Abstract
Aims
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly binds to ACE2 (angiotensin-converting enzyme 2) to facilitate cellular entry. Compared with the lung or respiratory tract, the human heart exhibits greater ACE2 expression. However, little substantial damage was found in the heart tissue, and no viral particles were observed in the cardiac myocytes. This study aims to analyse ACE2 and SARS-CoV-2 spike (S) protein proteases at the single-cell level, to explore the cardiac involvement in COVID-19 and improve our understanding of the potential cardiovascular implications of COVID-19.
Methods and Results
With meta-analysis, the prevalence of cardiac injury in COVID-19 patients varies from 2% [95% confidence interval (CI) 0–5%, I2 = 0%] in non-ICU patients to 59% (95% CI 48–71%, I2 = 85%) in non-survivors. With public single-cell sequence data analysis, ACE2 expression in the adult human heart is higher than that in the lung (adjusted P < 0.0001). Inversely, the most important S protein cleavage protease TMPRSS2 (transmembrane protease serine protease-2) in the heart exhibits an extremely lower expression than that in the lung (adjusted P < 0.0001), which may restrict entry of SARS-CoV-2 into cardiac cells. Furthermore, we discovered that other S protein proteases, CTSL (cathepsin L) and FURIN (furin, paired basic amino acid cleaving enzyme), were expressed in the adult heart at a similar level to that in the lung, which may compensate for TMPRSS2, mediating cardiac involvement in COVID-19.
Conclusion
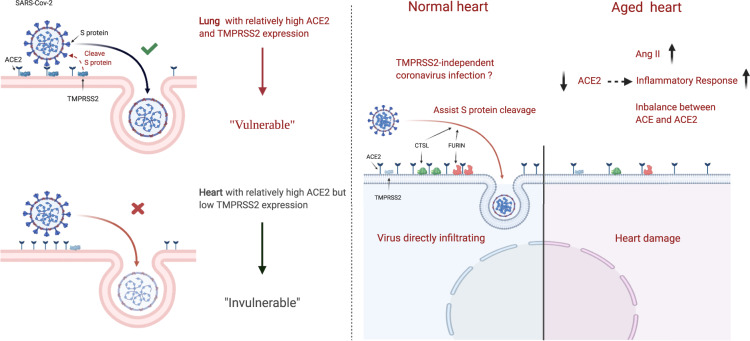
Compared with the lung, ACE2 is relatively more highly expressed in the human heart, while the key S protein priming protease, TMPRSS2, is rarely expressed. The low percentage of ACE2+/TMPRSS2+ cells reduced heart vulnerability to SARS-CoV-2 to some degree. CTSL and FURIN may compensate for S protein priming to mediate SARS-CoV-2 infection of the heart.
Keywords: COVID-19, SARS-CoV-2, Cardiac injury, ACE2, TMPRSS2
Graphical Abstract
Graphical Abstract.
Introduction
The continued spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has presented a pressing public challenge for the global community.1 Greater insights into the features of COVID-19 presentation and their pathogenesis remains of great importance. An increasing number of studies have shown that clinical manifestations of COVID-19 were not only primarily related to respiratory inflammation, but were also associated with cardiac injury.2–4 A large cohort study even found that cardiac injury was independently associated with increased in-hospital mortality.5 However, the degree of cardiovascular involvement in COVID-19 and mechanisms of cardiovascular tropism of SARS-CoV-2 virus are incompletely understood.
SARS-CoV-2 uses its spike (S) protein to directly bind with angiotensin-converting enzyme 2 (ACE2) on the host cell surface.6 Upon binding to ACE2, the S protein requires cleavage by proteases to fuse with the cell membrane and activate the process to enter the host cell.7 Several host proteases have the ability to cleave the S protein of SARS-CoV-2, including transmembrane protease serine protease-2 (TMPRSS2), cathepsins, and FURIN.8 Among these proteases, TMPRSS2 is essential for viral entry and spread in the host infected by SARS-CoV as well as SARS-CoV-2.8
According to a previous study, ACE2 exhibits greater expression in human foetal heart tissue (7.5%) than in the lung (<1%) and respiratory tract (∼2%),9 indicating that the human heart is vulnerable to SARS-CoV-2. However, pathological studies showed that little substantial damage was observed in the heart tissue and no viral particles were observed in the cardiac myocytes.10,11 The underlying mechanisms of this inconsistency between ACE2 expression and pathological manifestation remain to be elucidated.
In this study, we summarized the cardiac involvement of COVID-19 and analysed the expression of ACE2 and S protein cleavage proteases in human coronary artery, as well as in human foetal and adult heart tissue at the single-cell level, to identify mechanisms of cardiac injury in COVID-19 patients.
Methods
Study selection and data extraction for meta-analysis
Studies found on PubMed, Embase, a Chinese database (http://journal.yiigle.com/), and medRxiv were systematically reviewed by three authors (H.L., X.W., and S.G.) independently. The search was performed from its inception until 22 May 2020. Studies reporting data on complications of COVID-19 patients, including cardiac injury, liver injury (abnormal liver function), and renal injury, were eligible for inclusion. More detailed methods have been provided in the Supplementary material online.
Analysis of scRNA-seq data
Published single-cell RNA sequencing (scRNA-seq) datasets from human heart tissues, lung, and HBECs (human bronchial epithelial cells) were explored, and four types of scRNA-seq data were obtained: (i) foetal heart data were obtained from GSE106118, which contains 20 samples from different developmental stages, ranging from 5 weeks to 25 weeks of gestation;12 (ii) adult coronary artery data were obtained from GSE131778, which contains four cardiac transplant recipients;13 (iii) adult heart data were obtained from GSE109816 and GSE121893, which contain 14 normal heart samples from healthy organ donors, and six heart failure (HF) samples from patients undergoing heart transplantation;14 (iv) lung and HBEC data were obtained from a database (https://data.mendeley.com/datasets/7r2cwbw44m/1) which contains four endobronchial lining fluid-sampled patients and 12 lung cancer patients.8 We used criteria which are as similar to the original studies as possible when filtering cells; for adult cardiomyocytes, cells containing >72% mitochondrial genes were excluded.8,12–14 A Seurat object list was set up for each individual across different datasets. ‘SCTransform’ and ‘FindIntegrationAnchors’ functions in Seurat (v.3.1.5) were run to remove batch effects and integrate multiple samples across different datasets.15 Corrected UMIs (unique molecular identifiers) after integration were applied with ‘NormalizeData’ function (RC method, scale.factor = 1e6) to calculate CPM (counts per million), which was further transformed into log2(CPM + 1) to plot the expression level. Principal component analysis was used for dimensionality reduction, followed by clustering based on the K-nearest neighbour graph with optimal resolution. UMAP (https://github.com/lmcinnes/umap) was used for visualizing the results. The ‘FindAllMarkers’ function (min.pct = 0.25, logfc. threshold = 0.25) was applied to analyse differentially expressied genes. Cell identities were annotated based on data-derived marker genes and marker genes from the literature.12–14,16 Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, and visualization of results were performed using ClusterProfiler (version 3.14).17
Statistical analysis
Detailed statistical methods for the meta-analysis are shown in the Supplementary materials online. For analysis of scRNA-seq data, we used Wilcoxon rank-sum tests to detect differential genes, with P-values adjusted based on Bonferroni correction.18 For comparisons of expression values between two groups, the Wilcoxon rank sum test was used. For comparisons of multiple groups, the Kruskal–Wallis test was utilized,19 followed by a post-hoc test with Bonferroni-adjusted P-valuesy. The correlation between different cell types was presented with a Spearman similarity heat map. To explore the correlation between specific genes (e.g. ACE2) and clinical characteristics, gene average expression and percentage of positive cells per patient were calculated for further analysis. The relationship between different gene expression and age was analysed using a linear regression model, with R2 as model evaluation indices. Differences in the percentage of specific gene-positive cells between normal and HF tissues, as well as between male and female heart tissues, were tested using χ2 test, with P-values <0.05 considered as significant. Analyses were performed using R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria).
Results
Complications observed in COVID-19 patients
Forty studies were eventually included for analysis (Supplementary material online, Figure S1), and the majority of studies were from Wuhan, China (26/40, 65%) (Supplementary material online, Table S2). In total, 26, 11, and 23 cohorts reported the incidence of cardiac, liver, and renal injury, respectively, in COVID-19 patients (Supplementary material online, Table S3). The pooled prevalence estimate of cardiac injury varies from 2% [95% confidence interval (CI) 0–5%, I2 = 0%] in non-ICU patients to 59% (95% CI 48–71, I2 = 85%) in non-survivors (Supplementary material online, Figure S2). In studies where patients were not divided into specific groups, the overall prevalence of cardiac injury was 13% (95% CI 8–18%, I2 = 84%), intermediate between liver injury (23%, 95% CI 15–33%, I2 = 94.5%) and renal injury (9%, 95% CI 3–15%, I2 = 99%) (Supplementary material online, Figure S3). In addition, compared with their counterparts, high-severity patients showed a heavier burden of cardiovascular-related pre-existing conditions, with significantly increased hypertension [overall odds ratio (OR) 2.63, 95% CI 2.08–3.33, I2 = 39.9%), diabetes (overall OR 2.44, 95% CI 1.88–3.17, I2 = 26.9%), and cardiovascular disease (overall OR 4.54, 95% CI 2.69–7.64, I2 = 37.3%) (Supplementary material online, Figure S4). Moreover, compared with their counterparts, high-severity patients also exhibited a higher risk of cardiac injury (overall OR 4.72, 95% CI 2.48–8.97, I2 = 56.6%) (Supplementary material online, Figure S5).
Expression pattern of ACE2 and TMPRSS2 across different heart tissues
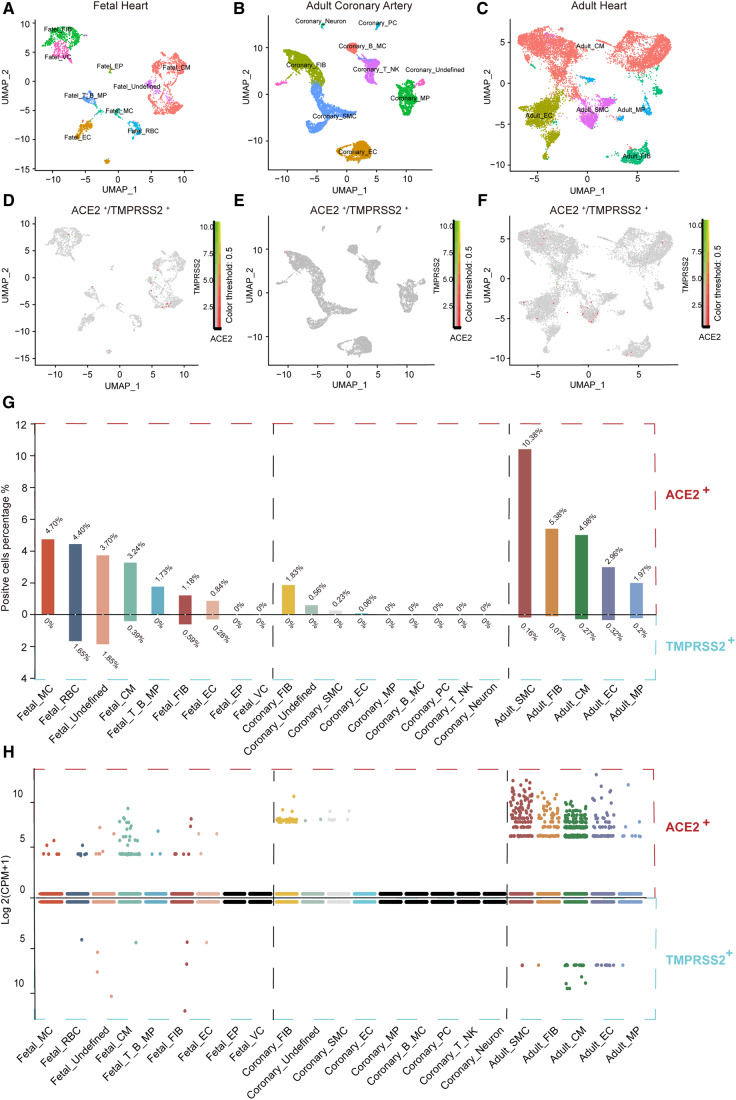
A total of 41 samples were included for analysis, among which adult heart accounted for 48.8% (20/41). After quality control, 3373 qualifying cells from foetal heart, 10 554 cells from adult coronary artery, and 12 554 cells from adult heart were obtained for subsequent analyses (Supplementary material online, Figure S1). Previous studies demonstrated that ACE2 and TMPRSS2 were important for SARS-CoV-2 S protein priming and cell entry.7 Thus, we explored the variation in ACE2 and TMPRSS2 expression across different heart tissues. Here, we present ‘gene expression’ from two perspectives, one is the percentage of gene-positive (UMI count >0) cells and the other is log2(CPM + 1).
As shown in Supplementary material online, Figure S1, the percentage of ACE2+ or TMPRSS2+ cells varied depending on sample source. Adult heart samples contained the largest number of ACE2+ cells (6.44%), followed by foetal heart samples (4.17%), and lastly adult coronary artery samples (0.46%). However, the percentages of TMPRSS2+ cells across different heart tissues were relatively low. Adult and foetal heart contained 0.36% and 0.47% TMPRSS2+ cells, respectively, with no TMPRSS2+ cells in adult coronary artery samples. After utilizing UMAP for dimensional reduction and visualization (Figure 1A–C), nine cell types in foetal heart, nine cell types in adult coronary artery, and five cell types in adult heart were identified. The distribution of ACE2+ or TMPRSS2+ cells among different cell types is shown in Figure 1D–F, and almost no cells expressed ACE2 and TMPRSS2 simultaneously. Thus, we further explored the percentage of ACE2+ or TMPRSS2+ cells across cell types and across heart tissues. As shown in Figure 1G, ACE2+ percentages across cell types in foetal heart and adult coronary artery samples were lower than 4.70% and 1.83%, respectively. In the adult heart, the percentage of ACE2+ cells was relatively high, with 10.38% in SMCs (smooth muscle cells). This raises a basic question: at what point can a specific cell type expressing ACE2 be considered vulnerable to COVID-19 infection? Several researchers have postulated that the heart is at high risk of COVID-19 infection, based on the findings that foetal myocardial cells have >7.5% ACE2+ cells, while type II alveolar cells (AT2) in the lung only have ∼1% ACE2+ cells.9 Here, we found that 62.5% (5/8) of cell types in the foetal heart (red blood cells were excluded), 11.1% (1/9) in the adult coronary artery, and 100% (5/5) in the adult heart display >1% ACE2+ cells, and among adult heart SMCs, ACE2+ cells reach 10.38% (Figure 1G). However, the percentage of ACE2+ cells may not be enough to draw this conclusion, especially since studies demonstrated that SARS-CoV-2 cell entry requires not only ACE2 binding but also cleavage by proteases such as TMPRSS2.7 We further explored the proportions of TMPRSS2+ cells across different cell types. Overall, percentages of TMPRSS2+ cells were <1.85% across all heart tissue cell types (Figure 1G).
Figure 1.
ACE2 and TMPRSS2 are expressed in specific cell types in the human heart. (A–C) Foetal heart cell atlas (A), adult coronary artery cell atlas (B), adult heart cell atlas (C) visualized by UMAP, coloured by the main cell groups. (D–F) Expression profile of ACE2 and TMPRSS2 in subset cell groups of the foetal heart (D), adult coronary artery (E), and adult heart (F); green represents the expression of TMPRSS2, red represents the expression of ACE2, and yellow represents the co-expression of ACE2 and TMPRSS2. (G) Percentage of ACE2- and TMPRSS2-positive cells in each subset cell group (nine cell groups in foetal heart, nine in adult coronary artery, and five in adult heart). (H) The expression of ACE2 and TMPRSS2 in each subset cell group. The expression level is displayed as log 2(CPM + 1). MC, mast cell; RBC, red blood cell; CM, cardiomyocyte; B, B cell; T, T cell; FIB, fibroblast; EC, endothelial cell; EP, epicardial cell; VC, valvular cell; SMC, smooth muscle cell; MP, macrophage; PC, pericyte; NK, natural killer cell; ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease serine 2; CPM, counts per million.
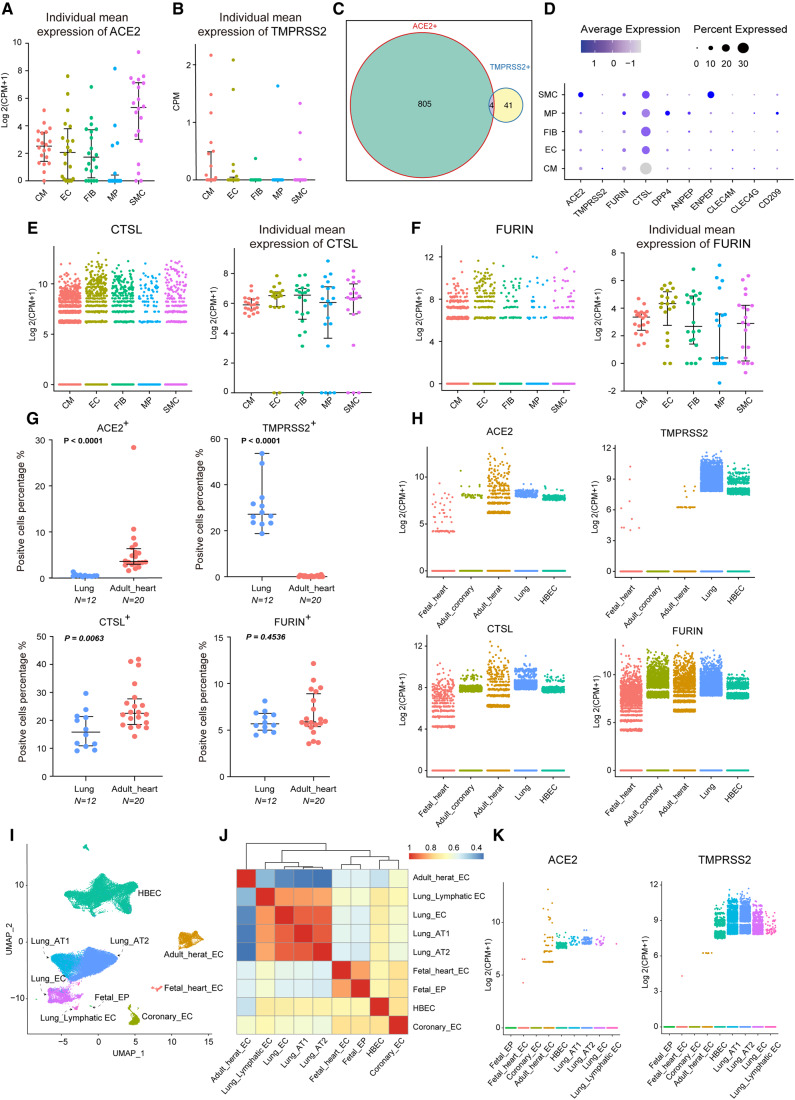
Considering that the percentages of ACE2+ or TMPRSS2+ cells were not the only parameters used to measure risk of SARS-CoV-2 infection, we further explored their gene expression levels (Figure 1H). The violin plot showed that ACE2 was highly expressed in the adult heart cell types; however, TMPRSS2 expression was still considered low across heart tissues. Considering the low expression of these genes and variation between individuals, we aggregated the mean expression of ACE2 and TMPRSS2 per cell type per patient (20 adult heart donors) (Figure2A andB). Adult heart SMCs show the highest ACE2 expression and no significant results in terms of TMPRSS2 expression (Supplementary material online, Figure S2A). Further subgroup analysis showed that in traditional SMCs (highly expressing MYH11), the SMC2 subgroup exhibited 3.5% ACE2+ cells, while the SMC1 subgroup highly expressing RGS5/ABCC9/CALD1 exhibited 15.7% ACE2+ cells (Supplementary material online, Figure S6). Further functional enrichment analysis revealed that both SMC1 and SMC2 displayed extracellular matrix (ECM)–receptor interaction, regulation of the actin cytoskeleton, vascular smooth muscle contraction, and other SMC-related functions, and the differential genes of SMC1 were more enriched in human papillomavirus infection, bacterial invasion of epithelial cells, Salmonella infection, and other viral and bacterial infection pathways (Supplementary material online, Figure S6E and F). Intercellular communication analysis revealed that SMC1 interacts with various cell groups in the heart (Supplementary material online, Figure S6G). TMPRSS2 is a transmembrane protease, thus ACE2 and TMPRSS2 dual-positive (ACE2+/TMPRSS2+) cells may be more permissive to SARS-CoV-2 infection.7,8,20 For this reason, we calculated ratios of ACE2+/TMPRSS2+ cells across heart tissues. ACE2+/TMPRSS2+ cells only exist in the adult heart, as shown in Figure 2C, and the percentage of these cells was rather small (4/12554, 0.03%).
Figure 2.
Expression pattern of ACE2, TMPRSS2, CTSL, and FURIN in the heart and lung. (A and B) Individual mean expression of ACE2 (A) and TMPRSS2 (B) in five major cell types; each point represents an individual sample (n = 20) and bars indicate the first and third quartile, with the median is shown as horizontal lines. The expression level is displayed as log 2(CPM + 1). (C) Venn plot showing the number of ACE2+ cells, TMPRSS2+ cells, and double positive cells in adult heart cells. (D) Dot plot representing the gene expression of the main coronavirus receptors and cofactors in major cell types of adult heart. (E and F) Expression of CTSL (E) and FURIN (F) in five major cell types; points represent cells (left) and individual samples (n = 20) (right). (G) Comparison of positive rates of ACE2, TMPRSS2, CTSL, and FURIN in individual samples between the lung and adult heart. Twelve patients are shown in lung and 20 patients in the heart. (H) Expression of ACE2, TMPRSS2, CTSL, and FURIN across five single-cell datasets. The expression level is displayed as log 2(CPM + 1). (I) Nine major endothelial and epithelial cell clusters in the lung and heart are visualized by UMAP, coloured by main cell groups. (J) Spearman similarity heat map of gene expression profiles of nine major endothelial and epithelial cell clusters in the lung and heart. (K) Expression of ACE2 (left) and TMPRSS2 (right) in nine major endothelial and epithelial cell clusters in the lung and heart; each point represents a cell. CTSL, cathepsin L; FURIN, furin (paired basic amino acid cleaving enzyme); HBEC, human bronchial epithelial cell. For comparison of positive rates of specific genes between the lung and heart, the Wilcoxon rank sum test was used. The P-values are shown in the figure.
Expression patterns of CTSL and FURIN in the adult heart
Although SARS-CoV-2 cell entry can be mainly inhibited by blocking TMPRSS2, there remained speculations as to whether any other protease can be used for virus priming.7,21 Prior studies have demonstrated that CTSL (cathepsin L) and FURIN (furin, paired basic amino acid cleaving enzyme) are expected to be efficient in facilitating cleavage of the SARS-CoV-2 S-protein.21,22 Thus, we explored expression patterns of CTSL and FURIN in the adult heart. CTSL+ and FURIN+ cells accounted for 28.6% and 8.7%, respectively, in the entire adult heart and appeared evenly distributed across different cell types (Figure 2D). In terms of CTSL and FURIN expression, there was no significant difference between cell types (Figure2E and F; Supplementary material online, Figure S2).
Comparison of specific gene expression between different tissues
The respiratory system is the major target of COVID-19 attack, and substantial damage can be seen in lung tissues from pathological findings.10 However, cardiac injury is common especially in high-severity patients, but little substantial damage was found from pathological findings.5,10 To learn more about this disparity, we compared the expression of ACE2 and TMPRSS2 between the heart and lung, also including HBECs. After integration as previously mentioned,15 we found that the percentage of ACE2+ cells in the adult heart was much higher than that in the lung (median: 3.6% vs. 0.5%, P < 0.0001), while the percentage of TMPRSS2+ cells was rather lower in the adult heart (median: 0% vs. 27.2%, P < 0.0001) (Figure 2G). Comparison of CTSL+ and FURIN+ cells between adult heart and lung isalso shown in Figure 2G. We further compared gene expression which showed that ACE2 expression in the adult heart is higher than that in the lung (P < 0.0001) and HBECs (P < 0.0001), while the TMPRSS2 expression level is considerably lower than that in the lung (P < 0.0001) and HBECs (P < 0.0001) (Figure 2H;Supplementary material online, Figure 2). These scRNA-seq-based results are also consistent with RNA-seq data from the NCBI Gene Database (Supplementary material online, Figure S7). Previous studies suggest that compared with adults, paediatric COVID-19 cases seemed less severe and symptoms experienced by children might be different.12,23 Thus, we compared ACE2 and TMPRSS2 expression between the adult and foetal heart. The results showed that ACE2 expression is lower in the foetal heart and TMPRSS2 expression exhibits no significant difference (Figure 2H;Supplementary material online, Figure S2). We speculate that a lower level of illness experienced by children might also exist in the cardiovascular system. However, these findings must be interpreted with caution.
Comparison of endothelium between heart and lung
After being released from the lung, SARS-CoV-2 circulates in the blood and may cause multiorgan damage through endothelial dysfunction.24,25 We further integrated endothelium from the heart and lung to compare their heterogeneity and correlations. Considering that diffuse alveolar damage was the primary finding in histology, epithelium-like type I and type II alveolar cells were also included in the analysis.24 Endothelium and epithelium from the heart and lung clustered separately in UMAP, but more closely among cells derived from the same tissue (Figure 2I). The Spearman similarity also shows a distinct gene expression of adult heart endothelium compared with the lung (Figure 2J). Interestingly, endothelium from the coronary artery and HBECs cluster together on the dendrogram, indicating partial overlap of the transcriptome signatures (Figure 2J). This is consistent with the opinion that epithelium in the airways plays a role similar to that of blood vessel endothelium.26 We also compared the expression of ACE2 and TMPRSS2 between heart and lung epithelium (Figure 2K). The expression pattern between these two genes is consistent with previous findings, which showed higher ACE2 but lower TMPRSS2 expression in the heart endothelium. Mean ranks and P-values of multiple group comparisons are shown in Supplementary material online, Figure S2.
Correlation between different genes and clinical characteristics
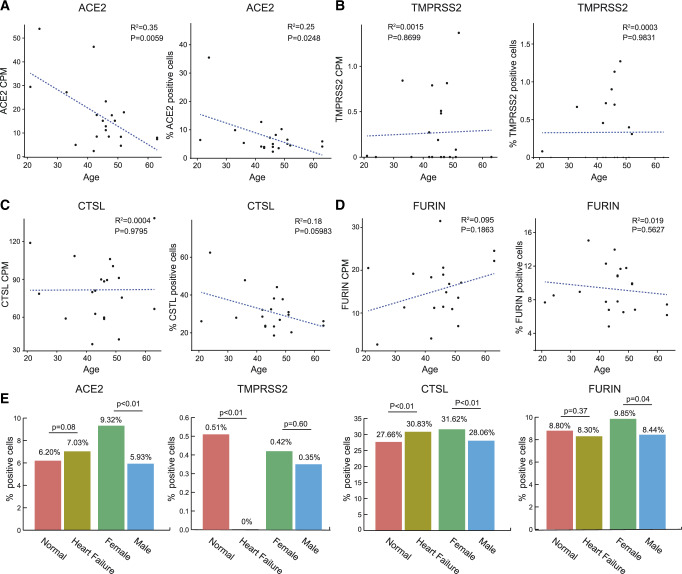
The clinical characteristics of adult heart donors in this study are shown in Supplementary material online, Table S8. A total of 14 healthy donors and 6 HF patients, ranging from 21 to 63 years of age, were included in this analysis. The average ACE2 expression value and percentage of ACE2+ cells per patient were calculated to assess the relationship between ACE2 and clinical characteristics, including age, sex, and heart failure. A negative correlation between overall expression of ACE2 and age (R2 = 0.35, P = 0.006) is shown in Figure 3A, which is consistent with previous studies observing that ACE2 is age related.27,28 In terms of the percentage of ACE2+ cells, a moderate inverse correlation with age still existed (R2 = 0.25, P = 0.025) (Figure 3A). There was no significant correlation between gene (TMPRSS2, CTSL, and FURIN) expression and age (Figure3C andD). Cells in the male adult heart showed a lower ACE2+ rate than in females (5.93% vs. 9.32%, P < 0.01) (Figure 3E). HF patients tended to exhibit a higher number of ACE2+ cells, but the difference was not statistically significant (7.03% vs. 6.20%, P = 0.08). HF cells showed a lower TMPRSS2+ rate (0% vs. 0.51%, P < 0.01) and higher CTSL+ rate (30.83% vs. 27.66%, P < 0.01) than cells in the normal heart. The male heart tended to have a lower ACE2+ rate (5.93% vs. 9.32%, P < 0.01), lower CTSL+ rate (28.06% vs. 31.62%, P < 0.01), and lower FURIN+ rate (8.44% vs. 9.85%, P = 0.04) (Figure 3E).
Figure 3.
Age, sex, and disease as possible factors influencing ACE2 expression. (A–D) Average ACE2 (A), TMPRSS2 (B), CTSL (C), and FURIN (D) expression value (left) and percentage of the above positive genes (right) over all individual samples vs. age in adult heart cells; points represent individual samples (n = 20). (E) Bar plot showing the comparison of the percentage of positive ACE2, TMPRSS2, CTSL, and FURIN cells in normal and heart failure hearts, as well as in female and male hearts. The significance of correlations was tested by using linear regression. Differences in the percentage of specific gene-positive cells between the two groups were tested using χ2 test. The P-values are shown in the figure.
Discussion
Clinical manifestations of COVID-19 were primarily related to respiratory inflammation, while cardiac complications are rapidly emerging as an important threat.5,29,30 In the present study, we also found that the incidence of cardiac injury increased with COVID-19 severity. However, regarding pathological findings, little substantial damage was seen in the heart tissue and no viral particles were observed in cardiac myocytes.10,11 This raises questions regarding cardiovascular involvement in COVID-19. Further, mechanisms of cardiovascular tropism of SARS-CoV-2 virus are incompletely understood. We explored the expression of ACE2 and SARS-CoV-2 spike (S) protein proteases at single-cell resolution and expect to improve our understanding of the cardiovascular implications of COVID-19.
Previous in vitro experimentation demonstrated that ACE2 can be used as the host cellular receptor for the SARS-CoV-2 S protein.22 Thus, several studies focusing on ACE2 expression in different tissues have been published;9,31 however, a comprehensive integration of cardiovascular datasets is lacking. We systematically analysed data from foetal heart, adult coronary artery, and adult heart tissues, and compared these with data from lung tissues and HBECs to explore the cardiovascular involvement in COVID-19. Our results showed a disparity in the expression of ACE2 and TMPRSS2 between the heart and lung, not only at the expression level, but also as regards the rates of positivity. The remarkable contrast in gene expression may reflect a difference between heart and lung in susceptibility to SARS-CoV-2 infection. Given that dual-positive ACE2 and TMPRSS2 cells are key conditions for SARS-CoV-2 cell entry, the heart may suffer less owing to the reduced expression of TMPRSS2, which may explain why little substantial damage occurs in the heart tissue compared with the lung. Evidence from other organs also supports our hypothesis. Research showed that ACE2 is highly expressed in the digestive system,32 and expression of TMPRSS2 in digestive system organs was considerably high (Supplementary material online, Figure S7), which indicates that the digestive system is at high risk of SARS-CoV-2 infection. This has been further proven by the clinical observation of gastrointestinal symptoms, such as diarrhoea, as well as the detection of viral RNA in digestive tissues.33 Thus, ACE2 and TMPRSS2 may determine SARS-CoV-2 infection to a large extent and, although cardiac cells exhibit abundant ACE2 as viral receptors, the possibility of infection could be low.10,11 Notably, several published COVID-19 cases exhibited dominant cardiac involvement (Supplementary material online, Table S9),11,34–36 which indicates that there may be a small number of people with severe heart infection. In this condition, we assume that CTSL and FURIN, which are considerably expressed in the adult heart, may play a potential role in mediating SARS-CoV-2 infection of the heart.
From the analysis above, cardiac injury seems more likely to be a concomitant phenomenon of systemic inflammatory responses.30,37 In this biological process, endothelial cells could be an important correlation between lung infection and other organ damage.25 We analysed features and correlations of endothelium from the heart and lung. The gene expression pattern of ACE2 and TMPRSS2 is heterogeneous between them, with higher ACE2 but lower TMPRSS2 expression in the heart endothelium. This indicates that the endothelium could be an abluminal barrier protecting cardiac tissue from circulating SARS-CoV-2 infection. From the recent autopsies performed on 67 COVID-19-positive patients, we can also see that little substantial endothelial dysfunction was found in 25 heart samples.24
Several limitations of this study should be mentioned. First, heterogeneity of our meta-analysis results is high, although we restricted criteria for study enrolment and performed subgroup analysis. An external validation of this estimate of cardiac injury should be considered. Secondly, we studied only single-cell transcriptome data of human heart and lung tissues, representing characteristics of the single-cell gene expression level. However, this analytical method therefore has an inherent bias because it is unclear what level of expression is considered biologically significant. In addition, confounders of sample source and depth of sequencing must be considered as a possible explanation for the results herein. However, the large number of cells (83 780) included in our study might dilute this impact. Although different data collaborations provided us with new insights into the possible explanation for the susceptibility heterogeneity in SARS-CoV-2 infection, our results and conclusions are data driven, which requires further laboratory verification. Lastly, limited sample size restricts determination of further implications of the correlation between different genes and clinical characteristics.
In conclusion, compared with lung tissues, ACE2 is relatively highly expressed in the human heart, while the key S protein priming protease, TMPRSS2, is barely expressed. The low percentage of ACE2+/TMPRSS2+ cells to some degree makes the heart less vulnerable to SARS-CoV-2 infection. The other S protein priming proteases, CTSL and FURIN, are relatively highly expressed in the human heart, which may compensate for mediating SARS-CoV-2 infection of the heart.
Supplementary Material
Acknowledgements
We thank the authors who uploaded their single-cell sequence data for others to perform further research. Our graphical abstract was created with BioRender.com.
Data availability
The heart data underlying this article are available in the Gene Expression Omnibus (GEO) at https://www.ncbi.nlm.nih.gov/geo/, and can be accessed with GEO accession numbers GSE106118 (foetal heart), GSE131778 (adult coronary artery), and GSE109816 and GSE121893 (adult heart). The lung and HBEC data underlying this article are available in Mendeley Data, v1 at https://data.mendeley.com/datasets/7r2cwbw44m/1.
Conflict of interest: none declared.
Funding
This work was supported by the National Natural Science Foundation of China [Key Program: grant no. 81830072].
Translational perspective
Understanding the mechanisms of cardiovascular tropism of SARS-CoV-2 is important for treating patients in a timely and effective way. We show that SARS-CoV-2 receptor ACE2 is relatively highly expressed in the human heart, while the key S protein priming protease, TMPRSS2, is barely expressed compared with lung tissues. The low percentage of ACE2+/TMPRSS2+ cells may indicate that the heart is less vulnerable to SARS-CoV-2 infection, and cardiac injury seems more likely to be a concomitant phenomenon of systemic inflammatory responses. These findings provide a new perspective for interpreting cardiac involvement in COVID-19 infection and highlight the importance of anti-inflammatory therapy.
References
- 1. Stower H. Spread of SARS-CoV-2. Nat Med 2020;26:465. [DOI] [PubMed] [Google Scholar]
- 2. Madjid M, Safavi-Naeini P, Solomon SD, Vardeny O.. Potential effects of coronaviruses on the cardiovascular system. JAMA Cardiol 2020;doi: 10.1001/jamacardio.2020.1286 [DOI] [PubMed] [Google Scholar]
- 3. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B.. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, Shi J, Zhou M, Wu B, Yang Z, Zhang C, Yue J, Zhang Z, Renz H, Liu X, Xie J, Xie M, Zhao J.. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol 2020;doi: 10.1016/j.jaci.2020.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi S, Qin M, Shen B, Cai Y, Liu T, Yang F, Gong W, Liu X, Liang J, Zhao Q, Huang H, Yang B, Huang C.. Association of cardiac injury with mortality in hospitalized patients with COVID-19 in Wuhan, China. JAMA Cardiol 2020;doi: 10.1001/jamacardio.2020.0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS.. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med 2020;46:586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N,, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S.. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lukassen S, Lorenz Chua R, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C, Eils R.. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J 2020;39:e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zou X, Chen K, Zou J, Han P, Hao J, Han Z.. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med 2020;14:185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS.. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8:420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tavazzi G, Pellegrini C, Maurelli M, Belliato M, Sciutti F, Bottazzi A, Sepe PA, Resasco T, Camporotondo R, Bruno R, Baldanti F, Paolucci S, Pelenghi S, Iotti GA, Mojoli F, Arbustini E.. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur J Heart Fail 2020;22:911–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cui Y, Zheng Y, Liu X, Yan L, Fan X, Yong J, Hu Y, Dong J, Li Q, Wu X, Gao S, Li J, Wen L, Qiao J, Tang F.. Single-cell transcriptome analysis maps the developmental track of the human heart. Cell Rep 2019;26:1934–1950.e1935. [DOI] [PubMed] [Google Scholar]
- 13. Wirka RC, Wagh D, Paik DT, Pjanic M, Nguyen T, Miller CL, Kundu R, Nagao M, Coller J, Koyano TK, Fong R, Woo YJ, Liu B, Montgomery SB, Wu JC, Zhu K, Chang R, Alamprese M, Tallquist MD, Kim JB, Quertermous T.. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell analysis. Nat Med 2019;25:1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang L, Yu P, Zhou B, Song J, Li Z, Zhang M, Guo G, Wang Y, Chen X, Han L, Hu S.. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat Cell Biol 2020;22:108–119. [DOI] [PubMed] [Google Scholar]
- 15. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R.. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang X, Lan Y, Xu J, Quan F, Zhao E, Deng C, Luo T, Xu L, Liao G, Yan M, Ping Y, Li F, Shi A, Bai J, Zhao T, Li X, Xiao Y.. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res 2018;47:D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu G, Wang LG, Han Y, He QY.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Curtin F, Schulz P.. Multiple correlations and Bonferroni’s correction. Biol Psychiatry 1998;44:775–777. [DOI] [PubMed] [Google Scholar]
- 19. Kruskal WH, Wallis WA.. Use of ranks in one‐criterion variance analysis. J Am Stat Assoc 1952;47:583–621. [Google Scholar]
- 20. Ziegler CGK, Allon SJ, Nyquist SK,, Mbano IM, Miao VN, Tzouanas CN, Cao Y, Yousif AS,, Bals J, Hauser BM, Feldman J, Muus C,, Wadsworth MH 2nd, Kazer SW, Hughes TK, Doran B, Gatter GJ, Vukovic M, Taliaferro F, Mead BE, Guo Z, Wang JP, Gras D, Plaisant M, Ansari M, Angelidis I, Adler H, Sucre JMS, Taylor CJ,, Lin B, Waghray A, Mitsialis V, Dwyer DF, Buchheit KM, Boyce JA, Barrett NA, Laidlaw TM, Carroll SL, Colonna L, Tkachev V, Peterson CW, Yu A, Zheng HB, Gideon HP, Winchell CG, Lin PL, Bingle CD, Snapper SB, Kropski JA, Theis FJ, Schiller HB, Zaragosi LE, Barbry P,, Leslie A,, Kiem HP, Flynn JL, Fortune SM, Berger B, Finberg RW, Kean LS, Garber M, Schmidt AG, Lingwood D, Shalek AK, Ordovas-Montanes J, Lung HCA Biological Network. Electronic address: lung-network@humancellatlas.org; HCA Lung Biological Network. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020;181:1016–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E.. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 2020;176:104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D.. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181:281–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu X, Zhang L, Du H, Zhang J, Li YY, Qu J, Zhang W, Wang Y, Bao S, Li Y, Wu C, Liu H, Liu D, Shao J, Peng X, Yang Y, Liu Z, Xiang Y, Zhang F, Silva RM, Pinkerton KE, Shen K, Xiao H, Xu S, Wong GWK.. SARS-CoV-2 infection in children. N Engl J Med 2020;382:1663–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bryce C, Grimes Z, Pujadas E, Ahuja S, Beasley MB, Albrecht R, Hernandez T, Stock A, Zhao Z, Al Rasheed M, Li Chen J, Wang L, Corben D, Haines A, Westra K, Umphlett W, Gordon M, Reidy RE, Petersen J, Salem B, Fiel F, El Jamal SM M, Tsankova NM, Houldsworth J, Mussa Z, Liu W-C, Veremis B, Sordillo E, Gitman M, Nowak M,, Brody R, Harpaz N,, Merad M, Gnjatic S, Donnelly R, Seigler P, Keys C, Cameron J, Moultrie I, Washington K-L, Treatman J, Sebra R, Jhang J, Firpo A, Lednicky J, Paniz-Mondolfi A, Cordon-Cardo C, Fowkes M. Pathophysiology of SARS-CoV-2: targeting of endothelial cells renders a complex disease with thrombotic microangiopathy and aberrant immune response. The Mount Sinai COVID-19 autopsy experience. medRxiv 2020:2020.2005.2018.20099960; doi: 10.1101/2020.05.18.20099960.
- 25. Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, Mehra MR, Schuepbach RA, Ruschitzka F, Moch H.. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020;395:1417–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Luscher TF, Vanhoutte PM.. The Endothelium: Modulator of Cardiovascular Function. Boca Raton, FL: CRC Press; 2020. [Google Scholar]
- 27. Xie X, Chen J, Wang X, Zhang F, Liu Y.. Age- and gender-related difference of ACE2 expression in rat lung. Life Sci 2006;78:2166–2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. AlGhatrif M, Cingolani O, Lakatta EG.. The dilemma of coronavirus disease 2019, aging, and cardiovascular disease: insights from cardiovascular aging science. JAMA Cardiol 2020;doi: 10.1001/jamacardio.2020.1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xiong TY, Redwood S, Prendergast B, Chen M.. Coronaviruses and the cardiovascular system: acute and long-term implications. Eur Heart J 2020;41:1798–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng YY, Ma YT, Zhang JY, Xie X.. COVID-19 and the cardiovascular system. Nat Rev Cardiol 2020;17:259–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qi F, Qian S, Zhang S, Zhang Z.. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem Biophys Res Commun 2020;526:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liang W, Feng Z, Rao S, Xiao C, Xue X, Lin Z, Zhang Q, Qi W.. Diarrhoea may be underestimated: a missing link in 2019 novel coronavirus. Gut 2020;69:1141–1143. [DOI] [PubMed] [Google Scholar]
- 33. Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H.. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020;158:1831–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu H, Ma F, Wei X, Fang Y.. Coronavirus fulminant myocarditis saved with glucocorticoid and human immunoglobulin. Eur Heart J 2020;41:doi: 10.1093/eurheartj/ehaa190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hua A, O’Gallagher K, Sado D, Byrne J.. Life-threatening cardiac tamponade complicating myo-pericarditis in COVID-19. Eur Heart J 2020;41:2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Inciardi RM, Lupi L, Zaccone G, Italia L, Raffo M, Tomasoni D, Cani DS, Cerini M, Farina D, Gavazzi E, Maroldi R, Adamo M, Ammirati E, Sinagra G, Lombardi CM, Metra M.. Cardiac involvement in a patient with coronavirus disease 2019 (COVID-19). JAMA Cardiol 2020;doi: 10.1001/jamacardio.2020.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen L, Hao G.. The role of angiotensin-converting enzyme 2 in coronaviruses/influenza viruses and cardiovascular disease. Cardiovasc Res 2020;doi: 10.1093/cvr/cvaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The heart data underlying this article are available in the Gene Expression Omnibus (GEO) at https://www.ncbi.nlm.nih.gov/geo/, and can be accessed with GEO accession numbers GSE106118 (foetal heart), GSE131778 (adult coronary artery), and GSE109816 and GSE121893 (adult heart). The lung and HBEC data underlying this article are available in Mendeley Data, v1 at https://data.mendeley.com/datasets/7r2cwbw44m/1.
Conflict of interest: none declared.