2013). Pan-genomic binding of hypoxia-inducible transcription factors. Biol. Chem 394, 507–517. [DOI] [PubMed] [Google Scholar]
- Semenza GL (2012). Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]


