Abstract
Background
The ideal severe acute respiratory syndrome coronavirus 2 (SARs-CoV-2) testing method would be accurate and also be patient-performed to reduce exposure to healthcare workers. The aim of this study was to compare patient-performed testing based on a morning saliva sample with the current standard testing method, healthcare worker-collected sampling via a nasopharyngeal swab (NPS).
Methods
This was a prospective single center study which recruited 217 asymptomatic adult male participants in a coronavirus disease 2019 (COVID-19) quarantine center who had tested positive for SARS-CoV-2 8–10 days prior to isolation. Paired NPS and saliva specimens were collected and processed within 5 hours of sample collection. Real time reverse transcription polymerase chain reaction (RT-PCR) targeting Envelope (E) and RNA-dependent RNA polymerase (RdRp) genes was performed and the results were compared.
Results
Overall, 160 of the 217 (74%) participants tested positive for COVID-19 based on saliva, NPS, or both testing methods. The detection rate for SARS-CoV-2 was higher in saliva compared to NPS testing (93.1%, 149/160 vs 52.5%, 84/160, P < .001). The concordance between the 2 tests was 45.6% (virus was detected in both saliva and NPS in 73/160), whereas 47.5% were discordant (87/160 tested positive for 1 whereas negative for the other). The cycle threshold (Ct) values for E and RdRp genes were significantly lower in saliva specimens compared to NP swab specimens.
Conclusions
Our findings demonstrate that saliva is a better alternative specimen for detection of SARS-CoV-2. Taking into consideration, the simplicity of specimen collection, shortage of PPE and the transmissibility of the virus, saliva could enable self-collection for an accurate SARS-CoV-2 surveillance testing.
Keywords: COVID-19, SARS-CoV-2, saliva, nasopharyngeal swab, pandemic
Saliva has better detection rate of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) via reverse transcription polymerase chain reaction (RT-PCR) assay in comparison to nasopharyngeal swab. Saliva specimen collection is noninvasive and reduce hazards exposure among healthcare workers during sampling. Saliva could enable self-collection for accurate SARS-CoV-2 surveillance testing.
(See the Editorial Commentary by Ali and Sweeney on pages e357–8.)
A pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is spreading across the world and human to human spread has been confirmed [1, 2]. The number of infected SARS-CoV-2 cases in Malaysia has been rising dramatically since the beginning of the spread despite mitigation measures. Healthcare worker are at the frontline of the coronavirus disease 2019 (COVID-19) outbreak response, making them vulnerable to the infection. Up until 23 April 2020, 325 healthcare workers have been confirmed to have contracted SARS-CoV-2. Although none of their source of infection has been linked to the management or treatment of SARS-CoV-2 patient, source of infection among 30% these healthcare workers are yet to be discovered [3].
The virus has been detected in various clinical specimen such as bronchoalveolar lavage [4], sputum [5], saliva [6], throat [7], stool, nasopharyngeal (NPS), oropharyngeal (OPS) swabs, and blood [8]. The current standard sampling techniques such as NPS and OPS used for surveillance and serial monitoring of infected patients are exposing healthcare workers to SARS-CoV-2 virus and other unknown pathogens via aerosols from swabbing and jeopardizing physical distancing. At the same time, the collection of these specimen types causes discomfort and minor injuries such as bleeding and ulceration of mucosal layer, especially in patients with predisposing factors.
Saliva specimens have demonstrated high concordance rate of >90% with NPS in the detection of coronavirus. In addition to that, some studies have used saliva specimen in surveillance (screening coronavirus) and serial monitoring of viral load [6, 9] has demonstrated the present of coronavirus in saliva but not in nasopharyngeal aspirate. Moreover, saliva specimens can be obtained noninvasively, simply by spitting into a sterile container. Saliva collection is regarded as a safer noninvasive specimen alternative to NPS or OPS. Other than minimizing exposure of the healthcare workers to hazards, it is self-collected, requires no special chilled media for transportation of samples, and less specimen degradation from delay in processing. Thus, this would maintain physical distancing and minimizing the chance of exposing front-liners to the virus.
The exploration on comparability of saliva versus NPS will assist in sampling protocol, the management of patients and reduce hazards exposure among healthcare workers. Therefore, we assessed the comparability between saliva and NPS specimens for SARS-CoV-2 detection via reverse transcription polymerase chain reaction (RT-PCR).
MATERIALS AND METHODS
Study Design, Participants and Setting
This prospective single center diagnostic study was conducted among 217 individuals who were tested positive for SARS-CoV-2 via NPS at a COVID-19 quarantine center, MAEPS. These selected individuals were on days 8–10 of isolation during the sampling. The inclusion criteria were those participants above 18 years old and able to obey commands. Individuals with respiratory aid were excluded. Assent and written informed consent were obtained from study participants.
Participants’ sociodemographic and symptoms at the time of sampling were collected. As a standard protocol, NPS from individuals were collected using sterile flocked swab and placed in sterile tube containing viral transport medium (VTM). Before collecting swabs, individuals were asked to provide self-collected deep throat saliva sample in a sterile collection container. Instruction on self-collected deep throat saliva was announced to participants a day prior. Briefly, upon waking up, the individuals were instructed to avoid food, water, and brushing of teeth before the collection of 2 mL of saliva. The time span taken between saliva collection and NPS was approximately 3 hours. All samples were stored at room temperature and transported to research lab at Institute for Medical Research, Kuala Lumpur, within 5 hours of sample collection for further processing.
SARS-CoV-2 Detection via RT-PCR Assay
On arrival at the research lab, all clinical specimens were inactivated at 65ºC for 1 hour in the Biosafety cabinet. Total nucleic acid extraction was performed using the MagNA Pure 96 system with the MagNA Pure 96 DNA and Viral NA Small Volume extraction kit (Roche Diagnostic GmBH, Germany), from 200 μL of viral transport medium containing the NPS or 200 μL of saliva. Extracted RNA was eluted in 50 μL of elution buffer. For SARS-CoV-2 RNA detection, 5 μL of RNA template was tested using 1-step RT-PCR of Real-Q 2019 nCoV detection kit (Biosewoom, Inc, South Korea), which has been certified for in vitro diagnostic product (IVD) use by International Medical Device Regulators Forums (IMDRF) jurisdiction. This kit uses the SARS-CoV-2 probe and primer targeting E-gene and RdRp of SARS-CoV-2 and human RNase gene as an internal control. Samples were classified as positive for SARS-CoV-2 when both E-gene and RdRp primer-probe sets were detected at cycle threshold (Ct) value of <38.
Data Processing and Analysis
Data were analyzed for normality and descriptive statistics were presented as a number (%) for categorical variables and mean ± standard deviation (SD) or median (interquartile range [IQR]) for continuous variables. McNemar’s test was used to compare the detection rate for two sampling methods in terms of the number of patients. Agreement between NPS and saliva was performed using κ statistics. Ct values were compared using paired t test. The correlation of Ct values between NPS and saliva was assessed using Pearson correlation coefficient. A P value < .05 was considered statistically significant. All statistical analysis was performed using jamovi software, version 1.2.22.0 [10].
RESULTS
Patients’ Characteristics
All participants were male recruits and asymptomatic at the time of sampling. The median age of the participant was 27 (IQR: 18–36) years.
Comparison of SARS-CoV-2 Detection Between Saliva and NPS
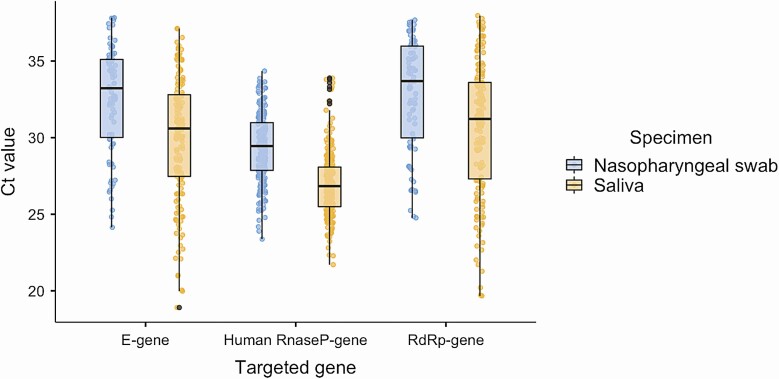
Among the 217 eligible participants, 73.7% (160/217) participants were tested positive for SARS-CoV-2 either from saliva, NPS, or both. There was an overall significant fair agreement between saliva and NPS (73.7%; 160/217, κ 0.260, 95% CI .158–.363, P < .001). Among patients with concordant results, 45.6% (73/160) had virus detected in both saliva and NPS. Eighty-seven patients had discordant results between saliva and NPS via SARS-CoV-2 RT-PCR assay, by which 47.5% (76/160) patients with virus detected in saliva but not in NPS and 6.9% (11/160) patients with virus detected in NPS but not in saliva. The detection rate of SARS-CoV-2 virus in saliva was higher than of NPS and was statistically significant (93.1%; 149/160 vs 52.5%; 84/160; P < .001). The Human RNase P gene was detectable in all specimens. The median (IQR) Ct values of E and RdRp genes were 30.6 (27.5–32.8) and 31.2 (27.3–33.6), respectively in saliva specimens, and 33.2 (30.0–35.1) and 33.7 (30.0–36.0), respectively, in nasopharyngeal swabs (Figure 1).
Figure 1.
Boxplots of SARS-CoV-2 Ct value (mean and interquartile range) of both E-gene and RdRp-gene of all the positive specimens. Ct values of both genes were lower in the saliva than nasopharyngeal swabs of studied asymptomatic individuals. Also included are Ct values of internal control (Human RnaseP-gene) of all the samples in both specimens. Abbreviations: Ct, cycle threshold; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Viral Load Analysis
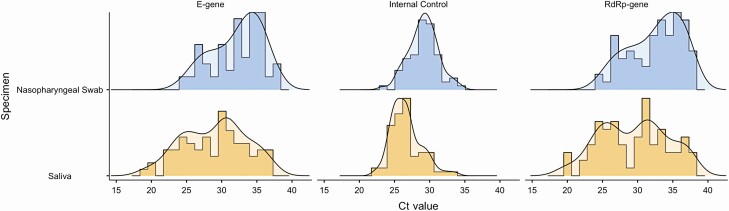
Among the 73 individuals with concordant results, the Ct values were significantly lower in saliva than those in NPS for both E-gene and RdRp-gene of SARS-CoV-2 virus (P < .05). There was significant correlation between saliva and NPS for the Ct values for SARS-CoV-2 (E-gene, r2 0.329; RdRp-gene, r2 0.352 with P < .05). Saliva shares large distribution of Ct value in comparison to NPS. Meanwhile, most NPS Ct values were edging the upper limit (Figure 2).
Figure 2.
Histogram represents distribution of Ct value of concordant (matched) specimens (n = 73) compared between specimen types and the targeted genes. Saliva has large distribution of Ct value of all the targeted genes in comparison to NPS. Meanwhile, nasopharyngeal swab shares most density of the histogram edging the upper limit. Abbreviation: Ct, cycle threshold.
DISCUSSION
Obtaining an optimal clinical specimen for detection of SARS-CoV-2 is central in controlling the global pandemic. Less invasive, good safety measure, amicable, cost effective, timely, and brief are among the major principles of clinical specimen collection during the global pandemic [11, 12]. In this study, we showed a comparable detection rate of SARS-CoV-2 between saliva and nasopharyngeal swabs as recommended by interim guidelines for clinical specimens of COVID-19 testing [13].
This study showed the value of testing a saliva sample as a noninvasive method of detection of SARS-CoV-2. The RT-PCR assay demonstrates high detection rate of SARS-CoV-2 in saliva and comparable performance to the current standard of nasopharyngeal swab. The Cohen’s κ coefficient value showed a fair (0.21–0.40) agreement of the diagnosis between standard nasopharyngeal swab and the saliva sample [14]. However, the Cohen’s κ value in this study is weaker than in a previous study [15]. The lower agreement of these 2 sampling methods could have several reasons. It can be attributed to larger size of discordant results between sampling methods. At the same time, the methodology (study design) and disease prevalence could ascribe the lower Cohen’s κ value in this study [16].
The overall detection rate from saliva samples was higher and significant (P < .05) than the reference standard for SARS-CoV-2 testing via RT-PCR assay. This finding reflects that salivary gland and tongue are possibly the major sites for SARS-CoV-2 viral replication and shedding as these tissues express ACE2 receptor for the viral attachment [17]. It is also possible that the virus migrates from the nasopharynx or lower respiratory tract to the oral cavity as described in a previous study [18]. In addition, it can be hypothesized that the inoculum size of nasopharyngeal swab may not be sufficient for viral transfer. But this hypothesis is yet to be investigated.
A few studies have compared the viral load between nasopharyngeal swab and saliva specimens Wyllie et al [19] has demonstrated that viral loads in saliva is higher than nasopharyngeal swab. Although Williams et al [20] and Becker et al [21] have demonstrated that saliva is less sensitive in comparison to nasopharyngeal swab. Meanwhile, Chau et al [22] and Jamal et al [23] demonstrated equivalent viral load between nasopharyngeal swab and saliva among symptomatic patients but lower in saliva of asymptomatic individuals. Contrary to that, our results showed significant difference in detection of SARS-CoV-2 between these 2 sampling methods among asymptomatic individuals. Saliva had a higher detection rate and lower Ct value (high viral load) than nasopharyngeal swab among the concordant results. However, we speculate that these different findings between studies are possibly due to distinct sampling techniques, detection kits, and study population.
Nevertheless, we had 72 individuals with their saliva specimen tested positive for SARS-CoV-2, although they tested negative for nasopharyngeal swab. This could be due to the property of saliva that acts as wide resource for genomic information by preventing RNA decomposition [24]. It is noteworthy that these inconsistent results may be related to timing of sampling, methodology of sample processing, and severity of disease [25]. Nonetheless, this finding has raised concerns on management of the individuals and the transmissibility of SARS-CoV-2 via saliva or other body fluid as demonstrated by Chen et al [26].
Earlier studies have demonstrated that bronchoalveolar lavage fluid and sputum specimens have the highest detection rate for SARS-CoV-2 testing [27]. However, these specimens are not easy to obtain as 80% of infected individuals commonly present with a dry cough at the onset of illness, and they are not suitable for surveillance as large proportion of the population is asymptomatic [28, 29]. Conversely, saliva can be easily obtained, either by self-collected or under guidance for pediatrics population regardless of illness manifestations. Moreover, saliva collection is noninvasive, patient friendly, and applicable for surveillance testing.
In the face of shortages of both swabs and personal protective equipment as described by Ranney et al [30], saliva is an alternative diagnostic specimen for the detection of SARS-CoV-2. Self-collecting saliva can negate the need for direct healthcare worker-patient interaction, reduce the overall nosocomial infection risk, reduce the waiting time in a busy clinical setting and cost efficient by easing the supply demands on swabs, personal protective equipment and manpower. In addition to that, the use of saliva instead of nasopharyngeal swab also enhance recruitment of individual for community surveillance studies [31]. Taking into consideration, the simplicity of specimen collection, shortage of PPE and the transmissibility of the virus, saliva could enable self-collection for accurate large-scale SARS-CoV-2 surveillance testing.
Limitations of This Study
First, we only recruited adult patients. Further evaluation should be conducted in the pediatric population. Second, the spectrum of the disease ranges from asymptomatic to severely ill patients, but our study only focused on homogenously composed of asymptomatic individuals. Therefore, performance of the saliva test for the detection of SARS-CoV-2 among symptomatic remains unknown. The saliva sample collections were not screened microscopically to assess saliva quality. Third, saliva was collected prior nasopharyngeal swab sampling. This is because longer duration required for collection of 2 mL of saliva. For that reason, we believe that walk-in patients’ saliva may not meet the specified standards for saliva collection. Further study is required to assess the detection of SARS-CoV-2 in random saliva collection. And finally, the Ct value in this study portrays a trend in viral load but not the viral copies per mL. This is due to lack of a reliable quantified positive control in our laboratory.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors are grateful to the Institute for Medical Research, Kuala Lumpur and MAEPS Centre for their support and facilities. The authors would like to thank the Director General of Health Malaysia for allowing us to publish our findings. This study obtained approval from the National Health Institute Human Research Ethics Committee (KKM/NIHSEC/P20-1045). Although this study did not involve any types of intervention in diagnosis and treatment, written informed consent was considered necessary. It involves the use of anonymized patient medical data. Only demographic data were obtained. Therefore, no breach of privacy or confidentiality of patient. This study reviewed human patients only, and no animals were involved in any aspect of the study.
Financial support. This study was supported by a grant from the National Institute of Malaysia, Ministry of Health, Malaysia (NMRR-20-860-54884).
Potential conflicts of Interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Phan T. Novel coronavirus: from discovery to clinical diagnostics. Infect Genet Evol 2020; 79:104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishiura H, Linton NM, Akhmetzhanov AR, et al. . Initial cluster of novel coronavirus (2019-nCoV) infections in Wuhan, China, is consistent with substantial human-to-human transmission. J Clin Med 2020; 9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tharanya Arumugam RK. 325 medical workers test positive for Covid-19. News Strait Times, NST [Internet]. 2020. [cited 2020 Jul 29]. Available at: https://www.nst.com.my/news/nation/2020/04/586972/325-medical-workers-test-positive-covid-19 [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, et al. ; China Novel Coronavirus Investigating and Research Team . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020; 382:727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin X, Gong Z, Xiao Z, Xiong J, Fan B, Liu J. Novel coronavirus pneumonia outbreak in 2019: computed tomographic findings in two cases. Korean J Radiol 2020; 21:365–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To KK-WW, Tsang OT-YY, Yip CC-Y, et al. . Consistent detection of 2019 novel coronavirus in Saliva. Clin Infect Dis. 2020:4–6. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7108139/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastola A, Sah R, Rodriguez-Morales AJ, et al. . The first 2019 novel coronavirus case in Nepal. Lancet Infect Dis 2020; 20:279–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Du RH, Li B, et al. . Molecular and serological investigation of 2019 nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect 2020; 9:386–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.To KKW, Yip CCY, Lai CYW, et al. . Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect 2019; 25:372–8. [DOI] [PubMed] [Google Scholar]
- 10.The jamovi project. jamovi - Stats. Open. Now. [Internet]. Version 1.2.2020. [cited 2020 Jun 22]. Available at: https://www.jamovi.org/.
- 11.World Health Organization. Guidelines for the collection of clinical specimens during field investigation of outbreaks WHO/CDS/CSR/EDC/2000.4. World Health Organization, 2000:1–51. Available at: https://www.who.int/ihr/publications/WHO_CDS_CSR_EDC_2000_4/en/. Accessed 22 June 2020. [Google Scholar]
- 12.Wilson ML. General principles of specimen collection and transport. Clin Infect Dis 1996; 22:766–77. [DOI] [PubMed] [Google Scholar]
- 13.CDC. Interim guidelines for clinical specimens for COVID-19 | CDC. Centers for Disease Control and Prevention. 2020. [cited 2020 Jun 22]. Available at: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. [Google Scholar]
- 14.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977; 33:159. [PubMed] [Google Scholar]
- 15.Pasomsub E, Watcharananan SP, Boonyawat K, et al. . Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect 2020.. doi: 10.1016/j.cmi.2020.05.001. Available at: http://www.clinicalmicrobiologyandinfection.com/article/S1198743X20302780/fulltext. [DOI] [PMC free article] [PubMed]
- 16.Thompson WD, Walter SD. A reappraisal of the kappa coefficient. J Clin Epidemiol 1988; 41:949–58. [DOI] [PubMed] [Google Scholar]
- 17.Xu R, Cui B, Duan X, Zhang P, Zhou X, Yuan Q. Saliva: potential diagnostic value and transmission of 2019-nCoV. Int J Oral Sci 2020; 12:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azzi L, Carcano G, Gianfagna F, et al. . Saliva is a reliable tool to detect SARS-CoV-2. J Infect 2020; 1–6. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyllie AL, Fournier J, Casanovas-Massana A, et al. . Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv. 2020. doi:10.11012020.04.16.20067835. Available at: https://www.medrxiv.org/content/10.1101/2020.04.16.20067835v1?fbclid=IwAR19q_Mv8NnVtKwnCdZGOpvaKGuOUxNMPhS04tf08gS7vbLq1vkOcsufNIs. [Google Scholar]
- 20.Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol 2020;50. Available at: https://jcm.asm.org/content/early/2020/04/17/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker D, Sandoval E, Amin A, et al. . Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv. 2020. doi: 10.1101/2020.05.11.20092338. Available at: https://www.medrxiv.org/content/10.1101/2020.05.11.20092338v2. [DOI] [Google Scholar]
- 22.Chau NVV, Lam VT, Dung NT, et al. . The natural history and transmission potential of asymptomatic SARS-CoV-2 infection. medRxiv [Internet]. 2020. doi: 10.1101/2020.04.27.20082347. Available at: https://www.medrxiv.org/content/10.1101/2020.04.27.20082347v1. [DOI] [Google Scholar]
- 23.Jamal AJ, Mozafarihashjin M, Coomes E, et al. . Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiwari M. Science behind human saliva. J Natural Sci Biol Med 2; 2011:53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwasaki S, Fujisawa S, Nakakubo S, et al. . Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J Infect 2020; 81:e145–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, Chen L, Deng Q, et al. . The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol 2020; 92:833–40. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Xu Y, Gao R, et al. . Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 2020;323:2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quilty BJ, Clifford S, Flasche S, Eggo RM. Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019-nCoV). Eurosurveillance 2020; 25:2000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, Zheng Z, Zhang C, et al. . Clinical characteristics of 145 patients with corona virus disease 2019 (COVID-19) in Taizhou, Zhejiang, China. Infection 2020;1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranney ML, Griffeth V, Jha AK. Critical supply shortages: the need for ventilators and personal protective equipment during the Covid-19 pandemic. N Engl J Med; 2020; 382:E41. [DOI] [PubMed] [Google Scholar]
- 31.To KKWW, Yip CCYY, Lai CYWW, et al. Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect; 2019; 25:372–8. Available at: http://www.ncbi.nlm.nih.gov/pubmed/29906597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.