Abstract
The first total synthesis of the pro-resolving lipid mediator 7(S),12(R),13(S)-Resolvin T2 [7(S), 12(R), 13(S)-RvT2] and its 13(R)-epimer, derived from n-3 docosapentaenoic acid (n-3 DPA), are described. 7(S), 12(R), 13(S)-RvT2 and its 13(R)-epimer were obtained by total synthesis using a chiral pool strategy to introduce the chiral centers. C7 was generated from S-(−)-1,2,4-butanetriol in both molecules and the C12 and C13 centers were generated from L-(+)-ribose and D-(−)-arabinose respectively. Cis and trans-selective Wittig reactions, selective deprotections, and Dess-Martin periodinane oxidation were the key steps in the syntheses.
Keywords: 7(S),12(R),13(S)-Resolvin T2; 7(S),12(R),13(R)-Resolvin T2; Chiral pool; Specialized pro-resolving mediators (SPMs)
Graphical Abstract
To create your abstract, type over the instructions in the template box below. Fonts or abstract dimensions should not be changed or altered.

The self-resolving acute inflammation is an active process to protect against infection and tissue damage, and it involves the biosynthesis of the specialized pro-resolving mediators (SPMs) to achieve final homeostasis.1-5 These SPMs include the lipoxins derived from arachidonic acid (AA), the E-series resolvins derived from eicosapentaenoic acid (EPA), and the D-series resolvins, maresins, protectins and their sulfido-conjugates derived from docosahexaenoic acid (DHA).6-19 In addition docosapentaenoic acid (n-3 DPA) has been shown to produce SPMs similar to DHA.20 More recently Dalli and collaborators discovered the 13-series resolvins (RvT1, RvT2, RvT3 and RvT4) that are formed from 13(R)-hydroxy-docosapentaenoic acid.21 The precursor 13-hydroxy-docosapentaenoic acid is produced in endothelial cells with 90% R-selectivity, but 10% of the 13S-epimer was observed by chiral-HPLC. These resolvins are early (< 4h) produced during infection and have shown to be protective against lethal doses of E-coli in animals.21-24 Due to the increasing bacterial resistance towards currently used antibiotics these novel SPMs could provide an alternative way to clear infections utilizing the immune system.
The proposed biosynthesis of RvT1-RvT4 is outlined in Figure 1. Dalli and collaborators showed that n-3 DPA is converted via endothelial COX-2 to the 13(R)-hydroperoxy-docosapentaenoic acid that after reduction gives 13(R)-hydroxy-docosapentaenoic acid.21 The synthesis of this precursor has been recently described by Hansen and collaborators.22 13(R)-hydroxy-docosapentaenoic acid is converted by neutrophils to 7-hydroperoxy-13(R)-hydroxy-docosapentaenoic acid that after reduction of the hydroperoxy-group gives RvT4. Enzymatic lipoxidase reaction transforms RvT4 into RvT1. 18O2 incorporation experiments confirmed that the hydroxyl groups at C-7 and C-20 are derived from enzymatic lipoxidase reaction, known to introduce these centers in polyunsaturated fatty acids with (S)-chirality. The 7-hydroperoxy-13(R)-hydroxy-docosapentaenoic acid can form an allylic epoxide intermediate [7,8-epoxy], that is enzymatically hydrolyzed to RvT2 and RvT3 as shown in Figure 1. It is worthwhile to note that due to the change in priority of the groups surrounding chiral center C13 in RvT2 and RvT3, according to the Cahn, Ingold, Prelog rules of nomenclature, this center has now to be assigned as S.
Figure 1.
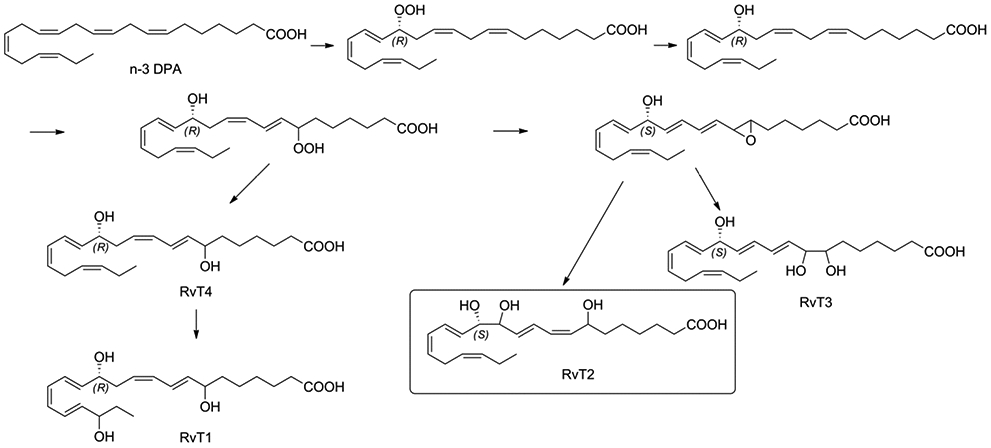
Proposed biosynthesis of RvT1, RvT2, RvT3 and RvT4.
In order to explore the biological and pharmacological properties and due to the limited amounts available from biological sources these products need to be prepared by total syntheses. Based on biosynthetic considerations we recently reported the total syntheses of 7(S),13(R),20(S)-RvTl and 7(S),13(R)-RvT4,25 and herein we describe the total syntheses of 7(S), 12(R), 13(S)-RvT2 [(7S,8Z, 10E, 12R, 13S, 14E, 16Z, 19Z)-7,12,13-trihydroxy-8,10,14,16,19-docosapentaenoic acid (1)] and 7(S), 12(R), 13(R)-RvT2 [(7S,8Z, 10E, 12R, 13R, 14E, 16Z, 19Z)-7,12,13-trihydroxy-8,10,14,16,19-docosapentaenoic acid (2)].
As shown in the retrosynthetic scheme (Figure 2) compound 1 and 2 were synthesized using the same strategy utilizing a final cis-selective Wittig olefination reaction. The key intermediate 3 was obtained from S-(−)-1,2,4-butanetriol (9) and intermediates 4 and 5 were generated from L-(+)-ribose (6) and D-(−)-arabinose (8) respectively.
Figure 2.

Retrosynthetic approach to 7(S),12(R),13(S)-RvT2 and 7(S),12(R),13(R)-RvT2.
The synthesis of the C1–C8 fragment was achieved starting from the crystalline phosphonium iodide 7 (Scheme 1) readily available from S-(−)-1,2,4-butanetriol (9) similar as described by Taber and collaborators.20,27 The ylide generated with n-BuLi was reacted with aldehyde-ester 10 to give the isopropylidene protected intermediate 11 in 66% yield. Hydrogenation of 11 with 10% Pt/C in hexane containing 10% methyl acetate gave 12 in quantitative yield.28 The isopropylidene group was cleaved with 1 N HCl m CH3OH to give 13 in 87% yield.28 The diol 13 was converted to the di-TBS derivative 14 in 93% yield. Selective deprotection of the primary TBS-group with 0.5 equiv of 10-camphorsulfonic acid (CSA) in CH2Cl2/CH3OH at 0 °C gave 15 in 49% isolated yield. Compound 15 was converted to the key intermediate 3 via Dess-Martin periodinane oxidation in CH2Cl2.29
Scheme 1. Reagents and conditions:

(a) n-BuLi, THF, −78 °C to −20 °C, 66%; (b) H2, Pt/C, hexane/methyl acetate 9/1, rt, quantitative; (c) 1 N HCl, CH3OH, 0 °C, 87%; (d) TBSCl, imidazole, 4-DMAP, CH2Cl2, 0 °C to rt, 93%; (e) CSA, CH2Cl2/CH3OH 1/1, 0 °C, 49%; (f) Dess-Martin periodinane, CH2Cl2, rt, 60%.
As described in Scheme 2 the C9–C22 key intermediate 4 was obtained from L-(+)-ribose (6) in nine steps. Wittig reaction of methyl (triphenylphosphoranylidene)acetate (16) with 6 in the presence of catalytic benzoic acid in 1,4-dioxane provided crude tetrol-ester 17 (a small sample was purified by flash chromatography SiO2/ethyl acetate for analytical purpose). Crude 17 was treated with acetone/2,2-dimethoxypropane and catalytic p-toluenesulfonic acid to give the diprotected-ester 18 in 39% yield over two steps.30 Cleavage of the terminal acetonide in 18 was accomplished with amberlyst® 15 in CH3OH/H2O to give 19 in 69% yield. Diol cleavage with NaIO4 in CH3OH/H2O gave the aldehyde 20 that was treated with 1 equiv of (triphenylphosphoranylidene)acetaldehyde (21) in benzene at rt to give the α,β-unsaturated aldehyde 22 in 58% yield over two steps.25 The use of CHCl3 and CH3CN31 as solvent gave substantial amounts of the cis-aldehyde with this type of substrate. Wittig coupling of 22 with 1.5 equivalents of the ylide prepared from the crystalline phosphonium iodide 2332 and n-BuLi at −78 °C in THF gave cleanly the cis-coupled product 24 in 69% yield. The geometry of the 14E,16Z-diene unit in 24 was confirmed by the 1H-1H coupling constants (J14,15 = 15.0 Hz and J16,17 = 11.1 Hz).33 Compound 24 was converted to alcohol 25 via DIBAL reduction in 85% yield followed by CBr4/PPh3 reaction to give bromide 26 in 86% yield.34 The key intermediate 4 was obtained in quantitative yield by slow addition of 5 equiv of PPh3 dissolved in CH2Cl2 to 26 at 0 °C to rt after purification by flash chromatography (SiO2, CH2Cl2/CH3OH 9/1).
Scheme 2. Reagents and conditions:

(a) 16, benzoic acid, 1,4-dioxane, reflux; (b) acetone/2,2-dimethoxypropane (2/1), p-toluenesulfonic acid, rt, 39% (over two steps); (c) amberlyst® 15, CH3OH/H2O 9/1, rt, 69%; (d) NaIO4, CH3OH/H2O 10/4, 0 °C to rt; (e) 21, benzene, rt, 58% (over two steps); (f) 23, n-BuLi, THF, −78 °C to −20 °C, 69%; (g) DIBAL-H, CH2Cl2, −78 °C to −20 °C, 85%; (h) CBr4, Ph3P, CH2Cl2, 0 °C, 86%; (i) Ph3P, CH2Cl2, 0 °C to rt, quantitative.
The skeleton of 7(S),12(R),13(S)-RvT2 (1) was assembled from the key intermediates 3 and 4 employing a cis-selective Wittig reaction using KHMDS as a base in THF at −78 °C to give 27 (Scheme 3). The geometry of the 8Z,10E-diene unit in 27 was confirmed by the 1H-1H coupling constants (J8,9 = 11.1 Hz and J10,11 = 15.0 Hz).33 Deprotection of the isopropylidene and TBS groups was achieved in one step with 2 N HCl in CH3OH at 0 °C to rt to give 2835 that was purified by HPLC [Zorbax SB-C18, 250 × 21.2 mm, 235 nm, CH3OH/H2O 80/20]. Mild alkaline hydrolysis of 28 with 1 N LiOH in CH3OH/H2O at 0 °C to rt gave, after HPLC purification [Zorbax SB-C18 250 × 21.2 mm, 235 nm, CH3OH/H2O (0.1% NH4OAc, pH 5.6, 0.05% EDTA disodium) 58/42] and desalting, 7(S), 12(R), 13(R)-RvT2 (1) in 58% yield. The 1H NMR, 13C NMR, UV, and HPLC/UV/MS analysis were consistent with the structure of 1.33
Scheme 3. Reagents and conditions:

(a) KHMDS, THF, −78 °C to −20 °C, 29%; (b) 2 N HCl, CH3OH, 0 °C to rt, 76%; (c) 1 N LiOH, CH3OH/H2O 2/1, 0 °C to rt, 79% and after HPLC and desalting 58%.
7(S),12(R),13(R)-RvT2 methyl ester (29) was prepared from D-(−)-arabinose (8) in a similar manner as outlined for 7(S),12(R),13(S)-RvT2 methyl ester (28). Mild alkaline hydrolysis of 29 with 1 N LiOH in CH3OH/H2O at 0 °C to rt gave, after HPLC purification [Zorbax SB-C18 250 × 21.2 mm, 235 nm, CH3OH/H2O (0.1% NH4OAc, pH 5.6, 0.05% EDTA disodium) 60/40] and desalting, 7(S),12(R),13(R)-RvT2 (2) in 52% yield (Scheme 4). The 1H NMR, 13C NMR, UV, and HPLC/UV/MS analysis were consistent with the structure of 2.33
Scheme 4. Reagents and conditions:
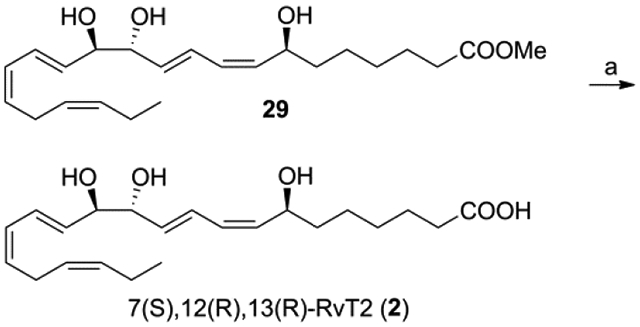
(a) 1 N LiOH, CH3OH/H2O 2/1, 0 °C to rt, 52% (after HPLC and desalting).
HPLC analysis of 1 and 2 was performed [Zorbax SB-C18, 1.8 μm, 50 x 2.1 mm, 235 nm, CH3OH/H2O (0.1% formic acid) 50:50–70:30, 0.2 mL/min] where tR [7(S),12(R),13(S)-RvT2 (1)]= 13.3 min and tR [7(S), 12(R), 13(R)-RvT2 (2)]= 13.9 mm.
In summary, the total syntheses of 7(S),12(R),13(S)-RvT2 (1) and its 13(R)-epimer (2) have been achieved,33 making these pro-resolving lipid mediators from n-3 DPA available for further biological testing. The synthesis of RvT3 will be reported in due course.
Supplementary Material
Highlights.
First total syntheses 7(S),12(R),13(S)-Resolvin T2 and its 13(R)-epimer have been achieved
Chiral center C7 was generated by chiral pool strategy from S-(−)-1,2,4-butanetriol
Chiral centers C12 and C13 were generated from L-(+)-ribose and D-(−)-arabinose
Key steps: Cis and trans-selective Wittig reactions, selective deprotections, Dess-Martin oxidation.
Acknowledgments
Financial support of this research by the NIH (R01AI128202) is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References and notes
- 1.Serhan CN, Levy BD, J. Clin. Invest 128 (2018) 2657–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serhan CN, de la Rosa X, Jouvene C, J. Intern. Med 286 (2018) 240–258. [DOI] [PubMed] [Google Scholar]
- 3.Serhan CN, Nature 510 (2014) 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serhan CN, FASEB J. 31 (2017) 1273–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Serhan CN, Chiang N, J. Dalli. Mol. Aspects Med. 64 (2018) 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serhan CN, Petasis NA, Chem. Rev 111 (2011) 5922–5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serhan CN, Dalli J, Karamnov S, Choi A, Park CK, Xu ZZ, Ji RR, Zhu M, Petasis NA, FASEB J. 26 (2012) 1755–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serhan CN, Gotlinger K, Hong S, Lu Y, Siegelman J, Baer T, Yang R, Colgan SP, Petasis NA, J. Immunol 176 (2006) 1848–1859. [DOI] [PubMed] [Google Scholar]
- 9.Sasaki K, Urabe D, Arai H, Arita M, Inoue M, Chem. Asian J 6(2011) 534–543. [DOI] [PubMed] [Google Scholar]
- 10.Dalli J, Ramon S, Norris PC, Colas RA, Serhan CN, FASEB J. 29 (2015). 2120–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalli J, Chiang N, Serhan CN, Proc. Natl. Acad. Sci. USA 111 (2014) E4753–E4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez AR, Spur BW, Tetrahedron Lett. 56 (2015) 256–259. [Google Scholar]
- 13.Rodriguez AR, Spur BW, Tetrahedron Lett. 56 (2015) 3936–3940. [Google Scholar]
- 14.Rodriguez AR, Spur BW, Tetrahedron Lett. 56 (2015) 5811–5815. [Google Scholar]
- 15.Dalli J, Sanger JM, Rodriguez AR, Chiang N, Spur BW, Serhan CN, PLoS One (2016) 11(2):e0149319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalli J, Vlasakov I, Riley IR, Rodriguez AR, Spur BW, Petasis NA, Chiang N, Serhan CN, Proc. Natl. Acad. Sci. USA 113 (2016) 12232–12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez AR, Spur BW, Tetrahedron Lett. 58 (2017) 1662–1668. [Google Scholar]
- 18.Chiang N, Riley IR, Dalli J, Rodriguez AR, Spur BW, Serhan CN, FASEB J. 32 (2018) 4043–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Rosa X, Norris PC, Chiang N, Rodriguez AR, Spur BW, Serhan CN, Am. J. Pathol 188 (2018) 950–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vik A, Dalli J, Hansen TV, Bioorg. Med. Chem. Lett 27 (2017) 2259–2266. [DOI] [PubMed] [Google Scholar]
- 21.Dalli J, Chiang N, Serhan CN, Nat. Med 21 (2015) 1071–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Primdahl KG, Aursnes M, Walker ME, Colas RA, Serhan CN, Dalli J, Hansen TV, Vik A, J. Nat. Prod 79 (2016) 2693–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker ME, Souza PR, Colas RA, Dalli J, FASEB J. 31 (2017)3636–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen TV, Dalli J, Serhan CN, Prostaglandins Other Lipid Mediat. 133 (2017) 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez AR, Spur BW, Tetrahedron Lett. 61 (2020) 151473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taber DF, M. Xu. J.C. Hartnett. J. Am. Chem. Soc. 124 (2002) 13121–13126. [DOI] [PubMed] [Google Scholar]
- 27.Deredas D, Skowron M, Salomon E, Tarnus C, Tschamber T, Wolf WM, Frankowski A, Tetrahedron 63 (2007) 2915–2922. [Google Scholar]
- 28.Lees WJ, Whitesides GM, J. Org. Chem 58 (1993) 1887–1894. [Google Scholar]
- 29.Dess DB, Martin JC, J. Am. Chem. Soc 113 (1991) 7277–7287. [Google Scholar]
- 30.Koh D, Agric. Chem. Biotechnol 47 (2004) 81–85. [Google Scholar]
- 31.Allard M, Barnes K, Chen X, Cheung Y-Y, Duffy B, Heap C, Inthavongsay J, Johnson M, Krishnamoorthy R, Manley C, Steffke S, Varughese D, Wang R, Wang Y, Schwartz CE, Tetrahedron Lett. 52 (2011) 2623–2626. [Google Scholar]
- 32.Oger C, Bultel-Ponce V, Guy A, Durand T, Galano J-M, Eur. J. Org. Chem (2012) 2621–2634. [Google Scholar]
- 33.Satisfactory spectroscopic data were obtained for all compounds. Selected physical data: Compound 7: 1H NMR (CDCl3, 300 MHz): δ 7.8–7.6 (m, 15H). 4.5–4.4 (m. 1H). 4.1–4.0 (dd. J = 8.4. 6.0 Hz, 1H). 4.1–3.9 (m, 1H). 3.6–3.4 (dd, J = 8.4. 6.0 Hz, 1H), 3.5–3.2 (m, 1H). 2.1—1.9 (m, 1H). 1.8–1.6 (m, 1H). 1.2 (s, 3H), 1.2 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 135.08 (d, J = 2.9 Hz, 3C). 133.39 (d, J = 9.8 Hz. 6C), 130.36 (d, J = 12.7 Hz, 6C), 117.55 (d, J = 86.4 Hz, 3C), 108.93. 74.38 (d, J = 15.5 Hz). 68.61, 27.07 (d, J = 3.4 Hz), 26.69, 24.96, 19.41 (d, J = 53.0 Hz); [α]D20 = +2 (c 0.37, CHCl3). Compound 11: 1H NMR (CDCl3, 300 MHz): δ 5.6–5.4 (m, 2H). 4.1 (m, 1H), 4.0 (dd, J = 7.8. 6.0 Hz, 1H), 3.6 (s, 3H), 3.6–3.5 (dd, J = 7.8, 6.9 Hz, 1H), 2.5–2.2 (m, 6H), 1.4 (s, 3H), 1.3 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 173.30, 130.34, 125.78, 108.99, 75.57, 69.05, 51.40, 33.93, 31.55, 26.90, 25.64, 23.03; [α]D20 = +20 (c 0.30, CHCl3), Compound 12: 1H NMR (CDCl3, 300 MHz): δ 4.1–4.0 (m, 2H), 3.6 (s, 3H), 3.5–3.4 (m, 1H), 2.3 (t, J = 7.5 Hz, 2H), 1.7–1.2 (m, 8H), 1.4 (s, 3H), 1.3 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 174.15, 108.65, 75.98, 69.45, 51.47, 33.94, 33.38, 29.11, 26.94, 25.73, 25.43, 24.79; [α]D20 = +16 (c 0.32, CHCl3), Compound 13: 1H NMR (CDCl3, 300 MHz): δ 3.8–3.6 (m, 1H), 3.6 (s, 3H), 3.6 (dd, J = 10.8, 3.0 Hz, 1H), 3.4 (dd, J = 10.8, 7.5 Hz, 1H), 2.3 (t, J = 7.5 Hz, 2H), 2.1–2.0 (br s, 2H), 1.7–1.6 (m, 2H), 1.5–1.3 (m, 6H); 13C NMR (CDCl3, 75.5 MHz): δ 174.24, 72.11, 66.79, 51.50, 33.92, 32.87, 29.02, 25.14, 24.73; [α]D20 = −0.8 (c 1.8, CHCl3), Compound 14: 1H NMR (CDCl3, 300 MHz): δ 3.7–3.6 (m, 1H), 3.6 (s, 3H), 3.5 (dd, J = 9.7, 5.5 Hz, 1H), 3.4–3.3 (dd, J = 9.7, 6.4 Hz, 1H), 2.3–2.2 (t, J = 7.5 Hz, 2H), 1.7–1.5 (m, 2H), 1.5–1.2 (m, 6H), 0.87 (s, 9H), 0.86 (s, 9H), 0.03 (s, 6H), 0.02 (s, 3H), 0.02 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 174.28, 73.02, 67.40, 51.43, 34.13, 34.06, 29.37, 25.98 (3C), 25.89 (3C), 24.95, 24.74, 18.38, 18.15, −4.25, −4.73, −5.31, −5.37; [α]D20 = −14 (c 0.13, CHCl3), Compound 15: 1H NMR (CDCl3, 300 MHz): δ 3.8–3.6 (m, 1H), 3.6 (s, 3H), 3.6–3.5 (dd, J = 11.1, 3.6 Hz, 1H), 3.4 (dd, J = 11.1, 5.1 Hz, 1H), 2.3–2.2 (t, J= 7.5 Hz, 2H), 1.7–1.5 (m, 2H), 1.6–1.4 (m, 2H), 1.4–1.2 (m, 4H), 0.88 (s, 9H), 0.06 (s, 6H); 13C NMR (CDCl3, 75.5 MHz): δ 174.16, 72.76, 66.24, 51.46, 33.97, 33.75, 29.28, 25.82 (3C), 25.01, 24.84, 18.07, −4.47, −4.57; [α]D20 = +7.5 (c 0.3, CHCl3), Compound 3: 1H NMR (CDCl3, 300 MHz): δ 9.6 (d, J = 1.8 Hz, 1H), 4.0–3.8 (td, J = 6.0, 1.8 Hz, 1H), 3.6 (s, 3H), 2.3–2.2 (t, J = 7.5 Hz, 2H), 1.7–1.5 (m, 4H), 1.5–1.2 (m, 4H), 0.89 (s, 9H), 0.05 (s, 3H), 0.04 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 204.29, 174.05, 77.52, 51.45, 33.87, 32.36, 28.92, 25.70 (3C), 24.69, 24.27, 18.15, −4.66, −4.98, Compound 17: 1H NMR (CD3OD, 300 MHz): δ 7.2–7.1 (dd, J = 15.9, 4.9 Hz, 1H), 6.1 (dd, J = 15.9, 1.8 Hz, 1H), 4.5–4.4 (td, J = 4.9, 1.8 Hz, 1H), 3.8–3.7 (m, 1H), 3.7 (s, 3H), 3.7–3.5 (m, 3H); 13C NMR (CD3OD, 75.5 MHz): δ 168.63, 149.45, 122.12, 75.72, 73.98, 73.02, 64.66, 52.01; [α]D20 = +23 (c 0.21, H2O). Compound 18: 1H NMR (CDCl3, 300 MHz): δ 7.1–7.0 (dd, J = 15.6, 4.5 Hz, 1H), 6.2–6.1 (dd, J = 15.6, 1.8 Hz, 1H), 4.9–4.7 (ddd, J = 6.6, 4.5, 1.8 Hz, 1H), 4.2–4.1 (dd, J = 8.4, 6.6 Hz, 1H), 4.1–4.0 (m, 1H), 4.0–3.8 (m, 2H), 3.7 (s, 3H), 1.45 (s, 3H), 1.37 (s, 3H), 1.34 (s, 3H), 1.27 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 166.53, 143.78, 121.88, 109.67, 109.61, 78.79, 76.53, 73.92, 67.48, 51.59, 27.46, 26.81, 25.24, 25.19; [α]D20 = +43 (c 0.37, CHCl3), Compound 19: 1H NMR (CDCl3, 300 MHz): δ 7.1 (dd, J = 15.6, 4.8 Hz, 1H), 6.2–6.1 (dd, J = 15.6, 1.8 Hz, 1H), 4.9–4.8 (ddd, J = 6.6, 4.8, 1.8 Hz, 1H), 4.2–4.1 (dd, J = 8.8, 6.6 Hz, 1H), 3.8–3.7 (m, 1H), 3.7 (s, 3H), 3.7–3.5 (m, 2H), 3.0–2.8 (br s, 2H), 1.5 (s, 3H), 1.3 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 167.09, 144.15, 121.73, 109.59, 77.85, 76.53, 69.91, 64.48, 51.79, 27.51, 25.15, Compound 22: 1H NMR (CDCl3, 300 MHz): δ 9.5 (d, J = 7.8 Hz, 1H), 6.7 (dd, J = 15.4, 5.5 Hz, 1H), 6.6–6.5 (dd, J = 15.7, 5.2 Hz, 1H), 6.3 (ddd, J = 15.7, 7.8, 1.2 Hz, 1H), 6.1–6.0 (dd, J = 15.4, 1.2 Hz, 1H), 5.0–4.8 (m, 2H), 3.7 (s, 3H), 1.5 (s, 3H), 1.4 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 192.65, 165.87, 150.29, 141.77, 133.46, 123.29, 110.56, 77.19 (2C), 51.77, 27.49, 25.19, Compound 24: 1H NMR (CDCl3, 300 MHz): δ 6.8–6.7 (dd, J = 15.6, 4.9 Hz, 1H), 6.6 (dd, J = 15.0, 11.1 Hz, 1H), 6.1–6.0 (dd, J = 15.6, 1.2 Hz, 1H), 6.0–5.9 (t, J= 11.1 Hz, 1H), 5.5–5.2 (m, 4H), 4.8–47 (m, 2H), 3.7 (s, 3H), 3.0–2.8 (brt, J = 7.3 Hz, 2H), 2.1–2.0 (m, 2H), 1.5 (s, 3H), 1.4 (s, 3H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 166.33, 143.94, 132.57, 132.24, 129.82, 127.33, 127.23, 126.20, 122.35, 109.50, 79.50, 77.69, 51.65, 27.80, 25.98, 25.40, 20.56, 14.20, Compound 25: 1H NMR (CDCl3, 300 MHz): δ 6.6–6.5 (br dd, J = 15.3, 11.2 Hz, 1H), 6.0–5.9 (t, J = 11.2 Hz, 1H), 6.0–5.8 (dt, J = 15.4, 5.4 Hz, 1H), 5.7–5.6 (ddt, J = 15.4, 7.5, 1.5 Hz, 1H), 5.6–5.5 (dd, J = 15.3, 7.8 Hz, 1H), 5.5–5.2 (m, 3H), 4.7–4.5 (m, 2H), 4.2–4.1 (dd, J = 5.4, 1.5 Hz, 2H), 3.0–2.8 (br t, J = 7.3 Hz, 2H), 2.1–2.0 (m, 2H), 1.5 (s, 3H), 1.4 (s, 3H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 133.66, 132.48, 131.62, 128.93, 128.45, 127.44, 127.05, 126.30, 108.78, 79.49, 79.07, 62.78, 28.02, 25.97, 25.50, 20.53, 14.20, Compound 26: 1H NMR (CDCl3, 300 MHz): δ 6.6–6.5 (br dd, J = 15.0, 11.1 Hz, 1H), 6.0 (brt, J = 11.1 Hz, 1H), 6.0–5.9 (m, 1H), 5.7–5.6 (dd, J = 15.3, 6.9 Hz, 1H), 5.6–5.4 (dd, J = 15.3, 7.8 Hz, 1H), 5.5–5.2 (m, 3H), 47–4.5 (m, 2H), 4.1 (d, J = 7.8 Hz, 2H), 3.0–2.8 (br t, J = 7.3 Hz, 2H), 2.1–2.0 (m, 2H), 1.5 (s, 3H), 1.4 (s, 3H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 132.49, 131.74, 131.27, 129.65, 129.01, 128.18, 127.39, 126.31, 109.04, 79.53, 78.30, 31.41, 27.94 25.99, 25.45, 20.55, 14.22, Compound 4: 1H NMR (CDCl3, 300 MHz): δ 7.9–7.7 (m, 9H), 7.7–7.6 (m, 6H), 6.3 (dd, J = 15.0, 11.1 Hz, 1H), 6.0–5.8 (dt, J = 15.0, 5.7 Hz, 1H), 5.7 (t, J = 11.1 Hz, 1H), 5.6–5.5 (m, 1H), 5.4–5.2 (m, 3H), 5.2–5.1 (dd, J = 15.0, 7.5 Hz, 1H), 4.9–4.6 (m, 2H), 4.6.–4.5 (m, 1H), 4.5–4.4 (m, 1H), 3.1–2.8 (br t, J = 7.5 Hz, 2H), 2.1–1.9 (m, 2H), 1.3 (s, 6H), 0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 137.46 (d, J = 13.1 Hz), 134.99 (d, J = 2.9 Hz, 3C), 133.95 (d, J = 9.8 Hz, 6C), 132.49, 131.85, 130.33 (d, J = 12.7 Hz, 6C), 129.33, 127.50, 127.35, 126.18, 117.92 (d, J = 85.8 Hz, 3C), 117.74 (d, J = 9.2 Hz), 108.75, 79.09 (d, J = 2.9 Hz), 78.30 (d, J = 2.3 Hz), 27.75, 27.35 (d, J = 49.5 Hz), 25.92, 25.36, 20.51, 14.17, Compound 27: 1H NMR (CDCl3, 300 MHz): δ 6.6–6.5 (dd, J = 15.0, 11.1 Hz, 1H), 6.5–6.4 (dd, J = 15.0, 11.1 Hz, 1H), 6.0–5.9 (t, J = 11.1 Hz, 1H), 5.9 (t, J = 11.1 Hz, 1H), 5.6–5.4 (m, 2H), 5.4–5.1 (m, 4H), 47–4.6 (m, 2H), 4.50.4 (m, 1H), 3.6 (s, 3H), 2.9 (br t, J = 7.4 Hz, 2H), 2.3–2.2 (t, J = 7.5 Hz, 2H), 2.1–2.0 (m, 2H), 1.6–1.2 (m, 8H), 1.5 (s, 3H), 1.4 (s, 3H), 1.0–0.9 (t, J = 7.5 Hz, 3H), 0.83 (s, 9H), −0.01 (s, 3H), −0.04 (s, 3H); 13C NMR (CDCl3, 75.5 MHz): δ 174.20, 136.55, 132.46, 131.46, 130.18, 128.83, 128.52, 128.50, 127.53, 126.42, 126.35, 108.88, 79.75, 79.59, 68.79, 51.43, 38.16, 34.01, 29.06, 28.04, 25.97, 25.82 (3C), 25.56, 24.96, 24.90, 20.54, 18.16, 14.22, −4.27, −4.79, 7(S),12(R),13(S)-Resolvin T2 methyl ester (28): 1H NMR (CD3OD, 300 MHz): δ 6.7–6.5 (2 dd, J = 14.7, 11.1 Hz, 2H), 6.1–6.0 (t, J = 11.1 Hz, 1H), 6.0 (t, J = 11.1 Hz, 1H), 5.9–5.7 (2 dd, J = 15.3, 5.4 and 15.3, 6.0 Hz, 2H), 5.5–5.2 (m, 4H), 4.6–4.5 (m, 1H), 4.1–40 (m, 2H), 3.6 (s, 3H), 3.0–2.9 (br t, J = 7.2 Hz, 2H), 2.3 (t, J = 7.5 Hz, 2H), 2.2–2.0 (m, 2H), 1.7–1.2 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CD3OD, 75.5 MHz): δ 175.99, 135.49, 135.28, 133.83, 133.15, 131.20, 129.98, 129.24, 128.14, 127.85, 127.80, 76.61, 76.46, 68.34, 51.98, 38.45, 34.74, 30.13, 26.85, 26.07, 25.98, 21.47, 14.63, UV (CH3OH) λmax 240 nm, HPLC/UV: Zorbax SB-08, 1.8 μm, 50 x 2.1 mm, 235 nm, CH3OH/H2O (0.1% formic acid) 50:50–85:15, 0.2 mL/min, tR = 9.4 min; HPLC/MS (m/z): 415.2 [M+Na]+. 7(S),12(R),13(R)-Resolvin T2 (1): 1H NMR (CD3OD, 300 MHz): δ 6.7–6.5 (2 dd, J = 14.7, 11.1 Hz, 2H), 6.1–6.0 (t, J = 11.1 Hz, 1H), 6.0 (t, J = 11.1 Hz, 1H), 5.9–5.7 (2 dd, J = 15.3, 5.4 and 15.3, 6.3 Hz, 2H), 5.5–5.2 (m, 4H), 4.6–45 (m, 1H), 4.1–4.0 (m, 2H), 3.0–2.9 (br t, J = 7.2 Hz, 2H), 2.3–2.2 (t, J = 7.5 Hz, 2H), 2.2–2.0 (m, 2H), 1.7–1.3 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CD3OD, 75.5 MHz): δ 178.63, 135.46, 135.31, 133.83, 133.14, 131.20, 129.98, 129.24, 128.12, 127.94, 127.81, 76.60, 76.50, 68.35, 38.51, 35.68, 30.28, 26.85, 26.36, 26.16, 21.47, 14.63. UV (CH3OH) λmax 240 nm, HPLC/UV: Zorbax SB-C18, 1.8 μm, 50 x 2.1 mm, 235 nm, CH3OH/H2O (0.1% formic acid) 50:50–70:30, 0.2 mL/min, tR = 13.3 min; HPLC/MS (m/z): 377.2 [M-H]−. 7(S),12(R),13(R)-Resolvin T2 methyl ester (29): 1H NMR (CD3OD, 300 MHz): δ 6.7–6.5 (2 dd, J = 15.0, 11.1 Hz, 2H), 6.1–6.0 (t, J = 11.1 Hz, 1H), 6.0–5.9 (t, J = 11.1 Hz, 1H), 5.8–5.6 (2 dd, J = 15.0, 6.0 Hz, 2H), 5.5–5.2 (m, 4H), 4.6–4.5 (m, 1H), 4.1–4.0 (m, 2H), 3.6 (s, 3H), 3.0–2.9 (brt, J = 7.3 Hz, 2H), 2.3 (t, J = 7.5 Hz, 2H), 2.2–2.0 (m, 2H), 1.7–1.3 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CD3OD, 75.5 MHz): δ 175.98, 135.39, 135.27, 133.63, 133.16, 131.32, 129.91, 129.17, 128.27, 127.98, 127.78, 76.69, 76.46, 68.33, 51.98, 38.44, 34.74, 30.13, 26.85, 26.07, 25.98, 21.47, 14.64. UV (CH3OH) λmax 240 nm, HPLC/UV: Zorbax SB-C18, 1.8 μm, 50 x 2.1 mm, 235 nm, CH3OH/H2O (0.1% formic acid) 50:50–85:15, 0.2 mL/min, tR = 9.4 min; HPLC/MS (m/z): 415.2 [M+Na]+, 7(S),12(R),13(R)-Resolvin T2 (2): 1H NMR (CD3OD, 300 MHz): δ 6.7–6.5 (2 dd, J = 15.3, 11.1 Hz, 2H), 6.1–6.0 (t, J = 11.1 Hz, 1H), 6.0–5.9 (t, J = 11.1 Hz, 1H), 5.8–5.6 (m, 2H), 5.5–5.2 (m, 4H), 4.6–45 (m, 1H), 41–4.0 (m, 2H), 3.0–2.9 (brt, J = 7.2 Hz, 2H), 2.3–2.2 (t, J = 7.5 Hz, 2H), 2.2–2.0 (m, 2H), 1.7–1.3 (m, 8H), 1.0–0.9 (t, J = 7.5 Hz, 3H); 13C NMR (CD3OD, 75.5 MHz): δ 178.56, 135.42, 135.24, 133.62, 133.15, 131.32, 129.91, 129.17, 128.26, 128.06, 127.78, 76.67, 76.50, 68.35, 38.51, 35.64, 30.27, 26.85, 26.35, 26.16, 21.47, 14.63. UV (CH3OH) λmax 240 nm, HPLC/UV: Zorbax SB-08, 1.8 μm, 50 x 2.1 mm, 235 nm, CH3OH/H2O (0.1% formic acid) 50:50–70:30, 0.2 mL/min, tR = 13.9 min; HPLC/MS (m/z):377.2 [M-H]−.
- 34.Guindon Y, Zamboni R, Lau C-K, Rokach J, Tetrahedron Lett. 23 (1982) 739–742. [Google Scholar]
- 35.Rodriguez AR, Spur BW, Tetrahedron Lett. 45 (2004) 8717–8720. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


