Abstract
Background
Coronavirus disease 2019 (COVID-19) is a global pandemic with no licensed vaccine or specific antiviral agents for therapy. Little is known about the longitudinal dynamics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–specific neutralizing antibodies (NAbs) in patients with COVID-19.
Methods
Blood samples (n = 173) were collected from 30 patients with COVID-19 over a 3-month period after symptom onset and analyzed for SARS-CoV-2–specific NAbs using the lentiviral pseudotype assay, coincident with the levels of IgG and proinflammatory cytokines.
Results
SARS-CoV-2–specific NAb titers were low for the first 7–10 days after symptom onset and increased after 2–3 weeks. The median peak time for NAbs was 33 days (interquartile range [IQR], 24–59 days) after symptom onset. NAb titers in 93.3% (28/30) of the patients declined gradually over the 3-month study period, with a median decrease of 34.8% (IQR, 19.6–42.4%). NAb titers increased over time in parallel with the rise in immunoglobulin G (IgG) antibody levels, correlating well at week 3 (r = 0.41, P < .05). The NAb titers also demonstrated a significant positive correlation with levels of plasma proinflammatory cytokines, including stem cell factor (SCF), TNF-related apoptosis-inducing ligand (TRAIL), and macrophage colony-stimulating factor (M-CSF).
Conclusions
These data provide useful information regarding dynamic changes in NAbs in patients with COVID-19 during the acute and convalescent phases.
Keywords: SARS-CoV-2, neutralizing antibodies, longitudinal dynamics, COVID-19, serological immune response
NAbs were detectable at the early stage of COVID-19, peaked around week 4–5, and declined gradually during the 3 months following symptom onset. A positive correlation between NAb titers and IgG levels was observed at 3 weeks after symptom onset.
Coronavirus disease 2019 (COVID-19) is a novel respiratory disease that is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Since the outbreak of SARS-CoV-2 last year, it has spread rapidly and caused a global pandemic [1]. As of 28 July 2020, over 16 million people worldwide have been reportedly infected and more than 650 800 individuals have died of COVID-19 [2]. Currently, considerable progress is being made to understand SARS-CoV-2 pathogenesis, epidemiology, antiviral drug development, and vaccine design. However, no licensed specific antiviral drugs or prophylactic vaccines are available. Developing effective viral inhibitors and antibody-based therapeutics to prevent or treat COVID-19 infection is a high global priority.
The SARS-CoV-2 RNA genome encodes 29 structural and nonstructural proteins, including spike (S), envelope (E), membrane (M), nucleocapsid (N) proteins, and the ORF1a/b polyprotein [3]. The S glycoprotein is responsible for SARS-CoV-2 attachment and entry into target host cells via its binding to the angiotensin-converting enzyme 2 (ACE-2) receptor [4]. Virus-specific neutralizing antibodies (NAbs) play a key role in reducing viral replication and increasing viral clearance [5, 6]. Neutralizing antibodies mainly act against the receptor-binding domain (RBD) of the SARS-CoV-2 S protein [7–9], effectively blocking viral entry. Thus, serological testing, especially to detect NAbs, is essential in determining the onset of the serological immune response, evaluating the potential capacity of the host body for viral clearance, and identifying donors for passive antibody therapy trials. In patients with COVID-19, NAbs can be detected within 2 weeks of symptom onset [10, 11]. The serological antibody response continues for at least 3 weeks and, in some cases, substantially longer [12, 13]. However, the dynamics and roles of SARS-CoV-2–specific NAbs and their correlation with antibody responses have not been explored in patients with COVID-19 more than 2 months after symptom onset.
Previous studies of severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) demonstrated that the most immunogenic antigens are the S and N proteins, and the development of serological tests (such as enzyme-linked immunosorbent assay and magnetic chemiluminescence enzyme immunoassay) for SARS-CoV-2 immunoglobulin (Ig) G or IgM antibodies has focused on these viral proteins. However, it is still unknown whether serological antibodies predict neutralizing activities or protection against viral reinfection [14]. For enveloped viruses, NAbs blocking virus infection are mainly associated with the envelop glycoprotein, thus playing a key role in viral clearance. Neutralizing antibody levels can be either determined using authentic or pseudotype virus in cellular bioassays.
Due to the highly pathogenic nature of SARS-CoV-2, infectious SARS-CoV-2 must be handled in a biosafety level 3 (BSL-3) facility. Moreover, recent studies indicated that pseudotype neutralization tests displayed consistent results with plaque-reduction neutralization testing assay with authentic virus [15, 16]. In this study, by utilizing SARS-CoV-2 pseudovirus, we first analyzed the 3-month longitudinal dynamics of in vitro NAb titers in 30 patients who recovered from COVID-19. Second, we evaluated the correlation between the dynamics of NAb titers and serological immunoglobulin G (IgG) levels, as well as inflammatory cytokine levels. Our study may provide useful information regarding dynamic changes in NAbs in patients with COVID-19 during the acute and convalescent phases and aid in the development of vaccines against SARS-CoV-2.
METHODS
Clinical Characteristics
A total of 30 patients who had recovered from COVID-19 and were discharged from the Yongchuan Hospital of Chongqing Medical University were included in our cohort. A confirmed case of COVID-19 was defined as an individual with nasopharyngeal swabs that were positive for laboratory-based polymerase chain reaction (PCR) testing. Patients with COVID-19 who meet following criteria can be discharged: 2 consecutive negative reverse transcriptase (RT)-PCR results on respiratory tract samples, body temperature is back to normal for more than 3 days, respiratory symptoms are obviously improved, and pulmonary imaging shows obvious absorption of inflammation. On 2 April and 8 May 2020 (follow-up point 1 and follow-up point 2, respectively) 2 follow-up visits were conducted. Sequential serum samples were collected from patients in the acute phase (3 or 4 samples per patient) and the convalescent phase (2 follow-up points: 60 [54–63] days and 96 [90–99] days after symptom onset) to measure and characterize the dynamic changes in virus-specific IgG and NAb titers. The acute phase was defined as the period when the viral RNA can be found in a respiratory specimen.
Ethical Approval
The study was approved by the Ethics Commission of Chongqing Medical University (reference number 2020003). Written informed consent was waived by the Ethics Commission of the designated hospital for emerging infectious diseases.
Plasmids
The codon-optimized genes encoding the SARS-CoV S protein (AAP13567.1) and SARS-CoV-2 S protein (QHD43416) with the 19 C-terminal amino acids deleted were synthesized by Sino Biological Inc (Beijing, China) and cloned into the pCMV3 vector, respectively. The HIV-1 NL4-3 ΔEnv Vpr luciferase reporter vector (pNL4-3.Luc.R-E-), constructed by N. Landau [17], was provided by Cheguo Cai, Wuhan University (Wuhan, China). The vesicular stomatitis virus G (VSV-G)–expressing plasmid pMD2.G was provided by Ding Xue, Tsinghua University (Beijing, China).
Cell Lines
HEK293T cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). Cells were maintained in Dulbecco’s modified Eagle medium (DMEM; Hyclone, Waltham, MA) supplemented with 10% fetal bovine serum (Gibco, Rockville, MD), 100 mg/mL streptomycin, and 100 U/mL of penicillin at 37°C in 5% CO2. HEK293T cells transfected with human ACE2 (293T-ACE2) were cultured under the same conditions, with the addition of G418 (0.5 mg/mL) to the medium.
Production and Titration of SARS-CoV-2 S Pseudoviruses
The SARS-CoV and SARS-CoV-2 pseudoviruses were generated as previously described, with some modifications [18]. Briefly, HEK293T cells (5 × 106) were co-transfected with pNL4-3.Luc.R-E- and recombinant SARS-CoV S or SARS-CoV-2 S plasmid using the Lipofectamine 3000 transfection reagent (Invitrogen, Rockville, MD), according to the manufacturer’s instructions. The cells were transferred to fresh DMEM 12 hours later. The supernatant-containing pseudovirions were harvested 48 hours after transfection and passed through a 0.45-μm filter. To construct the VSV-G pseudovirus, pMD2.G was co-transfected with the pNL4-3.Luc.R-E- plasmid. Viral titers (RNA copy number, copies/mL) were determined as described previously by real-time RT-quantitative PCR (-qPCR) using primers targeting the long terminal repeat (LTR) region [19].
Neutralization Assays
The 293T-ACE2 cells (2 × 104 cells/well) were seeded in 96-well plates. For the neutralization assay, 50 μL of pseudovirus (3.8 × 104 copies) was incubated with serial dilutions of serum samples from patients and human control serum as a negative control for 1 hour at 37°C and then added to the 96-well 293T-ACE2 plates. After 12 hours of infection, the culture medium was refreshed. After 72 hours postinfection, the 293T-ACE2 cells were lysed with 30 μL lysis buffer (Promega, Madison, WI) to measure pseudoviral transduction. Relative luminescence units of Luc activity were determined using the Luciferase Assay Kit (Promega). The titers of NAbs were calculated as 50% inhibitory dose (ID50), expressed as the highest dilution of plasma that resulted in a 50% reduction of luciferase luminescence compared with virus control, using a cutoff titer of 1:20.
Detection of IgG Against SARS-CoV-2
All serum samples were inactivated at 56°C for 30 minutes and stored at −20°C before testing. IgG against SARS-CoV-2 in plasma samples was tested using magnetic chemiluminescence enzyme immunoassay kits supplied by Bioscience Co (approved by the China National Medical Products Administration; approval number 20203400183), according to the manufacturer’s instructions. IgG levels are presented as the measured chemiluminescence values divided by the cutoff (S/CO).
Statistical Analysis
Continuous variables are expressed as median (interquartile range [IQR]) and categorical variables are expressed as number (%). Comparisons between 2 groups were performed using the Mann-Whitney U test or Fisher’s exact test. A 2-sided α of less than .05 was considered statistically significant. Statistical analyses were performed using R software, version 3.6.0 (R Foundation for Statistical Computing). Two-tailed Pearson correlation test was used to calculate the correlation coefficient of NAb to IgG levels or cytokines.
RESULTS
Clinical Characteristics
Of the total 30 patients in the cohort, 60.0% (18/30) were female and 10.0% (3/30) were categorized as severe based on the COVID-19 treatment guidelines (National Health Commission of the People’s Republic of China) (Table 1) who met any of the following criteria: (1) respiratory distress (≥30 breaths/minutes), (2) oxygen saturation of 93% or less at rest, (3) arterial partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) of 300 mmHg or less. The median length of the hospital stay was 22 days (IQR, 15–26 days).
Table 1.
Clinical Characteristics of 30 Patients Enrolled in the Study
| Characteristics | Patients (N = 30) |
|---|---|
| Age, median (IQR), years | 52 (45–67) |
| Sex, n (%) | |
| Male | 12 (40.0) |
| Female | 18 (60.0) |
| Exposure, n (%) | |
| From Wuhan | 4 (13.3) |
| Close contacts | 26 (76.7) |
| Severity, n (%) | |
| Nonsevere | 3 (10.0) |
| Severea | 27 (90.0) |
| Comorbidities, n (%) | |
| Hypertension | 9 (30.0) |
| Cardiovascular disease | 2 (6.7) |
| Diabetes | 2 (6.7) |
| COPD | 1 (3.3) |
| Chronic kidney disease | 1 (3.3) |
| Chronic liver disease | 2 (6.7) |
| Any | 13 (43.3) |
| Signs and symptoms, n (%) | |
| Fever | 11 (36.7) |
| Fatigue | 2 (6.7) |
| Dry cough | 10 (33.3) |
| Lack of appetite | 2 (6.7) |
| Myalgia | 3 (10.0) |
| Dyspnea | 5 (16.7) |
| Expectoration | 7 (23.3) |
| Pharyngalgia | 5 (16.7) |
| Diarrhea | 1 (3.3) |
| Nausea | 1 (3.3) |
| Dizziness | 2 (6.7) |
| Vomiting | 1 (3.3) |
| Chills | 3 (10.0) |
| Rhinorrhea | 1 (3.3) |
| Chest stuffiness | 1 (3.3) |
Abbreviations: COPD, chronic obstructive pulmonary disease; IQR, interquartile range (range from lower quartile to upper quartile).
aSevere was defined according to the Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial version 7), released by the National Health Commission and State Administration of Traditional Chinese Medicine.
Specific Serum Response to SARS-CoV-2 Pseudovirus Infection
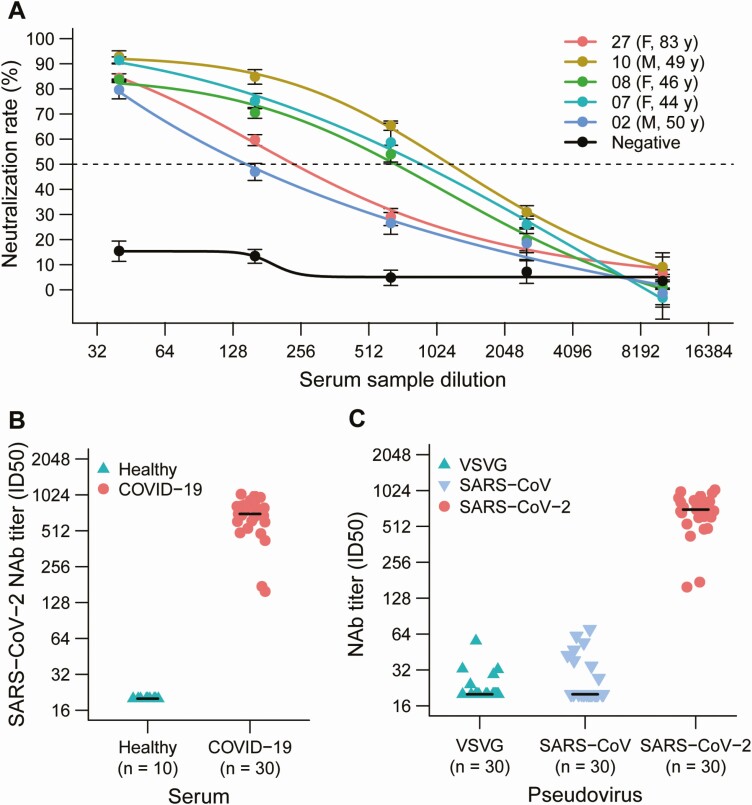
Sera from 5 representative convalescent patients with COVID-19, collected during the fourth week following symptom onset, were analyzed for their NAb titers against SARS-CoV-2 pseudovirus infection of 293T-ACE2 cells. All 5 serum samples demonstrated neutralizing activity against SARS-CoV-2 pseudovirus infection while the control serum from healthy individuals showed no neutralizing activity (Figure 1A and 1B). Furthermore, 30 plasma samples with strong SARS-CoV-2 neutralizing activity were evaluated for neutralization of VSV-G, SARS-CoV, and SARS-CoV-2 pseudoviruses. Weak cross-reactivity was detected between SARS-CoV-2 and SARS-CoV or VSV-G pseudovirus control (Figure 1C). These results suggest that SARS-CoV-2 infection could not stimulate strong cross-neutralizing antibodies against SARS-CoV.
Figure 1.
Analysis of the plasma response to SARS-CoV-2 infection. A, Sera from 5 convalescent patients with COVID-19 neutralized the SARS-CoV-2 pseudovirus. A serum sample from a healthy individual served as the negative control. The assay was performed in triplicate, and the median percentage of neutralization is shown. B, SARS-CoV-2 NAb titers of 20 plasma samples from convalescent patients with COVID-19 and 10 plasma samples from healthy donors. C, NAbs against VSVG, SARS-CoV, and SARS-CoV-2 pseudovirus in the sera from 30 convalescent patients with COVID-19. Abbreviations: COVID-19, coronavirus disease 2019; F, female; ID50, 50% inhibitory dose; M, male; NAb, neutralizing antibody; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; VSVG, vesicular stomatitis virus G.
Dynamic Changes in Neutralizing Antibodies Against SARS-CoV-2
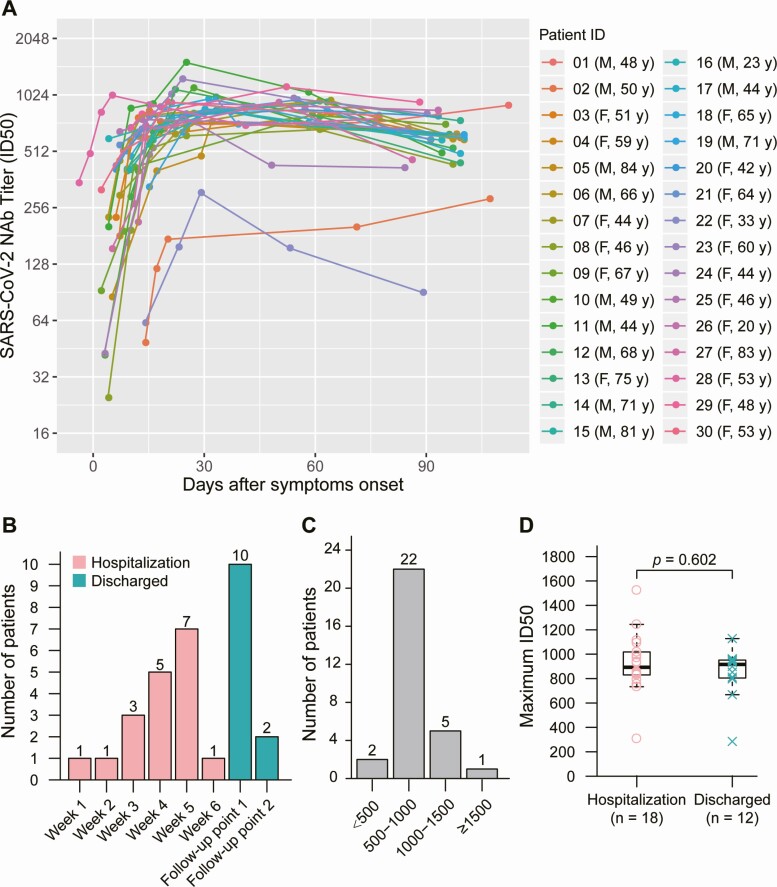
We analyzed the longitudinal dynamics of virus-specific IgG and NAb levels in 30 patients who were positive for SARS-CoV-2 using real-time RT-qPCR. SARS-CoV-2–specific NAb titers were low before day 7–10 and increased at week 2–3 after symptom onset. The highest NAb levels were detected 3 months after symptom onset in 28 of 30 (93.3%) patients with COVID-19 (Figure 2A). The median time for peak NAb levels was 33 days (IQR, 24–59 days) after symptom onset, and the NAb levels plateaued in 60% (18/30) of the patients with COVID-19 during hospitalization (Figure 2B). The peak NAb levels varied among the patients; 6.7%, 73.3%, and 20% of patients showed low (ID50 <500), medium-low (ID50, 500–999), and medium-high (ID50, 1000–2500) NAb titers, respectively (Figure 2C). There was no statistical difference among peak NAb titers that occurred during hospitalization and convalescence (Figure 2D).
Figure 2.
Dynamic changes in NAbs against SARS-CoV-2. A, Kinetics of SARS-CoV-2 NAb levels in 30 patients with COVID-19. B, Number of patients experiencing peak NAb levels during hospitalization or after discharge. Follow-up point 1 represents the follow-up study conducted on 2 April while follow-up point 2 represents the follow-up study conducted on 8 May 2020. C, Peak NAb levels in the patients. D, Comparison of peak NAb levels between patients with COVID-19 experiencing peak NAb levels during hospitalization and patients with COVID-19 experiencing peak NAb levels after discharge. Abbreviations: COVID-19, coronavirus disease 2019; F, female; ID50, 50% inhibitory dose; M, male; NAb, neutralizing antibody; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
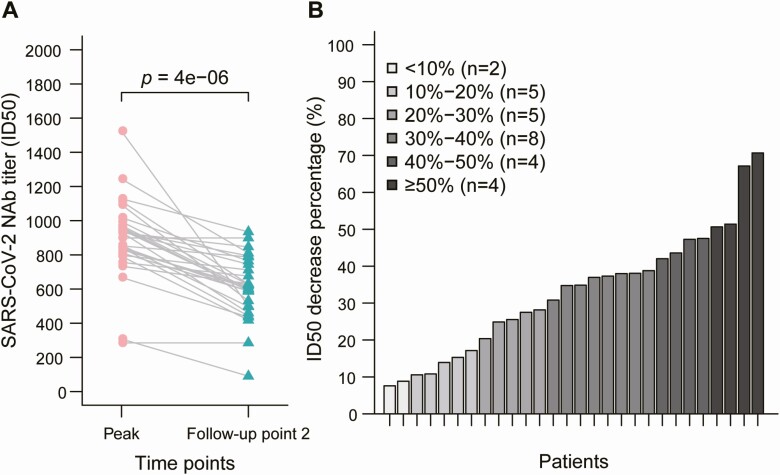
The duration and maintenance of peak of NAb levels in patients with COVID-19 are of great concern. Thus, we compared NAb levels between the peak time point and the final follow-up time point. A decline in NAb levels was observed in 93.3% (28/30) of SARS-CoV-2–infected patients, with a median decrease of 34.8% (IQR, 19.6–42.4%) (Figure 3A). Patients were also grouped according to their rate of decrease in NAb levels; more than 20% of the patients showed a more than 70% decrease in NAb levels during this time period (21/30) (Figure 3B).
Figure 3.
Decrease in NAb levels in patients with COVID-19. A, Comparison of NAb levels between the peak point and the follow-up time point 2. B, Percentage decrease in NAbs in patients with COVID-19. Abbreviations: COVID-19, coronavirus disease 2019; ID50, 50% inhibitory dose; NAb, neutralizing antibody; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Correlation Between Dynamics of Neutralizing Antibody and IgG Levels in Patients With COVID-19
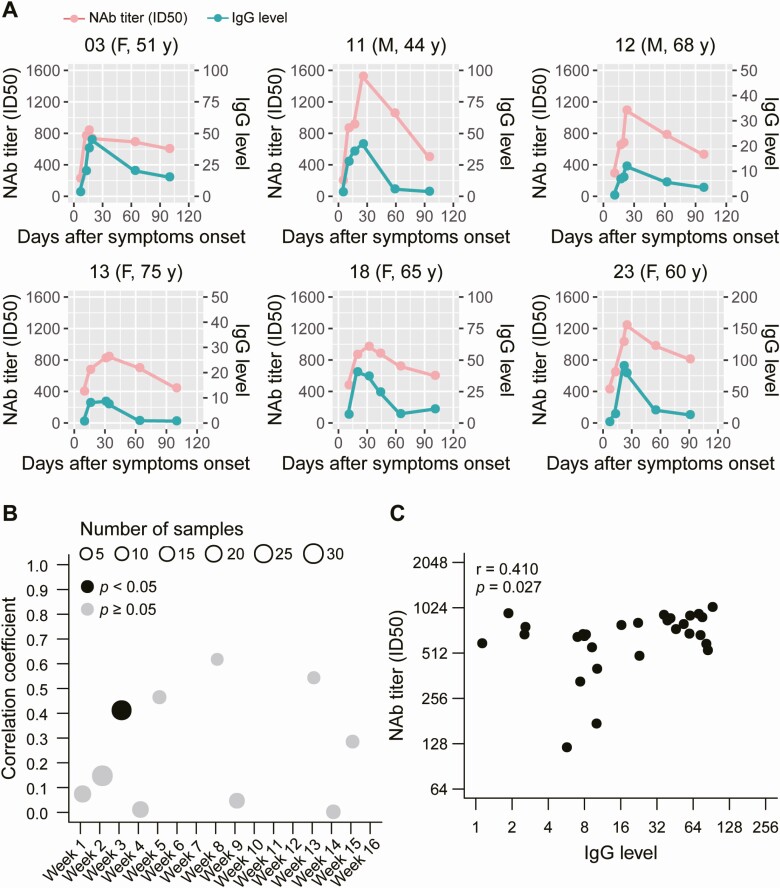
The kinetic levels of NAbs and virus-specific IgG over time in patients with COVID-19 are still unknown. To address this, we first determined the relationship between the NAb levels and virus-specific IgG levels in individual patients (Figure 4A, Supplementary Figure 1); similar dynamic changes were observed for the NAbs and virus-specific IgG levels in some patients. Furthermore, to determine if there was a statistical correlation between NAb levels and virus-specific IgG levels in patients with COVID-19, serum samples were grouped by time (weeks) after symptom onset. A statistically significant positive correlation was only observed in samples obtained 3 weeks after symptom onset (P = .027, r = 0.410) (Figure 4B).
Figure 4.
Correlation between the dynamics of NAb and virus-specific IgG levels. A, Kinetics of NAb and IgG levels in 6 patients. Plasma samples were collected at different time points after symptom onset. B, A total of 152 serum samples were grouped by time of collection after symptom onset; correlations were analyzed between NAb levels and IgG levels in each group. C, Correlations between NAb levels and IgG levels from serum samples collected 3 weeks after symptom onset. Abbreviations: F, female; ID50, 50% inhibitory dose; IgG, immunoglobulin G; M, male; NAb, neutralizing antibody.
Roles of Cytokines in Antibody Production
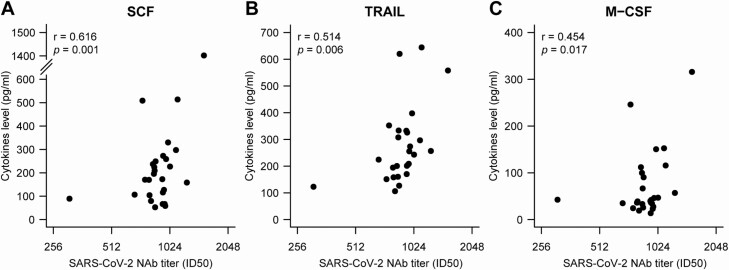
We analyzed the correlation between cytokine and chemokine levels and NAb levels in patients with COVID-19 during the acute phase. Interestingly, we observed that NAb levels were positively correlated with stem cell factor (SCF) (r = 0.616, P = .001), tumor necrosis factor (TNF)–related apoptosis-inducing ligand (TRAIL) (r = 0.514, P = .008), and macrophage colony-stimulating factor (M-CSF) (r = 0.454, P = .017) levels (Figure 5).
Figure 5.
A–C, Correlation between peak NAb levels and cytokines in sera. Serum samples with the highest NAb levels for all individuals were collected during hospitalization. Cytokine or levels were measured using the Bio-Plex Human Cytokine Screening Panel (48-Plex no. 12007283; Bio-Rad) on a Luminex 200 (Luminex Multiplexing Instrument, Merck Millipore), following the manufacturer’s instructions. Pearson’s correlation was used to analyze differences between NAb levels and cytokine levels. Abbreviations: ID50, 50% inhibitory dose; M-CSF, macrophage colony-stimulating factor; NAb, neutralizing antibody; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SCF, stem cell factor; TRAIL, tumor necrosis factor (TNF)–related apoptosis-inducing ligand.
Discussion
Virus-specific NAbs have been considered an important determinant for viral clearance. The pseudovirus-based assay is suitable for the high-throughput screening of SARS-CoV-2 NAbs in plasma donors without the requirement of BSL-3 laboratories. The assay has been widely used for evaluating NAbs in highly pathogenic viruses, such as Ebola, SARS-CoV, MERS-CoV, and highly pathogenic influenza viruses [20]. Herein, we described the dynamics of SARS-CoV-2–specific NAbs generated during both the acute and convalescent phases of SARS-CoV-2 infection using a pseudovirus-based neutralization assay. We found that SARS-CoV-2–specific NAb titers were low before day 7–10, peaked at approximately day 33 after symptom onset, and then gradually declined over a 3-month period. Meanwhile, SARS-CoV-2–specific NAbs were detected concurrently with and positively correlated with IgG antibodies in our cohort, indicating that the NAb response may play an important role in viral clearance.
Our understanding of the duration and nature of protective immunity to SARS-CoV-2 is currently very limited. The kinetics of antibody-mediated immunity to SARS-CoV-2 infection and how long this immunity lasts are unknown. Our data suggest that NAb titers in patients were variable, and the protective humoral immune response to SARS-CoV-2 may abate over time, which is in accordance with findings in patients infected with other human coronaviruses, such as HCoV-229E [21, 22]. The short-term humoral immune response in patients with COVID-19 is also highly consistent with that observed in patients infected with SARS-CoV and MERS-CoV [23, 24], who show a rapid decrease in virus-specific antibody titers within 3–4 months. Among the 30 recovered patients in our study, 2 patients showed very low NAb titers during the acute phase and 3-month follow-up, indicating that other immune responses, involving T cells and inflammatory cytokines, may have contributed to viral clearance. These data suggest that the antibody titers may diminish with time or some recovered patients may not produce a high-titer response during SARS-CoV-2 infection.
Recently, in a rhesus macaque model, SARS-CoV-2 infection evoked a robust protective immune response when the animals were re-exposed to SARS-CoV-2 at 1 month after the initial viral infection [25]. However, natural infection and volunteer challenge studies hint that coronavirus infections, including those with HCoV-229E and HCoV-OE43, cannot induce stable protective immunity; thus, reinfection occurs frequently. Moreover, an SARS-CoV antigen-specific memory B-cell response was not detectable in patients who had recovered from SARS at 6 years after disease onset, whereas SARS-CoV–specific memory T cells persisted in patients who had recovered from SARS [26, 27]. Although the role of memory T cells in the protective immune response to SARS-CoV-2 needs further evaluation, a robust T-cell response is required for viral clearance.
We also described here the dynamic correlation between SARS-CoV-2–specific NAbs and serological total IgG levels. Neutralizing antibody titers appeared concomitantly and correlated moderately with IgG levels at week 3 after symptom onset, which is consistent with other reports regarding patients who recovered from COVID-19 [13, 28]. The antigen epitope used for IgG detection in our study contained the nucleoprotein peptide, as well as the RBD domain of the S protein, which partially explains the discrepancy in NAb titers and IgG levels at weeks 4, 9, and 14 after symptom onset. The nucleoprotein is one of the major antigens of SARS-CoV-2 [29]. The binding antibodies detected by the total IgG assay may also be involved in viral clearance through antibody-dependent cytotoxicity. Therefore, the roles of binding antibodies and NAbs in disease progression need further evaluation.
Currently, adaptive immunotherapy using convalescent plasma (CP) from patients who recovered from COVID-19 is being used as a potential therapeutic approach to confer antiviral protection [30]. Several preliminary clinical trials have proven its effectiveness in treating SARS-CoV-2 [5, 6]. The efficacy of CP transfusion is attributed to the neutralizing effect of antibodies; thus, the NAb titer is the major determinant for CP therapy. Monitoring NAb levels and their duration will provide valuable data for evaluating the effectiveness of CP therapy. In our study, the levels of NAbs declined gradually over the 3-month follow-up period, with a median decrease of 34.8%. Thus, CP samples with high titers of NAbs from patients in the early stage of convalescence will be more suitable for clinical use.
There are some limitations to this study, which should be addressed. Due to the small sample size, we could not find any correlation between the dynamics of NAb titers and clinical characteristics contributing to different clinical outcomes. Serological blood samples were collected up to 3 months after symptom onset; data collected over longer follow-up times should be obtained to demonstrate the duration of humoral immunity after SARS-CoV-2 infection. The lack of data to determine an anamnestic immune response, such as tests for SARS-CoV-2–specific memory B cells, memory T cells, and specific cytokine-dependent memory cells, hampered the evaluation of the immune response, especially protective immunity against viral reinfection. These are major issues that should be investigated in future studies.
In summary, we determined the dynamics of NAb titers within 3 months after symptom onset in 30 SARS-CoV-2–infected patients and found a positive correlation between NAb titers and IgG antibodies. Our work provides valuable insight into the humoral immunity against SARS-CoV-2 infection. We also described a pseudotype system for measuring NAb titers, which could be expanded to antiviral drug screening and vaccine development.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank Professor Cheguo Cai (Wuhan University, Wuhan, China) for providing the pNL4-3.Luc.R-E- plasmid.
Financial support. This work was supported by the Emergency Project from the Science and Technology Commission of Chongqing (cstc2020jscx-fyzx0053), the Emergency Project for Novel Coronavirus Pneumonia from the Chongqing Medical University (CQMUNCP0302, CQMUNCP0304), the Leading Talent Program of Chongqing Science and Technology Commission (CSTCCXLJRC201719), and a Major National Science and Technology Program grant (2017ZX10202203) from the Science and Technology Commission of China.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020; 579:270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Coronavirus disease (COVID-19) situation reports. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/. Accessed 28 July 2020.
- 3. Sun J, He WT, Wang L, et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med 2020; 26:483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol 2020; 20:339–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duan K, Liu B, Li C, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci USA 2020; 117:9490–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shen C, Wang Z, Zhao F, et al. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. JAMA 2020; 323: 1582–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Y, Wang F, Shen C, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020; 368:1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of conva lescent patients’ B cells. Cell 2020; 182:73–84, e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-bind ing site of SARS-CoV-2. Nature 2020; 584:120–4. Available at: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 10. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 11. van der Heide V. Neutralizing antibody response in mild COVID-19. Nat Rev Immunol 2020; 20:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harvala H, Robb M, Watkins N, et al. Convalescent plasma therapy for the treatment of patients with COVID-19: assessment of methods available for antibody detection and their correlation with neutralising antibody levels. medRxiv 2020. doi: 10.1101/2020.05.20.20091694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ng D, Goldgof G, Shy B, et al. SARS-CoV-2 seroprevalence and neutralizing activity in donor and patient bloo d from the San Francisco Bay Area. medRxiv 2020. doi: 10.1101/2020.05.19.20107482. Available at: https://www.medrxiv.org/content/10.1101/2020.05.19.20107482v2. Accessed 8 July 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Luchsinger LL, Ransegnola B, Jin D, et al. Serological analysis of New York City COVID19 convalescent plasma donors. medRxiv 2020. doi: 10.1101/2020.06.08.20124792. Available at: https://www.medrxiv.org/content/10.1101/2020.06.08.20124792v1. Accessed 8 July 2020. [DOI] [Google Scholar]
- 15. Wang Y, Zhang L, Sang L, et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest 7 July 2020. doi: 10.1172/JCI138759. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jackson LA, Anderson EJ, Rouphael NG, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med 2020; NEJMoa2022483. doi: 10.1056/NEJMoa2022483. Epub 14 July 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Connor RI, Chen BK, Choe S, Landau NR. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 1995; 206:935–44. [DOI] [PubMed] [Google Scholar]
- 18. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020; 11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geraerts M, Willems S, Baekelandt V, Debyser Z, Gijsbers R. Comparison of lentiviral vector titration methods. BMC Biotechnol 2006; 6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Steffen I, Simmons G. Pseudotyping viral vectors with emerging virus envelope proteins. Curr Gene Ther 2016; 16:47–55. [DOI] [PubMed] [Google Scholar]
- 21. Callow KA, Parry HF, Sergeant M, Tyrrell DA. The time course of the immune response to experimental coronavirus infection of man. Epidemiol Infect 1990; 105:435–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cao WC, Liu W, Zhang PH, Zhang F, Richardus JH. Disappearance of antibodies to SARS-associated coronavirus after recovery. N Engl J Med 2007; 357:1162–3. [DOI] [PubMed] [Google Scholar]
- 23. Zhang JS, Chen JT, Liu YX, et al. A serological survey on neutralizing antibody titer of SARS convalescent sera. J Med Virol 2005; 77:147–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ko JH, Seok H, Cho SY, et al. Challenges of convalescent plasma infusion therapy in Middle East respiratory coronavirus infection: a single centre experience. Antivir Ther 2018; 23:617–22. [DOI] [PubMed] [Google Scholar]
- 25. Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020; 369:812–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang F, Quan Y, Xin ZT, et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol 2011; 186:7264–8. [DOI] [PubMed] [Google Scholar]
- 27. Yang LT, Peng H, Zhu ZL, et al. Long-lived effector/central memory T-cell responses to severe acute respiratory syndrome coronavirus (SARS-CoV) S antigen in recovered SARS patients. Clin Immunol 2006; 120:171–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu F, Wang A, Liu M, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv 2020. doi: 10.1101/2020.03.30.20047365. Available at: https://www.medrxiv.org/content/10.1101/2020.03.30.20047365v2. Accessed 8 July 2020. [DOI] [Google Scholar]
- 29. Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS–CoV-2: a target for vaccine development. J Virol 2020; 94:e00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis 2020; 20:398–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.