Abstract
The novel coronavirus (2019-nCoV) has recently emerged, causing COVID-19 outbreaks and significant societal/global disruption. Importantly, COVID-19 infection resembles SARS-like complications. However, the lack of knowledge about the underlying genetic mechanisms of COVID-19 warrants the development of prospective control measures. In this study, we employed whole-genome alignment and digital DNA–DNA hybridization analyses to assess genomic linkage between 2019-nCoV and other coronaviruses. To understand the pathogenetic behavior of 2019-nCoV, we compared gene expression datasets of viral infections closest to 2019-nCoV with four COVID-19 clinical presentations followed by functional enrichment of shared dysregulated genes. Potential chemical antagonists were also identified using protein–chemical interaction analysis. Based on phylogram analysis, the 2019-nCoV was found genetically closest to SARS-CoVs. In addition, we identified 562 upregulated and 738 downregulated genes (adj. P ≤ 0.05) with SARS-CoV infection. Among the dysregulated genes, SARS-CoV shared ≤19 upregulated and ≤22 downregulated genes with each of different COVID-19 complications. Notably, upregulation of BCL6 and PFKFB3 genes was common to SARS-CoV, pneumonia and severe acute respiratory syndrome, while they shared CRIP2, NSG1 and TNFRSF21 genes in downregulation. Besides, 14 genes were common to different SARS-CoV comorbidities that might influence COVID-19 disease. We also observed similarities in pathways that can lead to COVID-19 and SARS-CoV diseases. Finally, protein–chemical interactions suggest cyclosporine, resveratrol and quercetin as promising drug candidates against COVID-19 as well as other SARS-like viral infections. The pathogenetic analyses, along with identified biomarkers, signaling pathways and chemical antagonists, could prove useful for novel drug development in the fight against the current global 2019-nCoV pandemic.
Keywords: 2019-nCoV, coronavirus, COVID-19, microarray, SARS-CoV-2, comorbidities
Background
During the past century, several human coronaviruses have emerged, causing severe respiratory illness and global outbreaks in humans [1]. Coronaviruses (CoVs) are a large group of positive-sense single-stranded RNA viruses belonging to the Coronaviridae family [1, 2]. Six species of coronavirus have been identified as human pathogens [1, 3]. The spectrum of clinical manifestations ranges from mild to severe infection of the respiratory tract [3]. Four species OC43, HKU1, NL63 and 229E cause common cold in immune-compromised patients [1, 3]. Notably, two other coronaviruses, namely, severe acute the respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV), have plagued the general public and caused global outbreaks in 2003 and 2012, respectively [1, 2]. In the late December 2019, however, a novel coronavirus (2019-nCoV), which may have evolved through host-induced natural selection [4], was first identified in patients with viral pneumonia in Wuhan City, China [1, 2]. As of 30 April 2020, there has been a total of 3 200 414 confirmed 2019-nCoV cases and a death toll of 226 893 across 210 countries and territories (https://worldometers.info/coronavirus/). At the time of article writing, Bangladesh had 7103 confirmed cases with 163 deaths (http://corona.gov.bd/). The number of cases is still increasing worldwide with 4.5% global mortality rate [5, 6]. On 7 January 2020, newly evolved SARS-CoV-2 was first identified in Wuhan City, China [1]. The virus was initially named as 2019 novel coronavirus (2019-nCoV) by the World Health Organization [7]. However, based on established taxonomic methods, the virus has been recognized as a sister to SARS-CoV, hence, formally designated as SARS-CoV-2 by the Coronavirus Study Group (CSG) of the International Committee on Taxonomy of Viruses. The disease outbreak is named as coronavirus disease 19 (COVID-19) [7].
The SARS-CoV-2 genome encodes for four major proteins, i.e. spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins. The S-protein of SARS-CoVs facilitates the viral entry upon binding to angiotensin-converting enzyme 2 (ACE2) expressed in human lung cells [8, 9]. However, the binding affinity of S-protein towards ACE2 is higher in SARS-CoV-2 due to the translational alteration of five out of six vital amino acids in the active site of its S-protein allowing improved hydrophobic and salt–bridge interactions [8]. Upon binding, viral membrane fuses with host cells. After fusion occurs, transmembrane serine protease 2 (TMPRSS2) present on the host cell surface activates the ACE2-S-protein complex followed by the conformational changes that allow the virus to enter the cells [10].
The viral entry induces the host’s immune response by exposing itself to antigen-presenting cells (APCs) which have different types of pattern recognition receptors (PRRs) such as toll-like receptors (TLRs) [11]. For instance, TLR4 recognizes the S-protein and activate NF-κB transcription factors and pathogen-activated protein kinase (MAPK) pathway to induce proinflammatory proteins [10]. Meanwhile, other TLRs such as TLR3 could detect an RNA genome that eventually ends up activating the IRF3 and NF-κB transcription factors to induce proinflammatory cytokines such as IFN-α which in turn form complexes with its receptor, IFNAR, thereby, initiating the JAK-STAT pathways [10]. Further, JAK1 and TYK2 kinases phosphorylate STAT1 and 2 followed by its complexation with IRF9, and together they migrate into the nucleus to initiate the transcription of IFN-stimulated genes (ISGs) and lead to suppression of viral replication and prevent the severity of the disease [12]. However, excess releasing of pro-inflammatory cytokines (i.e. IFN-α, IFN-γ, IL-6, IL-18, TNF-α, TGFβ, etc.) and chemokines (i.e. CCL2, CCL5, CXCL8, CXCL10, etc.) from immune effector cells causes cytokine storms which will eventually lead to acute respiratory distress syndrome (ARDS) [10, 13]. However, for better survival and host infections, SARS-CoVs developed immune invasion strategies such as the formation of double vesicles during APC recognition by PRRs. In addition, it has several other proteins such as Nsp1 which can suppress INF-α activity through host translational machinery inactivation and inhibition of STAT1 phosphorylation [12].
Human coronavirus disease is a zoonosis, infection of animal origins. Most coronaviruses are originated from bats where they are non-virulent [14]. SARS-CoV-2 shares 88–92% genomic similarities with the most bat SARS-like coronaviruses, except W1V1 which showed ⁓96% sequence homology [14]. This bat coronavirus (W1V1) can use bat, civet and human ACE2 as a receptor for cell entry. Intriguingly, convalescent sera from SARS patients were able to neutralize WIV1 [14]. Therefore, it has been proposed that the bat is the original host of SARS-CoV-2 [15]. However, viral transmission from bats to humans requires several intermediate reservoirs [14]. In support, several studies have linked pangolins, cats, dogs and hamsters with SARS-CoV-2 infection and transmission [16–20]. These indicate the widespread prevalence of SARS-CoV-2 across animals as well as the potential threats to humans [21]. Preliminary evidence indicated that the COVID-19 outbreak started at the Wuhan seafood market. However, several recent studies suggest otherwise. For instance, among the first 41 cases, 14 had no prior contact with the market. In another study, five out of seven cases had no link with the market. Therefore, the virus could have amplified in the market, but it might neither have been the site of origin nor the only source of the outbreak [15].
The COVID-19 outbreak in China reiterates the significant and continued threat of zoonotic diseases caused by coronaviruses to the global population [1]. Unlike most other strains, the newly emerged 2019-nCoV causes SARS-like severe respiratory illness [2]. Like SARS-CoV infection, most infected patients develop pneumonia with prominent fever, shortness of breath, invasive lung lesions, anosmia and sometimes diarrhea [22–25]. The rate of severity and recovery from the COVID-19 is involved with the age, biological sex and other health conditions of the infected patients [6]. For instance, the majority of the COVID-19 patients are male (54.3%), while the fatality is more frequent (15%) in older age groups (above 80 years old) than in the younger population [6, 26]. In most cases, however, symptoms are similar to seasonal flu [27]. Nevertheless, in a recent Chinese study, 80% of cases were identified as asymptomatic, making early diagnosis difficult [28]. As a result, these undetected asymptomatic patients could be a major source of contagion [28]. Furthermore, the transmission of SARS-CoV-2 from humans to humans is highly frequent in family, health care and community setting [15]. The transmission occurs via respiratory tract droplets or fomites while fecal–oral spread is also possible. The estimated basic reproduction number (RO) of SARS-CoV-2 ranged between 2.0 and 2.8 [15]. Therefore, early detection is crucial for reducing viral shedding as well as for effective treatment [28]. Despite the knowledge and experience related to SARS-CoV and MERS-CoV, major gaps related to the epidemiology, clinical manifestations and pathogenetic behaviors of this emerging 2019-nCoV remain to be fulfilled [2]. Since no vaccine or antiviral drugs are available, present treatments are based on managing symptoms and complications. Therefore, insight into biomarkers and molecular mechanisms that transform the 2019-nCoV infection into COVID-19 disease and related complications is necessary to evaluate the clinical significance and in assessing the severity and prognosis of the disease.
In this study, we aimed to explore the molecular pathogenesis of COVID-19. As the 2019-nCoV shares characteristics with SARS-CoV in terms of genomic and pathogenic features, existing SARS-CoV experimental data is used to study molecular aspects of 2019-nCoV. To accomplish this, we first validated the genetic linkage among 2019-nCoV and other coronaviruses using phylogenetic analysis. We employed a multistep statistical method to determine differentially expressed genes (DEGs), regulatory networks and gene–disease associations as illustrated in Figure 1. Using the shared DEGs, we identified hub-proteins and regulatory biomarkers, i.e. transcription factors (TFs) and microRNAs (miRNAs). Furthermore, gene ontology and pathway analyses were performed to determine significant regulatory checkpoints related to disease complications. Finally, we screened potential chemical antagonists useful in the fight against COVID-19 disease.
Figure 1.
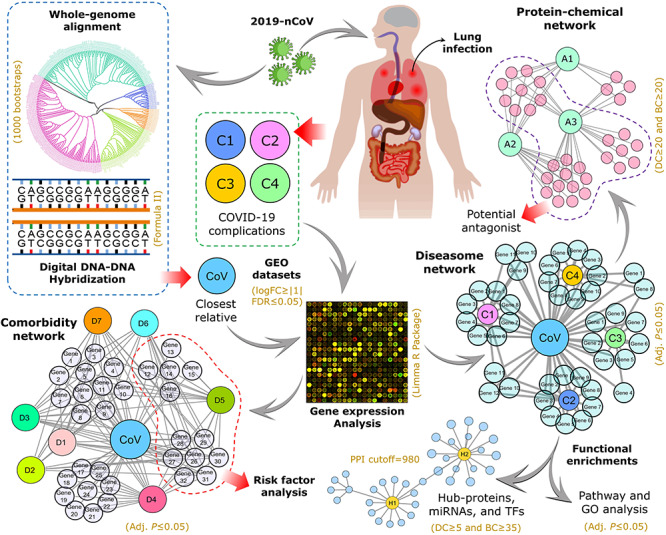
The overall experimental workflow applied in this study. Following establishing SARS-CoV as the closest relative of 2019-nCoV through whole-genome alignment (1000 bootstrap tests), the microarray datasets related to SARS-CoV infection and four COVID-19 complications (i.e. SARS, PNA, SHOB and DRA) were retrieved and analyzed to identify differentially expressed genes (DEGs). Herein, Limma R package was employed where logFC≥|1| was used to distinguish differential expression, and adjusted P-value ≤0.05 was set as cutoff for statistical significance. Then, the DEGs from SARS-CoV infection were subjected to validation with gold-standard databases (i.e. OMIM disease, OMIM expanded and dbGaP) for comorbidity and risk factor assessment followed by comparative analysis with the disease datasets. Finally, we used the shared DEGs for network-based interactome analysis and functional enrichments in terms of protein–protein interactions (PPI confidence cutoff = 980), gene–biomarker associations (DC≥5 and BC≥35) and protein–chemical interactions (DC≥20 and BC≥20). For pathway analysis, we considered BioCarta (2016), Reactome (2016), KEGG (human; 2019) and WikiPathways (human; 2019) databases, while Biological Process (2018), Molecular Function (2018) and Human Phenotype Ontology databases were used for gene ontological evaluation. In both cases, adjusted P-value ≤0.05 was considered as significant.
Materials and methods
Datasets
The complete genomes used in this study were collected from the Virus Pathogen Database and Analysis Resource (ViPR; https://viprbrc.org/). To investigate the impact of COVID-19 at the molecular level, we employed gene expression microarray datasets that were obtained from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Due to the genetic closeness and unavailability of COVID-19-related datasets, however, we used microarray datasets related to SARS-CoV infection and four COVID-19 complications, i.e. pneumonia (PNA), severe acute respiratory syndrome (SARS), shortness of breath (SHOB) and diarrhea (DRA). In this study, five different datasets with accession numbers GSE30589, GSE103119, GSE14841, GSE137268 and GSE1739 were used [29–33]. The SARS-CoV dataset (GSE30589) is an Affymetrix microarray data involving 33 infected and 9 mock-infected patients with 3 hybridizations. The SARS dataset (GSE1739) is a gene expression profile derived from peripheral blood mononuclear cells (PBMCs) of 10 patients and 4 healthy controls using Affymetrix HG-Focus target array platform. The PNA dataset (GSE103119) is an Illumina humanHT-12 v4.0 gene expression array of 152 patients with community-acquired pneumonia and 20 healthy patients. The SHOB dataset (GSE137268) is a gene expression array of 54 asthma patients and 15 healthy individuals developed using Illumina humanRef-8 v2.0 bead chip. The DRA dataset (GSE14841) is an Affymetrix human genome U133 plus 2.0 array from jejunal mucosa of five patients with diarrhea-irritable bowel syndrome and four healthy volunteers.
Methods
To determine the relationship between 2019-nCoV and other coronaviruses, we created a comprehensive phylogram that includes the complete genomes of 389 human beta-coronaviruses (Supplementary File S1). The tree was inferred with the FastME program integrated with ViPR database using genomic data with 1000 bootstrap replications. Then, Newick-formatted phylogram was redesigned with the interactive tree of life (iTOL) online tool (https://itol.embl.de/). In addition, the genomic distance and G+C difference were calculated through digital DNA–DNA hybridization (dDDH) using Genome-to-Genome Distance Calculator (GGDC) server v2.1 with default settings [34]. For this analysis, we considered reference genomes of 2019-nCoV (NC_045512.2), SARS-CoV (NC_004718.3), MERS-CoV (NC_019843.3), CoV-OC43 (NC_006213.1) and CoV-HKU1 (NC_006577.2); and formula-2 derived dDDH and G+C difference scores were emphasized as per the server’s recommendation [34].
Evaluation of gene expression microarray is very efficient when it comes to determining underlying molecular mechanisms that lead to the development of disease [35]. Using the Limma R package [36], we analyzed the selected microarray datasets as described earlier [35, 37]. Briefly, we categorized the samples in each dataset into two groups (treatment and control) followed by log transformation with a series of statistical methods (i.e. B- and t-statistics) and P-value adjustment with Benjamini and Hochberg method (false discovery rate). Herein, genes were considered as significantly upregulated or downregulated when expressed at adjusted P-value ≤0.05 with logFC score greater or lesser than (or equal to) +1 or −1, respectively.
To determine gene–disease associations, we applied neighborhood-based benchmark and topological methods. Thus, we built a comorbidity network from the gene–disease association where the node in the network can be either a disease or a gene. This network is a type of bipartite graph in which diseases are connected when they share at least 1 significant dysregulated gene. Let a particular set of human diseases be D and a set of human genes be G; a comorbidity network then attempts to find whether gene g ϵ G is associated with disease d ϵ D. If Gi and Gj, the sets of significant up- and downregulated genes, are associated with diseases i and j, respectively, then the number of shared dysregulated genes (ngij) associated with both diseases i and j is as follows:
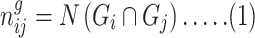 |
Co-occurrence indicates the number of common genes in the comorbidity network. Based on the Jaccard similarity index, the common neighbors are found. For each node pair with the edge (E), the prediction point is as follows:
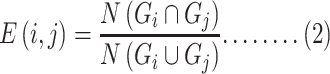 |
The protein–protein interaction (PPI), regulatory biomarkers (i.e. TFs and miRNAs), protein–drug interaction (PDI) and protein–chemical interaction (PCI) networks were constructed using NetworkAnalyst tool [38]. Experimentally validated STRING interactome with 980 confidence cutoff was used for PPI network. For TFs and miRNAs, we employed experimentally verified TF target genes from the JASPAR database [39] and miRNA target genes from TarBase [40] and miRTarBase [41] via NetworkAnalyst [38]. Similarly, PDI and PCI were evaluated based on DrugBank v5.0 and Comparative Toxicogenomics Database, respectively [42, 43]. All interactions were visualized and customized in Cytoscape v3.7.2 [44].
The degree centrality (DC) and betweenness centrality (BC) were applied to filter out potential regulatory biomarkers and chemical antagonists. Nodes with a high degree act as important ‘hubs’ in a network, while high betweenness indicates nodes as important ‘bottlenecks’ in a network. Briefly, the DC of a node v in a network can be defined as the total number of nodes that are directly connected to node v in that network which can also be expressed as follows:
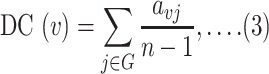 |
where n represents the total number of nodes in the network and avj represents that node v and j are directly connected. BC represents the total number of times node v appears in the shortest path between other nodes. It is also defined as follows:
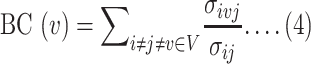 |
where σij is the total number of shortest paths from node i to node j and σivj is the total number of paths through node v.
Furthermore, we analyzed the pathways and gene ontology (GO) of the shared DEGs with the Enrichr web application [45] to understand how genetic determinants transform gene expressions in developing SARS-CoV complications. For pathways, we considered BioCarta (2016), Reactome (2016), KEGG (human; 2019) and WikiPathways (human; 2019) databases and GO Biological Process (2018), GO Molecular Function (2018) and Human Phenotype Ontology databases for GO analysis. Herein, an adjusted P-value ≤0.05 was considered as statistically significant.
Results
Human coronavirus phylogram
Whole-genome phylogenetic analysis revealed the relation among the different human beta-coronaviruses as shown in Figure 2. Importantly, the recent epidemic 2019-nCoV formed a sister clade with human SARS-CoV indicating their closest relationship with SARS-CoV at the genomic level. Moreover, as per the phylogenetic position, both SARS-CoV and 2019-nCoV share a common ancestral origin with MERS-CoV, which happened to be closely related to the CoV-HKU1 (Figure 2). This result coincides with the estimated genomic distance and G+C content difference. In our study, the taxonomic distance between SARS-CoV and 2019-nCoV was 0.1985 rendering 22.10% of DNA–DNA hybridization. Moreover, the G+C content of 2019-nCoV was different from SARS-CoV, MERS-CoV and CoV-HKU1 by 2.79, 3.26 and 5.91%, respectively. Interestingly, the G+C difference between 2019-nCoV and CoV-OC43 was lower (1.18%) compared to the closest neighbors. Since the G+C difference is bound to ≤1% within the same species, all these results suggest 2019-nCoV as a distinct coronavirus species.
Figure 2.
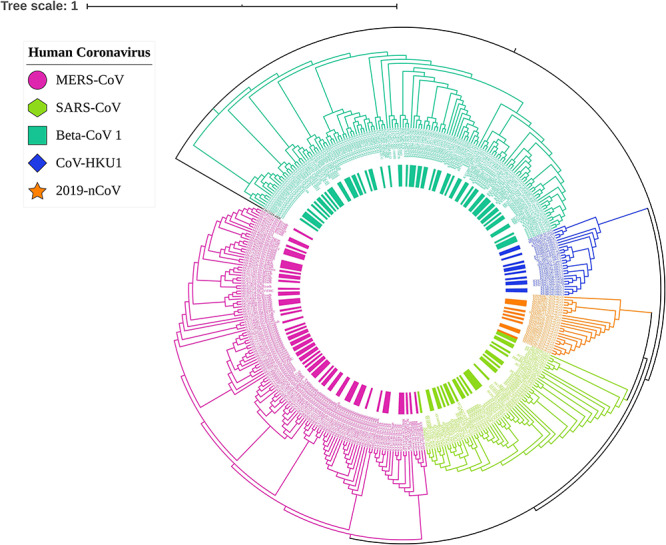
Phylogram of human coronavirus shows the respective position of newly emerged 2019-nCoV among the 389 beta-coronavirus. Different colors indicate the clades of different coronavirus where 2019-nCoV (orange) shared the same branch with SARS-CoV (lime). The whole-genome phylogram was inferred with 1000 bootstrap values in ViPR database and customized with the interactive tree of life (iTOL) web tool.
Infectome and diseasome
To investigate the host response upon 2019-nCoV infection, we analyzed the gene expression pattern of SARS-CoV-infected tissues and healthy tissues using microarray data. We found that 1300 genes were differentially expressed as compared to healthy controls in which 562 genes were upregulated while 738 genes were downregulated significantly with infection (adj. P ≤ 0.05). Similarly, we also analyzed the diseasome of four COVID-19 complications to observe their association with SARS-CoV infection. We identified a large number of DEGs for each condition, i.e. 427 in DRA, 581 in SHOB, 997 in PNA and 728 in SARS (Supplementary File S2). Then, we performed a cross-comparative analysis to find common significant DEGs between each condition and SARS-CoV infection. We found a total of 87 unique shared DEGs in which SARS-CoV infection shares 18, 15, 21 and 40 genes with DRA, SHOB, PNA and SARS, respectively. To visualize their association, we construct infectome–diseasome relationship networks centered on the SARS-CoV infection, in which two diseases are comorbid if there exists one or more genes that are associated with both diseases (Figure 3).
Figure 3.
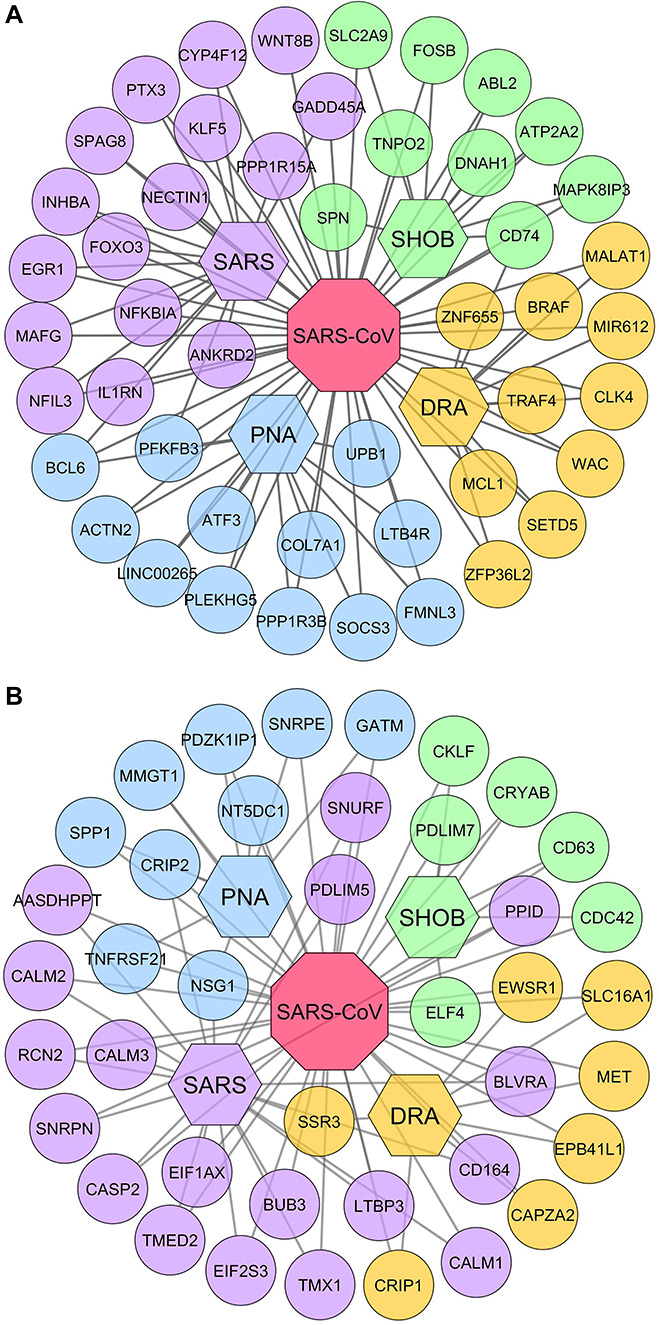
Infectome–diseasome network. The figure showing the interconnection of the shared DEGs: (A) upregulated genes and (B) downregulated genes. The octagon- and hexagon-shaped nodes represent viral infection (SARS-CoV) and four COVID-19 diseases (i.e. SARS, PNA, SHOB and DRA), respectively, while circular nodes delineate the genes involved. Clusters of different colored nodes indicate the disease-wise gene group.
Table 1 includes comprehensive details of important DEGs in terms of their expression pattern, associated conditions and pathogenic roles in disease development. Interestingly, SARS-CoV infection shared ≤19 upregulated and ≤22 downregulated genes with each different COVID-19 complication. Notably, upregulation of BCL6 and PFKFB3 genes was common to SARS-CoV, PNA and SARS (Figure 3A) while they shared CRIP2, NSG1 and TNFRSF21 genes in downregulation (Figure 3B). Nevertheless, less frequent COVID-19 conditions such as DRA and SHOB shared upregulated CD74 with SARS-CoV while the SSR3 gene was downregulated in DRA, in SARS as well as in SARS-CoV infection. However, no downregulated gene related to SHOB was found in the three other conditions.
Table 1.
List of shared DEGs with their expression pattern and pathogenetic mechanisms involved in the disease progression
| Gene symbol | Name | Category | Expression pattern | Associated conditions | Pathogenetic mechanisms | Infectious agents | Reference |
|---|---|---|---|---|---|---|---|
| SSR3 | Signal sequence receptor subunit 3 | Protein | Down | VAD, SARS | Ssr3 is a B-cell activation-related gene and was found to express in plasma and memory B cells in the early stage of COVID-19 recovery. It also has the most complex interacting network with viral proteins. | SARS-CoV-2 | [46, 47] |
| ABL2 | Abelson tyrosine-protein kinase 2 | Protein | Up | SHOB | Abl2 kinase activity is important for SARS-like viral entry and efficient replication. | SARS-CoV MERS-CoV SARS-CoV2 |
[48, 49] |
| CRIP2 | Cysteine-rich intestinal protein 2 | Protein | Down | PNA, SARS | Crip2 functions in the differentiation of airway smooth muscle and correlated negatively with ACE2 protein. | – | [50] |
| ATF3 | Activating transcription factor 3 | Protein | Up | PNA | Atf3 is involved in apoptosis, especially in renal tissue damage and upregulated in MERS and COVID-19 infection. | MERS-CoV SARS-CoV-2 |
[51, 52] |
| SPP1 | Secreted phosphoprotein 1 | Protein | Down | PNA | Spp1 is a significant pro-fibrotic gene which is involved in M2-like macrophage infiltration correlated to COVID-19 severity. | SARS-CoV-2 | [53] |
| BCL6 | B-cell lymphoma 6 | Protein | Up | PNA | Bcl6 regulates the inhibition of antiviral resistance in follicular Th cells, thereby increasing the susceptibility to viral infections. | HIV | [54] |
| TNFRSF21 | TNF receptor superfamily member 21 | Protein | Down | PNA, SARS | Tnfrsf21 activates the JNK and NF-κB pathways that eventually lead to a series of inflammatory reactions, and it also promotes cellular apoptosis. | – | [55] |
| COL7A1 | Collagen-type VII alpha 1 chain | Protein | Up | PNA | Col7a1 is upregulated in idiopathic interstitial pneumonia and involved in cell–matrix interactions | – | [56] |
| FMNL3 | Formin-like 3 | Protein | Up | PNA | Fmnl3 controls cytoskeleton organization and cell morphogenesis and is implicated in various diseases. | PRRSV | [57, 58] |
| LINC00265 | Long intergenic nonprotein coding RNA 265 | lncRNA | Up | PNA | Linc00265 activates PI3K-AKT pathway which is required for viral entry into cells and regulates inflammation. | HSV VACV |
[60–61] |
| LTB4R | Leukotriene B4 receptor | Protein | Up | PNA | Ltb4 overexpression triggers acute asthma attack, and it is important in inflammatory pathways and is highly prevalent in type 1 diabetic patients. | – | [62, 63] |
| CALM1 | Calmodulin 1 | Protein | Down | SARS | Calm1 regulates and modulates the cardiac ion channels and renin secretion. It is also involved in viral replication. | HIV | [64, 65] |
| SOCS3 | Suppressor of cytokine signaling 3 | Protein | Up | PNA | Socs3 controls the negative feedback mechanism of IL-6 which is highly prevalent in COVID-19 patients. | SARS-CoV-2 | [66] |
| LTBP3 | Latent-transforming growth factor beta binding protein 3 | Protein | Down | SARS | Ltbp3 enhances lung alveolarization, but nsp12 protein of SARS-CoV-2 can modulate it otherwise. | SARS-CoV-2 | [67] |
| EGR1 | Early growth response 1 | Protein | Up | SARS | Egr1 activates TGF-β1 promoter via ROS/p38 MAPK/STAT3 pathway by papain-like protease (PLpro) of SARS-like virus. | SARS-CoV | [68] |
| FOXO3 | Forkhead box O3 | Protein | Up | SARS | Foxo3 increases tissue-specific polyclonal cytotoxic T-cell expansion by reducing apoptosis and targeted by miR-223 for type 1 IFN production. | SARS-CoV-2 VSV |
[69, 70] |
| GADD45A | Growth arrest and DNA-damage-inducible 45 alpha | Protein | Up | SARS | Gadd45a is a key modulator of the MAPK signaling pathway which promotes cell death and asthma in SARS-like disease. | SARS-CoV-2 SARS-CoV |
[71, 72] |
| IL1RN | Interleukin-1 receptor antagonist | Protein | Up | SARS | Il1rn inhibits IL-1α and IL-1β, thereby modulating inflammatory responses in COVID-19, and remains elevated even after viral clearance. | SARS-CoV-2 MERS-CoV | [73] |
| INHBA | Inhibin subunit beta A | Protein | Up | SARS | Inhba overexpression induced by influenza virus causes acute respiratory distress-like syndrome in the lung. | H1N1 | [75] |
| KLF5 | Kruppel-like factor 5 | Protein | Up | SARS | Klf5 regulates IFN-induced transmembrane proteins (IFITM1, 2 and 3) in A549 cells, and IFITM1 is involved in inflammatory bowel disease. | H5N1 | [75, 76] |
| NECTIN1 | Nectin cell adhesion molecule 1 | Protein | Up | SARS | Nectin1 is a receptor for several viruses and also expressed in COVID-19. | SARS-CoV-2 | [77] |
| NFIL3 | Nuclear factor, interleukin-3 regulated | Protein | Up | SARS | Nfil3 plays a critical role in NK cell development and overexpressed in SARS-CoV infection. | SARS-CoV | [78, 79] |
| NFKBIA | NF-κB inhibitor alpha | Protein | Up | SARS | Nfkbia is a key modulator of IL-1 and TNF-α-related chemokine signaling, thereby contributing to infectious and inflammatory diseases. | SARS-CoV MERS-CoV |
[80–82] |
| PPP1R15A | Protein phosphatase 1 regulatory subunit 15A | Protein | Up | SARS | Ppp1r15a is a translation regulator that controls cytokine production and is upregulated by coronavirus replication and also found in cells with high levels of SARS-CoV-2. | SARS-CoV-2 HCoV-229E |
[83, 84] |
| CD74 | Cluster of differentiation 74 | Protein | Up | SHOB | Cd74 induces p44/p42 MAPK activation and MIF-mediated pulmonary inflammation. It contributes to the adaptive immunity impairment in COVID-19 patients. | SARS-CoV-2 | [85, 86] |
| ELF4 | E74-like ETS transcription factor 4 | Protein | Down | SHOB | Elf4 contributes in NK cell development and function as well as cell cycle arrest in naive CD8+ cells upon viral infection. | RSV | [87] |
| SLC16A1 | Solute carrier family 16 member 1 | Protein | Down | VAD | Slc16a1 is downregulated in patients with inflammatory bowel disease in response to cytokines TNF-α and IFN-γ. It also plays an important role in SARS-CoV-2 cell entry. | SARS-CoV-2 | [88, 89] |
| BRAF | B-Raf proto-oncogene | Protein | Up | VAD | Braf regulates MAP kinase/ERKs signaling pathway affecting cell division, differentiation and secretion. | – | [90] |
| MALAT1 | Metastasis-associated lung adenocarcinoma transcript 1 | lncRNA | Up | VAD | Malat1 targets miR-155 to promote GATA3 and Th2 cytokine production that leads to severe asthma and loss of lung function. | – | [91] |
| MCL1 | Myeloid cell leukemia sequence 1 | Protein | Up | VAD | Mcl1 interacts with orf7a of coronavirus and initiates apoptosis process as antiviral defense, and it is upregulated in SARS-like coronaviruses. | SARS-CoV-2 SARS-CoV MERS-CoV |
[92] |
| MIR612 | microRNA 612 | MiRNA | Up | VAD | miR-612 represses the transcription of IL-8 and ADARB2 and inhibits HIV production in concert with lncRNAs. | HIV | [93] |
| TRAF4 | TNF receptor-associated factor 4 | Protein | Up | VAD | Traf is upregulated by coronaviruses, and it connects IL-1R/Toll and TNF receptors activating of NF-κB and MAPK pathways. | SARS-CoV CoV-229E |
[94] |
| ZNF655 | Zinc finger protein 655 | Protein | Up | VAD | Znf655 may be involved in cell cycle phase transition to facilitate delayed and lack of several antiviral mechanisms observed in COVID-19. | SARS-CoV-2 | [95] |
| CRIP1 | Cysteine-rich protein 1 | Protein | Down | VAD | Crip1 helps TGF-β to inhibit the Th1 and Th2 differentiation, is involved in ACE2 expression and is downregulated by coronavirus. | SARS-CoV-2 | [96, 97] |
| CALM2 | Calmodulin 2 | Protein | Down | SARS | Calm2 is involved in calcium signaling which is significantly downregulated in virus-infected ciliated cells. | SARS-CoV-2 | [52] |
| EIF1AX | Eukaryotic translation initiation factor 1A X-linked | Protein | Down | SARS | Eif1ax enhances ribosome dissociation and stabilizes the binding of the initiator Met-tRNA to 40S ribosomal subunit. | SARS-CoV-2 | [98] |
| MAFG | MAF BZIP transcription factor G | Protein | Up | SARS | Mafg is an important transcription factor and a hub-gene in COVID-19 and upregulated upon viral infection. | SARS-CoV-2 | [99] |
| PTX3 | Pentraxin 3 | Protein | Up | SARS | Ptx3 plays an important role in host defense and inflammatory response in the lung and a protective role in pulmonary infection. | MHV-1 SARS-CoV |
[100] |
| SPAG8 | Sperm-associated antigen 8 | Protein | Up | SARS | Spag8 is involved in fertilization and compromised in ACE2-positive cells which is targeted by SARS-CoV-2 leading to the disruption of spermatogenesis. | SARS-CoV-2 | [101] |
| EWSR1 | Ewing sarcoma breakpoint region 1 | Protein | Down | VAD | Ewsr1 interacts with viral cis-acting replication element (CRE) for efficient replication and regulates cell cycle progression and DNA damage. | HCV | [102] |
| ATP2A2 | ATPase sarcoplasmic/ER Ca2+ transporting 2 | Protein | Up | SHOB | Atp2a2 is associated with cellular processes important in coronavirus replication. | SARS-CoV MERS-CoV |
[103] |
| FOSB | FosB proto-oncogene | Protein | Up | SHOB | Fosb is a part of activator protein 1 which is central to cytokine storms inflicted by SARS-like viruses and upregulated by their N and 3b proteins. | SARS-CoV MERS-CoV |
[104, 105] |
| MAPK8IP3 | Mitogen-activated protein kinase 8-interacting protein 3 | Protein | UP | SHOB | Mapk8ip3 plays an essential role in MAPK pathway to facilitate viral replication and can also trigger airway inflammation. | H7N9 | [106] |
| PFKFB3 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | Protein | Up | PNA | Pfkfb3 enhances glycolysis in macrophage glycolysis to promote its antiviral defense (removal and engulfment) but can be resulted in accelerated sepsis while being accompanied by zhx2 protein. | RSV Lentivirus |
[107, 108] |
*Genes that are studied for their involvement in viral diseases, especially in SARS-like viral infection, were considered, and their roles in respective pathogenesis are tabulated in light of the cited literatures and GeneCards (https://www.genecards.org/), a human gene database.
Abbreviations: DRA, diarrhea; PNA, pneumonia; SHOB, shortness of breath; SARS, severe acute respiratory syndrome; Down, downregulation; Up, upregulation; RSV, respiratory syncytial virus; H7N9, influenza A strain H7N9; MHV-1, coronavirus murine hepatitis virus strain 1; HCV, hepatitis C virus; HIV, human immunodeficiency virus; H5N1, influenza A strain H5N1; H1N1, influenza A strain H5N1; VSV, vesicular stomatitis virus; VACV, vaccinia virus; PRRSV, porcine reproductive and respiratory syndrome virus; ACE2, angiotensin-converting enzyme 2; COVID-19, coronavirus disease 2019.
Comorbidities of SARS-CoV infection
We used genes that are significantly dysregulated upon SARS-CoV infection to construct gene–disease association networks (GDN) to explore the shared pathogenetic projections and comorbidities in COVID-19, as shown in Figure 4. This multi-relational GDN is a bipartite graph consisting of two disjointed sets of nodes in which one set represents the known genetic disorders while others correspond to the significantly altered genes for SARS-CoV infection. The information related to disorders, genes and their possible association that we embedded into the GDN was obtained from OMIM and dbGaP databases (Supplementary File S3). Based on the physiological system affected, we classified the associated disorders into 13 categories including SARS-CoV. Herein, the nodes in the GDN indicate either disease categories or involved genes, and two diseases are related to each other if they have at least one gene in common (Figure 4).
Figure 4.
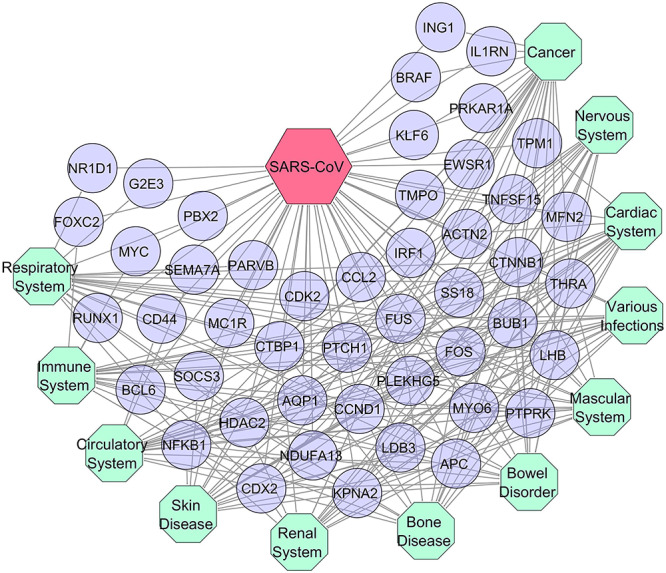
Comorbidity profile of SARS-CoV infection. Dysregulated genes showing association with various types of diseases and comorbidities. Herein, octagon-shaped nodes indicate the different affiliation of SARS-CoV-associated diseases (turquoise), while circular-shaped nodes represent different genes (violet) that are common with the other categories of disorders.
The number of interconnected genes between SARS-CoV and other diseases suggested that cardiac (20), immunological (18), cancer (23) and respiratory (19) diseases are greatly influenced by SARS-CoV and vice versa due to their higher number of shared genes (Figure 4). Interestingly, 14 genes were also common to all included categories of disease such as APC, KPNA2, CTNNB1, CCND1, NFKB1, FOS, HDAC2, PTPRK, LHB, FUS, CTBP1, THRA, CDX2 and BUB1 which further indicate their strong association with these diseases.
Functional enrichment
In complex pathological conditions, analysis of signaling pathways can shed light on the underlying molecular mechanisms linking multiple diseases to each other. Therefore, we analyzed a total of 87 unique shared DEGs from four COVID-19 presentations to determine the significantly involved pathways (Supplementary File S4). Table 2 shows a disease-wise total of 22 significant pathways that are relevant and shared among 4 COVID-19 presentations and SARS-CoV disease with their associated genes and adjusted P-values. For example, we identified five pathways including chemokine signaling pathway NRF2 pathway, non-small cell lung cancer, FoxO signaling pathway and hypoxia and p53 in the cardiovascular system that are significantly expressed in SARS condition (Table 2). In addition, we observed five pathways related to SHOB that are FOSB gene expression and drug abuse, EGFR downregulation, MAPK signaling pathway, TGF-β signaling pathway and oxidative damage. In this study, PNA-related significant pathways were mostly associated with cytokine signaling (Table 2); and DRA shared five pathways with SARS-CoV disease.
Table 2.
Signaling pathways involved with the shared dysregulated genes
| Pathways | Adj. P-value * | Associated genes |
|---|---|---|
| DRA | ||
| Negative feedback regulation of MAPK pathway | 5.38 × 10−3 | BRAF, MET |
| Bile salt and organic anion SLC transporters | 1.25 × 10−2 | SLC16A1 |
| Serotonin receptor 4/6/7 and NR3C signaling | 1.69 × 10−2 | BRAF |
| Hemostasis | 1.25 × 10−2 | CD74, SLC16A1, CAPZA2 |
| Focal adhesion | 1.34 × 10−2 | BRAF, MET |
| SHOB | ||
| FOSB gene expression and drug abuse | 3.74 × 10−3 | FOSB |
| EGFR downregulation | 2.01 × 10−2 | CDC42 |
| MAPK signaling pathway | 2.01 × 10−2 | CDC42, MAPK8IP3 |
| TGF-beta signaling pathway | 4.29 × 10−3 | CDC42, FOSB |
| Oxidative damage | 2.96 × 10−2 | CDC42 |
| PNA | ||
| Leukotriene receptors | 5.24 × 10−3 | LTB4R |
| Interleukin-6 signaling | 1.15 × 10−2 | SOCS3 |
| PERK regulates gene expression | 2.90 × 10−2 | ATF3 |
| Osteopontin signaling | 1.36 × 10−2 | SPP1 |
| IL22 soluble receptor signaling pathway | 1.15 × 10−2 | SOCS3 |
| Regulation of IFN-γ signaling | 1.46 × 10−2 | SOCS3 |
| Interleukin-11 signaling pathway | 4.52 × 10−2 | SOCS3 |
| SARS | ||
| Chemokine signaling pathway | 4.25 × 10−2 | NFKBIA, FOXO3 |
| NRF2 pathway | 3.45 × 10−2 | EGR1, MAFG |
| Non-small cell lung cancer | 7.72 × 10−3 | GADD45A, FOXO3 |
| FoxO signaling pathway | 2.32 × 10−3 | BCL6, GADD45A, FOXO3 |
| Hypoxia and p53 in the cardiovascular system | 4.12 × 10−2 | GADD45A |
*Pathways that are relevant to the respective conditions and have adjusted P-value ≤0.05 were considered.
Abbreviations: DRA, diarrhea; PNA, pneumonia; SHOB, shortness of breath; and SARS, severe acute respiratory syndrome.
In addition to pathways, GO properties were anticipated to determine the overrepresented ontological groups among the DEGs (Supplementary File S5). We found 25 significant GO groups that are linked to the highly expressed genes shared by SARS-CoV and COVID-19 diseases. The genes associated with their GO terms and adjusted P-values are presented in Table 3. Each of the complication PNA, SHOB and DRA shared six significant pathways with SARS-CoV, while SARS had seven pathways related to SARS-CoV disease (Table 3).
Table 3.
Gene ontological features associated with the shared dysregulated genes
| Terms | Pathways | Adj. P-value * | Associated genes |
|---|---|---|---|
| DRA | |||
| GO:0033674 | Positive regulation of kinase activity | 1.35 × 10−4 | CD74, TRAF4, MET |
| GO:0070372 | Regulation of ERK1 and ERK2 cascade | 1.32 × 10−3 | CD74, BRAF, MET, ZFP36L2 |
| GO:0000165 | MAPK cascade | 1.86 × 10−3 | BRAF, MET, ZFP36L2, CD74 |
| GO:0071277 | Cellular response to calcium ion | 3.71 × 10−2 | BRAF |
| GO:0035023 | Regulation of Rho protein signal transduction | 5.01 × 10−2 | MET |
| HP:0002020 | Gastroesophageal reflux | 3.98 × 10−2 | BRAF |
| SHOB | |||
| GO:0070296 | Sarcoplasmic reticulum calcium ion transport | 6.73 × 10−3 | ATP2A2 |
| GO:0042058 | Regulation of epidermal growth factor receptor signaling pathway | 4.26 × 10−2 | CDC42 |
| GO:0030544 | Hsp70 protein binding | 2.01 × 10−2 | SPN |
| GO:0008009 | Chemokine activity | 3.39 × 10−2 | CKLF |
| HP:0002747 | Respiratory insufficiency due to muscle weakness | 3.54 × 10−2 | CRYAB |
| HP:0002878 | Respiratory failure | 4.26 × 10−2 | CRYAB |
| PNA | |||
| GO:2000665 | Regulation of interleukin-10/13 secretion | 6.28 × 10−3 | TNFRSF21 |
| GO:0071345 | Cellular response to cytokine stimulus | 1.15 × 10−2 | SOCS3, BCL6, TNFRSF21 |
| GO:0006954 | Inflammatory response | 2.84 × 10−2 | LTB4R, TNFRSF21 |
| GO:0046426 | Negative regulation of JAK-STAT cascade | 4.82 × 10−2 | SOCS3 |
| GO:0004974 | Leukotriene receptor activity | 6.28 × 10−3 | LTB4R |
| HP:0002747 | Respiratory insufficiency due to muscle weakness | 4.92 × 10−2 | PLEKHG5 |
| SARS | |||
| GO:0043068 | Positive regulation of programmed cell death | 1.54 × 10−4 | BCL6, GADD45A, CASP2, FOXO3, PPID |
| GO:0071345 | Cellular response to cytokine stimulus | 2.05 × 10−3 | EGR1, IL1RN, BCL6, FOXO3, TNFRSF21 |
| GO:0034599 | Cellular response to oxidative stress | 2.22 × 10−2 | ANKRD2, FOXO3 |
| GO:1900744 | Regulation of p38MAPK cascade | 5.08 × 10−2 | GADD45A |
| GO:0042345 | Regulation of NF-κB import into nucleus | 5.08 × 10−2 | NFKBIA |
| GO:0019901 | Protein kinase binding | 5.01 × 10−5 | PPP1R15A, CALM3, ANKRD2, FOXO3, PDLIM5, CALM1, CALM2 |
| HP:0006530 | Interstitial pulmonary disease | 2.18 × 10−2 | IL1RN |
*Pathways that are highly relevant to the respective conditions and have adjusted P-value ≤0.05 were considered.
Abbreviations: DRA, diarrhea; PNA, pneumonia; SHOB, shortness of breath; and SARS, severe acute respiratory syndrome.
Among the 25 ontological features, notable GO terms were regulation of p38MAPK cascade (GO:1900744), regulation of NF-κB import into the nucleus (GO:0042345), protein kinase binding (GO:0019901), negative regulation of JAK-STAT cascade (GO:0046426), leukotriene receptor activity (GO:0004974), respiratory insufficiency due to muscle weakness (HP:0002747), respiratory failure (HP:0002878), regulation of epidermal growth factor receptor signaling pathway (GO:0042058), MAPK cascade (GO:0000165) and regulation of ERK1 and ERK2 cascade (GO:0070372). These ontological features were common in SARS-CoV disease and COVID-19 complications. Therefore, they could either be risk factors or regulatory checkpoints in COVID-19 disease.
Hub-proteins and their interactions
Using the shared DEGs, 12 proteins having experimental evidence were characterized as seed and formed an interaction network with the other 122 proteins. These proteins are SOCS3, BUB3, EIF2S3, MET, MCL1, NFKBIA, GADD45A, ATF3, EGR1, FOSB, ATP2A and KLF5. Among these 12 proteins, 7 proteins have interactions with 8 or more other proteins and are identified as hub-proteins (Figure 5). In our study, the majority of hub-proteins were related to SARS such as NFKBIA, BUB3, EIF2S3 and GADD45A, while two proteins (MET and MCL1) belonged to DRA and a single protein, SOCS3, was associated with PNA. However, no hub-protein was found in the shared DEGs for SHOB complication. From Figure 5, we can see the interrelationships among the different diseases, hub-proteins and their protein networks. However, EGR1 of SARS and ATF3 of PNA were intermediately connected, while FOSB, KLF5 and ATP2A2 were non-hub proteins.
Figure 5.
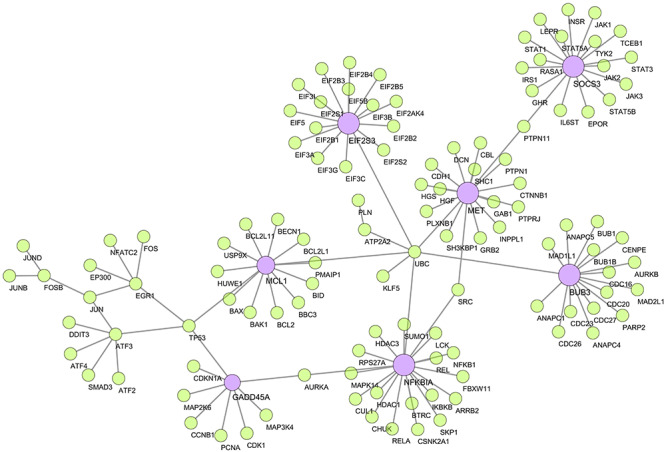
Protein–protein interaction network. The network was created using the experimentally validated STRING interactome with 980 as confidence cutoff. The seven large nodes (violet) indicate the hub-proteins, while smaller nodes (lime) and edges are the associated genes and interactions, respectively.
Transcription factors and microRNAs
We found 47 miRNAs and 28 TFs that might influence the differential expression of these genes thereby regulating disease progression. Among the 47 miRNAs, four human miRNAs (i.e. let-7c-5p, miR-27b-3p, miR-98-5p and miR-125a-5p) were found to be targeting the SARS-CoV-2 genome, while many others were associated with the development of COVID-19 symptoms, i.e. let-7b-5p, miR-155-5p, miR-186-5p, miR-16-5p, miR-27b-3p, miR-29a-3p, miR-30a-5p, etc., as depicted in Figure 6A. Among the 28 TFs, notable TFs are RELA, E2F1, STAT3, TP53, NFKB1, GATA3 and CREB1 as shown in Figure 6B.
Figure 6.
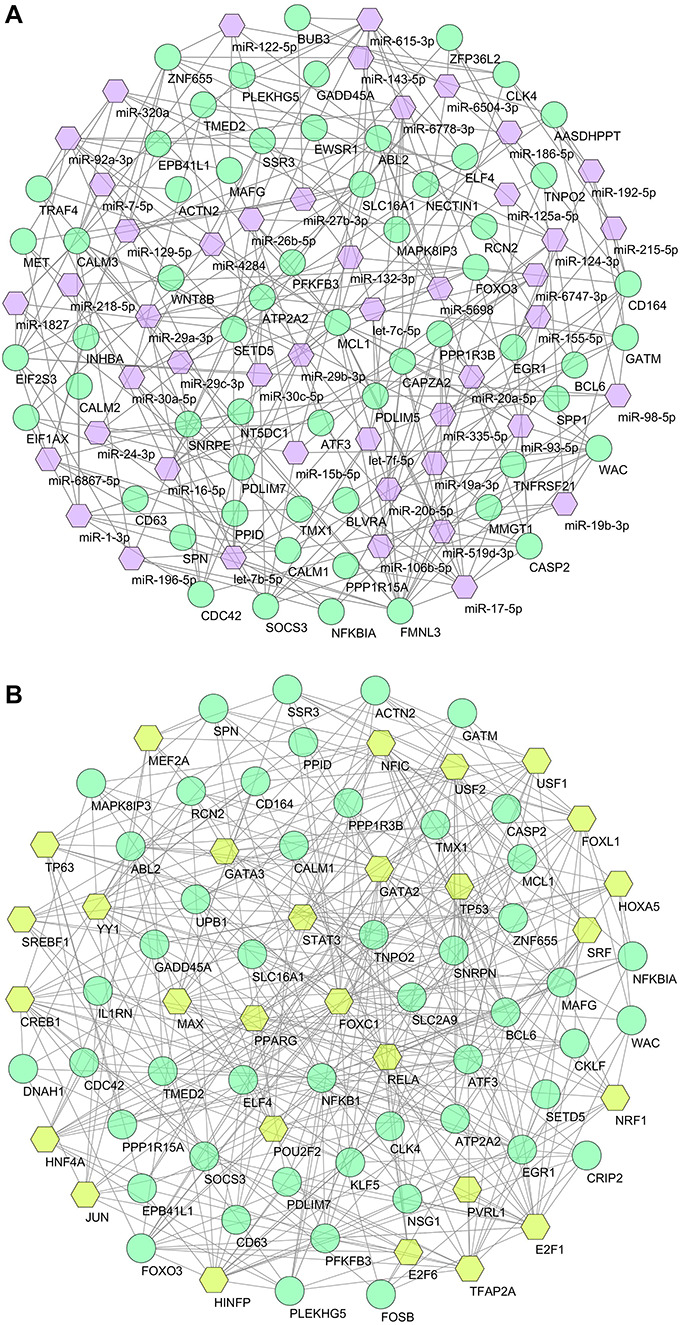
Regulatory networks of gene–biomarker interactions. (A) Interacting network of miRNAs and (B) interacting network of TFs. Both networks were filtered with degree centrality ≥5 and betweenness centrality ≥35, where circles are dysregulated genes (turquoise) and hexagons are associated miRNAs (violet) or TFs (lime).
Potential drugs and chemical agents
To identify potent drugs and antiviral agents, we used shared DEGs to analyze their interactions with different drugs and chemical agents. Using PDI approach, we found 13 drugs that act against the MET gene (Supplementary File S6). Besides, the PCI method predicted 31 potential chemical agents related to 52 genes as shown in Figure 7. Among the 13 drugs, most of them were kinase inhibitors such as antibiotic K-252A, cabozantinib, amuvatinib, crizotinib and SGX-523. Out of the total 31 chemicals, relevant chemical agents were cyclosporine, quercetin, resveratrol, (+)-JQ1 compound, SB-431542 and dorsomorphin.
Figure 7.
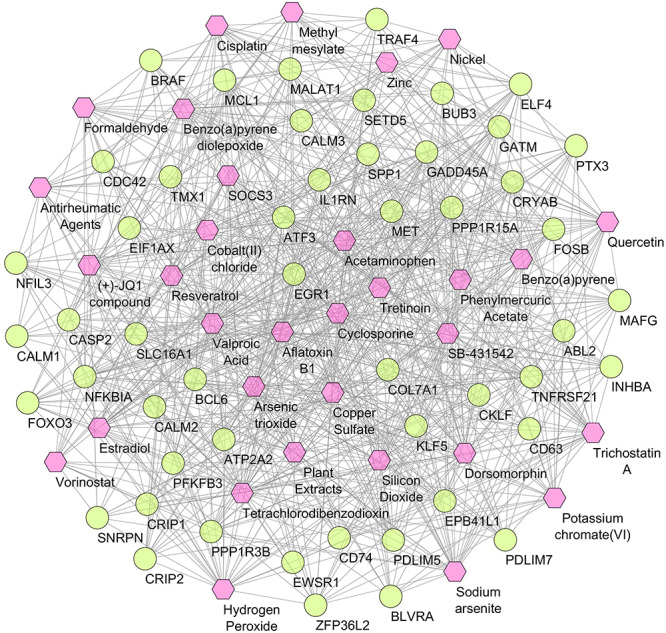
Protein–chemical interaction network. Of the total 83 nodes, circles represent the shared dysregulated genes (lime), while hexagons indicate the interacting chemical agents (pink). Degree centrality ≥20 and betweenness centrality ≥20 were considered for this network.
Discussion
To understand the genetics of COVID-19, we developed a network-based framework where intra- and interconnections among genes and proteins may provide valuable information on their roles in developing certain diseases or conditions. In this study, we first established the genomic closeness and position of 2019-nCoV within the Coronaviridae family using the phylogenic approach. In the phylogram, both 2019-nCoV and SARS-CoV formed two clades in a single node which confirms their close relationship. However, it also revealed that 2019-nCoV is not descended from SARS-CoV; rather they have a common ancestral origin. This observation is in line with another recent study that led to the naming of 2019-nCoV as SARS-CoV-2 [7]. Due to higher genetic similarity, all SARS-like coronaviruses (i.e. 2019-nCoV, SARS-CoV, MERS-CoV and CoV-HKU1) formed a separate branch from all other coronaviruses included in Beta-CoV 1, such as HCoV-OC43. This relationship is in line with an earlier report [108]. Furthermore, a recent study reveals that 2019-nCoV shares the highest (79.7%) nucleotide sequence identity with SARS-CoV [109]. This high degree of genomic similarity could be responsible for their analogous pathogenic behavior and clinical disease presentation. Therefore, SARS-CoV might share pathogenic signatures and pathways involved in the progression of COVID-19 upon 2019-nCoV infection, thereby providing the basis for further investigation.
Based on this guiding principle, we identified 87 significant DEGs shared among SARS-CoV infection and 4 COVID-19-related disorders. These genes could be associated with 2019-nCoV infections as well. Therefore, we reviewed the molecular pathogenesis of 44 genes (out of 87 DEGs) that are involved in COVID-19 and other viral infections, especially SARS-like viruses. Intriguingly, the expression pattern and pathogenetic roles of most genes are indifferent to COVID-19 and other viral infections, except for few. In a recent study, for example, the increased level of inflammatory chemokines such as IL-1 and TNF-α has been observed in COVID-19 patients [22]. As we speculated, the upregulation of NFKBIA and FOXO3 could be involved in this process by being the key modulators of IL-1 and TNF-α-related chemokine signaling [81, 110, 111]. In addition, the association of NFKBIA gene in SARS-CoV and MERS-CoV infections was found in a previous study [79]. Another gene Abl2 is important for cell entry and replication of SARS-like viruses [47, 48]. Furthermore, LTB4 is highly prevalent in type 1 diabetic patients, and overexpression of this gene triggers acute asthma attacks in SARS-related diseases [61, 62]. This also explains the increased susceptibility of diabetic patients to COVID-19. Therefore, they are not merely associated with SARS-CoV rather may have broader roles that might include their involvement in COVID-19 pathogenesis. In addition, we have characterized GO and pathways that are regulated by these genes. About 22 pathways were identified that are linked to SARS-CoV infection and could possibly be involved in COVID-19 as well. For instance, high expression of IL-6 signaling [22, 112] and IFN-γ production [113] was observed in COVID-19 sufferers. Therefore, these pathways could be linked to COVID-19 and may reveal important checkpoints for drug targets. We also identified 25 important GO terms that are related to the clinical conditions of COVID-19. Therefore, the regulation of these pathways might be important in COVID-19 progression. For example, interferon-MAPK pathway-mediated adaptive immune response against COVID-19 was observed through blood single-cell immune profiling [2]. Therefore, it is probable that COVID-19 disease progression might be mediated by those and other significant pathways.
To validate our findings, significant DEGs were employed to predict gene–disease association using benchmark databases. In doing so, we created a GDN network as comorbidities of SARS-CoV, to understand the possible COVID-19 projections and risk factors. Comorbidities may result from pathogenic relationships or common risk factors among concomitant disorders [114]. Apart from the respiratory disease, the anticipated comorbid profiles suggested a wide array of diseases involved in 2019-nCoV infection including cardiac, renal, brain and blood-related problems as well as different types of cancer. For example, 17 genes were found to be associated with blood diseases including thalassemia, thrombocytopenia, hypertension, thrombophilia and leukemia. Strikingly, a recent meta-analysis included thrombocytopenia as a comorbidity of COVID-19 [115]. Very recently, unusual blood clots have been found in multiple organs causing strokes and organ failures [116, 117]. This could be due to thrombophilia, an abnormal blood clotting disorder caused by altered expression of the associated genes upon 2019-nCoV infection. On the other hand, comorbidities related to brain diseases include encephalopathy, dementia and Leigh syndrome. Interestingly, a recent study suggested that COVID-19 could lead to acute hemorrhagic necrotizing encephalopathy, which is in line with our findings [118].
Furthermore, a study suggested hypertension as a potential risk factor in 2019-nCoV infection [119]. Two recent studies from China have shown a strong association between hypertension and COVID-19 deaths (Hazard ratio 1.70 to 3.05), raising concerns that hypertension is confounded by treatment with specific antihypertensive medications such as ACE inhibitors (ACEIs) and angiotensin receptor blockers (ARBs) [120, 121]. In response, the European Society of Cardiology Council on Hypertension made the following statement, ‘The Council on Hypertension strongly recommends that physicians and patients should continue treatment with their usual anti-hypertensive therapy because there is no clinical or scientific evidence to suggest that treatment with ACEIs or ARBs should be discontinued because of the COVID-19 infection [123]’. According to the American College of Cardiology (ACC) clinical bulletin, the fatality rate in COVID-19 is higher in patients with preexisting conditions such as cancer (5.6%), hypertension (6.0%), cardiovascular diseases (10.5%) and diabetes (7.3%) [123–125]. Furthermore, about 16.7% of patients developed arrhythmia, and 7.2% experienced an acute cardiac injury in addition to 2019-nCoV-associated complications [123]. These cardiovascular complications may result from various mechanisms, including relative ischemia, systemic inflammation and pathogen-mediated damage [126]. Interestingly, we identified 20 genes that are involved in different cardiovascular diseases and 16 genes related to diabetes mellitus (type 1 and 2). Evidence to date suggests that people who died of COVID-19 were more likely to have comorbidities such as hypertension, diabetes, cardiovascular disease or chronic lung disease [127, 128]. Further research on these genes could reveal long-term cardiovascular implications of viral infection, thus closing the significant knowledge gaps in the recent COVID-19 outbreak.
Using the shared genes, a PPI network was constructed to pinpoint the key disease-modifying agents in COVID-19 in light of SARS-CoV pathobiology. Hubs are defined as proteins with eight or more interactions, while proteins with less than four interactions are named non-hubs and the rest are intermediately connected [129]. We identified seven hub-proteins (MCL1, NFKBIA, BUB3, MET, EIF2S3, GADD45A and SOCS3) involved in SARS-CoV and COVID-19 diseases. The protein NFKBIA regulates inflammatory responses and plays an important role in the pathogenesis of acute respiratory distress syndrome [130]. Another hub-protein GADD45A is known to play a significant role in lung injury due to inflammation [131, 132]. Furthermore, the expression of SOCS3 downregulated JAK2/STAT3 pathway to promote macrophage polarization that plays a key role in lung inflammation [133]. Therefore, these hub-proteins can be regarded as candidate biomarkers or, if their biological role in COVID-19 is confirmed, as potential drug targets. Furthermore, TFs control the rate of transcription [134], while miRNAs are key players in RNA silencing and regulation of gene expression at the posttranscription level [135]. Hence, both are essential to understand particular disease development. In this regard, this study uncovered relationships among the shared DEGs and their respective TFs and regulatory miRNAs. Herein, we identified several TFs, such as STAT3, NFKB1A, E2F1, TP53, GATA3 CREB1, which are known to involve in viral-mediated acute respiratory diseases [86, 136, 137]. Further, some miRNAs associated with lung diseases (i.e. let-7b-5p and let-7c-5p), asthma (i.e. miR-155-5p, miR-186-5p, miR-16-5p, miR-27b-3p) and pneumonia (i.e. miR-29a-3p and miR-30a-5p) were also found highly prevalent in COVID-19 [86, 138–140].
TFs and miRNAs usually target host proteins to alter their expression during the development of specific diseases. For coronavirus, for instance, miR-98 modulates the host inflammatory response by interacting with STAT3. For other respiratory viruses, let-7b and let-7c target STAT3, while miR-30a influences TP53 [86]. Both are regulatory TFs that are identified as important biomarkers in this study. Furthermore, miR-16 and miR-155 modulate inflammatory responses by targeting the MCL1 and NFKBIA hub-proteins [86]. Interestingly, we also identified four miRNAs (let-7c-5p, miR-125a-5p, miR-27b-3p and miR-98-5p) that are known to target different genes of 2019-nCoV [141, 142]. For instance, let-7c-5p targets ORF1ab gene of 2019-nCoV and inhibits viral replication [142]. Conversely, the spike protein from SARS-CoV may downregulate the expression of miR-98 in bronchoalveolar stem cells to control their differentiation and inflammatory cytokine production. Moreover, we identified two long noncoding RNA (lncRNA) in our shared DEGs, i.e. MALAT1 and LINC00265. These lncRNAs may act in concert with miRNAs to orchestrate the competing endogenous RNA (ceRNA) network in disease progression. For instance, upregulation of MALAT1 ceases miR-155 expression which promotes GATA3 and Th2 cytokine production leading to severe asthma and loss of lung function [90]. On the other hand, LINC00265 inhibits let-7a activity using ceRNA mechanism, which results in the upregulation of IL6 [143]. All these eventually lead to the deterioration of lung tissues and successful viral replication [144].
In addition to regulatory molecules, we identified 13 MET gene-targeting drugs using PDI method that are mostly kinase inhibitors. Kinases play a key role in the airway muscle contractions and in the release of pro-inflammatory mediators [145]. Therefore, our results suggest plausible mechanisms, since most of the proteins are related to inflammatory responses. Nevertheless, we excluded these drugs since they have multiple known side effects including hypertension, diarrhea, anorexia and nausea [146]. Furthermore, we anticipated PCI network to identify the chemical agents that could act as potential antagonists. Among the total 31 chemical agents, we could find the potent antagonists by targeting the hub-proteins that are linked to COVID-19 progression as stated above. Previously, several drugs and chemical agents showed potent therapeutic action against 2019-nCoV. For example, remdesivir and chloroquine have been reported to inhibit 2019-nCoV and other SARS-like viruses [147]. Furthermore, a Japanese flu–drug favipiravir showed a significant protective effect against COVID-19 [148]. Moreover, a recent nonrandomized clinical trial showed a promising effect of hydroxychloroquine in concert with azithromycin against 2019-nCoV by inhibiting their genomic replication [149, 150]. Although hydroxychloroquine possesses less toxicity, prolonged and overdose usage of this anti-inflammatory agent can still cause poisoning [151]. Alternatively, the chemical agents identified in this study can be checked against 2019-nCoV or its disease form COVID-19. For example, we identified quercetin which is a potent antiviral and anti-inflammatory agent active against different strains of influenza virus (i.e. H1N1 and H3N2) [152]. Another identified chemical antagonist was resveratrol in our study. Interestingly, resveratrol is a prominent antiviral agent which showed antiviral activity against several human and animal viruses [153, 154]. Most importantly, Lin et al. showed resveratrol-mediated effective inhibition of MERS-CoV due to the downregulation of apoptosis [155]. As we stated earlier, overexpression of NF-κB is central to asthma and other pulmonary diseases [156]. Resveratrol inhibits the expression of NF-κB and related pathways, thereby subduing the conditions [154]. Another major identified compound is cyclosporine, which is known to inhibit the replication of RNA viruses including SARS-CoV, MERS-CoV and CoV-229E by targeting cyclophilins [157, 158]. Cyclophilins regulate the replication of RNA viruses and are usually overexpressed in viral infections as well as in inflammatory diseases such as asthma [157–159]. Importantly, a recent study includes resveratrol and cyclosporine A in the list of therapeutic options for 2019-nCoV [160]. Overall, these data are of clinical interest and may shed light on the cause and progression of COVID-19, as well as any new prospective therapeutic strategies.
Conclusion
The present study highlights molecular insight on potential biomarkers, regulatory components and molecular checkpoints that may help in developing therapeutics to combat the current 2019-nCoV pandemic. Based on gene expression analysis, we highlight significantly expressed genes with possible projections and risk factors associated with COVID-19, in the form of comorbidities. These identified biomarkers could be potential drug targets based on their role in regulatory checkpoints and disease progression. Furthermore, potent chemical agents that are identified could be used to target-specific disease-modifying factors. The comorbidity analysis presented here provides disease mechanistic insight and prognostic features relevant in COVID-19 disease. These data will aid in unlocking new opportunities for novel therapeutics to control this, as well as future viral outbreaks.
Key Points
The 2019-nCoV and SARS-CoV have a common ancestral origin, and their pathobiological profiles could have a number of significantly dysregulated genes in common. These could be compared with upcoming COVID-19 transcriptomes to assess the distinct genetic mechanisms involved in a disease.
Comorbidity profiles and associated genes could be evaluated for their roles in recently observed clinical presentations and future implications of COVID-19.
In protein–protein network analyses, identified hub-proteins can lead us to co-expression partners related to normal and disease states, which can help in the assessment of risk factors related to COVID-19 complications.
TFs and miRNAs identified as biomarkers can be used as prognostic signatures to differentiate patients and healthy individuals. Their role as disease-modifying traits can further be explored.
Signaling pathways that are common to SARS-CoV and COVID-19 presentations can be used as molecular checkpoints to develop therapeutics against 2019-nCoV infection.
Protein–chemical interactome analysis suggests cyclosporine, resveratrol and quercetin as potential chemical antagonists against COVID-19 as well as SARS-like viral infections.
Supplementary Material
Acknowledgements
We thank all those who have contributed in 2019-nCoV genome sequences deposited to the GISAID database (https://www.gisaid.org/) and provided ideas to the Virological website (http://virological.org/).
Zulkar Nain is a Research Fellow at the Department of Genetic Engineering and Biotechnology, East West University, Bangladesh. His research interests are bioinformatics, functional genomics, molecular modelling as well as computer-aided drug and vaccine design.
Humayan Kabir Rana is a Senior Lecturer at the Department of Computer Science and Engineering, Green University of Bangladesh. His research interest includes bioinformatics, machine learning, digital image and signal processing.
Pietro Liò is a Full Professor of Computational Biology and a member of the Artificial Intelligence Group at the University of Cambridge. His research interest focuses on bioinformatics and machine learning related to biological complexity.
Sheikh Mohammed Shariful Islam is a Physician Scientist at the Institute for Physical Activity and Nutrition, Deakin University. His research interest is focused on understanding diabetes and cardiovascular diseases using innovative information technologies.
Matthew A. Summers is a Research Scientist at the Garvan Institute of Medical Research, Australia. His research interests include understanding genetic and molecular mechanisms of rare muscle and bone diseases using cutting-edge single-cell genomic and transcriptomic methods.
Mohammad Ali Moni is a Research Fellow and Conjoint Lecturer at the University of New South Wales, Australia. He received his PhD degree from the University of Cambridge. His research interest encompasses artificial intelligence, machine learning, data science and clinical bioinformatics.
Authors’ contributions
M.A.M. and Z.N. conceived and designed the study; Z.N. performed the computational analyses, prepared the illustrations and wrote the manuscript; H.K.R. helped in the experimental procedure and draft preparation; P.L., M.A.S., S.M.S.I. and M.A.M. contributed in the critical analysis and revision; M.A.M. supervised the whole study. All authors approved the final version of the manuscript.
Conflict of interest
The authors declare no conflict of interests.
Ethical statement
Not applicable.
References
- 1. Zhu N, Zhang D, Wang W, et al. . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, et al. . Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang SF, Tuo JL, Huang XB, et al. . Epidemiology characteristics of human coronaviruses in patients with respiratory infection symptoms and phylogenetic analysis of HCoV-OC43 during 2010-2015 in Guangzhou. PLoS One 2018;13(1):e0191789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andersen KG, Rambaut A, Lipkin WI, et al. . The proximal origin of SARS-CoV-2. Nat Med 2020;89:44–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu JT, Leung K, Bushman M, et al. . Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med 2020;26:506–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Syal K. COVID-19: herd immunity and convalescent plasma transfer therapy. J Med Virol 2020;1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorbalenya AE, Baker SC, Baric RS, et al. . The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 2020;5:536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gheblawi M, Wang K, Viveiros A, et al. . Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system. Circ Res 2020;126:1456–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ge XY, Li JL, Lou YX, et al. . Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013;503(7477):535–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Astuti I, Ysrafil. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab Syndr Clin Res Rev 2020;14:407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rabi FA, Al Zoubi MS, Kasasbeh GA, et al. . SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens 2020;9:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol 2020;38:1–9. [DOI] [PubMed] [Google Scholar]
- 13. Li X, Geng M, Peng Y, et al. . Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal 2020;10:102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ye Z-W, Yuan S, Yuen K-S, et al. . Zoonotic origins of human coronaviruses. Int J Biol Sci 2020;16:1686–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mackenzie JS, Smith DW. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol Aust 2020;41:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lam TT-Y, Jia N, Zhang Y-W, et al. . Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 2020;583:282–5. [DOI] [PubMed] [Google Scholar]
- 17. Shi J, Wen Z, Zhong G, et al. . Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 2020;368(6494):1016–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sia SF, Yan L-M, Chin AWH, et al. . Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 2020;583:834–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sit THC, Brackman CJ, Ip SM, et al. . Infection of dogs with SARS-CoV-2. Nature 2020;1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Halfmann PJ, Hatta M, Chiba S, et al. . Transmission of SARS-CoV-2 in domestic cats. N Engl J Med 2020; NEJMc2013400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen D, Sun J, Zhu J, et al. . Single-cell screening of SARS-CoV-2 target cells in pets, livestock, poultry and wildlife. bioRxiv 2020; 2020.06.13.149690. [Google Scholar]
- 22. Guo Y-R, Cao Q-D, Hong Z-S, et al. . The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil Med Res 2020;7(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Q, Guan X, Wu P, et al. . Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med 2020;382:1199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan JFW, Yuan S, Kok KH, et al. . A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395:514–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brann D, Tsukahara T, Weinreb C, et al. . Non-neural expression of SARS-CoV-2 entry genes in the olfactory epithelium suggests mechanisms underlying anosmia in COVID-19 patients. bioRxiv 2020; 2020.03.25.009084. [DOI] [PMC free article] [PubMed]
- 26. Zhang J-J, Dong X, Cao Y-Y, et al. . Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020;75:1730–41. [DOI] [PubMed] [Google Scholar]
- 27. Bordi L, Nicastri E, Scorzolini L, et al. . Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS-CoV-2), Italy, February 2020. Euro Surveill 2020;25(8):2000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Day M. Covid-19: four fifths of cases are asymptomatic, China figures indicate. BMJ 2020;369:m1375. [DOI] [PubMed] [Google Scholar]
- 29. DeDiego ML, Nieto-Torres JL, Jiménez-Guardeño JM, et al. . Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog 2011;7(10):e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wallihan RG, Suárez NM, Cohen DM, et al. . Molecular distance to health transcriptional score and disease severity in children hospitalized with community-acquired pneumonia. Front Cell Infect Microbiol 2018;8:382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodiño-Janeiro BK, Martínez C, Fortea M, et al. . Decreased TESK1-mediated cofilin 1 phosphorylation in the jejunum of IBS-D patients may explain increased female predisposition to epithelial dysfunction. Sci Rep 2018;8:2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baines KJ, Fricker M, McDonald VM, et al. . Sputum transcriptomics implicates increased p38 signalling activity in severe asthma. Respirology 2019;25(7):709–18. [DOI] [PubMed] [Google Scholar]
- 33. Reghunathan R, Jayapal M, Hsu LY, et al. . Expression profile of immune response genes in patients with severe acute respiratory syndrome. BMC Immunol 2005;6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meier-Kolthoff JP, Klenk HP, Göker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol 2014;64(Pt 2):352–6. [DOI] [PubMed] [Google Scholar]
- 35. Rana HK, Akhtar MR, Islam MB, et al. . Machine learning and bioinformatics models to identify pathways that mediate influences of welding fumes on cancer progression. Sci Rep 2020;10:2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005;21(9):2067–75. [DOI] [PubMed] [Google Scholar]
- 37. Moni MA, Lio P. Genetic profiling and comorbidities of Zika infection. J Infect Dis 2017;216(6):703–12. [DOI] [PubMed] [Google Scholar]
- 38. Zhou G, Soufan O, Ewald J, et al. . NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019;47(W1):W234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Khan A, Fornes O, Stigliani A, et al. . JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 2018;46(D1):D260–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA 2006;12(2):192–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Da HS, Lin FM, Wu WY, et al. . MiRTarBase: A database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 2011;39:D163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wishart DS, Feunang YD, Guo AC, et al. . DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46(D1):D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davis AP, Grondin CJ, Johnson RJ, et al. . The comparative Toxicogenomics database: update 2019. Nucleic Acids Res 2019;47:D948–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shannon P. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuleshov MV, Jones MR, Rouillard AD, et al. . Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim Y, Pierce CM, Robinson LA. Impact of viral presence in tumor on gene expression in non-small cell lung cancer. BMC Cancer 2018;18(1):843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rameshrad M, Ghafoori M, Mohammadpour AH, et al. . A comprehensive review on drug repositioning against coronavirus disease 2019 (COVID19). Naunyn Schmiedebergs Arch Pharmacol 2020;393:1137–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Coleman CM, Sisk JM, Mingo RM, et al. . Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J Virol 2016;90:8924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cava C, Bertoli G, Castiglioni I. In Silico discovery of candidate drugs against Covid-19. Viruses 2020;12(4):404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yeung ML, Yao Y, Jia L, et al. . MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat Microbiol 2016;1(3):16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ravindra NG, Alfajaro MM, Gasque V, et al. . Single-cell longitudinal analysis of SARS-CoV-2 infection in human bronchial epithelial cells. bioRxiv 2020; 2020.05.06.081695. [Google Scholar]
- 52. Liao M, Liu Y, Yuan J, et al. . Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020;26:842–4. [DOI] [PubMed] [Google Scholar]
- 53. Amet T, Son YM, Jiang L, et al. . BCL6 represses antiviral resistance in follicular T helper cells. J Leukoc Biol 2017;102(2):527–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pan G, Bauer JH, Haridas V, et al. . Identification and functional characterization of DR6, a novel death domain-containing TNF receptor. FEBS Lett 1998;431(3):351–6. [DOI] [PubMed] [Google Scholar]
- 55. Horimasu Y, Ishikawa N, Taniwaki M, et al. . Gene expression profiling of idiopathic interstitial pneumonias (IIPs): identification of potential diagnostic markers and therapeutic targets. BMC Med Genet 2017;18(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang Z, Zhou X, Michal JJ, et al. . Reactomes of porcine alveolar macrophages infected with porcine reproductive and respiratory syndrome virus. PLoS One 2013;8(3):e59229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kumar R, Corbett MA, Smith NJC, et al. . Homozygous mutation of STXBP5L explains an autosomal recessive infantile-onset neurodegenerative disorder. Hum Mol Genet 2014;24(7):2000–10. [DOI] [PubMed] [Google Scholar]
- 58. Liu XQ, Cohen JI. The role of PI3K/Akt in human herpesvirus infection: from the bench to the bedside. Virology 2015;479–480:568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soares JAP, Leite FGG, Andrade LG, et al. . Activation of the PI3K/Akt pathway early during Vaccinia and cowpox virus infections is required for both host survival and viral replication. J Virol 2009;83:6883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ma L, Kuai WX, Sun XZ, et al. . Long noncoding RNA LINC00265 predicts the prognosis of acute myeloid leukemia PATIENTS and functions as a promoter by activating pi3k-akt pathway. Eur Rev Med Pharmacol Sci 2018;22(22):7867–76. [DOI] [PubMed] [Google Scholar]
- 61. Pal K, Feng X, Steinke JW, et al. . Leukotriene A4 hydrolase activation and leukotriene B4 production by eosinophils in severe asthma. Am J Respir Cell Mol Biol 2019;60:413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Santos-Bezerra DP, Filgueiras LR, Monteiro MB, et al. . Leukotriene pathway activation associates with poor glycemic control and with cardiovascular autonomic neuropathy in type 1 diabetes. Mediators Inflamm 2020;2020:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Broto C, Dibyajyoti D, Gopalakrishnan B, et al. . Network-based analysis of fatal comorbidities of COVID-19 and potential therapeutics. ChemRxiv 2020. [Google Scholar]
- 64. Srinivas RV, Bernstein H, Oliver C, et al. . Calmodulin antagonists inhibit human immunodeficiency virus-induced cell fusion but not virus replication. AIDS Res Hum Retroviruses 1994;10:1489–96. [DOI] [PubMed] [Google Scholar]
- 65. Naqvi AAT, Fatima K, Mohammad T, et al. . Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim Biophys Acta Mol basis Dis 2020;1866:165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Islam ABMMK, MA-A- KK. Lung biopsy cells transcriptional landscape from COVID-19 patient stratified lung injury in SARS-CoV-2 infection through impaired pulmonary surfactant metabolism. BioRxiv 2020; 2020.05.07.082297. [Google Scholar]
- 67. Li SW, Wang CY, Jou YJ, et al. . SARS coronavirus papain-like protease induces Egr-1-dependent up-regulation of TGF-β1 via ROS/p38 MAPK/STAT3 pathway. Sci Rep 2016;6:25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Salimi S, Hamlyn JM. COVID-19 and crosstalk between the hallmarks of aging. J Gerontol 2020; Series A: glaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chen L, Song Y, He L, et al. . MicroRNA-223 promotes type I interferon production in antiviral innate immunity by targeting forkhead box protein O3 (FOXO3). J Biol Chem 2016;291:14706–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. He B, Garmire L. Prediction of repurposed drugs for treating lung injury in COVID-19. F1000Research 2020;9:609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mizutani T. Signaling pathways of SARS-CoV in vitro and in vivo. Mol Biol SARS-Coronavirus 2010;305–22. [Google Scholar]
- 72. Blanco-Melo D, Nilsson-Payant BE, Liu W-C, et al. . Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020;181(5):1036–45e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Linko R, Hedger MP, Pettilä V, et al. . Serum activin A and B, and follistatin in critically ill patients with influenza A(H1N1) infection. BMC Infect Dis 2014;14:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang HF, Chen L, Luo J, et al. . KLF5 is involved in regulation of IFITM1, 2, and 3 genes during H5N1 virus infection in A549 cells. Cell Mol Biol 2016;62(13):65–70. [DOI] [PubMed] [Google Scholar]
- 75. Kim SH, In Choi H, Choi MR, et al. . Epigenetic regulation of IFITM1 expression in lipopolysaccharide-stimulated human mesenchymal stromal cells. Stem Cell Res Ther 2020;11:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Prates ET, Garvin MR, Pavicic M, et al. . Functional immune deficiency syndrome via intestinal infection in COVID-19. BioRxiv 2020; 2020.04.06.028712. [Google Scholar]
- 77. Kostrzewski T, Borg AJ, Meng Y, et al. . Multiple levels of control determine how E4bp4/Nfil3 regulates NK cell development. J Immunol 2018;200:1370–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kong SL, Chui P, Lim B, et al. . Elucidating the molecular physiopathology of acute respiratory distress syndrome in severe acute respiratory syndrome patients. Virus Res 2009;145:260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moni MA, Liò P. Network-based analysis of comorbidities risk during an infection: SARS and HIV case studies. BMC Bioinformatics 2014;15(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tourniaire F, Romier-Crouzet B, Lee JH, et al. . Chemokine expression in inflamed adipose tissue is mainly mediated by NF-κB. PLoS One 2013;8(6):e66515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sandell LJ, Xing X, Franz C, et al. . Exuberant expression of chemokine genes by adult human articular chondrocytes in response to IL-1β. Osteoarthr Cartil 2008;16:1560–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Poppe M, Wittig S, Jurida L, et al. . The NF-κB-dependent and -independent transcriptome and chromatin landscapes of human coronavirus 229E-infected cells. PLoS Pathog 2017;13:e1006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Emanuel W, Kirstin M, Vedran F, et al. . Bulk and single-cell gene expression profiling of SARS-CoV-2 infected human cell lines identifies molecular targets for therapeutic intervention. BioRxiv 2020; 2020.05.05.079194. [Google Scholar]
- 84. Villena J, Kitazawa H. The modulation of mucosal antiviral immunity by Immunobiotics: could they offer any benefit in the SARS-CoV-2 pandemic? Front Physiol 2020;11:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Takahashi K, Koga K, Linge HM, et al. . Macrophage CD74 contributes to MIF-induced pulmonary inflammation. Respir Res 2009;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Leon-Icaza SA, Zeng M. Rosas-Taraco AG. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA 2019;1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Saksena S, Theegala S, Bansal N, et al. . Mechanisms underlying modulation of monocarboxylate transporter 1 (MCT1) by somatostatin in human intestinal epithelial cells. Am J Physiol Liver Physiol 2009;297:G878–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Radzikowska U, Ding M, Tan G, et al. . Distribution of ACE2, CD147, CD26 and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sumimoto H, Imabayashi F, Iwata T, et al. . The BRAF-MAPK signaling pathway is essential for cancer-immune evasion in human melanoma cells. J Exp Med 2006;203(7):1651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Liang Z, Tang F. The potency of lncRNA MALAT1/miR-155/CTLA4 axis in altering Th1/Th2 balance of asthma. Biosci Rep 2020;40(2):BSR20190397. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 91. Guzzi PH, Mercatelli D, Ceraolo C, et al. . Master regulator analysis of the SARS-CoV-2/human Interactome. J Clin Med 2020;9(4):982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ouyang J, Hu J, Chen JL. lncRNAs regulate the innate immune response to viral infection. Wiley Interdiscip Rev RNA 2016;7(1):129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hu W, Yen YT, Singh S, et al. . SARS-CoV regulates immune function-related gene expression in human monocytic cells. Viral Immunol 2012;25(4):277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Karunakaran KB, Balakrishnan N, Ganapathiraju MK. Interactome of SARS-CoV-2 / nCoV19 modulated host proteins with computationally predicted PPIs, Research Square 2020.
- 95. Lund RJ, Löytömäki M, Naumanen T, et al. . Genome-wide identification of novel genes involved in early Th1 and Th2 cell differentiation. J Immunol 2007;178(6):3648–60. [DOI] [PubMed] [Google Scholar]
- 96. Wang J, Luo Q, Chen R, et al. Susceptibility analysis of COVID-19 in smokers based on ACE2. 2020, 1–8.
- 97. Hachim IY, Hachim MY, Talaat IM, et al. . The molecular basis of gender variations in mortality rates associated with the novel coronavirus (COVID-19) outbreak. 2020. (Preprint); 2020050364. [DOI] [PMC free article] [PubMed]
- 98. Hemmat N, Derakhshani A, Bannazadeh Baghi H, et al. . Neutrophils, crucial, or harmful immune cells involved in coronavirus infection: a bioinformatics study. Front Genet 2020;11:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Han B, Ma X, Zhang J, et al. . Protective effects of long pentraxin PTX3 on lung injury in a severe acute respiratory syndrome model in mice. Lab Invest 2012;92:1285–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang Z, Xu X. scRNA-seq profiling of human testes reveals the presence of the ACE2 receptor, A target for SARS-CoV-2 infection in spermatogonia, leydig and sertoli cells. Cells 2020;9(4):920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Oakland TE, Haselton KJ, Randall G. EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication. J Virol 2013;87(12):6625–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wong HH, Kumar P, Tay FPL, et al. . Genome-wide screen reveals valosin-containing protein requirement for coronavirus exit from endosomes. J Virol 2015;89(21):11116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Coperchini F, Chiovato L, Croce L, et al. . The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev 2020;53:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM, et al. . Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res 2014;194:124–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Chen Y, Zhou J, Cheng Z, et al. . Functional variants regulating LGALS1 (galectin 1) expression affect human susceptibility to influenza A(H7N9). Sci Rep 2015;5:8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Z, Kong L, Tan S, et al. . Zhx2 accelerates sepsis by promoting macrophage glycolysis via Pfkfb3. J Immunol 2020;204(8):2232–41. [DOI] [PubMed] [Google Scholar]
- 107. Jiang H, Shi H, Sun M, et al. . PFKFB3-driven macrophage glycolytic metabolism is a crucial component of innate antiviral defense. J Immunol 2016;197(7):2880–90. [DOI] [PubMed] [Google Scholar]
- 108. Liò P. Phylogenomics and bioinformatics of SARS-CoV. Trends Microbiol 2004;12:106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhou Y, Hou Y, Shen J, et al. . Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 2020;6:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Joseph J, Ametepe ES, Haribabu N, et al. . Inhibition of ROS and upregulation of inflammatory cytokines by FoxO3a promotes survival against salmonella typhimurium. Nat Commun 2016;7:12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tourniaire F, Romier-Crouzet B, Lee JH, et al. . Chemokine expression in inflamed adipose tissue is mainly mediated by NF-κB. PLoS One 2013;8:e66515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhou Y, Fu B, Zheng X, et al. . Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci Rev 2020;7(6):998–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Liu C, Zhou Q, Li Y, et al. . Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci 2020;6(3):315–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hossain MA, Asa TA, Rahman MR, et al. . Network-based approach to identify key candidate genes and pathways shared by thyroid cancer and chronic kidney disease. Informatics Med Unlocked 2019;16:100240. [Google Scholar]
- 115. Lippi G, Plebani M, Michael Henry B. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin Chim Acta 2020;506:145–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Oxley TJ, Mocco J, Majidi S, et al. . Large-vessel stroke as a presenting feature of Covid-19 in the young. N Engl J Med 2020;382(20):e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Klok FA, Kruip MJHA, Meer NJM, et al. . Incidence of thrombotic complications in critically ill ICU patients with COVID-19. Thromb Res 2020;20:30120–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Poyiadji N, Shahin G, Noujaim D, et al. . COVID-19–associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology 2020;296(2):E119–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir 2020;2600:30116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wu C, Chen X, Cai Y, et al. . Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020;180(7):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhou F, Yu T, Du R, et al. . Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Simone G. Position statement of the ESC Council on hypertension on ACE-inhibitors and angiotensin receptor blockers. Eur Soc Cardiol 2020. [Google Scholar]
- 123. Wang D, Hu B, Hu C, et al. . Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J Am Med Assoc 2020;323:1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Chen N, Zhou M, Dong X, et al. . Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395(10223):507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med 2020;8(4):e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Xiong T-Y, Redwood S, Prendergast B, et al. . Coronaviruses and the cardiovascular system: acute and long-term implications. Eur Heart J 2020;41(19):1798–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Jordan RE, Adab P, Cheng KK. Covid-19: risk factors for severe disease and death. BMJ 2020;368:m1198. [DOI] [PubMed] [Google Scholar]
- 128. Guan W, Liang W, Zhao Y, et al. . Comorbidity and its impact on 1590 patients with Covid-19 in China: a nationwide analysis. Eur Respir J 2020;55(5):2000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Ekman D, Light S, Björklund ÅK, et al. . What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol 2006;7:R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Zhai R, Zhou W, Gong MN, et al. . Inhibitor κB-α haplotype GTC is associated with susceptibility to acute respiratory distress syndrome in Caucasians. Crit Care Med 2007;35:893–8. [DOI] [PubMed] [Google Scholar]
- 131. Meyer NJ, Huang Y, Singleton PA, et al. . GADD45a is a novel candidate gene in inflammatory lung injury via influences on Akt signaling. FASEB J 2009;23(5):1325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Mitra S, Wade MS, Sun X, et al. . GADD45a promoter regulation by a functional genetic variant associated with acute lung injury. PLoS One 2014;9:e100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chi X, Ding B, Zhang L, et al. . lncRNA GAS5 promotes M1 macrophage polarization via miR-455-5p/SOCS3 pathway in childhood pneumonia. J Cell Physiol 2019;234:13242–51. [DOI] [PubMed] [Google Scholar]
- 134. Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol 1997;29:1305–12. [DOI] [PubMed] [Google Scholar]
- 135. Ambros V. The functions of animal microRNAs. Nature 2004;431(7006):350–5. [DOI] [PubMed] [Google Scholar]
- 136. Krug N, Hohlfeld JM, Kirsten AM, et al. . Allergen-induced asthmatic responses modified by a GATA3-specific DNAzyme. N Engl J Med 2015;372:1987–95. [DOI] [PubMed] [Google Scholar]
- 137. Bartel S, Schulz N, Alessandrini F, et al. . Pulmonary microRNA profiles identify involvement of Creb1 and Sec14l3 in bronchial epithelial changes in allergic asthma. Sci Rep 2017;7:46026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Herrera-Rivero M, Zhang R, Heilmann-Heimbach S, et al. . Circulating microRNAs are associated with pulmonary hypertension and development of chronic lung disease in congenital diaphragmatic hernia. Sci Rep 2018;8:10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kho AT, Sharma S, Davis JS, et al. . Circulating MicroRNAs: association with lung function in asthma. PLoS One 2016;11(6):e0157998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Huang F, Zhang J, Yang D, et al. . MicroRNA expression profile of whole blood is altered in adenovirus-infected pneumonia children. Mediators Inflamm 2018;2018:2320640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Sardar R, Satish D, Birla S, et al. . Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. BioRxiv 2020; 2020.03.21.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Demirci MDS, Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ 2020;8:e9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Cai B, Yang B, Huang D, et al. . STAT3-induced up-regulation of lncRNA NEAT1 as a ceRNA facilitates abdominal aortic aneurysm formation by elevating TULP3. Biosci Rep 2020;40(1): BSR20193299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Mallick B, Ghosh Z, Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in Bronchoalveolar stem cells. PLoS One 2009;4:e7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Barnes PJ. Kinases as novel therapeutic targets in asthma and chronic obstructive pulmonary disease. Pharmacol Rev 2016;68:788–815. [DOI] [PubMed] [Google Scholar]
- 146. Ariyawutyakorn W, Saichaemchan S, Varella-Garcia M. Understanding and targeting MET Signaling in solid Tumors - are we there yet? J Cancer 2016;7:633–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Wang M, Cao R, Zhang L, et al. . Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res 2020;30:269–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Cai Q, Yang M, Liu D, et al. . Experimental treatment with Favipiravir for COVID-19: an open-label control study. Engineering 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Liu J, Cao R, Xu M, et al. . Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 2020;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Gautret P, Lagier J-C, Parola P, et al. . Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents 2020;105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Liu J, Cao R, Xu M, et al. . Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov 2020;6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Wu W, Li R, Li X, et al. . Quercetin as an antiviral agent inhibits influenza a virus (IAV) entry. Viruses 2015;8(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Abba Y, Hassim H, Hamzah H, et al. . Antiviral activity of resveratrol against human and animal viruses. Adv Virol 2015;2015:184241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Zhao X, Cui Q, Fu Q, et al. . Antiviral properties of resveratrol against pseudorabies virus are associated with the inhibition of IκB kinase activation. Sci Rep 2017;7:8782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Lin SC, Ho CT, Chuo WH, et al. . Effective inhibition of MERS-CoV infection by resveratrol. BMC Infect Dis 2017;17(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Schuliga M. NF-kappaB signaling in chronic inflammatory airway disease. Biomolecules 2015;5(3):1266–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Wilde AH, Zevenhoven-Dobbe JC, Meer Y, et al. . Cyclosporin A inhibits the replication of diverse coronaviruses. J Gen Virol 2011;92:2542–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Dyall J, Gross R, Kindrachuk J, et al. . Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs 2017;77(18):1935–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Nigro P, Pompilio G, Capogrossi MC. Cyclophilin A: a key player for human disease. Cell Death Dis 2013;4(10):e888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov 2020;19:149–50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


